Abstract
More than a passive effector of gene expression, mRNA translation (protein synthesis) by the ribosome is a rapidly tunable and dynamic molecular mechanism. Neurodevelopmental disorders are associated with abnormalities in the mRNA translation, protein synthesis, and neocortical development; yet, we know little about the molecular mechanisms behind these abnormalities. Furthermore, our understanding of the regulation of the ribosome and mRNA translation during normal brain development is poor. mRNA translation is emerging as a key driver of the rapid and timed regulation of spatiotemporal gene expression in the developing nervous system, including the neocortex. In this review, we focus on the regulatory role of the ribosome in neocortical development, and construct a current understanding of how ribosomal complex specificity may contribute to the development of the neocortex. We also present a microarray analysis of ribosomal protein-coding mRNAs across the neurogenic phase of neocortical development, in addition to the dynamic enrichment of these mRNAs in actively translating neocortical polysomal ribosomes. Understanding the multivariate control of mRNA translation by ribosomal complex specificity will be critical to reveal the intricate mechanisms of normal brain development and pathologies of neurodevelopmental disorders.
Neocortical development and post-transcriptional processing
The neocortex forms the central neuronal circuit of higher cognitive function and voluntary motor behavior in evolutionarily advanced species (Rakic, 2009; Lui et al., 2011). Development of this complex laminar structure from neural stem cells requires an intricate progression of spatially and temporally controlled molecular events (Molyneaux et al., 2007; Leone et al., 2008, Kriegstein and Alvarez-Buylla, 2009; DeBoer et al., 2013). During neocortical development, neocortical neural stem cells initially divide symmetrically, then transition to multipotent radial glia (RG) progenitors dividing asymmetrically to acquire progressively specific neuronal fates first, followed by glial fates. During the neurogenic phase of neocortical development, the uninterrupted progressive specification of RG is essential to generate a great diversity of glutamatergic pyramidal projection neuron subpopulations, with lower layers born first and upper layers born second.
Each layer’s pyramidal neurons assume a characteristic molecular profile and form specific axonal projections with different targets throughout gestation, laying the foundation for extensive refinements lasting into early adulthood (Molyneaux et al., 2007; Thomson et al., 2007; Petreanu et al., 2009; Rakic, 2009; DeBoer et al., 2013). Lower layer 5 and 6 pyramidal neurons will predominantly project to subcortical structures, including the thalamus and spinal cord. In contrast, upper layers 2 and 3 will solely project intracortically within the ipsilateral hemisphere, or to contralateral targets via the corpus callosum. Thus, the intricate development of neocortical structure lays the foundation for the functional output of its circuits.
Differential transcription factor expression was shown to be a major driver of neuronal fate during RG differentiation (Molyneaux et al., 2007; Leone et al., 2008; Kriegstein and Alvarez-Buylla, 2009; DeBoer et al., 2013). A focus on regulation at the transcriptional level, combined with advances in nucleic acid-based technologies, has led to a series of studies analyzing the genome and transcriptome of the neocortex. The transcriptional signature of the neocortex has been studied throughout pre- and post-natal development (Ayoub et al., 2011; Kang et al., 2011; Hawrylycz et al., 2012; Pletikos et al., 2013; Fertuzinhos et al., 2014; Jaffe et al., 2014; Miller et al., 2014), across neocortical layers (Belgard et al., 2011; Hoerder-Suabedissen et al., 2013), and even at the neuronal-subtype level (Molyneaux et al., 2015). Research on neurological disorders involving the neocortex has also highlighted the transcriptional framework, with many studies on autism spectrum disorders focusing on abnormalities at the genomic (Bartlett et al., 2005; Morrow et al., 2008; Weiss et al., 2009; Pinto et al., 2010; Sanders et al., 2011; Michaelson et al., 2012; O’Roak et al., 2012) and transcriptomic (Voineagu et al., 2011; Parikshak et al., 2013) levels. Other studies have begun to reveal epigenetic signatures in different cortical areas during development, across evolution, aging, and disease (Hirabayashi and Gotoh, 2010; Numata et al., 2012; Petanjek and Kostovic, 2012; Hata et al., 2013; Reilly et al., 2015). Taken together, these excellent studies support the observation that transcriptional regulation determines neuronal subtype differentiation.
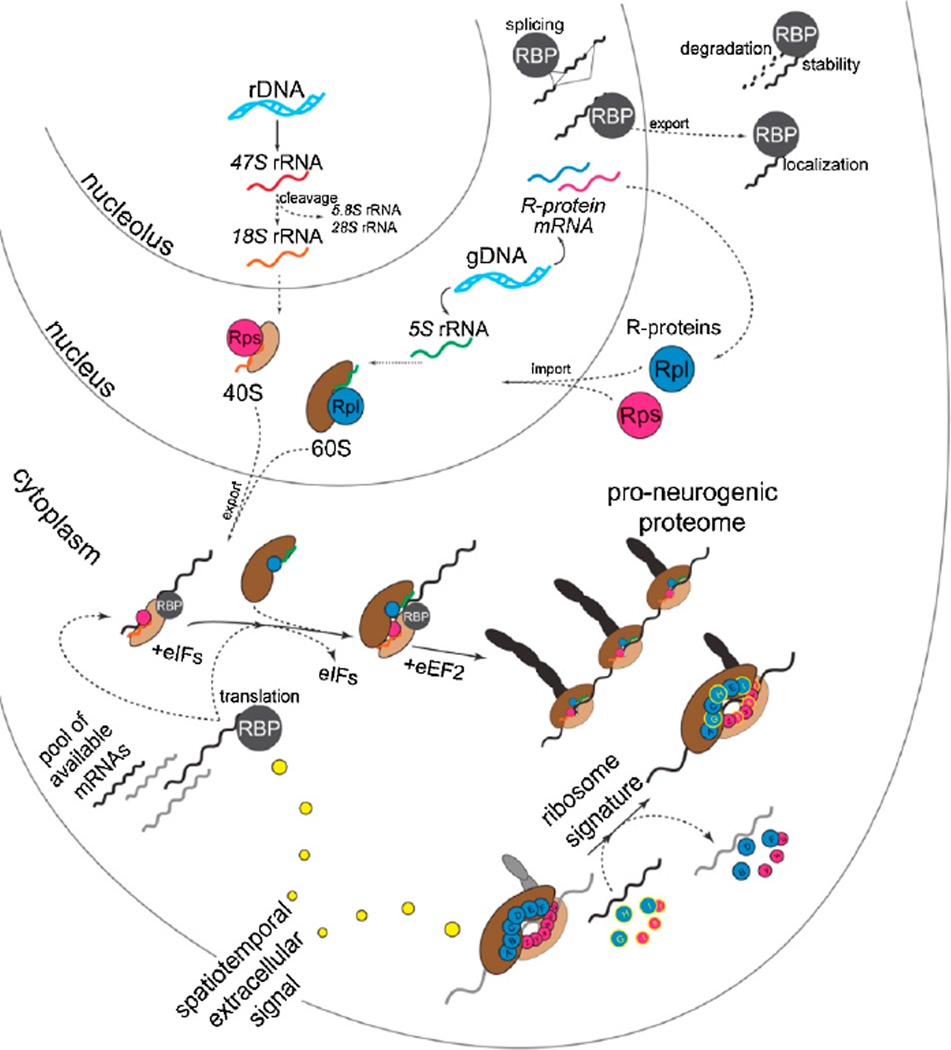
