Abstract
Genomic instability is a hallmark of cancer and aging. Premature aging (progeroid) syndromes are often caused by mutations in genes whose function is to ensure genomic integrity. The RecQ family of DNA helicases is highly conserved and plays crucial roles as genome caretakers. In human, mutations in three RecQ genes — BLM, WRN, and RECQL4 — give rise to Bloom’s syndrome (BS), Werner syndrome (WS), and Rothmund-Thomson’s syndrome (RTS), respectively. WS is a prototypic premature aging disorder; however, the clinical features present in BS and RTS do not indicate accelerated aging. The BLM helicase has pivotal functions at the crossroads of DNA replication, recombination, and repair. BS cells exhibit a characteristic form of genomic instability that includes excessive recombination. The excessive homologous recombination drives the development of the many types of cancers that affect persons in the normal population. Replication delay and slower cell turnover rates have been proposed to explain many features of Bloom’s syndrome, such as short stature. More recently, aberrant transcriptional regulation of growth and survival genes has been proposed as a hypothesis.
Keywords: Bloom’s syndrome, RecQ helicases, BLM, genomic instability, cancer susceptibility, aging
1. Introduction
Genomic instability is a biological condition in which mutations accumulate in the genome at a higher rate than in normal cells. The distinction between rate and frequency is an important one, because differences in mutation frequency can be explained by differences in mutation rate or by differences in cell turnover (i.e., proliferation and apoptosis rates). Genomic instability can be environmentally determined by mutagenic exposure or genetically determined by mutation of DNA damage response or DNA repair genes. It is a common feature of cancers where both mechanisms play an important role in its generation. The theoretical framework linking mutation and cancer initiation and progression is well established because somatic mutations in oncogenes and tumor suppressors are major drivers of cancer. Moreover, there are a wide variety of clinical entities associated with cancer susceptibility and caused by mutations in genes that maintain genomic integrity. There are also prominent examples of associations between genomic instability syndromes and both aging and neurological disorders. The theory linking mutation with aging or neuronal cell death dysfunction is not as well understood.
Bloom’s syndrome (BS; OMIM #210900) is one of a group of rare autosomal recessive disorders characterized by chromosomal instability that includes Fanconi anemia (FA) and ataxia telangiectasia (German, 1995). As molecular analysis of mutation and gene function has advanced, clinical entities with defects in DNA damage response or DNA repair have been grouped under the rubric of genomic instability syndromes. From the clinical standpoint, the genomic instability syndromes are highly heterogeneous. The ones that are inherited as dominant traits, such as Hereditary Breast and Ovarian Cancer syndrome and Lynch syndrome, have no clinical manifestations other than cancer in the proband and in the proband’s family. The genomic instability syndromes that are inherited as autosomal recessives are associated with a wide variety of clinical features of varying severity. This heterogeneity is nowhere more prominent than in the syndromes defined by mutations in the RecQ helicase family.
Genes in the RecQ helicase family are known for evolutionarily conserved regions that encode the helicase domains of the proteins. These proteins are DNA-dependent ATPases and ATP-dependent DNA unwinding enzymes. The first RecQ gene was identified in Escherichia coli with isolation of the recQ1 mutant, which is resistant to thymineless death (Nakayama et al., 1984). Single RecQ genes were identified in Saccharomyces cerevisiae and in Schizosaccharomyces pombe. Isolation of the S. cerevisiae gene SGS1, for slow growth suppressor, yielded important insights because sgs1 mutants arose as suppressors to topoisomerase 3 (top3) mutations and the protein products topoisomerase III and SGS1 physically interact (Bennett et al., 2000; Gangloff et al., 1994). Besides slow growth, top3 mutants exhibit excessive homologous recombination [HR; (Wallis et al., 1989)] as do sgs1 mutants (Watt et al., 1996). Topoisomerase III is a type one topoisomerase that breaks and religates single-stranded DNA (ssDNA) but differs from topoisomerase I in that it acts on ssDNA more efficiently than double-stranded DNA (dsDNA). Jim Wang postulated that E. coli topoisomerase III could have a function in unraveling complementary DNA strands as opposing replication forks approach each other for replication termination (Wang, 1991). This early suggestion has been supported more recently with the discovery of a role for the topoisomerase IIIα-RecQ complex in preventing ultrafine DNA bridges (UFBs; see Section 4.3) In addition to a function in replication termination, the topoisomerase IIIα-RecQ complex also possesses a unique activity that can resolve recombination intermediates without crossing-over (see Section 4.1). Physical or functional interactions between topoisomerase IIIs and many RecQs are evolutionarily conserved.
In humans, there are five RecQ helicase genes, presented here in the order of their discovery, namely RECQL, BLM, WRN, RECQL4, and RECQL51 (Croteau et al., 2014). During the radiation of metazoans, gene duplication produced three RecQ genes in worms and flies and five in essentially all vertebrates. Evolution has innovated distinct functions for the RecQ genes. Mutations in human BLM result in BS (Ellis et al., 1995). Mutations in human WRN and RECQL4 are linked to Werner syndrome (WS) and Rothmund-Thomson syndrome (RTS), respectively (Kitao et al., 1999; Yu et al., 1996). Each of these syndromes features genomic instability and susceptibility to cancer, but the characteristics of the genomic instability varies and the sites and types of cancers associated with each syndrome are different. BS and RTS are early developmental disorders characterized by small size. WS is characterized by premature development of features associated with aging. While WS is classified as a segmental progeroid syndrome (Martin, 1997), the other two syndromes are not progeroid. In this review, we will present BS and its clinical features, review BLM functions in the various aspects of DNA metabolism, and we will conclude with some considerations about BS and aging.
2. Clinical features of Bloom’s syndrome
The most striking clinical feature of BS is small but proportional body size (German, 1969). Birth weight for persons with BS ranges from 0.7 to 3.2 kg with mean term weights of 1.89 kg ± 0.35 kg for BS boys and 1.87 kg ± 0.35 kg for BS girls compared to 3.27 kg ± 0.44 kg and 3.23 kg ± 0.53 kg for normal boys and girls, respectively (Keller et al., 1999)2. Mean birth lengths are similarly reduced with 43.4 cm ± 4.4 cm for BS boys and 43.8 cm ± 2.8 cm for BS girls compared to 50.5 cm ± 2.5 cm and 49.9 cm ± 2.7 cm for normal boys and girls, respectively. Means for both birth weight and birth length in BS are more than two standard deviations below normal, demonstrating that the growth retardation in BS is prenatal. With the advent of fetal ultra-sound, the small size has proven evident in early fetal development. The pathognomonic genomic instability of BS, namely elevated sister chromatid exchanges (SCEs; see Section 3.1), can be detected in fetal cells and used for prenatal diagnosis; molecular analysis of the BS gene BLM can also be used (Sanz and German, 2006).
Growth is retarded throughout early life and into maturity. Mean adult height is 145.5 cm ± 7.6 cm for BS males and 141.5 cm ± 6.6 cm for BS females compared to 176.8 cm ± 6.7 cm and 163.7 cm ± 6.1 cm for normal males and females, respectively (Keller et al., 1999). Current data from the Bloom’s Syndrome Registry is similar, with an average adult height of 149 cm (range 128–164) for BS males and 138 cm (range 115–160) for BS females (http://weill.cornell.edu/bsr/data_from_registry/). Mean adult weights are correspondingly below normal at 41.3 kg ± 8.8 kg for BS males and 36.6 kg ± 8.6 kg for BS females compared to 68.9 kg ± 9.1 kg and 58.3 kg ± 8.1 kg for normal males and females, respectively. Body mass index at birth and during post-natal development is also well below normal but the deficit decreases into adulthood.
Although the body overall is well proportioned, the head is somewhat small and narrow relative to BS body size. The malar and lower mandibular areas are underdeveloped. The voice is high-pitched (Sanz and German, 2006).
The skin in persons with BS appears normal at birth, but in the first or second year of life and in response to sun exposure, children develop an erythematous rash on the nose and cheeks and around the mouth. It sometimes involves the backs of the hands and forearms but it does not develop on the arms and legs, the trunk, or buttocks. The rash can include telangiectasia. The severity of the rash varies with some persons losing eyelashes and blistering around the mouth, and some others where it is very faint. Many persons with BS have café-au-lait spots and hypopigmented areas of skin; the hypopigmented areas are often associated with contiguous hyperpigmented areas.
The majority of newborns, infants, and young children with BS show a lack of interest in food. Severe gastroesophageal reflux is common in children and diarrhea is also a frequent complaint. The increased gastroesophageal reflux could be the source of the increased frequency of pneumonia and otitis media in BS due to increased exposure to gastric contents due to aspiration (Sanz and German, 2006). The feeding problems go hand in hand with a paucity of sub-cutaneous fat in many persons with BS. Gastric feeding tubes have been placed in some children, resulting in an increase in adiposity but not height.
The syndrome is associated with a variety of other clinical features, including a mild immunodeficiency manifested by reduced levels of one or more plasma immunoglobulins, azoospermia or severe oligospermia in males and premature cessation of menstruation in females, and minor anatomic defects. Intellectual abilities are limited in some persons and normal in others; some have achieved graduate degrees. Major anatomic defects are not increased in frequency.
The main cause of death in BS is from complications from cancer. There are three main characteristics of cancer in BS: (i) for each tumor type, cancer development occurs on average at an earlier age than normal, (ii) many persons have been diagnosed with multiple independent primary cancers, and (iii) the type and sites of cancer are very broad, including the major epithelial cancers (lung, colorectal, breast, skin, esophageal, genital-urinary tract, and liver), lymphoma, leukemia, and rare pediatric tumors such as retinoblastoma, Wilm’s tumor, and osteosarcoma. In the most recent update from the Bloom’s Syndrome Registry (Sanz and German, 2006), there have been 205 malignancies in 129 persons of the 265 BS persons followed. Analysis of standardized incidence ratios in BS showed that persons with BS are 99 times (95% confidence interval 83-117) more likely to be diagnosed with any cancer relative to the general population (Shi et al., 2006). The most common epithelial cancer is colorectal (n=30), for which cancer the standardized incidence ratio is 521 (95% confidence interval 318-804). The standardized incidence ratio for breast cancer (n=16) is 90. Skin cancer (n=27) is also common. Lymphoma (n=35), acute lymphoblastic leukemia (n=12), and acute myelogenous leukemia (n=25) are common, and standardized incidence ratios for lymphoid malignancy is 166 and for leukemia is 152. Myelodysplasia (n=22), which is a precursor of myelogenous leukemia, is relatively frequent. The mean age of diagnosis of any cancer is 23.
Another major medical complication that is common in BS is diabetes mellitus (n=47). It resembles the adult-onset type of diabetes and is associated with abnormalities in insulin release and glucose tolerance (Diaz et al., 2006). The mean age of diagnosis is 26.6 years. Chronic obstructive pulmonary disease is also common. Pulmonary failure has been the cause of death in five persons with BS.
Table 1 compares the clinical features and major medical complications of the RecQ syndromes. Quite a few of the clinical features of BS and RTS are shared, including growth retardation, a prominent skin rash, gastrointestinal reflux and diarrhea during infancy, and hypogonadism; but the syndromes also present some striking differences. The skin rash in RTS is striking. It is described as poikiloderma, in which there is redness, telangiectasia, spots of atrophy, and alternating areas of hypo- and hyper-pigmented skin, giving an overall mottled appearance (Wang and Plon, 1999). The rash begins on the face then spreads to the arms and legs and sometimes involves the buttocks but spares the trunk, and it persists throughout life. The skin rash in BS is mild by comparison and generally limited to the face. Effects of RECQL4 mutation on skeletal and dental development are a prominent part of RTS, whereas in persons with BLM mutation skeletal and dental abnormalities are infrequent. Similarly, hematologic anomalies, juvenile cataracts, and sparse scalp hair are common in RTS but not BS. On the other hand, persons with BS have more frequent infections, have deficits in one or more immunoglobulins and in the delayed hypersensitivity reaction, and have a more severe hypogonadism. Although the cancer diatheses in BS and RTS are similar in some ways, carcinomas of epithelial tissue are very common in BS but not in RTS, and melanoma and osteosarcoma are relatively rare in BS but quite common in RTS.
Table 1.
Comparison of clinical features and complications in Bloom’s, Rothmund-Thomson, and Werner syndromes.
| Anatomic feature | Bloom’s syndrome | Rothmund-Thomson syndrome1 | Werner syndrome |
|---|---|---|---|
| Growth | < 3rd percentile | < 5th percentile | Absence of adolescent growth spurt2 |
| Skin | Sun-sensitive erythema on butterfly area of face, telangiectasias, mouth fissures, café-au-lait and hypopigmented spots | Poikiloderma – erythema on face that spreads to buttocks and extremities, telangiectasias, blistering, developing into atrophic and keratotic skin; café-au-lait and hypopigmented spots | Deep, chronic ulcers around the ankles; scleroderma-like changes2 |
| Gastrointestinal | Increased reflux, vomiting, and diarrhea in children | Increased reflux, vomiting, and diarrhea in children | Normal |
| Immune system | Increased otitis media and pneumonia, deficit of one or more immunoglogulins | Normal3 | Normal |
| Hematologic | Normal | Anemia, neutropenia, aplastic anemia | Normal |
| Cardiovascular | Normal | Normal | Normal anatomy. Early onset coronary artery atherosclerosis and arteriosclerosis |
| Bone | No major skeletal or anatomic abnormalities4 | Frequent, major skeletal abnormalities | Normal anatomy. Early onset of osteoporosis |
| Teeth | Occasional absence of lateral incisors | Frequent abnormalities including hypoplastic teeth, microdontia, delayed eruption, missing and supernumerary teeth, ectopic eruption | Normal |
| Eyes | Normal | Juvenile cataracts | Bilateral cataracts2 |
| Hair | Normal | Sparse scalp hair or total alopecia in children and adults | Premature greying and thinning of scalp hair2 |
| Gonads | Infertility in males, sub-fertility in females | Subfertility | Early decline in fertility |
| Intelligence | Normal intelligence with some limitations | Normal | Normal |
| Mean lifespan | 26 | Not reported | 54 |
| Most common cause of death | Cancer | Cancer | Myocardial infarction |
| Cancer | Adenocarcinoma of many epithelial tissues, lymphoma, myelodysplasia and leukemia5 | Osteosarcoma, skin (basal and squamous), melanoma, lymphoma, myelodysplasia and leukemia | Soft-tissue sarcoma, osteosarcoma, melanoma6, thyroid carcinoma |
| Diabetes | 17% of persons | Not reported as elevated | 71% of persons |
Mutations in RECQL4 are associated with many but not all persons with Rothmund-Thomson syndrome (RTS). Other genes linked to this syndrome may exist. Mutations in RECQL4 are also associated with RAPADILINO and Baller Gold syndrome. RAPADILINO was described in Finland, and it overlaps clinically with RTS but without the poikiloderma. Baller-Gold syndrome also overlaps with RTS but also exhibits craniostenosis.
