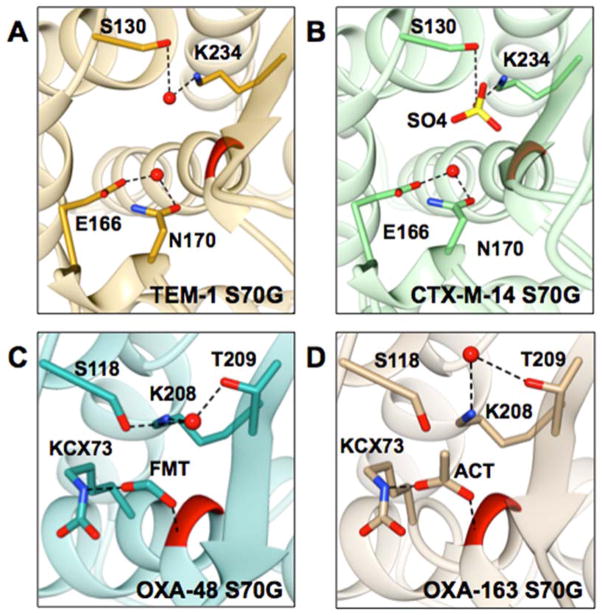
Figure 7.
Representations of the active sites of class A and class D S70G mutants. Position 70 is colored in red, selected residues and respective anions are represented in stick and labeled; water molecules are represented as red balls, and dashed lines represent selected interactions. A) TEM-1 S70G (PDB ID: 1ZG6) is shown in gold, the deacylating water is coordinated by E166 and N170. Stec et al. proposed that the water molecule coordinated by S130 and K234 is involved in the hydrolysis mechanism of the S70G mutant.57 B) CTX-M-14 S70G (PDB ID: 4PM6) is shown in green, the deacylating water is coordinated by E166 and N170. S130 and K234 coordinate a sulfate ion that is shown in yellow. C) OXA-48 S70G (PDB ID: 5HAQ) is shown in dark cyan. S118, K208, and T209 coordinate a water molecule. Additionally, a formate ion is coordinated by the N-carbamylated K73 and the –NH of G70. D) OXA-163 S70G (PDB ID: 5HAR) is shown in tan. K208 and T209 coordinate a water molecule. Also, an acetate ion is coordinated by N-carbamylated K73 and the –NH of G70.