Abstract
Objective
To investigate the significance of functional polymorphisms of inflammatory response genes by analysis of a large population of patients, both with and without severe sepsis, and representative of the diverse populations (geographic diversity, physician diversity, clinical treatment diversity) that would be encountered in critical care clinical practice.
Design
Collaborative case-control study conducted from July 2001 to December 2005.
Setting
A heterogeneous population of patients from 12 USA intensive care units (ICUs) represented by the Genetic Predisposition to Severe Sepsis (GenPSS) archive.
Patients
Eight hundred and fifty-four patients with severe sepsis and an equal number of mortality, age, gender, and race-matched patients also admitted to the ICU without evidence of any infection (matched nonseptic controls).
Measurements and Main Results
We developed assays for six functional single nucleotide polymorphisms (SNPs) present before the first codon of TNF at −308, IL1B at −511, IL6 at −174, IL10 at −819, and CD14 at −159, and in the first intron of LTA (also known as TNF-β) at +252 (LTA(+252)). The Project IMPACT™ critical care clinical database information management system developed by the Society of Critical Care Medicine and managed by Tri-Analytics, Inc. and Cerner Corporation was utilized. Template-directed dye-terminator incorporation assay with fluorescence polarization detection was used as a high-throughput genotyping strategy. Fifty-three percent of the patients were male with 87.3 % and 6.4 % of Caucasian and African American racial types, respectively. Overall mortality was 35.1 % in both severe sepsis (SS) and matched nonseptic control (MC) patients group. Average ages (SD) of the SS and MC patients were 63.0 (16.05) and 65.0 (15.58) years old, respectively. Among the 6 SNPs, LTA(+252) was most over-represented in the septic patient group (% severe sepsis; AA 45.6: AG 51.1: GG 56.7, P = .005). Moreover, the genetic risk effect was most pronounced in males, age > 60 yrs (P = .005).
Conclusions
LTA(+252) may influence predisposition to severe sepsis, a predisposition that is modulated by gender and age. Although the genetic influences can be overwhelmed by both comorbid factors and acute illness in individual cases, population studies suggest that this is an influential biological pathway modulating risk of critical illnesses.
Keywords: Genetic Predisposition to Disease, Sepsis, Lymphotoxin-alpha, Gender, Aging
Sepsis remains a critical and costly public health problem. In the United States, more than 750,000 patients become septic each year, and more than 25% of these will die [1]. The annual total cost for care of sepsis in 1995 exceeded $16 billion [1] and patients suspected with severe sepsis are reported to account for 500,000 emergency department visits annually [2]. Recently, an evidence-based system for assessing quality of evidence of sepsis diagnosis and strength of recommendations for treatment [3, 4] has been used to develop an aggressive strategy for intervention for patients with this complication (Surviving Sepsis Campaign [www.survivingsepsis.com]). It is difficult, however, to identify prognostic factors that may predict response to the therapies implemented for severe sepsis. Advances in diagnosis, intervention, and prognostication of sepsis increasingly rely on tools that were recently confined to research laboratories much as molecular DNA typing, quantitative reverse transcriptase PCR and ELISA assays.
Predisposition is complementary to diagnosis, intervention and prognostication. ‘Human Genetic Variation’ was selected as the ‘Breakthrough of the year 2007′ [5] and has been promoted as a strategy to identify patients predisposed to sepsis and other critical illnesses [6-11]. It is believed that the mortality of patients with severe sepsis correlates with markers of inflammatory responses [12-14] expressed as inflammatory cytokine levels in the blood. The inflammatory cytokine production of intensive care unit (ICU) patients with sepsis has been reported to be largely affected by interleukin-1 (IL-1) and tumor necrosis factor (TNF) related genetic polymorphisms in a single population study [10].
Assessing genetic predisposition is more challenging in population studies than the investigation of inheritance of Mendelian traits within specific kindreds. The challenges are related to the underlying composition of the populations in population association studies as well as the specific multigenic disease, trait or response under investigation. Sample sizes are often inadequate to achieve significant statistical power for predictive purposes and there is often reluctance to attribute seemingly significant results to chance [15, 16]. Further complicating the analyses is the fact that many genetic polymorphisms demonstrate ethnic differences in allele distributions [17]. This may be a factor in sepsis research because African American patients have been reported to have a higher population-based incidence of severe sepsis than either Caucasians or Hispanics, possibly attributable to differences in clinical, social, geographic, or access to healthcare services in the United States ICUs [18]. Gender of the patient with sepsis is also implicated as a prognostic factor for severe sepsis in both clinical [1, 19] and basic science studies [20]. Beneficial effects of estrogen through cytokine modulations of female patients with septic insults have been examined by experimental studies [20, 21]. Meanwhile, human [22, 23] and animal [24] studies have also demonstrated that the elderly experience more severe inflammatory responses and are more vulnerable to severe sepsis than the young due to dysregulation of cytokine storm. However, the biological basis of these differences is more complex than simply the presence or absence of sex hormones and may be modulated by genetic background and inflammatory gene expression [25, 26]. According to experts [15, 27, 28], issues in population genetics studies that complicate the correlation with clinical events in addition to inadequate statistical power include the heterogeneity of population subgroups, difficulty in acquiring matched control groups, genetic factors obscured by overwhelming disease pathophysiology, genetic variants that have undefined pathogenic consequences on gene function, and the low allele frequency of some polymorphic variants.
Herein, we report on functional polymorphisms of six inflammatory response genes following analysis of a large and diverse population of patients both with and without severe sepsis from 12 medical centers in the USA. In this project, we used the Project IMPACT™ (PI) critical care clinical database information management system developed by the Society of Critical Care Medicine (SCCM) to facilitate the prospective collection of clinical samples for genetic analysis using a case-control design. Control patients were selected from those admitted to the same ICUs and having the same gender, age, race and outcome but absent severe sepsis. Simultaneously, we established a high-throughput, cost-effective genotyping of samples by introducing the template-directed dye-terminator incorporation assay with fluorescence polarization (TDI-FP) detection [29]. On the basis of the results of population association, we performed further assessment on the joint association between heritable and acquired factors with conventional tools and with recursive partitioning analysis.
[Materials and Methods]
Design and data source of ‘Genetic Predisposition to Severe Sepsis’ Project
Critically ill patients from 12 intensive care units across the United States were studied as part of the Genetic Predisposition to Severe Sepsis (GenPSS) project. The study was designed to assess the significance of functional polymorphisms of inflammatory response genes by analysis of a large, diverse population of patients both with and without severe sepsis from geographically distinct medical centers with different clinicians and operating procedures. All participating clinical centers utilized the Project IMPACT (PI) database information management system developed by the Society for Critical Care Medicine (SCCM) and managed by Tri-Analytics, Inc. (http://www.trianalytics.com/, accessed 2Jan2009) and Cerner Corporation (Kansas City, MO). This database provided the separation of the patients’ identity and personal health information (PHI) from the central laboratory where genotyping was performed. PHI was scrubbed or masked from patient records by Tri-Analytics prior to delivery of clinical outcome data to the investigators. As the intended consequence of this study design (which also relied upon the use of waste blood, vide infra), the study was approved for conduct with waiver of written informed consent. The project design also meets the confidentiality and specimen access controls required of a typical NIGMS Human Genetic Cell Repository (http://ccr.coriell.org/Sections/Collections/NIGMS/?SsId=8 , accessed 2Jan2009).
Patient Selection and Definitions
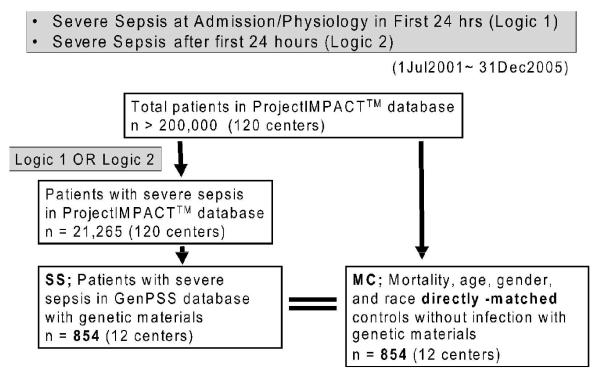
The PI database was queried across the study period of July 2001 to December 2005 to identify specimens of blood submitted from 854 patients with unambiguous severe sepsis (SS) and an equal number of similar mortality, age (matched within a 10-yr margin), gender, and race matched controls (MC) without evidence of any infection during their ICU stay. The diagnosis of severe sepsis was made when a patient met the criteria proposed by the American College of Chest Physicians/Society of Critical Care Medicine Consensus Conference [30]. We used a previously validated approach to the identification of severe sepsis involving the co-occurrence of International Classification of Diseases, Ninth Revision (ICD-9) codes for a bacterial or fungal infectious process and acute organ dysfunction using the following Boolean logic criteria (Fig. 1)[31]:
Figure 1.
