Abstract
Linkage disequilibrium — the nonrandom association of alleles at different loci — is a sensitive indicator of the population genetic forces that structure a genome. Because of the explosive growth of methods for assessing genetic variation at a fine scale, evolutionary biologists and human geneticists are increasingly exploiting linkage disequilibrium in order to understand past evolutionary and demographic events, to map genes that are associated with quantitative characters and inherited diseases, and to understand the joint evolution of linked sets of genes. This article introduces linkage disequilibrium, reviews the population genetic processes that affect it and describes some of its uses. At present, linkage disequilibrium is used much more extensively in the study of humans than in non-humans, but that is changing as technological advances make extensive genomic studies feasible in other species.
Linkage disequilibrium (LD) is one of those unfortunate terms that does not reveal its meaning. As every instructor of population genetics knows, the term is a barrier not an aid to understanding. LD means simply a nonrandom association of alleles at two or more loci, and detecting LD does not ensure either linkage or a lack of equilibrium. The term was first used in 1960 by Lewontin and Kojima1 and it persists because LD was initially the concern of population geneticists who were not picky about terminology as long as the mathematical definition was clear. At first, there were few data with which to study LD, and its importance to evolutionary biology and human genetics was unrecognized outside of population genetics. However, interest in LD grew rapidly in the 1980s once the usefulness of LD for gene mapping became evident and large-scale surveys of closely linked loci became feasible. By then, the term was too well established to be replaced.
LD is of importance in evolutionary biology and human genetics because so many factors affect it and are affected by it. LD provides information about past events and it constrains the potential response to both natural and artificial selection. LD throughout the genome reflects the population history, the breeding system and the pattern of geographic subdivision, whereas LD in each genomic region reflects the history of natural selection, gene conversion, mutation and other forces that cause gene-frequency evolution. How these factors affect LD between a particular pair of loci or in a genomic region depends on local recombination rates. The population genetics theory of LD is well developed and is being widely used to provide insight into evolutionary history and as the basis for mapping genes in humans and in other species.
In this article, I will review the definitions of LD and the problems with assessing it, then outline the basic population genetics of LD that tells us how natural selection, genetic drift, recombination and mutation all affect levels of LD, and finally discuss some recent applications of LD to mapping genes, inferring the intensity of selection in the genome and estimating allele age.
Definitions
One pair of loci
LD between alleles at two loci has been defined in many ways (BOX 1), but all definitions depend on the quantity:
| (1) |
which is the difference between the frequency of gametes carrying the pair of alleles A and B at two loci (pAB) and the product of the frequencies of those alleles (pA and pB). Originally, the definition was in terms of gamete frequencies because that allows for the possibility that the loci are on different chromosomes. The usual application now is to loci on the same chromosome, in which case the allele pair AB is called a haplotype and pAB is the haplotype frequency. As defined, DAB characterizes a population; in practice, DAB is estimated from allele and haplotype frequencies in a sample. Standard sampling theory has to be applied to find the confidence intervals of estimated values2.
The quantity DAB is the coefficient of linkage disequilibrium. It is defined for a specific pair of alleles, A and B, and does not depend on how many other alleles are at the two loci — each pair of alleles has its own D. The values for different pairs of alleles are constrained by the fact that the allele frequencies at both loci and the haplotype frequencies have to add up to 1. If both loci are diallelic, as is the case with virtually all SNPs, the constraint is strong enough that only one value of D is needed to characterize LD between those loci. In fact, DAB = −DAb = −DaB = Dab, where a and b are the other alleles. In this case, the D is used without a subscript. The sign of D is arbitrary and depends on which pair of alleles one starts with.
If either locus has more than two alleles, no single statistic quantifies the overall LD between them. Although several have been suggested3,4, none has gained wide acceptance. Such a statistic is needed when both loci have numerous alleles, as is the case for many loci in the major histocompatibility complex in vertebrates, which have dozens or even hundreds of alleles, or for microsatellite loci, which often have 10 to 20 alleles. If there is no one pair of alleles of particular interest, the question is often whether there is more LD between one pair of loci than another pair, or more LD between a pair of loci in one species than in another5,6.
Linkage equilibrium
If D = 0 there is linkage equilibrium (LE), which has similarities to the Hardy–Weinberg equilibrium (HWE) in implying statistical independence. When genotypes at a single locus are at HWE, whether an allele is present on one chromosome is independent of whether it is present on the homologue. Consequently, the frequency of the AA homozygote is the square of the frequency of A () and the frequency of the Aa heterozygote is twice the product of pA and pa, the two being necessary to allow for both Aa and aA. The essential feature of HWE is that, regardless of the initial genotype frequencies, HWE is established in one generation of random mating. Any initial deviation from HWE disappears immediately. Significant departures from HWE indicate something interesting is going on, for example, extensive inbreeding, strong selection or genotyping error.
LE is similar to HWE because it implies that alleles at different loci are randomly associated. The frequency of the AB haplotype is the product of the allele frequencies (pApB). LE differs from HWE, however, because it is not established in one generation of random mating. Instead, D decreases at a rate that depends on the recombination frequency, c, between the two loci:
| (2) |
where t is time in generations. Even for unlinked loci (c = 0.5), D decreases only by a factor of a half each generation, something proved by Weinberg7 in 1909. The general formula was obtained first by Jennings8.
Although LE will eventually be reached, it will occur slowly for closely linked loci. That is the basis for the uses of LD discussed in later sections. Other population genetic forces, including selection, gene flow, genetic drift and mutation, all affect D, so substantial LD will persist under many conditions. Now that very large numbers of polymorphic loci can be surveyed, the extent of LD in a genome can be quantified with great precision, allowing a fine-scale analysis of forces governing genomic variation.
The coefficient of LD and related quantities are descriptive statistics. Their magnitude does not indicate whether or not there is a statistically significant association between alleles in haplotypes. Standard statistical tests, including the chi squared and Fisher's exact test, are commonly used to test for significance2.
Haplotype phase
D and related statistics implicitly assume that haploid individuals or gametes can be typed. But often, only diploid genotypes and not haplotypes can be determined. That is the case with all SNP surveys, other than those of the X chromosome in males (assuming males are the heterogametic sex) or when haploids can be typed. The problem is sketched in BOX 2. The extent of LD in genotypic data2,9 can be quantified, but the lack of information about the haplotype phase weakens the signal of nonrandom association sufficiently that this approach is not often taken. It is more common to use a statistical method based on population genetics theory (BOX 2) to infer haplotype phase from genotypic data and then to treat the inferred haplotypes as if they were data. Although this procedure is intuitively appealing and usually leads to reasonable results, especially for common haplotypes, it ignores the uncertainty that is inherent in the inference step and that might be important in some cases. Often, genotypes can be resolved into several possible haplotypes, and inferred frequencies of rare haplotypes can be quite wrong10. It is preferable, although sometimes difficult, to use methods that account for the uncertainly in the inferred haplotype frequencies, as is done in likelihood analysis with Metropolis algorithm using random coalescence (LAMARC)11 and some other computer program packages.
