Abstract
The plant hormone auxin regulates numerous aspects of plant growth and development. Early auxin response genes mediate its genomic effects on plant growth and development. Discovered in 1987, SMALL AUXIN UP RNAs (SAURs) are the largest family of early auxin response genes. SAUR functions have remained elusive, however, presumably due to extensive genetic redundancy. However, recent molecular, genetic, biochemical, and genomic studies have implicated SAURs in the regulation of a wide range of cellular, physiological, and developmental processes. Recently, crucial mechanistic insight into SAUR function was provided by the demonstration that SAURs inhibit PP2C.D phosphatases to activate plasma membrane (PM) H+-ATPases and promote cell expansion. In addition to auxin, several other hormones and environmental factors also regulate SAUR gene expression. We propose that SAURs are key effector outputs of hormonal and environmental signals that regulate plant growth and development.
Keywords: auxin, SAURs, plant growth and development, cell expansion, acid growth, PP2C.D phosphatases, PM H+-ATPases
INTRODUCTION
Auxin, derived from the Greek word αυξειν (auxein, meaning to grow or increase), was the first identified plant hormone. Since its discovery in the 1930s, auxin has fascinated plant biologists, who have sought to understand its biosynthesis, metabolism, transport, perception, signaling, and responses. Auxin influences nearly all aspects of plant growth and development through regulating cell division, expansion, differentiation, and patterning. In the nucleus, a co-receptor complex that is composed of TRANSPORT INHIBITOR RESPONSE1/AUXIN SIGNALING F-BOX PROTEINS (TIR1/AFBs) and AUXIN/INDOLE ACETIC ACID (AUX/IAA) transcriptional repressors perceives auxin to regulate gene transcription (reviewed in Wang and Estelle, 2014). These auxin-regulated genes then control downstream auxin responses.
Early or primary auxin response genes are transcriptionally induced by auxin within minutes, and this induction does not require de novo protein synthesis (Abel et al., 1994). Most early auxin response genes are classified into three families: AUX/IAAs, GRETCHEN HAGEN3s (GH3s), and SAURs (reviewed in Hagen and Guilfoyle, 2002). SAURs were originally identified in a differential hybridization screen for genes that were rapidly induced by auxin in elongating soybean hypocotyl sections (McClure and Guilfoyle, 1987). Since then, SAURs have been identified in diverse plant species, including mung bean (Yamamoto et al., 1992), pea (Guilfoyle et al., 1993), tomato (Zurek et al., 1994), Arabidopsis (Gil et al., 1994), apple (Watillon et al., 1998), radish (Anai et al., 1998), maize (Yang and Poovaiah, 2000), rice (Jain et al., 2006), moss (Rensing et al., 2008), sorghum (Wang et al., 2010), potato (Wu et al., 2012), cotton (Yang et al., 2012), litchi (Kuang et al., 2012), tobacco (Wu et al., 2012), pepper (Wu et al., 2012), petunia (Wu et al., 2012), peach (Tatsuki et al., 2013), and poplar (Wang et al., 2014a). While substantial progress has been made toward understanding the functions of both Aux/IAA and GH3 proteins in auxin responses (Tiwari et al., 2001; Staswick et al., 2002; Tiwari et al., 2004; Calderón Villalobos et al., 2012), functional studies on SAURs have lagged behind. Nearly 30 years after their discovery, we have only just begun to unlock the secrets of the SAURs. In this review, we describe recent advances that implicate SAURs in regulating a wide range of cellular, physiological, and developmental processes.
SAURs, the Largest Family of Early Auxin Response Genes
Among early auxin response genes, the SAUR gene family is the most numerous. Genomic bioinformatic analyses have revealed that there are 81 SAURs (including two pseudogenes) in Arabidopsis (Hagen and Guilfoyle, 2002), 58 SAURs (including two pseudogenes) in rice (Jain et al., 2006), 18 SAURs in moss (Rensing et al., 2008), 71 SAURs in sorghum (Wang et al., 2010), 134 SAURs in potato (Wu et al., 2012), 99 SAURs in tomato (Wu et al., 2012), and 79 SAURs in maize (Chen et al., 2014). Typically, SAUR genes are not randomly distributed in the genome, as many of them are found in tandem arrays of extremely highly related genes in soybean (McClure et al., 1989), Arabidopsis (Hagen and Guilfoyle, 2002), rice (Jain et al., 2006), tomato (Wu et al., 2012), and maize (Chen et al., 2014). Tandem and segmental duplication events likely contributed to the expansion of the SAUR gene family (Wu et al., 2012; Chen et al., 2014). The genomic structures of SAUR genes show similar features. The vast majority of SAUR genes lack introns. Many also contain one or more auxin response elements (AuxREs) within their promoter region, and possess a downstream destabilizing (DST) element in the 3’ untranslated region (UTR) (Hagen and Guilfoyle, 2002; Jain et al., 2006; Wu et al., 2012; Chen et al., 2014). The DST consists of three conserved elements separated by non-conserved bases of variable length (GGA(N)xATAGAT(N)xGTA) (McClure et al., 1989; Newman et al., 1993; Sullivan and Green, 1996). This sequence is found in 30 of the 79 Arabidopsis SAUR genes (Supplementary Table 1). For at least some SAUR transcripts, the DST element confers instability (Sullivan and Green, 1996). However, the functional significance of these DST elements is uncertain, as Arabidopsis mutants defective in DST-mediated mRNA degradation exhibit no apparent phenotype (Johnson et al., 2000).
SAUR genes encode small proteins that are unique to plants and contain no obvious characterized motifs indicative of a biochemical function. The predicted molecular weights of Arabidopsis SAUR proteins range from 9.3 to 21.4 kDa. SAUR proteins have been predicted to reside in the nucleus, cytosol, mitochondrion, chloroplast, and on the plasma membrane (Wu et al., 2012; Chen et al., 2014). Studies employing SAUR fusion proteins have provided evidence for SAUR localization to the nucleus (ZmSAUR2, Knauss et al., 2003; SAUR32, Park et al., 2007; SAUR36, Narsai et al., 2011), cytosol (OsSAUR39, Kant et al., 2009; SAUR55, Narsai et al., 2011; SAUR41, Kong et al., 2013; SAUR40 and SAUR71, Qiu et al., 2013), and plasma membrane (SAUR63, Chae et al., 2012; SAUR19, Spartz et al., 2012). While such findings should be interpreted with caution, as most of these studies were conducted with SAUR overexpression constructs and sometimes in heterologous systems, the findings suggest that different SAURs may localize to distinct cellular compartments.
Protein multiple sequence alignment revealed that SAURs from different plant species contain a central domain specific to SAUR proteins (CDD superfamily cI03633, Marchler-Bauer et al., 2013). This ∼ 60 amino acid domain, referred to here as the SAUR domain, is highly conserved (Figure 1), suggesting that this domain is essential for SAUR protein function. The SAUR domain is dominated by hydrophobic amino acids and also contains short, highly conserved, charged patches and a nearly invariant cysteine residue. Several proline and aromatic amino acids are also exceptionally highly conserved. Outside of the SAUR domain, SAUR proteins have variable length N- and C-terminal extensions, which exhibit much lower degrees of conservation (Supplementary Figure 1).
Figure 1. The SAUR domain.
Multiple sequence alignment of the SAUR domain of 247 SAUR proteins from Arabidopsis thaliana, Oryza sativa, Populus trichocarpa, and Physcomitrella patens was performed using Clustal Omega. The consensus residue(s) at each position are color coded. Dotted lines indicate the positions of short, nonconserved insertions present in a few SAUR family members that were removed from the alignment.
SAUR expression is regulated at multiple levels. Early studies revealed that several SAUR genes are transcriptionally induced by auxin in mung bean, pea, Arabidopsis, radish, and maize (reviewed in Hagen and Guilfoyle, 2002). More recent genomic methods of assessing gene expression have greatly expanded the number of known auxin-induced SAUR genes in Arabidopsis (Tian et al., 2002; Zhao et al., 2003; Goda et al., 2004; Nemhauser et al., 2004; Redman et al., 2004; Okushima et al., 2005; Chapman et al., 2012). However, this list likely remains incomplete as many SAUR genes are not represented on the Arabidopsis ATH1 gene chip and several of the SAUR probe sets that are on this chip are not specific to individual SAUR genes. With these caveats, it is estimated that about one-half of the Arabidopsis SAUR genes are upregulated to some extent by auxin treatment (Supplementary Figure 2). Interestingly, a small number of SAUR genes appear to be repressed by auxin (Supplementary Figure 2). In general, auxin-induced SAURs tend to be most highly expressed in shoots, while many auxin-repressed and non-responsive SAURs are preferentially expressed in roots (Paponov et al., 2008).
A second layer of regulation of SAUR expression involves mRNA stability. At least some SAUR transcripts are highly unstable (McClure and Guilfoyle, 1989; Franco et al., 1990). The DST element in the 3’-UTR of SAUR genes functions as an mRNA instability determinant, responsible for the rapid turnover of SAUR mRNAs (Newman et al., 1993). In the cases examined, auxin does not appear to regulate SAUR transcript stability (Gil and Green, 1996). Interestingly, some SAUR transcripts have also been shown to be specifically polyadenylated by PAPS1, one of the four canonical nuclear poly(A) polymerases of Arabidopsis (Vi et al., 2013). This appears to be functionally significant, as overexpression of a GFP-SAUR19 transgene containing a heterologous 3’-UTR could suppress a subset of paps1 mutant phenotypes.
In addition to regulation at the transcriptional and posttranscriptional levels, SAUR expression is also subject to posttranslational control. Several SAUR proteins have been found to be highly unstable. The first such report was with the maize SAUR2 (ZmSAUR2) protein (Knauss et al., 2003). 35S-methionine pulse-chase labeling experiments revealed that ZmSAUR2 is a short-lived protein with a half-life of about 7 minutes. More recently, the Arabidopsis SAUR19 and SAUR63 proteins were found to be highly unstable. Treatment with the 26S proteasome inhibitor MG132 causes accumulation of SAUR19 and SAUR63, suggesting the involvement of the ubiquitin-26S proteasome pathway in regulating SAUR degradation (Chae et al., 2012; Spartz et al., 2012). However, neither the mechanism nor regulation of SAUR ubiquitylation has been elucidated, although in the case of SAUR63, the degradation rate was reported to be faster under dim light than bright light (Chae et al., 2012). Interestingly, the SAUR19 and SAUR63 proteins are dramatically stabilized when expressed as fusions with GFP, GUS, or even some small epitope tags (StrepII) (Spartz et al, 2012; Chae et al., 2012).
