Abstract
Acetic acid-induced inhibition of yeast growth and metabolism limits the productivity of industrial fermentation processes, especially when lignocellulosic hydrolysates are used as feedstock in industrial biotechnology. Tolerance to acetic acid of food spoilage yeasts is also a problem in the preservation of acidic foods and beverages. Thus, understanding the molecular mechanisms underlying adaptation and tolerance to acetic acid stress is increasingly important in industrial biotechnology and the food industry. Prior genetic screens for S. cerevisiae mutants with increased sensitivity to acetic acid identified loss-of-function mutations in the YPK1 gene, which encodes a protein kinase activated by the Target of Rapamycin (TOR) Complex 2 (TORC2). We show here by several independent criteria that TORC2-Ypk1 signaling is stimulated in response to acetic acid stress. Moreover, we demonstrate that TORC2-mediated Ypk1 phosphorylation and activation is necessary for acetic acid tolerance, and occurs independently of Hrk1, a protein kinase previously implicated in the cellular response to acetic acid. In addition, we show that TORC2-Ypk1-mediated activation of L-serine: palmitoyl-CoA acyltransferase, the enzyme complex that catalyzes the first committed step of sphingolipid biosynthesis, is required for acetic acid tolerance. Furthermore, analysis of the sphingolipid pathway using inhibitors and mutants indicates that it is production of certain complex sphingolipids that contributes to conferring acetic acid tolerance. Consistent with that conclusion, promoting sphingolipid synthesis by adding exogenous long-chain base precursor phytosphingosine to the growth medium enhanced acetic acid tolerance. Thus, appropriate modulation of the TORC2-Ypk1-sphingolipid axis in industrial yeast strains may have utility in improving fermentations of acetic acid-containing feedstocks.
Keywords: budding yeast, weak acid, adaptation, lipid metabolism, protein kinases, protein phosphorylation
INTRODUCTION
Understanding the cellular response to acetic acid stress, and the ability to manipulate it, are increasingly important in two major industries. First, large-scale fermentations by the yeast Saccharomyces cerevisiae are contributing to an ever greater extent to the production of ethanol, other biofuels and bulk chemicals, and specialty chemicals and biologicals [1]. In this context, use of plant biomass as feedstock in place of simple sugars will have considerable economic impact [2, 3]. However, one impediment to using plant biomass-derived feedstocks is the presence of acetic acid in lignocellulosic hydrolysates, which inhibits yeast growth and hinders fermentation [4, 5]. Second, in the food industry, weak acids (in particular, acetic acid) are often used as preservatives. However, certain microbes are acetic acid-tolerant and, thus, able to survive and proliferate in foods and beverages preserved with this weak acid, causing spoilage that results in major economic loss [6].
At the low pH values reached in S. cerevisiae cultures, significantly below the pKa of acetic acid (~4.75), this weak acid is mainly in its undissociated state and, hence, uncharged, permitting its diffusion across the plasma membrane (PM), a process also facilitated through the aquaglyceroporin Fps1 [7, 8]. Upon entering the more basic environment of the cytosol, the acid dissociates into acetate anion and H+, which accumulate, thereby decreasing cytosolic pH, increasing membrane stretch (brought on by an increase in turgor pressure), and elevating reactive oxygen species [9, 10]. Ultimately, these stresses cause decreased metabolic activity [11], decreased cell growth and viability [12, 13], and decreased survival in stationary phase [14].
A number of adaptive responses of S. cerevisiae to acetic acid stress have been identified. Yeast cells challenged with acetic acid were found to increase vacuolar and plasma membrane (PM) H+-ATPase activity, presumably to allow recovery of the cytosolic pH to more physiological values [15, 16]. Exposure to high levels of acetic acid (≥100 mM) activates the Hog1 MAPK, which phosphorylates Fps1, thus promoting its endocytosis and thereby reducing acetic acid influx [7]. Exposure to acetic acid also induces the expression of the PM-localized members of the major facilitator superfamily Tpo2 and Tpo3 [17], via activation of the transcription factor Haa1 helping to reduce the concentration of the accumulated intracellular acetate. Genome-wide transcriptome analysis and functional genomics studies have revealed additional loci under Haa1 control, including the HRK1 gene, which encodes a H+-ATPase-activating protein kinase [17, 18], and other genes whose products contribute globally to yeast cell adaptation to acetic acid [19]. Also, phenotypic analysis of the S. cerevisiae genome-wide deletion collection identified a number of additional genes important for survival in response to acetic acid stress, including the protein kinase Ypk1 [20, 21]. The knowledge gathered so far on the understanding of molecular factors and cellular pathways involved in acetic acid tolerance has been used in the development of acetic acid-tolerant strains, either through the deletion or overexpression of single genes [22, 23], manipulation of genes that play a crucial role in the regulatory cascades that control acetic acid tolerance, such as manipulation of the Haa1-regulon [24–26], or the use of evolutionary engineering strategies to select strains that possess increased tolerance to the acid [27]. Manipulation of the composition of the growth medium in order to increase yeast tolerance to acetic acid has also been reported [20].
Ypk1 is a member of the AGC subfamily of protein kinases [28] and the ortholog of mammalian SGK1 [29]. Ypk1 activity requires phosphorylation of Thr504 in its activation loop by the protein kinases Pkh1 and Pkh2, which are associated with PM-localized structures called eisosomes [30, 31]. Ypk1 activity can be substantially increased by further phosphorylation at sites in its C-terminal domain (especially Ser644 and Thr662) by the PM-associated Target of Rapamycin (TOR) Complex 2 (TORC2) [31–33]. Certain membrane stresses, including sphingolipid depletion [32], heat shock [34], and hypotonic conditions [35], elevate TORC2 activity and consequently activate Ypk1, whereas other stresses, such as hypertonic conditions [36, 37], diminish TORC2 and Ypk1 activity.
Identification of physiologically-relevant substrates of Ypk1 has shed light on the molecular mechanisms downstream of TORC2-Ypk1 activation that promote tolerance to these various membrane stresses [32, 38–40]. For example, upon sphingolipid depletion or in hypo-osmotic medium (which causes membrane stretch due to the increased turgor pressure), Ypk1 phosphorylates Orm1 and Orm2 [32], thus blocking the inhibition that these two proteins normally exert on L-serine:palmitoyl-CoA acyltransferase (SPT), the enzyme complex catalyzing the first step in sphingolipid biosynthesis [41], thus increasing flux into this pathway. Moreover, Ypk1 phosphorylates and stimulates Lac1 and Lag1, the catalytic subunits of the ceramide synthase complex [40], thereby ensuring that the SPT-generated long-chain (sphingoid) base (LCB), mainly phytosphingosine (PHS), and fatty acyl-CoA [typically very long chain fatty acids (VLCFA), mainly C26] are diverted more efficiently into ceramides for use in the production of complex sphingolipids [42]. Furthermore, TORC2-Ypk1-dependent phosphorylation of both Orm1 and Orm2 and Lac1 and Lag1 are required for yeast cell survival under the stresses of sphingolipid depletion and hypotonic shock [32, 40].
Given that TORC2-Ypk1 signaling is known to respond to membrane stretch, like that elicited by acetic acid treatment, and that functional genomics suggested a role for Ypk1 in the adaptive response to acetic acid, we investigated whether TORC2-Ypk1 activity responds to challenging yeast cells with an inhibitory sub-lethal exogenous acetic acid concentration and whether TORC2-Ypk1 function contribute to the ability of yeast cells to survive the stress imposed by exposure to this weak acid. If so, we sought to determine whether it is changes in the production of sphingolipids per se that is responsible for the role that TORC2-Ypk1 signaling plays in the maintenance of acetic acid tolerance. Our findings provide previously uncharacterized mechanistic insights about how yeast cells adapt to exposure to acetic acid.
RESULTS
TORC2-dependent phosphorylation of Ypk1 is activated upon challenge with acetic acid
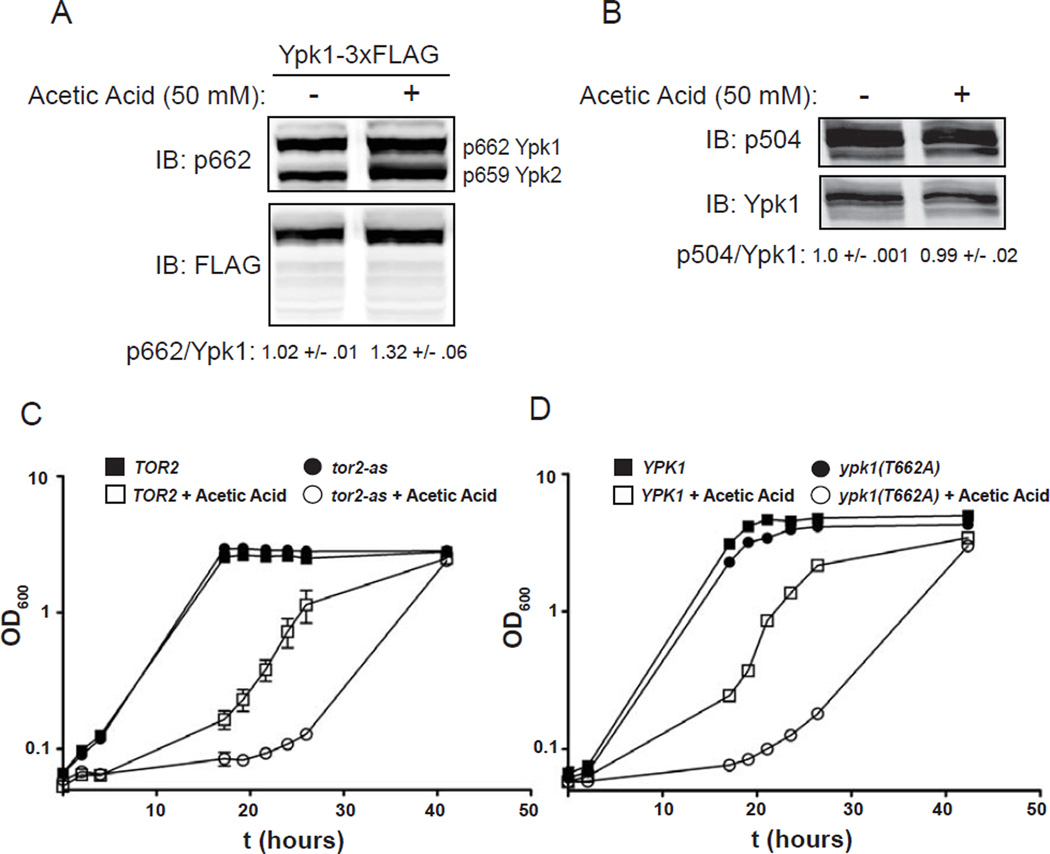
We first examined whether TORC2-Ypk1 signaling is modulated under acetic acid stress using a phosphosite-specific antibody that recognizes P-T662 in Ypk1 [33]. We have found (our unpublished results) that this antibody cross-reacts with an equivalent residue, P-T659, in the paralog Ypk2, which is also known to be phosphorylated in a TORC2-dependent manner [31]. Upon exposure of cells to 50 mM acetic acid, we observed a readily detectable and reproducible increase in the fraction of Ypk1 molecules phosphorylated at T622 (and a similar increase in P-T659 in Ypk2) (Fig. 1A). By contrast, under the exact same conditions, there was no observable increase in phosphorylation at T504 (Fig. 1B), the site regulated by Pkh1 and Pkh2 [30]. Thus, TORC2-dependent phosphorylation of Ypk1 and Ypk2 was specifically increased in response to acetic acid treatment, whereas Pkh1- and Pkh2-dependent phosphorylation was not.
