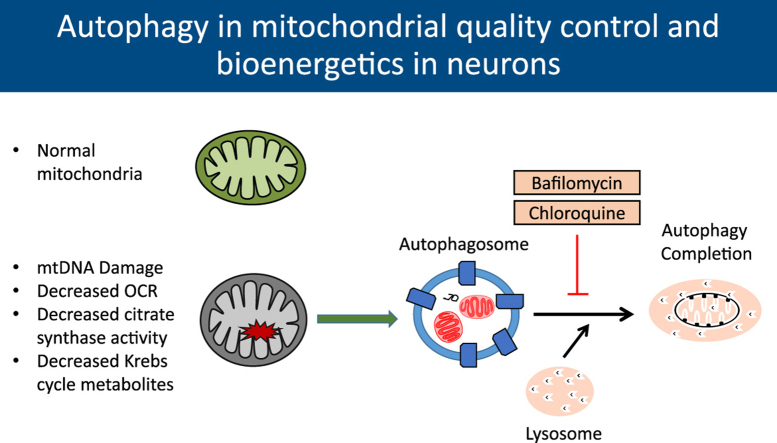
Abstract
Autophagy is an important cell recycling program responsible for the clearance of damaged or long-lived proteins and organelles. Pharmacological modulators of this pathway have been extensively utilized in a wide range of basic research and pre-clinical studies. Bafilomycin A1 and chloroquine are commonly used compounds that inhibit autophagy by targeting the lysosomes but through distinct mechanisms. Since it is now clear that mitochondrial quality control, particularly in neurons, is dependent on autophagy, it is important to determine whether these compounds modify cellular bioenergetics. To address this, we cultured primary rat cortical neurons from E18 embryos and used the Seahorse XF96 analyzer and a targeted metabolomics approach to measure the effects of bafilomycin A1 and chloroquine on bioenergetics and metabolism. We found that both bafilomycin and chloroquine could significantly increase the autophagosome marker LC3-II and inhibit key parameters of mitochondrial function, and increase mtDNA damage. Furthermore, we observed significant alterations in TCA cycle intermediates, particularly those downstream of citrate synthase and those linked to glutaminolysis. Taken together, these data demonstrate a significant impact of bafilomycin and chloroquine on cellular bioenergetics and metabolism consistent with decreased mitochondrial quality associated with inhibition of autophagy.
Highlights
-
•
Autophagy inhibition decreased mitochondrial bioenergetics in intact neurons.
-
•
Autophagy inhibition decreased mitochondrial complexes I, II or IV substrate linked respiration.
-
•
Autophagy inhibition increased mitochondrial DNA damage.
-
•
Autophagy inhibition decreased major components of the Krebs cycle.
-
•
Autophagy inhibition resulted in decreased citrate synthase activities.
Graphical abstract
1. Introduction
The autophagy-lysosomal pathway plays an important role in protein and organelle homeostasis [1], [2], [3], [4]. This pathway involves the engulfment of proteins or organelles by autophagosomes and subsequent degradation by lysosomes. This is a multi-step, dynamic process involving greater than 32 autophagy related proteins and lysosomal components [1], [2], [3], [4], [5], [6]. Pharmacological inhibitors of autophagy either at initiation or completion have been used widely in both normal and pathologic states in a variety of cells and tissues to provide insights into the protective or deleterious roles of autophagy. Their use has been indispensable in measuring autophagy and lysosomal activities and in some instances these inhibitors have been used in the clinic as well [7], [8], [9], [10], [11], [12], [13]. However, whether these compounds have off target effects on cellular bioenergetics is not clear. Interestingly, targeting the autophagy-lysosomal pathway would be expected to inhibit mitophagy and result in decreased mitochondrial quality. This view is supported by data in mitochondria isolated from ATG7 knockout mouse skeletal muscle that shows decreased mitochondrial respiration. Furthermore, MEFs isolated from these mice also show decreased basal and maximal oxygen consumption rates [14]. In this study we hypothesized that two structurally diverse and commonly used pharmacological agents, which both inhibit autophagy at the level of the lysosome through distinct mechanisms, should exhibit convergent effects on mitochondrial quality and cellular bioenergetics. This was tested using bafilomycin and chloroquine by assessing their effects on LC3-II accumulation, bioenergetics, and metabolism in primary neurons.
The macrolide antibiotic bafilomycin A1 was among the first of this class isolated from Streptomyces gresius, and has been shown to be a potent inhibitor of the Vacuolar H+ATPase which controls pH in the lysosome (V-ATPase) [15], [16], [17]. Through this mechanism bafilomycin inhibits autophagic flux by preventing the acidification of endosomes and lysosomes [18], [19]. The mechanisms of action of bafilomycin are complex. In one study, it was shown that it does not affect the E. coli F1,Fo-ATPase or the N. crassa mitochondrial F1,Fo-ATPase in vitro over a wide range of concentrations [15]. On the other hand, bafilomycin at low nM concentrations was reported to act as a potassium ionophore and at 30–100 nM decreases mitochondrial membrane potential and O2 consumption, and at ~300 nM induces swelling in isolated mitochondria from rat liver [20]. In differentiated PC12 cells, SH-SY5Y cells and cerebellar granule neurons, bafilomycin (at 50–250 nM after 45 min) partially uncouples mitochondria due to a decrease in the proportion of polarized mitochondria, i.e. stochastic flickering. Furthermore, it was shown to decrease the mitochondrial pH, Ca2+ and ΔΨm. This was associated with an elevation of mitochondrial respiration as assayed by MitoXpress and PtCPTE-CFR9 fluorescent probes both dose and time dependently [21]. These data suggest that bafilomycin has a number of off-target effects on mitochondria directly, making it difficult to determine which are a consequence of inhibiting autophagy. This is potentially important as the translational capability of bafilomycin is being explored in a wide range of models. For example, there is evidence that bafilomycin inhibits viral replication of Influenza A and B in canine kidney cells [22], [23]. In cancer, bafilomycin alone or as a co-treatment appears to be effective in enhancing the efficacy of other therapies [24], [25], [26]. Given the complex interactions between metabolism and autophagy we reasoned additional insight could be gained by comparing the impact of bafilomycin on cellular bioenergetics. This is an integrated measure of mitochondrial metabolism, with the cell providing its own substrates compared to measurements of individual components of the respiratory chain under conditions where substrates are not limiting. This can then be compared with parameters of mitochondrial quality to determine mechanism.
Similar to bafilomycin, the former malaria drug chloroquine (CQ) is now widely used as an inhibitor of autophagy in both cell culture and in vivo [13]. Chloroquine has a long history of human use and is currently being tested as a sensitizing agent for certain cancers, making understanding its mechanisms of action both topical and important [7], [8], [9]. Chloroquine is a lysosomotropic weak base, which in the monoprotonated form diffuses into the lysosome, where it becomes diprotonated and becomes trapped. Protonated chloroquine then changes the lysosomal pH, thereby inhibiting autophagic degradation in the lysosomes [27]. And while the direct effects of chloroquine on mitochondria have not been, to our knowledge, extensively studied, there is evidence to suggest that in already compromised mitochondria, as is the case in cardiac pressure overload, chloroquine exacerbates the decrease in mitochondrial quality and function [28].
Autophagy plays a critical role in maintaining healthy mitochondrial populations through mitochondrial specific recycling termed mitophagy. Mitochondria are key components in the cell responsible for not only energy production but for varied and diverse signaling pathways [29], [30]. Failure to maintain a population of healthy mitochondria can lead to decreased energy production, increased reactive oxygen species production and eventually to cytotoxicity. These outcomes are especially evident in chronic diseases including neurodegenerative and heart diseases [1], [2], [5], [30], [31], [32]. Since pharmacological inhibitors of autophagy are frequently used and combined with measurements of mitochondrial function, it is important to determine their effects in a defined cellular system. In addition to oxidative phosphorylation, mitochondria metabolism is also essential for multiple cellular functions beyond energetics [33], [34], [35].
In the present study, we have used primary cortical rat neurons to evaluate how the autophagy inhibitors chloroquine and bafilomycin affect mitochondrial bioenergetics and metabolism. We used the Seahorse extracellular flux analyzer and a targeted metabolomics approach to determine parameters of mitochondrial bioenergetics and metabolism in intact neurons providing a cellular context for understanding the autophagy pathway and its inhibitors. We have found that both chloroquine and bafilomycin decrease mitochondrial quality to a similar extent when levels are titrated to achieve a comparable level of autophagy inhibition.