Roles of SAURs in Plant Growth and Development
Since their discovery, SAURs have puzzled plant biologists. All plant genomes appear to contain large SAUR gene families. Because of likely genetic redundancy and the difficulties in generating higher order saur mutants due to the tight linkage of tandemly arrayed SAUR genes, it is challenging to employ loss-of-function genetic approaches to elucidate SAUR functions in plant growth and development. To circumvent this problem, gain-of-function genetic approaches that overexpress SAURs or stabilized SAUR fusion proteins have been utilized. Such studies have implicated SAURs in a wide range of cellular, physiological, and developmental processes involving hormonal and environmental control of plant growth and development. While the findings of overexpression studies should always be interpreted with caution, many of these findings have been supported by RNA silencing strategies that target multiple SAUR subfamily members, as well as additional biochemical and genetic findings. Below we review the evidence for SAUR-mediated regulation of several aspects of plant growth and development.
Cell Expansion
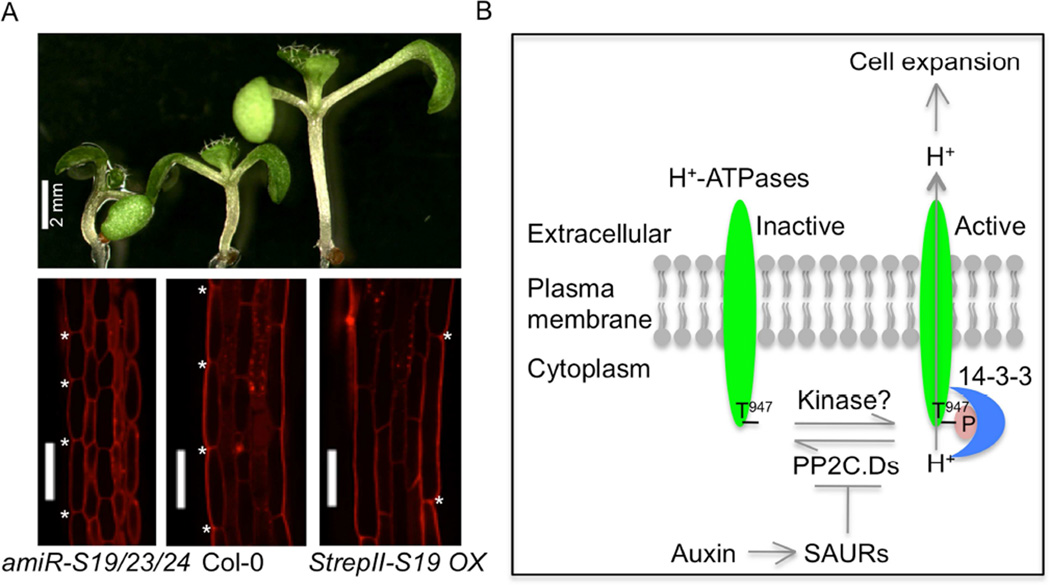
Cell expansion is a fundamental process essential for plant growth and development (reviewed in Braidwood et al., 2013). Hypocotyl growth is mainly caused by cell expansion, making it an excellent study system (Gendreau et al., 1997). Auxin plays a major role in regulating hypocotyl growth (Gray et al. 1998; Zhao et al. 2001). Many SAURs are highly expressed in elongating hypocotyls (McClure and Guilfoyle, 1987; Chae et al., 2012; Spartz et al., 2012; Stamm and Kumar, 2013), and it has long been hypothesized that SAURs may be involved in auxin-regulated cell expansion. Experimental results supporting a positive role for SAURs in cell expansion were obtained from several recent studies in Arabidopsis. Overexpression of SAUR36 (Stamm and Kumar, 2013), SAUR41 (Kong et al., 2013), or stabilized fusion proteins of SAUR19 (Spartz et al., 2012) or SAUR63 (Chae et al., 2012) promotes hypocotyl elongation as a result of increased cell expansion. By contrast, seedlings expressing artificial microRNAs (amiRNAs) targeting multiple members of the SAUR19 (Spartz et al., 2012) or SAUR63 (Chae et al., 2012) subfamilies have slightly shorter hypocotyls with smaller epidermal cells (Figure 2A). These findings suggest that these SAURs positively regulate cell expansion to promote hypocotyl growth. In addition to hypocotyl phenotypes, SAUR63 overexpression and amiRNA knockdown plants also exhibit longer and shorter stamen filaments, respectively. Likewise, SAUR19 and additional SAURs have also been implicated in leaf cell expansion, which we review below.
Figure 2. SAURs activate PM H+-ATPases via inhibiting PP2C.D phosphatases to promote cell expansion.
(A) Seven-day-old light-grown Arabidopsis seedlings. Seedlings were stained in 10 µg /ml propidium iodide, and hypocotyl epidermal cells were observed under a Nikon A1 spectral confocal microscope. Asterisks (*) indicate the intercellular space of two adjacent epidermal cells. Scale bar = 100 µm.
(B) A model for SAUR-regulated cell expansion. Adapted from Spartz et al. (2014).
What is the mechanism underlying SAUR-mediated cell expansion? Spartz et al. (2014) recently proposed that SAURs promote cell expansion via an acid growth mechanism. The acid growth theory was proposed in the 1970s to explain auxin-mediated cell expansion (Rayle and Cleland, 1970, 1980, 1992; Hager, 2003). According to this theory, auxin stimulates susceptible cells to excrete protons, lowering apoplastic pH and elevating membrane potential. Plasma membrane (PM) H+-ATPases are responsible for pumping protons into the apoplast. The resulting reduction in apoplastic pH activates cell wall-loosening enzymes such as expansins and hyperpolarizes the plasma membrane, thus increasing cell wall extensibility and promoting solute and water uptake to generate the turgor increase required for cell expansion. Until recently, however, the acid growth theory has lacked strong genetic support, and the underlying molecular mechanism(s) by which auxin activates PM H+-ATPase activity were uncertain.
The Arabidopsis genome encodes 11 PM H+-ATPases called AHAs (ARABIDOPSIS H+-ATPases), with AHA1 and AHA2 being the most highly expressed isoforms (Baxter et al., 2003; Haruta et al., 2010). Takahashi et al. (2012) provided key insight into auxin-mediated PM H+-ATPase activation with the demonstration that auxin rapidly induces phosphorylation of the penultimate threonine residue of the C-terminal autoinhibitory domain of AHAs (Thr947 in AHA2). Phosphorylation of this residue coincides with 14-3-3 protein binding and PM H+-ATPase activation (Fuglsang et al., 1999; Kinoshita and Shimazaki, 1999). SAUR19 activates PM H+-ATPases by promoting AHA Thr947 phosphorylation (Spartz et al., 2014). This phosphorylation increase results in elevated PM H+-ATPase activity. Consistent with these findings, plants overexpressing stabilized SAUR19 fusion proteins exhibit remarkable phenotypic similarity to open stomata2 (ost2) mutants. ost2 is a gain-of-function allele of AHA1 that results in a constitutively active enzyme (Merlot et al., 2007). Like ost2 mutants, seedlings expressing SAUR19 fusion proteins exhibit increased elongation growth, reduced aploplastic pH, drought hypersensitivity due to improper guard cell regulation, and constitutive expression of pathogen defense genes (Spartz et al., 2014). These phenotypes are also elicited by fusicoccin, a fungal wilting toxin that specifically activates PM H+-ATPases (Marre, 1979; Singh and Roberts, 2004).
SAUR19, as well as several additional SAURs, interact with several members of the 9-member PP2C.D family of protein phosphatases (Schweighofer et al., 2004; Spartz et al., 2014). Enzymatic assays revealed that SAUR binding inhibits phosphatase activity. Spartz et al. (2014) employed gain- and loss-of-function genetic studies to demonstrate that SAUR19 and PP2C.D phosphatases function antagonistically to regulate cell expansion by controlling AHA Thr947 phosphorylation and activity. For example, whereas seedlings overexpressing GFP-SAUR19 exhibit increased cell expansion, Thr947 phosphorylation, and PM H+-ATPase activity, PP2C.D1 overexpression confers reductions in all three processes. Importantly, both SAUR19-mediated growth promotion and PP2C.D1-mediated growth repression occur in a PM H+-ATPase-dependent manner.
Consistent with these gain-of-function findings, seedlings expressing a PP2C.D2/5/7/8/9 amiRNA knockdown construct partially phenocopy GFP-SAUR19 overexpression seedlings. Furthermore, PP2C.D1 interacts with AHA2 on the plasma membrane and can catalyze AHA Thr947 dephosphorylation both in vitro and when heterologously expressed in yeast. Based on these results, the authors proposed that auxin induction of SAUR expression results in repression of PP2C.D-mediated dephosphorylation of PM H+-ATPase Thr947. Consequently, PM H+-ATPase equilibrium is shifted to the phosphorylated, active state, resulting in increased apoplastic acidification and cell expansion (Figure 2B).
Curiously, in contrast to the above findings, some SAURs have been suggested to negatively regulate cell expansion. Transgenic seedlings overexpressing SAUR32 (also known as AAM1, ABOLISHED APICAL HOOK MAINTENANCE1) exhibit reduced hypocotyl elongation when grown either in the dark or under red light (Park et al., 2007). Likewise, saur36 T-DNA loss-of-function plants have larger leaves containing larger epidermal cells, suggesting that SAUR36 may negatively regulate cell expansion to inhibit leaf growth (Hou et al., 2013). However, Stamm and Kumar (2013) reported that SAUR36 overexpressing seedlings display a long hypocotyl phenotype. At this point, it is unclear whether these disparate findings with SAUR36 in leaves and hypocotyls are due to organ-specific regulation or some other factor. Future studies are required to elucidate the potential mechanisms of SAUR32 and SAUR36 inhibition of cell expansion. Nonetheless, it is tempting to speculate that different SAUR family members might act in an antagonistic fashion, a phenomenon frequently observed in other gene families, such as AUXIN RESPONSE FACTORS (ARFs) and ARABIDOPSIS RESPONSE REGULATORS (ARRs) (reviewed in Guilfoyle and Hagen, 2007; Argueso et al., 2010).