Figure 1.
Exposure to an inhibitory sub-lethal acetic acid concentration stimulates TORC2-dependent Ypk1 activation and both enzymes are important for cellular adaptation to this stress. (A) A Ypk1-3xFLAG expressing strain (YDB379) was grown to mid-exponential phase in MM4 medium and then cells were switched into control MM4 medium of MM4 medium supplemented with 50 mM acetic acid for 60 min. Cells were harvested and protein extracted. Extracts were resolved by SDS-PAGE, and blotted as in Materials and Methods. (B) Extracts from control or acetic acid treated wild-type (BY4741) cells were resolved by SDS-PAGE, and blotted as in Materials and Methods. (C) TOR2 (yKL4) or hypomorphic tor2-as (yKL5) strains were grown to exponential phase in MM4 medium and then diluted to OD600=0.05 in control MM4 medium or MM4 medium supplemented with 60 mM acetic acid and grown in batch culture for the indicated times. For liquid cultures, each was grown in triplicate and the error bars indicate the S.E.M. of replicates at each time point. (D) ypk1Δ (JTY6142) cells were transformed with plasmids expressing Ypk1 (pAM20) or Ypk1(T662A) (pFR221), which prevents TORC2 phosphorylation of Ypk1 at this residue and thus prevents full activation of Ypk1 by TORC2. Growth of cells expressing these constructs in MM4 medium containing acetic acid was determined as in (C).
TORC2 and Ypk1 action are important for cells to cope with acetic acid stress
We next investigated whether TORC2 activity and, more specifically, its activation of Ypk1 are important for cellular adaptation to acetic acid. First, growth of a tor2-as strain, which has chronically diminished TORC2 activity [43], was compared to growth of an otherwise isogenic TOR2+ strain on a defined medium (MM4) at pH 4 in the absence or presence of 60 mM acetic acid. Although both TOR2+ and tor2-as strains grew at very similar rates in the control medium lacking acetic acid, the tor2-as cells exhibited a much more protracted lag phase and a slower overall growth rate than the TOR2+ cells in the medium containing acetic acid (Fig. 1C). Second, and similarly, we used a Ypk1 mutant, Ypk1(T662A), that also displays diminished functionality [44]. Again, we observed that, although both YPK1+ and ypk1(T662A) strains grew at very similar rates in the control medium lacking acetic acid, the ypk1(T662A) cells exhibited a more protracted lag phase and a slower overall growth rate than the YPK1+ cells in the medium containing acetic acid (Fig. 1D). Thus, the stimulation of TORC2 in response to acetic acid, and its subsequent activation of Ypk1 by phosphorylation at T662, are physiologically important for the ability of yeast cells to adapt to sudden exposure to acetic acid. Thus, Ypk1 is the major downstream effector of TORC2 in this stress response.
Phosphorylation of Ypk1 substrates is upregulated in response to acetic acid stress
When Ypk1 becomes activated, among its well-documented in vivo substrates are Orm1 and Orm2, two protein inhibitors of SPT activity. It has been amply demonstrated that Ypk1-mediated phosphorylation of Orm1 occurs specifically on residues S51, S52 and S53 and, likewise, that Ypk1-mediated phosphorylation of Orm2 occurs at the equivalent residues (S45 S47 S48) [32]. Similarly, Ypk1 phosphorylates Lac1 and Lag1, the two highly related catalytic subunits of the ceramide synthase complex, at equivalent sites (S23 and S24 in both proteins) [40].
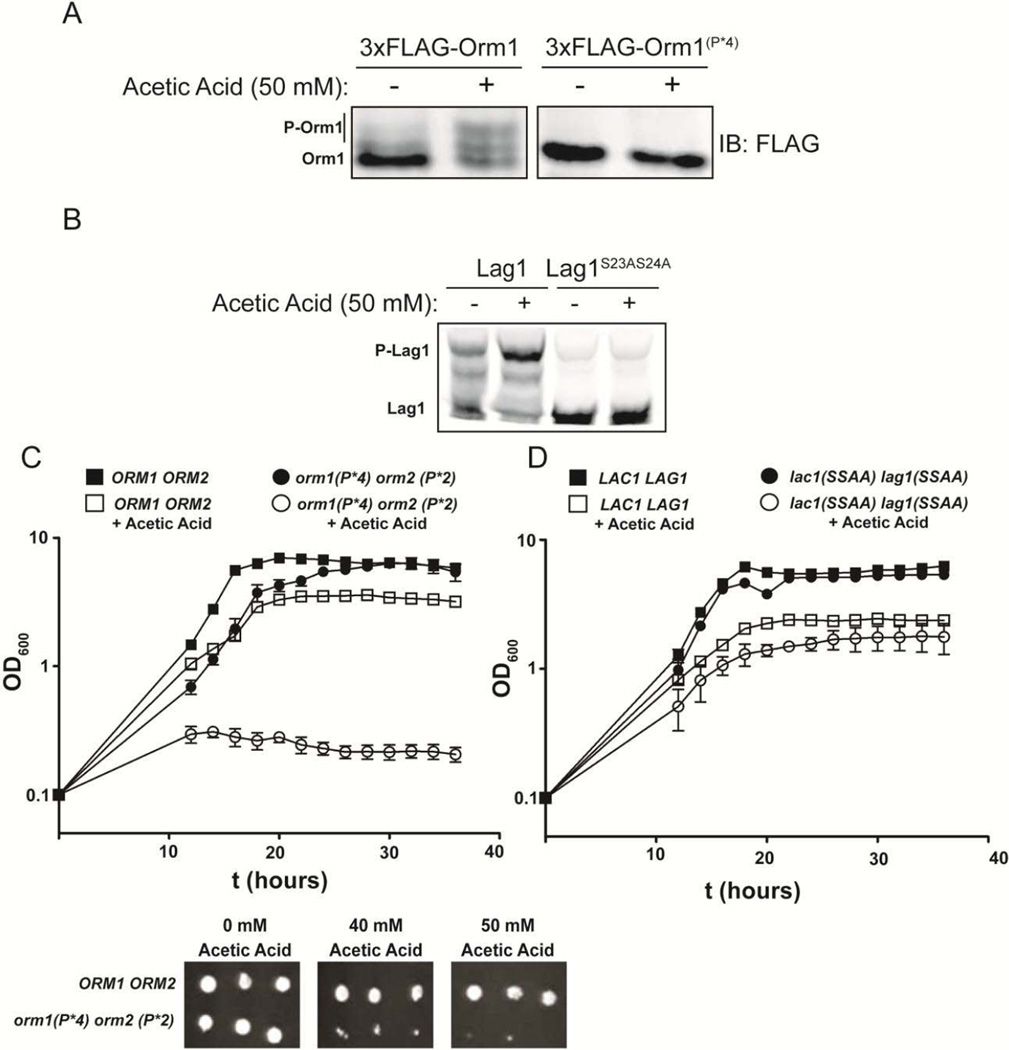
Therefore, to further verify that acetic acid stress activates Ypk1, we tested whether acetic treatment induced the Ypk1-mediated phosphorylation of these substrates. Modification of these proteins is most conveniently monitored by the fact that the mobility of the phosphorylated species is retarded, compared to the unphosphorylated state, when the proteins are resolved by SDS-PAGE or using the Phos-tag™ gel method. Indeed, we found that acetic acid treatment caused a prominent increase in the slower mobility Orm1 species (Fig. 2A, left panel), indicative of greatly enhanced phosphorylation. The appearance of these species was totally abrogated when cells expressed (as the sole source of Orm1) a mutant (orm1-P*4) [41] in which the known Ypk1 phosphorylation sites are mutated to Ala (Fig. 2A, right panel), consistent with the conclusion that Ypk1-dependent phosphorylation is likely responsible for the observed mobility shift. However, in addition to S51A S52A S53A, the orm1-P*4 allele also carries five other mutations (S29A S32A S34A S35A S36A) [41]. Hence, we could not rule out that some (or all) of the observed phosphorylated species might arise from modifications occurring at these other positions. For this reason, we also examined Lag1 and Lac1. Reassuringly, we found that acetic acid treatment also caused a marked increase in the slower mobility species for both Lag1 (Fig. 2B) and Lac1 (data not shown); moreover, these mobility shifts were completely abolished by mutation of just the two known Ypk1 sites in both proteins (S23A S24A), confirming that Ypk1 was responsible for the observed acetic acid-induced modifications.