2. Methods
2.1. Chemicals
Bafilomycin A1 (B1793), chloroquine (C6628), oligomycin (75351), FCCP (C2920), rotenone (R8875), antimycin-A (A8674), pyruvate (P5280), malate (M6413), ADP (A2754), succinate (S2378) were all obtained from Sigma. Neurobasal medium (21103-049), B-27 supplement (17504044-044), L-glutamine (25030-081) and penicillin-streptomycin (15140-122) were obtained from Life Technologies. PMP (102504) was obtained from Seahorse Bioscience.
2.2. Cell Culture
Primary cortical rat neurons were isolated at embryonic day E18. Dissections were performed in HBSS media and plated on poly-L-Lysine coated plates using Neural Basal media supplemented with L-glutamine, B27, and Penicillin/Streptomycin. Experiments were performed on DIV7, when half of the media was replaced with each compound diluted in Neural Basal supplemented with L-glutamine and Penicillin/Streptomycin only.
2.3. Cell viability
Cells were plated at 80k on XF96 plates and exposed for 24 h to bafilomycin or chloroquine. Cell viability was measured 24 h after the exposure by the Trypan Blue exclusion method.
2.4. Western blot analysis
Neurons were plated on 24-well plates seeded at 480k cells per well. After exposures to the pharmacological compounds, medium was aspirated. Cells were then washed once with ice cold PBS, and 40 µL of RIPA lysis buffer (50 mM Tris pH 7.8, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100%, and 0.1% SDS) with 1x Roche protease inhibitors (4693132001) was added to each well and cells were scraped. Two wells were then combined and centrifuged at 16,800×g for 15 min at 4 °C. The supernatant was collected and protein was measured using the BCA protein assay. 5x loading buffer was added to samples and then separated by SDS-PAGE using 12% or 15% polyacrylamide gels. Membranes were then immunoblotted for LC3 (Sigma L8918 1:2000), Citrate synthase (AB96600 1:1000), PGC1-α (SC-13067 1:1000), p62 (Abnova H00008878-M01 1:2000), aconitase (affinity purified as previously described [36], was a generous gift from Scott Ballinger at UAB) and actin (Sigma A5441 1:2500). Subunits of mitochondrial oxidative phosphorylation complexes I-V were blotted for by using Abcam's total OXPHOS rodent antibody cocktail (AB110413). Images were collected using an Amersham™ Imager600. Images were then analyzed using ImageJ [37].
2.5. Mitochondrial DNA copy number and DNA damage assays
Cells were plated at 480k cells per well on 24 well plates and were treated for 24 h. After treatment, DNA was extracted. Quantitative real-time PCR was performed using SYBR green in an ABI 7500 machine. Mitochondrial DNA copy number was determined by normalizing results from primers targeted to mtDNA (Forward: 5'-CCAAGGAATTCCCCTACACA-3' and Reverse: 5′-GAAATTGCGAGAATGGTGGT-3′) against results from primers targeted to nuclear 18S DNA (Forward: 5'-CGAAAGCATTTGCCAAGAAT-3' and Reverse: [38], [39]. Mitochondrial DNA damage was determined by PCR as previously described [40]. Briefly, mtDNA long segments (Forward: 5′-GGACAAATATCATTCTGAGGAGCT-3′ Reverse: (5′-GGATTAGTCAGCCGTAGTTTACGT-3′) and short segments (Forward: 5′-CCAAGGAATTCCCCTACACA-3′ Reverse: 5′-GAAATTGCGAGAATGGTGGT-3′) were amplified using an AccuPrimeTM Taq DNA Polymerase High Fidelity kit (Life Tech Corp). The products were separated on agarose gels, visualized by ethidium bromide, and analyzed by ImageJ. mtDNA Long PCR conditions were: 94 °C for 11 s, 25 cycles of denaturation at 94 °C for 15 s, annealing and extension at 67 °C for 12 min, and final extension at 72 °C for 10 min mtDNA Short PCR conditions were: 94 °C for 6 s, 18 cycles of denaturation at 94 °C for 20 s, annealing and extension at 65 °C for 1 min, and final extension at 72 °C for 10 min. Lesion frequency was calculated as follows [41]:
2.6. Bioenergetic analysis
Bioenergetic profiles were determined using the Seahorse Bioscience Extracellular Flux Analyzer (XF96) [42]. Cells were isolated as above and plated at 80k cells per well on XF96 plates. For mitochondrial stress test in intact neurons, after exposure to autophagy inhibitors, cells were switched to XF media 30 min prior to measurement of oxygen consumption rate (OCR), followed by sequential injection of oligomycin (1 µg/mL), FCCP (1 µM), and rotenone (1 µM)+antimycin A (10 µM), with 2 OCR measurements after each injection [42]. Total protein per well was measured by Lowry DC protein assay and OCR normalized to protein. Basal OCR was calculated as OCR before oligomycin minus OCR after Antimycin. Maximal OCR was calculated as OCR after FCCP minus OCR after Antimycin. Reserve Capacity was calculated as OCR after FCCP minus OCR before oligomycin. ATP-linked OCR was calculated as OCR before oligomycin minus OCR after oligomycin. Proton leak OCR was calculated as OCR after oligomycin minus OCR after Antimycin. Non-mitochondrial OCR is the OCR after antimycin injection.
For mitochondrial activity assay in permeabilized cells, 5 min prior to assay, cells were switched to MAS buffer (70 mM sucrose, 220 mM mannitol, 10 mM KH2PO4, 5 mM MgCl2, 2 mM HEPES, 1 mM EGTA, adjusted pH to 7.2) [43]. OCR was measured after injection 1 nM PMP (Seahorse Plasma Membrane Permeabilizer) plus complex I modulators (10 mM pyruvate, 1 mM malate and 1 mM ADP or 1 µM FCCP) for complex I activity assays, then injected with 1 μM rotenone to inhibit complex I activity, followed by injection of complex II modulators (10 mM succinate) to assay for complex II activity, then injected with 10 µM Antimycin. Complex IV activity was measured in PMP permeabilized neurons in MAS buffer by injection of PMP (1 nM) plus TMPD 0.5 mM, ascorbate 2 mM, and 1 mM ADP; after 2 measurements the inhibitor azide 20 mM was injected. Complex I activity was calculated as the difference between before and after rotenone injection. Complex II activity was calculated as the difference between before and after antimycin injection. Complex IV activity was calculated as the difference after ascorbate injection and after azide injection. The experiments were repeated with >3 independent cultures. Because of significant protein loss during assays utilizing MAS buffer, accurate measure of total protein was unobtainable, thus raw data is represented.
2.7. Metabolomics
Cells were plated on 24 well plates at 480k per well and treated with autophagy modulators for 24 h. Upon completion of treatment the cells were washed with 1 mL of ice cold PBS. The cells were then lysed and scraped in 500 µL of HPLC grade ice cold methanol and incubated at −80 °C for 2 h. Plates were scraped again and cell lysates were transferred to glass tubes where they were centrifuged at ~1000×g for 20 min at 4 °C. Supernatants were transferred to new glass tubes and stored at −80 °C until further processing [44].
Standards were generated as a master mix of all compounds at 100 μg/mL in H2O and serial diluted to 10x of the final concentrations (0.05–10 μg/mL, 9 standards). Standards were further diluted to 1x in methanol to a total volume of 1 mL, and dried by a gentle stream of N2. For cell extracts, 1 mL of each were transferred to a glass tube and dried under a gentle stream of N2. Standards and samples were resuspended in 50 µL of 5% acetic acid and vortexed for 15 s. Amplifex™ Keto Reagent (SCIEX, Concord, Ontario, Canada) (50 µL) was added to each sample and allowed to react for 1 h at room temperature. Standards and samples were then dried under a gentle stream of N2 and resuspended in 1 mL and 200 µL of 0.1% formic acid, respectively.