Flow chart of patients’ selection in the GenPSS project
GenPSS, Genetic Predisposition to Severe Sepsis
Logic 1—Severe Sepsis at Admission/Physiology Within First 24 hrs: Criteria 1 AND 2 fulfilled
- Severe Sepsis at Admission
-
1a. Acute ICU admission diagnosis (ICD-9 codes); sepsis (septicemia-bacteremia) 038 OR Septic Shock 785.59 OR Shock-Septic 785.59 OR Bacteremia 790. (excludes anaphylaxis)OR
-
1b. APACHE 2 diagnostic criteria; SepsisWITH
-
- Physiology, at least one of
-
2a. Systolic Blood Pressure; < 90mmHg as the lowest recorded systolic blood pressure in the first 24 hours OR use of vasopressor other than dopamine, such as phenylephrine; norepinephrine; and epinephrine. Dopamine is excluded because it continues to be used for purposes other than sepsis therapy.OR
-
2b. Acute renal failure as defined by APACHE 2 [32]; If a new rise in serum creatinine to 1.5 mg/dL or greater with oliguria (<135 mL urine any 8 hour period in the last 48 hours). The patient has to have had a recent period of oliguria. Discharge ICD coding was examined as a validity check to include ARF (584) with oliguria/anuria (788.5).OR
-
2c. Bilirubin > 2.0 mg/dLOR
- 2d. PaO2<90 torr for nonventilated patients receiving supplemental oxygen OR P/F ratio (PaO2/FIO2) < 225 for mechanical ventilated patients.
-
Logic 2—Severe Sepsis developing after admission (beyond 24 hours)
Project IMPACT™ coding as Severe Sepsis as a “complication not related to procedure”, with verified (a) Sepsis, severe (b) Sepsis-induced hypotension OR (c) Septic Shock.
Selection of Matched Controls
Controls were matched as follows: Participating centers submitted similar blood samples from critically ill patients who were not known to be septic, recognizing that some would become septic during the ICU stay. Most who were not septic on admission did not become septic during the ICU stay. All patients who did not meet severe sepsis criteria were placed into the matching pool, and those who had any evidence of infection were then excluded. This left a pool of potential non-sepsis match candidates who had no evidence of infection during their hospitalization as recorded in the PI database. Each septic patient was tentatively matched with all patients in the pool of the same gender, reported race/ethnicity and hospital discharge status (alive versus dead). Age matches were allowed +/− 10 years. Then, each severely septic patient was matched with one patient from its set of potential non-sepsis matches. If there was only one possible match for the septic patient, the match was assigned and then the match was removed from the pool. If more than a single matching patient was identified, then the match was made by selecting one of the potential matches at random, and then removing that match from the pool. The algorithm ran recursively until a match was made for each septic patient. In the event that a match corresponded to a sample that failed to yield DNA that could be analyzed, the algorithm was re-run against remaining matches in the control pool.
Occasionally, patients would have more than one ICU admission during a single hospitalization. If the patient was severely septic on any ICU admission, the patient was judged severely septic for that hospitalization and therefore could not also be used as a matched control.
Specimen Collection and Patient Confidentiality
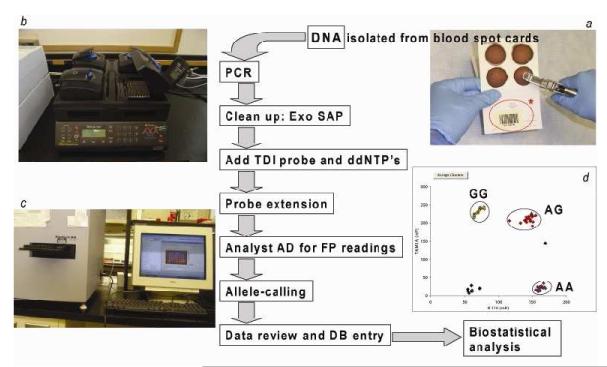
Fig. 2 illustrates our strategy for high-throughput, cost-effective genotyping of samples in this GenPSS project. Waste blood was collected from of ICU patients by participant ICU staff on blood spot cards (FTA®, Cat. No. WB120205, Whatman) as part of routine care. Typically, this waste blood was left in a syringe after sampling from an invasive arterial or central venous catheter. Cards were identified only by a bar code label. An identical bar code label was attached to the PI data sheet for each patient. The filter paper card was air-dried, inserted into a foil pouch (Cat. No. WB100037, Whatman), sealed in a cardboard mailing envelope (Cat. No. WB100016, Whatman), mailed to the genotyping laboratory via regular mail and received at a post office box for retrieval by lab personnel. Upon receipt at the GenPSS study center, the bar codes of the blood spot cards were scanned, entered into the local GenPSS database and then relabeled with a separate S/PAIR number. This S/PAIR re-coded blood spot card was forwarded to the genotyping lab.
Figure 2.
Workflow of genotyping in the GenPSS project
Figure 2 illustrates our strategy for high-throughput, cost-effective genotyping of samples in this GenPSS project. ICUs sent waste blood samples collected on a filter paper card (a). On this card, randomly- generated Bar-coded numbers (in the red circle*) were prepared to protect the confidentiality of individual patient. DNA for genotyping was isolated from these blood stains. First, we amplified the target region of DNA by PCR with primers specific for the sequences of each of our 6 selected markers. After a clean-up step of the PCR products to remove unused primers and nucleotides, we added a unique oligonucleotide probe molecule for the sequence-specific, single nucleotide extension step using dideoxyterminator nucleotides (ddNTPs). This is the TDI-FP as the high-throughput method for SNP analysis. PCR, Exo SAP, TDI reaction procedures were performed using a tetrad thermal cycler (b). FP values were directly measured using an Analyst AD fluorescence reader (c). In the result view which appears in the excel sheet (d), small dots indicate FP values obtained for individual patients’ samples and identification of specific genotype clusters from the TDI reaction.
GenPSS, Genetic Predisposition to Severe Sepsis; PCR, polymerase chain reaction; Exo SAP, E. coli exonuclease 1 and shrimp alkaline phosphatase; TDI, template-directed dye-terminator incorporation; ddNTP, dideoxynucleotide triphosphate; FP, fluorescence polarization; DB, database; SNP, single nucleotide polymorphism.
To promote confidentiality, genotyping lab personnel did not have information about the medical center, physician or patient from which the blood spot card originated. The PI personnel did not have access to genotyping determinations or the local GenPSS database at any time. The information management personnel queried the clinical PI database to identify specimens submitted from patients enrolled in GenPSS at participating ICUs without revealing any protected health information (PHI) to the genotyping lab personnel. Bar code identifiers were selected for PI records that corresponded to patients with severe sepsis and the matched control patients who did not have evidence of any infection at any time during their hospital course.
DNA Isolation
Using a 3 mm paper hole punch (McGill Inc., Marengo, IL), 2 FTA paper discs from the dried blood stain were punched out and placed in each tube of 8-strip thin wall tubes (MIDSCI, St. Louis, MO). DNA Elution Solution (PureGene, Qiagen (formerly Gentra Systems, Inc.) , MD) was added (200 μL per tube) and incubated overnight at 4°C. After incubation, the DNA elution solution was discarded. DNA purification solution (Gentra, Minneapolis, MN) 150 μL was added, incubated for 15 minutes at room temperature, mixed and discarded. This step was repeated 2 to 4 times until a colorless wash was obtained. DNA elution solution was added to the washed paper punches in each tube and incubated for 15 minutes. After removing as much solution as possible, DNA elution buffer was finally added to each tube. The sealed tubes were incubated for 15 minutes at 99°C within a MJ Research 225 Tetrad Thermal Cycler (GMI, Ramsey, MN) to facilitate release of the bound DNA from the paper discs. The eluted DNA was transferred to a fresh tube and stored at 4°C.
Polymerase Chain Reaction
We developed assays for 6 functional SNPs present in inflammation-related genes implicated as mediators of the sepsis cascade. These include SNPs present before the first codon of TNF at −308, IL1B at −511, IL6 at −174, IL10 at −819, and CD14 at −159 (TNF(−308)) [6-8], IL1B(−511) [33-35], IL6 (−174) [36, 37], IL10 (−819) [38, 39], and CD14 (−159) [40, 41]), and in the first intron of lymphotoxin-α (formerly known as TNF-β) at +252 (LTA(+252) [42, 43]. These 6 polymorphisms were selected from 22 candidate markers related to systemic inflammation based on our preliminary analysis of a separate pilot patient cohort. We focused on genes whose products had a known functional role in the inflammatory response [35, 39, 43-47].
Two microliters (μL) of genomic DNA (5 to 10 nanograms) was amplified in a mixture containing 2.5 μL of AmpliTaq Gold® PCR Master Mix (Applied Biosystems, Foster City, CA), 0.05 μL 50 mmol/L mixture of forward and reverse primer (Integrated DNA Technologies, Inc., Coralville, IA), 1.45 μL double-distilled water according to the conditions listed in Table 1a. To optimize the TDI-FP technique, PCR primers and conditions were designed to generate a homogeneous PCR product of 400 bp or smaller using Primer Premier ver. 5.0 software (Biosoft International, Palo Alto, CA). All reactions were performed in 0.2 mL black-skirted 96-well reaction plates (ABgene, Surrey, UK) with the MJ Research 225 Tetrad Thermal Cycler (BioRad, Hercules, CA).
Table 1a.