As research continues on the molecular basis of the vast complexity and diversity encompassing neocortical development, it is clear that a large-scale analysis of regulation at the post-transcriptional level is on the horizon. Regulation at the post-transcriptional level is well positioned to amplify the complexity and spatiotemporal specification of neuronal subtypes generated from RG in the developing neocortex (DeBoer et al., 2013; Pilaz and Silver, 2015) (Figure 1). RNA-binding proteins (RBPs) have been shown to play crucial roles in neocortical development, regulating all steps of post-transcriptional processing (splicing, localization, stability/decay, translation), and have been described in detail previously (DeBoer et al., 2013; Pilaz and Silver, 2015). Here, we focus on the final and essential step in functional gene expression, mRNA translation into proteins at the level of the ribosome, during neural development. We propose that the ribosome and its associated factors, constituting a highly dynamic and complex macromolecular machine, drive the spatiotemporal development of the neocortex.
Figure 1. The landscape of post-transcriptional regulation and the ribosome signature in neocortical development.
(Adapted from Kraushar et al., 2015).
Ribosomal complex assembly and diversity
The ribosome is a massive macromolecular complex of 4 rRNA species and at least 79 ribosomal proteins (R-proteins) in eukaryotes, where the intricate process of ribosome assembly – largely studied in yeast – involves over 150 non-ribosomal factors (Fromont-Racine et al., 2003; Henras et al., 2008; Kressler et al., 2010) (Figure 1). The eukaryotic 80S ribosome capable of mRNA translation and found in the cytoplasm is composed of two main subunits, a small 40S subunit and a large 60S subunit. The pathway assembling these subunits with their constituent rRNAs and R-proteins begins in the nucleolus and nucleoplasma with three separate RNA polymerases I-III (Pol-I, Pol-II, Pol-III). In the nucleolus, Pol-I transcribes 3 of the 4 rRNAs from a polycistronic rDNA locus to initially generate a 45S pre-rRNA transcript. The 45S pre-rRNA undergoes extensive site-specific processing, including sequential cleavage, pseudouridylation, and methylation via the concerted action of small nucleolar RNAs (snoRNAs) and ribonucleolar proteins (Tschochner and Hurt, 2003; Henras et al., 2008; Tafforeau et al., 2013), ultimately generating 18S, 5.8S, and 28S rRNAs. The fourth 5S rRNA is transcribed by Pol-III in the nucleoplasma from gDNA at a different locus.
R-proteins are transcribed by Pol-II in the nucleoplasma followed by translation in the cytoplasm, and then imported into the nucleus (Figure 1). It is worth noting that there are ~2,000 genomic sequences encoding at least 79 mammalian R-proteins; while most of these are predicted to be pseudogenes (Balasubramanian et al. 2009; Zhang et al., 2002), in lower-order eukaryotes Rproteins are often encoded by multiple genes acting as functionally distinct paralogues (Xue and Barna, 2012). Pseudogenes themselves have been implicated in the post-transcriptional regulatory framework (Muro et al., 2011), and indeed have been shown to regulate gene expression in the brain (Korneev et al., 1999). In the nucleolus and nucleoplasma, R-proteins complex with the 4 folded rRNAs (18S, 5.8S, 28S, and 5S) to progressively mature pre-ribosomal species. Ultimately, the small 40S subunit is composed of 18S rRNA and 32 R-proteins (Rproteins small; Rps), while the large 60S is composed of 5S, 5.8S, and 28S rRNAs with an additional 47 R-proteins (R-proteins large; Rpl). These mature 40S and 60S subunits exit the nucleoplasma through nuclear pore complexes via facilitated diffusion by a Ran-GTPase cycle.
Once in the cytoplasm, 40S subunits initiate mRNA translation via 5’ cap-dependent or cap-independent mechanisms (Jackson et al., 2010) (Figure 1). Subsequent 60S joining forms 80S ribosomes (monosomes), capable of active peptide elongation predominantly in complexes of multiple 80S ribosomes (polysomes) per mRNA, with monosomes translating a subset of mRNAs (Heyer and Moore, 2016). Active translation occurs until termination complex formation and subunit recycling (Dever and Green, 2012). Ribosomal function during the cycle of mRNA translation is itself facilitated by numerous eukaryotic initiation (eIF) and elongation (eEF) factors, while adopting a highly dynamic range of structural intermediates (Behrmann et al., 2015) within a landscape of energetic favorability (Munro et al., 2009). The activity of translation eIF and eEF co-factors is extensively regulated, largely by post-translational modifications like phosphorylation (Jackson et al., 2010; Dever and Green, 2012). Ribosome biosynthesis to actively translating complexes is an intricate assembly process involving hundreds of non-ribosomal molecules and substantial cellular energy resources. The numerous and complex steps of ribosome assembly afford many opportunities for regulation, modification, or dysfunction – many of which occur between core ribosomal components themselves for quality control and feedback mechanisms (Strunk et al., 2011, 2012; Karbstein, 2013).
