DNA sequencing technologies continue to make progress in increased throughput and quality, and decreased cost. As we transition from whole exome capture sequencing to whole genome sequencing (WGS), our ability to convert machine-generated variant calls, including single nucleotide variant (SNV) and insertion-deletion variants (indels), into human-interpretable knowledge has lagged far behind the ability to obtain enormous amounts of variants. To help narrow this gap, here we present WGSA (WGS annotator), a functional annotation pipeline for human genome sequencing studies, which is runnable out of the box on the Amazon Compute Cloud and is freely downloadable at https://sites.google.com/site/jpopgen/wgsa/.
Functional annotation is a key step in a WGS analysis. In one way, annotation helps the analyst filter to a subset of elements of particular interest (eg, cell type specific enhancers), in another way annotation helps the investigators to increase the power of identifying phenotype-associated loci (eg, association test using functional prediction score as a weight) and interpret potentially interesting findings. Currently, there are several popular gene model based annotation tools, including ANNOVAR [1], SnpEff [2] and the Ensembl Variant Effect Predictor (VEP) [3]. These can annotate a variety of protein coding and non-coding gene models from a range of species. It is well known among practitioners that different databases (eg, RefSeq [4] and Ensembl [5]) use different models for the same gene. Even when the same gene structure is implemented, predicted consequences of a given variant from different annotation tools may not be in agreement [6]. Therefore, it has been suggested to obtain annotation from tools across multiple databases for a more complete interpretation of the variants discovered in WGS [6]. Annotations of coding and non-coding variants include scores pertaining to functionality, conservation, population allele frequencies and disease-related annotations, that is, known disease-causing variants and disease-associated variants identified in genome-wide association analyses. Recent large-scale epigenomics projects provide rich datasets of cell-specific regulatory elements. Unfortunately, there are currently few tools available to integrate all those functional annotation resources and provide a convenient and efficient pipeline for annotating millions of variants discovered in a WGS study.
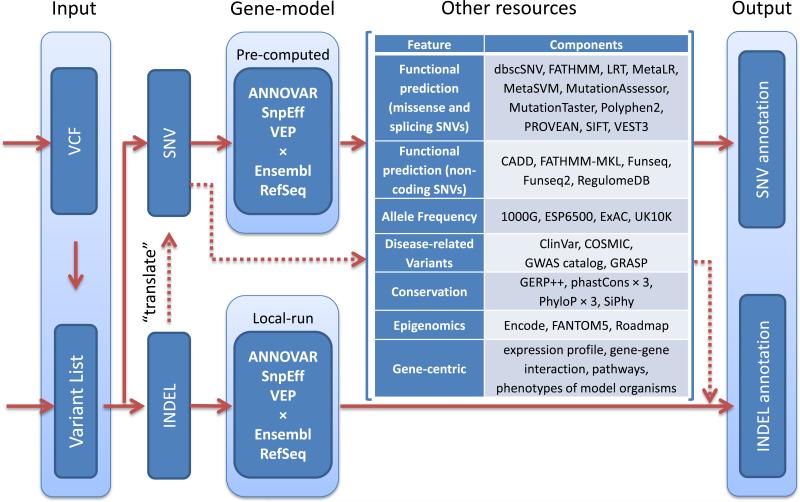
To facilitate the functional annotation step of WGS, we developed WGSA. Currently WGSA supports the annotation of SNVs and indels locally without remote database requests, allowing it to scale up for large WGS studies. The overview of the WGSA pipeline is presented in figure 1. The complete list of the resources (and their references) contained in WGSA can be found in online supplementary table S1. For gene-model based annotation, WGSA integrates the outputs from three annotation tools (ANNOVAR, SnpEff and VEP) versus two databases (RefSeq and Ensembl), and provides a summary of variant consequences from the six annotation results. To further speed up the process for large-scale WGS studies, we have precomputed annotations for all potential human SNVs (a total of 8,584,031,106) based on human reference hg19 non-N bases and use it as a local database. For SNV-centric resources, WGSA integrates five functional prediction scores, eight conservation scores, allele frequencies from four large-scale sequencing studies, variants in four disease-related databases, among others (figure 1 and online supplementary table S1). For regulatory region-centric resources, WGSA includes cell type specific transcription factor binding sites, DNAse I hypersensitivity regions and chromosome activity predictions from three epigenomics projects (figure 1 and online supplementary table S1). WGSA also contains rich functional annotations for non-synonymous SNVs and genes from our dbNSFP database [7, 8].
Figure 1.
Flowchart of the whole genome sequencing annotator (WGSA) annotation pipeline. Dotted lines show the “detour” of the indel annotation via the single nucleotide variant (SNV) annotation pipes.