Samples were analyzed by LC-multiple reaction ion monitoring-mass spectrometry. Liquid chromatography was performed by LC20AC HPLC system (Shimadzu, Columbia, MD) with a Synergi Hydro-RP 4 µm 80 A 250×2 mm ID column (Phenomenex, Torrance, CA). Mobile phases were: A) 0.1% formic acid and B) methanol/0.1% formic acid. Compounds were eluted using a 5–40% linear gradient of B from 1 to 7 min, followed by a column wash of 40–100% B from 7 to 10 min, and re-equilibrated at 5% B from 10.5 to 15 min. Column eluant was passed into an electrospray ionization interface of an API 4000 triple-quadrupole mass spectrometer (SCIEX). The following mass transitions were monitored in the positive ion mode: m/z 261/118 for α-ketoglutarate, m/z 247/144 for oxaloacetate and m/z 204/144, 204/118 and 204/60 for pyruvate. In the negative mode, the following transitions are monitored: m/z 115/71 for fumarate, m/z 89/43 for lactate, m/z 117/73 for succinate, m/z 133/115 for malate, m/z 173/85 for cis-aconitate, m/z 191/87 for citrate, m/z 191/73 for isocitrate, m/z 147/129 for 2-hydroxyglutarate, m/z 146/102 for glutamate, m/z 145/42 for glutamine and m/z 132/88 for aspartate. The 16 transitions were each monitored for 35 ms, with a total cycle time of 560 ms. MS parameters were CAD 4, CUR 15, GS1 60, GS2 30, TEM 600, IS −3500 V for negative polarity mode and IS 4500 for positive polarity mode. Peak areas of metabolites in the sample extracts are compared in MultiQuant software (SCIEX) to those of the known standards to calculate metabolite concentrations.
2.8. Citrate activity assay
Neurons were plated at 480k/well on 24 well plates. Cells were scraped into RIPA lysis buffer as before and frozen until assayed. Activity was measured using a Beckman Coulter DU800 spectrophotometer. 945 µL of 100 mM Tris (pH 8.0 at 37 °C) with 0.1% Triton X-100 was added to cuvette followed by 25 µL of sample, 10 µL of 10 mM acetyl-CoA, 10 µL 5,5′-dithiobis(2,4-nitrobenzoic acid), and 10 µL 20 mM oxaloacetate. Activity was measured for 4 min. The rate was calculated from the linear range and was normalized to total protein.
2.9. Normalized activity calculations
Citrate synthase activity (nmol/min/mg protein) was expressed as the ratio of enzyme activity to relative levels of citrate synthase protein as determined by western blotting. Complex I, II and IV-linked OCR were expressed as the ratio of their respective OCR normalized to relative levels of specific subunits within each complex as determined using western blotting.
2.10. Statistics
All results were expressed as mean±SEM. Statistical analysis of data were performed by using one-way ANOVA followed by Tukey's post hoc test. Values of p<0.05 were considered statistically significant.
3. Results
3.1. Effects of autophagy inhibitors on LC3-II accumulation and viability
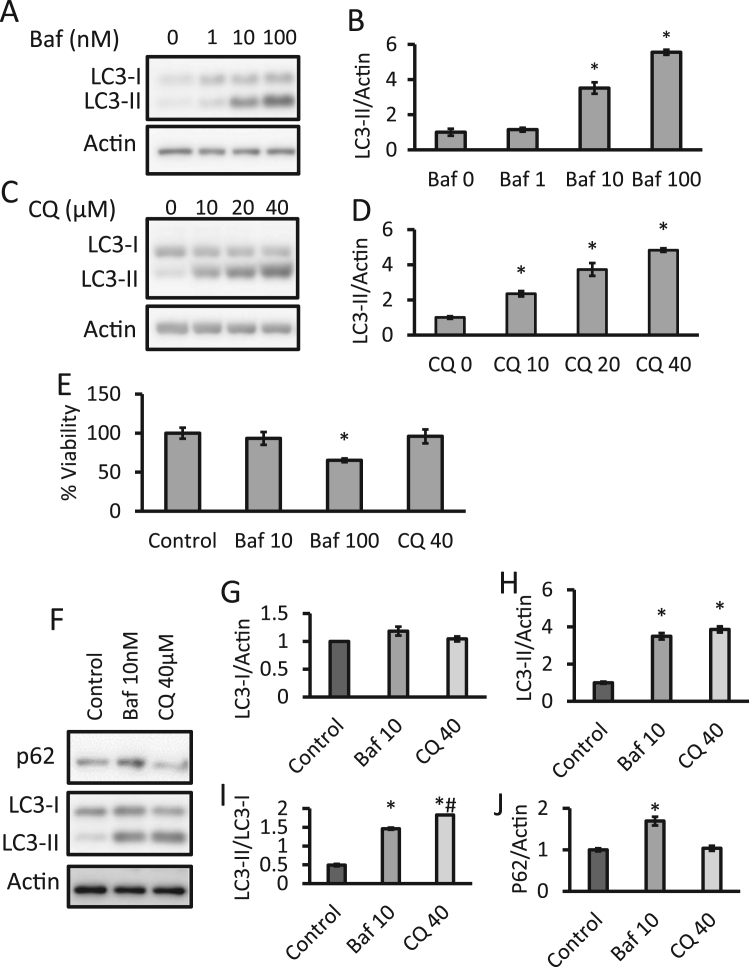
During the autophagy process, LC3-I is converted to LC3-II by lipidation, resulting in LC3-II migrating differently on SDS/PAGE. LC3-II is used as a quantitative marker of autophagy since it is required for the formation of the autophagosome and its level is proportional to the amount of autophagosomes in the cell [13], [45], [46]. To determine concentrations of bafilomycin and chloroquine which result in an approximately equivalent level of autophagy inhibition, cultured primary cortical rat neurons at 7 d in vitro (DIV7) were exposed to a range of concentrations for both compounds for 24 h. Significant increases in LC3-II were observed at 10 and 100 nM but not at 1 nM bafilomycin exposure (Fig. 1A, B). Significant increases in LC3-II were observed at 10, 20, and 40 µM concentrations of CQ (Fig. 1C, D). No significant changes in LC3-I were observed. There were no significant changes in the cell viability after exposure to chloroquine for 24 h. However, exposure to 100 nM but not 10 nM bafilomycin, decreased cell viability by approximately 35% (Fig. 1E). In a direct comparison of these two autophagy modulators, we observed no significant changes in LC3-I, but statistically similar increases in LC3-II for both bafilomycin and chloroquine exposures. The LC3-II/I ratio reflects the conversion of the protein from the cytosolic form to the autophagosome associated form upon autophagy activation and was also calculated and showed significant increases for both bafilomycin and chloroquine treatments, although chloroquine was statistically further increased, albeit modestly, from bafilomycin. The autophagy substrate p62 was also measured and showed increased levels for bafilomycin (Fig. 1F-J). Given the toxicity of 100 nM bafilomycin and the similar accumulation of LC3-II with 10 nM bafilomycin and 40 µM chloroquine treatments, all subsequent 24 h time point experiments were performed with these concentrations.
Fig. 1.
: Effects of bafilomycin and chloroquine on autophagy and cell survival. Primary rat cortical neurons at DIV7 were used for experiments. (A-D) Western blot analyses of LC3-I and LC3-II in lysates in neurons exposed to increasing concentrations of bafilomycin (baf) or chloroquine (CQ) for 24 h. (E) Trypan blue exclusion assay of cell viability after 24 h exposure to baf or CQ at indicated concentrations. (F-J) Western blot analyses of p62, LC3-I and LC3-II in lysates in neurons exposed to 10 nM baf and 40 µM CQ. *p<0.05 compared to control, #p<0.05 between baf and CQ, Data=mean±SEM, 3 independent experiments, and each experiment contain 3 independent samples.