While the extensive reviews cited above cover key steps in this process, many aspects of the ribosome biosynthetic pathway remain a mystery. This is particularly the case in higher-order eukaryotes, since the vast majority of previous analyses were performed in yeast and bacteria-derived in vitro systems. This was not previously a matter of contention, as ribosome biosynthesis and mRNA translation mechanisms were considered to be highly conserved. However, recent work strongly points towards system-specific complexity in ribosomal structure and function (Roberts et al., 2008). At the forefront of this research is emerging evidence that tissue patterning in mammalian development, including the neocortex, is specified at the level of the ribosome and mRNA translation (Mauro and Edelman, 2007; Kondrashov et al., 2011; Xue and Barna, 2012; Kraushar et al., 2014, 2015; Xue et al., 2015). Heterogeneity in core ribosomal components and associated co-factors may be a major driving force in the specification of stem cell fate during development (Brombin et al., 2015; Sanchez et al., 2016).
Dynamics of the riboproteome and RNA binding proteins – the ribosome signature
An increasing number of independent groups are providing evidence that ribosomes have differential protein compositions based on localization, developmental-stage, cell-type specificity, and responding to changing environmental conditions (Kondrashov et al., 2011; Xue and Barna, 2012; Reschke et al., 2013; Kraushar et al., 2014, 2015; Brombin et al., 2015; Slavov et al., 2015; Xue et al., 2015). We recently described the developmentally dynamic composition of R-proteins in ribosomal complexes during neocortical development as the neocortical “ribosome signature” (Kraushar et al., 2015).
The working hypothesis behind the ribosome signature is that it is determined by heterogeneity in ribosomal protein composition and the activity of ribosome-associated factors. This is supported by several unbiased mass-spectrometry analyses that identified the existence of ribosomes with distinct protein compositions (Reschke et al., 2013; Kraushar et al., 2015; Slavov et al., 2015). The differential incorporation of R-proteins may have specific physiologic functions, impacting the translation of specific mRNAs via their 5’ and 3’ untranslated regions (UTRs). This was shown in the developing axial skeleton, where differential Rpl38 expression was shown to regulate tissue patterning and mRNA translation by the Hox gene family via an IRES-dependent mechanism in their 5’-UTRs (Xue et al., 2015). Likewise, Rpl13a regulates Ceruloplasmin mRNA translation via its 3’-UTR in response to interferon-γ signaling in the immune system (Mazumder et al., 2003; Kapasi et al., 2007). Related mechanisms may exist in the developing neocortex, as we previously found that the same morphogen (Wnt3) regulating Rpl7 enrichment in neocortical polysomes stimulates the translation of Forkhead box protein P2 (Foxp2) mRNA via its 3’-UTR, driving lower-layer neuron differentiation (Kraushar et al., 2015). The exact mechanisms defining the differential incorporation of R-proteins into heterogeneous ribosome populations remain unclear, but the tracking of R-protein exchange between ribosomes has been described under stress conditions (Pulk et al., 2010). The combinatorial composition and stoichiometry of R-proteins in distinct ribosome species may act in concert with other modifications to core ribosome structure (Vesper et al., 2011; Byrgazov et al., 2013) to define the ribosome signature.
Of the proteins constituting ribosomal complexes, also known as the riboproteome, RBPs were dynamically enriched in a variety of analyzed mouse and human-derived cell types by Reschke and colleagues using stable isotope labeling by amino acids in cell culture (SILAC)-based mass spectrometry (Reschke et al., 2013). This study found a remarkable array of RBPs in complex with the ribosome, which may themselves serve to regulate the ribosome signature. The study of a prominent family of RBPs, the Hu antigens, has provided insight into the precise developmental role that such regulatory molecules can play in mRNA translation (Antic and Keene, 1997; Okano and Darnell, 1997; Lebedeva et al., 2001, Ince-Dunn et al., 2012; Darnell, 2013). Hu antigen R (HuR) was found to regulate the mRNA composition of actively translating ribosomes in neocortical development (Kraushar et al. 2014). Furthermore, deletion of HuR in the developing neocortex impacts the protein components of the translation machinery. Conditional HuR deletion (HuR-cKO) in all neocortical projection neurons affects the post-translational modification of core mRNA translation initiation and elongation factors. In addition, HuR-cKO depletes polysomes of R-proteins, like Rpl7, and subsets of other translation co-factors. Thus, HuR impacts mRNA translation on many levels, including the riboproteome and ribosome signature. HuR further results in agenesis of the corpus collosum and a smaller neocortex. A small neocortex is generally associated with microcephaly and neurodevelopmental dysfunction, reflecting abnormal development of neocortical projection neurons and consequently circuit level aberrations.