Figure 2.
Exposure to an inhibitory sub-lethal acetic acid concentration triggers Ypk1-mediated phosphorylation of proteins whose modification stimulates sphingolipid production and is important for survival under this stress. (A) A strain (JTY5125) expressing either 3xFLAG-Orm1 (YDB146) or 3xFLAG-Orm1(P*4), as indicated, was grown to mid-exponential phase in MM4 medium and then cells were switched into control MM4 medium or MM4 medium supplemented with 50 mM acetic acid for 60 min. Cells were harvested and protein extracted. Extracts were resolved by SDS-PAGE, and blotted as in Materials and Methods. (B) Wild-type (BY4741) cells expressing 3xFLAG-Lag1 (yAM159-A) or 3xFLAG-Lag1(S23AS24A) (yAM163-A) from their genomic loci were exposed to acetic acid, harvested and protein extracted as in (A). Extracts were resolved by SDS-PAGE, and blotted as in Materials and Methods. (C) Cells (JTY5215) expressing either 3xFLAG-Orm1 and 3xHA-Orm2 (JTY6140) or the 3xFLAG-Orm1(P*4) and 3xHA-Orm2(P*2) (JTY5215) mutants (which cannot be phosphorylated by Ypk1) were grown to mid-exponential phase in MM4 medium and then diluted to OD600=0.1 and grown in the wells of microtiter plates in either control MM4 medium or MM4 medium containing 50 mM acetic acid as described in Materials and Methods. For liquid cultures, each was grown in at least quadruplicate and the error bars indicate the SEM of replicates at each time point. (D) Liquid growth assays were performed as in (C) for LAC1 LAG1 (yAM205 - A) and Lac1(S23AS24A) Lag1(S23AS24A) (yAM207 - B) strains, as indicated. (E) Cells expressing either 3xFLAG-Orm1 and 3xHA-Orm2 (JTY6140) or 3xFLAG-Orm1(P*4) and 3xHA-Orm2(P*2) (JTY5215) were grown to mid-exponential phase and then serial dilutions were made and spotted onto MM4 agar plates containing the indicated acetic acid concentrations. Plates were imaged after 3 days of growth at 30 °C.
Ypk1 stimulation of the sphingolipid pathway is required for acetic acid tolerance
Having found that TORC2-Ypk1 activation in response to acetic acid triggers the phosphorylation of targets that should result in an increase in both SPT and ceramide synthase activity raised the possibility that the role that TORC2 and Ypk1 play in promoting adaptation to acetic acid stress is upregulation of sphingolipid biosynthesis. This hypothesis is consistent with two additional observations. First, other enzymes involved in sphingolipid metabolism have been implicated genetically in the response to acetic acid stress [20, 45]. Second, lipidomic analysis has shown that readily detectable increases in sphingolipid species occur in response to acetic acid treatment [46].
If TORC2-Ypk1-dependent upregulation of sphingolipid synthesis is critical for adaptation to acetic acid stress, we reasoned that preventing TORC2-Ypk1-mediated phosphorylation of Orm1 and Orm2, or Lac1 and Lag1, or both, would compromise the ability of yeast cells to grow when challenged with acetic acid. In agreement with this conclusion, we found that an ORM1+ ORM2+ strain and otherwise isogenic cells carrying the orm1-P*4 orm2-P*2 alleles, in which all of the Ypk1 sites (in bold) in both proteins have been mutated to Ala [P*4 = S29A S32A S34A S35A S36A S51A S52A S53A; P*2 = S9A S15A T18A S46A S47A S48A] grew at very similar rates in the control medium lacking acetic acid; but, in marked contrast, and unlike the parental ORM1+ ORM2+ strain, the orm1-P*4 orm2-P*2 cells were unable to grow in the medium containing acetic acid (Fig. 2C). It has been shown in prior work that orm1-P*4 orm2-P*2 cells have decreased SPT activity and are hypersensitive to myriocin, an antibiotic that is a potent SPT inhibitor [41], and that TORC2-dependent and Ypk1-mediated phosphorylation of Orm1 and Orm2 is required to derepress sphingolipid biosynthetic activity [32]. Thus, the inability of orm1-P*4 orm2-P*2 cells to grow in medium containing high acetic acid is consistent with the need for Ypk1-evoked stimulation of LCB production to cope with this stress.
It has been observed previously that basal ceramide synthase activity is quite high and that Ypk1-mediated phosphorylation stimulates this enzyme complex just 2-fold [40]. Moreover, cells carrying the lac1(S23A S24A) lag1(S23A S24A) alleles (lacking the Ypk1 sites in both proteins) are more sensitive to myriocin than otherwise isogenic LAC1+ LAG1+ cells, but only modestly so [40]. Nonetheless, although LAC1+ LAG1+ and lac1(S23A S24A) lag1(S23A S24A) cells grew quite comparably in the control medium lacking acetic acid, there was detectably poorer growth of the lac1(S23A S24A) lag1(S23A S24A) cells, compared to the LAC1+ LAG1+ control, in the medium containing acetic acid (Fig. 2D). Again, however, this effect was modest, compared to the severe growth inhibition observed upon challenge with an inhibitory acetic acid concentration when the blockade of SPT activity imposed by Orm1 and Orm2 cannot be alleviated by their Ypk1-mediated phosphorylation (Fig. 2E). Thus, elevating flux into the sphingolipid biosynthetic pathway via TORC2-dependent Ypk1-mediated activation of SPT is a prerequisite for achieving cell survival and adaptation in response to acetic acid stress.
TORC2-Ypk1 activation is not dependent on protein kinase Hrk1
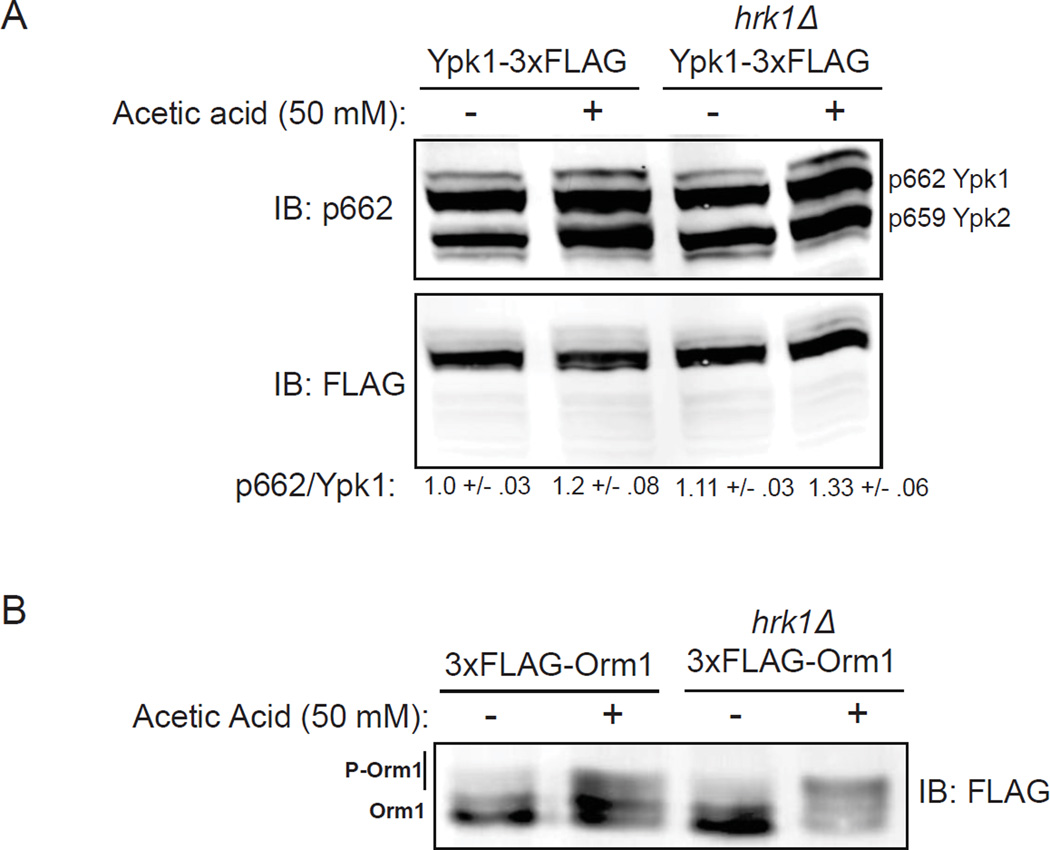
We next sought to determine whether any previously described acetic acid-evoked signaling system might be responsible for the activation of TORC2-Ypk1 by acetic acid stress that we observed. We focused our attention on the protein kinase Hrk1, an enzyme implicated in the regulation of the PM H+-ATPase (Pma1) [18] and expressed under the control of the transcription factor Haa1 in response to acetic acid stress [17], but otherwise a very poorly characterized member of the Npr1 sub-family of yeast protein kinases [47]. When cultivated in the presence of a mildly inhibitory concentration of acetic acid, hrk1Δ cells exhibit a protracted lag phase and an increase in the intracellular concentration of this weak acid, compared to HRK1+ cells [17]. Also, effects on the biosynthesis of complex sphingolipids are among the many cellular roles ascribed to the related protein kinase Npr1, a more well studied enzyme that is negatively regulated by TORC1 [48, 49]. Therefore, we examined whether Hrk1 is necessary for the TORC2-Ypk1 activation that occurs under acetic acid stress. However, upon challenge with 50 mM acetic, we found that TORC2-mediated phosphorylation of Ypk1 and Ypk2 was unaffected in hrk1Δ cells, compared to HRK1+ control cells (Fig, 3A) and, likewise, that Ypk1-mediated Orm1 phosphorylation was not affected (Fig. 3B). Thus, Hrk1 is not required for either TORC2 activation or Ypk1 signaling to the sphingolipid biosynthetic pathway upon acetic acid stress.