The “cardinal signs” of Werner syndrome appear in the second and third decades of life.
Isolated reports of immune dysfunction.
Minor anatomic anomalies have been noted.
46% of all persons followed in the Bloom’s Syndrome Registry have been diagnosed with a cancer.
Acral lentiginous melanoma, a very rare tumor, has a high relative incidence in Werner syndrome.
The clinical features of WS contrast sharply with those of BS and RTS. The four “cardinal signs” of WS — bilateral ocular cataracts, premature greying and thinning of scalp hair, deep and chronic ulcers around the ankles and scleroderma-like changes of the skin, and short stature — all appear in adolescence or after reaching adulthood. Many of the acquired clinical features in WS resemble aging including the scleroderma-like skin changes, progressive loss and greying hair, osteoporosis, arteriosclerosis, and coronary artery atherosclerosis. With the exception of sparse hair and cataracts in RTS, these features are absent from BS and RTS. The loss of hair and ocular cataracts in RTS appear early in life and are likely linked to defects in development of these tissues. The skin changes in BS arise after sun exposure in early life. In RTS, it is not clear that the skin changes result due to sun exposure, but they also arise in the first year of life. In both BS and RTS, the skin changes begin as an erythematous rash. There have been enough persons with BS who have lived past the age of 40 so that, if progeroid features had developed in BS, they should have come to the attention of the registrars of the Bloom’s Syndrome Registry. These considerations indicate that BS is not a progeroid syndrome, and it seems reasonable to await further information on the natural history of RTS before suggesting it as a progeroid syndrome.
3. Molecular genetics
3.1. Cellular phenotype
BS cells are characterized by increases in chromosomal aberrations, including chromatid gaps and breaks, telomere associations, and quadriradial (Qr) chromosomes, which result from unresolved recombination between homologous chromosomes (German et al., 1965; German et al., 1974; Rosin and German, 1985). Other common abnormalities at metaphase include chromosome fragmentation, lagging chromosomes at anaphase, and anaphase bridges. There is an excess of telophase cells with a chromatid strand stretched between them and increased micronuclei formation. BS cells exhibit increased mutation rates (Warren et al., 1981). In molecular genetic analysis, the genomic instability in BS includes elevated mitotic HR and unequal sister-chromatid exchange (Groden et al., 1990; Groden and German, 1992; Killen et al., 2009). By flow-cytometric analysis of red cells from persons heterozygous for the M and N blood group variants of the glycophorin A locus, there is a 50 fold increase over normal of M/M and N/N red cells, presumably generated by a mitotic HR event in a precursor cell; but there is also an increase in M/null and N/null cells, indicating a higher frequency of mutational inactivation (Kyoizumi et al., 1989; Langlois et al., 1989). Perhaps, the most characteristic feature is the more than ten fold increase in SCEs, which are the result of crossover events during HR repair of damaged replication forks (Chaganti et al., 1974).
As a comparison, cells derived from persons with WS do not exhibit these characteristics. Although the rate of spontaneous chromosomal breaks is higher in WS lymphocytes compared to normal ones, it is less than the rate in BS cells (Gebhart et al., 1988). WS cells also exhibit an increased frequency of mutations and of chromosomal aberrations, including variegated translocation mosaicism and large chromosomal deletions (Fukuchi et al., 1989; Hoehn et al., 1975; Salk et al., 1981). In contrast to BS cells, the levels of SCEs in WS cells are normal (Bartram et al., 1976; Yu et al., 1996), and WS cells do not exhibit elevated HR-mediated genomic instability (Killen et al., 2009), but have instead a deficit in HR repair (Chen et al., 2003; Prince et al., 2001; Saintigny et al., 2002). The presumptive genomic instability in RTS is not well understood and reports are often contradictory. Spontaneous or induced chromosome breakage in RTS is generally reported to be normal in lymphocytes and fibroblasts, and increased crossing-over has not been a consistent finding; SCEs are normal. Instead, there are characteristic structural chromosomal abnormalities, often involving chromosome 8, arising from misdivision of the centromere; for example, i(8p), i(8q), i(12p), and i(12q) are common abnormalities found in cultures of RTS cells (Larizza et al., 2006). Whether the rates of these events are increased has not been shown. Consequently, the genomic instabilities present in these three syndromes are distinct. Although the BLM, WRN, and RECQL4 genes are related, the different clinical and cellular features of the associated syndromes, and the fact that loss of either one cannot be compensated by the presence of other RecQ members, indicate that they have specific non-overlapping functions.
3.2. Structure and biochemical properties
BS is caused by homozygous or compound heterozygous mutations in the BLM gene, which is localized to chromosome 15q26.1. More than sixty different mutations have been identified, all resulting in BLM inactivation either by premature protein translation termination or by missense mutations that abrogate its helicase activity (German et al., 2007). The BLM gene encodes a 1417 amino-acid protein that localizes in the nucleus to PML nuclear bodies, which are discrete nuclear foci (Bischof et al., 2001; Zhong et al., 1999). BLM protein expression peaks in the S and G2/M phases of the cell cycle consistent with its role in DNA replication and recombination (Dutertre et al., 2000; Sanz et al., 2000).
BLM hydrolyzes ATP to unwind DNA by traversing ssDNA in a 3′ to 5′ direction (Karow et al., 1997). The RecQ helicases as a group are DNA structure-specific helicases. They play a critical role in the maintenance of genome stability by acting at the interface between DNA replication, recombination, and repair. Thus, it is not surprising that many substrates preferred by the RecQ helicases model DNA recombination intermediates, such as Holliday Junctions (HJs), displacement loops (D-loops), replication forks, and G-quadruplex (G4) DNA (Croteau et al., 2014; Larsen and Hickson, 2013). G4 DNA is a highly stable structure formed by Hoogsteen base pairing between four adjacent guanines arranged in a square planar configuration. Sequences that can form G4 DNA in vitro are found throughout the genome, including telomeres and the rDNA loci (Maizels and Gray, 2013) and mounting evidence indicates that G4 DNAs are biologically relevant structures (Rhodes and Lipps, 2015). G4 DNA formation can indeed be detrimental to the progression of replication and transcription machineries. In vitro, BLM is unable to unwind dsDNA from a blunt-ended terminus or from an internal nick and shows weak unwinding activity of duplex molecules with a single strand 3′-tail. However, BLM preferentially unwinds more elaborate structures such as bubbles, forked DNA duplexes, X-shaped molecules mimicking HJs and G4 DNA (Mohaghegh et al., 2001; Popuri et al., 2008; Sun et al., 1998). BLM specifically binds HJs and is able to catalyze ATP-dependent branch migration of HJs (Karow et al., 2000). BLM is also able to bind and efficiently disrupt D-loops formed during the initial step of HR (Bachrati et al., 2006; van Brabant et al., 2000). Additionally BLM can efficiently unwind RNA-DNA heteroduplexes and R-loops (a structure that can be formed by hybridization of a processed mRNA to the genomic DNA or structures similar to this structure), which could impede replication fork progression (Popuri et al., 2008).
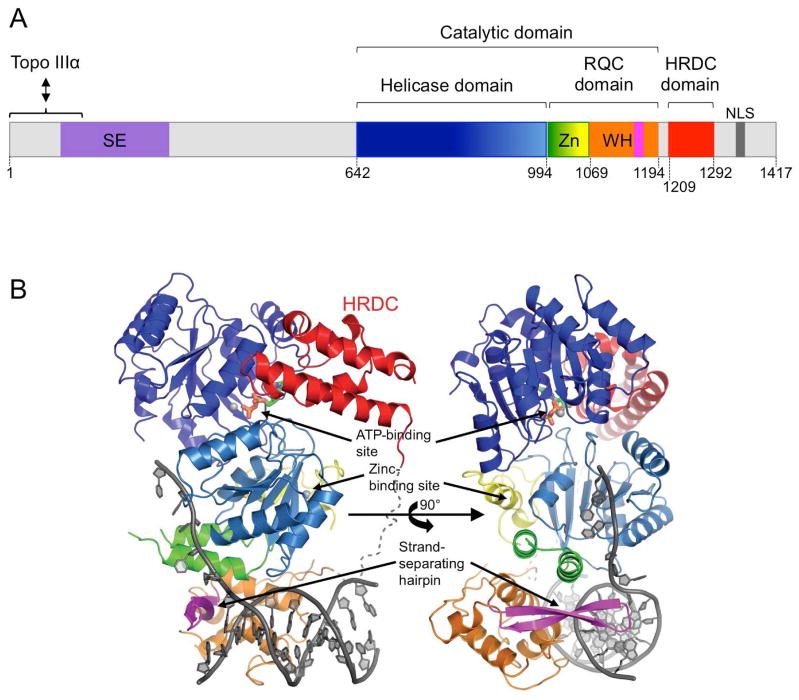
Preferential activity toward specific substrates distinguishes RecQ helicases from other helicases and can be associated with unique structural features. BLM possesses three typical domains shared by the other RecQ members (Fig. 1): the helicase domain, the RecQ carboxy-terminal (RQC) domain, and the helicase and ribonuclease D C-terminal (HRDC) domain (Kitano, 2014). The helicase and RQC domains form the catalytic core of the BLM helicase. The helicase domain is the most highly conserved component among the RecQ family and is characteristic of the SF2 superfamily of helicases. This domain is responsible for ATP binding and hydrolysis and acts as an ATP-dependent DNA translocation module that supplies a driving force for the helicase reaction (Swan et al., 2014). The second most conserved domain is the RQC, which is unique to the RecQ family of proteins. It is composed of a Zn2+-binding motif and a winged-helix (WH) domain that together act as a dsDNA-binding site. The -hairpin located in the WH domain acts as a scalpel, which is responsible for the duplex separation (Fig. 1B) (Kim et al., 2013; Kitano et al., 2010). The HRDC domain is the least conserved domain, and in human, it is only present in BLM and WRN proteins. Its function remains unclear. Neither WRN nor BLM HRDC domains directly bind to DNA (Kitano et al., 2007; Sato et al., 2010), but this domain may play a key role in the mechanism coupling the ATPase and helicase activities (Swan et al., 2014). The crystal structure of BLM in complex with ADP and a 3′-overhang DNA duplex reveals that the HRDC domain interacts with the helicase domain allowing the optimal configuration required to strongly couple ATP hydrolysis with DNA unwinding (Fig. 1B) (Swan et al., 2014). Additionally the HRDC domain may confer some DNA substrate specificity. Specifically, the HRDC domain of BLM is required for double HJ (dHJ) dissolution (Wu et al., 2005). Study of structural conformation of BLM suggests that the HRDC may act as a wedge required to open the HJ arms by electrostatic repulsion (Kim et al., 2013). Interestingly the WRN HRDC cannot substitute for the BLM HRDC in dHJ dissolution, indicating a evolutionary differentiation of the structure possibly mediating a different function or substrate specificity for the WRN HRDC (Wu et al., 2005).
Fig. 1.
Domain structure of human BLM protein. (A) Schematic representation of BLM domains. The functional domains are the helicase domain; the RecQ C-terminal domain (RQC) comprising the Zn2+-binding motif (Zn) and winged helix (WH) with the -hairpin in pink; and the helicase and ribonuclease D C-terminal (HRDC) domain. The N-terminal strand exchange (SE) domain is in purple and the interaction domain with topoisomerase III (TopoIII ) is indicated. The nuclear localization signal (NLS) is in dark grey. (B) Crystal structure of human BLM640-1298 in complex with ADP and a 3′-overhang DNA duplex. The colors are as in (A): the helicase domain is in blue, the Zn2+-binding motif is in green and yellow, the winged-helix (WH) domain is in orange with the -hairpin in pink, and the HRDC domain is in red. The ADP is shown in stick form and the DNA in cartoon form with the phosphate backbone in dark grey. The calcium and zinc ions are shown as light grey spheres. Reproduced with permission of the International Union of Crystallography (Swan et al., 2014).
In addition to their DNA unwinding activity, RecQ proteins catalyze ssDNA annealing (Wu, 2012). In BLM the N-terminus part of the protein has been shown to present annealing activity (Chen and Brill, 2010; Cheok et al., 2005; Machwe et al., 2005). The combination of unwinding and annealing activities catalyzes strand exchange, which might be required to promote either branch migration of HJs or the regression of stalled replication forks (see Sections 4.1 and 4.2). Strikingly, WRN is the only RecQ member to have an additional 3′ to 5′ exonuclease domain (Huang et al., 1998; Perry et al., 2006). This novel enzymatic activity distinguishes WRN from the other RecQ helicases; however, its exact role in WRN’s function is still unknown. The exonuclease may cooperate with DNA polymerase δ in removing damaged bases that might prevent elongation during lagging strand synthesis (Kamath-Loeb et al., 2012). It may also be important in WRN functions in translesion synthesis, non-homologous end joining (NHEJ), or DNA synthesis at telomeres. For example, exonuclease activity seems to play a role in DSB repair in coordination with Ku (Orren et al., 2001; Bukowy et al., 2008) and exonuclease activity is thought to cooperate with WRN helicase activity to promote replication fork regression (Machwe et al., 2007).
4. BLM functions in DNA transactions
Human RecQ helicases are involved in various aspects of DNA metabolism (Croteau et al., 2014). BLM is playing a critical role in HR, in DNA replication, and in stabilization and repair of damaged replication forks. BLM is also important for proper sister chromatid segregation in mitosis, and telomere maintenance.