3.2. Effects of bafilomycin and chloroquine on cellular bioenergetics in intact neurons
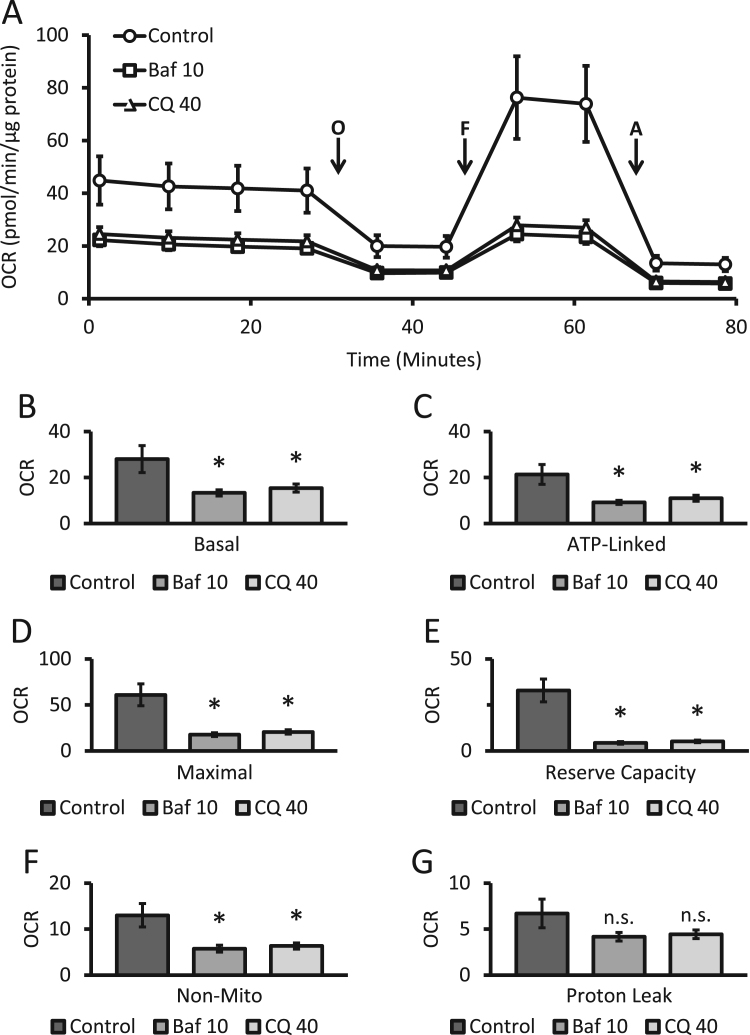
Primary cortical rat neurons were exposed to autophagy modulators for 24 h as before and medium was replaced by XF media free of autophagy modulators, followed by a mitochondrial stress test in intact cells using XF media. These cells exhibited the typical profile for the mitochondrial stress test we have previously reported for primary neurons [38], [47]. As expected, after establishing a stable baseline measurement of oxygen consumption rate (OCR) the injection of oligomycin resulted in a decrease in OCR due to the inhibition of ATP synthase. Next, FCCP, a mitochondrial uncoupler was found to stimulate OCR by depleting the proton gradient. Finally, antimycin was injected to inhibit consumption of oxygen by the mitochondrial electron transport chain [32], [42]. Basal, ATP-linked, maximal, reserve capacity, proton leak, and non-mitochondrial OCR were determined from this trace (Fig. 2A). Both bafilomycin exposure at 10 nM and chloroquine exposure at 40 µM resulted in similar decreases in overall OCR for basal, ATP-linked, maximal, reserve capacity and non-mitochondrial related parameters of oxygen consumption. No significant changes in proton leak were noted (Fig. 2B-G). Extracellular acidification rate (ECAR), a measurement of proton production from glycolysis and the TCA cycle, was also recorded using the same injection protocol as above. Statistically significant decreases in ECAR were observed for both bafilomycin and chloroquine and in the plot of OCR vs ECAR the cells a show a similar and a less energetic status (Supplemental Figure 1A-C).
Fig. 2.
Effects of bafilomycin and chloroquine on parameters of bioenergetics in intact neurons. (A) Oxygen consumption rate (OCR) from primary cortical neurons exposed to bafilomycin (10 nM) and chloroquine (40 µM) for 24 h was determined by the mitochondrial stress test. OCR was measured before and after sequential injection of Oligomycin (O, 1 µg/mL), FCCP (F, 1 µM), and Antimycin A (A, 10 µM). (B-G) Basal, maximal, reserve capacity, ATP linked, proton leak, and non-mitochondrial OCR were calculated from (A) as described in the Methods. *p<0.05 compared to control, from 3 independent experiments. Shown is the representative experiment with, n≥3 independent samples, Data=mean±SEM, n.s.: not significant.
Since bafilomycin has been reported to inhibit the mitochondrial ATP synthase and chloroquine can modulate organellar pH, either compound could acutely inhibit mitochondrial function. To test this, we measured basal OCR for 4 consecutive readings and then directly injected the compounds, recorded 4 more basal readings and then proceeded with a mitochondrial stress test. No discernable decreases of OCR were observed during the 30 min immediately after direct injection of the compounds or during the stress test (Supplemental Figure 2A). No significant alterations in p62, LC3-I, or LC3-II protein levels nor any changes in viability were observed (Supplemental Figure 2B-F).
3.3. Effects of bafilomycin and chloroquine on mitochondrial complex I, II and IV substrate–linked respiration
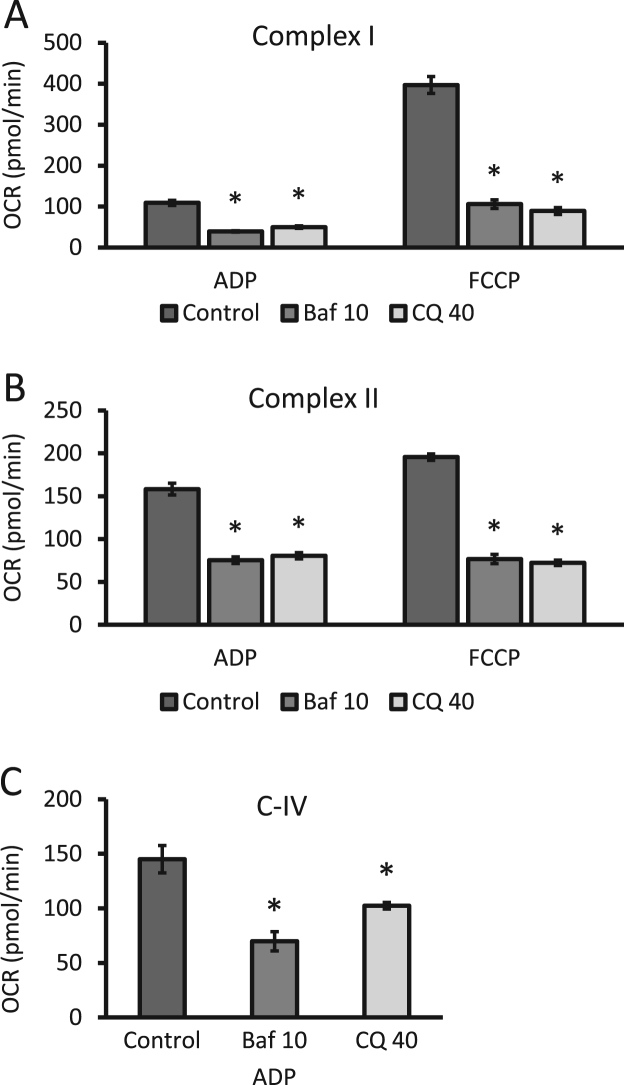
To assess the effect of the autophagy inhibitors on mitochondrial complexes, cells were exposed to 10 nM bafilomycin or 40 µM chloroquine (24 h) under the conditions described in Fig. 2 and then switched to MAS buffer before permeabilization with PMP and OCR measurements with complex I, II or IV-linked substrates. Complex I linked respiration was diminished by approximately 65% and 55% for bafilomycin and chloroquine treatment in the presence of ADP, respectively. In the presence of FCCP, decreases in OCR between 70% and 80% were observed after autophagy inhibitor treatment (Fig. 3A). Complex II linked activities were diminished by 50% in the presence of ADP and by 60% in the presence of FCCP after either bafilomycin or chloroquine exposure (Fig. 3B). Complex IV substrate-linked OCR in the presence of ADP was decreased by ~47% with bafilomycin and by ~30% with chloroquine (Fig. 3C).
Fig. 3.
Effects of bafilomycin and chloroquine on complexes I, II and IV substrate-linked OCR in permeabilized neurons. Neurons were exposed to 10 nM bafilomycin (baf) or 40 µM chloroquine (CQ) for 24 h. (A) The activities of complex I were determined in neurons permeabilized with PMP in the presence of complex I substrates (pyruvate and malate) utilizing either ADP or FCCP. (B) Complex II linked activities were determined in similar fashion utilizing complex II substrate succinate in the presence of either ADP or FCCP. (C) Complex IV linked activities were again measured using ascorbate and TMPD as substrates with ADP in permeabilized cells. *p<0.05 compared to control, from 3 independent experiments. Shown is the representative experiment with n≥3 independent samples, Data=mean±SEM.