PCR primers, Product Size, and Conditions
| Gene symbol |
SNP loci | dbSNP rs# clustered ID |
Primersa | PCR conditionsb |
|---|---|---|---|---|
| TNF | TNF(−308) | rs1800629 | TTTCTGAAGCCCCTCCCAGTT (F) CCCAAGGTGAGCAGAGGGAGA (R) |
95°C, 10 min; 44 cycles of (95°C, 30 sec, 60°C, 30 sec, 72°C, 45 sec), 72°C, 5 min |
| LTA | LTA(+252) | rs909253 | CGTGCTTCGTGCTTTGGACTA (F) CCCAAGGTGAGCAGAGGGAGA (R) |
95°C, 10 min; 40 cycles of (95°C, 30 sec, 60°C, 30 sec, 72°C, 45 sec), 72°C, 5 min |
| IL1B | IL1B(−511) | rs16944 | TGGCATTGATCTGGTTCATC (F) GTTTAGGAATCTTCCCACTT (R) |
95°C, 10 min; 40 cycles of (95°C, 30 sec, 60°C, 30 sec, 72°C, 45 sec), 72°C, 5 min |
| IL6 | IL6(−174) | rs1800795 | GCGCTAGCCTCAATGACGACC (F) ATCTTTGTTGGAGGGTGAGGG (R) |
95°C, 10 min; 40 cycles of (95°C, 30 sec, 64°C, 30 sec, 72°C, 45 sec), 72°C, 5 min |
| IL10 | IL10(−819) | rs1800871 | TACAGTAGGGTGAGGAAACC (F) GGTAGTGCTCACCATGACCC (R) |
95°C, 10 min; 40 cycles of (95°C, 30 sec, 62°C, 30 sec, 72°C, 45 sec), 72°C, 5 min |
| CD14 | CD14(−159) | rs2569190 | GCTTAGGCTCCCGAGTCAACA (F) TGTCATTCAGTTCCCTCCTC (R) |
95°C, 10 min; 40 cycles of (95°C, 30 sec, 63°C, 30 sec, 72°C, 45 sec), 72°C, 5 min |
Primers are listed in a 5′-3′ orientation (F, forward primer; R, reverse primer).
Following amplification, PCR products were stored at 4°.
dbSNP rs# clustered IDs are identified in NCBI, National Center for Biotechnology Information. SNP, single nucleotide polymorphism; TNF(−308), SNP at position −308 nucleotides 5′ of the first exon of the tumor necrosis factor-α gene; LTA(+252), SNP at position 252 site of the first intron of the lymphotoxin-α gene; IL1B(−511), SNP at position −511 nucleotides 5′ of the first exon of the interleukin-1β gene; IL6(−174), SNP at position −174 nucleotides before the first exon of the IL-6 gene; IL10(−819), SNP at position −819 nucleotides before the first exon of the IL-10 gene; CD14(−159), SNP at position −159 nucleotides before the first exon of the CD14 gene.
Amplicon Purification
To eliminate unincorporated single-stranded PCR primers and dNTPs which may reduce the specificity of the TDI-FP reaction, E. coli exonuclease 1 (New England Biolabs, Inc., Ipswich, MA) (0.1 μL), shrimp alkaline phosphatase (SAP; Roche Molecular Biochemicals, Indianapolis, IN) (1.0 μL), 10× SAP reaction buffer (0.2 μL), pyrophosphatase (Perkin Elmer Life Sciences, Inc, Boston, MA) (0.15 μL), and double-distilled water (0.55 μL) were added to the total volume of PCR product (6.0 μL) and incubated at 37°C for 60 minutes, followed by 85°C for 25 minutes for enzyme inactivation. Pyrophosphatase was omitted from the purification of the PCR products of TNF(−308) and LTA(+252).
Template-directed Dye-terminator Incorporation Reaction (TDI)
For all assays, we used the single-base extension SNP genotyping kit AcycloPrime™ FP SNP Detection from Perkin Elmer Life Sciences, Inc, Boston, MA. The purified PCR product was combined with 0.05 μL AcycloPol™ thermostabile polymerase, 2.0 μL of 10 × AcycloPrime™ Reaction Buffer, 1.0 μL of specific AcycloTerminator™ mix consisting of R110/TAMRA-labeled dideoxynucleotide pairs for each SNP variant, 1.0 μL of the site-specific TDI oligonucleotide probe, 10 μmol/L (Table 1a; Integrated DNA Technologies, Inc., Coralville, IA), and 8.95 μL double-distilled water. The final reaction volume was 21 μL. The reactions were incubated according to the conditions specified in Table 1b. The TDI oligonucleotide probes were 20 to 30 nucleotides designed to complement the sequence immediately adjacent to the SNP position. Annealing temperatures were selected to be approximately 10°C below the melting temperature of the oligonucleotide probe DNA sequence.
Table 1b.
TDI-FP Oligonucleotide Probes, Dye-Terminator Combinations, and Reaction Conditions
| Gene symbol |
SNP loci | Base change |
TDI-FP probesa | Dye terminator |
TDI-FP reaction conditionsb |
|---|---|---|---|---|---|
| TNF | TNF(−308) | G → A | GAGGCAATAGGTTTTGA GGGGCATG (F) |
G/A | 95°C, 2 min; 30 cycles of (95°C, 15 sec, 62°C, 30 sec) |
| LTA | LTA(+252) | A → G | TGTCACACATTCTCTGTT TCTGCCATG (F) |
G/A | 95°C, 2 min; 25 cycles of (95°C, 15 sec, 61°C, 30 sec) |
| IL1B | IL1B(−511) | C → T | GTCTCTACCTTGGGTGCT GTTCTCTGCCTC (R) |
G/A | 95°C, 2 min; 20 cycles of (95°C, 15 sec, 61°C, 30 sec) |
| IL6 | IL6(−174) | G → C | GTGCAATGTGACGTCCTT TAGCAT (R) |
G/C | 95°C, 2 min; 30 cycles of (95°C, 15 sec, 62°C, 30 sec) |
| IL10 | IL10(−819) | C → T | TGTACCCTTGTACAGGTG ATGTAA (F) |
C/T | 95°C, 2 min; 15 cycles of (95°C, 15 sec, 55°C, 30 sec) |
| CD14 | CD14(−159) | C → T | AATGAAGGATGTTTCAGG GAGGGGG |
G/A | 95°C, 2 min; 25 cycles of (95°C, 15 sec, 55°C, 30 sec) |
Probes are listed in a 5′-3′ orientation (F, forward primer; R, reverse primer).
All reaction products were stored at 4°.
TDI-FP, template-directed dye-terminator incorporation with fluorescence polarization detection; SNP, single nucleotide polymorphism; TNF(−308), SNP at position −308 nucleotides 5′ of the first exon of tumor necrosis factor-α; LTA(+252), SNP at position 252 site of the first intron of lymphotoxin-α; IL1B(−511), SNP at position −511 nucleotides 5′ of the first exon of interleukin-1β; IL6(−174), SNP at position −174 nucleotides before the first exon of IL-6; IL10(−819), SNP at position −819 nucleotides before the first exon of IL-10, CD14(−159), SNP at position −159 nucleotides before the first exon of CD14.
Fluorescence Polarization Determination
Fluorescence polarization (FP) values were directly measured using an Analyst AD fluorescence reader (Molecular Devices, Sunnyvale, CA) with excitation and emission wavelengths of 580 nm and 605 nm for R110, and 552 nm and 575 nm for TAMRA, respectively. For LTA(+252), IL6(−174), and CD14(−159), 0.1 μL of single-stranded DNA binding protein (Epicentre Biotechnologies, Madison, WI), 0.2 μL of 10× SAP reaction buffer, and 1.7 μL water were added to each TDI reaction product and incubated at 37°C for 60 minutes before FP determination. For the remainder of the SNPs analyzed, FP was determined immediately following the TDI reaction.
Between reaction steps (i.e., PCR, amplicon purification, TDI reaction, and incubation with single-stranded DNA binding protein) reaction plates were thermally sealed with adhesive sealing sheets (Brinkmann Instruments, Inc., Westbury, NY) and pulse centrifuged to minimize evaporative loss and cross contamination.
Allele Assignment
Allele assignment was objective and used SNPscorer software (Perkin Elmer Life Science, Inc at www.snpscoring.com, Samples that remained ambiguous following the first analysis were re-analyzed using identical methodology or direct DNA sequencing. Samples were considered of indeterminate genotype if allele assignment could not be made after two repetitions of TDI-FP methodology and sequencing. To confirm the accuracy of our approach, sequence-verified controls for each genotype were incubated in every TDI-FP assay performed. As a validity check, five percent of the TDI-FP calls were verified by conventional DNA sequencing.
Gene and Clinical Data Analysis
In addition to standard strategies, we employed recursive partitioning (RP) as a tool to identify relevant genetic and non-genetic predictors. We used HelixTree™ software (Golden Helix Inc, Bozeman, MT) for RP analysis. RP can be most easily viewed as conditional feature finding. Once a split is made based upon one gene/clinical feature, then the subsequent analysis is conditional on the presence or absence of that gene or clinical feature. Examination of a particular clinical outcome suggests that the effect of any feature will be, at least in part, dependent on the presence/absence of other features.
Statistical Analysis
Hardy-Weinberg equilibrium for the population distribution of the variant alleles was determined according to the approach described by Guo and Thompson [48]. Allelic chi-squares were also examined for each SNP. Strict Bonferroni correction is considered to be overly conservative, therefore a Bayesian formula was applied to obtain 0.95 posterior probability of a correct influence of association to a particular gene. In preliminary analysis, we considered differences significant at P < .05. In order to improve the likelihood that observed differences were not the result of a Type 1 error, we set final significance at P < .01.