4.1. Functions in homologous recombination
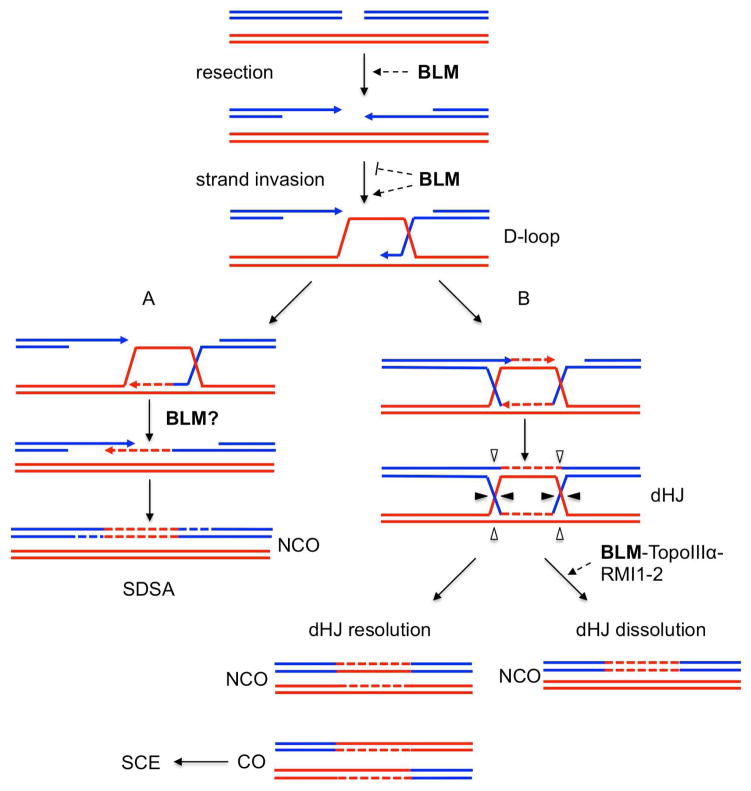
HR is a major high-fidelity repair pathway that in mammalian cells occurs in S and G2 phases of the cell cycle, when a homologous template is available. HR can repair various DNA lesions including double-strand breaks (DSBs), single-strand gaps, collapsed replication forks, and eroded telomeres (Kowalczykowski, 2015). For the repair of DSBs, the initial step after DSB formation is the 5′ to 3′ nucleolytic resection of both ends of the break by specialized exonucleases (Fig. 2). Single-strand binding protein RPA binds to the 3′ ssDNA-overhang generated, after which it is exchanged for the RAD51 recombinase by mediator proteins such as BRCA2 and RAD52. The ssDNA-RAD51 nucleoprotein filament searches for a complementary DNA sequence in the intact homologous DNA duplex, resulting in the invasion of the duplex by the 3′ ssDNA tail. This strand exchange process results in the formation of the D-loop, which is then extended by DNA synthesis using the 3′ invading end as a primer. Two distinct pathways can then resolve the D-loop. In the synthesis-dependent strand-annealing (SDSA) pathway (Fig. 2, pathway A), the polymerase-extended, invading strand is unwound from the intact duplex and is annealed to the complementary ssDNA on the other side of the break. Subsequent gap-filling DNA synthesis and ligation generate exclusively non-crossing-over (NCO) products. Alternatively, the ssDNA on the other side of the break can anneal to the displaced strand from the donor duplex, a process referred to as second-end capture. This annealing primes DNA synthesis in the other direction followed by rejoining of the two ends to produce the dHJ structure (Fig. 2, pathway B). dHJs are removed either by dissolution, producing only NCO products, or by resolution performed by structure-specific endonucleases and ligation to produce NCO or CO products.
Fig. 2.
Homologous recombination repair of double strand breaks (DSB). DSB is resected from 5′ to 3′ to produce a 3′-single strand overhang, which invades the intact homologous chromatid, to form the D-loop. (A) In synthesis-dependent strand-annealing (SDSA), the invading strand dissociates and re-anneals with the complementary 3′-tail to produce non-crossing over (NCO) products. (B) Second-end capture results in the formation of double Holliday Junction (dHJ). dHJ can be resolved by BLM-TopoIII -RMI-mediated dissolution to give NCO or can be cleaved by HJ resolvases (triangles) to produce both NCO and crossing-over (CO), depending on the cleavage plan (plain or empty triangles). CO products will manifest as sister-chromatid exchanges. Newly synthesized DNA is depicted as dashed line in the same color as the template. The established and potential roles of BLM in this pathway are indicated (dashed arrows). See text for details.
Various in vitro assays have revealed a potential role for BLM at most steps of the HR process, wherein BLM can have both pro- and anti-recombinogenic functions (Fig. 2). BLM promotes DSB end resection by stimulating the exonucleases DNA2 and EXO1 (Gravel et al., 2008; Nimonkar et al., 2011; Nimonkar et al., 2008). In the second step of HR, BLM can regulate D-loop formation. In vitro, BLM can disrupt the Rad51-ssDNA filament by removing RAD51 from ssDNA, when RAD51 is in its inactive ADP-bound form, thereby preventing D-loop formation (Bugreev et al., 2007). However, when RAD51 is in its active ATP-bound form, BLM stimulates RAD51 homology search and strand invasion, promoting D-loop formation (Bugreev et al., 2009). BLM is also involved in the later steps of HR after D-loop formation and DNA synthesis. In the SDSA pathway, BLM may play a role by disrupting the D-loop (Bachrati et al., 2006; van Brabant et al., 2000). Additionally, BLM together with topoisomerase III, RMI1, and RMI2, form the “dissolvasome” complex, which is critical for dHJs dissolution (Raynard et al., 2006; Singh et al., 2008; Wu et al., 2006; Xu et al., 2008). In vitro analyses suggest that BLM branch migrates the two HJ toward each other, converting the duplexes into hemicatenanes that are decatenated by topoisomerase III, leading to NCO products (Wu and Hickson, 2003). BLM’s dHJ dissolution function in the topoisomerase III -BLM complex cannot be replaced by any other RecQ helicase in the family (Wu et al., 2005). This conserved function limits crossing-overs during HR. In absence of dissolution, dHJs are processed through endonucleolytic cleavage and ligation resulting in equal numbers of NCO and CO products. These are wonderful biochemical insights, but they do not resolve an important question raised by the recombination repair model in Fig. 1, namely, the question of pathway choice. If, in the presence of BLM, most substrates are repaired by pathway A, then the dissolvasome does not have much work to do in DSB repair. In the absence of BLM, the substrates will flow through pathway B where the dHJ could only be repaired by breakage and rejoining because the dissolvasome is dependent on the presence of BLM. The temptation is to suggest that pathway B, then, is the source of SCEs in BS cells. The problem with this idea is that DSBs exogenously induced by irradiation or radiomimetics are poor SCE-inducing agents. SCEs are most strongly induced by agents that cause replication-associated damage (UV, crosslinking agents, replication inhibitors, etc.), and they derive from HR repair of multiple types replication damage, including post-replication gap repair and repair of DSBs at advancing or stalled forks. Post-replication gap repair can generate a dHJ that could be processed by the dissolvasome, but HR repair of DSBs generated at advancing or stalled replication forks leads to a single HJ and the dissolvasome does not resolve these structures (see Section 4.2). It is not known whether there are single or multiple sources for the SCEs in BS cells.
Several studies have highlighted functional and physical interactions between the FA core complex and the BLM dissolvasome. Five of the FA proteins were found in a complex with BLM-TopoIII -RMI1-2 (Meetei et al., 2003) and deficiencies of FANCC and FANCM in DT40 cells and of FANCJ in mammalian cells are associated with an approximately two fold elevation of SCE levels (Hirano et al., 2005; Rosado et al., 2009; Suhasini et al., 2011). FANCM has been shown to physically connect the dissolvasome and FA complex and this interaction is required for recruiting BLM to sites of DNA damage (Deans and West, 2009; Hoadley et al., 2012). The interaction between BLM and FANCJ is also important for response to replication stress (Suhasini et al., 2011; Suhasini and Brosh, 2012). Consequently, there is crosstalk between the BLM dissolvasome and the FA core complex and the coordination of their activities is critical for suppressing potentially harmful SCE events that can arise during DNA-damage repair. On the clinical side, there is an intriguing observation that BLM mutation can be associated with a more severe presentation of BS that includes Fanconi-like features (Saal et al., 2001). On the other hand, it should be noted that elevated SCEs is not a cytogenetic feature in FA.
BLM pro- and anti-recombinogenic activities are subtly regulated by post-translational modifications, including phosphorylation, sumoylation, and ubiquitination (Bohm and Bernstein, 2014). Of interest, BLM is modified by small ubiquitin-related modifiers (SUMO), particularly SUMO-2, and BLM contains a SUMO-binding motif that is required for BLM’s sumoylation and for localization of BLM to the PML nuclear bodies (Eladad et al., 2005; Zhu et al., 2008). Cells expressing full-length BLM proteins that contain mutations in the SUMO-acceptor sites (SUMO-deficient BLM) exhibited accumulations of DNA damage repair foci both in the presence or absence of exogenous genotoxic agents, excess DSBs, and hypersensitivity to replication stress (Ouyang et al., 2009; Ouyang et al., 2013). Most strikingly, there was in impairment of recruitment of RAD51 and a failure in HR repair at stalled replication forks in SUMO-deficient cells, suggesting that sumoylation regulates the switch between BLM’s pro- and anti-recombinogenic functions.
WRN is also involved in HR, but, whereas BLM deficiency induces an increase in HR, WRN-deficient cells show defects in HR (Chen et al., 2003; Prince et al., 2001; Saintigny et al., 2002). WRN can act early in the HR pathway and is redundant with BLM in stimulating DNA resection (Sturzenegger et al., 2014). In the later steps of HR, although WRN can catalyze branch migration of HJs in vitro (Constantinou et al., 2000), it cannot promote dHJ dissolution (Wu et al., 2005). The NHEJ pathway, which repairs DSBs by direct rejoining of the two extremities of the DSB, is normal in BS cells. NHEJ is error-prone compared to HR when the extremities of the DSB are processed by resection or when the pathway devolves to micro-homology searches. WRN, but not BLM, is involved in NHEJ repair (Chen et al., 2003; Perry et al., 2006), and a defect in NHEJ may explain the variegated translocation mosaicism and large chromosomal deletions observed in WS cells. WRN interacts with both Ku and DNA-PKcs (Cooper et al., 2000; Li and Comai, 2000), and it is a substrate for DNA-PKcs (Yannone et al., 2001).
4.2. Functions in DNA replication and in stabilization and repair of stalled forks
Cells derived from persons with BS exhibits defects in DNA replication that include slower progression of replication forks (Hand and German, 1975; Rao et al., 2007) and the accumulation of aberrant replication intermediates (Lonn et al., 1990). Additionally, BLM interacts with the p12 subunit of Pol (Selak et al., 2008), and large-scale analysis confirmed that BLM is present at the advancing replication fork (Alabert et al., 2014; Dungrawala et al., 2015). BLM may act during unperturbed DNA replication by removing DNA structures that could impede the progression of the replication machinery, such as G4 DNA. In vitro, BLM and WRN can efficiently bind and unwind G4, which could allow the replication machinery to progress without stalling, especially through DNA structures that form on the lagging strand (Chatterjee et al., 2014; Sun et al., 1998). Studies in chicken DT40 cells have shown that WRN and BLM cooperate with FANCJ to facilitate DNA replication through G4 structures (Sarkies et al., 2011) and BLM and WRN are also required for removal of G4 DNA during DNA replication at telomeres (see Section 4.4). There is evidence that BLM interacts with flap endonuclease 1 (FEN1) and may assist in the maturation of Okazaki fragments, especially when long flaps are generated (Bartos et al., 2006; Sharma et al., 2004). WRN also cooperates with FEN1 (Brosh et al., 2001), indicating possibly that these functions are overlapping.
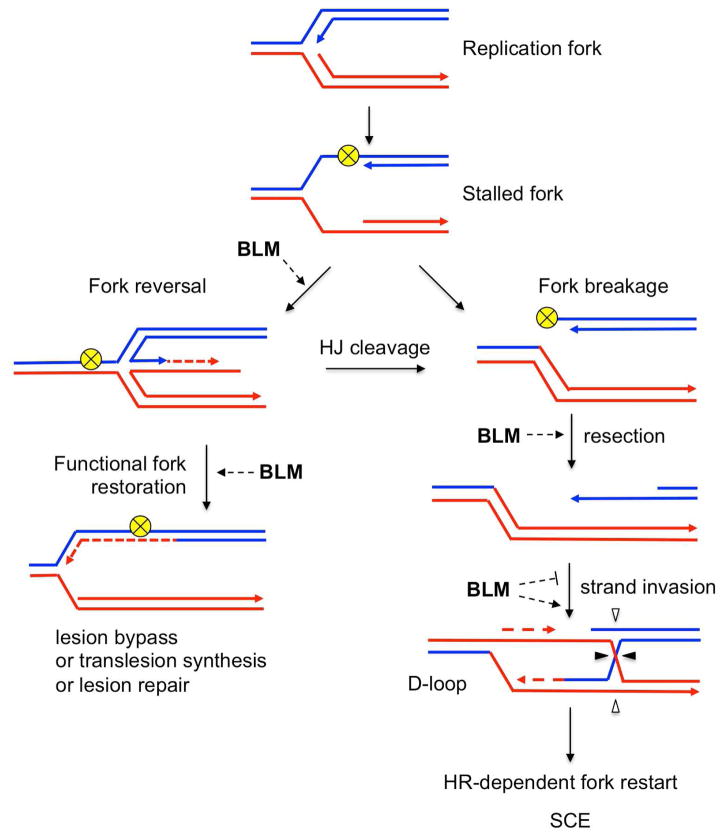
BLM is crucial during DNA replication stress, as evidenced by the hypersensitivity of BS cells to replication inhibitors (Davies et al., 2004). As replication forks progress, they encounter various obstacles, such as damage of the DNA template, unusual secondary structures, DNA-bound proteins, they collide with the transcription machinery, or they can be limited by depletion of the nucleotide pool. These impediments may result in fork stalling. The DNA replication checkpoint is activated to ensure that stalled forks are stabilized and to promote the efficient resumption of DNA synthesis once the damage has been repaired. Failure to stabilize stalled forks causes their collapse, as defined by their inability to complete replicon synthesis (Branzei and Foiani, 2010; Cortez, 2015). In that case, the completion of genome duplication relies on the firing of nearby backup origins, referred to as dormant origins. BLM-deficient cells show an increase of fork stalling, suggesting a role for BLM in the stabilization and processing of stalled forks (Rao et al., 2007). Consistent with this function, following replication stress, BLM is phosphorylated by the replication checkpoint kinases ATR and ATM and is recruited to stalled replication forks where focal concentrations of BLM protein accumulate (Beamish et al., 2002; Davalos et al., 2004; Davies et al., 2004; Dungrawala et al., 2015; Sengupta et al., 2003). BLM is required for the efficient restart of stalled forks after replicative stress and the reduced fork restart observed in BLM-deficient cells is accompanied by an increase in dormant origin firing (Davies et al., 2007). Recruitment of RAD51 to stalled forks and the HR pathway are crucial for the restart of stalled replication forks and the repair of collapsed forks (Petermann and Helleday, 2010). It has been proposed that RAD51-mediated fork remodeling occurs at stalled forks, involving the coordinated annealing of the newly synthesized strands and the re-annealing of the parental strands to generate a regressed fork (also known as a chicken foot), similar to a HJ structure (Fig. 3) (Neelsen and Lopes, 2015; Zellweger et al., 2015). A potential role for BLM at stalled forks could be to facilitate fork regression. In vitro, BLM can use its branch migration activity to mediate fork regression, generating arms greater than 250 bp (Karow et al., 2000; Ralf et al., 2006). Alternatively, subsequent to fork regression, BLM could promote branch migration of the regressed fork to restore a functional replication fork, allowing replication restart and preventing excessive recombination events (Machwe et al., 2011). Reversed forks can also be processed by structure-specific endonucleases into one-ended DSBs (Fig. 3). The recovery of these collapsed forks depends on RAD51-mediated HR. In absence of BLM there is a bias toward resolving these recombination intermediates by crossing over, as evidenced by elevated SCEs. The generation of SCEs in BS cells is dependent on the activities of the structure-specific nucleases MUS81 and SLX4, that is, depletion of either of these proteins in BS cells suppresses SCEs (Wechsler et al., 2011). These data suggest that cleavage of a stalled or regressed fork directs repair down an SCE-generating path (the right side of Fig. 3). BLM’s role in fork regression may not be unique or in fact, in the presence of BLM, the replication fork is stabilized and the regressed fork is not formed at all! Clearly, new approaches are needed to resolve all these different possibilities.