LD at more than two loci
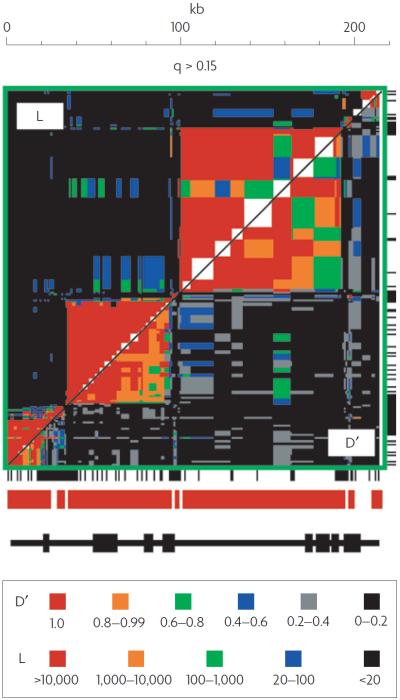
When more than two loci are considered together, a common practice is to distinguish graphically those pairs that have high levels of LD from those that do not3. The result is a graph of the type introduced by Miyashita and Langley12 to describe patterns of LD in Drosophila melanogaster. A more recent example is shown in FIG. 1. This figure indicates that a 216 kb segment in the class II region of the major histocompatibility complex in humans is made up of non-overlapping sets of loci in strong LD with each other. Each group is called a `haplotype block' and boundaries were shown to be associated with hot spots of recombination. Similar patterns were found in other genomic regions in humans13,14, leading to the hypothesis that most of the human genome had a block-like pattern of LD. Haplotype blocks in humans vary in size from a few kb to more than 100 kb15.
Figure 1. Haplotype blocks.
This graph provides some of the first evidence of haplotype blocks and their association with recombination hot spots. The figure shows the pattern of pairwise linkage disequilibrium (LD) in a 216 kb region of the class II region of the major histocompatibility complex in humans for all pairs of SNPs in the region for which the frequency (q) of the minor (that is, less common) allele exceeded 0.15. The region above the diagonal shows levels of L obtained when using D′ = 0 as the null hypothesis (L is the likelihood ratio from the test of linkage equilibrium). D′ is the ratio of D (a measure of LD) to its maximum possible absolute value, given the allele frequencies. This figure is reproduced, with permission, from Nature Genetics REF. 130 © (2001) Macmillan Publishers Ltd.
Haplotype blocks were a surprising discovery that was of great practical importance for the mapping of inherited diseases. Before their discovery, the prevailing view of LD in humans was represented by results from the simulation study of Kruglyak16, which showed that, under assumptions that were intended to approximate the history of modern humans, little LD would be expected beyond 3 kb. The discovery of haplotype blocks showed that LD usually extended over much longer chromosomal distances and suggested that testing one SNP within each block for significant association with a disease might be sufficient to indicate association with every SNP in that block, thus reducing the number of SNPs that need to be tested in case–control studies of disease association17. The situation turned out to be more complex both because some genomic regions were found to not have a block-like structure18 and because different ways of defining haplotype blocks resulted in different block boundaries19. Nevertheless, the observation that LD in humans extended over relatively large chromosomal distances provided a major part of the impetus for the International HapMap Project, which in its first generation identified over 1 million SNPs in humans and characterized the LD in 269 individuals in four ethnically different populations (European, Han Chinese, Japanese and Yoruban)20. The second generation HapMap published recently characterized 3.1 million SNPs in the same group of individuals21.
Haplotype blocks vary somewhat among human populations — they tend to be somewhat shorter in African populations15,20,21. Haplotype blocks have been studied in other species as well, both model organisms, including the mouse and rat22, and domesticated species, including cows23 and dogs24. The isolation of strains and breeds in these species results in much longer block lengths than are found in humans.
Variance in heterozygosity
A simple and often useful statistic describing the overall extent of LD in a genomic region is the variance in heterozygosity across loci, which increases as a linear function of D−2 (REF. 25). This statistic is useful when the density of polymorphic loci is low and the goal is to obtain a general idea of the importance of recombination. Maynard Smith et al.26 used this statistic to assess the overall degree of clonality of various pathogenic bacteria.
Higher-order disequilibria
When considering more than two loci, equation 1 can be generalized to define higher-order coefficients of LD. For alleles at three loci (A, B, and C) the third-order coefficient is:
| (3) |
where DAB, DBC and DAC are the pairwise disequilibrium coefficients. DABC is analogous to the three-way interaction term in an analysis of variance and can be interpreted as the non-independence among these alleles that is not accounted for by the pairwise coefficients. The decay of these higher-order coefficients under random mating was studied by Geiringer27 and has been worked out in some detail by later authors. Little practical use of these higher-order coefficients has been made, other than in the analysis of variation of human leukocyte antigen loci in humans, which suggested that two loci that are closely linked to a selected locus would display unusual patterns of LD28. It is worth considering whether higher-order disequilibrium coefficients can help to understand the patterns found in the HapMap and other large data sets.
LD within and between populations
When data for more than one population are available, LD between a pair of loci can be partitioned into contributions within and between populations. This partitioning, first suggested by Ohta29,30, is similar to Wright's31 partitioning of deviations from HWE frequencies into FIS, the average deviation within populations, and FST, the average deviation that is attributable to differences in allele frequency among populations31. Ohta30 partitioned DT, the total disequilibrium in a subdivided population, into DIS, the average disequilibrium within subpopulations, and DST, the contribution to the overall disequilibrium caused by differences in allele frequencies among subpopulations. Computer programs such as Genepop32 are available to calculate DIS and DST.
These statistics are used widely in the analysis of data from non-human populations but only rarely for human populations, probably because the focus in humans is on each population whereas the focus in other species is often on the overall pattern of LD. Natural selection favouring adaptations to local conditions will increase DST whenever alleles at different loci are favoured. Partitioning overall LD is an appropriate first step when trying to determine whether differences in LD result only from differences in allele frequency or from other factors that vary among populations.
Bypassing LD
Both D and measures based on D are descriptive statistics that quantify deviations from random association of alleles. The statistics themselves provide no information about why alleles at different loci are nonrandomly associated. There is no agreement about which is the best or most useful statistic2,33,34, in part because different statistics are sensitive to different population genetic processes that can cause nonrandom association. An alternative to using one or even several statistics is to ignore the coefficient of LD altogether and estimate parameters of the population genetic models as discussed in the following sections. Several methods to estimate recombination rates directly from haplotypes have been proposed35–39 and they have been used successfully to estimate local rates of recombination in the human genome and to identify DNA sequence motifs associated with hot spots of recombination40.
Bypassing descriptive statistics has the advantage of not having to decide which statistic best captures the underlying signal that is sought, but it has disadvantage of not providing a summary of the data independently of the model used.