Results
Characteristics of Severely Septic and Matched Control Patients
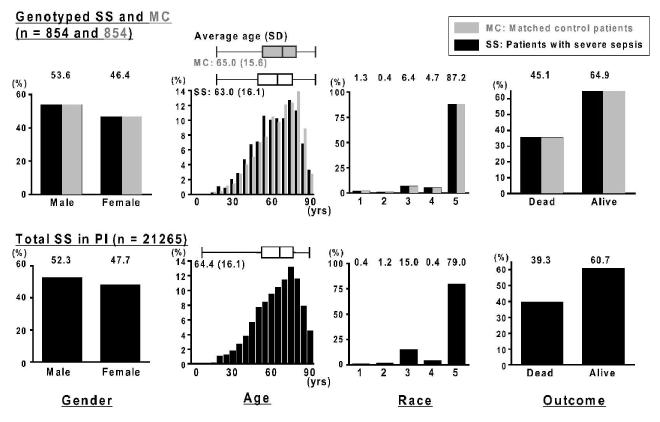
The characteristics of 854 severely septic (SS) and matched non-septic control (MC) patients are listed in Table 2. In both groups, 53.6 % were male, 87.2 % and 6.4 % were Caucasian and African American patients, respectively, and overall survival was 64.9 % (Fig. 3). In spite of their similar mortality, the MC patients had no evidence of infection at any time during their hospitalization. Average ages (SD) of SS and MC groups were 63.0 (16.05) and 65.0 (15.58) years old, respectively (Fig. 3) and the distribution of the age in each cohort was similar (F = 1.06, P = 0.389). The non-septic patients were selected as directed matches in terms of gender, age race and survival from the same Project IMPACT ™ database (Fig 1). By design, the cohort of control ICU patients are well-matched on these selected variables to the patients with severe sepsis.
Table 2.
Patient Characteristics (Severe Sepsis and Matched Controls)
| Genotyped patients with severe sepsis; SS (n = 854) |
Matched control patients; MC (n = 854) |
P values | |
|---|---|---|---|
| SAPS2 Prob. of Survival (mean±s.d.) | 57.0 ± 28.8 | 70.5 ± 28.1 | < .0001a |
| Septic Shock (%) | 7.5 | NA | NA |
| Pre-existing conditions (%) | |||
| Cardiovascular diseases | 94.0 | 89.2 | .0003b |
| Diabetes mellitus | 75.8 | 65.2 | < .0001b |
| COPD | 70.5 | 58.2 | < .0001b |
| Malignant neoplasm | 46.5 | 32.2 | < .0001b |
| Trauma | 2.2 | 9.4 | < .0001b |
| Obesity | 41.0 | 33.3 | .0009 b |
|
Acute organ dysfunctions and sequelae (%) |
|||
| Respiratory (Ventilated) | 63.1 | 48.6 | < .0001b |
| Cardiovascular | 13.1 | 4.3 | < .0001b |
| Renal | 25.6 | 6.9 | < .0001b |
| Hematologic | 6.3 | 1.5 | < .0001b |
| Neurologic | 3.2 | 3.0 | .8890b |
| Hepatic | 2.5 | .6 | .0016b |
| Number of acute organ dysfunctions (mean ±s.e.m) |
1.14 ± .03 | .65 ± .02 | < .0001a |
| DIC | 4.0 | 2.3 | .0529b |
| DVT | 6.6 | 4.7 | .0928b |
| GI bleeding | 1.6 | .7 | .0720b |
| Treatments (%) | |||
| Renal replacement therapy | 14.2 | 4.0 | < .0001b |
| Vasopressin | 7.5 | 1.2 | < .0001b |
| Blood product administrations | 18.5 | 11.2 | < .0001b |
| Enteral feeding | 46.8 | 18.7 | < .0001b |
, P value of unpaired t-test (two-tailed);
, P value with Fisher’s exact test.
Figure 3.
Representativeness of total patients with severe sepsis in Project IMPACT ™ critical care clinical database
Top:Gender, age race and outcome of patients with severe sepsis (SS; solid black bars) who were genotyped in the present study (n = 865). Gender, age race and outcome of matched controls without infection (MC; gray shadow bars) who were genotyped in the present study (n = 865). Bottom: Gender, age race and outcome of patients with severe sepsis who appeared in the Project IMPACT™ (PI) critical care clinical database (n = 21265).
Y-axes for all graphs show the relative frequencies of patients in each group. The boxplots shown above age distributions on each group indicate the median values and the inter-quartile range (sides of boxes).
SS, patients with severe sepsis; MC, mortality, age, gender, and race-matched controls without evidence of any infection; PI, the Project IMPACT™ critical care clinical database; Race #1, American Indian/Alaska native/Australian Aborigine; Race #2, Asian/Pacific islander; Race #3, African American/African European/Haitian; Race #4, Latin/Hispanic; Race #5, Caucasian.
Genotyped Specimens are Representative of the Total Severe Sepsis Population
Distributions of gender, age, race and outcome between the genotyped SS and the total SS were compared (Fig. 3). The mean age (SD) was 64.4 (16.12) years old and 52.3 % were men in total SS of PI database (n = 21265; Fig. 3). In this total population, 79.0 % and 15.0 % were Caucasian and African-American, respectively. Caucasians were slightly overrepresented in the genotyped population, most likely the result of the patient populations in the institutions that chose to participate in the study. The distribution of age between the total SS and the genotyped SS was similar (F = 1.01, P = .885). The survival rate in total SS population (60 %) was slightly lower than the genotyped SS population (64%). Overall, as illustrated in Fig. 3, genotyped specimens are fairly representative of the total severe sepsis population.
Genotypic Distributions of the Six Inflammation Gene Polymorphisms and Frequency of Severe Sepsis in the Genotype Categories
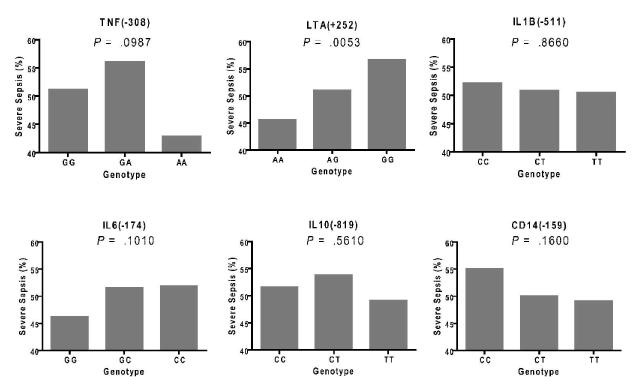
Genotype call rate of the six SNPs was 91.4 - 99.0 % although genotypic distributions in TNF (−308), LTA(+252) and IL1B(−511) diverged from Hardy-Weinberg equilibrium (HWE) in the studied subjects (P = .0013, < .0001 and .0065, respectively; Table 3). As expected given their collocation on chromosome 6, TNF(−308) and LTA(+252) were in linkage disequilibrium (LD correlation R = .4523, D’ = .7819, P < .0001). LTA(+252) was the marker among the 6 SNPs which was the most over-represented in the septic patient group with codominant (additive) model analysis and therefore significantly associated with the sepsis susceptibility in terms of dose effect of alleles (P = .005; Fig 4). IL6(−174), CD14(−159) and IL1B(−511) also maintained the allele dose effect although their splits trend did not reach statistical significance. As for TNF(−308), the genotype distribution of variant homozygotes (AA) was too low (3.59 %; Table 3) to evaluate the frequency of severe sepsis occurrence. It appears that, in this large and geographically diverse population of the critically ill patients, LTA(+252) genotype influences the risk of developing severe sepsis.
Table 3.
Genotypic distributions of the six SNPs
| SNP locus | Genotype; n (%) | Genoty pe call rate |
Hardy- Weinberg equilibrium test P value All/Septics/ Matched Controls |
||
|---|---|---|---|---|---|
| TNF(−308) | GG | GA | AA | 0.001 | |
| Total genotyped patients n = 1561 |
1123 (71.94 %) |
382 (24.27 %) | 56 (3.59 % ) |
.9139 | 0.453 <0.0001 |
|
| |||||
| LTA(+252) | AA | AG | GG | < 0.0001 | |
| Total genotyped patients n = 1690 |
730 (43.20 %) |
697 (41.24 %) | 263 (15.56 %) |
.9895 | 0.002 0.003 |
|
| |||||
| IL1B(−511) | CC | CT | TT | 0.006 | |
| Total genotyped patients n = 1571 |
683 (43.48 %) |
668 (42.52 %) | 220 (14.00 %) |
.9198 | 0.041 0.071 |
|
| |||||
| IL6(−174) | GG | GC | CC | 0.169 | |
| Total genotyped patients n = 1684 |
655 (38.90 %) |
769 (45.66 %) | 260 (15.44 %) |
.9859 | 0.768 0.109 |
|
| |||||
| IL10(−819) | CC | CT | TT | 0.141 | |
| Total genotyped patients n = 158 |
897 (56.59 %) |
576 (36.34 %) | 112 (7.07 % ) |
.9280 | 0.749 0.071 |
|
| |||||
| CD14(−159) | CC | CT | TT | 0.703 | |
| Total genotyped patients n = 1570 |
443 (28.22 %) |
789 (50.25 %) | 338 (21.53 %) |
.9192 | 0.791 0.385 |
SNP, single nucleotide polymorphism; TNF(−308), SNP at position −308 nucleotides 5′ of the first exon of tumor necrosis factor-α; LTA(+252), SNP at position 252 site of lymphotoxin-α; IL1B(−511), SNP at position −511 nucleotides 5′ of the first exon of interleukin-1β; IL6(−174), SNP at position −174 nucleotides before the first exon of IL-6; IL10(−819), SNP at position −819 nucleotides before the first exon of IL-10; CD14(−159), SNP at position −159 nucleotides before the first exon of CD14.