Fig. 3.
Model for replication fork stalling and restart. Replication fork stalls when it encounters an obstacle (yellow ball) leading to single-strand DNA exposure. Newly synthesized DNA strands can anneal to each other to produce a four-way Holliday junction (HJ) or reversed fork. Reversed fork can be converted back to a functional replication fork, allowing lesion bypass, or translesion synthesis, or repair. Reversed fork can also be cleaved, leading to the formation of one-ended DSB. The sister chromatid is used as a template for recombination-mediated restart of the replication fork. Depending on the HJ cleavage (triangles), homologous recombination (HR) process can lead to crossing-over (CO), visualized as sister chromatid exchanges (SCEs). Replication forks can also break directly when encountering DNA damage without fork regression. Newly synthesized DNA is depicted as dashed line in the same color as the template. The potential roles of BLM in stabilization and restart of stalled replication forks are indicated (dashed arrows). See text for details.
WRN is also involved in DNA replication and response to replicative stress (Pichierri et al., 2011; Sidorova, 2008). Several studies reported that WRN-deficient cells have an extended S phase (Poot et al., 1992; Takeuchi et al., 1982), an increase of spontaneous fork stalling (Rodriguez-Lopez et al., 2002), and a defect in fork progression especially after replication stress (Sidorova et al., 2013). During replicative stress, WRN prevents stalled fork to collapse through formation of DSBs (Franchitto et al., 2008). WRN and BLM seem to have redundant and non-redundant functions in DNA replication and stalled fork recovery (Sidorova et al., 2013). For instance, depletion of both WRN and BLM reduces fork progression more than depletion of either BLM or WRN on its own. Similar to BLM, WRN is recruited at stalled replication forks, and it is able to catalyze in vitro fork regression as well as conversion of regressed HJs to functional forks (Constantinou et al., 2000; Machwe et al., 2011). Additionally, WRN is able to stimulate the bypass of DNA damage by translesion polymerases, which are specialized polymerases that are able to replicate across damaged DNA, a function that BLM has not been reported to have (Kamath-Loeb et al., 2007; Maddukuri et al., 2014; Maddukuri et al., 2012; Phillips and Sale, 2010).
4.3. Functions in sister chromatid segregation
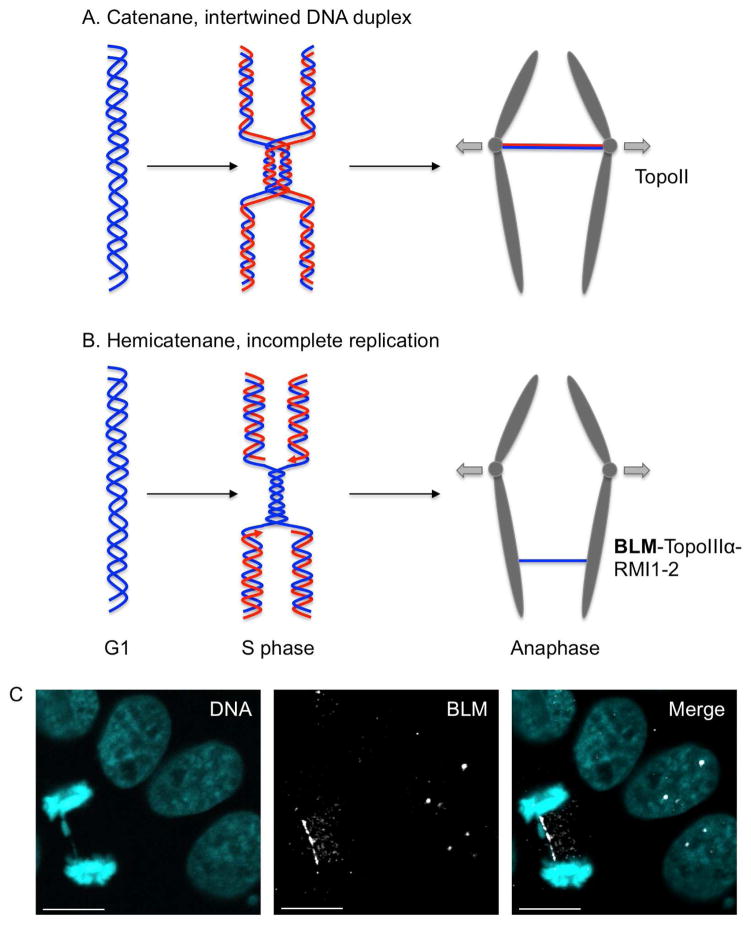
BLM facilitates sister chromatid segregation by processing unresolved replication intermediates that manifest in mitosis as UFBs linking the two daughter nuclei (Chan and Hickson, 2011; Liu et al., 2014). BLM together with topoisomerase III, RMI1, and RMI2 localize to UFBs in anaphase (Fig. 4C), and BLM-deficient cells exhibit an increased frequency of UFBs, suggesting a role for BLM in the resolution of these structures (Chan et al., 2007). Most UFBs detected in normal cells link the centromeres and are predominantly thought to contain unresolved dsDNA catenanes, which are resolved by topoisomerase II (Fig. 4A). Replication stress induces a second class of UFBs arising from fragile sites, which are regions intrinsically challenging for DNA replication (Fig. 4B) (Chan et al., 2009). These UFBs are characterized by the presence of Fanconi Anemia proteins FANCD2 and FANCI at each extremity of the DNA thread (Chan et al., 2009; Naim and Rosselli, 2009). These UFBs most likely correspond to under-replicated DNA, which is coated with RPA and fails to incorporate the thymidine analog EdU (Chan et al., 2009; Gemble et al., 2015). The FANC pathway and topoisomerase III -BLM complex cooperate to resolve the resulting hemicatenane structures. Failure to resolve these UFBs leads to DNA breakage as mitosis proceeds. The broken chromosomes are transmitted to daughter cells, where they become sequestered by p53-binding protein 1 (53BP1), a known mediator of DSB repair. 53BP1 is thought to shield the breaks against excessive resection until the next S phase when they can be repaired by recombination (Lukas et al., 2011). Breakage of the UFBs can also lead to the formation of micronuclei. DNA replication is less efficient in micronuclei and can result in further chromosome breakage (Zhang et al., 2015a). Consistent with a role of BLM in preventing UFBs breakage, BLM-deficient cells exhibit an increased frequency of micronuclei (Ke et al., 2011; Rosin and German, 1985). A third class of UFBs can also arise from telomeres, as mentioned below (Barefield and Karlseder, 2012).
Fig. 4.
Model for the origin of ultra-fine bridges (UFBs). (A) Fully replicated but still intertwined DNA duplexes give rise to UFBs in anaphase and are resolved by topoisomerase II. These catenanes structures are commonly observed at centromeres. (B) Late replication intermediates lead to UFBs in anaphase. These hemicatenane structures arise predominantly from fragile sites under replicative stress and are resolved by BLM-TopoIII -RMI1-2. Parental DNA molecule is depicted in blue and in red is the nascent strand synthesized in S phase (C) Visualization of UFB in anaphase. HeLa cells were fixed and stained for BLM (white) and DAPI (cyan). In interphase, BLM localizes to distinct nuclear loci (PML nuclear bodies) seen as discreet foci. In anaphase, BLM localizes to UFB that appears as a thin thread between the two masses of sister chromatids. Scale bar represent 10μm.
This function of BLM in resolving UFBs is unique among the RecQ helicases, and WRN does not localize to UFBs (Chan et al., 2007). However, WRN-deficient cells exhibit telomere dysfunction associated with an increase of UFBs emanating form the telomeres. BLM localizes to these telomeric UFBs and is most likely involved in their resolution (Barefield and Karlseder, 2012).
4.4. Functions in telomere maintenance
Telomeres are nucleoprotein structures that cap and protect the ends of chromosomes, preventing them from degradation and from being recognized and processed as DSBs by DNA repair mechanisms. Mammalian telomeres consist in several kilobases (10–15 kb for human telomeres) of non-coding tandem TTAGGG repeats that provide a buffer against loss of genetic information. Telomeres present a 30–400 nucleotide 3′-overhang of the G-rich strand, which can fold back and invade the double-stranded telomere DNA to create a telomere D-loop, or T-loop. The T-loop structure protects the chromosome ends from degradation (de Lange, 2005a). Shelterin is a specialized complex of six proteins that specifically bind telomeres and is essential for telomere capping and repression of inopportune DNA damage repair. However, telomeres present a challenge for the replication machinery in part because of the topological interference of the T-loop and the propensity of the G-rich strand to form G4 structures. In addition, replicative polymerases lack the capacity to replicate completely the terminal ends leading to replication-dependent erosion of the telomeres. The loss of telomeric DNA is compensated by telomerase, a specialized reverse transcriptase that adds telomeric repeats onto the 3′-end of chromosomes (Martinez and Blasco, 2015). In humans, telomerase activity is restricted to pluripotent cells and is insufficient to maintain telomere length over a human lifespan. Consequently, telomeres loose about 30–150 nucleotides with each cell division. Past a critical threshold, telomeres can no longer cap the chromosome ends leading to a constitutive DNA damage response that induces the cells to enter a stable and irreversible state of growth arrest, referred to as cellular senescence. This mechanism explains the limited proliferative capacity of normal somatic cells that was first described in 1961, when Hayflick and Moorehead showed that in vitro-cultured primary human fibroblasts stopped dividing after 25–50 cell divisions and entered senescence – a phenomenon now known as the Hayflick limit or replicative senescence (Hayflick and Moorhead, 1961). Loss of normal p53, a deficiency present in approximately 50% of cancers, is one mechanism by which a cell can bypass the senescence barrier, but as telomeres shorten further the chromosome end is recognized as a DSB, which triggers DNA repair mechanisms and end-to-end chromatid fusions. When sister chromatids fuse end-to-end, bridge-breakage-fusion cycles are initiated (de Lange, 2005b). Tumors must ultimately activate a telomere maintenance mechanism, predominantly telomerase reactivation, in order to achieve indefinite replication capacity. Progressive telomere attrition has been proposed as one of the molecular hallmarks of aging (Lopez-Otin et al., 2013). There is no evidence that BS cells reach replicative senescence faster than normal cells. However, BLM-deficient cells do exhibit telomere defects such as associations between homologous telomeres and sister-telomere loss, indicating a role for BLM in telomere maintenance (Barefield and Karlseder, 2012; Lillard-Wetherell et al., 2004). One possible role for BLM is to facilitate telomere replication by removing topological structures, such as the T-loop or G4 structures, that present impediments to the replication progression. For instance, analysis of telomeric DNA replication dynamics in murine fibroblasts revealed that replication fork progression in the telomere is hindered by the presence of G4 structures and requires BLM and WRN, which functionally overlap to some extent, to resolve these structures (Drosopoulos et al., 2015). Another role for BLM in telomere maintenance could be the resolution of late replication intermediates. Indeed, telomeres pose an inherent challenge to the replication machinery, and they have been shown to behave like fragile sites (Sfeir et al., 2009). Similar to common fragile sites and other difficult to replicate regions, telomeres are prone to form UFBs, which, as noted above, require BLM for their resolution (Barefield and Karlseder, 2012).
BLM is also involved in a recombination-mediated mechanism of telomere maintenance, referred to as alternative lengthening of telomeres (ALT) (Rezazadeh, 2013). ALT is an homology-directed telomere-synthesis process that is used in 10–15% of human tumors to prevent telomere attrition in the absence of telomerase (Pickett and Reddel, 2015). ALT cells present an up-regulation of recombination at the telomeres, accompanied by misprocessing of the T-loops. Absence of BLM or WRN in ALT-cells increases telomeric-sister chromatid exchanges (T-SCEs), indicating an overlapping role for BLM and WRN in the regulation of the HR-mediated telomere maintenance mechanism used by ALT cells to evade telomere attrition (Hagelstrom et al., 2010). In addition to elevated T-SCEs, depletion of BLM induces telomere shortening in ALT cells but not in telomerase-positive cells (Bhattacharyya et al., 2009). Both WRN and BLM have been shown to localize to telomeres specifically in ALT cells and to interact with proteins of the shelterin complex, including TRF1, TRF2, and POT1. These interactions modulate BLM and WRN unwinding activities on structures mimicking the T-loop in vitro (Lillard-Wetherell et al., 2004; Opresko et al., 2005; Opresko et al., 2004; Opresko et al., 2002). Consequently, the role of BLM and WRN in disrupting recombination intermediates at telomeres could either prevent telomeric HR in normal cells or facilitate completion of the ALT pathway in cancer cells. Although BLM and WRN are both involved in telomere maintenance, they seem to have non-redundant functions as evidenced by the synergistic effects of simultaneous loss of BLM in WRN in human or mouse cells (Barefield and Karlseder, 2012; Du et al., 2004).
5. Bloom’s syndrome, cancer, and premature aging
Aging is a complex biological process defined as the time-dependent decline of almost all physiological functions, and it is associated with an increasing rate of mortality. There is still no clear understanding of the molecular basis of aging, but progressive compromise of genome integrity is a central part of the process (Lenart and Krejci, 2016). Indeed, accumulation of DNA damage, telomere shortening, and alterations of chromatin structure are among the hallmarks of aging (Lopez-Otin et al., 2013). Moreover, premature aging syndromes are often associated with DNA repair genes (Bohr, 2002; Burtner and Kennedy, 2010; Freitas et al., 2011). As noted above, BS is sometimes mentioned in non-primary literature sources as a progeroid syndrome, mainly because BLM and WRN are associated as RecQ genes, the lifespan of persons with BS is reduced, and cancer develops at an early age in BS. Early onset of cancer and death do not necessarily indicate premature aging—there are old people who do not have cancer, and cancer is also associated with certain childhood conditions. In contrast with WS, persons with BS do not develop prematurely the features associated with aging, such as gray hair, cataracts, osteoporosis, skin changes, arteriosclerosis, and atherosclerosis (Table 1). Yet, the population of persons with BS is relatively young. The oldest person in the Bloom Syndrome Registry is 53 years old, and the average age at death is below 30. The principal cause of death in BS is from the complications of cancer. Average lifespan has increased since the establishment of the Registry; a plausible explanation for this increase is that more BS persons survive their first cancer diagnosis due to improvements of cancer treatments. As more persons with BS reach an older age, we will gain more information about the aging process in BS.