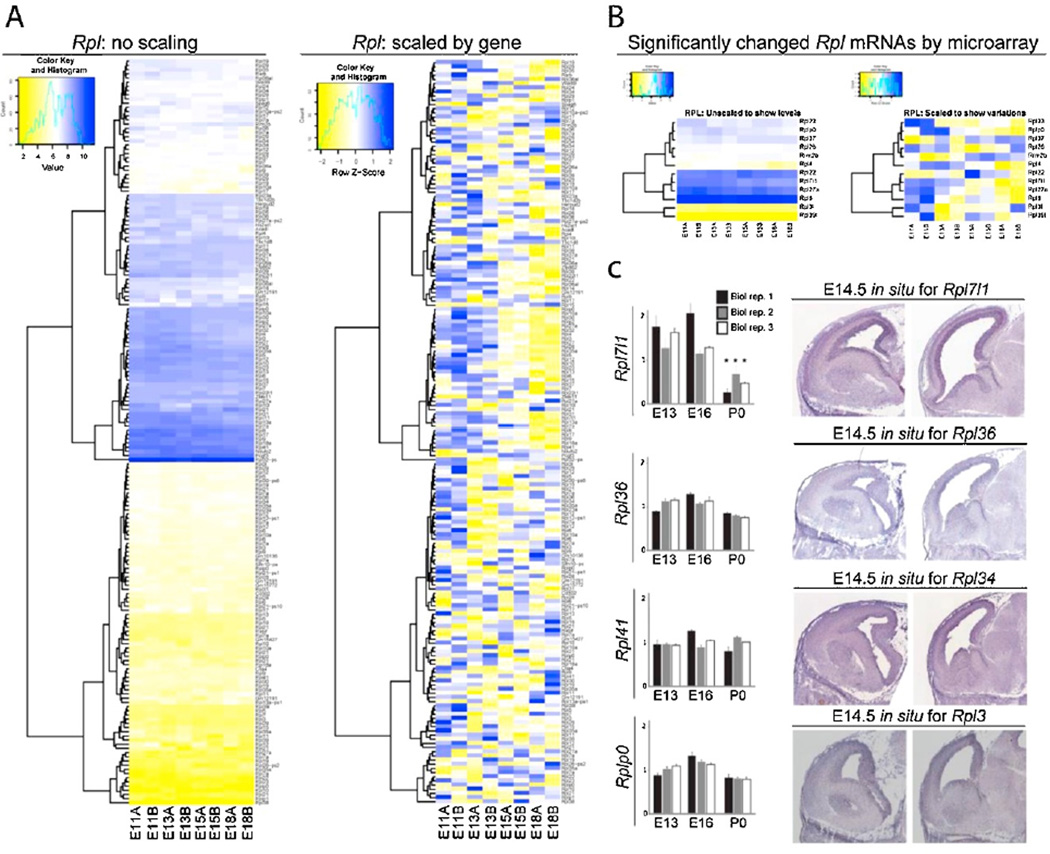
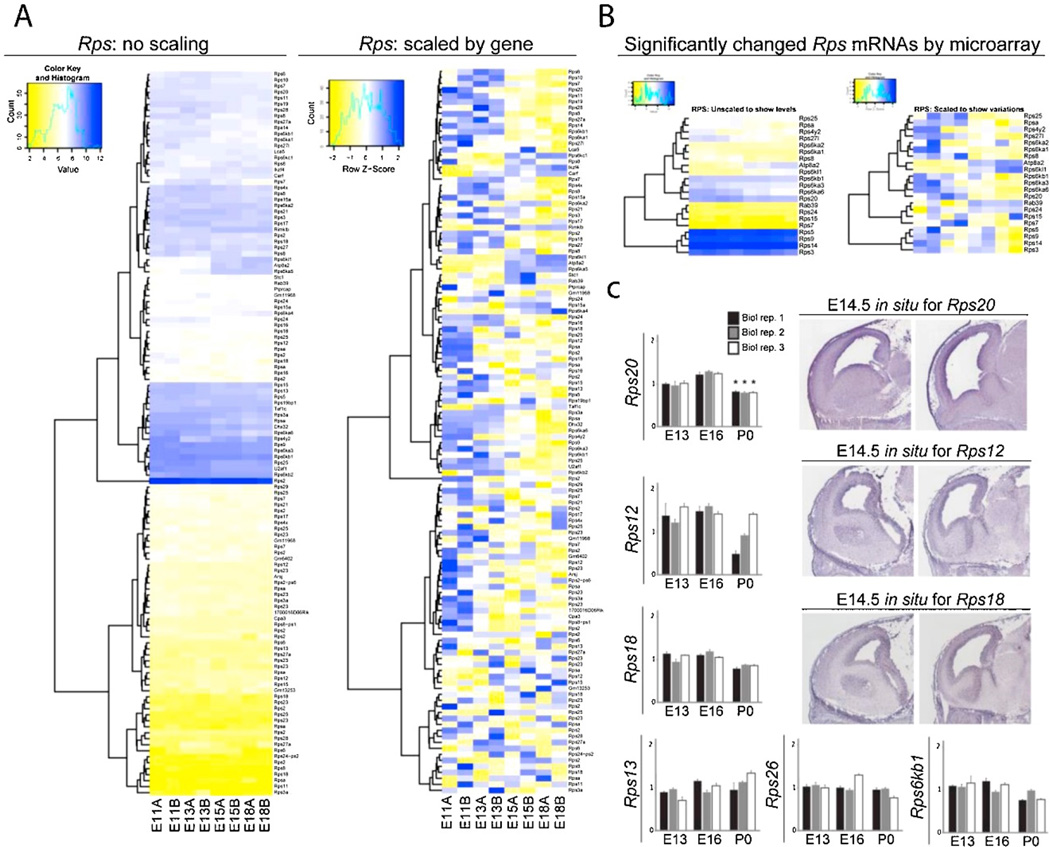
Bioinformatics pathway analysis of previously published microarray data (DeBoer et al., 2013) revealed that many mRNAs encoding R-proteins (R-mRNAs) are expressed across neocortical development (Figures 2A and 3A). The neocortex was analyzed at E11 (onset of neural stem cell proliferation), E13 (RG asymmetric divisions generating lower-layer neurons), E15 (switch to RG asymmetric divisions generating upper-layer neurons), and E18 (resolution of neurogenesis). Further bioinformatic analysis compared the dynamic temporal changes in steady state levels of mRNAs encoding ribosomal proteins between E11, E13, E15 and E18. Heat maps were scaled with respect to distinct gene transcript levels at each adjacent stage, revealing changes in the total expression levels of only a small number of Rpl (6.7%, 12/178) and Rps (17%, 21/119) R-mRNAs (Figures 2B and 3B, respectively). Changes in the levels of R-mRNAs such as Rpl7l1 and Rps20 were confirmed using quantitative real-time PCR (qRT-PCR; Figures 2C and 3C, left). In situ hybridization analysis using available online databases (www.genepaint.org as in DeBoer et al., 2013) showed particular R-mRNA enrichment in the neocortical ventricular zone (VZ) and cortical plate (CP) at E14.5 (Figures 2C and 3C, right). Most of the changes in total neocortical R-mRNA levels occurred during mid-neurogenesis between E15–E18 (Figures 2B and 3B), which reflected the observed dynamics for other post-transcriptional processing genes (DeBoer et al., 2013; Kraushar et al., 2015). While some R-proteins were specifically shown to change in their polysome association despite constant steady state levels (Kraushar et al., 2015), these microarray and qRT-PCR findings suggest that a small subset may be regulated in total levels transcriptionally.