Shade Avoidance Responses
Under shade (a low red/far-red ratio) conditions, many plants exhibit increased elongation growth of stems and petioles, hyponastic leaves, reduced branching, increased leaf senescence, and early flowering. These growth responses are known as shade avoidance responses. In Arabidopsis seedlings, the most dramatic shade avoidance response is hypocotyl elongation (reviewed in Casal, 2013; de Wit et al., 2014). Mutant seedlings defective in auxin biosynthesis, transport, signaling, and response exhibit reduced hypocotyl growth in response to shade, indicating that auxin is an important regulator of shade avoidance responses. In response to shade, the PHYTOCHROME INTERACTING FACTOR (PIF4, PIF5, and PIF7) transcription factors activate expression of auxin biosynthetic genes to upregulate IAA production.
A link between SAURs and shade avoidance response was suggested by Roig-Villanova et al. (2007). In Arabidopsis, PHYTOCHROME RAPIDLY REGULATED1 (PAR1) and PAR2 are atypical basic helix–loop–helix (bHLH) proteins, which lack the basic domain required for DNA-binding. Expression of PAR1 and PAR2 is upregulated by shade in seedlings, inhibiting shade avoidance responses. PAR1 and PAR2 repress the expression of SAUR15 (also known as SAUR-AC1), SAUR67 (misannotated as SAUR68), and several additional SAURs. PAR1 and PAR2 lack a DNA binding domain. Instead, PAR1 interacts with PIF4 to inhibit PIF4 DNA-binding activity (Hao et al., 2012). Many SAURs, including SAUR15, are potential direct targets of PIF4 (Oh et al., 2012). Therefore, PAR1-mediated transcriptional repression of SAUR15 likely involves PAR1 inhibition of PIF4-mediated transcriptional activation. Like PAR1 and PAR2, SAUR15 and SAUR67 are also induced by shade. However, this induction is transient, suggesting that PAR1 and PAR2 repression of SAUR expression may serve as a feedback mechanism for attenuating shade-induced SAUR expression.
Additional evidence supporting a role of SAURs in shade-stimulated elongation growth was provided by Hornitschek et al. (2012). PIF4 and PIF5 function redundantly to promote shade-induced hypocotyl growth. The authors used chromatin immunoprecipitation sequencing (ChIP-seq) to identify the genomic binding sites for PIF5 in plants exposed to simulated shade for 2 hours. The potential direct PIF5 targets include the SAUR19 subfamily genes (SAUR19, 20, 21, 22, 23, and 24) as well as additional SAUR genes. In response to shade, expression of SAUR19, SAUR21, SAUR23, and SAUR24 is rapidly induced in elongating hypocotyls (Spartz et al., 2012). Upregulation of SAUR gene expression by shade could be achieved both directly by PIF4 and PIF5-mediated gene activation and indirectly by increased auxin levels controlled by PIF4, PIF5, and PIF7. Presumably, these increases in SAUR expression promote the cell expansion underlying the increased hypocotyl and petiole growth characteristic of shade avoidance growth. However, genetic studies are clearly needed to support this hypothesis.
High Temperature-induced Growth
Similar to shade, high temperature also induces rapid elongation growth. Arabidopsis seedlings exhibit elongated hypocotyls and petioles in response to high temperature. High temperature promotes auxin biosynthesis in Arabidopsis seedlings (Gray et al., 1998). This upregulation of auxin biosynthesis is controlled by PIF4, the primary PIF family member that controls high temperature-mediated growth (Koini et al., 2009). PIF4 directly regulates the expression of auxin biosynthesis genes TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS1 (TAA1) and CYP79B2 (Franklin et al., 2011) and YUCCA8 (YUC8) (Sun et al., 2012) to increase auxin production in response to high temperature.
Recent findings indicate that SAURs contribute to PIF4-mediated elongation growth at high temperature. SAURs in the SAUR19 subfamily positively regulate cell expansion to promote hypocotyl growth (Spartz et al., 2012; Spartz et al., 2014). High temperature rapidly induces the expression of these genes in hypocotyls (Franklin et al., 2011). However, pif4 null mutants exhibit dramatically reduced expression of these genes at high temperature, indicating that high temperature-induced SAUR gene expression is PIF4-dependent. Furthermore, SAUR19 overexpression rescues the hypocotyl elongation defects of pif4 mutants at high temperature. These results suggest that the SAUR19 subfamily genes function downstream of PIF4 to regulate hypocotyl growth in response to high temperature (Franklin et al., 2011). PIF4-dependent SAUR gene expression could be explained by two distinct mechanisms. As described above, PIF4 upregulates auxin biosynthesis in response to elevated temperatures. Since the SAUR19 subfamily genes are auxin inducible (Spartz et al., 2012), PIF4-mediated increases in auxin levels would induce the expression of these genes. Additionally, PIF4 may directly regulate the expression of the SAUR19 subfamily genes. Oh et al. (2012) provided experimental evidence to support this hypothesis, as SAUR19, 20, 21, 22, 23, and 24 were all identified as high-confidence PIF4 target genes by ChIP-seq analysis. Although targeted follow up experiments are needed to confirm that PIF4 directly binds to the promoters of these genes, it seems likely that both mechanisms contribute to PIF4-dependent SAUR gene expression to promote elongation growth at high temperature.
Tropic growth
SAUR-mediated changes in cell expansion have also been implicated in tropic growth responses – directional growth in response to environmental stimuli such as light and gravity. Light and gravity signals cause an asymmetric auxin distribution, resulting in differential growth (bending) of shoots and roots during phototropic and gravitropic responses. Shoots exhibit positive phototropism, growing toward the light source. Auxin accumulates on the shaded side of plant stems, promoting the differential cell expansion that results in stem bending toward the light. In response to gravity, shoots exhibit negative gravitropism (grow upward), while roots exhibit positive gravitropism (grow downward). A higher concentration of auxin on the lower side of shoots causes increased growth, while a higher concentration of auxin on the lower side of roots inhibits growth (reviewed in Goyal et al., 2013; Vandenbrink et al., 2014).
Several of the first studies on SAUR genes reported increases in SAUR expression within the elongating cells of tropically stimulated stems. Using a tissue printing technique, McClure and Guilfoyle (1989) examined the distribution of SAUR transcripts in the elongating region of excised soybean hypocotyls during gravitropism. Upon gravity stimulation, SAUR transcripts changed from a symmetric distribution to an asymmetric distribution, accumulating on the lower side of hypocotyls. Likewise, increased GUS activity was observed on the lower side of gravistimulated tobacco stems expressing a pSAUR10A::GUS reporter (Li et al., 1991), and RNA in situ hybridization detected more abundant soybean SAUR transcripts in the epidermis and cortex on the lower side of hypocotyls during gravitropic response (Gee et al., 1991). In addition to gravitropism, asymmetric pSAUR10A::GUS activity was also observed in tobacco stems during phototropic growth, with the shaded side exhibiting increased GUS staining (Li et al., 1991). More recent transcriptomic studies have revealed that many SAUR genes are preferentially expressed on the lower, elongating sides of gravistimulated rice shoots and Arabidopsis stems (Hu et al., 2013; Taniguchi et al., 2014). In fact, of the 30 genes exhibiting differential expression between the upper and lower flanks of Arabidopsis stems following a 30-min gravistimulation, 14 were SAUR genes. Curiously, all 14 of these genes belong to the SAUR19 and SAUR63 clades of the phylogeny (Supplementary Figure 2).
Overexpression studies have provided some functional support for the involvement of SAURs in tropic growth responses. Arabidopsis seedlings expressing a GFP-SAUR19 transgene driven by the cauliflower mosaic virus 35S promoter exhibit a dramatic delay in phototropic bending and a wavy root growth habit (Spartz et al., 2012). SAUR41 is specifically expressed in the endodermis of the root elongation zone on the upper side of gravistimulated roots, and ectopic expression of SAUR41 resulted in altered gravitropic root growth (Kong et al., 2013). Together with the above gene expression studies correlating SAUR expression with differential growth of the two sides of tropically stimulated roots and shoots, it seems likely that asymmetric SAUR expression may promote the cell expansion underlying tropic growth. Disrupting the normal expression pattern by expressing SAUR genes from heterologous promoters results in the loss of the ability to control differential cell expansion. Interestingly, a recent modeling study (Hohm et al., 2014) found that apoplastic acidification is an important component of phototropic bending, suggesting that SAUR-mediated activation of PM H+-ATPases may drive the differential cell expansion associated with tropic growth.
Apical Hook Development
Like tropic growth, the development, maintenance, and opening of apical hooks involves the regulated formation of auxin gradients to control differential cell expansion (reviewed in Abbas et al., 2013). Dicotyledonous seedlings grown in the dark develop an apical hook structure caused by differential cell expansion. The apical hook protects the shoot apical meristem from damage when seedlings grow through soil. Upon reaching the soil surface and perceiving light, hook opening is triggered.
Gene expression and gain-of-function studies in Arabidopsis have suggested that SAURs are involved in apical hook development. The auxin regulated reporter DR5::GUS is preferentially expressed on the inner side of the apical hook (Abbas et al., 2013). Although SAUR32 expression does not appear to be auxin-inducible, SAUR32 expression in apical hooks closely resembles that of DR5::GUS (Park et al., 2007). While the apical hooks of a saur32 T-DNA null mutant appear wild-type, SAUR32 overexpression from the 35S promoter confers a hookless phenotype due to a defect in apical hook maintenance. Interestingly, SAUR32 overexpression seedlings also have short hypocotyls, suggesting that SAUR32 negatively regulates cell expansion. Together, these findings suggest that during apical hook development, SAUR32 might inhibit cell expansion of the inner side of the apical hook, which is important for hook maintenance. Paradoxically, however, overexpression of SAURs that promote hypocotyl cell expansion such as SAUR19 and SAUR36 also confers a hookless phenotype (Spartz et al., 2012; Stamm and Kumar, 2013). Although additional studies are clearly required, the above findings suggest that proper expression of both growth promoting and growth inhibiting SAUR genes is required to achieve the differential cell expansion necessary to achieve normal apical hook development. The challenge is to understand the molecular mechanisms behind SAUR-regulated differential growth in the apical hook. That said, initial genetic studies of the recently proposed SAUR-PP2C.D mediated regulation of PM H+-ATPase activity support a role for this regulatory module in apical hook development. When grown in the dark, pp2c.d1 and pp2c.d2 mutants, as well as the constitutively active ost2 PM H+-ATPase mutant, exhibit reduced apical hook angles and open cotyledon phenotypes (Spartz et al., 2014).