The development of cancer depends on the process of mutation. The probability of a particular cancer-causing mutation event or series of mutation events is generally low; consequently, as the number of cell generations increases, that is, as we age, the expectation that cancer-causing mutations will occur increases. In the realm of cancer genetics, it has long been known but poorly understood that syndromes that feature different types of genomic instability give rise to varied cancer susceptibilities. Lynch syndrome is caused by mismatch repair gene mutations, exhibits a genomic instability characterized by replication errors, and features mainly increased incidence of colon, endometrial, and stomach cancer. Broadly speaking, replication errors are a genomic instability that impact tissues with higher turnover rates. Mutations in BRCA1 and BRCA2 lead to defects in HR and feature increased incidence of breast and ovarian cancer; the specificity of the dependence of breast and ovarian tissue on HR is less obvious. It is relatively more obvious that a defect in the repair of UV damage, which is exhibited in the rare autosomal recessive syndrome xeroderma pigmentosum, is associated with increased skin cancer, because the sun is a cancer-inducing environmental exposure. More recently, connections have been made between repair of DNA damage due to naturally occurring aldehydes and the cancers that arise in FA (Langevin et al., 2011). Understanding the biology of environmental inducers for others cancers may provide insight to the varied cancer diatheses of these different genomic instabilities.
In the case of BS, we encounter a genomic instability that increases the probability of cancer-causing events in most organ systems, whereas in WS we encounter a genomic instability that gives rise to a more restricted cancer diathesis. One prominent difference in the genomic instabilities in these two syndromes is the frequency of HR, which is elevated in BS cells and depressed in WS cells. HR is a powerful accelerator of carcinogenesis because a cell that harbors a first-hit mutation in a tumor suppressor gene will be more likely to generate a homozygous, tumor-suppressor-negative successor cell through mitotic crossing-over between homologs followed by segregation at metaphase of the recombinant and non-recombinant chromatids. The frequency of mitotic crossing-over in BS cells is 20–50 times the frequency in normal cells, as evidenced by the frequency of Qr figures in metaphases from BS lymphocytes3 and the frequency of homozygous M/M or N/N red cells in glycophorin A heterozygotes (Langlois et al., 1989). Homozygous null blm mutations in the mouse are organismal lethal but cell viable; yet, a mouse model for BS was generated using a hypomorphic allele of Blm that produces greatly reduced levels of BLM protein. In this model, elevated mitotic crossing-over events drive cancer development (Luo et al., 2000). Elevated mitotic crossing-over has also been detailed in mouse embryonic stem (ES) cells that contain homozygous blm null mutations (LaRoque et al., 2011). The overall conclusion is that suppression of crossing-over between homologs is an important barrier to carcinogenesis in humans and other species and this suppression is partly achieved by preventing crossovers when interhomolog interactions do occur. This suppression does not appear to be important for organismal aging because aging is not driven by homozygous null mutations in “aging-suppressor genes.” Heterozygotes for WRN mutation ostensibly age at the same rate as normal persons whereas there have been reports that the rate of colorectal cancer is elevated in BLM heterozygotes (Gruber et al., 2002).
Another important difference between BLM and WRN that very likely impacts the cancer phenotypes in the two syndromes is in transcriptional regulation. BLM is expressed only in dividing cells; it is absent or greatly reduced in quiescent stem cells or in terminally differentiated non-dividing cells (Kawabe et al., 2000; Turley et al., 2001). All dividing cells depend on BLM, underscoring the primacy of BLM’s function during DNA synthesis. WRN on the other hand is expressed in both dividing and non-dividing cells (Gee et al., 2002; Szekely et al., 2005). Moreover, the tissues that are most affected by the absence of WRN are the slowly dividing mesenchymal cells, including the vasculature, fat, bone, and dermis. This association extends to the cancer diathesis in WS, where the sites and types of cancer that arise are soft-tissue sarcomas, osteosarcomas, thyroid cancer, and atypical melanomas—rare tumors that are derived from mesenchymal cells (Goto et al., 1996). This is not a tissue compartment that is normally predisposed to cancer young or old. Thus, these slowly dividing tissues are dependent on WRN to avoid cancer-inducing mutation events. WRN has important connections with non-S-phase-dependent DNA repair events such as NHEJ and excision repair. While quiescent stem cells up-regulate DNA repair pathways are they re-enter the cell cycle in order to repair accumulated DNA damage, WRN is expressed in the quiescent stem cell (Beerman et al., 2014). Because stem cells replenish the tissues and maintain homeostasis, genomic instability in this compartment has a large impact and cancerization and aging.
At the cellular level, one of the hallmarks of aging is telomere shortening, which is linked to replicative senescence or Hayflick limit (Hayflick and Moorhead, 1961). Cells derived from WS patients exhibit a limited replicative capacity and enter prematurely into senescence when cultured in vitro, compared to age-matched normal cells (Martin, 2005; Salk et al., 1981). The telomere dysfunction observed in WS cells may be the critical contributor to premature replicative senescence and aging in WS (Opresko, 2008). Mouse models with homozygous Wrn null mutations are relatively healthy, but telomeres in Mus musculus are many times longer than telomeres in Homo sapiens. When the Wrn null mutations were combined with null mutations in the RNA component of telomerase Terc and bred three to six generations, a WS-like aging phenotype was recapitulated in the mouse (Chang et al., 2004; Du et al., 2004). More recently, when induced pluripotent stem cells (iPSCs) were generated from skin fibroblast from persons with WS, cellular phenotypes associated with aging were reversed (Cheung et al., 2014; Shimamoto et al., 2015). iPSCs are generated by expression of key transcription factors that maintain embryonic stem cell status and telomerase is re-expressed during the transcriptional reprogamming. When the iPSCs were differentiated into mesenchymal stem cells, the senescence phenotypes returned, but senescence could be suppressed by expression of telomerase (Cheung et al., 2014). Differentiation to neuronal cells was not associated with return of the senescence phenotypes, modeling the situation in WS. In consideration of the connection between aging and telomerase mutations in the human syndrome dyskeratosis congenita, the premature aging in WS is very likely caused by the telomere dysfunction in cells derived from mesenchymal stem cells. BLM also plays a role in telomere replication, but there is no evidence that BS cells reach replicative senescence any faster than normal. Moreover, telomeres do not appear to shorten prematurely in BS cells (Yankiwski et al., 2000). It may be that whatever functions BLM performs at telomeres are largely overlapping with WRN or disruption of these functions leads to a DNA damage response and apoptosis instead of a senescence response. WRN on the other hand has unique functions at telomeres that BLM is unable to perform. The type of genomic instability at telomeres provides a central part of the explanation for why there is premature aging in WS but not in BS.
Another possibility that merits attention relates to genome structure and gene expression. Aging is associated with epigenetic alterations in DNA methylation and histones modifications associated with chromatin remodeling (Lenart and Krejci, 2016; Lopez-Otin et al., 2013). Interestingly, in a recent study, mesenchymal stem cells derived from WRN-deficient human embryonic stem cells presented disorganization of heterochromatin architecture associated with loss of H3K9 methylation that is thought to promote premature senescence (Zhang et al., 2015b). Heterochromatin alterations in BLM-deficient cells have not been reported. Microarray studies identifying genes and miRNAs that are differentially expressed in absence of BLM or WRN have shown that BS and WS are transcriptionally distinct diseases (Johnson et al., 2010; Nguyen et al., 2014; Tang et al., 2016). WRN mutation or depletion perturbs the expression of several senescence-associated gene programs, possibly contributing to the disease pathogenicity. Part of these differences may trace to differences in the structure of gene-expression regulatory G4 DNAs around the genome. The genes that are differentially expressed in the absence of BLM or WRN are in fact significantly enriched in G4 motifs around the transcription start site or at the 5′ end of the first intron. This observation provides strong evidence that G4 DNA is an in vivo target for WRN and BLM helicases. More than ten different human helicases have been shown to bind and unfold G4 DNA and from a structural standpoint, G4 DNA structures are very diverse, depending on the number of G4 motifs, the orientation of the strands, the nature and the length of interconnecting loops within the G4 DNA fold and between G4 DNA folds, and so on (Mendoza et al., 2016). Therefore, the fact that distinct helicases modulate different genes supports the view that these helicases exploit the structural diversity of G4 to differentially modulate human gene expression. These differences of action of WRN and BLM toward G4 DNA structures provide a different perspective on the explanations for differences observed in WS and BS pathogenicity.
The gene expression studies also provide an alternative view for some of the other prominent features in BS, in particular, the short stature, which is not due to substantial deficits in production of growth hormones. Because the size of cells in persons with BS is normal, the presumption is that there are fewer of them. Accordingly, to explain a lack of normal accumulations of cells, there is either a diminution in the rate of cell division or an increase in apoptosis or both. In the blm null mouse model, there is a wave of apoptosis in the hematopoietic compartment prior to, and most likely explaining, the embryonic death; but excess apoptosis does not appear to be the sole reason for small size. BS lymphocytes in short-term cultures proliferate at the same rate as normal cells whereas some primary BS fibroblasts proliferate more slowly; however, establishment of BS cells in culture can be challenge for unknown reasons and it is generally unsatisfying to extrapolate organismal size from proliferation rates of cells in culture. BS cells have trouble meeting the demands of the rapid cell divisions that constitute the tissues during embryonic development. The leading hypothesis has been that problems arising during DNA synthesis require longer S phases to handle the challenge, slowing the rate of cell division and increasing the rate of apoptosis. The gene expression studies, on the other hand, has shown that individual genes and gene networks whose expression is affected by BLM mutation include growth/proliferation and cell death/survival. Thus, the gene expression defect could contribute to the short stature, hypogonadism, and immune deficiency. Similarly, WRN mutation perturbs the expression of several senescence-associated gene programs, contributing to the aging phenotype.
In summary, although there are many similarities in gene function and activity between BLM and WRN, the clinical outcomes of mutations in these genes are very different. Differences in the genetic mechanisms by which the clinical features and major complications that develop due to mutations in BLM and WRN — the expressivity that connects genotype to phenotype — undeniably explain how BS and WS are so different clinically. Differences in particular genomic instabilities that occur in BS and WS cells impact the types of cancer that arise in the two syndromes. Defects in maintenance of stability at telomeres specific to the WRN are important to the aging process in WS. Further research into differential transcription programs in BS and WS will doubtless further our understanding of the various syndrome-specific phenotypes.
Acknowledgments
The authors thank Kelvin W. Pond for the confocal images in Figure 4. This work was supported by grants from the National Institutes of Health (R01-CA140804, U01-CA153060, and P30-CA023074 to N.A.E.) The content of this article is solely the responsibility of the authors and does not represent the official views of the National Institutes of Health.
Footnotes
Due to the vagaries of human activity, there are some especially unhappy aliases in the literature for human RecQ genes. We use here the HUGO gene nomenclature committee designations.
The clinical data discussed in this section originates from the Bloom’s Syndrome Registry, which was established by James L. German III, M.D., in the 1960s. The Registry has served as the repository for clinical, genetic, cytogenetic, and molecular information on BS and has provided the basis for a thorough and detailed natural history of the disorder.