Figure 4.
Frequency of severe sepsis in genotype categories of the six single nucleotide polymorphisms
SNP, single nucleotide polymorphism; TNF(−308), SNP at position −308 nucleotides 5’ of the first exon of tumor necrosis factor-α; LTA(+252), SNP at position 252 site of lymphotoxin-α; IL1B(−511), SNP at position −511 nucleotides 5’ of the first exon of interleukin-1β; IL6(−174), SNP at position −174 nucleotides before the first exon of IL-6; IL10(−819), SNP at position −819 nucleotides before the first exon of IL-10, CD14(−159), SNP at position −159 nucleotides before the first exon of CD14. Y-axes for all graphs show the frequencies of severe sepsis in each genotype category. P values in SNP were evaluated with chi-square test on codominant model analysis.
Lymphotoxin-α intron Polymorphism and Severe Sepsis after Gender, Age and Racial Stratification
Table 5 shows the association of the LTA(+252) genotype and the other factors, i.e., age, gender, and race, on severe sepsis risk. In addition to the codominant model analysis, LTA(+252) variant was more frequent in the septic patient group with both recessive and dominant model analysis (P = .0044 and .0126, respectively). Severe sepsis risk did not segregate with the LTA(+252) among female ICU patients, but did it among male patients. Likewise, severe sepsis risk was associated with the LTA(+252) only among older (> 60 yrs) ICU patients. Furthermore, the statistical significance (P = .0016; Table 5) and the allele dose effect of the LTA(+252) on susceptibility to severe sepsis was maintained only among Caucasian patients. In each category (Male SS v Male MC, Female SS v Female MC, SS > 60 yrs v MC > 60 yrs, SS ≤ 60 yrs v MC ≤ 60 yrs, and Caucasian SS v Caucasian MC), the odds ratios of ‘AA v GG’ were 1.90, 1.22, 1.68, 1.36 and 1.55, respectively and were higher than the comparison of ‘AG v GG’ (1.40, 1.08, 1.18, 1.32, and 1.22, respectively), demonstrating reasonable trends in terms of LTA(+252) allele dose effect.
Table 5.
Genotype distributions of the lymphotoxin alpha first intron polymorphism stratified by gender, age and race
| SNP loci | Genotype; n (%) | Genotyp e call rate |
Hardy- Weinberg equilibriu m test P value |
||
|---|---|---|---|---|---|
| LTA(+252) | AA | AG | GG | ||
|
| |||||
| Total genotyed patients (n = 1690) |
730 (43.20 %) |
697 (41.24 %) |
263 (15.56 %) |
.9895 | < .0001 |
|
| |||||
| Male (n= 908) | 388 (42.73 %) |
371 (40.86 %) |
149 (16.41 %) |
.9913 | .0002 |
| Female (n = 782) | 342 (43.73 %) |
326 (41.69 %) |
114 (14.58 %) |
.9874 | .0130 |
|
| |||||
| > 60 yrs (n = 1022) | 440 (43.05 %) |
434 (42.47 %) |
148 (14.48 %) |
.9894 | .0162 |
| ≤ 60 yrs (n = 668) | 290 (43.41 %) |
263 (39.37 %) |
115 (17.22 %) |
.9896 | < .0001 |
|
| |||||
| Caucasian (n = 1474) | 649 (44.03 %) |
612 (41.52 %) |
213 (14.45 %) |
.9893 | .0006 |
| Non-Caucasian (n = 216) |
81 (37.50 %) |
85 (39.35 %) |
50 (23.15 %) |
.9908 | .0039 |
LTA(+252), single nucleotide polymorphism at position 252 site of lymphotoxin-α.
Joint Association of the Lymphotoxin-α intron Polymorphism and Age and Gender on Susceptibility to Severe Sepsis
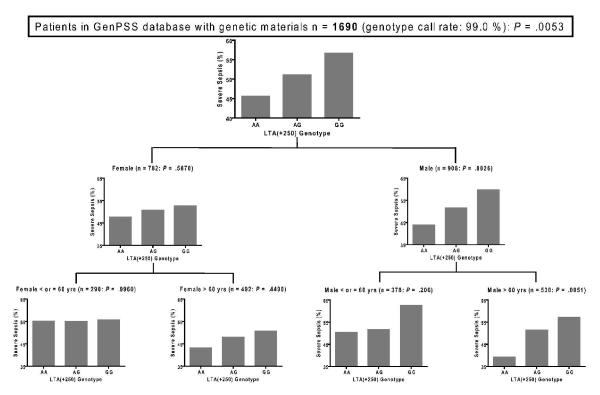
Fig. 5 shows a tree diagram with the RP method indicating joint association of LTA(+252) and age and gender on susceptibility to severe sepsis. As shown in Fig. 5, in male population (P = .0026), LTA(+252) had significantly larger influence on severe sepsis risk than female population (P = .5870). In both gender groups, older (> 60 yrs) patients were more susceptible to severe sepsis, which was consistent with the results in Table 4. Although all populations exhibited the allele dose effect, there was statistical significance only in the older male population (P = .0051). Recursive partitioning indicated that the effect of LTA(+252) on severe sepsis risk was concentrated primarily in the older male members of the patient population.
Figure 5.
Frequency of severe sepsis in the lymphotoxin alpha first intron single nucleotide polymorphism after gender and age stratification.
LTA(+252), single nucleotide polymorphism at position 252 site of the lymphotoxin-α gene, Y-axes for all graphs shows the frequencies of severe sepsis. P values in SNP were evaluated with chi-square test on codominant model analysis.
Table 4.
Chi-square test on allelic frequencies and genotype distributions with respect to the predisposition to severe sepsis in the lymphotoxin alpha first intron polymorphism
| Patients’ subgroups (n; genotyped patients’ numbers) |
Chi-square test on LTA(+252) |
Odds ratio for incidence of severe sepsis |
|||
|---|---|---|---|---|---|
| Allelic frequency |
Genotypic distribution | ||||
|
| |||||
| Chi-square (df=1) (P value) |
Chi-square of recessive model (AA v AG+GG) (df=1) (P value) |
Chi-square of dominant model (AA+AG v GG) (df=1) (P value) |
AG v GG [95% CI] |
AA v GG [95% CI] |
|
| Total SS (n = 838) v Total MC (n = 852) |
11.60 ( .0007) |
8.10 ( .0044) |
6.22 ( .0126) |
1.25 [ .94, 1.67] |
1.56 [1.17, 2.07] |
|
| |||||
| Male SS (n= 450) v Male MC (n = 458) |
13.28 ( .0003) |
8.95 ( .0028) |
7.38 ( .0066) |
1.40 [ .95, 2.06] |
1.90 [1.30, 2.79] |
| Female SS (n = 388) v Fe male MC (n = 394) |
1.15 ( .2843) |
.93 ( .3349) |
.49 ( .4860) |
1.08 [ .71, 1.66] |
1.22 [ .80, 1.87] |
|
| |||||
| SS > 60 yrs (n = 477) v MC > 60 yrs (n = 545) |
10.66 ( .0011) |
9.52 ( .0020) |
3.79 ( .0516) |
1.18 [ .82, 1.73] |
1.68 [1.16, 2.45] |
| SS ≤ 60 yrs (n = 361) v MC ≤ 60 yrs (n = 307) |
1.73 ( .1879) |
.55 ( .4596) |
1.99 ( .1588) |
1.32 [ .89, 1.96] |
1.36 [ .88, 2.11] |
|
| |||||
| Caucasian SS (n = 730) v Caucasian MC (n = 744) |
9.96 ( .0016) |
7.69 ( .0056) |
4.62 ( .0315) |
1.22 [ .89, 1.67] |
1.55 [1.13, 2.11] |
| Non-Caucasian SS (n = 108) v Non-Caucasian MC (n = 108) |
1.60 ( .2062) |
.49 ( .4822) |
1.67 ( .1969) |
1.48 [ .73, 3.00] |
1.56 [ .77, 3.18] |
LTA(+252), single nucleotide polymorphism at position 252 site of lymphotoxin-α; df, degree of freedom; CI, confidence interval; SS, patients with severe sepsis; MC, mortality, age, gender, and race-matched controls without evidence of any infection. P values are examined with Chi-square test.