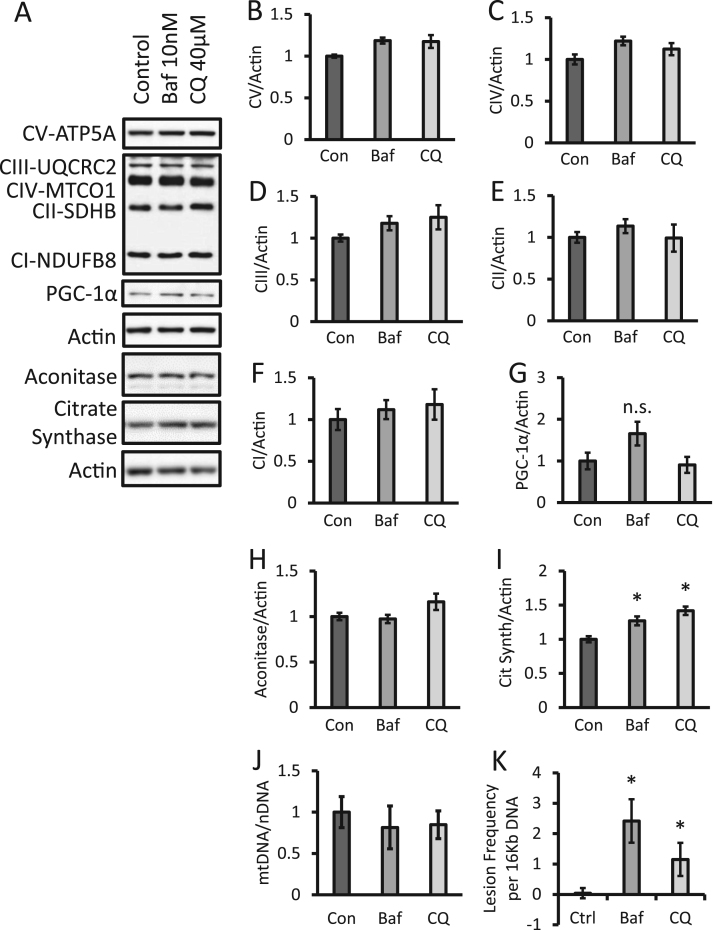
3.4. Effects autophagy inhibitors on levels of mitochondrial proteins and DNA
Given the significant decrease in all mitochondrial respiratory complexes after 24 h exposure we reasoned this could be due to either a decrease in mitochondrial quality or mitochondrial number. This was tested using a number of approaches. First, we determined if levels of selected mitochondrial electron transport chain complexes and Krebs cycle enzymes were altered by autophagy inhibitors after a 24 h exposure (Fig. 4A). No significant changes in representative subunits of mitochondrial complexes I-V were observed (Fig. 4B-F). Using these data and the respiratory chain complex activities (Fig. 3), a normalized activity was calculated for each complex. Supplemental figure 3 shows the complex I, II and IV activities normalized to protein and demonstrates that they are suppressed by bafilomycin or chloroquine by ~40–80% for all the respiratory chain complexes. It does not appear mitochondrial biogenesis is activated since we did not observe any changes on PGC-1α, (Fig. 4G). The levels of Krebs cycle enzyme aconitase did not change, while citrate synthase showed a modest increase in protein levels (Fig. 4H, I). Importantly, no significant changes in mtDNA copy number were observed; however, a significant increase in the amount of mitochondrial DNA damage occurred after bafilomycin or chloroquine exposure for 24 h, consistent with a decrease in mitochondrial quality (Fig. 4J, K).
Fig. 4.
Increased mitochondrial DNA damage without significant changes of PGC-1α, key electron transport chain proteins or mitochondrial DNA copy number in response to bafilomycin or chloroquine. (A) Western blot analysis of subunits of complex I-V, PGC-1α, aconitase, and citrate synthase, in lysates from cells exposed to 10 nM bafilomycin or 40 µM chloroquine for 24 h. (B-I) Relative quantification of western blots (A). (J) Analysis of mitochondrial DNA copy number normalized to nuclear DNA. (K) Analysis of mitochondrial DNA damage. *p<0.05 compared to control, n≥3 independent samples and treatment from 1 experiment, Data=mean±SEM.
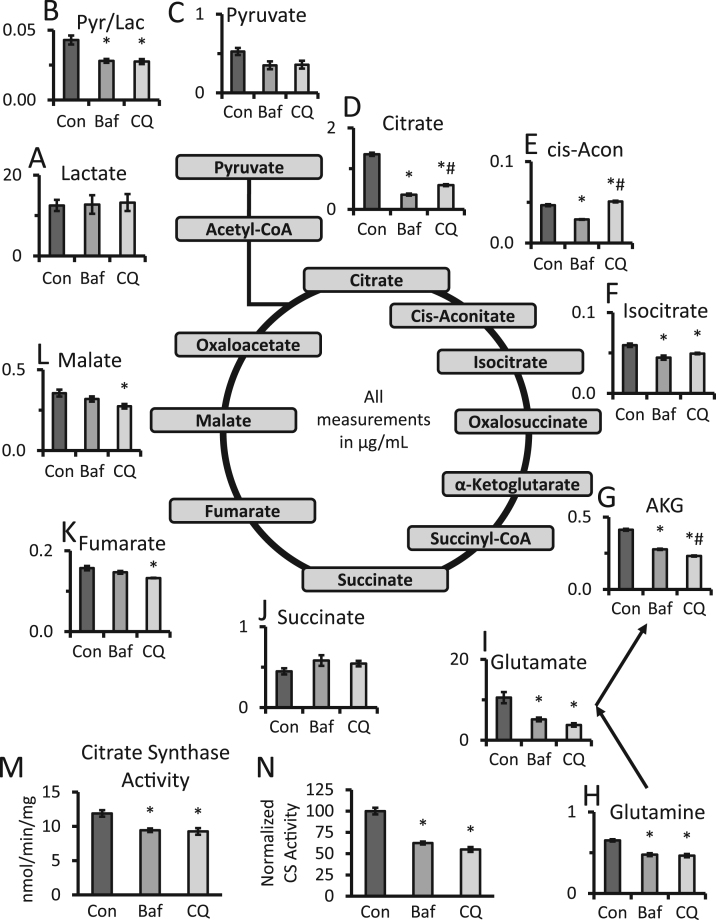
3.5. Effects autophagy inhibitors on Krebs cycle intermediates
We next determined the level of Krebs cycle metabolites and lactate using a targeted metabolomics approach (Fig. 5). We detected no significant changes in lactate levels but observed a trend towards decreased pyruvate levels after bafilomycin and chloroquine exposure. The pyruvate to lactate ratio was then calculated and significant decreases of similar levels were observed after both bafilomycin and chloroquine exposure (Fig. 5A-C). Citrate, cis-aconitate and isocitrate levels were also measured, with significant decreases of both citrate and isocitrate in response to both bafilomycin and chloroquine. Interestingly, cis-aconitate was decreased by bafilomycin but increased by chloroquine (Fig. 5D-F). Key components of glutaminolysis, including α-ketoglutarate, glutamate, and glutamine, were significantly decreased by both bafilomycin and chloroquine while succinate levels remained unchanged (Fig. 5G-J). Fumarate and malate were also decreased but achieved significance with chloroquine only (Fig. 5K, L). To determine the potential mechanism behind the decrease of Krebs cycle intermediates in response to bafilomycin or chloroquine, we measured citrate synthase activities and found that it significantly decreased by ~20% after either bafilomycin or chloroquine exposure (Fig. 5M). Furthermore, we normalized its activity to levels of citrate synthase protein present in the cells, and found an even further decrease of ~40%, consistent with a substantially lower level of active protein (Fig. 5N).
Fig. 5.