Population genetics of LD
Natural selection
Initial interest in LD arose from questions about the operation of natural selection. If alleles at two loci are in LD and they both affect reproductive fitness, the response to selection on one locus might be accelerated or impeded by selection affecting the other.
One line of research in this area concerns the effect of LD on long-term trends in evolution. Kimura41, Nagylaki42,43 and others showed that unless interacting loci are very closely linked or selection is very strong, recombination dominates and, to a good approximation, LD can be ignored. This theory supports Fisher's44 depiction of natural selection steadily increasing the average fitness of a population. This theory also shows that when selection is strong and fitness interactions among loci are complex, average fitness might not increase every generation because LD constrains the way in which haplotype frequencies respond to selection. In that case, linkage must be accounted for explicitly before even qualitative predictions can be made.
In some cases, selection alone can increase LD. This occurs when fitnesses are more than multiplicative, meaning that the average fitness of an individual carrying the AB haplotype exceeds the product of the average fitnesses of individuals carrying A alone or B alone45. The pattern is easiest to see with diallelic loci in haploid organisms. If the relative fitness (w) of the ab, Ab, and aB haplotypes are 1, wAb and waB, then selection will increase LD if wAB > wAbwaB.
If both A and B are maintained by balancing selection, then LD can persist indefinitely1,46,47. Furthermore, when more than two loci interact in this way, large blocks of LD can be maintained by selection, leading to the suggestion that an individual locus is not the appropriate unit of selection48,49. Interest in this kind of theory diminished in the 1970s when it was discovered that LD could not be detected between alleles that are distinguishable by protein electrophoresis50,51. This theory will become popular again or perhaps be reinvented as studies find increasing evidence of intragenic interactions that can create strong epistasis in fitness52.
Genetic drift
Genetic drift alone can create LD between closely linked loci — the effect is similar to taking a small sample from a large population. Even if two loci are in LE, sampling only a few individuals will create some LD. Results first obtained in the late 1960s suggested that genetic drift balanced by mutation and recombination would maintain only low levels of LD, and the expectation of D2 is small even if there is no recombination53,54 because the flux of mutations at both loci tends to eliminate most LD. For that reason, drift was largely ignored as a cause of LD. However, the expectation of D2 does not tell the whole story because it includes cases in which one or both loci are monomorphic (when D is necessarily 0). The expectation of D2 when both loci are polymorphic cannot be calculated analytically, but simulations show that much larger values are seen35,55,56.
Genetic drift interacts with selection in a surprising way. Selection affecting closely linked loci becomes slightly weakened because drift creates small amounts of LD that, on average, reduces the response to selection57. This effect, called the Hill–Robertson effect, is relatively weak when only two loci are considered but is much stronger per locus when many selected loci are closely linked58.
Felsenstein59 showed that the Hill–Robertson effect might have a crucial role in the evolution of recombination and sexual reproduction. The basic idea is that the Hill–Robertson effect causes selection to be inefficient in purging deleterious mutations in a species with a low recombination rate. Hence, natural selection will favour any mutation that increases recombination rates. This early result has been confirmed and extended by many others60,61. As interactions among intragenic SNPs become better understood, the Hill–Robertson effect will have to be taken into account when considering the evolution of gene function, especially in the first few generations of a new selective regime.
Population subdivision and population bottlenecks
Natural selection affects only one or a small number of loci. By contrast, population subdivision, changes in population size and the exchange of individuals among populations all affect LD throughout the genome. Consequently, genome-wide patterns of LD can help us understand the history of changes in population size and the patterns of gene exchange.
The intentional or unintentional mixing of individuals from subpopulations that have different allele frequencies creates LD62,63. The effect is obvious in an extreme case. Suppose that one subpopulation is fixed for A and B whereas another is fixed for a and b. Any mixture of individuals from the two subpopulations would contain only the AB and ab haplotypes, implying that there is perfect LD (D′ = 1; D′ is the ratio of D to its maximum possible absolute value, given the allele frequencies), when in fact there is no LD in either subpopulation. This effect is similar to the Wahlund effect — the inbreeding coefficient at a locus when subpopulations with different allele frequencies are mixed. The reason is the same: the inbreeding coefficient measures the covariance between alleles at a locus just as D measures the covariance between alleles at different loci2. Differences in allele frequencies among subpopulations create additional covariance in both cases.
The movement of individuals or gametes among subpopulations causes gene flow, which increases LD in each subpopulation whenever allele frequencies differ among subpopulations. The decay of LD under recombination alone can be greatly retarded62. If selection maintains differences in allele frequencies at two or more loci among subpopulations, LD in each subpopulation will persist64,65.
Changes in population size, particularly an extreme reduction in size (a population bottleneck), can increase LD. Colonizing species undergo repeated bottlenecks in size, and many models of the history of hominids assume a bottleneck occurred when modern humans first left Africa66. After a bottleneck, some haplotypes will be lost, generally resulting in increased LD. A subsequent period of small population size will augment LD by increasing the effect of genetic drift. Several studies of humans have argued that long-distance LD in humans is the results of a bottleneck early in human history67–69. Detecting higher levels of genome-wide LD in one population than in another can then indicate a past bottleneck68.
Inbreeding, inversions and gene conversion
Other forces create LD as well. Inbreeding creates LD for the same reason as population subdivision. Because of recent common ancestry, inbreeding augments the covariance between alleles at different loci70,71. Theory predicts that this effect is largest in selfing species, but the expected pattern is not evident in the most thoroughly studied selfing species, Arabidopsis thaliana72,73.
Genomic inversions greatly reduce recombination between the inverted and non-inverted segments because recombination produces aneuploid gametes. Consequently, the inverted and original segments become equivalent to almost completely isolated subpopulations between which LD accumulates. This fact has long been appreciated by Drosophila geneticists50.
Gene conversion affects LD at a pair of loci in the same way that reciprocal recombination does. The equivalence can be seen by considering a pair of diallelic loci A/a and B/b. Gene conversion at the B/b locus will result in an individual with haplotype phase AB/ab who will produce Ab or aB gametes depending on whether B converts b or the reverse. However, gene conversion differs from recombination when more than two loci are considered together. Reciprocal crossing over affects LD between all pairs of loci on opposite sides of where the crossing over took place. By contrast, gene conversion affects loci only within the conversion track, which is generally quite short. Loci that are not within the track are unaffected. For example, if three loci, A/a, B/b and C/c, are on a chromosome in that order and only B/b is within a conversion track, LD between A/a and B/b and between B/b and C/c is affected by conversion but the LD between A/a and C/c is not. Several methods for inferring the relative rates of gene conversion and recombination have been based on this idea74–78.