To verify the effects suggested by recursive partitioning, we formally tested the association between the sepsis case status and the marker a LTA(+252) adjusting for potential non-genetic covariates using a full versus reduced logistic regression approach based on an additive genotypic model. The full model consists of LTA(+252) allele counts along with the non-genetic covariates, and the reduced model consists only of the non-genetic covariates. This tests whether or not the allele count covariate adds significantly to the model. The non-genetic covariates considered for this test were age, pre-existing heart conditions, COPD, diabetes, malignant cancer, obesity, and whether the patient was admitted to the ICU due to trauma. The p-value of the full model was 4.53 × 10−22, and the p-value of the full versus reduced model test was 0.0016. Of the non-genetic covariates age, COPD, malignant cancer, and trauma were significant in the model with respective p-values of 0.00044, 0.00658, 0.00011, 2.42 × 10−11. When the same regression was run with only these significant covariates the p-value of the full model was 2.24× 10−22 and the p-value of the full versus reduced model was 0.001738. This indicates that there remains strong evidence that the LTA(+252) marker is associated with the sepsis response after accounting for these potential confounders.
Discussion
This study shows that in a large and heterogeneous population of critically ill patients who received medical care in diverse settings, a variant allele of the LTA gene is associated with increased risk of severe sepsis. This finding is important because prior studies have generally focused on care of a single ethnic group or care at a single medical center, and often report findings in relatively small study populations [49]. Moreover, this adverse effect of the variant allele of LTA(+252) on susceptibility to severe sepsis in this study is influenced by non-modifiable characteristics of gender and age (Fig. 5). The adverse effect of the LTA(+252)G on male patients is also reminiscent of the previous report by Schröder [19]. However, the surprising finding in the present study is that the influence of LTA(+252)G presence on the predisposition to severe sepsis is modified not only by gender (male) but also by age (elderly). Furthermore, the present study has more statistical power than the previous study plus the severe sepsis risk was evaluated with a completely matched control population among patients registered in the Project IMPACT™ (PI) database. This result suggests that inflammatory cytokine production is somewhat regulated by LTA(+252) in sepsis pathophysiology and the genetic risk association may be more pronounced among elderly male patients. Although the genotype distributions of LTA(+252) did not differ between young and elderly nor between male and female (Table 4), the largest genetic association appeared in older male patients. Interpretive caution is advised since a significant result in one subgroup and a lack of statistical significance in another does not necessarily signify a different effect according to subgroup. The interaction statistics between subgroups were not significant.
Stewart, et al. and Marik, et al. have consistently shown that TNF production by the elderly was higher in severe biliary infections [23] and more generally in septic shock [22]. As for sexual dimorphism, Moxley, et al. demonstrated that LPS-stimulated TNF levels in peripheral blood of healthy female patients were lower than in blood of male patients [21]. Although the study design did not provide for measurements of cytokines, we speculate that their concentration might be significantly stratified by LTA(+252) especially in older male ICU patients. Furthermore, a recent report suggested that gene expression variance in critically ill patients due to age, gender, and ethnic background is greater than that due to infecting organism [50]. Our study failed to recruit a sufficient number of African-Americans with severe sepsis to reach reliable statistical inferences concerning any influence that race/ethnicity might have on sepsis predisposition phenotype among carriers of the various LTA(+252) alleles. Many of the contributing centers were community-based. Of note, there was no statistical difference in the frequency of African-Americans in those centers who did and who did not have genetic specimens contributed.
Contrary to prior reports, many of the genetic polymorphisms besides LTA(+252) that had been already reported as predictors of severe sepsis did not demonstrate a statistically significant effect on sepsis risk. Thus variant allele carriers of CD14(−159) appeared somewhat less susceptible to severe sepsis in our result (Fig. 4), which is inconsistent with a published report [41] even if the influence of CD14(−159) in our result was not significant. This apparent discrepancy might be partially due to the larger size of our study or to differences in control selection. In our study, the US ICU patients from 12 the different centers who had no evidence of any infections were selected as control. In contrast, earlier studies that reported rare allele of CD14(−159) had adverse effect on sepsis risk chose healthy volunteers in a single center as their controls [40, 41, 51].
The early enthusiasm fueled by smaller studies that suggested the existence of a number of gene variants that might predispose to sepsis and/or an adverse outcome has been dampened by the realization that many of those studies were underpowered, their findings were likely falsely positive and their conclusions were unwarranted [49]. This concern applies to LTA (+252), which has previously been reported in smaller studies to both confer ( 5 studies) and not to infer (8 studies) an increased risk of sepsis [49]. One approach to minimizing the risk of falsely positive attributions is the calculation of a false positive report probability (FPRP) [52]. Even setting a conservative FPRP value of 0.2 still suggests that the observed influence of LTA(+252) on severe sepsis predisposition in our study to be noteworthy.
A sufficiently powered, carefully described whole genome study in sepsis genomics appears important to understanding both sepsis biology as well as the interplay among genetic and non-genetic influences upon susceptibility and outcome. The importance of comprehensive clinical databases that collect not only demographic data and pre-existing conditions but also describe the septic event in detail—including all causative organisms, putative sources, treatments and apparent success or failure—cannot be overstated. Such process variables allow for rigorous disease-based grouping (e.g., analysis of patients with pneumococcal pneumonia) as opposed to the current stopgap strategy of syndrome-based grouping (e.g. severe vs. mild sepsis).
Comprehensive clinical data contained in PI and similar databases enable not only close matching of cases and controls, but also some insight into selection bias that might affect the cases themselves. The specimens from patients with severe sepsis in the GenPSS archive appeared to be representative of the total severe sepsis population from the PI database (Fig. 3). Yet there was a difference in survival rate between ‘genotyped SS v total SS patients’ (64.87 v 60.72 %, P = .0102; Fig. 3) that might reflect differences in the approach of participating centers to the care of severely septic patients. Although the genotype failure rate approached 9% for some alleles, this appeared to be unrelated to the contributing center or to patient status (severe sepsis versus matched controls). Provided that the failure was unrelated to a specific allele, modeling suggests that the no additional SNPs would have become significant if genotype failures had not occurred
Alternatively, there may be more complex genetic effects. In our ICU patients’ cohort, there are deviations from Hardy-Weinberg equilibrium (HWE) on LTA(+252). The Fisher’s Exact HWE test p-value is 1.21 × 10−5 and the HWE correlation R value is 0.10696. When cases and controls are considered separately, the Fisher’s exact HWE p-value is 0.00226 for cases and 0.00346 for controls for the LTA(+252) marker. Thus there is evidence to suggest that the association between the septic shock response and this marker is not driven by departures from HWE. It is important to note that the samples were not randomly drawn from a population, but instead were taken from patients admitted to the ICU. The LTA(+252) marker has been shown in previous studies to be associated with both sepsis and respiratory failure [53]. The likelihood of cases and controls suffering from either sepsis, respiratory failure or both is greater than would normally be found in the general “healthy” population, plausibly accounting for the departures from HWE. Plausible is different from certain, thus one can only conclude that for some genotypes evaluated, the alleles were not assorted randomly in the population under study.
We believe that the intersection of clinical care and disease association genetics demands study of large (>100,000) populations whose clinical data are rigorously recorded and whose genomes are studied in their entirely. The costs of such an effort, previously prohibitive, are now mitigated by the US federal requirements for portable electronic records and advances in the efficiency of screening techniques. The risks of such an effort, primarily related to the potential for discrimination should genetic data leak to payors and insurers, have been mitigated by passage of the Genetic Insurability Nondiscrimination Act (GINA) signed into US law on April 24, 2008.
Our mid-scale population association study supports the hypothesis that genetic predisposition to severe sepsis exists and is further modulated by both genetic (gender) and non-genetic (age) factors. Variation in the LTA(+252) gene appears to explain, in part, the susceptibility to sepsis and the effect may be greater in older Caucasian men.
[Acknowledgments]
Maureen Stark, formerly of Tri-Analytics and presently with Cerner Corporation provided tireless support.
We thank the technicians and summer students who developed, qualified and performed the assays: Beverly Gibson, Brooke Stroup,Curtis Wilson, Xiaomin (May) Wu, Syamal (Dave) Bhattacharya, Michelle Mergler and Derek Bogdan.
We are pleased to acknowledge the dedication and participation of the following individuals and institutions: Jackie O’Brien, Robert W. Taylor, St. John’s Mercy Medical Center, St. Louis MO; Brenda Snyder, Ellie Blasco, Michael Schwarz, North Colorado Medical Center, Greeley CO; Leslie DeSouza, Lori-Ann Kozikowski, Tom Higgins, Baystate Medical Center, Springfield MA, Dee Dee Boss, Gerald Plost, St. John Medical Center, Tulsa OK; Angela Dickson, St. John Medical Center, Longview WA; Mary Katherine Blackburn, Daniel Trahan, Learnard Chabert Medical Center, Houma LA; Howard Corwin, Dartmouth Hitchcock Medical Center, Lebanon NH; Terri Conner, Seton Medical Canter, Austin TX; Sandralee Blosser, Penn State Hershey Medical Center, Hershey PA; Christopher Dunatov, Milt L. McPherson, Northeast Medical Center, Concord NC; Sandy Hartenstein, Eastern Idaho Regional Medical Canter, Idaho Falls ID; Dianne Gergely, Devendra Amin, Morton Plant Hospital, Clearwater FL; Richard Riker, Maine Medical Center, Portland ME; Joanne Kuszat, Amy Fraccola, Rex Healthcare, Raleigh NC.
An independent statistical opinion was provided by Christophe Lambert.
Source of funding: This research was supported by NIH/GM (062809) and the Uehara Memorial Foundation. The funding sources had no role in study design, collection, analysis, or interpretation of data.