In a typical culture of phytohemagglutanin-stimulated lymphocytes from a person with BS, one to two Qr figures is encountered in every 100 metaphases. The frequency in a similar culture from a normal person has not been properly measured, but it is a rare event well below one per thousand that a cytogeneticist will greet with astonishment and awe.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alabert C, Bukowski-Wills JC, Lee SB, Kustatscher G, Nakamura K, de Lima Alves F, Menard P, Mejlvang J, Rappsilber J, Groth A. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat Cell Biol. 2014;16:281–293. doi: 10.1038/ncb2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachrati CZ, Borts RH, Hickson ID. Mobile D-loops are a preferred substrate for the Bloom’s syndrome helicase. Nucleic Acids Res. 2006;34:2269–2279. doi: 10.1093/nar/gkl258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barefield C, Karlseder J. The BLM helicase contributes to telomere maintenance through processing of late-replicating intermediate structures. Nucleic Acids Res. 2012;40:7358–7367. doi: 10.1093/nar/gks407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartos JD, Wang W, Pike JE, Bambara RA. Mechanisms by which Bloom protein can disrupt recombination intermediates of Okazaki fragment maturation. J Biol Chem. 2006;281:32227–32239. doi: 10.1074/jbc.M606310200. [DOI] [PubMed] [Google Scholar]
- Bartram CR, Koske-Westphal T, Passarge E. Chromatid exchanges in ataxia telangiectasia, Bloom syndrome, Werner syndrome, and xeroderma pigmentosum. Ann Hum Genet. 1976;40:79–86. doi: 10.1111/j.1469-1809.1976.tb00166.x. [DOI] [PubMed] [Google Scholar]
- Beamish H, Kedar P, Kaneko H, Chen P, Fukao T, Peng C, Beresten S, Gueven N, Purdie D, Lees-Miller S, Ellis N, Kondo N, Lavin MF. Functional link between BLM defective in Bloom’s syndrome and the ataxia-telangiectasia-mutated protein, ATM. J Biol Chem. 2002;277:30515–30523. doi: 10.1074/jbc.M203801200. [DOI] [PubMed] [Google Scholar]
- Beerman I, Seita J, Inlay MA, Weissman IL, Rossi DJ. Quiescent hematopoietic stem cells accumulate DNA damage during aging that is repaired upon entry into cell cycle. Cell Stem Cell. 2014;15:37–50. doi: 10.1016/j.stem.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett RJ, Noirot-Gros MF, Wang JC. Interaction between yeast sgs1 helicase and DNA topoisomerase III. J Biol Chem. 2000;275:26898–26905. doi: 10.1074/jbc.M003137200. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya S, Keirsey J, Russell B, Kavecansky J, Lillard-Wetherell K, Tahmaseb K, Turchi JJ, Groden J. Telomerase-associated protein 1, HSP90, and topoisomerase IIIalpha associate directly with the BLM helicase in immortalized cells using ALT and modulate its helicase activity using telomeric DNA substrates. J Biol Chem. 2009;284:14966–14977. doi: 10.1074/jbc.M900195200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof O, Kim SH, Irving J, Beresten S, Ellis NA, Campisi J. Regulation and localization of the Bloom syndrome protein in response to DNA damage. J Cell Biol. 2001;153:367–380. doi: 10.1083/jcb.153.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohm S, Bernstein KA. The role of post-translational modifications in fine-tuning BLM helicase function during DNA repair. DNA Repair (Amst) 2014;22:123–132. doi: 10.1016/j.dnarep.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohr VA. Human premature aging syndromes and genomic instability. Mechanisms of ageing and development. 2002;123:987–993. doi: 10.1016/s0047-6374(02)00039-8. [DOI] [PubMed] [Google Scholar]
- Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol. 2010;11:208–219. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- Brosh RM, Jr, von Kobbe C, Sommers JA, Karmakar P, Opresko PL, Piotrowski J, Dianova I, Dianov GL, Bohr VA. Werner syndrome protein interacts with human flap endonuclease 1 and stimulates its cleavage activity. EMBO J. 2001;20:5791–5801. doi: 10.1093/emboj/20.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugreev DV, Mazina OM, Mazin AV. Bloom syndrome helicase stimulates RAD51 DNA strand exchange activity through a novel mechanism. J Biol Chem. 2009;284:26349–26359. doi: 10.1074/jbc.M109.029371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugreev DV, Yu X, Egelman EH, Mazin AV. Novel pro- and anti-recombination activities of the Bloom’s syndrome helicase. Genes Dev. 2007;21:3085–3094. doi: 10.1101/gad.1609007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukowy Z, Harrigan JA, Ramsden DA, Tudek B, Bohr VA, Stevnsner T. WRN Exonuclease activity is blocked by specific oxidatively induced base lesions positioned in either DNA strand. Nucleic Acids Res. 2008;36:4975–4987. doi: 10.1093/nar/gkn468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtner CR, Kennedy BK. Progeria syndromes and ageing: what is the connection? Nat Rev Mol Cell Biol. 2010;11:567–578. doi: 10.1038/nrm2944. [DOI] [PubMed] [Google Scholar]
- Chaganti RS, Schonberg S, German J. A manyfold increase in sister chromatid exchanges in Bloom’s syndrome lymphocytes. Proc Natl Acad Sci U S A. 1974;71:4508–4512. doi: 10.1073/pnas.71.11.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KL, Hickson ID. New insights into the formation and resolution of ultra-fine anaphase bridges. Semin Cell Dev Biol. 2011;22:906–912. doi: 10.1016/j.semcdb.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Chan KL, North PS, Hickson ID. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J. 2007;26:3397–3409. doi: 10.1038/sj.emboj.7601777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KL, Palmai-Pallag T, Ying S, Hickson ID. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol. 2009;11:753–760. doi: 10.1038/ncb1882. [DOI] [PubMed] [Google Scholar]
- Chang S, Multani AS, Cabrera NG, Naylor ML, Laud P, Lombard D, Pathak S, Guarente L, DePinho RA. Essential role of limiting telomeres in the pathogenesis of Werner syndrome. Nat Genet. 2004;36:877–882. doi: 10.1038/ng1389. [DOI] [PubMed] [Google Scholar]
- Chatterjee S, Zagelbaum J, Savitsky P, Sturzenegger A, Huttner D, Janscak P, Hickson ID, Gileadi O, Rothenberg E. Mechanistic insight into the interaction of BLM helicase with intra-strand G-quadruplex structures. Nature communications. 2014;5:5556. doi: 10.1038/ncomms6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CF, Brill SJ. An essential DNA strand-exchange activity is conserved in the divergent N-termini of BLM orthologs. EMBO J. 2010;29:1713–1725. doi: 10.1038/emboj.2010.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Huang S, Lee L, Davalos A, Schiestl RH, Campisi J, Oshima J. WRN, the protein deficient in Werner syndrome, plays a critical structural role in optimizing DNA repair. Aging Cell. 2003;2:191–199. doi: 10.1046/j.1474-9728.2003.00052.x. [DOI] [PubMed] [Google Scholar]
- Cheok CF, Wu L, Garcia PL, Janscak P, Hickson ID. The Bloom’s syndrome helicase promotes the annealing of complementary single-stranded DNA. Nucleic Acids Res. 2005;33:3932–3941. doi: 10.1093/nar/gki712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung HH, Liu X, Canterel-Thouennon L, Li L, Edmonson C, Rennert OM. Telomerase protects werner syndrome lineage-specific stem cells from premature aging. Stem Cell Reports. 2014;2:534–546. doi: 10.1016/j.stemcr.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinou A, Tarsounas M, Karow JK, Brosh RM, Bohr VA, Hickson ID, West SC. Werner’s syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep. 2000;1:80–84. doi: 10.1093/embo-reports/kvd004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper MP, Machwe A, Orren DK, Brosh RM, Ramsden D, Bohr VA. Ku complex interacts with and stimulates the Werner protein. Genes Dev. 2000;14:907–912. [PMC free article] [PubMed] [Google Scholar]
- Cortez D. Preventing replication fork collapse to maintain genome integrity. DNA Repair (Amst) 2015 doi: 10.1016/j.dnarep.2015.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croteau DL, Popuri V, Opresko PL, Bohr VA. Human RecQ helicases in DNA repair, recombination, and replication. Annu Rev Biochem. 2014;83:519–552. doi: 10.1146/annurev-biochem-060713-035428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davalos AR, Kaminker P, Hansen RK, Campisi J. ATR and ATM-dependent movement of BLM helicase during replication stress ensures optimal ATM activation and 53BP1 focus formation. Cell Cycle. 2004;3:1579–1586. doi: 10.4161/cc.3.12.1286. [DOI] [PubMed] [Google Scholar]
- Davies SL, North PS, Dart A, Lakin ND, Hickson ID. Phosphorylation of the Bloom’s syndrome helicase and its role in recovery from S-phase arrest. Mol Cell Biol. 2004;24:1279–1291. doi: 10.1128/MCB.24.3.1279-1291.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies SL, North PS, Hickson ID. Role for BLM in replication-fork restart and suppression of origin firing after replicative stress. Nat Struct Mol Biol. 2007;14:677–679. doi: 10.1038/nsmb1267. [DOI] [PubMed] [Google Scholar]
- Deans AJ, West SC. FANCM connects the genome instability disorders Bloom’s Syndrome and Fanconi Anemia. Mol Cell. 2009;36:943–953. doi: 10.1016/j.molcel.2009.12.006. [DOI] [PubMed] [Google Scholar]
- de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- de Lange T. Telomere-related genome instability in cancer. Cold Spring Harb Symp Quant Biol. 2005b;70:197–204. doi: 10.1101/sqb.2005.70.032. [DOI] [PubMed] [Google Scholar]
- Diaz A, Vogiatzi MG, Sanz MM, German J. Evaluation of short stature, carbohydrate metabolism and other endocrinopathies in Bloom’s syndrome. Horm Res. 2006;66:111–117. doi: 10.1159/000093826. [DOI] [PubMed] [Google Scholar]
- Drosopoulos WC, Kosiyatrakul ST, Schildkraut CL. BLM helicase facilitates telomere replication during leading strand synthesis of telomeres. J Cell Biol. 2015;210:191–208. doi: 10.1083/jcb.201410061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X, Shen J, Kugan N, Furth EE, Lombard DB, Cheung C, Pak S, Luo G, Pignolo RJ, DePinho RA, Guarente L, Johnson FB. Telomere shortening exposes functions for the mouse Werner and Bloom syndrome genes. Mol Cell Biol. 2004;24:8437–8446. doi: 10.1128/MCB.24.19.8437-8446.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dungrawala H, Rose KL, Bhat KP, Mohni KN, Glick GG, Couch FB, Cortez D. The replication checkpoint prevents two types of fork collapse without regulating replisome stability. Mol Cell. 2015;59:998–1010. doi: 10.1016/j.molcel.2015.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutertre S, Ababou M, Onclercq R, Delic J, Chatton B, Jaulin C, Amor-Gueret M. Cell cycle regulation of the endogenous wild type Bloom’s syndrome DNA helicase. Oncogene. 2000;19:2731–2738. doi: 10.1038/sj.onc.1203595. [DOI] [PubMed] [Google Scholar]
- Eladad S, Ye TZ, Hu P, Leversha M, Beresten S, Matunis MJ, Ellis NA. Intra-nuclear trafficking of the BLM helicase to DNA damage-induced foci is regulated by SUMO modification. Human molecular genetics. 2005;14:1351–1365. doi: 10.1093/hmg/ddi145. [DOI] [PubMed] [Google Scholar]
- Ellis NA, Groden J, Ye TZ, Straughen J, Lennon DJ, Ciocci S, Proytcheva M, German J. The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- Franchitto A, Pirzio LM, Prosperi E, Sapora O, Bignami M, Pichierri P. Replication fork stalling in WRN-deficient cells is overcome by prompt activation of a MUS81-dependent pathway. J Cell Biol. 2008;183:241–252. doi: 10.1083/jcb.200803173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas AA, Vasieva O, de Magalhaes JP. A data mining approach for classifying DNA repair genes into ageing-related or non-ageing-related. BMC Genomics. 2011;12:27. doi: 10.1186/1471-2164-12-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuchi K, Martin GM, Monnat RJ., Jr Mutator phenotype of Werner syndrome is characterized by extensive deletions. Proc Natl Acad Sci U S A. 1989;86:5893–5897. doi: 10.1073/pnas.86.15.5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff S, McDonald JP, Bendixen C, Arthur L, Rothstein R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol Cell Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhart E, Bauer R, Raub U, Schinzel M, Ruprecht KW, Jonas JB. Spontaneous and induced chromosomal instability in Werner syndrome. Hum Genet. 1988;80:135–139. doi: 10.1007/BF00702855. [DOI] [PubMed] [Google Scholar]
- Gee J, Ding Q, Keller JN. Analysis of Werner’s expression within the brain and primary neuronal culture. Brain Res. 2002;940:44–48. doi: 10.1016/s0006-8993(02)02588-x. [DOI] [PubMed] [Google Scholar]
- Gemble S, Ahuja A, Buhagiar-Labarchede G, Onclercq-Delic R, Dairou J, Biard DS, Lambert S, Lopes M, Amor-Gueret M. Pyrimidine pool disequilibrium induced by a cytidine deaminase deficiency inhibits PARP-1 activity, leading to the under replication of DNA. PLoS Genet. 2015;11:e1005384. doi: 10.1371/journal.pgen.1005384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German J. Bloom’s syndrome. I. Genetical and clinical observations in the first twenty-seven patients. American journal of human genetics. 1969;21:196–227. [PMC free article] [PubMed] [Google Scholar]
- German J. Bloom’s syndrome. Dermatol Clin. 1995;13:7–18. [PubMed] [Google Scholar]
- German J, Archibald R, Bloom D. Chromosomal breakage in a rare and probably genetically determined syndrome of man. Science. 1965;148:506–507. doi: 10.1126/science.148.3669.506. [DOI] [PubMed] [Google Scholar]
- German J, Crippa LP, Bloom D. Bloom’s syndrome. III. Analysis of the chromosome aberration characteristic of this disorder. Chromosoma. 1974;48:361–366. doi: 10.1007/BF00290993. [DOI] [PubMed] [Google Scholar]
- German J, Sanz MM, Ciocci S, Ye TZ, Ellis NA. Syndrome-causing mutations of the BLM gene in persons in the Bloom’s Syndrome Registry. Hum Mutat. 2007;28:743–753. doi: 10.1002/humu.20501. [DOI] [PubMed] [Google Scholar]
- Goto M, Miller RW, Ishikawa Y, Sugano H. Excess of rare cancers in Werner syndrome (adult progeria) Cancer Epidemiol Biomarkers Prev. 1996;5:239–246. [PubMed] [Google Scholar]
- Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groden J, German J. Bloom’s syndrome. XVIII. Hypermutability at a tandem-repeat locus. Hum Genet. 1992;90:360–367. doi: 10.1007/BF00220459. [DOI] [PubMed] [Google Scholar]
- Groden J, Nakamura Y, German J. Molecular evidence that homologous recombination occurs in proliferating human somatic cells. Proc Natl Acad Sci U S A. 1990;87:4315–4319. doi: 10.1073/pnas.87.11.4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber SB, Ellis NA, Scott KK, Almog R, Kolachana P, Bonner JD, Kirchhoff T, Tomsho LP, Nafa K, Pierce H, Low M, Satagopan J, Rennert H, Huang H, Greenson JK, Groden J, Rapaport B, Shia J, Johnson S, Gregersen PK, Harris CC, Boyd J, Rennert G, Offit K. BLM heterozygosity and the risk of colorectal cancer. Science. 2002;297:2013. doi: 10.1126/science.1074399. [DOI] [PubMed] [Google Scholar]