Applications of LD
Mutation and gene mapping
Mutation has a unique role in creating LD. When a mutant allele, M, first appears on a chromosome, it is in low frequency, pM = 1/(2N) (N is the population size) and is in perfect LD with the alleles at other loci that are on the chromosome carrying the first copy of M; perfect LD means that D′ = 1 (BOX 1). If D′ = 1, only three of the four possible haplotypes are present in the population (BOX 3). Perfect LD will persist until recombination involving an M-bearing chromosome creates a non-ancestral haplotype. Consequently, loci that are closely linked to M will remain in perfect LD for a long time and in strong LD for even longer.
The persistence of strong LD between a mutant allele and the loci closely linked to it has many practical implications. Rare marker alleles in strong LD with a monogenic disease locus have to be closely linked to the causative locus. Relatively simple mathematical theory indicates just how close. The resulting method, called LD mapping, has been successfully used with several diseases (BOX 4).
The same idea underlies association mapping of complex diseases. Closely linked polymorphic SNPs tend to be in strong LD with one another. The fine-scale pattern of LD in humans confirms that the human genome is comprised of haplotype blocks within which most or all SNPs are in high LD21. These high levels of LD among SNPs are assumed to be true for alleles that increase the risk of complex inherited diseases. This idea, combined with the development of efficient methods for surveying large numbers of SNPs, has led to the many recent genome-wide association (GWA) studies that have detected SNPs that are significantly associated with breast cancer79,80, colorectal cancer81,82, type 2 diabetes83–86 and heart disease87,88, among other diseases.
However, one potential problem in GWA studies is that, as mentioned above, LD can be created by unrecognized population subdivision. Several methods have been proposed to account for such LD89,90.
Although GWA studies have been successful in finding new causative alleles, the overall proportion of risk that is accounted for is often low. For example, Easton et al.79 found five new variants associated with familial breast cancer, but only 3.6% of familial breast cancer is accounted for by those alleles. Seventy percent of the genetic basis of familial breast cancer remains unaccounted for. Alleles accounting for a greater proportion of risk might be found in even larger studies, but it is unclear whether most causative variants will ultimately be found this way. The reason is that GWA studies are more effective in finding causative alleles that are in relative high frequency. Other methods might be needed for low-frequency causative alleles.
Detecting natural selection
Strong positive selection quickly increases the frequency of an advantageous allele, with the result that linked loci remain in unusually strong LD with that allele. This idea originated with Maynard Smith and Haigh91, who called it genetic hitch-hiking. Their paper focused on an advantageous allele that goes to fixation and causes a substantial reduction of heterozygosity at closely linked neutral loci. Recently, methods have been developed for detecting regions of unusually low heterozygosity that are indications of past hitch-hiking events92–94.
If an advantageous allele has not gone to fixation, variability at linked markers will be lower on chromosomes bearing that allele than on other chromosomes. Several tests of neutrality have been based on this idea. One class of methods assumes that a potentially advantageous allele at a locus has been identified and tests whether there is significantly more LD with that allele than with other alleles at the same locus95,96. A second class of methods assumes only that the potentially selected locus has been identified and tests whether patterns of haplotype variation at that locus are consistent with neutrality97,98. Recently, Sabeti et al.99,100 and Voight et al.101 developed computationally efficient methods for detecting evidence of selection in whole genomes and have applied those methods to the HapMap populations. These studies found several regions in the human genome that were previously not suspected to harbour selected variants.
Estimating allele age
Strong LD with an allele in a relatively large region indicates that not much time has passed since the allele arose by mutation. If the mutant allele has reached a relatively high frequency in a short time, it is likely to have done so under the effect of positive selection. This tendency provides the basis for the various tests of selection mentioned in the previous section. In addition to testing for selection, LD can indicate the point in time that the allele arose by mutation, that is, the allele age. The idea is based on equation 2 above. From an observed level of LD, allele age is estimate by solving for t (REFS 95,102). This approach is straightforward and leads to reasonable estimates of allele age, but it does not take account of the stochastic nature of recombination and genetic drift, and hence exaggerates the accuracy of the resulting estimates. Various statistical methods have been developed that provide more realistic confidence intervals of estimated ages103–106.
The future of LD studies
In human population genetics, the future of LD is now. Very large-scale GWA studies have already been carried out and many more are in progress. The technological problem of efficiently genotyping 500,000 or more SNPs has been solved, and costs of genotyping will continue to decline. And soon new technologies will allow large resequencing studies, including the 1000 Genomes Project107, to take place. The limiting factor will be the availability of people who are willing to participate in GWA studies and the resources needed for accurate clinical assessment.
The methods currently used for association mapping will be used even more extensively in the future to study the history of human populations. At present, most analysis is done on the four HapMap populations, but large-scale surveys of SNPs and resequencing studies will be complete for a much broader range of populations. Advances in understanding human history will be increasingly limited by people's willingness to participate in genetic studies, something that is influenced by political and ethical concerns in addition to scientific ones108–110.
With the increased resolution of LD patterns, the study of human history will shift in focus from understanding the average history of populations to understanding the history of different genomic regions. Unusually large variation in LD within the human genome already suggests that ancient human populations were subdivided111,112 and that some genomic regions of modern humans were brought by introgression from an extinct ancestor, possibly Neanderthals113.
In model organisms, large SNP and resequencing studies on the scale of human studies are now being done73,114. Other species will have to wait a technological generation or two before such large-scale surveys will be possible, in part because of the lower levels of funding available for studying non-model species. The range of possible selective regimes and population histories is vastly greater for non-humans than for humans, and new theoretical methods will no doubt be needed. Extensive studies of variation and examination of LD patterns will probably reveal levels of complexity not seen in humans. In some groups, widespread trans-species polymorphism and evidence of inter-specific gene transfer will be so apparent that some higher organisms might come to resemble bacteria in their genetic promiscuity.
At present, the emphasis is on LD between SNPs, which are diallelic and which mutate at such low rates that current patterns of LD are nearly unaffected by recent mutation. Less attention has been paid to studying LD between other kinds of genetic variants, including microsatellites, insertions, deletions and inversions. The relatively high rate of mutation in microsatellites makes possible the assessment of LD that was created recently115,116. The potential selective effect of indels and inversions, combined with more efficient means of their detection117,118, will provide additional rich sources of LD in humans and other species.
Box 1 | Definitions of LD.
Different definitions of linkage disequilibrium (LD) have been proposed because they capture different features of nonrandom association. All of them are related to D, which is defined in equation 1 in the text. Although D completely characterizes the extent to which two alleles, A and B, are nonrandomly associated, it is often not the best statistic to use when comparing LD at different pairs of loci because the range of possible values of D for each pair is constrained by the allele frequencies. The smallest possible value, Dmin, is the less negative value of −pApB and −(1 − pA)(1 − pB), where p is the frequency of the allele. The largest possible value, Dmax, is the smaller of pA(1 − pB) and pB(1 − pA). Lewontin119 defined D′ to be the ratio of D to its maximum possible absolute value, given the allele frequencies. This definition has the convenient property that when D′ = 1 it indicates that at least one of the four possible haplotypes is absent, regardless of the allele frequencies (BOX 2), a situation commonly described as a `perfect' disequilibrium.