Appendix: Report of power calculations
Our study suggested that three SNPs maintained an allele dose effect although the splits trend did not reach statistical significance. In order to help the reader interpret the data and this statement, we performed and provide power calculations that vary with posited genetic effect and also with minor allele frequency assuming 854 case-control pairs, a severe sepsis rate of 5% in the ICU population, and a two-tailed α = 0.01. The calculations were performed with Quanto, a freely downloadable application offered by the University of Southern California and available at http://hydra.usc.edu/gxe/.
Three models are presented. The first model, the dominant model, assumes that either one or two copies of the minor allele is sufficient to predispose to severe sepsis. The second model, the log-additive or coDominant model, assumes that two copies of the minor allele have double the probability of predisposing to severe sepsis. The third model, the recessive model, requires two copies of the minor allele to predispose to severe sepsis. Since there is no way of knowing how the minor allele might have its effect in advance, all three models are considered.
For each of the models, a range of minor allele frequencies ranging from 0.18 to 0.38 is illustrated. A minor allele frequency of 0.18 corresponds approximately to the lowest frequency observed among the six minor alleles, that of TNF(−308). The highest frequency shown corresponds to the frequency of the minor allele of LTA(+252).
The power calculation also depends on the degree of genetic predisposition. This is shown in the table as the RG, which can be thought of the increased odds of becoming septic if the model conditions are fulfilled. These are shown from 1.25 to 2.5 for each model.
Each combination of model, minor allele frequency, and odds ratio produces a unique power value. A value of 0.8 corresponds to 80% , meaning that the study has an 80% probability of detecting the effect if the effect exists. Again, we set the p-value at a conservative 0.01.
These data are calculated for single genes. They do not account either for gene-gene interactions or for multiple comparisons. Moreover they are power calculations only and have no influence on the analysis of the data.
The table shows that if two copies of the minor allele are required to predispose to severe sepsis(recessive model), then either the minor allele must be relatively frequent or else the predisposition must be substantial (e.g. doubling the risk).
| Power | ||||
|---|---|---|---|---|
| Minor Allele Frequency |
Effect (Odds Ratio) |
Recessive | coDominant | Dominant |
| 0.18 | 1.25 | 0.0429 | 0.5009 | 0.352 |
| 1.5 | 0.1671 | 0.9858 | 0.9258 | |
| 1.75 | 0.3868 | 0.9999 | 0.9986 | |
| 2 | 0.6284 | 0.9999 | 0.9999 | |
| 2.25 | 0.8136 | 0.9999 | 0.9999 | |
| 2.5 | 0.9209 | 0.9999 | 0.9999 | |
| 0.23 | 1.25 | 0.0673 | 0.592 | 0.3837 |
| 1.5 | 0.2914 | 0.9952 | 0.942 | |
| 1.75 | 0.6172 | 0.9999 | 0.9991 | |
| 2 | 0.8563 | 0.9999 | 0.9999 | |
| 2.25 | 0.961 | 0.9999 | 0.9999 | |
| 2.5 | 0.9919 | 0.9999 | 0.9999 | |
| 0.28 | 1.25 | 0.0995 | 0.6571 | 0.392 |
| 1.5 | 0.4383 | 0.998 | 0.9437 | |
| 1.75 | 0.8019 | 0.9999 | 0.9991 | |
| 2 | 0.96 | 0.9999 | 0.9999 | |
| 2.25 | 0.9949 | 0.9999 | 0.9999 | |
| 2.5 | 0.9995 | 0.9999 | 0.9999 | |
| 0.33 | 1.25 | 0.1391 | 0.7015 | 0.382 |
| 1.5 | 0.5838 | 0.999 | 0.9354 | |
| 1.75 | 0.9124 | 0.9999 | 0.9987 | |
| 2 | 0.9913 | 0.9999 | 0.9999 | |
| 2.25 | 0.9995 | 0.9999 | 0.9999 | |
| 2.5 | 0.9999 | 0.9999 | 0.9999 | |
| 0.38 | 1.25 | 0.1842 | 0.7299 | 0.3579 |
| 1.5 | 0.7076 | 0.9993 | 0.9168 | |
| 1.75 | 0.9651 | 0.9999 | 0.9975 | |
| 2 | 0.9984 | 0.9999 | 0.9999 | |
| 2.25 | 0.9999 | 0.9999 | 0.9999 | |
| 2.5 | 0.9999 | 0.9999 | 0.9999 | |
Footnotes
The authors have not disclosed any potential conflicts of interest.
Special Instructions: none
[References]
- 1.Angus DC, Linde-Zwirble WT, Lidicker J, et al. Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit Care Med. 2001;29:1303–1310. doi: 10.1097/00003246-200107000-00002. [DOI] [PubMed] [Google Scholar]
- 2.Wang HE, Shapiro NI, Angus DC, et al. National estimates of severe sepsis in United States emergency departments. Crit Care Med. 2007;35:1928–1936. doi: 10.1097/01.CCM.0000277043.85378.C1. [DOI] [PubMed] [Google Scholar]
- 3.Dellinger RP, Levy MM, Carlet JM, et al. Surviving Sepsis Campaign: international guidelines for management of severe sepsis and septic shock: 2008. Intensive Care Med. 2008;34:17–60. doi: 10.1007/s00134-007-0934-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dellinger RP, Levy MM, Carlet JM, et al. Surviving Sepsis Campaign: international guidelines for management of severe sepsis and septic shock: 2008. Crit Care Med. 2008;36:296–327. doi: 10.1097/01.CCM.0000298158.12101.41. [DOI] [PubMed] [Google Scholar]
- 5.Pennisi E. Breakthrough of the year. Human genetic variation. Science. 2007;318:1842–1843. doi: 10.1126/science.318.5858.1842. [DOI] [PubMed] [Google Scholar]
- 6.McGuire W, Hill AV, Allsopp CE, et al. Variation in the TNF-alpha promoter region associated with susceptibility to cerebral malaria. Nature. 1994;371:508–510. doi: 10.1038/371508a0. [DOI] [PubMed] [Google Scholar]
- 7.Stuber F, Udalova IA, Book M, et al. −308 tumor necrosis factor (TNF) polymorphism is not associated with survival in severe sepsis and is unrelated to lipopolysaccharide inducibility of the human TNF promoter. J Inflamm. 1995;46:42–50. [PubMed] [Google Scholar]
- 8.Mira JP, Cariou A, Grall F, et al. Association of TNF2, a TNF-alpha promoter polymorphism, with septic shock susceptibility and mortality: a multicenter study. JAMA. 1999;282:561–568. doi: 10.1001/jama.282.6.561. [DOI] [PubMed] [Google Scholar]
- 9.Tabrizi AR, Zehnbauer BA, Freeman BD, et al. Genetic markers in sepsis. J Am Coll Surg. 2001;192:106–117. doi: 10.1016/s1072-7515(00)00748-1. quiz 145-106. [DOI] [PubMed] [Google Scholar]
- 10.Watanabe E, Hirasawa H, Oda S, et al. Extremely high interleukin-6 blood levels and outcome in the critically ill are associated with tumor necrosis factor- and interleukin-1-related gene polymorphisms. Crit Care Med. 2005;33:89–97. doi: 10.1097/01.ccm.0000150025.79100.7d. discussion 242-243. [DOI] [PubMed] [Google Scholar]
- 11.Villar J, Perez-Mendez L, Flores C, et al. A CXCL2 polymorphism is associated with better outcomes in patients with severe sepsis. Crit Care Med. 2007;35:2292–2297. doi: 10.1097/01.ccm.0000284511.73556.59. [DOI] [PubMed] [Google Scholar]
- 12.Pinsky MR, Vincent JL, Deviere J, et al. Serum cytokine levels in human septic shock. Relation to multiple-system organ failure and mortality. Chest. 1993;103:565–575. doi: 10.1378/chest.103.2.565. [DOI] [PubMed] [Google Scholar]
- 13.Remick DG, Bolgos GR, Siddiqui J, et al. Six at six: interleukin-6 measured 6 h after the initiation of sepsis predicts mortality over 3 days. Shock. 2002;17:463–467. doi: 10.1097/00024382-200206000-00004. [DOI] [PubMed] [Google Scholar]
- 14.Oda S, Hirasawa H, Shiga H, et al. Sequential measurement of IL-6 blood levels in patients with systemic inflammatory response syndrome (SIRS)/sepsis. Cytokine. 2005;29:169–175. doi: 10.1016/j.cyto.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 15.Cardon LR, Bell JI. Association study designs for complex diseases. Nat Rev Genet. 2001;2:91–99. doi: 10.1038/35052543. [DOI] [PubMed] [Google Scholar]
- 16.Colhoun HM, McKeigue PM, Smith G Davey. Problems of reporting genetic associations with complex outcomes. Lancet. 2003;361:865–872. doi: 10.1016/s0140-6736(03)12715-8. [DOI] [PubMed] [Google Scholar]
- 17.Zehnbauer B. Population genetics in critical illness. Crit Care Med. 2005;33:242–243. doi: 10.1097/01.ccm.0000150763.21694.8a. [DOI] [PubMed] [Google Scholar]
- 18.Barnato AE, Alexander S, Linde-Zwirble WT, et al. Racial Variation in the Incidence, Care, and Outcomes of Severe Sepsis. Am J Respir Crit Care Med. 2007 doi: 10.1164/rccm.200703-480OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schroder J, Kahlke V, Book M, et al. Gender differences in sepsis: genetically determined? Shock. 2000;14:307–310. discussion 310-303. [PubMed] [Google Scholar]