Effects of bafilomycin and chloroquine on Krebs cycle metabolites. (A-L) Targeted metabolomics of primary cortical neurons exposed to 10 nM bafilomycin or 40 µM chloroquine for 24 h. Levels of metabolites were shown in μg/mL. *p<0.05 compared to control, #p<0.05 compared between bafilomycin and chloroquine, n=5 samples. (M) Measurement of citrate synthase activity after 24 h exposure to bafilomycin or chloroquine. Activities were normalized to total protein. (N) Citrate synthase activities were normalized to relative citrate synthase protein levels shown in Fig. 4A, I, and expressed as a percentage of untreated controls. *p<0.05 compared to control, n=3–5 independent samples and treatment from 1 experiment, Data=mean±SEM.
4. Discussion
Autophagy gene Atg7 knockout in skeletal muscle, mouse embryonic fibroblasts and pancreatic β cells has been previously found to exhibit decreased state IV, State III, or basal, ATP-linked, and maximal activities. Atg7 knockout mouse embryonic fibroblasts also exhibited increased DCFDA fluorescence suggesting increased production of reactive oxygen species, and Atg7 knockout pancreatic β cells exhibited accumulation of swollen dysmorphic mitochondria [14]. This suggests that autophagy failure leads to a decrease in mitochondrial quality. In this study we have provided the first direct comparison on the effects of two widely used autophagy inhibitors, bafilomycin and chloroquine, on bioenergetics.
Attenuated autophagy has been reported to accompany cellular bioenergetic dysfunction in various genetic and oxidative stress models [48], [49], [50], [51]. Previous studies suggested that bafilomycin has a direct effect on mitochondrial function [20], [21]. In our study, neither bafilomycin nor chloroquine for 32 min changed OCR in intact cells (Supplemental Figure 2A). These data indicate that the observed deficits in mitochondria function are not due to off target effects of the compounds on mitochondria but rather through autophagy failure resulting in accumulation of damaged mitochondria. Our studies demonstrate that in neurons, a 24 h exposure to autophagy inhibitors also inhibits parameters of mitochondrial function (Fig. 2), even in the absence of p62 accumulation, as the case in chloroquine exposure. Furthermore, decreases were observed for complex I, II, and IV substrate-linked respiration in permeabilized cells, indicating widespread deficiencies in mitochondrial electron transport chain function (Fig. 3). These deficiencies occurred without any changes in PGC-1α, mitochondrial electron transport chain proteins, or matrix proteins except for an increase in citrate synthase and a trend toward increased aconitase protein which did not reach significance (Fig. 4A-I). However, while citrate synthase protein levels were modestly increased, its activity was diminished and its normalized activity, was diminished even further (Fig. 4M, N). Mitochondrial DNA damage was also increased (Fig. 4J, K). Taken together, these observations are all consistent with a decrease in mitochondrial quality.
Cellular metabolic programs play important roles in health and disease [44], [52], [53], [54]. How metabolic pathways respond to autophagy inhibition have not been fully investigated. In the metabolomics analysis we observed that pyruvate or lactate levels themselves showed no significant change, the more sensitive measure of the relative activity of glycolysis to mitochondrial function, the pyruvate/lactate ratio, was decreased after bafilomycin or chloroquine exposure, consistent with a relative increase in glycolysis (Fig. 5A-C and Supplemental figure 1). Furthermore, Krebs cycle intermediates, including citrate, cis-aconitate, and isocitrate are diminished. This is likely resulting from the inhibition of citrate synthase and its inability to provide substrates for downstream processing which was confirmed by direct measurement of citrate synthase activity (Fig. 5D-F). The differences in cis-aconitate levels in response to bafilomycin and chloroquine also suggest a possible decrease in aconitase activity. Additionally, key components of glutaminolysis, α-ketoglutarate, glutamate, and glutamine, were decreased while succinate, fumarate and malate have approximately been restored (Fig. 5G-L). This observation would be consistent with an increase in glutaminolysis in an attempt to restore downstream intermediates.
In summary, at a concentration and duration of exposure that led to a similar level of accumulation of LC3-II, we found that both bafilomycin and chloroquine also decreased parameters of mitochondrial function in intact neurons. The levels of representative mitochondrial electron transport chain and mitochondrial matrix proteins were essentially unchanged by bafilomycin or chloroquine, but all showed a decrease in normalized activity suggesting multiple deficits in mitochondrial enzymes in oxidative phosphorylation and the TCA cycle. Furthermore, mtDNA levels were unchanged but damage was increased. These findings are consistent with a secondary effect on bioenergetics by bafilomycin and chloroquine due to autophagy inhibition and a resulting decrease in mitochondrial quality.
Acknowledgment
This work was supported by UAB Health Service Foundation General Endowment Fund, UAB-UCSD O′Brien Acute Kidney Injury Center (P30 DK079337), UAB Blue Sky Program, and NIH (NIHR01-NS064090).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2016.11.004.
Appendix A. Supplementary material
Supplementary material Supplemental Figure 1: Extracellular acidification rate is decreased after autophagy modulator treatment. (A) Extracellular acidification rate (ECAR) was determined from primary cortical neurons exposed to bafilomycin (10 nM) and chloroquine (40 µM) for 24 h and were then subjected to a mitochondrial stress test consisting of the sequential injection of Oligomycin (O, 1 µg/mL), FCCP (F, 1 µM), and Antimycin A (A, 10 µM). (B) ECAR from (A) versus OCR from Figure 2A were plotted. (C) The OCR and ECAR ratios were also determined in these conditions. *p<0.05 compared to control, from 3 independent experiments. Shown is the representative experiment with n≥3 independent samples as in Figure 2, Data=mean±SEM. Supplemental Figure 2: No change in parameters of bioenergetics in intact neurons after short-term exposure to bafilomycin (Baf) and chloroquine (CQ). (A) Mitochondrial oxygen consumption rate (OCR) was measured for 4 consecutive readings and then 100 nM baf and 40 µM CQ were injected. OCR was measured for an additional 4 readings. A mitochondrial stress test consisting of sequential injection of oligomycin (O), FCCP (F), and Antimycin A (A) was performed. No significant differences in OCR were observed, from 3 independent experiments. Shown is the representative of one experiment with data=mean±SEM n≥3 independent samples. (B) Western blot analysis of primary neurons exposed to baf and CQ 32 min after direct injection. (C-E) Quantification of western blots (B), n≥3 independent culture and treatment from 1 experiment. (F) Trypan blue exclusion assessment of cell viability of primary neurons exposed to baf or CQ for 32 min, n≥3 independent culture and treatment from 1 experiment, Data=mean±SEM. Supplemental Figure 3: Normalized activities of mitochondrial complexes are decreased after exposure to bafilomycin (Baf) and chloroquine (CQ). (A, B) Mitochondrial complex normalized activity was calculated by normalizing complex I and complex II substrate-linked OCRs in the presence of either ADP or FCCP (Figure 3) to subunits of mitochondrial complex protein levels determined in Figure 4. (C) Normalized activity of Complex IV was determined in the presence of ADP (Figure 3) and normalized to protein levels as in Figure 4. *p<0.05 compared to control, OCR data were from 3 independent experiments. Shown is the representative of one experiment with n≥3 independent samples as in Figure 3. Western blot analyses were from n≥3 independent culture and treatment from 1 experiment as in Figure 4, Data=mean±SEM.
.