Another commonly used way to quantify LD is with r2:
which is a correlation coefficient of 1/0 (all or none) indicator variables indicating the presence of A and B. In general, r2 is similar to D′ in that it can be nearly one even if one or both alleles are in low frequency.
Still another measure is δA, defined to be pA + D/pB. It is the conditional probability that a chromosome carries an A allele, given that it carries a B allele. It is useful for characterizing the extent to which a particular allele is associated with a genetic diease120.
Box 2 | Genotype data and haplotype phase.
When the genotype of a dipoid individual is determined, the result is a list of genotypes for each locus surveyed. If three diallelic loci are surveyed, the genotypes of four individuals might be AA bb CC, Aa BB cc, aa Bb Cc and Aa Bb Cc. The haplotypes of the first two individuals are immediately apparent. Individual 1 has two copies of AbC and individual 2 has ABc and aBc. There is no uncertainty if no more than one locus is heterozygous. Otherwise, haplotypes cannot be determined without further information. Individual 3 could have haplotypes aBC/abc or aBc/abC. The number of possible resolutions increases exponentially with the number of heterozygous loci. Individual 4 could have haplotypes ABC/abc, ABc/abC, aBc/AbC or aBC/Abc.
There are several ways to determine haplotyes from genotypes; this is commonly referred to as resolving haplotype phase. If the parental genotypes are known, the haplotype phase of the offspring can usually, but not always, be determined. If the parents of individual 3 have genotypes Aa BB Cc and Aa Bb cc, then the individual's haplotype phase has to be aBC/abc. However, if instead the parents' genotypes are Aa Bb Cc and Aa Bb cc, then the haplotype phase still cannot be resolved.
Another way to resolve haplotype phase is to use a biochemical method that separately amplifies each chromosome, allowing direct determination of haplotype phase121. Although such methods exist, they are currently too slow and costly to be used in large genomic surveys.
It is much more common to use a statistical method based on the assumption that haplotypes are randomly joined into genotypes. The basic idea is that individuals that are homozygous at all loci or all but one locus provide some information about haplotype frequencies that can then be used to infer the haplotype phase of the other individuals. Various methods — including those based on maximum likelihood122, parsimony123, combinatorial theory124 and a priori distribution derived from coalescent theory125 — have been developed. The last method is the basis for the program PHASE, which has performed the best in extensive simulation studies126. The emerging view of this problem is that inferring haplotype phase is similar to other cases in which missing data (in this case the haplotype phase of a diploid genotype) has to be imputed127.
Box 3 | Four-haplotype test.
If, initially, a locus is polymorphic for two alleles, A and a, and a linked locus is fixed for b, there are only two haplotypes present in the population — Ab and ab. When a new allele, B, appears by mutation, there are three haplotypes, the new one being AB or aB depending on which copy of b mutated. In both cases, D′ = 1 (D′ is the ratio of D (a measure of linkage disequilibrium) to its maximum possible absolute value, given the allele frequencies). If there is no recombination and no additional copy of B is created by mutation, the fourth haplotype will never appear. Therefore, if all four haplotypes are detected, there has to have been either recombination or recurrent mutation36. The four-haplotype test or equivalently detecting D′ < 1 provides a simple way to determine whether recombination has occurred, provided that recurrent mutation can be ignored.
Box 4 | LD mapping.
Linkage disequilibrium (LD) mapping of a disease-associated allele is based on the slow decay of LD with closely linked markers. One of the first successful examples of LD mapping was by Hästbacka et al.128, who mapped the locus associated with diastrophic dysplasia (DTD) in Finland. In this population a majority of cases seemed to be caused by a dominant mutation that had been mapped by conventional methods to 5q31–5q34. Hästbacka et al. surveyed several restriction site polymorphisms in that region and found two that showed strong LD with DTD. They found that on a sample of non-DTD chromosomes, the numbers of the four possible haplotypes (labelled 11, 12, 21 and 22) were 4, 28, 7, and 84, whereas on a sample of DTD chromosomes the numbers were 144, 1, 0, 2. These data indicated that the mutation causing DTD in Finland arose on a chromosome with haplotype 11, and that LD had decayed little in the 2,000 years (roughly 100 generations) since the founding of the Finnish population. Hästbacka et al. applied the Luria–Delbrück theory to approximate the history of the DTD mutation and concluded that the mutation was approximately 0.06 cM or 60 kb from these marker loci. It was later found at a distance of 70 kb129.
Acknowledgements
The writing of this paper was supported in part by a grant from the US National Institutes of Health, R01-GM40282. I thank J. Felsenstein for discussions of this topic and the translation of Weinberg's paper, and M. Kirkpatrick and the referees for comments on an earlier version of this paper.
References
- 1.Lewontin RC, Kojima K. The evolutionary dynamics of complex polymorphisms. Evolution. 1960;14:458–472. [Google Scholar]
- 2.Weir BS. Genetic Data Analysis II. Sinauer Assoc; Sunderland, Massachusetts: 1996. [Google Scholar]
- 3.Hedrick PW. Genetic disequilibrium measures: proceed with caution. Genetics. 1987;117:331–341. doi: 10.1093/genetics/117.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper and the reply by Lewontin (reference 33) point out many of the logical and statistical difficulties in attempting to define a `best' LD statistic.
- 4.Abecasis GR, Cookson WOC. GOLD — Graphical Overview of Linkage Disequilibrium. Bioinformatics. 2000;16:182–183. doi: 10.1093/bioinformatics/16.2.182. [DOI] [PubMed] [Google Scholar]
- 5.Zhao H, Nettleton D, Dekkers JCM. Evaluation of linkage disequilibrium measures between multi-allelic markers as predictors of linkage disequilibrium between single nucleotide polymorphisms. Genet. Res. 2007;89:1–6. doi: 10.1017/S0016672307008634. [DOI] [PubMed] [Google Scholar]
- 6.Meyer D, Single RM, Mack SJ, Erlich HA, Thomson G. Signatures of demographic history and natural selection in the human major histocompatibility complex loci. Genetics. 2006;173:2121–2142. doi: 10.1534/genetics.105.052837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weinberg W. Uber vererbungsgesetze beim menschen. Z. Abst V. Vererb. 1909;1:276–330. [Google Scholar]
- 8.Jennings HS. The numerical results of diverse systems of breeding, with respect to two pairs of characters, linked and independent, with special relation to the effects of linkage. Genetics. 1917;2:97–154. doi: 10.1093/genetics/2.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weir BS. Inferences about linkage disequilibrium. Biometrics. 1979;35:235–254. [PubMed] [Google Scholar]
- 10.Excoffier L, Slatkin M. Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol. Biol. Evol. 1995;12:921–927. doi: 10.1093/oxfordjournals.molbev.a040269. [DOI] [PubMed] [Google Scholar]
- 11.Kuhner MK. LAMARC 2.0: maximum likelihood and Bayesian estimation of population parameters. Bioinformatics. 2006;22:768–770. doi: 10.1093/bioinformatics/btk051. [DOI] [PubMed] [Google Scholar]
- 12.Miyashita N, Langley CH. Molecular and phenotypic variation of the white locus region in Drosophila melanogaster. Genetics. 1988;120:199–212. doi: 10.1093/genetics/120.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daly MJ, Rioux JD, Schaffner SF, Hudson TJ, Lander ES. High-resolution haplotype structure in the human genome. Nature. 2001;29:229–232. doi: 10.1038/ng1001-229. [DOI] [PubMed] [Google Scholar]; This paper presents the first clear evidence of haplotype blocks in the human genome and the first method for detecting block boundaries.