Figure 2. Microarray analysis of neocortical Rpl mRNA expression across development.
(A) Total expression levels of Rpl mRNAs measured in the E11, E13, E15, and E18 neocortex by microarray (DeBoer et al., 2013), shown as unscaled (global levels relative to other mRNAs, left) and scaled for each gene (relative levels over time for each mRNA, right). Unscaled heat maps have been scaled over the samples using the standard RMA method in limma. This was done to show comparative expression levels between genes. The scaled version divides the values in each row (each gene) by its own mean and is therefore expressed as a z-score (see scale). Microarray analysis was performed in duplicate for each developmental stage (A+B). (B) Subset of Rpl mRNAs significantly changing levels across development in the neocortex. (C) qRT-PCR confirmation of Rpl mRNAs measured by microarray to significantly change (Rpl7l1) or remain unchanged (Rpl36, Rpl41, Rplp0) across neocortical development (left, P < 0.05). In situ hybridization (right) in the E14.5 necortex for confirmed R-mRNAs (www.genepaint.org as in DeBoer et al., 2013).
Figure 3. Microarray analysis of neocortical Rps mRNA expression across development. (A–C).
Rps mRNA analysis, as shown in Figure 2 for Rpl mRNAs.
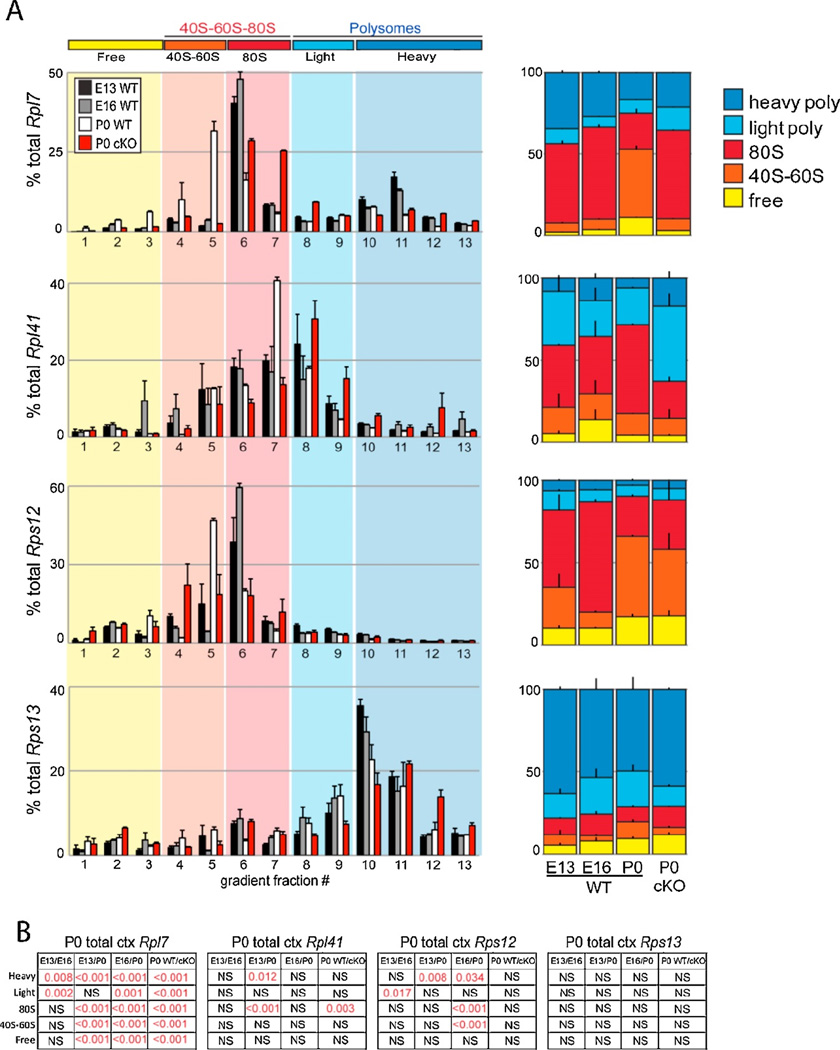
In addition to unbiased mass-spectrometry analysis of the riboproteome, ribosome profiling by RNA sequencing identifies the mRNA targets of dynamic ribosome complexes. Our previous analysis revealed many mRNAs are specifically dynamic in their association with actively translating polysomes across neocortical development, while their overall steady-state levels remain unchanged (Kraushar et al., 2014, 2015). These translationally regulated mRNAs largely code for either transcription or translation pathway genes, including R-mRNAs. We performed sucrose density gradient ultracentrifugation and fractionation of E13, E16, and P0 WT neocortex followed by RNA isolation and qRT-PCR measurement of R-mRNA levels in 40S-60S-80S and polysome complexes, selecting R-mRNAs whose steady-state levels were not found to change previously by RNAseq (Figure 4). The results confirmed the RNAseq data, showing developmentally dynamic shifts in R-mRNAs associated with 40S-60S-80S and polysomal fractions between E13, E16, and P0. The Rpl7 transcript was reduced in polysomes over time but unchanged in total levels; this was determined in both GO and KEGG analyses and was confirmed by qRT-PCR. In contrast, Rps13 did not vary significantly in GO analysis, and was shown to be unchanged in polysomes during development. Similar results for other Rpl and Rps mRNAs confirmed dynamic changes in association with 40S-60S-80S and polysomal fractions. Further more stringent statistical testing with a Monte Carlo-based simulation approach (Kraushar et al., 2014) was also performed, which reinforced the significance of Rpl7 and Rps12, while Rps13 and 18S rRNA (not shown) remained insignificant. Consistent with previous findings (Kruashar et al., 2015), these results suggest that temporally regulated shifts in the association of distinct mRNA populations with polysomes, including R-mRNAs, begin at E16 and continue to P0.