References
- 1.Dodson M., Darley-Usmar V., Zhang J. Cellular metabolic and autophagic pathways: traffic control by redox signaling. Free Radic. Biol. Med. 2013;63:207–221. doi: 10.1016/j.freeradbiomed.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee J., Giordano S., Zhang J. Autophagy, mitochondria and oxidative stress: cross-talk and redox signalling. Biochem. J. 2012;441(2):523–540. doi: 10.1042/BJ20111451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yorimitsu T., Klionsky D.J. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12(Suppl 2):1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klionsky D.J., Emr S.D. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290(5497):1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang J. Autophagy and mitophagy in cellular damage control. Redox Biol. 2013;1(1):19–23. doi: 10.1016/j.redox.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang J. Teaching the basics of autophagy and mitophagy to redox biologists--mechanisms and experimental approaches. Redox Biol. 2015;4:242–259. doi: 10.1016/j.redox.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Slater A.F., Cerami A. Inhibition by chloroquine of a novel haem polymerase enzyme activity in malaria trophozoites. Nature. 1992;355(6356):167–169. doi: 10.1038/355167a0. [DOI] [PubMed] [Google Scholar]
- 8.Trape J.F., Pison G., Preziosi M.P., Enel C., Desgrees du Lou A., Delaunay V., Samb B., Lagarde E., Molez J.F., Simondon F. Impact of chloroquine resistance on malaria mortality. C. R. Acad. Sci. III. 1998;321(8):689–697. doi: 10.1016/s0764-4469(98)80009-7. [DOI] [PubMed] [Google Scholar]
- 9.Kimura T., Takabatake Y., Takahashi A., Isaka Y. Chloroquine in cancer therapy: a double-edged sword of autophagy. Cancer Res. 2013;73(1):3–7. doi: 10.1158/0008-5472.CAN-12-2464. [DOI] [PubMed] [Google Scholar]
- 10.Hook G., Jacobsen J.S., Grabstein K., Kindy M., Hook V. Cathepsin B is a new drug target for traumatic brain injury therapeutics: evidence for E64d as a promising lead drug candidate. Front. Neurol. 2015;6:178. doi: 10.3389/fneur.2015.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hook G., Hook V., Kindy M. The cysteine protease inhibitor, E64d, reduces brain amyloid-beta and improves memory deficits in Alzheimer's disease animal models by inhibiting cathepsin B, but not BACE1, beta-secretase activity. J. Alzheimers Dis. 2011;26(2):387–408. doi: 10.3233/JAD-2011-110101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hook G., Yu J., Toneff T., Kindy M., Hook V. Brain pyroglutamate amyloid-beta is produced by cathepsin B and is reduced by the cysteine p inhibitor E64d, representing a potential Alzheimer's disease therapeutic. J. Alzheimers Dis. 2014;41(1):129–149. doi: 10.3233/JAD-131370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klionsky D.J., Abdalla F.C., Abeliovich H., Abraham R.T., Acevedo-Arozena A., Adeli K., Agholme L., Agnello M., Agostinis P., Aguirre-Ghiso J.A. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8(4):445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu J.J., Quijano C., Chen E., Liu H., Cao L., Fergusson M.M., Rovira I.I., Gutkind S., Daniels M.P., Komatsu M. Mitochondrial dysfunction and oxidative stress mediate the physiological impairment induced by the disruption of autophagy. Aging. 2009;1(4):425–437. doi: 10.18632/aging.100038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bowman E.J., Siebers A., Altendorf K. Bafilomycins: a class of inhibitors of membrane ATPases from microorganisms, animal cells, and plant cells. Proc. Natl. Acad. Sci. U.S.A. 1988;85(21):7972–7976. doi: 10.1073/pnas.85.21.7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Werner G., Hagenmaier H., Drautz H., Baumgartner A., Zahner H. Metabolic products of microorganisms. 224. Bafilomycins, a new group of macrolide antibiotics. Production, isolation, chemical structure and biological activity. J. Antibiot. 1984;37(2) doi: 10.7164/antibiotics.37.110. (110–17) [DOI] [PubMed] [Google Scholar]
- 17.Yoshimori T., Yamamoto A., Moriyama Y., Futai M., Tashiro Y. Bafilomycin A1, a specific inhibitor of vacuolar-type H(+)-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J. Biol. Chem. 1991;266(26):17707–17712. [PubMed] [Google Scholar]
- 18.Yamamoto A., Tagawa Y., Yoshimori T., Moriyama Y., Masaki R., Tashiro Y. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct. Funct. 1998;23(1):33–42. doi: 10.1247/csf.23.33. [DOI] [PubMed] [Google Scholar]
- 19.Klionsky D.J., Elazar Z., Seglen P.O., Rubinsztein D.C. Does bafilomycin A1 block the fusion of autophagosomes with lysosomes? Autophagy. 2008;4(7):849–850. doi: 10.4161/auto.6845. [DOI] [PubMed] [Google Scholar]
- 20.Teplova V.V., Tonshin A.A., Grigoriev P.A., Saris N.E., Salkinoja-Salonen M.S. Bafilomycin A1 is a potassium ionophore that impairs mitochondrial functions. J. Bioenerg. Biomembr. 2007;39(4):321–329. doi: 10.1007/s10863-007-9095-9. [DOI] [PubMed] [Google Scholar]
- 21.Zhdanov A.V., Dmitriev R.I., Papkovsky D.B. Bafilomycin A1 activates respiration of neuronal cells via uncoupling associated with flickering depolarization of mitochondria. Cell Mol. Life Sci. 2011;68(5):903–917. doi: 10.1007/s00018-010-0502-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ochiai H., Sakai S., Hirabayashi T., Shimizu Y., Terasawa K. Inhibitory effect of bafilomycin A1, a specific inhibitor of vacuolar-type proton pump, on the growth of influenza A and B viruses in MDCK cells. Antivir. Res. 1995;27(4):425–430. doi: 10.1016/0166-3542(95)00040-s. [DOI] [PubMed] [Google Scholar]
- 23.Yeganeh B., Ghavami S., Kroeker A.L., Mahood T.H., Stelmack G.L., Klonisch T., Coombs K.M., Halayko A.J. Suppression of influenza A virus replication in human lung epithelial cells by noncytotoxic concentrations bafilomycin A1. Am. J. Physiol. Lung Cell. Mol. Physiol. 2015;308(3):L270–L286. doi: 10.1152/ajplung.00011.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hernandez-Breijo B., Monserrat J., Roman I.D., Gonzalez-Rodriguez A., Fernandez-Moreno M.D., Lobo M.V., Valverde A.M., Gisbert J.P., Guijarro L.G. Azathioprine desensitizes liver cancer cells to insulin-like growth factor 1 and causes apoptosis when it is combined with bafilomycin A1. Toxicol. Appl. Pharmacol. 2013;272(3):568–578. doi: 10.1016/j.taap.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 25.Ohta T., Arakawa H., Futagami F., Fushida S., Kitagawa H., Kayahara M., Nagakawa T., Miwa K., Kurashima K., Numata M. Bafilomycin A1 induces apoptosis in the human pancreatic cancer cell line Capan-1. J. Pathol. 1998;185(3):324–330. doi: 10.1002/(SICI)1096-9896(199807)185:3<324::AID-PATH72>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 26.Lim J.H., Park J.W., Kim M.S., Park S.K., Johnson R.S., Chun Y.S. Bafilomycin induces the p21-mediated growth inhibition of cancer cells under hypoxic conditions by expressing hypoxia-inducible factor-1alpha. Mol. Pharmacol. 2006;70(6):1856–1865. doi: 10.1124/mol.106.028076. [DOI] [PubMed] [Google Scholar]
- 27.Homewood C.A., Warhurst D.C., Peters W., Baggaley V.C. Lysosomes, pH and the anti-malarial action of chloroquine. Nature. 1972;235(5332):50–52. doi: 10.1038/235050a0. [DOI] [PubMed] [Google Scholar]
- 28.Chaanine A.H., Gordon R.E., Nonnenmacher M., Kohlbrenner E., Benard L., Hajjar R.J. High-dose chloroquine is metabolically cardiotoxic by inducing lysosomes and mitochondria dysfunction in a rat model of pressure overload hypertrophy. Physiol. Rep. 2015;3:7. doi: 10.14814/phy2.12413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J., Darley-Usmar V. In: Mitochondrial Dysfunction in Neurodegenerative Disorders. Reeve A.K., Krishnan K.J., Duchen M.R., Turnbull D.M., editors. Springer; London: 2012. pp. 95–111. [Google Scholar]
- 30.Mitchell T., Chacko B., Ballinger S.W., Bailey S.M., Zhang J., Darley-Usmar V. Convergent mechanisms for dysregulation of mitochondrial quality control in metabolic disease: implications for mitochondrial therapeutics. Biochem. Soc. Trans. 2013;41(1):127–133. doi: 10.1042/BST20120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Redmann M., Dodson M., Boyer-Guittaut M., Darley-Usmar V., Zhang J. Mitophagy mechanisms and role in human diseases. Int. J. Biochem. Cell Biol. 2014;53:127–133. doi: 10.1016/j.biocel.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hill B.G., Benavides G.A., Lancaster J.R., Jr., Ballinger S., Dell'Italia L., Jianhua Z., Darley-Usmar V.M. Integration of cellular bioenergetics with mitochondrial quality control and autophagy. Biol. Chem. 2012;393(12):1485–1512. doi: 10.1515/hsz-2012-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Butow R.A., Avadhani N.G. Mitochondrial signaling: the retrograde response. Mol. Cell. 2004;14(1):1–15. doi: 10.1016/s1097-2765(04)00179-0. [DOI] [PubMed] [Google Scholar]