- 20.Choudhry MA, Bland KI, Chaudry IH. Trauma and immune response--effect of gender differences. Injury. 2007;38:1382–1391. doi: 10.1016/j.injury.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moxley G, Posthuma D, Carlson P, et al. Sexual dimorphism in innate immunity. Arthritis Rheum. 2002;46:250–258. doi: 10.1002/1529-0131(200201)46:1<250::AID-ART10064>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 22.Marik PE, Zaloga GP. The effect of aging on circulating levels of proinflammatory cytokines during septic shock. Norasept II Study Investigators. J Am Geriatr Soc. 2001;49:5–9. doi: 10.1046/j.1532-5415.2001.49003.x. [DOI] [PubMed] [Google Scholar]
- 23.Stewart L, Grifiss JM, Jarvis GA, et al. Elderly patients have more severe biliary infections: influence of complement-killing and induction of TNFalpha production. Surgery. 2008;143:103–112. doi: 10.1016/j.surg.2007.06.035. [DOI] [PubMed] [Google Scholar]
- 24.Turnbull IR, Wlzorek JJ, Osborne D, et al. Effects of age on mortality and antibiotic efficacy in cecal ligation and puncture. Shock. 2003;19:310–313. doi: 10.1097/00024382-200304000-00003. [DOI] [PubMed] [Google Scholar]
- 25.Suffredini AF. Systemic inflammation and sexual dimorphism: more than meets the eye. Crit Care Med. 2007;35:1610–1612. doi: 10.1097/01.CCM.0000266793.09378.22. [DOI] [PubMed] [Google Scholar]
- 26.Trentzsch H, Stewart D, De Maio A. Genetic background conditions the effect of sex steroids on the inflammatory response during endotoxic shock. Crit Care Med. 2003;31:232–236. doi: 10.1097/00003246-200301000-00036. [DOI] [PubMed] [Google Scholar]
- 27.Risch NJ. Searching for genetic determinants in the new millennium. Nature. 2000;405:847–856. doi: 10.1038/35015718. [DOI] [PubMed] [Google Scholar]
- 28.Terwilliger JD, Haghighi F, Hiekkalinna TS, et al. A bias-ed assessment of the use of SNPs in human complex traits. Curr Opin Genet Dev. 2002;12:726–734. doi: 10.1016/s0959-437x(02)00357-x. [DOI] [PubMed] [Google Scholar]
- 29.Chen X, Levine L, Kwok PY. Fluorescence polarization in homogeneous nucleic acid analysis. Genome Res. 1999;9:492–498. [PMC free article] [PubMed] [Google Scholar]
- 30.American College of Chest Physicians/Society of Critical Care Medicine Consensus Conference: definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. Crit Care Med. 1992;20:864–874. [PubMed] [Google Scholar]
- 31.Weycker D, Akhras KS, Edelsberg J, Angus DC, Oster G. Long-term mortality and medical care charges in patients with severe sepsis. Crit Care Med. 2003;31:2316–23. doi: 10.1097/01.CCM.0000085178.80226.0B. [DOI] [PubMed] [Google Scholar]
- 32.Knaus WA, Draper EA, Wagner DP, et al. APACHE II: a severity of disease classification system. Crit Care Med. 1985;13:818–829. [PubMed] [Google Scholar]
- 33.Watanabe E, Hirasawa H, Oda S, et al. Cytokine-related genotypic differences in peak interleukin-6 blood levels of patients with SIRS and septic complications. J Trauma. 2005;59:1181–1189. doi: 10.1097/00005373-200511000-00025. discussion 1189-1190. [DOI] [PubMed] [Google Scholar]
- 34.Ma P, Chen D, Pan J, et al. Genomic polymorphism within interleukin-1 family cytokines influences the outcome of septic patients. Crit Care Med. 2002;30:1046–1050. doi: 10.1097/00003246-200205000-00015. [DOI] [PubMed] [Google Scholar]
- 35.Wen AQ, Wang J, Feng K, et al. Effects of haplotypes in the interleukin 1beta promoter on lipopolysaccharide-induced interleukin 1beta expression. Shock. 2006;26:25–30. doi: 10.1097/01.shk.0000223125.56888.c7. [DOI] [PubMed] [Google Scholar]
- 36.Schluter B, Raufhake C, Erren M, et al. Effect of the interleukin-6 promoter polymorphism (−174 G/C) on the incidence and outcome of sepsis. Crit Care Med. 2002;30:32–37. doi: 10.1097/00003246-200201000-00005. [DOI] [PubMed] [Google Scholar]
- 37.Sutherland AM, Walley KR, Manocha S, et al. The association of interleukin 6 haplotype clades with mortality in critically ill adults. Arch Intern Med. 2005;165:75–82. doi: 10.1001/archinte.165.1.75. [DOI] [PubMed] [Google Scholar]
- 38.Lowe PR, Galley HF, Abdel-Fattah A, et al. Influence of interleukin-10 polymorphisms on interleukin-10 expression and survival in critically ill patients. Crit Care Med. 2003;31:34–38. doi: 10.1097/00003246-200301000-00005. [DOI] [PubMed] [Google Scholar]
- 39.Allen ML, Hoschtitzky JA, Peters MJ, et al. Interleukin-10 and its role in clinical immunoparalysis following pediatric cardiac surgery. Crit Care Med. 2006;34:2658–2665. doi: 10.1097/01.CCM.0000240243.28129.36. [DOI] [PubMed] [Google Scholar]
- 40.Hubacek JA, Stuber F, Frohlich D, et al. The common functional C(−159)T polymorphism within the promoter region of the lipopolysaccharide receptor CD14 is not associated with sepsis development or mortality. Genes Immun. 2000;1:405–407. doi: 10.1038/sj.gene.6363691. [DOI] [PubMed] [Google Scholar]
- 41.Gibot S, Cariou A, Drouet L, et al. Association between a genomic polymorphism within the CD14 locus and septic shock susceptibility and mortality rate. Crit Care Med. 2002;30:969–973. doi: 10.1097/00003246-200205000-00003. [DOI] [PubMed] [Google Scholar]
- 42.Majetschak M, Flohe S, Obertacke U, et al. Relation of a TNF gene polymorphism to severe sepsis in trauma patients. Ann Surg. 1999;230:207–214. doi: 10.1097/00000658-199908000-00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stuber F, Petersen M, Bokelmann F, et al. A genomic polymorphism within the tumor necrosis factor locus influences plasma tumor necrosis factor-alpha concentrations and outcome of patients with severe sepsis. Crit Care Med. 1996;24:381–384. doi: 10.1097/00003246-199603000-00004. [DOI] [PubMed] [Google Scholar]
- 44.Wilson AG, Symons JA, McDowell TL, et al. Effects of a polymorphism in the human tumor necrosis factor alpha promoter on transcriptional activation. Proc Natl Acad Sci U S A. 1997;94:3195–3199. doi: 10.1073/pnas.94.7.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fishman D, Faulds G, Jeffery R, et al. The effect of novel polymorphisms in the interleukin-6 (IL-6) gene on IL-6 transcription and plasma IL-6 levels, and an association with systemic-onset juvenile chronic arthritis. J Clin Invest. 1998;102:1369–1376. doi: 10.1172/JCI2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Terry CF, Loukaci V, Green FR. Cooperative influence of genetic polymorphisms on interleukin 6 transcriptional regulation. J Biol Chem. 2000;275:18138–18144. doi: 10.1074/jbc.M000379200. [DOI] [PubMed] [Google Scholar]
- 47.Lin J, Yao YM, Yu Y, et al. Effects of CD14-159 C/T polymorphism on CD14 expression and the balance between proinflammatory and anti-inflammatory cytokines in whole blood culture. Shock. 2007;28:148–153. doi: 10.1097/SHK.0b013e3180341d35. [DOI] [PubMed] [Google Scholar]
- 48.Guo SW, Thompson EA. Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics. 1992;48:361–372. [PubMed] [Google Scholar]
- 49.Clark MF, Baudouin SV. A systematic review of the quality of genetic association studies in human sepsis. Intensive Care Med. 2006;32:1706–1712. doi: 10.1007/s00134-006-0327-y. [DOI] [PubMed] [Google Scholar]
- 50.McDunn JE, Husain KD, Polpitiya AD, et al. Plasticity of the systemic inflammatory response to acute infection during critical illness: development of the riboleukogram. PLoS ONE. 2008;3:e1564. doi: 10.1371/journal.pone.0001564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Agnese DM, Calvano JE, Hahm SJ, et al. Human toll-like receptor 4 mutations but not CD14 polymorphisms are associated with an increased risk of gram-negative infections. J Infect Dis. 2002;186:1522–1525. doi: 10.1086/344893. [DOI] [PubMed] [Google Scholar]
- 52.Wacholder S, Chanock S, Garcia-Closas M, El Ghormli L, Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst. 2004;96:434–42. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Waterer GW, Quasney MW, Cantor RM, et al. Septic shock and respiratory failure in community-acquired pneumonia have different TNF polymorphism associations. Am J Respir Crit Care Med. 2001;163:1599–1604. doi: 10.1164/ajrccm.163.7.2011088. [DOI] [PubMed] [Google Scholar]