Leaf Growth and Senescence
Auxin plays a critical role in leaf growth and development, regulating leaf initiation, lamina formation, vein development, and leaf shape and size (reviewed in Scarpella et al., 2010; Byrne, 2012). Several lines of evidence have suggested that SAURs regulate leaf growth through controlling cell expansion or division, contributing to auxin-regulated leaf growth and development.
Spartz et al. (2012) found that the SAUR19 subfamily genes positively regulate leaf growth. Plants expressing an artificial miRNA targeting SAUR19, 23, and 24 display reduced leaf area, while plants expressing stabilized SAUR19 fusion proteins exhibit larger leaves than wild-type. These changes in leaf size appear to be entirely due to altered cell size, suggesting that the SAUR19 subfamily genes positively regulate cell expansion to promote leaf growth. Strikingly, SAUR19 appears quite unique among leaf growth promoting genes. For example, a recent study examining 13 genes that confer increased leaf size when either overexpressed or mutated revealed that SAUR19 and EXPANSIN10 were the only ones that specifically affected cell expansion rather than cell division (Vanhaeren et al., 2014). Furthermore, when tested in pairwise combinations with the other growth promoting mutations/transgenes, SAUR19 overexpression resulted in synergistic increases in leaf size. These findings suggest that the SAUR19 subfamily genes are attractive targets for future genetic engineering efforts aimed at enhancing plant biomass.
Unlike the SAUR19 subfamily genes, a few SAURs have been implicated as negative regulators of leaf growth. SAUR76 is highly expressed in roots, but only weakly expressed in leaves. Interestingly, while auxin treatment resulted in a dramatic upregulation in roots, no increase in SAUR76 expression was observed in leaves (Markakis et al., 2013). When overexpressed from the 35S promoter, SAUR76 conferred a reduction in leaf size. This effect appears to be the result of a reduction in cell number rather than cell size, suggesting that SAUR76 might negatively regulate cell division to inhibit leaf growth (Markakis et al., 2013). Similarly, SAUR36 also appears to negatively regulate leaf growth, as saur36 null mutants exhibit enlarged leaves (Hou et al., 2013). However, unlike SAUR76 overexpression, saur36 mutants exhibit an increase in leaf epidermal cell size, suggesting that SAUR36 negatively regulates leaf cell expansion rather than cell division.
In addition to regulating leaf growth and development, auxin has also been implicated in leaf senescence, a process eventually leading to leaf death. However, the precise functions of auxin in leaf senescence are unclear, as contradictory results have been reported as to whether auxin negatively or positively regulates leaf senescence (reviewed in Jibran et al., 2013; Khan et al., 2014). Recent studies of auxin induced SAUR genes in Arabidopsis and rice suggest that auxin may promote leaf senescence through the expression of SAURs. Arabidopsis SAUR36, also known as SAG201 (SENESCENCE-ASSOCIATED GENE201), is highly upregulated during leaf senescence (Hou et al., 2013), and leaf senescence of two independent saur36 T-DNA mutants was significantly delayed. Furthermore, inducible overexpression of SAUR36 promoted premature leaf senescence. Interestingly, only SAUR36 expression constructs that lacked the DST element within the 3’-UTR conferred an early senescence phenotype, suggesting that post-transcriptional regulation of SAUR36 transcript levels may play an important functional role. Together, these findings convincingly demonstrate that SAUR36 positively regulates leaf senescence. Similar findings were obtained with OsSAUR39 in rice. Though loss-of-function support is lacking, OsSAUR39 expression is elevated in older leaves, and overexpression of OsSAUR39 promoted early senescence (Kant et al., 2009). Further studies are needed to provide mechanistic insight and to determine whether the upregulation of SAUR36 and OsSAUR39 expression during senescence is auxin-dependent.
Root Growth and Development
Auxin is essential for root growth and development, regulating stem cell specification and division, meristem size, cell elongation, and differentiation (reviewed in Sozzani and Iyer-Pascuzzi, 2014; Takatsuka and Umeda, 2014). In addition to the primary roots, auxin also regulates the initiation and growth and development of lateral roots. While the majority of SAUR studies discussed above have focused on expression patterns and functional studies in shoots, several of the aforementioned SAUR overexpression lines also exhibit root phenotypes.
SAUR76 is expressed in the pericycle and endodermal layers of the root elongation zone and at lateral root initiation sites (Markakis et al., 2013). Auxin treatment strongly upregulates SAUR76 expression in roots. While saur76 null mutants exhibited no apparent phenotype, overexpression of SAUR76 conferred increases in root meristem size and primary root growth, suggesting that SAUR76 may positively regulate root growth. SAUR41, on the other hand, is specifically expressed in the quiescent center and cortical/endodermal initials of the stem cell niche of primary and lateral roots. Its expression pattern is expanded to the root endodermis in seedlings treated with auxin for 1 hour. Plants overexpressing SAUR41 exhibited increased primary root growth and lateral root numbers (Kong et al., 2013). Interestingly, expression of SAUR41 under the control of tissue/cell type-specific promoters (PIN1, WOX5, PLT2, and others) caused defects in root meristem patterning and auxin distribution within root tips (as assayed using the DR5rev::GFP reporter). As in shoots, there is some evidence for positive and negative acting SAURs in root growth and development. In contrast to the above findings with Arabidopsis SAUR41 and SAUR76, overexpression of OsSAUR39 in rice resulted in reductions in root elongation and lateral root development (Kant et al., 2009). To date, no molecular or biochemical mechanism has been elucidated to explain how SAUR proteins might regulate root growth and development.
Calcium Signaling
In response to cytosolic free Ca2+ increases, calcium-binding proteins, such as calmodulins (CaM) and CaM-like proteins (CML), sense Ca2+ signals and initiate signaling cascades to regulate many cellular, physiological, and developmental processes (reviewed in Dodd et al., 2010; Kudla et al., 2010). Numerous studies have suggested that Ca2+ is involved in auxin signaling or responses (reviewed in Vanneste and Friml, 2013). Auxin induces a rapid and transient increase of cytosolic Ca2+ concentration in various plant species. However, the functions of Ca2+ in auxin signaling or responses are poorly understood. SAUR proteins may provide a functional link between Ca2+ and auxin responses.
Using recombinant 35S-labeled CaM to screen a maize root cDNA expression library, Yang and Poovaiah (2000) isolated ZmSAUR1 as a CaM-interacting protein that binds to CaM in a calcium-dependent manner. Deletion analyses revealed that the CaM-binding motif is located in the N-terminal region of ZmSAUR1. This study and subsequent studies have identified additional SAUR proteins (maize ZmSAUR2, Arabidopsis SAUR15 and SAUR70, and soybean SAUR10A5) capable of binding CaM (Knauss et al., 2003; Popescu et al., 2007). Our search for potential CaM-binding sites within the 79 Arabidopsis SAUR proteins using CALMODULIN TARGET DATABASE (http://calcium.uhnres.utoronto.ca/ctdb/ctdb/sequence.html) identified potential CaM-binding motifs in all family members except SAUR4, 6, 10, 18, 30, 33, 40, 41, 52, 57, 58, 67, and 74. To date, however, only SAUR70 has been shown to bind CaM or CaM-related proteins in planta (Popescu et al., 2007) and no functional significance has been ascribed to this potential activity of SAUR proteins.
Auxin transport
Several studies have indicated that SAURs can modulate polar auxin transport. In Arabidopsis, SAUR19 overexpression and SAUR19/23/24 amiRNA knockdown seedlings conferred increased and decreased basipetal indole-3-acetic acid (IAA) transport, respectively, in hypocotyls (Spartz et al., 2012). Similar findings have been obtained with SAUR41 and SAUR63 (Chae et al., 2012; Kong et al., 2013). In contrast, overexpression of OsSAUR39 in rice resulted in reduced basipetal IAA transport (Kant et al., 2009), again raising the possibility that different SAUR proteins may function antagonistically. The simplest explanation for increased auxin transport is the elevated PM H+-ATPase activity resulting from SAUR overexpression. As per the chemiosmotic model for IAA transport (Blakeslee et al., 2005), the increase in apoplastic acidification and the predicted increase in plasma membrane potential associated with SAUR overexpression would be expected to promote IAA transport. However, other mechanisms are also possible, including the aforementioned CaM binding activity of SAURs, as polar auxin transport is highly dependent on calcium (Vanneste and Friml, 2013). Regardless of the mechanism, the fact that SAUR proteins are capable of modulating IAA transport provides a potential explanation for the diversity of effects, including altered cell expansion, division, and patterning that result from ectopic SAUR expression.
Hormonal Regulation of SAUR Expression - more than just Auxin
In addition to auxin, brassinosteroids (BR), gibberellins (GA), jasmonate (JA), and abscisic acid (ABA) have been reported to regulate the expression of some SAUR genes, indicating that SAURs likely contribute to other hormone-regulated aspects of plant growth and development. Similar to auxin, BR is a growth-promoting hormone that regulates many physiological and developmental processes (reviewed in Fàbregas and Caño-Delgado, 2014; Wang et al., 2014b). A large body of evidence suggests that the BR and auxin pathways exhibit extensive crosstalk and converge at the transcriptional level to regulate many shared target genes (reviewed in Vert and Chory, 2011; Wang et al., 2012), including many SAUR genes (Goda et al., 2004; Nemhauser et al., 2004). In the BR signaling pathway, the BRI1-EMS-SUPPRESSOR1 (BES1)/BRASSINAZOLE RESISTANT1 (BZR1) family transcription factors regulate gene expression (He et al., 2005; Yin et al., 2005). Chromatin immunoprecipitation – DNA microarray (ChIP-chip) studies identified many SAUR genes as potential direct targets of BZR1 and BES1 (Supplementary Table 2) (Sun et al., 2010; Yu et al., 2011). Indeed, ChIP assays have confirmed that BZR1 and BES1 bind to the SAUR15 promoter (Yin et al., 2005; Sun et al., 2010; Walcher and Nemhauser, 2012), and BES1 binds to SAUR36 and SAUR59 promoters (Yu et al., 2011). While the precise roles of SAUR proteins in BR action remain unclear, given the well-established role of BR in promoting cell expansion, it seems likely that SAURs are downstream effectors that mediate at least some aspects of BR-mediated expansion growth.