- 14.Gabriel SB, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 15.Wall JD, Pritchard JK. Haplotype blocks and linkage disequilibrium in the human genome. Nature Rev. Genet. 2003;4:587–597. doi: 10.1038/nrg1123. [DOI] [PubMed] [Google Scholar]
- 16.Kruglyak L. Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nature Genet. 1999;22:139–144. doi: 10.1038/9642. [DOI] [PubMed] [Google Scholar]
- 17.Carlson CS, et al. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am. J. Hum. Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phillips MS, et al. Chromosome-wide distribution of haplotype blocks and the role of recombination hot spots. Nature Genet. 2003;33:382–387. doi: 10.1038/ng1100. [DOI] [PubMed] [Google Scholar]
- 19.Anderson EC, Novembre J. Finding haplotype block boundaries by using the minimum-description-length principle. Am. J. Hum. Genet. 2003;73:336–354. doi: 10.1086/377106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.International HapMap Consortium A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guryev V, et al. Haplotype block structure is conserved across mammals. PLoS Genet. 2006;2:1111–1118. doi: 10.1371/journal.pgen.0020121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gautier M, et al. Genetic and haplotypic structure in 14 European and African cattle breeds. Genetics. 2007;177:1059–1070. doi: 10.1534/genetics.107.075804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindblad-Toh K, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 25.Brown AHD, Feldman MW, Nevo E. Multilocus structure of natural populations of Hordeum spontaneum. Genetics. 1980;96:523–536. doi: 10.1093/genetics/96.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maynard Smith J, Smith NH, O'Rourke M, Spratt BG. How clonal are bacteria? Proc. Natl Acad. Sci. USA. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geiringer H. On the probability theory of linkage in Mendelian heredity. Annals of Mathematical Statistics. 1944;15:25–57. [Google Scholar]
- 28.Grote MN, Klitz W, Thomson G. Constrained disequilibrium values and hitchhiking in a three-locus system. Genetics. 1998;150:1295–1307. doi: 10.1093/genetics/150.3.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohta T. Linkage disequilibrium due to random genetic drift in finite subdivided populations. Proc. Natl. Acad. Sci. USA. 1982;79:1940–1944. doi: 10.1073/pnas.79.6.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohta T. Linkage disequilibrium with the island model. Genetics. 1982;101:139–155. doi: 10.1093/genetics/101.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wright S. Breeding structure of populations in relation to speciation. Am. Nat. 1940;74:232–248. [Google Scholar]
- 32.Raymond M, Rousset F. Genepop (Version 1.2) — population-genetics software for exact tests and ecumenicism. J. Hered. 1995;86:248–249. [Google Scholar]
- 33.Lewontin RC. On measures of gametic disequilibrium. Genetics. 1988;120:849–852. doi: 10.1093/genetics/120.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maniatis N, Morton NE, Xu CF, Hosking LK, Collins A. The optimal measure of linkage disequilibrium reduces error in association mapping of affection status. Hum. Mol. Genet. 2005;14:145–153. doi: 10.1093/hmg/ddi019. [DOI] [PubMed] [Google Scholar]
- 35.Hudson RR. Properties of a neutral allele model with intragenic recombination. Theor. Popul. Biol. 1983;23:183–201. doi: 10.1016/0040-5809(83)90013-8. [DOI] [PubMed] [Google Scholar]; This paper presents the first coalescent model with recombination.
- 36.Hudson RR, Kaplan NL. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics. 1985;111:147–164. doi: 10.1093/genetics/111.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuhner MK, Yamato J, Felsenstein J. Maximum likelihood estimation of recombination rates from population data. Genetics. 2000;156:1393–1401. doi: 10.1093/genetics/156.3.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hudson RR. Two-locus sampling distributions and their applications. Genetics. 2001;159:1805–1817. doi: 10.1093/genetics/159.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McVean G, Awadalla P, Fearnhead P. A coalescent-based method for detecting and estimating recombination from gene sequences. Genetics. 2002;160:1231–1241. doi: 10.1093/genetics/160.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]; This paper applies the method described in reference 39 to human HapMap data and demonstrates the ubiquity of recombinational hot spots and identifies a DNA sequence motif that is associated with elevated recombination rates.
- 41.Kimura M. Attainment of quasi linkage equilibrium when gene frequencies are changing by natural selection. Genetics. 1965;52:875–890. doi: 10.1093/genetics/52.5.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagylaki T. Quasilinkage equilibrium and the evolution of two-locus systems. Proc. Natl. Acad. Sci. USA. 1974;71:526–530. doi: 10.1073/pnas.71.2.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagylaki T. The evolution of one and two-locus systems. Genetics. 1976;83:583–600. [PMC free article] [PubMed] [Google Scholar]
- 44.Fisher RA. The Genetical Theory of Natural Selection. Clarendon; Oxford: 1930. [Google Scholar]
- 45.Felsenstein J. The effect of linkage on directional selection. Genetics. 1965;52:349–363. doi: 10.1093/genetics/52.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karlin S, Feldman MW. Linkage and selection: two locus symmetric viability model. Theor. Popul. Biol. 1970;1:39–71. doi: 10.1016/0040-5809(70)90041-9. [DOI] [PubMed] [Google Scholar]
- 47.Feldman MW, Franklin I, Thomson GJ. Selection in complex genetic systems I. The symmetric equilibria of the three-locus symmetric viability model. Genetics. 1974;76:135–162. doi: 10.1093/genetics/76.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Franklin I, Lewontin RC. Is the gene the unit of selection? Genetics. 1970;65:707–734. doi: 10.1093/genetics/65.4.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Slatkin M. On treating the chromosome as the unit of selection. Genetics. 1972;72:157–168. doi: 10.1093/genetics/72.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Charlesworth B, Charlesworth D. Study of linkage disequilibrium in populations of Drosophila melanogaster. Genetics. 1973;73:351–359. doi: 10.1093/genetics/73.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Langley CH, Tobari YN, Kojima KI. Linkage disequilibrium in natural populations of Drosophila melanogaster. Genetics. 1974;78:921–936. doi: 10.1093/genetics/78.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hamon SC, et al. Evidence for consistent intragenic and intergenic interactions between SNP effects in the APOA1/C3/A4/A5 gene cluster. Hum. Hered. 2006;61:87–96. doi: 10.1159/000093384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hill WG, Robertson A. Linkage disequilibrium in finite populations. Theor. Appl. Genet. 1968;38:226–231. doi: 10.1007/BF01245622. [DOI] [PubMed] [Google Scholar]
- 54.Ohta T, Kimura M. Linkage disequilibrium at steady state determined by random genetic drift and recurrent mutation. Genetics. 1969;63:229–238. doi: 10.1093/genetics/63.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hudson RR. The sampling distribution of linkage disequilibrium under an infinite allele model without selection. Genetics. 1985;109:611–631. doi: 10.1093/genetics/109.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Slatkin M. Linkage disequilibrium in growing and stable populations. Genetics. 1994;137:331–336. doi: 10.1093/genetics/137.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hill WG, Robertson A. The effect of linkage on limits to artificial selection. Genet. Res. 1966;8:269–294. [PubMed] [Google Scholar]
- 58.McVean GAT, Charlesworth B. The effects of Hill–Robertson interference between weakly selected mutations on patterns of molecular evolution and variation. Genetics. 2000;155:929–944. doi: 10.1093/genetics/155.2.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Felsenstein J. The evolutionary advantage of recombination. Genetics. 1974;78:737–756. doi: 10.1093/genetics/78.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is the first paper to recognize the Hill–Robertson effect and its implications for the evolution of sex and recombination.