Figure 4. R-mRNA translation across neocortical development.
(A) qRT-PCR analysis of Rpl7, Rpl41, Rps12, and Rps13 R-mRNAs in WT E13, E16, P0 vs. P0 Kcnc2-Cre/Wnt3flf (cKO) sucrose density gradient fractionations. These R-mRNAs were found to have unchanged steady-state levels across neocortical development, with a subset changing levels specifically in 40S-60S-80S and/or polysome complexes, as measured by RNAseq previously (Kraushar et al., 2015). mRNA levels in individual fractions (left), or grouped by free (1–3), 40S–60S (4–5), 80S (6–7), light (8–9), and heavy (10–13) polysome associated fractions (right) are shown for each RmRNA (n=2–6 neocortices, 1–3 fractionations, in duplicate/condition). (B) Statistical analysis of (A), significant comparisons highlighted in red (ANOVA, Tukey’s HSD post hoc).
Recently, Cho and colleagues performed a similar analysis of translational efficiency in the hippocampus during the acute and long-term phases of memory encoding in the hippocampus (Cho et al., 2015). Ribosome profiling of translationally active mRNAs normalized to the steady-state transcriptome measured by RNAseq revealed the translational regulation of mRNAs in response to a memory task. Surprisingly, the predominant trend was towards translational suppression in the acute phase of memory encoding in the hippocampus, and particularly suppression of R-mRNA expression. While the regulation of R-mRNA translation has been described to occur in both 5’-terminal oligopyrimidine tract (5’-TOP) (Meyuhas, 2000) and 3’-polyA dependent (Subtelny et al., 2014) mechanisms in other systems, its role in the brain is unclear. Intriguingly, early work on the regulation of balanced R-protein expression in prokaryotes found that R-proteins are capable of regulating the translation of their own R-mRNAs (Nomura et al., 1984), suggesting a translationally-based feeback mechanism regulating ribosome assembly. This regulation may further include the splicing of pre-R-mRNA transcripts influenced by R-proteins in eukaryotes (Mitrovich and Anderson, 2000; Ivanov et al., 2006; Malygin et al., 2007).
While the picture in eukaryotes is less clear, our previous findings of increasing Rpl7 protein in neocortical polysomes at later developmental stages (Kraushar et al., 2015) in concert with decreasing polysomal Rpl7 R-mRNA (Figure 4) may reflect similar feedback mechanisms. Furthermore, there may be an overlap between translationally-specific regulation in neocortical development (Kraushar et al., 2015) (Figure 4) and memory formation (Cho et al., 2015), also evidenced by the regulation of eIF2 phosphorylation in both contexts (Costa-Mattioli et al., 2005, 2007; Kraushar et al., 2014). Taken together, these findings point to a mechanism of regulation that is tightly controlled in two intricate processes: brain development and memory formation. The result of this is profound: an exceptionally complex organ that can encode and recall its experiences - no small feat for a group of translation-specific molecules.
The neocortical ribosome signature and extracellular factors
Timed extracellular signaling events determine normal neocortical development. One of these major developmental events is the ingrowth of thalamic axons at mid-neurogenesis, which in mouse occurs around embryonic days 15–16 (López-Bendito and Molnár, 2003), and may further impact both human neurodevelopment (Kostović and Judaš, 2007; Fair et al., 2010; Toulmin et al., 2015) and consequently disorders like autism (Nair et al., 2013) and schizophrenia (Woodward et al., 2012; Woodward and Heckers, 2014). The timed ingrowth of thalamocortical axons is accompanied by secretion of extracellular factors into the developing neocortex.
Extracellular factors have been shown to regulate mRNA translation specificity in multiple neuronal systems (Campbell and Holt, 2001; Petroulakis and Wang, 2002; Schratt, 2004). For example, the BDNF trophic factor regulates translation of a select group of mRNAs during neuronal development, specifically within neuronal dendrites in a mammalian target of rapamycin (mTOR)-dependent pathway (Schratt, 2004). Thalamocortical axons secrete a member of the Wnt morphogen family, Wingless-related MMTV integration site 3 (Wnt3), which regulates the neocortical ribosome signature by targeted mRNA translation and specifying the R-protein composition neocortical polysomes (Kraushar et al., 2015). Thalamocortical Wnt3 stimulates the enrichment of Rpl7 in polysomes of the sensorimotor neocortex, and further promotes neocortical neuronal differentiation while suppressing oligodendrocyte differentiation, in a translation-dependent manner. Specifically, Wnt3 drives the differentiation of Foxp2+ lower-layer neocortical neurons by the translation of Foxp2 mRNA via its 3’-UTR. Furthermore, thalamic Wnt3-cKO influences the translation of R-mRNAs, including Rpl7 (Figure 4, “cKO”). Taken together, these data suggest that timed signaling during neocortical development acts at multiple steps to regulate the neocortical riboproteome and specific mRNA translation, thus regulating the neocortical ribosome signature.
Ribosomes and mRNA translation in neurodevelopmental disorders
Ribosomopathies are associated with mRNA translation deficiency, cell cycle arrest, and neurodevelopmental disorders (Narla and Ebert, 2011; Teng et al., 2013; Brooks et al., 2014). In particular, an X-linked missense mutation in a single R-protein, Rpl10, results in microcephaly, cognitive deficits, and seizures in humans (Brooks et al., 2014). The developmental impact of Rpl10 deficiency was confirmed in this study with an Rpl10-targeting morpholino in zebrafish, resulting in microcephaly and a decrease in global mRNA translation. Rpl10 is enriched in actively translating ribosomes in the early developing neocortex and decreases by the postnatal period (Kraushar et al., 2015). As neurodevelopmental disorders have been linked to both abnormal thalamocortical connectivity (Kostović and Judaš, 2007; Woodward et al., 2012; Woodward and Heckers, 2014) and RBPs, such as dysmorphic dendrites, cognitive dysfunction, and seizure susceptibility in HuD KO mice (DeBoer et al., 2014), it is conceivable that even slight alterations in regulatory signaling to the ribosome signature and mRNA translation during neocortical development could lead to life-long structural and functional brain abnormalities.