Figure 3.
The acetic acid-tolerance determinant protein kinase Hrk1 is not required for either TORC2 activation or Ypk1 action upon acetic acid stress. (A) Ypk1-3xFLAG (YDB379) or hrk1Δ Ypk1-3xFLAG (yAM336) strains were treated as in Fig. 1A. Ypk1 T662 (and Ypk2 T659) phosphorylation was determined by immunoblotting as in Fig. 1A. (B) 3xFLAG-Orm1 (YDB146) or hrk1Δ 3xFLAG-Orm1 (yAM336) strains were treated as above. Orm1 phosphorylation was determined by immunoblotting as in Fig. 2A.
Acetic acid tolerance requires production of complex sphingolipids
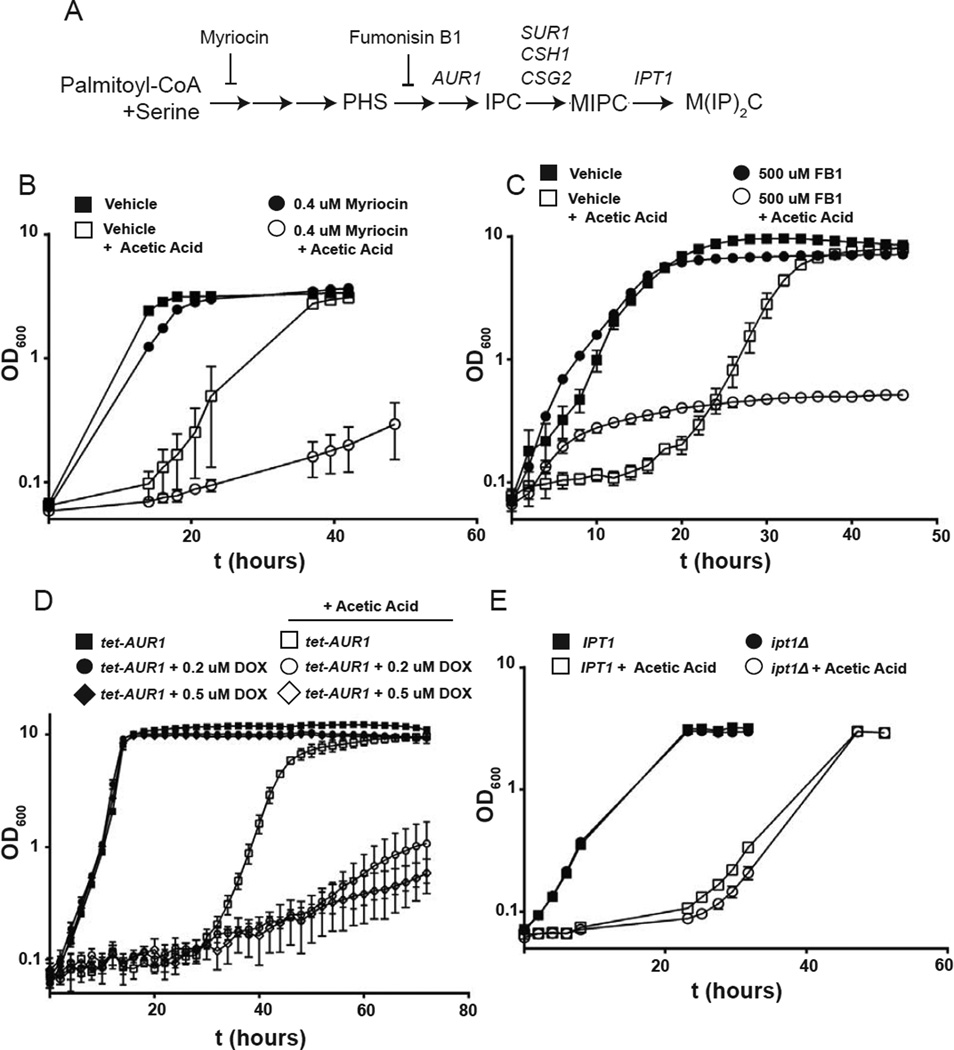
Activation of TORC2-Ypk1 signaling increases metabolic flux into the sphingolipid pathway, which commences with the generation of LCBs, which are converted to ceramides and then to more complex sphingolipids. Therefore, we next investigated which of the intermediates or end-products of the sphingolipid biosynthetic pathway must be produced for cells to acquire acetic acid tolerance. Toward this end, we used either chemical and/or genetic means to block the sphingolipid pathway at discrete steps (Fig. 4A) and assessed the ability of the cells subjected to these blocks to cope with the challenge of acetic acid stress, as judged by their growth rate. We found, first, that a sublethal dose of myriocin, which causes only a minor effect on the specific growth rate and final biomass concentration of wild-type cells in medium lacking acetic acid, greatly exacerbated the growth inhibitory effect of exposure to 60 mM acetic acid at pH 4 (Fig. 4B), consistent with the need to upregulate flux into the sphingolipid pathway via generation of LCBs to adapt to acetic acid stress. Likewise, preventing utilization of the primary yeast LCB, PHS, by addition of fumonisin B1, a specific inhibitor of ceramide synthase [50], also greatly potentiated the growth inhibitory effect of acetic acid treatment (Fig. 4C). Similarly, using the Tet-Off system to block production of phosphatidylinositol:ceramide phosphoinositol transferase (Aur1), the enzyme responsible for inositolphosphorylceramide (IPC) synthesis [51], while causing no obvious growth defect in medium lacking acetic acid, caused a severe growth debility in the medium containing acetic acid (Fig. 4D). At last, we found in our work that ipt1Δ cells, which lack the enzyme (inositolphosphotransferase) necessary for mannose di(inositolphosphoryl)ceramide [M(IP)2C] formation, are no more acetic acid sensitive than IPT1+ control cells (Fig. 4E), suggesting that either IPC or mannosylinositol phosphorylceramide (MIPC), but not the M(IP)2C derived from it, are required for adaptation to acetic acid stress.
Figure 4.
Perturbation of complex sphingolipid synthesis causes increased acetic acid sensitivity. (A) Schematic diagram of sphingolipid biosynthesis in S. cerevisiae. Adapted from (52). Pharmacological or genetic perturbations used in this study and their points of inhibition are shown. (B) Growth of wild-type (BY4741) yeasts in MM4 medium supplemented with or without acetic acid in the presence of vehicle (methanol) or 0.4 µM myriocin in the same solvent was determined by liquid growth assay as in Fig. 1C. (C) Wild-type (BY4741) yeasts were grown to exponential phase in MM4 medium and then diluted to OD600=0.05 and grown in MM4 medium supplemented with or without acetic acid in the presence of vehicle (methanol) or 500 µM fumonisin B1 in the same solvent. (D) Growth of a tet-AUR1 strain (MTY175) in MM4 medium supplemented with or without acetic acid in the presence of vehicle (50% v/v ethanol) or either 0.2 or 0.5 µM doxycycline in the same solvent, as indicated, was determined by liquid growth assay as in (C). (E) Growth of IPT1 (BY4741) or ipt1Δ (M24) yeast strains in MM4 medium supplemented with or without acetic acid was measured as in (B).
Supplementation with phytosphingosine increases acetic acid tolerance
If elevated intracellular LCB production in response to acetic acid challenge is required to drive higher levels of complex sphingolipids to permit cellular adaptation to this stress, then we reasoned that perhaps the simple expedient of supplementing the growth medium with the cell-permeable LCB PHS might be sufficient to increase flux into the sphingolipid biosynthetic pathway and thereby confer greater acetic acid tolerance. Supplementing the MM4 medium with 20–30 µM PHS did not impair the growth of wild-type (ORM1+ OMR2+) cells in medium lacking acetic, nor did it markedly improve the growth of the cells in the presence of 40 mM acetic acid (Fig. 5, top row). However, in otherwise isogenic cells carrying the orm1-P*4 orm2-P*2 alleles (where upregulation of flux into the sphingolipid pathway in response to acetic acid-evoked TORC2-Ypk1 signaling cannot occur), which had significantly impaired growth in the presence of 40 mM acetic acid, the presence of PHS in this concentration range (20–30 mM) permitted detectable improvement in the level of survival (Fig. 5, bottom rows). Thus, in the presence of an inhibitory concentration of acetic acid, LCBs (and the sphingolipids derived from them, based on the other results we have presented here) are indeed limiting factors for adaptation to this stressful condition.
Figure 5.
Exogenously-supplied PHS enhances acetic acid tolerance of S. cerevisiae. Serial dilution of otherwise wild-type cells expressing either +93xFLAG-Orm1 and 3xHA-Orm2 (JTY6140) or the 3xFLAG-Orm1(P*4) and 3xHA-Orm2(P*2) mutants were grown to mid-exponential phase and then serial dilutions were made and spotted onto MM4 agar plates containing the indicated acetic acid and PHS concentrations. Plates were imaged after 3 days of growth at 30 °C.