- 60.Keightley PD, Otto SP. Interference among deleterious mutations favours sex and recombination in finite populations. Nature. 2006;443:89–92. doi: 10.1038/nature05049. [DOI] [PubMed] [Google Scholar]
- 61.Barton NH. A general model for the evolution of recombination. Genet. Res. 1995;65:123–144. doi: 10.1017/s0016672300033140. [DOI] [PubMed] [Google Scholar]
- 62.Nei M, Li W. Linkage disequilibrium in subdivided populations. Genetics. 1973;75:213–219. doi: 10.1093/genetics/75.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mitton JB, Koehn RK, Prout T. Population genetics of marine pelecypods. III. Epistasis between functionally related isoenzymes of Mytilus edulis. Genetics. 1973;73:487–496. doi: 10.1093/genetics/73.3.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li WH. Stable linkage disequilibrium without epistasis in subdivided populations. Theor. Popul. Biol. 1974;6:173–183. doi: 10.1016/0040-5809(74)90022-7. [DOI] [PubMed] [Google Scholar]
- 65.Slatkin M. Gene flow and selection in a 2-locus system. Genetics. 1975;81:787–802. doi: 10.1093/genetics/81.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Noonan JP, et al. Sequencing and analysis of Neanderthal genomic DNA. Science. 2006;314:1113–1118. doi: 10.1126/science.1131412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schmegner C, Hoegel J, Vogel W, Assum G. Genetic variability in a genomic region with long-range linkage disequilibrium reveals traces of a bottleneck in the history of the European population. Hum. Genet. 2005;118:276–286. doi: 10.1007/s00439-005-0056-2. [DOI] [PubMed] [Google Scholar]
- 68.Zhang WH, et al. Impact of population structure, effective bottleneck time, and allele frequency on linkage disequilibrium maps. Proc. Natl. Acad. Sci. USA. 2004;101:18075–18080. doi: 10.1073/pnas.0408251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thornton K, Andolfatto P. Approximate Bayesian inference reveals evidence for a recent, severe bottleneck in a Netherlands population of Drosophila melanogaster. Genetics. 2006;172:1607–1619. doi: 10.1534/genetics.105.048223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weir BS, Cockerham CC. Group inbreeding with 2 linked loci. Genetics. 1969;63:711–742. doi: 10.1093/genetics/63.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Golding GB, Strobeck C. Linkage disequilibrium in a finite population that is partially selfing. Genetics. 1980;94:777–789. doi: 10.1093/genetics/94.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nordborg M, et al. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 2005;3:1289–1299. doi: 10.1371/journal.pbio.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim S, et al. Recombination and linkage disequilibrium in Arabidopsis thaliana. Nature Genet. 2007;39:1151–1155. doi: 10.1038/ng2115. [DOI] [PubMed] [Google Scholar]
- 74.Wiehe T, Mountain J, Parham P, Slatkin M. Distinguishing recombination and intragenic gene conversion by linkage disequilibrium patterns. Genet. Res. 2000;75:61–73. doi: 10.1017/s0016672399004036. [DOI] [PubMed] [Google Scholar]
- 75.Ardlie K, et al. Lower-than-expected linkage disequilibrium between tightly linked markers in humans suggests a role for gene conversion. Am. J. Hum. Genet. 2001;69:582–589. doi: 10.1086/323251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Padhukasahasram B, Marjoram P, Nordborg M. Estimating the rate of gene conversion on human chromosome 21. Am. J. Hum. Genet. 2004;75:386–397. doi: 10.1086/423451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gay J, Myers S, McVean G. Estimating meiotic gene conversion rates from population genetic data. Genetics. 2007;177:881–894. doi: 10.1534/genetics.107.078907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Frisse L, et al. Gene conversion and different population histories may explain the contrast between polymorphism and linkage disequilibrium levels. Am. J. Hum. Genet. 2001;69:831–843. doi: 10.1086/323612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Easton DF, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stacey SN, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nature Genet. 2007;39:865–869. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 81.Tomlinson I, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nature Genet. 2007;39:984–988. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 82.Zanke BW, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nature Genet. 2007;39:989–994. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 83.Diabetes Genetics Initiative, Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 84.Zeggini E, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Scott LJ, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sladek R, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 87.Gudbjartsson DF, et al. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature. 2007;448:353–357. doi: 10.1038/nature06007. [DOI] [PubMed] [Google Scholar]
- 88.McPherson R, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kohler K, Bickeboller H. Case–control association tests correcting for population stratification. Ann. Hum. Genet. 2006;70:98–115. doi: 10.1111/j.1529-8817.2005.00214.x. [DOI] [PubMed] [Google Scholar]
- 90.Pritchard JK, Donnelly P. Case–control studies of association in structured or admixed populations. Theor. Popul. Biol. 2001;60:227–237. doi: 10.1006/tpbi.2001.1543. [DOI] [PubMed] [Google Scholar]
- 91.Maynard Smith J, Haigh J. The hitch-hiking effect of a favourable gene. Genet. Res. 1974;23:23–35. [PubMed] [Google Scholar]
- 92.Kim Y, Stephan W. Detecting a local signature of genetic hitchhiking along a recombining chromosome. Genetics. 2002;160:765–777. doi: 10.1093/genetics/160.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Przeworski M. Estimating the time since the fixation of a beneficial allele. Genetics. 2003;164:1667–1676. doi: 10.1093/genetics/164.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nielsen R, et al. Genomic scans for selective sweeps using SNP data. Genome Res. 2005;15:1566–1575. doi: 10.1101/gr.4252305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Stephens JC, et al. Dating the origin of the CCR5–Delta32 AIDS-resistance allele by the coalescence of haplotypes. Am. J. Hum. Genet. 1998;62:1507–1515. doi: 10.1086/301867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Slatkin M, Bertorelle G. The use of intra-allelic variability for testing neutrality and estimating population growth rate. Genetics. 2001;158:865–874. doi: 10.1093/genetics/158.2.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hudson RR, Bailey K, Skarecky D, Kwiatowski J, Ayala FJ. Evidence for positive selection in the superoxide dismutase (Sod) region of Drosophila melanogaster. Genetics. 1994;136:1329–1340. doi: 10.1093/genetics/136.4.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Depaulis F, Veuille M. Neutrality tests based on the distribution of haplotypes under an infinite-site model. Mol. Biol. Evol. 1998;15:1788–1790. doi: 10.1093/oxfordjournals.molbev.a025905. [DOI] [PubMed] [Google Scholar]
- 99.Sabeti PC, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 100.Sabeti PC, et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper and reference 101 are among the first to show the feasibility of testing for selection on a genome-wide scale.