- 34.Goldenthal M.J., Marin-Garcia J. Mitochondrial signaling pathways: a receiver/integrator organelle. Mol. Cell. Biochem. 2004;262(1–2):1–16. doi: 10.1023/b:mcbi.0000038228.85494.3b. [DOI] [PubMed] [Google Scholar]
- 35.Gupta S. Molecular signaling in death receptor and mitochondrial pathways of apoptosis (Review) Int. J. Oncol. 2003;22(1):15–20. [PubMed] [Google Scholar]
- 36.Fetterman J.L., Pompilius M., Westbrook D.G., Uyeminami D., Brown J., Pinkerton K.E., Ballinger S.W. Developmental exposure to second-hand smoke increases adult atherogenesis and alters mitochondrial DNA copy number and deletions in apoE(-/-) mice. PloS One. 2013;8(6):e66835. doi: 10.1371/journal.pone.0066835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giordano S., Dodson M., Ravi S., Redmann M., Ouyang X., Darley Usmar V.M., Zhang J. Bioenergetic adaptation in response to autophagy regulators during rotenone exposure. J. Neurochem. 2014 doi: 10.1111/jnc.12844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boyer-Guittaut M., Poillet L., Liang Q., Bole-Richard E., Ouyang X., Benavides G.A., Chakrama F.Z., Fraichard A., Darley-Usmar V.M., Despouy G. The role of GABARAPL1/GEC1 in autophagic flux and mitochondrial quality control in MDA-MB-436 breast cancer cells. Autophagy. 2014;10(6):986–1003. doi: 10.4161/auto.28390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schapira A.H., Gegg M. Mitochondrial contribution to Parkinson's disease pathogenesis. Park. Dis. 2011;2011:159160. doi: 10.4061/2011/159160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Knight-Lozano C.A., Young C.G., Burow D.L., Hu Z.Y., Uyeminami D., Pinkerton K.E., Ischiropoulos H., Ballinger S.W. Cigarette smoke exposure and hypercholesterolemia increase mitochondrial damage in cardiovascular tissues. Circulation. 2002;105(7):849–854. doi: 10.1161/hc0702.103977. [DOI] [PubMed] [Google Scholar]
- 42.Dranka B.P., Benavides G.A., Diers A.R., Giordano S., Zelickson B.R., Reily C., Zou L., Chatham J.C., Hill B.G., Zhang J. Assessing bioenergetic function in response to oxidative stress by metabolic profiling. Free Radic. Biol. Med. 2011;51(9):1621–1635. doi: 10.1016/j.freeradbiomed.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salabei J.K., Gibb A.A., Hill B.G. Comprehensive measurement of respiratory activity in permeabilized cells using extracellular flux analysis. Nat. Protoc. 2014;9(2):421–438. doi: 10.1038/nprot.2014.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith M.R., Vayalil P.K., Zhou F., Benavides G.A., Beggs R.R., Golzarian H., Nijampatnam B., Oliver P.G., Smith R.A., Murphy M.P. Mitochondrial thiol modification by a targeted electrophile inhibits metabolism in breast adenocarcinoma cells by inhibiting enzyme activity and protein levels. Redox Biol. 2016;8:136–148. doi: 10.1016/j.redox.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mizushima N., Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3(6):542–545. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- 46.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19(21):5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benavides G.A., Liang Q., Dodson M., Darley-Usmar V., Zhang J. Inhibition of autophagy and glycolysis by nitric oxide during hypoxia-reoxygenation impairs cellular bioenergetics and promotes cell death in primary neurons. Free Radic. Biol. Med. 2013;65:1215–1228. doi: 10.1016/j.freeradbiomed.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dodson M., Liang Q., Johnson M.S., Redmann M., Fineberg N., Darley-Usmar V.M., Zhang J. Inhibition of glycolysis attenuates 4-hydroxynonenal-dependent autophagy and exacerbates apoptosis in differentiated SH-SY5Y neuroblastoma cells. Autophagy. 2013;9:12. doi: 10.4161/auto.26094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hjelmeland A., Zhang J. Metabolic, autophagic, and mitophagic activities in cancer initiation and progression. Biomed. J. 2016;39(2):98–106. doi: 10.1016/j.bj.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liang Q., Benavides G.A., Vassilopoulos A., Gius D., Darley-Usmar V., Zhang J. Bioenergetic and autophagic control by Sirt3 in response to nutrient deprivation in mouse embryonic fibroblasts. Biochem. J. 2013;454(2):249–257. doi: 10.1042/BJ20130414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Redmann M., Darley-Usmar V., Zhang J. The role of autophagy, mitophagy and lysosomal functions in modulating bioenergetics and survival in the context of redox and proteotoxic damage: implications for neurodegenerative diseases. Aging Dis. 2016;7(2):150–162. doi: 10.14336/AD.2015.0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Z., Klipfell E., Bennett B.J., Koeth R., Levison B.S., Dugar B., Feldstein A.E., Britt E.B., Fu X., Chung Y.M. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472(7341):57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Z., Tang W.H., Cho L., Brennan D.M., Hazen S.L. Targeted metabolomic evaluation of arginine methylation and cardiovascular risks: potential mechanisms beyond nitric oxide synthase inhibition. Arterioscler Thromb. Vasc. Biol. 2009;29(9):1383–1391. doi: 10.1161/ATVBAHA.109.185645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chouchani E.T., Pell V.R., Gaude E., Aksentijevic D., Sundier S.Y., Robb E.L., Logan A., Nadtochiy S.M., Ord E.N., Smith A.C. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature. 2014;515(7527):431–435. doi: 10.1038/nature13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material Supplemental Figure 1: Extracellular acidification rate is decreased after autophagy modulator treatment. (A) Extracellular acidification rate (ECAR) was determined from primary cortical neurons exposed to bafilomycin (10 nM) and chloroquine (40 µM) for 24 h and were then subjected to a mitochondrial stress test consisting of the sequential injection of Oligomycin (O, 1 µg/mL), FCCP (F, 1 µM), and Antimycin A (A, 10 µM). (B) ECAR from (A) versus OCR from Figure 2A were plotted. (C) The OCR and ECAR ratios were also determined in these conditions. *p<0.05 compared to control, from 3 independent experiments. Shown is the representative experiment with n≥3 independent samples as in Figure 2, Data=mean±SEM. Supplemental Figure 2: No change in parameters of bioenergetics in intact neurons after short-term exposure to bafilomycin (Baf) and chloroquine (CQ). (A) Mitochondrial oxygen consumption rate (OCR) was measured for 4 consecutive readings and then 100 nM baf and 40 µM CQ were injected. OCR was measured for an additional 4 readings. A mitochondrial stress test consisting of sequential injection of oligomycin (O), FCCP (F), and Antimycin A (A) was performed. No significant differences in OCR were observed, from 3 independent experiments. Shown is the representative of one experiment with data=mean±SEM n≥3 independent samples. (B) Western blot analysis of primary neurons exposed to baf and CQ 32 min after direct injection. (C-E) Quantification of western blots (B), n≥3 independent culture and treatment from 1 experiment. (F) Trypan blue exclusion assessment of cell viability of primary neurons exposed to baf or CQ for 32 min, n≥3 independent culture and treatment from 1 experiment, Data=mean±SEM. Supplemental Figure 3: Normalized activities of mitochondrial complexes are decreased after exposure to bafilomycin (Baf) and chloroquine (CQ). (A, B) Mitochondrial complex normalized activity was calculated by normalizing complex I and complex II substrate-linked OCRs in the presence of either ADP or FCCP (Figure 3) to subunits of mitochondrial complex protein levels determined in Figure 4. (C) Normalized activity of Complex IV was determined in the presence of ADP (Figure 3) and normalized to protein levels as in Figure 4. *p<0.05 compared to control, OCR data were from 3 independent experiments. Shown is the representative of one experiment with n≥3 independent samples as in Figure 3. Western blot analyses were from n≥3 independent culture and treatment from 1 experiment as in Figure 4, Data=mean±SEM.