Many human and animal model studies of Autism Spectrum Disorders (ASDs) have implicated genes and pathways that converge on mRNA translation in the cortex as a target in ASD etiology (Kelleher and Bear, 2008; Park et al., 2008; Gkogkas et al., 2013; Santini et al., 2013). This is reinforced by ASD subtypes that are caused by defective genes associated with mRNA translation (Kelleher and Bear, 2008), which are expressed in the developing neocortex. In parallel, abnormal neocortical development is strongly implicated in ASDs (State and Šestan, 2012). However, while the regulation of core translation components has been implicated in ASDs (Park et al., 2008; Darnell et al., 2011; Gkogkas et al., 2013), how their dysfunction leads to abnormal neocortical development remains unanswered and a key direction for future research.
Conclusions and future directions
Recent research has revealed the centrality of post-transcriptional regulation in brain development and function. RBPs, for example, have been shown to play a major role in neocortical development. It has also begun to hint at the heterogeneity of the ribosome and the particular role that it plays in development. However, there is still a significant gap in knowledge regarding the role that post-transcriptional regulation plays in normal and abnormal brain development. The interplay between heterogeneous ribosome complexes and their capability to both promote and suppress translation of a specific mRNA in a given place and time will be particularly critical to understand brain development and disease. This may act in concert with the regulation by translation co-factors like eIF2 as described above, and eIF4E which was shown to regulate the pool of pro-neurogenic mRNAs bound in granules with the repressor 4E-T in the neocortex (Yang et al., 2014). Furthermore, modulation of mRNA translation rate itself regulates neocortical neurogenesis of upper layer neurons derived from intermediate progenitor cells, via a mechanism involving addition of 5-methoxycarbonylmethyl (mcm5) and 5-carbamoylmethyl (ncm5) groups to U34 in 11 tRNA species (Laguesse et al., 2015). Conditional deletion of the Elongator (Elp3) enzyme responsible for these tRNA modifications in neural progenitors triggers the unfolded protein response (UPR) and inhibition of indirect upper-layer neurogenesis. Thus, future work will be required to elucidate both specific targeting and general regulation by the mRNA translation apparatus in neocortical neurogenesis. As the findings of our group and others have shown, many changes to mRNA translation complexes occur at mid-neurogenesis, overlapping with the switch in lower- to upper-layer neurogenesis. How this timed posttranscriptional regulation of gene expression serves to specify neuronal subtypes will be the focus of future efforts. Overall, the field should strive towards as deep an understanding of posttranscriptional regulation as we have of transcriptional regulation.
Future studies will determine ribosome signature specificity in developing complex systems, such as the neocortex with its many circuits, neuronal subpopulations, and sub-compartments. Along these lines, one interesting question to consider is the specificity of the ribosome signature and mRNA translation regulation in dendritic compartments, since local mRNA translation is a critical mechanism in developing neurites and mature dendrites (Bramham and Wells, 2007; Kong and Lasko, 2012). The ribosome signature provides an unexpected and intriguing level of complexity in the rapid and timed regulation of functional gene expression. From transcription, splicing, localization, stability/degradation, and finally translation, the journey of mRNA transcripts between the genome to functional proteome is a long one (Figure 1). At each of these steps, regulation multiplies the possible outcomes, amplifying complexity in the molecular specification of neural stem cells to differentiated functional neuronal circuits in the neocortex.
Highlights.
-
-
mRNA translation (protein synthesis) by the ribosome is a rapidly tunable and dynamic molecular mechanism in developing nervous system
-
-
ribosomes have differential protein compositions based on localization, developmental-stage, and cell-type specificity
-
-
timed extracellular signals dictate ribosome and mRNA translation specificity
Acknowledgments
This work was supported by National Institutes of Health (NIH) grants (NS064303 and NS075367) to M.R.R. M.L.K. was the recipient of a Kirschstein National Research Service Award F30 NIH Fellowship (1F30MH106220). We thank many individuals at our institution for their critical input to the manuscript and without whom this work would have been incomplete. A special thank you to Dr. Ronald P. Hart (Rutgers University) for his continuous support and bioinformatics analysis across many years of our work, part of which we included here.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Antic D, Keene JD. Embryonic Lethal Abnormal Visual RNA-Binding Proteins Involved in Growth, Differentiation, and Posttranscriptional Gene Expression. Am J Hum Genet. 1997;61:273–278. doi: 10.1086/514866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayoub A, Oh S, Xie Y, Leng J, Cotney J, Dominguez MH, Noonan JP, Rakic P. Transcriptional programs in transient embryonic zones of the cerebral cortex defined by high-resolution mRNA sequencing. Proc Natl Acad Sci USA. 2011;108:14950–14955. doi: 10.1073/pnas.1112213108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S, Zheng D, Liu Y-J, Fang G, Frankish A, Carriero N, Robilotto R, Cayting P, Gerstein M. Comparative analysis of processed ribosomal protein pseudogenes in four mammalian genomes. Genome Biol. 2009;10:R2–R2.10. doi: 10.1186/gb-2009-10-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett CW, Gharani N, Millonig JH, Brzustowicz LM. Three autism candidate genes: A synthesis of human genetic analysis with other disciplines. Int J Dev Neurosci. 2005;23:221–234. doi: 10.1016/j.ijdevneu.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Behrmann E, Loerke J, Budkevich TV, Yamamoto K, Schmidt A, Penczek PA, Vos MR, Bürger J, Mielke T, Scheerer P, Spahn CMT. Structural Snapshots of Actively Translating Human Ribosomes. Cell. 2015;161:845–857. doi: 10.1016/j.cell.2015.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgard TG, Marques AC, Oliver PL, Abaan HO, Sirey TM, Hoerder-suabedissen A, Garcia-Moreno F, Molnár Z, Margulies EH, Ponting CP. NeuroResource: A Transcriptomic Atlas of Mouse Neocortical Layers. Neuron. 2011;71:605–616. doi: 10.1016/j.neuron.2011.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramham CR, Wells DG. Dendritic mRNA: transport, translation and function. Nat Rev Neurosci. 2007;8:776–789. doi: 10.1038/nrn2150. [DOI] [PubMed] [Google Scholar]