- Hagelstrom RT, Blagoev KB, Niedernhofer LJ, Goodwin EH, Bailey SM. Hyper telomere recombination accelerates replicative senescence and may promote premature aging. Proc Natl Acad Sci U S A. 2010;107:15768–15773. doi: 10.1073/pnas.1006338107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand R, German J. A retarded rate of DNA chain growth in Bloom’s syndrome. Proc Natl Acad Sci U S A. 1975;72:758–762. doi: 10.1073/pnas.72.2.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- Hirano S, Yamamoto K, Ishiai M, Yamazoe M, Seki M, Matsushita N, Ohzeki M, Yamashita YM, Arakawa H, Buerstedde JM, Enomoto T, Takeda S, Thompson LH, Takata M. Functional relationships of FANCC to homologous recombination, translesion synthesis, and BLM. EMBO J. 2004;24:418–427. doi: 10.1038/sj.emboj.7600534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoadley KA, Xue Y, Ling C, Takata M, Wang W, Keck JL. Defining the molecular interface that connects the Fanconi anemia protein FANCM to the Bloom syndrome dissolvasome. Proc Natl Acad Sci U S A. 2012;109:4437–4442. doi: 10.1073/pnas.1117279109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoehn H, Bryant EM, Au K, Norwood TH, Boman H, Martin GM. Variegated translocation mosaicism in human skin fibroblast cultures. Cytogenet Cell Genet. 1975;15:282–298. doi: 10.1159/000130526. [DOI] [PubMed] [Google Scholar]
- Huang S, Li B, Gray MD, Oshima J, Mian IS, Campisi J. The premature ageing syndrome protein, WRN, is a 3′-->5′ exonuclease. Nat Genet. 1998;20:114–116. doi: 10.1038/2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JE, Cao K, Ryvkin P, Wang LS, Johnson FB. Altered gene expression in the Werner and Bloom syndromes is associated with sequences having G-quadruplex forming potential. Nucleic Acids Res. 2010;38:1114–1122. doi: 10.1093/nar/gkp1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath-Loeb AS, Lan L, Nakajima S, Yasui A, Loeb LA. Werner syndrome protein interacts functionally with translesion DNA polymerases. Proc Natl Acad Sci U S A. 2007;104:10394–10399. doi: 10.1073/pnas.0702513104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath-Loeb AS, Shen JC, Schmitt MW, Loeb LA. The Werner syndrome exonuclease facilitates DNA degradation and high fidelity DNA polymerization by human DNA polymerase. J Biol Chem. 2012;287:12480–12490. doi: 10.1074/jbc.M111.332577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karow JK, Chakraverty RK, Hickson ID. The Bloom’s syndrome gene product is a 3′-5′ DNA helicase. J Biol Chem. 1997;272:30611–30614. doi: 10.1074/jbc.272.49.30611. [DOI] [PubMed] [Google Scholar]
- Karow JK, Constantinou A, Li JL, West SC, Hickson ID. The Bloom’s syndrome gene product promotes branch migration of holliday junctions. Proc Natl Acad Sci U S A. 2000;97:6504–6508. doi: 10.1073/pnas.100448097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe T, Tsuyama N, Kitao S, Nishikawa K, Shimamoto A, Shiratori M, Matsumoto T, Anno K, Sato T, Mitsui Y, Seki M, Enomoto T, Goto M, Ellis NA, Ide T, Furuichi Y, Sugimoto M. Differential regulation of human RecQ family helicases in cell transformation and cell cycle. Oncogene. 2000;19:4764–4772. doi: 10.1038/sj.onc.1203841. [DOI] [PubMed] [Google Scholar]
- Ke Y, Huh JW, Warrington R, Li B, Wu N, Leng M, Zhang J, Ball HL, Li B, Yu H. PICH and BLM limit histone association with anaphase centromeric DNA threads and promote their resolution. EMBO J. 2011;30:3309–3321. doi: 10.1038/emboj.2011.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller C, Keller KR, Shew SB, Plon SE. Growth deficiency and malnutrition in Bloom syndrome. J Pediatr. 1999;134:472–479. doi: 10.1016/s0022-3476(99)70206-4. [DOI] [PubMed] [Google Scholar]
- Killen MW, Stults DM, Adachi N, Hanakahi L, Pierce AJ. Loss of Bloom syndrome protein destabilizes human gene cluster architecture. Human molecular genetics. 2009;18:3417–3428. doi: 10.1093/hmg/ddp282. [DOI] [PubMed] [Google Scholar]
- Kim SY, Hakoshima T, Kitano K. Structure of the RecQ C-terminal domain of human Bloom syndrome protein. Sci Rep. 2013;3:3294. doi: 10.1038/srep03294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano K. Structural mechanisms of human RecQ helicases WRN and BLM. Frontiers in genetics. 2014;5:366. doi: 10.3389/fgene.2014.00366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano K, Kim SY, Hakoshima T. Structural basis for DNA strand separation by the unconventional winged-helix domain of RecQ helicase WRN. Structure. 2010;18:177–187. doi: 10.1016/j.str.2009.12.011. [DOI] [PubMed] [Google Scholar]
- Kitano K, Yoshihara N, Hakoshima T. Crystal structure of the HRDC domain of human Werner syndrome protein, WRN. J Biol Chem. 2007;282:2717–2728. doi: 10.1074/jbc.M610142200. [DOI] [PubMed] [Google Scholar]
- Kitao S, Shimamoto A, Goto M, Miller RW, Smithson WA, Lindor NM, Furuichi Y. Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nat Genet. 1999;22:82–84. doi: 10.1038/8788. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski SC. An overview of the molecular mechanisms of recombinational DNA repair. Cold Spring Harbor Perspect Biol. 2015;7 doi: 10.1101/cshperspect.a016410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyoizumi S, Nakamura N, Takebe H, Tatsumi K, German J, Akiyama M. Frequency of variant erythrocytes at the glycophorin-A locus in two Bloom’s syndrome patients. Mutat Res. 1989;214:215–222. doi: 10.1016/0027-5107(89)90166-8. [DOI] [PubMed] [Google Scholar]
- Langevin F, Crossan GP, Rosado IV, Arends MJ, Patel KJ. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature. 2011;475:53–58. doi: 10.1038/nature10192. [DOI] [PubMed] [Google Scholar]
- Langlois RG, Bigbee WL, Jensen RH, German J. Evidence for increased in vivo mutation and somatic recombination in Bloom’s syndrome. Proc Natl Acad Sci U S A. 1989;86:670–674. doi: 10.1073/pnas.86.2.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larizza L, Magnani I, Roversi G. Rothmund-Thomson syndrome and RECQL4 defect: splitting and lumping. Cancer Lett. 2006;232:107–120. doi: 10.1016/j.canlet.2005.07.042. [DOI] [PubMed] [Google Scholar]
- LaRocque JR, Stark JM, Oh J, Bojilova E, Yusa K, Horie K, Takeda J, Jasin M. Interhomolog recombination and loss of heterozygosity in wild-type and Bloom syndrome helicase (BLM)-deficient mammalian cells. Proc Natl Acad Sci U S A. 2011;108:11971–11976. doi: 10.1073/pnas.1104421108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen NB, Hickson ID. RecQ helicases: conserved guardians of genomic integrity. Adv Exp Med Biol. 2013;767:161–184. doi: 10.1007/978-1-4614-5037-5_8. [DOI] [PubMed] [Google Scholar]
- Lenart P, Krejci L. DNA, the central molecule of aging. Mutat Res. 2016;786:1–7. doi: 10.1016/j.mrfmmm.2016.01.007. [DOI] [PubMed] [Google Scholar]
- Li B, Comai L. Functional interaction between Ku and the werner syndrome protein in DNA end processing. J Biol Chem. 2000;275:28349–28352. doi: 10.1074/jbc.C000289200. [DOI] [PubMed] [Google Scholar]
- Lillard-Wetherell K, Machwe A, Langland GT, Combs KA, Behbehani GK, Schonberg SA, German J, Turchi JJ, Orren DK, Groden J. Association and regulation of the BLM helicase by the telomere proteins TRF1 and TRF2. Human molecular genetics. 2004;13:1919–1932. doi: 10.1093/hmg/ddh193. [DOI] [PubMed] [Google Scholar]
- Liu Y, Nielsen CF, Yao Q, Hickson ID. The origins and processing of ultra fine anaphase DNA bridges. Current opinion in genetics & development. 2014;26:1–5. doi: 10.1016/j.gde.2014.03.003. [DOI] [PubMed] [Google Scholar]
- Lonn U, Lonn S, Nylen U, Winblad G, German J. An abnormal profile of DNA replication intermediates in Bloom’s syndrome. Cancer Res. 1990;50:3141–3145. [PubMed] [Google Scholar]
- Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas C, Savic V, Bekker-Jensen S, Doil C, Neumann B, Pedersen RS, Grofte M, Chan KL, Hickson ID, Bartek J, Lukas J. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol. 2011;13:243–253. doi: 10.1038/ncb2201. [DOI] [PubMed] [Google Scholar]
- Luo G, Santoro IM, McDaniel LD, Nishijima I, Mills M, Youssoufian H, Vogel H, Schultz RA, Bradley A. Cancer predisposition caused by elevated mitotic recombination in Bloom mice. Nat Genet. 2000;26:424–429. doi: 10.1038/82548. [DOI] [PubMed] [Google Scholar]
- Machwe A, Karale R, Xu X, Liu Y, Orren DK. The Werner and Bloom syndrome proteins help resolve replication blockage by converting (regressed) holliday junctions to functional replication forks. Biochemistry. 2011;50:6774–6788. doi: 10.1021/bi2001054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machwe A, Xiao L, Groden J, Matson SW, Orren DK. RecQ family members combine strand pairing and unwinding activities to catalyze strand exchange. J Biol Chem. 2005;280:23397–23407. doi: 10.1074/jbc.M414130200. [DOI] [PubMed] [Google Scholar]
- Machwe A, Xiao L, Lloyd RG, Bolt E, Orren DK. Replication fork regression in vitro by the Werner syndrome protein (WRN): holliday junction formation, the effect of leading arm structure and a potential role for WRN exonuclease activity. Nucleic Acids Res. 2007;35:5729–5747. doi: 10.1093/nar/gkm561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddukuri L, Ketkar A, Eddy S, Zafar MK, Eoff RL. The Werner syndrome protein limits the error-prone 8-oxo-dG lesion bypass activity of human DNA polymerase kappa. Nucleic Acids Res. 2014;42:12027–12040. doi: 10.1093/nar/gku913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddukuri L, Ketkar A, Eddy S, Zafar MK, Griffin WC, Eoff RL. Enhancement of human DNA polymerase eta activity and fidelity is dependent upon a bipartite interaction with the Werner syndrome protein. J Biol Chem. 2012;287:42312–42323. doi: 10.1074/jbc.M112.410332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizels N, Gray LT. The G4 genome. PLoS Genet. 2013;9:e1003468. doi: 10.1371/journal.pgen.1003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GM. The Werner mutation: does it lead to a “public” or “private” mechanism of aging? Mol Med. 1997;3:356–358. [PMC free article] [PubMed] [Google Scholar]
- Martin GM. Genetic modulation of senescent phenotypes in Homo sapiens. Cell. 2005;120:523–532. doi: 10.1016/j.cell.2005.01.031. [DOI] [PubMed] [Google Scholar]
- Martinez P, Blasco MA. Replicating through telomeres: a means to an end. Trends Biochem Sci. 2015 doi: 10.1016/j.tibs.2015.06.003. [DOI] [PubMed] [Google Scholar]
- Meetei AR, Sechi S, Wallisch M, Yang D, Young MK, Joenje H, Hoatlin ME, Wang W. A multiprotein nuclear complex connects Fanconi anemia and Bloom syndrome. Mol Cell Biol. 2003;23:3417–3426. doi: 10.1128/MCB.23.10.3417-3426.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza O, Bourdoncle A, Boulé JB, Brosh RM, Jr, Mergny JL. G-quadruplexes and helicases. Nucleic Acids Res. 2016;44:1989–2006. doi: 10.1093/nar/gkw079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohaghegh P, Karow JK, Brosh RM, Jr, Bohr VA, Hickson ID. The Bloom’s and Werner’s syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naim V, Rosselli F. The FANC pathway and BLM collaborate during mitosis to prevent micro-nucleation and chromosome abnormalities. Nat Cell Biol. 2009;11:761–768. doi: 10.1038/ncb1883. [DOI] [PubMed] [Google Scholar]
- Nakayama H, Nakayama K, Nakayama R, Irino N, Nakayama Y, Hanawalt PC. Isolation and genetic characterization of a thymineless death-resistant mutant of Escherichia coli K12: identification of a new mutation (recQ1) that blocks the RecF recombination pathway. Mol Gen Genet. 1984;195:474–480. doi: 10.1007/BF00341449. [DOI] [PubMed] [Google Scholar]
- Neelsen KJ, Lopes M. Replication fork reversal in eukaryotes: from dead end to dynamic response. Nat Rev Mol Cell Biol. 2015;16:207–220. doi: 10.1038/nrm3935. [DOI] [PubMed] [Google Scholar]
- Nguyen GH, Tang W, Robles AI, Beyer RP, Gray LT, Welsh JA, Schetter AJ, Kumamoto K, Wang XW, Hickson ID, Maizels N, Monnat RJ, Jr, Harris CC. Regulation of gene expression by the BLM helicase correlates with the presence of G-quadruplex DNA motifs. Proc Natl Acad Sci U S A. 2014;111:9905–9910. doi: 10.1073/pnas.1404807111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011;25:350–362. doi: 10.1101/gad.2003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimonkar AV, Ozsoy AZ, Genschel J, Modrich P, Kowalczykowski SC. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc Natl Acad Sci U S A. 2008;105:16906–16911. doi: 10.1073/pnas.0809380105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orren DK, Machwe A, Karmakar P, Piotrowski J, Cooper MP, Bohr VA. A functional interaction of Ku with Werner exonuclease facilitates digestion of damaged DNA. Nucleic Acids Res. 2001;29:1926–1934. doi: 10.1093/nar/29.9.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opresko PL. Telomere ResQue and preservation--roles for the Werner syndrome protein and other RecQ helicases. Mechanisms of ageing and development. 2008;129:79–90. doi: 10.1016/j.mad.2007.10.007. [DOI] [PubMed] [Google Scholar]
- Opresko PL, Mason PA, Podell ER, Lei M, Hickson ID, Cech TR, Bohr VA. POT1 stimulates RecQ helicases WRN and BLM to unwind telomeric DNA substrates. J Biol Chem. 2005;280:32069–32080. doi: 10.1074/jbc.M505211200. [DOI] [PubMed] [Google Scholar]
- Opresko PL, Otterlei M, Graakjaer J, Bruheim P, Dawut L, Kolvraa S, May A, Seidman MM, Bohr VA. The Werner syndrome helicase and exonuclease cooperate to resolve telomeric D loops in a manner regulated by TRF1 and TRF2. Mol Cell. 2004;14:763–774. doi: 10.1016/j.molcel.2004.05.023. [DOI] [PubMed] [Google Scholar]
- Opresko PL, von Kobbe C, Laine JP, Harrigan J, Hickson ID, Bohr VA. Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J Biol Chem. 2002;277:41110–41119. doi: 10.1074/jbc.M205396200. [DOI] [PubMed] [Google Scholar]
- Ouyang KJ, Woo LL, Zhu J, Huo D, Matunis MJ, Ellis NA. SUMO modification regulates BLM and RAD51 interaction at damaged replication forks. PLoS biology. 2009;7:e1000252. doi: 10.1371/journal.pbio.1000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang KJ, Yagle MK, Matunis MJ, Ellis NA. BLM SUMOylation regulates ssDNA accumulation at stalled replication forks. Front Genet. 2013;4:167. doi: 10.3389/fgene.2013.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry JJ, Yannone SM, Holden LG, Hitomi C, Asaithamby A, Han S, Cooper PK, Chen DJ, Tainer JA. WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing. Nat Struct Mol Biol. 2006;13:414–422. doi: 10.1038/nsmb1088. [DOI] [PubMed] [Google Scholar]