GA is also a growth-promoting hormone that regulates diverse physiological and developmental processes, such as seed germination and stem elongation. The DELLA repressors function as central regulators in the GA signaling pathway, and GA promotes DELLA protein degradation by the 26S proteasome to stimulate growth (reviewed in Claeys et al., 2014; Xu et al., 2014). Consistent with a possible role for SAURs in GA-dependent growth, Bai et al. (2012) recently found that 27 SAUR genes were upregulated following a 12h GA treatment (Supplementary Table 3). This increase in SAUR expression is likely the result of enhanced ARF, PIF, and BZR1/BES1 transcription factor binding to SAUR promoters following DELLA proteolysis, as DELLAs interact with, and inhibit the binding of each of these transcription factors (de Lucas et al., 2008; Feng et al., 2008; Bai et al., 2012; Oh et al., 2014).
A second connection between SAUR proteins and GA-regulated growth is in seed germination. GA promotes germination, and RGA-LIKE2 (RGL2) is a major DELLA protein that represses seed germination in Arabidopsis. Stamm et al. (2012) reported that RGL2 upregulates SAUR36, which is highly expressed in dry and imbibed seeds, and ChIP-qPCR analysis detected RGL2 binding to the SAUR36 promoter. Consistent with the upregulation of SAUR36 by RGL2, GA represses SAUR36 expression (Stamm and Kumar, 2013). Genetic studies support a role for SAUR36 in regulating seed germination, although curiously, germination of both saur36 null mutants and SAUR36 overexpression lines was hypersensitive to paclobutrazol (PAC, a gibberellin biosynthesis inhibitor) and ABA treatments. Further research is required to understand the precise role of SAUR36 in seed germination.
In contrast to the general trend of increased SAUR expression elicited by the above growth promoting hormones, many SAUR genes are repressed by the stress-related hormones jasmonate and ABA (Supplementary Table 4) (Nemhauser et al., 2006). ABA is a major regulator of the adaptive responses of plants to stresses, such as drought and high salt, which cause plant growth inhibition (reviewed in Yoshida et al., 2014). Many SAUR genes exhibit reduced expression in response to ABA treatment, as well as drought and osmotic stresses (Kodaira et al., 2011). To investigate the mechanisms underlying abiotic stress-regulated growth inhibition, Kodaira et al. (2011) studied two transcriptional repressors AZF1 (ARABIDOPSIS ZINC-FINGER PROTEIN1) and AZF2. Expression of AZF1 and AZF2 is induced by ABA, drought, and high salt, and AZF overexpression confers growth repression. Transcriptome analyses of DEX-inducible AZF1 and AZF2 overexpression plants identified 27 SAUR genes, whose expression is downregulated. These genes included nearly all members of the SAUR19 and SAUR63 subfamilies, and gel shift assays demonstrated that recombinant AZF1 and AZF2 proteins could bind to the promoters of SAUR20 and SAUR63, suggesting that at least these SAURs are direct targets of AZF-mediated repression. Consistent with these findings, expression of SAUR16 and SAUR63 is increased in the azf1 azf2 double mutant in response to high salt stress. As several studies have found that the SAUR19 and SAUR63 subfamily genes positively regulate cell expansion to promote plant growth (Chae et al., 2012; Spartz et al., 2012; Spartz et al., 2014), these findings suggest that AZF-mediated down-regulation of SAUR expression may contribute to the growth repression characteristic of ABA and abiotic stress treatments. Potentially, the molecular basis for this regulation could be the aforementioned SAUR regulation of PP2C.D phosphatase activity to modulate PM H+-ATPase activity. While both auxin treatment and SAUR19 overexpression promote increases in PM H+-ATPase C-terminal Thr947 phosphorylation and enzymatic activity, ABA treatment results in reduced Thr947 phosphorylation and ATPase activity (Hayashi et al., 2014).
CONCLUSIONS AND FUTURE PERSPECTIVES
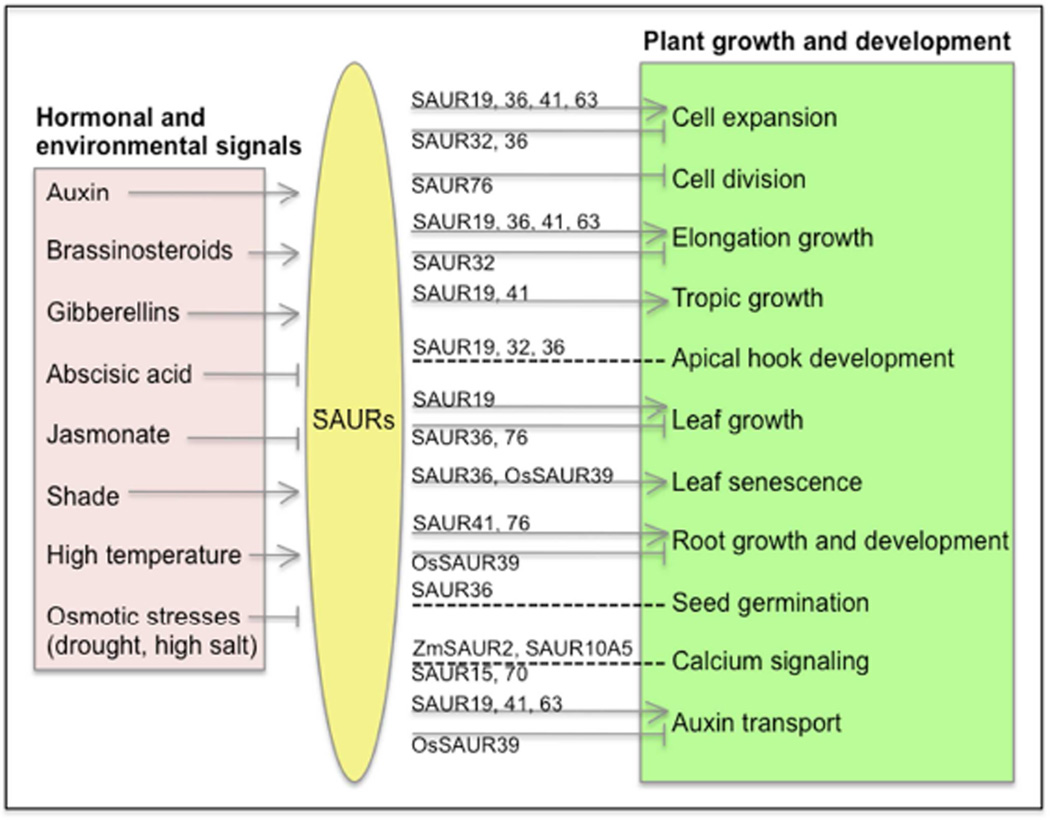
Following a protracted lag phase, the functions of the large, highly conserved, and plant specific SAUR gene family are beginning to be elucidated. Data obtained from molecular, genetic, biochemical, and genomic studies have suggested that SAURs are involved in regulating a wide range of cellular, physiological, and developmental processes in response to hormonal and environmental signals. We suggest that SAURs play key roles in integrating hormonal and environmental signals into distinct growth and developmental responses (Figure 3). Based on our current knowledge, this regulation is accomplished largely through the induction or repression of SAUR gene transcription. In particular, the SAUR19 and SAUR63 subfamilies, both of which have been intimately linked with cell expansion, appear to be especially responsive to hormonal and environmental signals that regulate plant growth. That said, there is strong evidence that SAUR activities are also controlled post-transcriptionally, via the regulation of mRNA and protein stability. Determining the extent and mechanisms by which hormonal and environmental signals contribute to these modes of regulation is a crucial area for future research.
Figure 3. SAURs integrate hormonal and environmental signals to regulate plant growth and development.
Arrows and blunted lines indicate positive and negative regulation, respectively. Dashed lines indicate either positive or negative regulation. Hormones and environmental factors confer changes in SAUR expression. Particular SAURs then positively or negatively regulate specific cellular, physiological, and developmental processes. The indicated effects of the various hormones and environmental factors on SAUR expression reflect the overall trend.
Although considerable progress has been made in recent years, we still have only a rudimentary understanding of the functions and molecular mechanisms of SAUR-regulated plant growth and development. Much of our current knowledge is based on expression patterns and gain-of-function studies, which, at best, can only suggest potential functions. While SAUR loss-of-function studies are complicated by likely extensive genetic redundancy and tight linkage arrangements, modern genome editing techniques such as TALEN and CRISPR/Cas9 nucleases offer potential solutions.
Molecular and biochemical studies are also desperately needed to provide additional mechanistic insight into SAUR protein function. The recent demonstration that SAUR19 inhibits PP2C.D phosphatase activity to control PM H+-ATPase activity is an exciting first step in this direction (Spartz et al., 2014). And indeed, the crucial role that PM H+-ATPases play in establishing electrochemical gradients across the plasma membrane could potentially control, either directly or indirectly, many of the growth responses that appear to be influenced by SAURs. However, the observations that different SAUR proteins exhibit distinct subcellular localizations strongly suggest that PM H+-ATPase regulation is only one of several SAUR functions. That said, the possibility that PP2C.D regulation is the primary function of SAUR proteins remains an intriguing prospect. There are 9 PP2C.D family members in Arabidopsis, and initial work suggests that these proteins are differentially localized (Tovar-Mendez et al., 2014). Thus, a SAUR-PP2C.D “code” may exist, whereby distinct SAUR-PP2C.D combinations in different tissues and subcellular compartments could modulate the phosphorylation status and activities of many downstream effectors to control diverse aspects of growth and development. Detailed PP2C.D genetic studies, characterization of PP2C.D expression patterns, and the identification of additional PP2C.D substrates will likely be informative in testing this possibility and potentially forging new links with SAUR proteins. In addition, the CaM binding activity exhibited by several SAUR proteins also warrants additional studies that may lead to the identification of distinct biochemical functions for SAUR proteins.
It now seems apparent that SAUR proteins are crucial regulators of diverse aspects of plant growth and development. Unraveling the molecular networks that link specific hormonal and environmental signals to particular SAUR proteins and cellular/developmental outputs will undoubtedly improve our understanding of SAUR function, and may lead to novel strategies for manipulating plant growth. We expect many new and exciting findings on this long neglected family of plant proteins in the years to come.
Supplementary Material
Acknowledgments
We thank Angela K. Spartz and Mee Yeon Park for critical reading and comments on the manuscript, and thank the University of Minnesota - University Imaging Center for the Nikon A1 spectral confocal microscope.