DISCUSSION
In this work, we obtained several important new insights about the mechanisms by which S. cerevisiae cells combat exposure to an inhibitory but sub-lethal acetic acid concentration. We documented, for the first time, that under this stress TORC2-Ypk1 signaling is activated. Moreover, we demonstrated that TORC2-Ypk1-mediated stimulation of sphingolipid biosynthesis makes a major contribution to the ability of the cells to endure acetic acid stress because inhibition of either TORC2 or Ypk1, or blockade of sphingolipid production, substantially exacerbates the growth inhibitory effect of exposure to acetic acid. Conversely, we showed that even a simple manipulation to increase through-put through the sphingolipid pathway, such as supplying exogenously the specific pathway precursor PHS, improves the tolerance of a susceptible yeast strain to acetic acid. Our findings greatly extend previous genomewide studies that implicated genes involved in sphingolipid metabolism, among many other genes, in acquisition of tolerance to acetic acid [17, 20, 21]. Our results also explain the underlying biochemical basis for a prior observation, revealed by lipidomic profiling. It was found that exponentially-growing yeast cells adapted to a concentration of acetic acid that reduced their doubling time by 50% exhibited an increase in their total content of complex sphingolipids and extensive changes in the profile of the complex sphingolipids present, compared to cells cultivated in the same way, but not exposed to acetic acid [46]. The conclusions of our study are also in agreement with a recent report [52] providing evidence that the intrinsically high sphingolipid content in the PM of Zygosaccharomyces bailii, a hemiascomycete distantly related to S. cerevisiae, is an important contributor to the high acetic acid tolerance of this organism. In this regard, we demonstrated that, at least in S. cerevisiae, it is IPC and MIPC, and not M(IP)2C, that are required for acetic acid tolerance. Three observations suggest this. First, it has been observed in prior work that a mutation (csg2Δ) that impairs MIPC formation causes growth impairment at low pH values [21, 53], like the pH 4 MM4 medium that we used to test the effect of exogenous acetic acid concentration. Second, and more tellingly, it has been reported previously that sur1Δ cells, which also have reduced MIPC production, are more susceptible to the growth inhibitory effect of acetic acid [20]. Third, we observed in this work that a mutant with impaired M(IP)2C formation, exhibited no increase in acetic acid sensitivity when compared with the corresponding parental strain. Collectively, the results presented are consistent with the conclusion that it is the early complex sphingolipids IPC and MIPC that are the species primarily responsible for conferring acetic acid tolerance. In agreement with this scenario, loss-of-function mutations in SUR2 (an enzyme that generates PHS de novo) [53] and in YPC1 (an enzyme that can produce PHS from the breakdown of phytoceramides) [54] have lower levels of this LCB, and both genes are transcriptionally upregulated in a Haa1-dependent manner in response to acetic acid stress [17, 20]. This upregulation presumably leads to an increase in the rate of synthesis of those sphingolipid species most needed to best resist the effects of acetic acid-induced stress. In this same regard, a scs7Δ mutant, which lacks an enzyme necessary to alpha-hydroxylate the VLCFA in sphingolipids, contains an altered spectrum of IPC species and is more sensitive to acetic acid than the corresponding parental strain [20]. Given that TORC2, Ypk1 and the mechanism by which they regulate sphingolipid production appear to be largely conserved throughout the fungal clade [40], the response we have described here likely plays an important role in the tolerance to acetic acid of many other yeast species, such as Z. bailii [52].
Given what we have now established, several questions arise that are worthy of further investigation. First, it is unclear why elevation of complex sphingolipids permits growth on medium containing inhibitory concentrations of acetic acid. One suggestion is that an increase in the content of VLCFA-containing sphingolipids leads to a thicker and more dense PM, thereby increasing the free energy barrier for permeation of acetic acid [52], or enhancing cohesion among the lipids in the outer leaflet, which would be important to counteract the hypotonic shock reported to be induced by this weak acid [55]. Moreover, inadequate sphingolipid biosynthesis is known to deleteriously influence the trafficking of integral membrane proteins from the Golgi body to the PM [56, 57], and several proteins required for this vesicle-mediated protein sorting (Sur2, Sur4 and Ypc1) have known roles in sphingolipid metabolism [58]. Since several PM proteins (e.g., Fps1, Pma1, Tpo2, Tpo3), as well as Yro2 and its paralog Mrh1 [59], have been implicated in yeast adaptive response to acetic acid, it is possible that increasing sphingolipid production may increase the efficiency of the delivery to the PM of these proteins required for acetic acid tolerance.
Another open question of interest is how acetic acid stress stimulates TORC2 activity. TORC2 could be coupled physically to a PM protein that serves as a direct sensor of acetic acid or its influx, in agreement with the recent observation that Tor2 (the catalytic component of TORC2) interacts physically with Sho1, an integral PM protein that is a putative osmosensor [60]. Additionally, TORC2 level or activity could be increased indirectly by changes elicited by acetic acid-sensing pathways or by changes in cellular properties brought on by the effects of this weak acid. Neither the transcription of TOR2, nor any of the genes encoding the other subunits of the TORC2 complex (AVO1, AVO2, AVO3/TSC11, BIT2, BIT61, SLM1 or SLM2), nor YPK1, is induced by acetic acid stress or dependent for their expression under acetic acid stress on the Haa1 transcription factor [17]. Thus, as we showed by immunoblotting for Ypk1 itself, the levels of the components of TORC2 are likely not affected when cells are challenged with acetic acid. Similarly, we found that the protein kinase Hkr1 that is upregulated by acetic acid treatment in a Haa1-dependent manner, is not required for the observed increase in TORC2-Ypk1 signaling that occurs in response to acetic acid stress. So, given that TORC2 is a PM-associated protein complex [61], TORC2 may instead sense the presence of acetic acid indirectly by responding to the ensuing membrane stretch, akin to the activation of TORC2 elicited by other hypo-osmotic stress conditions, which appears to depend on changes in the localization of the Slm1 and Slm2 subunits of TORC2 [35]. Alternatively, given that the MAPK Slt2/Mpk1 is also activated under hypo-osmotic conditions [62], it may be involved, directly or indirectly, in stimulating TORC2 activity.
A third lingering question is whether TORC2-activated Ypk1 also contributes to acetic acid tolerance in other ways, aside from elevating the production of IPC and MIPC per se. For example, sphingolipids are largely found in the outer leaflet of the PM [41, 56, 57], and Ypk1 also affects the composition of the PM bilayer by negatively regulating the protein kinases Fpk1 and Fpk1, whose function is, in turn, to stimulate the flippases (mainly Dnf1 and Dnf2) that translocate aminoglycerophospholipids from the outer to the inner leaflet [38]. Thus, Ypk1-mediated down-modulation of flippase function may be physiologically important in altering the properties of the PM in response to acetic acid stress. One group has reported that Lag1-dependent de novo ceramide production (as well as the production of ceramides by the mitochondrially-localized complex sphingolipid hydrolase Isc1) is causal for the mitochondrial damage and increased cell inviability induced by acetic acid [45]. However, as we have shown before, activated Ypk1 phosphorylates Lag1 and Lac1 and stimulates ceramide production [40], which, as we have documented here, is protective for cells exposed to acetic acid. In this same vein, the apparent damage to mitochondria caused by acetic acid treatment reportedly increases cellular ROS levels to a harmful level [63, 64] and, consistent with the findings we present here, others have shown that TORC2-Ypk1 signaling and sphingolipid production are important for maintaining cellular ROS levels in a tolerable range [65], although the mechanism by which TORC2-Ypk1 signaling does so is unclear.
Finally, from a practical standpoint, we have shown here that just promoting sphingolipid production by adding PHS to the growth medium provides a clear survival benefit to cells during adaptation to acetic acid stress. Thus, appropriate engineering to upregulate the sphingolipid pathway in situ may provide a readily accessible and rational means to enhance the acetic acid tolerance of industrially valuable S. cerevisiae strains, either alone or in combination with other genetic strategies that appear to improve the acetic acid tolerance of this organism [66–68].
MATERIALS AND METHODS
Construction of yeast strains and growth conditions
S. cerevisiae strains used in this study are listed in Table 1. Strains were constructed using standard yeast genetic manipulations [69]. When generating strains with chromosomal modification, integration of the DNA fragment into the correct genomic loci was confirmed by PCR using genomic DNA from isolated colonies as template and with oligonucleotides complementary to the integrated DNA fragment and complementary to genomic sequence at least 150 bases away from the integration site as primers for the reaction.
Table 1.
Yeast strains used in this study.
| Strain | Genotype | Source/reference |
|---|---|---|
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Research Genetics, Inc. |
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Research Genetics, Inc. |
| YDB379 | BY4741 Ypk1-3xFLAG::natNT2 | J.S. Weissman, Univ. of California, San Francisco |
| yKL4 | BY4741 TOR2+::Hygr | [40] |
| yKL5z | BY4741 Tor2(L2178A)::Hygr | [40] |
| JTY6142 | BY4741 ypk1Δ::KanMX4 | Research Genetics, Inc. |
| YDB146 | BY4741 3xFlag-Orm1 | [41] |
| yAM159-A | BY4741 3xFLAG-Lag1::LEU2 | [40] |
| yAM163-A | 3xFLAG-Lag1(S23A S24A)::LEU2 | [40] |
| JTY6140 | BY4741 MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 can1Δ::pSte2-His5 lyp1Δ::pSte3- Leu5 3xFLAG-Orm1 3xHA-Orm2 |
[41] |
| JTY5215 | BY4741 MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 can1Δ::pSte2-His5 lyp1Δ::pSte3- Leu5 3xFLAG-Orm1-P*4 3xHA-Orm2-P*2 |
[41] |
| yAM205-A | BY4742 Lac1::LEU2 Lag1::LEU2 | [40] |
| yAM207-B | BY4742 Lac1(S23A S24A)::LEU2 Lag1(S23A S24A)::LEU2 |
[40] |
| yAM327 | BY4741 Ypk1-3xFLAG hrk1Δ::kanMX4 | This study |
| yAM336 | BY4741 3xFlag-Orm1 hrk1Δ::kanMX4 | This study |
| M24 | BY4741 ipt1Δ::kanMX4 | Research Genetics, Inc. |
| MTY175 | BY4741, tetO7-AUR1::kanMX4 URA3 | M. Tani, Kyushu University, Japan |
Yeast cultures were grown in either Yeast Peptone Dextrose medium (YPD) (containing per liter: 10 g Bacto yeast extract, 20 g Bacto Peptone, and 20 g glucose) or MM4 medium adjusted to pH 4.0 (containing per liter: 1.7 g yeast nitrogen base without amino acids or ammonium sulfate, 20 g glucose, 2.65 g ammonium sulfate and 60 mg leucine, and further supplemented with 20 mg methionine, 20 mg histidine, 60 mg leucine, 20 mg uracil, or 30 mg lysine according to the strains' autotrophy).