- Petermann E, Helleday T. Pathways of mammalian replication fork restart. Nat Rev Mol Cell Biol. 2010;11:683–687. doi: 10.1038/nrm2974. [DOI] [PubMed] [Google Scholar]
- Phillips LG, Sale JE. The Werner’s Syndrome protein collaborates with REV1 to promote replication fork progression on damaged DNA. DNA Repair (Amst) 2010;9:1064–1072. doi: 10.1016/j.dnarep.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichierri P, Ammazzalorso F, Bignami M, Franchitto A. The Werner syndrome protein: linking the replication checkpoint response to genome stability. Aging (Albany NY) 2011;3:311–318. doi: 10.18632/aging.100293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett HA, Reddel RR. Molecular mechanisms of activity and derepression of alternative lengthening of telomeres. Nat Struct Mol Biol. 2015;22:875–880. doi: 10.1038/nsmb.3106. [DOI] [PubMed] [Google Scholar]
- Poot M, Hoehn H, Runger TM, Martin GM. Impaired S-phase transit of Werner syndrome cells expressed in lymphoblastoid cell lines. Exp Cell Res. 1992;202:267–273. doi: 10.1016/0014-4827(92)90074-i. [DOI] [PubMed] [Google Scholar]
- Popuri V, Bachrati CZ, Muzzolini L, Mosedale G, Costantini S, Giacomini E, Hickson ID, Vindigni A. The human RecQ helicases, BLM and RECQ1, display distinct DNA substrate specificities. J Biol Chem. 2008;283:17766–17776. doi: 10.1074/jbc.M709749200. [DOI] [PubMed] [Google Scholar]
- Prince PR, Emond MJ, Monnat RJ., Jr Loss of Werner syndrome protein function promotes aberrant mitotic recombination. Genes Dev. 2001;15:933–938. doi: 10.1101/gad.877001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralf C, Hickson ID, Wu L. The Bloom’s syndrome helicase can promote the regression of a model replication fork. J Biol Chem. 2006;281:22839–22846. doi: 10.1074/jbc.M604268200. [DOI] [PubMed] [Google Scholar]
- Rao VA, Conti C, Guirouilh-Barbat J, Nakamura A, Miao ZH, Davies SL, Sacca B, Hickson ID, Bensimon A, Pommier Y. Endogenous gamma-H2AX-ATMChk2 checkpoint activation in Bloom’s syndrome helicase deficient cells is related to DNA replication arrested forks. Mol Cancer Res. 2007;5:713–724. doi: 10.1158/1541-7786.MCR-07-0028. [DOI] [PubMed] [Google Scholar]
- Raynard S, Bussen W, Sung P. A double Holliday junction dissolvasome comprising BLM, topoisomerase IIIalpha, and BLAP75. J Biol Chem. 2006;281:13861–13864. doi: 10.1074/jbc.C600051200. [DOI] [PubMed] [Google Scholar]
- Rezazadeh S. On BLM helicase in recombination-mediated telomere maintenance. Mol Biol Rep. 2013;40:3049–3064. doi: 10.1007/s11033-012-2379-0. [DOI] [PubMed] [Google Scholar]
- Rhodes D, Lipps HJ. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015;43:8627–8637. doi: 10.1093/nar/gkv862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Lopez AM, Jackson DA, Iborra F, Cox LS. Asymmetry of DNA replication fork progression in Werner’s syndrome. Aging Cell. 2002;1:30–39. doi: 10.1046/j.1474-9728.2002.00002.x. [DOI] [PubMed] [Google Scholar]
- Rosado IV, Niedzwiedz W, Alpi AF, Patel KJ. The Walker B motif in avian FANCM is required to limit sister chromatid exchanges but is dispensable for DNA crosslink repair. Nucleic Acids Res. 2009;37:4360–4370. doi: 10.1093/nar/gkp365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosin MP, German J. Evidence for chromosome instability in vivo in Bloom syndrome: increased numbers of micronuclei in exfoliated cells. Hum Genet. 1985;71:187–191. doi: 10.1007/BF00284570. [DOI] [PubMed] [Google Scholar]
- Saintigny Y, Makienko K, Swanson C, Emond MJ, Monnat RJ., Jr Homologous recombination resolution defect in werner syndrome. Mol Cell Biol. 2002;22:6971–6978. doi: 10.1128/MCB.22.20.6971-6978.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salk D, Au K, Hoehn H, Martin GM. Cytogenetics of Werner’s syndrome cultured skin fibroblasts: variegated translocation mosaicism. Cytogenet Cell Genet. 1981;30:92–107. doi: 10.1159/000131596. [DOI] [PubMed] [Google Scholar]
- Sanz MM, German J. Bloom’s Syndrome. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD, Fong CT, Mefford HC, Smith RJH, Stephens K, editors. GeneReviews(R) Seattle (WA): 2006. [Google Scholar]
- Sanz MM, Proytcheva M, Ellis NA, Holloman WK, German J. BLM, the Bloom’s syndrome protein, varies during the cell cycle in its amount, distribution, and co-localization with other nuclear proteins. Cytogenet Cell Genet. 2000;91:217–223. doi: 10.1159/000056848. [DOI] [PubMed] [Google Scholar]
- Sato A, Mishima M, Nagai A, Kim SY, Ito Y, Hakoshima T, Jee JG, Kitano K. Solution structure of the HRDC domain of human Bloom syndrome protein BLM. J Biochem. 2010;148:517–525. doi: 10.1093/jb/mvq097. [DOI] [PubMed] [Google Scholar]
- Sato A, Mishima M, Nagai A, Kim SY, Ito Y, Hakoshima T, Jee JG, Kitano K. Solution structure of the HRDC domain of human Bloom syndrome protein BLM. J Biochem. 2010;148:517–525. doi: 10.1093/jb/mvq097. [DOI] [PubMed] [Google Scholar]
- Szekely AM, Bleichert F, Nümann A, Van Komen S, Manasanch E, Ben Nasr A, Canaan A, Weissman SM. Werner protein protects nonproliferating cells from oxidative DNA damage. Mol Cell Biol. 2005;25:10492–10506. doi: 10.1128/MCB.25.23.10492-10506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selak N, Bachrati CZ, Shevelev I, Dietschy T, van Loon B, Jacob A, Hubscher U, Hoheisel JD, Hickson ID, Stagljar I. The Bloom’s syndrome helicase (BLM) interacts physically and functionally with p12, the smallest subunit of human DNA polymerase delta. Nucleic Acids Res. 2008;36:5166–5179. doi: 10.1093/nar/gkn498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Linke SP, Pedeux R, Yang Q, Farnsworth J, Garfield SH, Valerie K, Shay JW, Ellis NA, Wasylyk B, Harris CC. BLM helicase-dependent transport of p53 to sites of stalled DNA replication forks modulates homologous recombination. EMBO J. 2003;22:1210–1222. doi: 10.1093/emboj/cdg114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Sommers JA, Wu L, Bohr VA, Hickson ID, Brosh RM., Jr Stimulation of flap endonuclease-1 by the Bloom’s syndrome protein. J Biol Chem. 2004;279:9847–9856. doi: 10.1074/jbc.M309898200. [DOI] [PubMed] [Google Scholar]
- Shi W, Zauber A, Sanz M, German J. Analysis of the markedly increased incidence of cancer in individuals with Bloom’s syndrome (Abstract). 56th annual meeting of the American Society of Human Genetics; New Orleans. 2006. [Google Scholar]
- Shimamoto A, Kagawa H, Zensho K, Sera Y, Kazuki Y, Osaki M, Oshimura M, Ishigaki Y, Hamasaki K, Kodama Y, Yuasa S, Fukuda K, Hirashima K, Seimiya H, Koyama H, Shimizu T, Takemoto M, Yokote K, Goto M, Tahara H. Reprogramming suppresses premature senescence phenotypes of Werner syndrome cells and maintains chromosomal stability over long-term culture. PLoS One. 2014;9:e112900. doi: 10.1371/journal.pone.0112900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorova JM. Roles of the Werner syndrome RecQ helicase in DNA replication. DNA Repair (Amst) 2008;7:1776–1786. doi: 10.1016/j.dnarep.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorova JM, Kehrli K, Mao F, Monnat R., Jr Distinct functions of human RECQ helicases WRN and BLM in replication fork recovery and progression after hydroxyurea-induced stalling. DNA Repair (Amst) 2013;12:128–139. doi: 10.1016/j.dnarep.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh TR, Ali AM, Busygina V, Raynard S, Fan Q, Du CH, Andreassen PR, Sung P, Meetei AR. BLAP18/RMI2, a novel OB-fold-containing protein, is an essential component of the Bloom helicase-double Holliday junction dissolvasome. Genes Dev. 2008;22:2856–2868. doi: 10.1101/gad.1725108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturzenegger A, Burdova K, Kanagaraj R, Levikova M, Pinto C, Cejka P, Janscak P. DNA2 cooperates with the WRN and BLM RecQ helicases to mediate long-range DNA end resection in human cells. J Biol Chem. 2014;289:27314–27326. doi: 10.1074/jbc.M114.578823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhasini AN, Brosh RM., Jr Fanconi anemia and Bloom’s syndrome crosstalk through FANCJ-BLM helicase interaction. Trends Genet. 2012;28:7–13. doi: 10.1016/j.tig.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhasini AN, Rawtani NA, Wu Y, Sommers JA, Sharma S, Mosedale G, North PS, Cantor SB, Hickson ID, Brosh RM., Jr Interaction between the helicases genetically linked to Fanconi anemia group J and Bloom’s syndrome. EMBO J. 2011;30:692–705. doi: 10.1038/emboj.2010.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Karow JK, Hickson ID, Maizels N. The Bloom’s syndrome helicase unwinds G4 DNA. J Biol Chem. 1998;273:27587–27592. doi: 10.1074/jbc.273.42.27587. [DOI] [PubMed] [Google Scholar]
- Swan MK, Legris V, Tanner A, Reaper PM, Vial S, Bordas R, Pollard JR, Charlton PA, Golec JM, Bertrand JA. Structure of human Bloom’s syndrome helicase in complex with ADP and duplex DNA. Acta Crystallogr D Biol Crystallogr. 2014;70:1465–1475. doi: 10.1107/S139900471400501X. [DOI] [PubMed] [Google Scholar]
- Takeuchi F, Hanaoka F, Goto M, Yamada M, Miyamoto T. Prolongation of S phase and whole cell cycle in Werner’s syndrome fibroblasts. Exp Gerontol. 1982;17:473–480. doi: 10.1016/s0531-5565(82)80009-0. [DOI] [PubMed] [Google Scholar]
- Tang W, Robles AI, Beyer RP, Gray LT, Nguyen GH, Oshima J, Maizels N, Harris CC, Monnat RJ., Jr The Werner syndrome RECQ helicase targets G4 DNA in human cells to modulate transcription. Human molecular genetics. 2016 doi: 10.1093/hmg/ddw079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Brabant AJ, Ye T, Sanz M, German IJ, Ellis NA, Holloman WK. Binding and melting of D-loops by the Bloom syndrome helicase. Biochemistry. 2000;39:14617–14625. doi: 10.1021/bi0018640. [DOI] [PubMed] [Google Scholar]
- Wallis JW, Chrebet G, Brodsky G, Rolfe M, Rothstein R. A hyper-recombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell. 1989;58:409–419. doi: 10.1016/0092-8674(89)90855-6. [DOI] [PubMed] [Google Scholar]
- Wang JC. DNA topoisomerases: why so many? J Biol Chem. 1991;266:6659–6662. [PubMed] [Google Scholar]
- Wang LL, Plon SE. Rothmund-Thomson Syndrome. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD, Fong CT, Mefford HC, Smith RJH, Stephens K, editors. GeneReviews(R) Seattle (WA): 1999. [Google Scholar]
- Warren ST, Schultz RA, Chang CC, Wade MH, Trosko JE. Elevated spontaneous mutation rate in Bloom syndrome fibroblasts. Proc Natl Acad Sci U S A. 1981;78:3133–3137. doi: 10.1073/pnas.78.5.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt PM, Hickson ID, Borts RH, Louis EJ. SGS1, a homologue of the Bloom’s and Werner’s syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics. 1996;144:935–945. doi: 10.1093/genetics/144.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler T, Newman S, West SC. Aberrant chromosome morphology in human cells defective for Holliday junction resolution. Nature. 2011;471:642–646. doi: 10.1038/nature09790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Bachrati CZ, Ou J, Xu C, Yin J, Chang M, Wang W, Li L, Brown GW, Hickson ID. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc Natl Acad Sci U S A. 2006;103:4068–4073. doi: 10.1073/pnas.0508295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Chan KL, Ralf C, Bernstein DA, Garcia PL, Bohr VA, Vindigni A, Janscak P, Keck JL, Hickson ID. The HRDC domain of BLM is required for the dissolution of double Holliday junctions. EMBO J. 2005;24:2679–2687. doi: 10.1038/sj.emboj.7600740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Hickson ID. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- Wu Y. Unwinding and rewinding: double faces of helicase? J Nucleic Acids. 2012;2012:140601. doi: 10.1155/2012/140601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Guo R, Sobeck A, Bachrati CZ, Yang J, Enomoto T, Brown GW, Hoatlin ME, Hickson ID, Wang W. RMI, a new OB-fold complex essential for Bloom syndrome protein to maintain genome stability. Genes Dev. 2008;22:2843–2855. doi: 10.1101/gad.1708608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yankiwski V, Marciniak RA, Guarente L, Neff NF. Nuclear structure in normal and Bloom syndrome cells. Proc Natl Acad Sci U S A. 2000;97:5214–5219. doi: 10.1073/pnas.090525897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yannone SM, Roy S, Chan DW, Murphy MB, Huang S, Campisi J, Chen DJ. Werner syndrome protein is regulated and phosphorylated by DNA-dependent protein kinase. J Biol Chem. 2001;276:38242–38248. doi: 10.1074/jbc.M101913200. [DOI] [PubMed] [Google Scholar]
- Yu CE, Oshima J, Fu YH, Wijsman EM, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, Martin GM, Mulligan J, Schellenberg GD. Positional cloning of the Werner’s syndrome gene. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- Zellweger R, Dalcher D, Mutreja K, Berti M, Schmid JA, Herrador R, Vindigni A, Lopes M. Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. J Cell Biol. 2015;208:563–579. doi: 10.1083/jcb.201406099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CZ, Spektor A, Cornils H, Francis JM, Jackson EK, Liu S, Meyerson M, Pellman D. Chromothripsis from DNA damage in micronuclei. Nature. 2015a;522:179–184. doi: 10.1038/nature14493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Li J, Suzuki K, Qu J, Wang P, Zhou J, Liu X, Ren R, Xu X, Ocampo A, Yuan T, Yang J, Li Y, Shi L, Guan D, Pan H, Duan S, Ding Z, Li M, Yi F, Bai R, Wang Y, Chen C, Yang F, Li X, Wang Z, Aizawa E, Goebl A, Soligalla RD, Reddy P, Esteban CR, Tang F, Liu GH, Belmonte JC. Aging stem cells. A Werner syndrome stem cell model unveils heterochromatin alterations as a driver of human aging. Science. 2015b;348:1160–1163. doi: 10.1126/science.aaa1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Hopkin RJ, Saal HM. Atypical presentations of Bloom syndrome. 548th annual meeting of the American Society of Human Genetics; San Diego, CA. 2001. Abstract 635. [Google Scholar]
- Zhong S, Hu P, Ye TZ, Stan R, Ellis NA, Pandolfi PP. A role for PML and the nuclear body in genomic stability. Oncogene. 1999;18:7941–7947. doi: 10.1038/sj.onc.1203367. [DOI] [PubMed] [Google Scholar]
- Zhu J, Zhu S, Guzzo CM, Ellis NA, Sung KS, Choi CY, Matunis MJ. Small ubiquitin-related modifier (SUMO) binding determines substrate recognition and paralog-selective SUMO modification. J Biol Chem. 2008;283:29405–29415. doi: 10.1074/jbc.M803632200. [DOI] [PMC free article] [PubMed] [Google Scholar]