Annotating the consequences of indels raises special challenges. In addition to the allele frequencies and consequences predicted by the three annotation tools (ANNOVAR, SnpEff and VEP), we take an approach to first “translate” an indel to local SNVs by incorporating their effect (insertion, deletion, replacement) on nearby flanking sequences, annotating those SNVs with other available resources, and then summarizing these results for the indel (figure 1, online supplementary figure S1 and details in online supplementary notes). Although this approach is a simplification of the potentially complicated impact of an indel, it will provide additional information on an indel regarding the focal region it resides in, such as whether it is a short tandem repeat mutation, whether it breaks a transcription factor binding site, local functional prediction scores, local conservation scores, etc.
To provide convenient access for a broad community, we have built an Amazon Machine Image (AMI) for running WGSA on the cloud via Amazon Web Services (AWS). To run WGSA, the user only needs to upload a variant list file (or a .vcf file) and a configuration file, containing a list of the resources to be included in the annotation. The annotation pipeline can be run with just two command line calls. We also provide WGSA as a downloadable version at https://sites.google.com/site/jpopgen/wgsa for bioinformaticians who prefer to build WGSA locally. It can be used as a foundation resource for customized annotation pipelines, as is the case for Baylor College of Medicine's Human Genome Sequencing Center annotation software, Cassandra (https://www.hgsc.bcm.edu/software/cassandra). The runtime of two experimental runs with 46.6 million variants is shown in online supplementary table S2. WGSA was written in Java so that most of its annotation modules (the whole SNV annotation and most of the indel annotation except VEP) can easily be run across different platforms. Detailed protocols for using WGSA in the cloud and building it locally can be found in online supplementary notes and at https://sites.google.com/site/jpopgen/wgsa/. As changes inevitably occur in the source databases and as new annotation resources emerge, WGSA will be updated in response. Users will be able to receive update notices and detailed instruction on updating steps (for the local version).
Supplementary Material
Acknowledgments
We gratefully acknowledge contributions from the researchers of the CHARGE sequencing project, especially Drs. L Adrienne Cupples and Fuli Yu and other members of the CHARGE Analysis & Bioinformatics Committee. We thank Mike Dahdouli for providing technical support. We thank Jin Yu for advising on AWS usage.
Funding This study was supported by the US National Institutes of Health (5RC2HL102419 and U54HG003273).
Footnotes
Contributors X.L., A.D.J., J.A.B., A.H.L., A.C., P.W., Z.H., R.J.K. and E.B. designed the study. X.L. collected the annotation resources and developed the tool. S.W. tested the pipeline. B.P. provided tools for retrieving the RegulomeDB data set. E.B. and R.G. supervised the study. X.L., S.W. and E.B. wrote the draft manuscript and all authors provided critical edits.
Competing interests The authors declare that they have no competing interests.
Reference
- 1.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–70. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pruitt KD, Brown GR, Hiatt SM, Thibaud-Nissen F, Astashyn A, Ermolaeva O, Farrell CM, Hart J, Landrum MJ, McGarvey KM, Murphy MR, O'Leary NA, Pujar S, Rajput B, Rangwala SH, Riddick LD, Shkeda A, Sun H, Tamez P, Tully RE, Wallin C, Webb D, Weber J, Wu W, DiCuccio M, Kitts P, Maglott DR, Murphy TD, Ostell JM. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res. 2014;42:D756–63. doi: 10.1093/nar/gkt1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cunningham F, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, Gil L, Girón CG, Gordon L, Hourlier T, Hunt SE, Janacek SH, Johnson N, Juettemann T, Kähäri AK, Keenan S, Martin FJ, Maurel T, McLaren W, Murphy DN, Nag R, Overduin B, Parker A, Patricio M, Perry E, Pignatelli M, Riat HS, Sheppard D, Taylor K, Thormann A, Vullo A, Wilder SP, Zadissa A, Aken BL, Birney E, Harrow J, Kinsella R, Muffato M, Ruffier M, Searle SMJ, Spudich G, Trevanion SJ, Yates A, Zerbino DR, Flicek P. Ensembl 2015. Nucleic Acids Res. 2015;43:D662–9. doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCarthy DJ, Humburg P, Kanapin A, Rivas MA, Gaulton K, The WGS500 Consortium. Cazier J-B, Donnelly P. Choice of transcripts and software has a large effect on variant annotation. Genome Med. 2014;6:26. doi: 10.1186/gm543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu X, Jian X, Boerwinkle E. dbNSFP: A lightweight database of human nonsynonymous SNPs and their functional predictions. Hum Mutat. 2011;32:894–9. doi: 10.1002/humu.21517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu X, Jian X, Boerwinkle E. dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum Mutat. 2013;34:E2393–402. doi: 10.1002/humu.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.