FUNDING
This work was supported by grants to WMG from the National Institutes of Health (GM067203) and National Science Foundation (MCB-0817205).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTARY DATA
Supplementary data are available at Molecular Plant Online.
No conflict of interest declared.
REFERENCES
- Abbas M, Alabadí D, Blázquez MA. Differential growth at the apical hook: all roads lead to auxin. Front. Plant Sci. 2013;4:441. doi: 10.3389/fpls.2013.00441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel S, Oeller PW, Theologis A. Early auxin-induced genes encode short-lived nuclear proteins. Proc. Natl. Acad. Sci. U S A. 1994;91:326–330. doi: 10.1073/pnas.91.1.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anai T, Kono N, Kosemura S, Yamamura S, Hasegawa K. Isolation and characterization of an auxin-inducible SAUR gene from radish seedlings. DNA Seq. 1998;9:329–333. [PubMed] [Google Scholar]
- Argueso CT, Raines T, Kieber JJ. Cytokinin signaling and transcriptional networks. Curr. Opin. Plant Biol. 2010;13:533–539. doi: 10.1016/j.pbi.2010.08.006. [DOI] [PubMed] [Google Scholar]
- Bai MY, Shang JX, Oh E, Fan M, Bai Y, Zentella R, Sun TP, Wang ZY. Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat. Cell Biol. 2012;14:810–817. doi: 10.1038/ncb2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter I, Tchieu J, Sussman MR, Boutry M, Palmgren MG, Gribskov M, Harper JF, Axelsen KB. Genomic comparison of P-type ATPase ion pumps in Arabidopsis and rice. Plant Physiol. 2003;132:618–628. doi: 10.1104/pp.103.021923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakeslee JJ, Peer WA, Murphy AS. Auxin transport. Curr. Opin. Plant Biol. 2005;8:494–500. doi: 10.1016/j.pbi.2005.07.014. [DOI] [PubMed] [Google Scholar]
- Braidwood L, Breuer C, Sugimoto K. My body is a cage: mechanisms and modulation of plant cell growth. New Phytol. 2013;201:388–402. doi: 10.1111/nph.12473. [DOI] [PubMed] [Google Scholar]
- Byrne ME. Making leaves. Curr. Opin. Plant Biol. 2012;15:24–30. doi: 10.1016/j.pbi.2011.10.009. [DOI] [PubMed] [Google Scholar]
- Calderón Villalobos LI, Lee S, De Oliveira C, Ivetac A, Brandt W, Armitage L, Sheard LB, Tan X, Parry G, Mao H, Zheng N, Napier R, Kepinski S, Estelle M. A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat. Chem. Biol. 2012;8:477–485. doi: 10.1038/nchembio.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ. Photoreceptor signaling networks in plant responses to shade. Annu. Rev. Plant Biol. 2013;64:403–427. doi: 10.1146/annurev-arplant-050312-120221. [DOI] [PubMed] [Google Scholar]
- Chae K, Isaacs CG, Reeves PH, Maloney GS, Muday GK, Nagpal P, Reed JW. Arabidopsis SMALL AUXIN UP RNA63 promotes hypocotyl and stamen filament elongation. Plant J. 2012;71:684–697. doi: 10.1111/j.1365-313X.2012.05024.x. [DOI] [PubMed] [Google Scholar]
- Chapman EJ, Greenham K, Castillejo C, Sartor R, Bialy A, Sun TP, Estelle M. Hypocotyl transcriptome reveals auxin regulation of growth-promoting genes through GA-dependent and -independent pathways. PLoS One. 2012;7:e36210. doi: 10.1371/journal.pone.0036210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Hao X, Cao J. Small auxin upregulated RNA (SAUR) gene family in maize: identification, evolution, and its phylogenetic comparison with Arabidopsis, rice, and sorghum. J. Integr. Plant Biol. 2014;56:133–150. doi: 10.1111/jipb.12127. [DOI] [PubMed] [Google Scholar]
- Claeys H, De Bodt S, Inzé D. Gibberellins and DELLAs: central nodes in growth regulatory networks. Trends Plant Sci. 2014;19:231–239. doi: 10.1016/j.tplants.2013.10.001. [DOI] [PubMed] [Google Scholar]
- de Lucas M, Daviere JM, Rodriguez-Falcon M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blazquez MA, Titarenko E, Prat S. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451:480–484. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- de Wit M, Lorrain S, Fankhauser C. Auxin-mediated plant architectural changes in response to shade and high temperature. Physiol. Plant. 2014;151:13–24. doi: 10.1111/ppl.12099. [DOI] [PubMed] [Google Scholar]
- Dodd AN, Kudla J, Sanders D. The language of calcium signaling. Annu. Rev. Plant Biol. 2010;61:593–620. doi: 10.1146/annurev-arplant-070109-104628. [DOI] [PubMed] [Google Scholar]
- Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, Schafer E, Fu X, Fan LM, Deng XW. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature. 2008;451:475–479. doi: 10.1038/nature06448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco AR, Gee MA, Guilfoyle TJ. Induction and superinduction of auxin-responsive mRNAs with auxin and protein synthesis inhibitors. J. Biol. Chem. 1990;265:15845–15849. [PubMed] [Google Scholar]
- Franklin KA, Lee SH, Patel D, Kumar SV, Spartz AK, Gu C, Ye S, Yu P, Breen G, Cohen JD, Wigge PA, Gray WM. Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc. Natl. Acad. Sci. U S A. 2011;108:20231–20235. doi: 10.1073/pnas.1110682108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuglsang AT, Visconti S, Drumm K, Jahn T, Stensballe A, Mattei B, Jensen ON, Aducci P, Palmgren MG. Binding of 14-3-3 protein to the plasma membrane H+-ATPase AHA2 involves the three C-terminal residues Tyr946-Thr-Val and requires phosphorylation of Thr947. J. Biol. Chem. 1999;274:36774–36780. doi: 10.1074/jbc.274.51.36774. [DOI] [PubMed] [Google Scholar]
- Fàbregas N, Caño-Delgado AI. Turning on the microscope turret: a new view for the study of brassinosteroid signaling in plant development. Physiol. Plant. 2014;151:172–183. doi: 10.1111/ppl.12130. [DOI] [PubMed] [Google Scholar]
- Gee MA, Hagen G, Guilfoyle TJ. Tissue-specific and organ-specific expression of soybean auxin-responsive transcripts GH3 and SAURs. Plant Cell. 1991;3:419–430. doi: 10.1105/tpc.3.4.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendreau E, Traas J, Desnos T, Grandjean O, Caboche M, Hofte H. Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol. 1997;114:295–305. doi: 10.1104/pp.114.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil P, Green PJ. Multiple regions of the Arabidopsis SAUR-AC1 gene control transcript abundance: the 3’ untranslated region functions as an mRNA instability determinant. EMBO J. 1996;15:1678–1686. [PMC free article] [PubMed] [Google Scholar]
- Gil P, Liu Y, Orbović V, Verkamp E, Poff KL, Green PJ. Characterization of the auxin-inducible SAUR-AC1 gene for use as a molecular genetic tool in Arabidopsis. Plant Physiol. 1994;104:777–784. doi: 10.1104/pp.104.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Sawa S, Asami T, Fujioka S, Shimada Y, Yoshida S. Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 2004;134:1555–1573. doi: 10.1104/pp.103.034736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal A, Szarzynska B, Fankhauser C. Phototropism: at the crossroads of light-signaling pathways. Trends Plant Sci. 2013;18:393–401. doi: 10.1016/j.tplants.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Gray WM, Ostin A, Sandberg G, Romano CP, Estelle M. High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc. Natl. Acad. Sci. U S A. 1998;95:7197–7202. doi: 10.1073/pnas.95.12.7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle TJ, Hagen G. Auxin response factors. Curr. Opin. Plant Biol. 2007;10:453–460. doi: 10.1016/j.pbi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- Guilfoyle TJ, Hagen G, Li Y, Ulmasov TN, Liu Z, Strabala T, Gee M. Auxin-regulated Transcription. Aust. J. Plant Physiol. 1993;20:489–502. [Google Scholar]
- Hagen G, Guilfoyle T. Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol. Biol. 2002;49:373–385. [PubMed] [Google Scholar]
- Hager A. Role of the plasma membrane H+-ATPase in auxin-induced elongation growth: historical and new aspects. J. Plant Res. 2003;116:483–505. doi: 10.1007/s10265-003-0110-x. [DOI] [PubMed] [Google Scholar]
- Hao Y, Oh E, Choi G, Liang Z, Wang ZY. Interactions between HLH and bHLH factors modulate light-regulated plant development. Mol. Plant. 2012;5:688–697. doi: 10.1093/mp/sss011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta M, Burch HL, Nelson RB, Barrett-Wilt G, Kline KG, Mohsin SB, Young JC, Otegui MS, Sussman MR. Molecular characterization of mutant Arabidopsis plants with reduced plasma membrane proton pump activity. J. Biol. Chem. 2010;285:17918–17929. doi: 10.1074/jbc.M110.101733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y, Takahashi K, Inoue SI, Kinoshita T. Abscisic Acid Suppresses Hypocotyl Elongation by Dephosphorylating Plasma Membrane H+-ATPase in Arabidopsis thaliana. Plant Cell Physiol. 2014;55:845–853. doi: 10.1093/pcp/pcu028. [DOI] [PubMed] [Google Scholar]
- He JX, Gendron JM, Sun Y, Gampala SS, Gendron N, Sun CQ, Wang ZY. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science. 2005;307:1634–1638. doi: 10.1126/science.1107580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohm T, Demarsy E, Quan C, Allenbach Petrolati L, Preuten T, Vernoux T, Bergmann S, Fankhauser C. Plasma membrane H+-ATPase regulation is required for auxin gradient formation preceding phototropic growth. Mol. Syst. Biol. 2014;10:751. doi: 10.15252/msb.20145247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P, Kohnen MV, Lorrain S, Rougemont J, Ljung K, Lopez-Vidriero I, Franco-Zorrilla JM, Solano R, Trevisan M, Pradervand S, Xenarios I, Fankhauser C. Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 2012;71:699–711. doi: 10.1111/j.1365-313X.2012.05033.x. [DOI] [PubMed] [Google Scholar]