Plasmids and recombinant DNA methods
Plasmids used in this study are listed in Table 2. All plasmids were constructed by standard laboratory methods [70]. Constructs generated in this study had all promoter and coding regions sequences confirmed by sequencing analysis.
Table 2.
Plasmids used in this study.
Preparation of cell extracts and immunoblotting of phosphorylated proteins
Immunoblot analysis of phosphorylated Lac1, Lag1 and Ypk1 species was performed as previously described [37, 40]. Briefly, alkaline lysis and trichloroacetic acid precipitation [71] was used to make whole cell protein extracts. To detect Ypk1 phosphorylated species, 15 µL of the extract was resolved by standard SDS-PAGE using 8% acrylamide gels. To detect Lac1 and Lag1 phosphorylated species, 15 µL of TCA extract was resolved by Phostag SDS-PAGE [72] at 160 V using gels containing 8% acrylamide, 35 µM MnCl2, and 35 µM Phostag (Wako Chemicals USA, Inc.).
After SDS-PAGE resolution, protein extracts were transferred to nitrocellulose and incubated with primary antibody in Odyssey buffer (Licor Biosciences). Membranes were washed and then incubated with appropriate secondary antibodies [anti-mouse, anti-rabbit or anti-goat IRDye680LT or IRDye800 conjugated IgG (Licor Biosciences)] diluted 1:10000 in Odyssey buffer with 0.1% Tween-20 and 0.02% SDS). Blots were imaged using an Odyssey infrared scanner (Licor Biosciences).
Primary antibodies and dilutions used in this study were: 1:5000 mouse anti-FLAG (Sigma-Aldrich), 1:500 rabbit anti-pSGK (T256) (to recognize Ypk1 phosphorylated at T504) (Santa Cruz Biotechnology, Dallas, TX), 1:20000 rabbit anti-Ypk1(P-T662) (generous gift from Ted Powers, University of California, Davis) and 1:1000 goat anti-Ypk1 (Santa Cruz Biotechnology, Dallas, TX).
Immunopurification and immunodetection of Orm1
Exponentially growing cells expressing 3xFLAG-Orm1 were pelleted and transferred into MM4 or MM4 with 50 mM acetic acid for 1 h. 40 mL of each culture was then harvested and washed with 1 mL of media. The cells were then frozen in liquid nitrogen and stored at −80 °C. The pellets were resuspended in 0.5 mL of ice cold IP buffer (50 mM Hepes-KOH pH 7.5, 150 mM KOAc, 2 mM MgOAc, 1 mM CaCl2, 15% glycerol, 4 mM NaVO4, 50 mM NaF, 20 mM Na-PPi, and 1× complete protease inhibitor [Roche, Basel, Switzerland]) with 0.1% digitonin. An equal volume of glass beads were added to each cell slurry and cells were lysed by shaking using a FastPrep 120 (Thermo Fisher Scientific). 0.5 mL of cold IP buffer with 1.9% digitonin was added and lysates were rotated for 45 min. at 4 °C. Lysates were clarified by centrifugation for 30 min at 13,200 rpm (16,100×g) in a microfuge (Eppendorf 5415D) at 4 °C. Lysate protein concentration was measured by Pierce BCA protein assay (Thermo Fisher Scientific) and protein concentration of each lysate was normalized. Immunoprecipitation was carried out by adding 25 µl of anti-FLAG agarose (Sigma) washed with 1 ml cold IP buffer with 0.1% digitonin and resuspended in 25 µL of the same buffer to each sample and rotating this slurry at 4 °C for 2.5 h. After rotation, the resin was washed four times with 1 ml cold IP buffer with 0.1% digitonin. Protein was eluted by adding 25 µl of PBS and 10 µl of 5× Laemlli buffer, and boiling for 8 min. Five µl of the IP extract were resolved by SDS-PAGE using 10% 75:1 acrylamide:bis-acrylamide gels resolved at 120 V. Proteins were subsequently transferred from SDS–PAGE gels to nitrocellulose membranes and blotted as described above.
Yeast growth assays
Growth assays were carried out either in solid agar media or in liquid broth batch cultures in flasks or microtiter plates. For growth assays in agar, cells were grown in MM4 media at pH 4.5 at 30°C, with orbital agitation (250 rpm) until culture OD600 = 0.7 ± 0.07 was reached. These cells were resuspended in sterile water to obtain suspensions with OD600 = 0.05 ± 0.005. These cell suspensions and dilutions of 1:5 and 1:10 were applied as 3 µl spots onto the surface of either MM4 medium (pH 4.5) or MM4 medium (pH 4.5), supplemented with 0.05% Tergitol Type NP-40, and with adequate concentrations of acetic acid and solvent (methanol) or PHS, and incubated at 30 °C for 2 to 3 days, according to the severity of growth inhibition. Plates were then scanned on a flat bed scanner and growth phenotypes examined.
For liquid growth assays in flasks, cells cultivated until mid-exponential phase (OD600 = 0.8 ± 0.08) in MM4 growth medium (at pH 4.0) were used to inoculate cultures at an initial OD600 of 0.05 ± 0.005 in MM4 (at pH 4.0) or MM4 medium supplemented with 50 mM acetic acid and either solvent alone (methanol) or 0.4 µM myriocin. Cultures were then grown at 30 °C and the OD600 of the culture was determined at the indicated times.
For liquid growth assays in microtiter plates, exponentially growing cells in MM4 medium were diluted to OD600 = 0.1 or OD600 = 0.05 in either MM4 medium or MM4 medium with 40 or 50 mM acetic acid and solvent [methanol or 50% ethanol (v/v)] or 500 µM fumonisin B1 or 0.2–1 µg/ml doxycycline. Then, 100 µl or 150 µl of each culture was placed in a well of a 96 flat bottom well plate. Cultures were grown with orbital shaking at 30°C in a Tecan Infinite M-1000 PRO plate reader (Tecan Systems Inc., San Jose, CA) or FilterMax F5 Multi-Mode Microplate Readers (Molecular Devices) for 36 or 72 hr, respectively. Absorbance measurements were taken every 15 min. Absorbance values were converted to OD600 values using a standard curve of absorbance values of cultures at known OD600 taken on the same plate reader.
Summary Statement.
We document that during yeast adaptation to acetic acid stress, TORC2-Ypk1 signaling is activated, contributing to yeast cell ability to endure this stress. Manipulation of the sphingolipid pathway is proposed as a way to increase yeast fitness in industrial biotechnology.
Acknowledgments
We thank Jonathan Weissman and Motohiro Tani for the generous gift of certain yeast strains used in this study, Garrett Timmons for technical assistance, Françoise Roelants for technical assistance and critical discussion, and other members of the Thorner Lab for various reagents, plasmids, and helpful advice.
FUNDING
This work was supported by Ph.D. grant SFRH/BD/80065/2011 (to J.F.G.) from the Fundação para a Ciência e a Tecnologia (FCT), by NIH Predoctoral Training Grant GM07232 and a Predoctoral Fellowship from the UC Systemwide Cancer Research Coordinating Committee (to A.M.), by NIH R01 Research Grant GM21841 and Senior Investigator Award 11-0118 from the American Asthma Foundation (to J.T.), by FCT contracts PTDC/BBB-BEP/0385/2014 and UID/BIO/04565/2013 (to I.S.C.) and by funds (Project N. 007317) from Programa Operacional Regional de Lisboa 2020 to the iBB.
Abbreviations
- IPC
inositolphosphorylceramide
- LCB
long-chain base
- MIPC
mannosyl-inositol-phosphorylceramide
- M(IP)2C
mannosyl-diinositolphosphorylceramide
- PHS
phytosphingosine
- PM
plasma membrane
- TORC2
target of rapamycin complex 2
- VLCFA
very long chain fatty acids
- YPD
yeast extract peptone dextrose
Footnotes
AUTHOR CONTRIBUTIONS
J.F.G. and A.M. performed the experimental work, S.R. provided invaluable guidance, A.M., J.F.G., I.S.C. and J.T. designed the experiments, analyzed the data, and wrote the paper.