- 101.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:446–458. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Reich DE, Goldstein DB. In: Microsatellies: Evolution and Applications. Goldstein DB, Schlötterer C, editors. Oxford University Press; Oxford: 1999. pp. 129–138. [Google Scholar]
- 103.Kaplan NL, Lewis PO, Weir BS. Age of the ΔF508 cystic fibrosis mutation. Nature Genet. 1994;8:216. doi: 10.1038/ng1194-216a. [DOI] [PubMed] [Google Scholar]
- 104.Slatkin M, Rannala B. Estimating the age of alleles by use of intraallelic variability. Am. J. Hum. Genet. 1997;60:447–458. [PMC free article] [PubMed] [Google Scholar]
- 105.Guo SW, Xiong M. Estimating the age of mutant disease alleles based on linkage disequilibrium. Hum. Hered. 1997;47:315–337. doi: 10.1159/000154431. [DOI] [PubMed] [Google Scholar]
- 106.Slatkin M. A Bayesian method for jointly estimating allele age and selection intensity. Genet. Res. 2008;90:119–128. doi: 10.1017/S0016672307008944. [DOI] [PubMed] [Google Scholar]
- 107.Kaiser J. DNA sequencing: A plan to capture human diversity in 1000 Genomes. Science. 2008;319:395. doi: 10.1126/science.319.5862.395. [DOI] [PubMed] [Google Scholar]
- 108.Barker J. The human genome diversity project — `Peoples', `populations' and the cultural politics of identification. Cultural Studies. 2004;18:571–606. [Google Scholar]
- 109.Cunningham H. Colonial encounters in postcolonial contexts — patenting indigenous DNA and the Human Genome Diversity Project. Crit. Anthropol. 1998;18:205–233. [Google Scholar]
- 110.Kahn P. Genetic diversity project tries again. Science. 1994;266:720–722. doi: 10.1126/science.7973621. [DOI] [PubMed] [Google Scholar]
- 111.Wall JD. Detecting ancient admixture in humans using sequence polymorphism data. Genetics. 2000;154:1271–1279. doi: 10.1093/genetics/154.3.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Plagnol V, Wall JD. Possible ancestral structure in human populations. Plos Genet. 2006;2:972–979. doi: 10.1371/journal.pgen.0020105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Evans PD, Mekel-Bobrov N, Vallender EJ, Hudson RR, Lahn BT. Evidence that the adaptive allele of the brain size gene microcephalin introgressed into Homo sapiens from an archaic Homo lineage. Proc. Natl. Acad. Sci. USA. 2006;103:18178–18183. doi: 10.1073/pnas.0606966103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Begun DJ, et al. Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 2007;5:e310. doi: 10.1371/journal.pbio.0050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tishkoff SA, et al. Global patterns of linkage disequilibrium at the CD4 locus and modern human origins. Science. 1996;271:1380–1387. doi: 10.1126/science.271.5254.1380. [DOI] [PubMed] [Google Scholar]
- 116.Mountain JL, et al. SNPSTRs: empirically derived, rapidly typed, autosomal haplotypes for inference of population history and mutational processes. Genome Res. 2002;12:1766–1772. doi: 10.1101/gr.238602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tuzun E, et al. Fine-scale structural variation of the human genome. Nature Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 118.Conrad DF, Andrews TD, Carter NP, Hurles ME, Pritchard JK. A high-resolution survey of deletion polymorphism in the human genome. Nature Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- 119.Lewontin RC. The interaction of selection and linkage. I. General considerations; heterotic models. Genetics. 1964;49:49–67. doi: 10.1093/genetics/49.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bengtsson BO, Thomson G. Measuring the strength of associations between HLA antigens and diseases. Tissue Antigens. 1981;18:356–363. doi: 10.1111/j.1399-0039.1981.tb01404.x. [DOI] [PubMed] [Google Scholar]
- 121.Clark AG, et al. Haplotype structure and population genetic inferences from nucleotide-sequence variation in human lipoprotein lipase. Am. J. Hum. Genet. 1998;63:595–612. doi: 10.1086/301977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hill WG. Estimation of linkage disequilibrium in randomly mating populations. Heredity. 1974;33:229–239. doi: 10.1038/hdy.1974.89. [DOI] [PubMed] [Google Scholar]
- 123.Clark AG. Inference of haplotypes from PCR-amplified samples of diploid populations. Mol. Biol. Evol. 1990;7:111–122. doi: 10.1093/oxfordjournals.molbev.a040591. [DOI] [PubMed] [Google Scholar]
- 124.Eskin E, Halperin E, Karp RM. Efficient reconstruction of haplotype structure via perfect phylogeny. J. Bioinform. Comput. Biol. 2003;1:1–20. doi: 10.1142/s0219720003000174. [DOI] [PubMed] [Google Scholar]
- 125.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Marchini J, et al. A comparison of phasing algorithms for trios and unrelated individuals. Am. J. Hum. Genet. 2006;78:437–450. doi: 10.1086/500808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nature Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 128.Hästbacka J, et al. Linkage disequilibrium mapping in isolated founder populations: diastrophic dysplasia in Finland. Nature Genet. 1992;2:204–211. doi: 10.1038/ng1192-204. [DOI] [PubMed] [Google Scholar]
- 129.Hästbacka J, et al. The diastrophic dysplasia gene encodes a novel sulfate transporter — positional cloning by fine-structure linkage disequilibrium mapping. Cell. 1994;78:1073–1087. doi: 10.1016/0092-8674(94)90281-x. [DOI] [PubMed] [Google Scholar]
- 130.Jeffreys AJ, Kauppi L, Neumann R. Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nature Genet. 2001;29:217–222. doi: 10.1038/ng1001-217. [DOI] [PubMed] [Google Scholar]; This paper presents the first experimental demonstration of hot spots of recombination along with evidence of their association with haplotype blocks.