- Hou K, Wu W, Gan SS. SAUR36, a small auxin up RNA gene, is involved in the promotion of leaf senescence in Arabidopsis. Plant Physiol. 2013;161:1002–1009. doi: 10.1104/pp.112.212787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Mei Z, Zang A, Chen H, Dou X, Jin J, Cai W. Microarray analyses and comparisons of upper or lower flanks of rice shoot base preceding gravitropic bending. PLoS One. 2013;8:e74646. doi: 10.1371/journal.pone.0074646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Tyagi AK, Khurana JP. Genome-wide analysis, evolutionary expansion, and expression of early auxin-responsive SAUR gene family in rice (Oryza sativa) Genomics. 2006;88:360–371. doi: 10.1016/j.ygeno.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Jibran R, Hunter DA, Dijkwel PP. Hormonal regulation of leaf senescence through integration of developmental and stress signals. Plant Mol. Biol. 2013;82:547–561. doi: 10.1007/s11103-013-0043-2. [DOI] [PubMed] [Google Scholar]
- Johnson MA, Perez-Amador MA, Lidder P, Green PJ. Mutants of Arabidopsis defective in a sequence-specific mRNA degradation pathway. Proc. Natl. Acad. Sci. U S A. 2000;97:13991–13996. doi: 10.1073/pnas.240354097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant S, Bi YM, Zhu T, Rothstein SJ. SAUR39, a small auxin-up RNA gene, acts as a negative regulator of auxin synthesis and transport in rice. Plant Physiol. 2009;151:691–701. doi: 10.1104/pp.109.143875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M, Rozhon W, Poppenberger B. The role of hormones in the aging of plants - a mini-review. Gerontology. 2014;60:49–55. doi: 10.1159/000354334. [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Shimazaki K. Blue light activates the plasma membrane H+-ATPase by phosphorylation of the C-terminus in stomatal guard cells. EMBO J. 1999;18:5548–5558. doi: 10.1093/emboj/18.20.5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauss S, Rohrmeier T, Lehle L. The auxin-induced maize gene ZmSAUR2 encodes a short-lived nuclear protein expressed in elongating tissues. J. Biol. Chem. 2003;278:23936–23943. doi: 10.1074/jbc.M212585200. [DOI] [PubMed] [Google Scholar]
- Kodaira KS, Qin F, Tran LS, Maruyama K, Kidokoro S, Fujita Y, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis Cys2/His2 zinc-finger proteins AZF1 and AZF2 negatively regulate abscisic acid-repressive and auxin-inducible genes under abiotic stress conditions. Plant Physiol. 2011;157:742–756. doi: 10.1104/pp.111.182683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koini MA, Alvey L, Allen T, Tilley CA, Harberd NP, Whitelam GC, Franklin KA. High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr. Biol. 2009;19:408–413. doi: 10.1016/j.cub.2009.01.046. [DOI] [PubMed] [Google Scholar]
- Kong Y, Zhu Y, Gao C, She W, Lin W, Chen Y, Han N, Bian H, Zhu M, Wang J. Tissue-specific expression of SMALL AUXIN UP RNA41 differentially regulates cell expansion and root meristem patterning in Arabidopsis. Plant Cell Physiol. 2013;54:609–621. doi: 10.1093/pcp/pct028. [DOI] [PubMed] [Google Scholar]
- Kuang JF, Wu JY, Zhong HY, Li CQ, Chen JY, Lu WJ, Li JG. Carbohydrate stress affecting fruitlet abscission and expression of genes related to auxin signal transduction pathway in litchi. Int. J. Mol. Sci. 2012;13:16084–16103. doi: 10.3390/ijms131216084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla J, Batistic O, Hashimoto K. Calcium signals: the lead currency of plant information processing. Plant Cell. 2010;22:541–563. doi: 10.1105/tpc.109.072686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Hagen G, Guilfoyle TJ. An Auxin-Responsive Promoter Is Differentially Induced by Auxin Gradients during Tropisms. Plant Cell. 1991;3:1167–1175. doi: 10.1105/tpc.3.11.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Zheng C, Chitsaz F, Derbyshire MK, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Lu S, Marchler GH, Song JS, Thanki N, Yamashita RA, Zhang D, Bryant SH. CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res. 2013;41:D348–D352. doi: 10.1093/nar/gks1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markakis MN, Boron AK, Van Loock B, Saini K, Cirera S, Verbelen JP, Vissenberg K. Characterization of a small auxin-up RNA (SAUR)-like gene involved in Arabidopsis thaliana development. PLoS One. 2013;8:e82596. doi: 10.1371/journal.pone.0082596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marre E. Fusicoccin: A tool in plant physiology. Ann. Rev. Plant Physiol. 1979;30:273–288. [Google Scholar]
- McClure BA, Guilfoyle T. Characterization of a class of small auxin-inducible soybean polyadenylated RNAs. Plant Mol. Biol. 1987;9:611–623. doi: 10.1007/BF00020537. [DOI] [PubMed] [Google Scholar]
- McClure BA, Guilfoyle T. Rapid redistribution of auxin-regulated RNAs during gravitropism. Science. 1989;243:91–93. doi: 10.1126/science.11540631. [DOI] [PubMed] [Google Scholar]
- McClure BA, Hagen G, Brown CS, Gee MA, Guilfoyle TJ. Transcription, organization, and sequence of an auxin-regulated gene cluster in soybean. Plant Cell. 1989;1:229–239. doi: 10.1105/tpc.1.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlot S, Leonhardt N, Fenzi F, Valon C, Costa M, Piette L, Vavasseur A, Genty B, Boivin K, Müller A, Giraudat J, Leung J. Constitutive activation of a plasma membrane H+-ATPase prevents abscisic acid-mediated stomatal closure. EMBO J. 2007;26:3216–3226. doi: 10.1038/sj.emboj.7601750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narsai R, Law SR, Carrie C, Xu L, Whelan J. In-depth temporal transcriptome profiling reveals a crucial developmental switch with roles for RNA processing and organelle metabolism that are essential for germination in Arabidopsis. Plant Physiol. 2011;157:1342–1362. doi: 10.1104/pp.111.183129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser JL, Mockler TC, Chory J. Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol. 2004;2:E258. doi: 10.1371/journal.pbio.0020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser JL, Hong F, Chory J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell. 2006;126:467–475. doi: 10.1016/j.cell.2006.05.050. [DOI] [PubMed] [Google Scholar]
- Newman TC, Ohme-Takagi M, Taylor CB, Green PJ. DST sequences, highly conserved among plant SAUR genes, target reporter transcripts for rapid decay in tobacco. Plant Cell. 1993;5:701–714. doi: 10.1105/tpc.5.6.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Wang ZY. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 2012;14:802–809. doi: 10.1038/ncb2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Bai MY, Arenhart RA, Sun Y, Wang ZY. Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. Elife. 2014;3:e03031. doi: 10.7554/eLife.03031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Lui A, Nguyen D, Onodera C, Quach H, Smith A, Yu G, Theologis A. Functional Genomic Analysis of the AUXIN RESPONSE FACTOR Gene Family Members in Arabidopsis thaliana: Unique and Overlapping Functions of ARF7 and ARF19. Plant Cell. 2005;17:444–463. doi: 10.1105/tpc.104.028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paponov IA, Paponov M, Teale W, Menges M, Chakrabortee S, Murray JA, Palme K. Comprehensive transcriptome analysis of auxin responses in Arabidopsis. Mol. Plant. 2008;1:321–337. doi: 10.1093/mp/ssm021. [DOI] [PubMed] [Google Scholar]
- Park JE, Kim YS, Yoon HK, Park CM. Functional characterization of a small auxin-up RNA gene in apical hook development in Arabidopsis. Plant Sci. 2007;172:150–157. [Google Scholar]
- Popescu SC, Popescu GV, Bachan S, Zhang Z, Seay M, Gerstein M, Snyder M, Dinesh-Kumar SP. Differential binding of calmodulin-related proteins to their targets revealed through high-density Arabidopsis protein microarrays. Proc. Natl. Acad. Sci. U S A. 2007;104:4730–4735. doi: 10.1073/pnas.0611615104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu T, Chen Y, Li M, Kong Y, Zhu Y, Han N, Bian H, Zhu M, Wang J. The tissue-specific and developmentally regulated expression patterns of the SAUR41 subfamily of small auxin up RNA genes: potential implications. Plant Signal Behav. 2013;8:e25283. doi: 10.4161/psb.25283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayle DL, Cleland R. Enhancement of wall loosening and elongation by Acid solutions. Plant Physiol. 1970;46:250–253. doi: 10.1104/pp.46.2.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayle DL, Cleland RE. Evidence that Auxin-induced Growth of Soybean Hypocotyls Involves Proton Excretion. Plant Physiol. 1980;66:433–437. doi: 10.1104/pp.66.3.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayle DL, Cleland RE. The Acid Growth Theory of auxin-induced cell elongation is alive and well. Plant Physiol. 1992;99:1271–1274. doi: 10.1104/pp.99.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redman JC, Haas BJ, Tanimoto G, Town CD. Development and evaluation of an Arabidopsis whole genome Affymetrix probe array. Plant J. 2004;38:545–561. doi: 10.1111/j.1365-313X.2004.02061.x. [DOI] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud PF, Lindquist EA, Kamisugi Y, Tanahashi T, Sakakibara K, Fujita T, Oishi K, Shin IT, Kuroki Y, Toyoda A, Suzuki Y, Hashimoto S, Yamaguchi K, Sugano S, Kohara Y, Fujiyama A, Anterola A, Aoki S, Ashton N, Barbazuk WB, Barker E, Bennetzen JL, Blankenship R, Cho SH, Dutcher SK, Estelle M, Fawcett JA, Gundlach H, Hanada K, Heyl A, Hicks KA, Hughes J, Lohr M, Mayer K, Melkozernov A, Murata T, Nelson DR, Pils B, Prigge M, Reiss B, Renner T, Rombauts S, Rushton PJ, Sanderfoot A, Schween G, Shiu SH, Stueber K, Theodoulou FL, Tu H, Van de Peer Y, Verrier PJ, Waters E, Wood A, Yang L, Cove D, Cuming AC, Hasebe M, Lucas S, Mishler BD, Reski R, Grigoriev IV, Quatrano RS, Boore JL. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- Roig-Villanova I, Bou-Torrent J, Galstyan A, Carretero-Paulet L, Portoles S, Rodriguez-Concepcion M, Martinez-Garcia JF. Interaction of shade avoidance and auxin responses: a role for two novel atypical bHLH proteins. EMBO J. 2007;26:4756–4767. doi: 10.1038/sj.emboj.7601890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpella E, Barkoulas M, Tsiantis M. Control of leaf and vein development by auxin. Cold Spring Harb. Perspect. Biol. 2010;2:a001511. doi: 10.1101/cshperspect.a001511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweighofer A, Hirt H, Meskiene I. Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci. 2004;9:236–243. doi: 10.1016/j.tplants.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Singh J, Roberts MR. Fusicoccin activates pathogen-responsive gene expression independently of common resistance signalling pathways, but increases disease symptoms in Pseudomonas syringae-infected tomato plants. Planta. 2004;219:261–269. doi: 10.1007/s00425-004-1234-5. [DOI] [PubMed] [Google Scholar]