LITERATURE CITED
- 1.Erickson B, Nelson, Winters P. Perspective on opportunities in industrial biotechnology in renewable chemicals. Biotechnology journal. 2012;7:176–185. doi: 10.1002/biot.201100069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carroll A, Somerville C. Cellulosic biofuels. Annual review of plant biology. 2009;60:165–182. doi: 10.1146/annurev.arplant.043008.092125. [DOI] [PubMed] [Google Scholar]
- 3.Chundawat SP, Beckham GT, Himmel ME, Dale BE. Deconstruction of lignocellulosic biomass to fuels and chemicals. Annual review of chemical and biomolecular engineering. 2011;2:121–145. doi: 10.1146/annurev-chembioeng-061010-114205. [DOI] [PubMed] [Google Scholar]
- 4.Zha Y, Muilwijk B, Coulier L, Punt PJ. Inhibitory Compounds in Lignocellulosic Biomass Hydrolysates during Hydrolysate Fermentation Processes. J Bioprocess Biotechniq. 2012;2:1–11. [Google Scholar]
- 5.Franden MA, Pienkos PT, Zhang M. Development of a high-throughput method to evaluate the impact of inhibitory compounds from lignocellulosic hydrolysates on the growth of Zymomonas mobilis. Journal of biotechnology. 2009;144:259–267. doi: 10.1016/j.jbiotec.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 6.Fleet GH. Yeasts in foods and beverages: impact on product quality and safety. Current opinion in biotechnology. 2007;18:170–175. doi: 10.1016/j.copbio.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 7.Mollapour M, Piper PW. Hog1 mitogen-activated protein kinase phosphorylation targets the yeast Fps1 aquaglyceroporin for endocytosis, thereby rendering cells resistant to acetic acid. Molecular and cellular biology. 2007;27:6446–6456. doi: 10.1128/MCB.02205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casal M, Cardoso H, Leão C. Mechanisms regulating the transport of acetic acid in Saccharomyces cerevisiae. Microbiology. 1996;142:1385–1390. doi: 10.1099/13500872-142-6-1385. [DOI] [PubMed] [Google Scholar]
- 9.Piper P, Calderon CO, Hatzixanthis K, Mollapour M. Weak acid adaptation: the stress response that confers yeasts with resistance to organic acid food preservatives. Microbiology. 2001;147:2635–2642. doi: 10.1099/00221287-147-10-2635. [DOI] [PubMed] [Google Scholar]
- 10.Mira NP, Teixeira MC. Microbial mechanisms of tolerance to weak acid stress. Frontiers in microbiology. 2013;4:416. doi: 10.3389/fmicb.2013.00416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arneborg N, Jespersen L, Jakobsen M. Individual cells of Saccharomyces cerevisiae and Zygosaccharomyces bailii exhibit different short-term intracellular pH responses to acetic acid. Archives of microbiology. 2000;174:125–128. doi: 10.1007/s002030000185. [DOI] [PubMed] [Google Scholar]
- 12.Moon NJ. Inhibition of the growth of acid tolerant yeasts by acetate, lactate and propionate and their synergistic mixtures. Journal of Applied Microbiology. 1983;55:453–460. [Google Scholar]
- 13.Giannattasio S, Guaragnella N, Zdralevic M, Marra E. Molecular mechanisms of Saccharomyces cerevisiae stress adaptation and programmed cell death in response to acetic acid. Frontiers in microbiology. 2013;4:33. doi: 10.3389/fmicb.2013.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burtner CR, Murakami CJ, Kennedy BK, Kaeberlein M. A molecular mechanism of chronological aging in yeast. Cell Cycle. 2009;8:1256–1270. doi: 10.4161/cc.8.8.8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stratford M, Nebe-von-Caron G, Steels H, Novodvorska M, Ueckert J, Archer DB. Weak-acid preservatives: pH and proton movements in the yeast Saccharomyces cerevisiae. International journal of food microbiology. 2013;161:164–171. doi: 10.1016/j.ijfoodmicro.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 16.Carmelo V, Santos H, Sá-Correia I. Effect of extracellular acidification on the activity of plasma membrane ATPase and on the cytosolic and vacuolar pH of Saccharomyces cerevisiae. Biochimica et biophysica acta. 1997;1325:63–70. doi: 10.1016/s0005-2736(96)00245-3. [DOI] [PubMed] [Google Scholar]
- 17.Mira NP, Becker JD, Sá-Correia I. Genomic expression program involving the Haa1p-regulon in Saccharomyces cerevisiae response to acetic acid. Omics : a journal of integrative biology. 2010;14:587–601. doi: 10.1089/omi.2010.0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goossens A, de La Fuente N, Forment J, Serrano R, Portillo F. Regulation of yeast H(+)-ATPase by protein kinases belonging to a family dedicated to activation of plasma membrane transporters. Molecular and cellular biology. 2000;20:7654–7661. doi: 10.1128/mcb.20.20.7654-7661.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mira NP, Teixeira MC, Sá-Correia I. Adaptive response and tolerance to weak acids in Saccharomyces cerevisiae: a genome-wide view. Omics : a journal of integrative biology. 2010;14:525–540. doi: 10.1089/omi.2010.0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mira NP, Palma M, Guerreiro JF, Sá-Correia I. Genome-wide identification of Saccharomyces cerevisiae genes required for tolerance to acetic acid. Microbial cell factories. 2010;9:79. doi: 10.1186/1475-2859-9-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawahata M, Masaki K, Fujii T, Iefuji H. Yeast genes involved in response to lactic acid and acetic acid: acidic conditions caused by the organic acids in Saccharomyces cerevisiae cultures induce expression of intracellular metal metabolism genes regulated by Aft1p. FEMS yeast research. 2006;6:924–936. doi: 10.1111/j.1567-1364.2006.00089.x. [DOI] [PubMed] [Google Scholar]
- 22.Lee Y, Nasution O, Choi E, Choi IG, Kim W, Choi W. Transcriptome analysis of acetic-acid-treated yeast cells identifies a large set of genes whose overexpression or deletion enhances acetic acid tolerance. Applied microbiology and biotechnology. 2015;99:6391–6403. doi: 10.1007/s00253-015-6706-y. [DOI] [PubMed] [Google Scholar]
- 23.Chen YY, Stabryla L, Wei N. Improved Acetic Acid Resistance in Saccharomyces cerevisiae by Overexpression of the WHI2 Gene Identified through Inverse Metabolic Engineering. Appl Environ Microb. 2016;82:2156–2166. doi: 10.1128/AEM.03718-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen YY, Sheng JY, Jiang T, Stevens J, Feng XY, Wei N. Transcriptional profiling reveals molecular basis and novel genetic targets for improved resistance to multiple fermentation inhibitors in Saccharomyces cerevisiae. Biotechnol Biofuels. 2016;9 doi: 10.1186/s13068-015-0418-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inaba T, Watanabe D, Yoshiyama Y, Tanaka K, Ogawa J, Takagi H, Shimoi H, Shima J. An organic acid-tolerant HAA1-overexpression mutant of an industrial bioethanol strain of Saccharomyces cerevisiae and its application to the production of bioethanol from sugarcane molasses. Amb Express. 2013;3 doi: 10.1186/2191-0855-3-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakihama Y, Hasunuma T, Kondo A. Improved ethanol production from xylose in the presence of acetic acid by the overexpression of the HAA1 gene in Saccharomyces cerevisiae. Journal of bioscience and bioengineering. 2015;119:297–302. doi: 10.1016/j.jbiosc.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 27.Meijnen JP, Randazzo P, Foulquie-Moreno MR, van den Brink J, Vandecruys P, Stojiljkovic M, Dumortier F, Zalar P, Boekhout T, Gunde-Cimerman N, Kokosar J, Stajdohar M, Curk T, Petrovic U, Thevelein JM. Polygenic analysis and targeted improvement of the complex trait of high acetic acid tolerance in the yeast Saccharomyces cerevisiae. Biotechnol Biofuels. 2016;9:5. doi: 10.1186/s13068-015-0421-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pearce LR, Komander D, Alessi DR. The nuts and bolts of AGC protein kinases. Nature reviews. Molecular cell biology. 2010;11:9–22. doi: 10.1038/nrm2822. [DOI] [PubMed] [Google Scholar]
- 29.Casamayor A, Torrance PD, Kobayashi T, Thorner J, Alessi DR. Functional counterparts of mammalian protein kinases PDK1 and SGK in budding yeast. Current biology : CB. 1999;9:186–197. doi: 10.1016/s0960-9822(99)80088-8. [DOI] [PubMed] [Google Scholar]
- 30.Roelants FM, Torrance PD, Bezman N, Thorner J. Pkh1 and Pkh2 differentially phosphorylate and activate Ypk1 and Ykr2 and define protein kinase modules required for maintenance of cell wall integrity. Molecular biology of the cell. 2002;13:3005–3028. doi: 10.1091/mbc.E02-04-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kamada Y, Fujioka Y, Suzuki NN, Inagaki F, Wullschleger S, Loewith R, Hall MN, Ohsumi Y. Tor2 directly phosphorylates the AGC kinase Ypk2 to regulate actin polarization. Molecular and cellular biology. 2005;25:7239–7248. doi: 10.1128/MCB.25.16.7239-7248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roelants FM, Breslow DK, Muir A, Weissman JS, Thorner J. Protein kinase Ypk1 phosphorylates regulatory proteins Orm1 and Orm2 to control sphingolipid homeostasis in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:19222–19227. doi: 10.1073/pnas.1116948108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niles BJ, Mogri H, Hill A, Vlahakis A, Powers T. Plasma membrane recruitment and activation of the AGC kinase Ypk1 is mediated by target of rapamycin complex 2 (TORC2) and its effector proteins Slm1 and Slm2. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:1536–1541. doi: 10.1073/pnas.1117563109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun Y, Miao Y, Yamane Y, Zhang C, Shokat KM, Takematsu H, Kozutsumi Y, Drubin DG. Orm protein phosphoregulation mediates transient sphingolipid biosynthesis response to heat stress via the Pkh-Ypk and Cdc55-PP2A pathways. Molecular biology of the cell. 2012;23:2388–2398. doi: 10.1091/mbc.E12-03-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berchtold D, Piccolis M, Chiaruttini N, Riezman I, Riezman H, Roux A, Walther TC, Loewith R. Plasma membrane stress induces relocalization of Slm proteins and activation of TORC2 to promote sphingolipid synthesis. Nature cell biology. 2012;14:542–547. doi: 10.1038/ncb2480. [DOI] [PubMed] [Google Scholar]