- Sozzani R, Iyer-Pascuzzi A. Postembryonic control of root meristem growth and development. Curr. Opin. Plant Biol. 2014;17:7–12. doi: 10.1016/j.pbi.2013.10.005. [DOI] [PubMed] [Google Scholar]
- Spartz AK, Ren H, Park MY, Grandt KN, Lee SH, Murphy AS, Sussman MR, Overvoorde PJ, Gray WM. SAUR Inhibition of PP2C–D Phosphatases Activates Plasma Membrane H+-ATPases to Promote Cell Expansion in Arabidopsis. Plant Cell. 2014;26:2129–2142. doi: 10.1105/tpc.114.126037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spartz AK, Lee SH, Wenger JP, Gonzalez N, Itoh H, Inzé D, Peer WA, Murphy AS, Overvoorde PJ, Gray WM. The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J. 2012;70:978–990. doi: 10.1111/j.1365-313X.2012.04946.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm P, Kumar PP. Auxin and gibberellin responsive Arabidopsis SMALL AUXIN UP RNA36 regulates hypocotyl elongation in the light. Plant Cell Rep. 2013;32:759–769. doi: 10.1007/s00299-013-1406-5. [DOI] [PubMed] [Google Scholar]
- Stamm P, Ravindran P, Mohanty B, Tan EL, Yu H, Kumar PP. Insights into the molecular mechanism of RGL2-mediated inhibition of seed germination in Arabidopsis thaliana. BMC Plant Biol. 2012;12:179. doi: 10.1186/1471-2229-12-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Tiryaki I, Rowe ML. Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell. 2002;14:1405–1415. doi: 10.1105/tpc.000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan ML, Green PJ. Mutational analysis of the DST element in tobacco cells and transgenic plants: identification of residues critical for mRNA instability. RNA. 1996;2:308–315. [PMC free article] [PubMed] [Google Scholar]
- Sun J, Qi L, Li Y, Chu J, Li C. PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating arabidopsis hypocotyl growth. PLoS Genet. 2012;8:e1002594. doi: 10.1371/journal.pgen.1002594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Fan XY, Cao DM, Tang W, He K, Zhu JY, He JX, Bai MY, Zhu S, Oh E, Patil S, Kim TW, Ji H, Wong WH, Rhee SY, Wang ZY. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell. 2010;19:765–777. doi: 10.1016/j.devcel.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Hayashi K, Kinoshita T. Auxin activates the plasma membrane H+-ATPase by phosphorylation during hypocotyl elongation in Arabidopsis. Plant Physiol. 2012;159:632–641. doi: 10.1104/pp.112.196428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuka H, Umeda M. Hormonal control of cell division and elongation along differentiation trajectories in roots. J. Exp. Bot. 2014;65:2633–2643. doi: 10.1093/jxb/ert485. [DOI] [PubMed] [Google Scholar]
- Taniguchi M, Nakamura M, Tasaka M, Morita M. Identification of gravitropic response indicator genes in Arabidopsis inflorescence stems. Plant Signal Behav. 2014;9:e29570. doi: 10.4161/psb.29570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuki M, Nakajima N, Fujii H, Shimada T, Nakano M, Hayashi K, Hayama H, Yoshioka H, Nakamura Y. Increased levels of IAA are required for system 2 ethylene synthesis causing fruit softening in peach (Prunus persica L. Batsch) J. Exp. Bot. 2013;64:1049–1059. doi: 10.1093/jxb/ers381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Q, Uhlir NJ, Reed JW. Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. Plant Cell. 2002;14:301–319. doi: 10.1105/tpc.010283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle TJ. Aux/IAA Proteins Contain a Potent Transcriptional Repression Domain. Plant Cell. 2004;16:533–543. doi: 10.1105/tpc.017384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Wang XJ, Hagen G, Guilfoyle TJ. AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell. 2001;13:2809–2822. doi: 10.1105/tpc.010289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovar-Mendez A, Miernyk JA, Hoyos E, Randall DD. A functional genomic analysis of Arabidopsis thaliana PP2C clade D. Protoplasma. 2014;251:265–271. doi: 10.1007/s00709-013-0526-7. [DOI] [PubMed] [Google Scholar]
- Vandenbrink JP, Kiss JZ, Herranz R, Medina FJ. Light and gravity signals synergize in modulating plant development. Front. Plant Sci. 2014;5:563. doi: 10.3389/fpls.2014.00563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhaeren H, Gonzalez N, Coppens F, De Milde L, Van Daele T, Vermeersch M, Eloy NB, Storme V, Inzé D. Combining growth-promoting genes leads to positive epistasis in Arabidopsis thaliana. Elife. 2014;3:e02252. doi: 10.7554/eLife.02252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S, Friml J. Calcium: The Missing Link in Auxin Action. Plants. 2013;2:650–675. doi: 10.3390/plants2040650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Chory J. Crosstalk in cellular signaling: background noise or the real thing? Dev. Cell. 2011;21:985–991. doi: 10.1016/j.devcel.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vi SL, Trost G, Lange P, Czesnick H, Rao N, Lieber D, Laux T, Gray WM, Manley JL, Groth D, Kappel C, Lenhard M. Target specificity among canonical nuclear poly(A) polymerases in plants modulates organ growth and pathogen response. Proc. Natl. Acad. Sci. U S A. 2013;110:13994–13999. doi: 10.1073/pnas.1303967110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walcher CL, Nemhauser JL. Bipartite promoter element required for auxin response. Plant Physiol. 2012;158:273–282. doi: 10.1104/pp.111.187559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Du Q, Yang X, Zhang D. Identification and characterization of nuclear genes involved in photosynthesis in Populus. BMC Plant Biol. 2014a;14:81. doi: 10.1186/1471-2229-14-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Estelle M. Diversity and specificity: auxin perception and signaling through the TIR1/AFB pathway. Curr. Opin. Plant Biol. 2014;21:51–58. doi: 10.1016/j.pbi.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Bai Y, Shen C, Wu Y, Zhang S, Jiang D, Guilfoyle TJ, Chen M, Qi Y. Auxin-related gene families in abiotic stress response in Sorghum bicolor. Funct. Integr. Genomics. 2010;10:533–546. doi: 10.1007/s10142-010-0174-3. [DOI] [PubMed] [Google Scholar]
- Wang W, Bai MY, Wang ZY. The brassinosteroid signaling network-a paradigm of signal integration. Curr. Opin. Plant Biol. 2014b;21C:147–153. doi: 10.1016/j.pbi.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Bai MY, Oh E, Zhu JY. Brassinosteroid signaling network and regulation of photomorphogenesis. Annu. Rev. Genet. 2012;46:701–724. doi: 10.1146/annurev-genet-102209-163450. [DOI] [PubMed] [Google Scholar]
- Watillon B, Kettmann R, Arredouani A, Hecquet JF, Boxus P, Burny A. Apple messenger RNAs related to bacterial lignostilbene dioxygenase and plant SAUR genes are preferentially expressed in flowers. Plant Mol. Biol. 1998;36:909–915. doi: 10.1023/a:1005914506110. [DOI] [PubMed] [Google Scholar]
- Wu J, Liu S, He Y, Guan X, Zhu X, Cheng L, Wang J, Lu G. Genome-wide analysis of SAUR gene family in Solanaceae species. Gene. 2012;509:38–50. doi: 10.1016/j.gene.2012.08.002. [DOI] [PubMed] [Google Scholar]
- Xu H, Liu Q, Yao T, Fu X. Shedding light on integrative GA signaling. Curr. Opin. Plant Biol. 2014;21:89–95. doi: 10.1016/j.pbi.2014.06.010. [DOI] [PubMed] [Google Scholar]
- Yamamoto KT, Mori H, Imaseki H. cDNA Cloning of Indole-3-Acetic Acid-Regulated Genes: Aux22 and SAUR from Mung Bean (Vigna radiata) Hypocotyl Tissue. Plant Cell Physiol. 1992;33:93–97. [Google Scholar]
- Yang T, Poovaiah BW. Molecular and biochemical evidence for the involvement of calcium/calmodulin in auxin action. J. Biol. Chem. 2000;275:3137–3143. doi: 10.1074/jbc.275.5.3137. [DOI] [PubMed] [Google Scholar]
- Yang X, Zhang X, Yuan D, Jin F, Zhang Y, Xu J. Transcript profiling reveals complex auxin signalling pathway and transcription regulation involved in dedifferentiation and redifferentiation during somatic embryogenesis in cotton. BMC Plant Biol. 2012;12:110. doi: 10.1186/1471-2229-12-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell. 2005;120:249–259. doi: 10.1016/j.cell.2004.11.044. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Mogami J, Yamaguchi-Shinozaki K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 2014;21:133–139. doi: 10.1016/j.pbi.2014.07.009. [DOI] [PubMed] [Google Scholar]
- Yu X, Li L, Zola J, Aluru M, Ye H, Foudree A, Guo H, Anderson S, Aluru S, Liu P, Rodermel S, Yin Y. A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 2011;65:634–646. doi: 10.1111/j.1365-313X.2010.04449.x. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science. 2001;291:306–309. doi: 10.1126/science.291.5502.306. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Dai X, Blackwell HE, Schreiber SL, Chory J. SIR1, an upstream component in auxin signaling identified by chemical genetics. Science. 2003;301:1107–1110. doi: 10.1126/science.1084161. [DOI] [PubMed] [Google Scholar]
- Zurek DM, Rayle DL, McMorris TC, Clouse SD. Investigation of Gene Expression, Growth Kinetics, and Wall Extensibility during Brassinosteroid-Regulated Stem Elongation. Plant Physiol. 1994;104:505–513. doi: 10.1104/pp.104.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.