- 36.Lee YJ, Jeschke GR, Roelants FM, Thorner J, Turk BE. Reciprocal phosphorylation of yeast glycerol-3-phosphate dehydrogenases in adaptation to distinct types of stress. Molecular and cellular biology. 2012;32:4705–4717. doi: 10.1128/MCB.00897-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muir A, Roelants FM, Timmons G, Leskoske KL, Thorner J. Down-regulation of TORC2-Ypk1 signaling promotes MAPK-independent survival under hyperosmotic stress. eLif. 2015;4:e09336. doi: 10.7554/eLife.09336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roelants FM, Baltz AG, Trott AE, Fereres S, Thorner J. A protein kinase network regulates the function of aminophospholipid flippases. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:34–39. doi: 10.1073/pnas.0912497106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rispal D, Eltschinger S, Stahl M, Vaga S, Bodenmiller B, Abraham Y, Filipuzzi I, Movva NR, Aebersold R, Helliwell SB, Loewith R. Target of rapamycin complex 2 regulates actin polarization and endocytosis via multiple pathways. J Biol Chem. 2015 doi: 10.1074/jbc.M114.627794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muir A, Ramachandran S, Roelants FM, Timmons G, Thorner J. TORC2-dependent protein kinase Ypk1 phosphorylates ceramide synthase to stimulate synthesis of complex sphingolipids. eLife. 2014;3:e03779. doi: 10.7554/eLife.03779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Breslow DK, Collins SR, Bodenmiller B, Aebersold R, Simons K, Shevchenko A, Ejsing CS, Weissman JS. Orm family proteins mediate sphingolipid homeostasis. Nature. 2010;463:1048–1053. doi: 10.1038/nature08787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schorling S, Vallee B, Barz WP, Riezman H, Oesterhelt D. Lag1p and Lac1p are essential for the Acyl-CoA-dependent ceramide synthase reaction in Saccharomyces cerevisae. Molecular biology of the cell. 2001;12:3417–3427. doi: 10.1091/mbc.12.11.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kliegman JI, Fiedler D, Ryan CJ, Xu YF, Su XY, Thomas D, Caccese MC, Cheng A, Shales M, Rabinowitz JD, Krogan NJ, Shokat KM. Chemical genetics of rapamycin-insensitive TORC2 in S. cerevisiae. Cell reports. 2013;5:1725–1736. doi: 10.1016/j.celrep.2013.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roelants FM, Torrance PD, Thorner J. Differential roles of PDK1- and PDK2-phosphorylation sites in the yeast AGC kinases Ypk1, Pkc1 and Sch9. Microbiology. 2004;150:3289–3304. doi: 10.1099/mic.0.27286-0. [DOI] [PubMed] [Google Scholar]
- 45.Rego A, Costa M, Chaves SR, Matmati N, Pereira H, Sousa MJ, Moradas-Ferreira P, Hannun YA, Costa V, Corte-Real M. Modulation of mitochondrial outer membrane permeabilization and apoptosis by ceramide metabolism. PloS one. 2012;7:e48571. doi: 10.1371/journal.pone.0048571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lindberg L, Santos AX, Riezman H, Olsson L, Bettiga M. Lipidomic profiling of Saccharomyces cerevisiae and Zygosaccharomyces bailii reveals critical changes in lipid composition in response to acetic acid stress. PloS one. 2013;8:e73936. doi: 10.1371/journal.pone.0073936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hunter T, Plowman GD. The protein kinases of budding yeast: six score and more. Trends in biochemical sciences. 1997;22:18–22. doi: 10.1016/s0968-0004(96)10068-2. [DOI] [PubMed] [Google Scholar]
- 48.Shimobayashi M, Oppliger W, Moes S, Jeno P, Hall MN. TORC1-regulated protein kinase Npr1 phosphorylates Orm to stimulate complex sphingolipid synthesis. Molecular biology of the cell. 2013;24:870–881. doi: 10.1091/mbc.E12-10-0753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jin N, Mao K, Jin Y, Tevzadze G, Kauffman EJ, Park S, Bridges D, Loewith R, Saltiel AR, Klionsky DJ, Weisman LS. Roles for PI(3,5)P2 in nutrient sensing through TORC1. Molecular biology of the cell. 2014;25:1171–1185. doi: 10.1091/mbc.E14-01-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu WI, McDonough VM, Nickels JT, Jr, Ko J, Fischl AS, Vales TR, Merrill AH, Jr, Carman GM. Regulation of lipid biosynthesis in Saccharomyces cerevisiae by fumonisin B1. The Journal of biological chemistry. 1995;270:13171–13178. doi: 10.1074/jbc.270.22.13171. [DOI] [PubMed] [Google Scholar]
- 51.Tani M, Kuge O. Hydroxylation state of fatty acid and long-chain base moieties of sphingolipid determine the sensitivity to growth inhibition due to AUR1 repression in Saccharomyces cerevisiae. Biochemical and biophysical research communications. 2012;417:673–678. doi: 10.1016/j.bbrc.2011.11.138. [DOI] [PubMed] [Google Scholar]
- 52.Lindahl L, Genheden S, Eriksson LA, Olsson L, Bettiga M. Sphingolipids contribute to acetic acid resistance in Zygosaccharomyces bailii. Biotechnology and bioengineering. 2016;113:744–753. doi: 10.1002/bit.25845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dudley AM, Janse DM, Tanay A, Shamir R, Church GM. A global view of pleiotropy and phenotypically derived gene function in yeast. Molecular systems biology. 2005;1 doi: 10.1038/msb4100004. 2005 0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mao C, Xu R, Bielawska A, Obeid LM. Cloning of an alkaline ceramidase from Saccharomyces cerevisiae. An enzyme with reverse (CoA-independent) ceramide synthase activity. The Journal of biological chemistry. 2000;275:6876–6884. doi: 10.1074/jbc.275.10.6876. [DOI] [PubMed] [Google Scholar]
- 55.Mollapour M, Piper PW. Hog1p mitogen-activated protein kinase determines acetic acid resistance in Saccharomyces cerevisiae. FEMS yeast research. 2006;6:1274–1280. doi: 10.1111/j.1567-1364.2006.00118.x. [DOI] [PubMed] [Google Scholar]
- 56.Dickson RC. Thematic review series: sphingolipids. New insights into sphingolipid metabolism and function in budding yeast. Journal of lipid research. 2008;49:909–921. doi: 10.1194/jlr.R800003-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hannich JT, Umebayashi K, Riezman H. Distribution and functions of sterols and sphingolipids. Cold Spring Harbor perspectives in biology. 2011;3 doi: 10.1101/cshperspect.a004762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Proszynski TJ, Klemm RW, Gravert M, Hsu PP, Gloor Y, Wagner J, Kozak K, Grabner H, Walzer K, Bagnat M, Simons K, Walch-Solimena C. A genome-wide visual screen reveals a role for sphingolipids and ergosterol in cell surface delivery in yeast. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:17981–17986. doi: 10.1073/pnas.0509107102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takabatake A, Kawazoe N, Izawa S. Plasma membrane proteins Yro2 and Mrh1 are required for acetic acid tolerance in Saccharomyces cerevisiae. Applied microbiology and biotechnology. 2015;99:2805–2814. doi: 10.1007/s00253-014-6278-2. [DOI] [PubMed] [Google Scholar]
- 60.Lam MH, Snider J, Rehal M, Wong V, Aboualizadeh F, Drecun L, Wong O, Jubran B, Li M, Ali M, Jessulat M, Deineko V, Miller R, Lee M, Park HO, Davidson A, Babu M, Stagljar I. A Comprehensive Membrane Interactome Mapping of Sho1p Reveals Fps1p as a Novel Key Player in the Regulation of the HOG Pathway in S. cerevisiae. Journal of molecular biology. 2015;427:2088–2103. doi: 10.1016/j.jmb.2015.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Berchtold D, Walther TC. TORC2 plasma membrane localization is essential for cell viability and restricted to a distinct domain. Molecular biology of the cell. 2009;20:1565–1575. doi: 10.1091/mbc.E08-10-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Davenport KR, Sohaskey M, Kamada Y, Levin DE, Gustin MC. A second osmosensing signal transduction pathway in yeast. Hypotonic shock activates the PKC1 protein kinase-regulated cell integrity pathway. The Journal of biological chemistry. 1995;270:30157–30161. doi: 10.1074/jbc.270.50.30157. [DOI] [PubMed] [Google Scholar]
- 63.Ludovico P, Rodrigues F, Almeida A, Silva MT, Barrientos A, Côrte-Real M. Cytochrome c release and mitochondria involvement in programmed cell death induced by acetic acid in Saccharomyces cerevisiae. Molecular biology of the cell. 2002;13:2598–2606. doi: 10.1091/mbc.E01-12-0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Semchyshyn HM, Abrat OB, Miedzobrodzki J, Inoue Y, Lushchak VI. Acetate but not propionate induces oxidative stress in bakers' yeast Saccharomyces cerevisiae. Redox report : communications in free radical research. 2011;16:15–23. doi: 10.1179/174329211X12968219310954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Niles BJ, Joslin AC, Fresques T, Powers T. TOR complex 2-Ypk1 signaling maintains sphingolipid homeostasis by sensing and regulating ROS accumulation. Cell reports. 2014;6:541–552. doi: 10.1016/j.celrep.2013.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang JG, Liu XY, He XP, Guo XN, Lu Y, Zhang BR. Improvement of acetic acid tolerance and fermentation performance of Saccharomyces cerevisiae by disruption of the FPS1 aquaglyceroporin gene. Biotechnology letters. 2011;33:277–284. doi: 10.1007/s10529-010-0433-3. [DOI] [PubMed] [Google Scholar]
- 67.Zhang MM, Zhao XQ, Cheng C, Bai FW. Improved growth and ethanol fermentation of Saccharomyces cerevisiae in the presence of acetic acid by overexpression of SET5 and PPR1. Biotechnology journal. 2015;10:1903–1911. doi: 10.1002/biot.201500508. [DOI] [PubMed] [Google Scholar]
- 68.dos Santos SC, Sa-Correia I. Yeast toxicogenomics: lessons from a eukaryotic cell model and cell factory. Current opinion in biotechnology. 2015;33:183–191. doi: 10.1016/j.copbio.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 69.Burke D, Amberg D, Strathern J. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Laboratory Press. 2005 [Google Scholar]
- 70.Green MR, Sambrook J. Molecular cloning : a laboratory manual. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 2012. [Google Scholar]
- 71.Westfall PJ, Patterson JC, Chen RE, Thorner J. Stress resistance and signal fidelity independent of nuclear MAPK function. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:12212–12217. doi: 10.1073/pnas.0805797105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kinoshita E, Kinoshita-Kikuta E, Koike T. Advances in Phos-tag-based methodologies for separation and detection of the phosphoproteome. Biochimica et biophysica acta. 2015;1854:601–608. doi: 10.1016/j.bbapap.2014.10.004. [DOI] [PubMed] [Google Scholar]