- Brombin A, Joly J-S, Jamen F. New tricks for an old dog: ribosome biogenesis contributes to stem cell homeostasis. Curr Opin Genet Dev. 2015;34:61–70. doi: 10.1016/j.gde.2015.07.006. [DOI] [PubMed] [Google Scholar]
- Brooks SS, Wall AL, Golzio C, Reid DW, Kondyles A, Willer JR, Botti C, Nicchitta CV, Katsanis N, Davis EE. A Novel Ribosomopathy Caused by Dysfunction of X-Linked Microcephaly in Humans. Genetics. 2014;198:723–733. doi: 10.1534/genetics.114.168211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrgazov K, Vesper O, Moll I. Ribosome heterogeneity: Another level of complexity in bacterial translation regulation. Curr Opin Microbiol. 2013;16:133–139. doi: 10.1016/j.mib.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell DS, Holt CE. Chemotropic responses of retinal growth cones mediated by rapid local protein synthesis and degradation. Neuron. 2001;32:1013–1026. doi: 10.1016/s0896-6273(01)00551-7. [DOI] [PubMed] [Google Scholar]
- Cho J, Yu N-K, Choi J-H, Sim S-E, Kang SJ, Kwak C, Lee S-W, Kim J-i, Choi D, Il, Kim VN, Kaang B-K. Multiple repressive mechanisms in the hippocampus during memory formation. Science. 2015;350:82–87. doi: 10.1126/science.aac7368. [DOI] [PubMed] [Google Scholar]
- Costa-Mattioli M, Gobert D, Harding H, Herdy B, Azzi M, Bruno M, Bidinosti M, Ben Mamou C, Marcinkiewicz E, Yoshida M, Imataka H, Cuello AC, Seidah N, Sossin W, Lacaille J-C, Ron D, Nader K, Sonenberg N. Translational control of hippocampal synaptic plasticity and memory by the eIF2α kinase GCN2. Nature. 2005;436:1166–1173. doi: 10.1038/nature03897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Mattioli M, Gobert D, Stern E, Gamache K, Colina R, Cuello C, Sossin W, Kaufman R, Pelletier J, Rosenblum K, Krnjevic K, Lacaille JC, Nader K, Sonenberg N. eIF2alpha Phosphorylation Bidirectionally Regulates the Switch from Short- to Long-Term Synaptic Plasticity and Memory. Cell. 2007;129:195–206. doi: 10.1016/j.cell.2007.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KYS, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, Licatalosi DD, Richter JD, Darnell RB. FMRP Stalls Ribosomal Translocation on mRNAs Linked to Synaptic Function and Autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell RB. RNA protein interaction in neurons. Annu Rev Neurosci. 2013;36:243–270. doi: 10.1146/annurev-neuro-062912-114322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBoer EM, Azevedo R, Vega TA, Brodkin J, Akamatsu W, Okano H, Wagner GC, Rasin M-R. Prenatal Deletion of the RNA-Binding Protein HuD Disrupts Postnatal Cortical Circuit Maturation and Behavior. J Neurosci. 2014;34:3674–3686. doi: 10.1523/JNEUROSCI.3703-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBoer EM, Kraushar ML, Hart RP, Rasin M-R. Post-transcriptional regulatory elements and spatiotemporal specification of neocortical stem cells and projection neurons. Neuroscience. 2013;248:499–528. doi: 10.1016/j.neuroscience.2013.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE, Green R. The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb Perspect Biol. 2012;4:a013706. doi: 10.1101/cshperspect.a013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fair DA, Bathula D, Mills KL, Dias TGC, Blythe MS, Zhang D, Snyder AZ, Raichle ME, Stevens Aa, Nigg JT, Nagel BJ. Maturing thalamocortical functional connectivity across development. Front Syst Neurosci. 2010;4(10):1–10. doi: 10.3389/fnsys.2010.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fertuzinhos S, Li M, Kawasawa YI, Ivic V, Franjic D, Singh D, Crair M, Sestan N. Laminar and temporal expression dynamics of coding and noncoding RNAs in the mouse neocortex. Cell Rep. 2014;6:938–950. doi: 10.1016/j.celrep.2014.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromont-Racine M, Senger B, Saveanu C, Fasiolo F. Ribosome assembly in eukaryotes. Gene. 2003;313:17–42. doi: 10.1016/s0378-1119(03)00629-2. [DOI] [PubMed] [Google Scholar]
- Gkogkas CG, Khoutorsky A, Ran I, Rampakakis E, Nevarko T, Weatherill DB, Vasuta C, Yee S, Truitt M, Dallaire P, Major F, Lasko P, Ruggero D, Nader K, Lacaille J-C, Sonenberg N. Autism-related deficits via dysregulated eIF4E-dependent translational control. Nature. 2013;493:371–377. doi: 10.1038/nature11628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata K, Mizukami H, Sadakane O, Watakabe A, Ohtsuka M, Takaji M, Kinoshita M, Isa T, Ozawa K, Yamamori T. DNA methylation and methyl-binding proteins control differential gene expression in distinct cortical areas of macaque monkey. J Neurosci. 2013;33:19704–19714. doi: 10.1523/JNEUROSCI.2355-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawrylycz MJ, Lein ES, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras AK, Soudet J, Gérus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol life Sci. 2008;65:2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer EE, Moore MJ. Redefining the Translational Status of 80S Monosomes. Cell. 2016;164:757–769. doi: 10.1016/j.cell.2016.01.003. [DOI] [PubMed] [Google Scholar]
- Hirabayashi Y, Gotoh Y. Epigenetic control of neural precursor cell fate during development. Nat Rev Neurosci. 2010;11:377–388. doi: 10.1038/nrn2810. [DOI] [PubMed] [Google Scholar]
- Hoerder-Suabedissen A, Oeschger FM, Krishnan ML, Belgard TG, Wang WZ, Lee S, Webber C, Petretto E, Edwards AD, Molnár Z. Expression profiling of mouse subplate reveals a dynamic gene network and disease association with autism and schizophrenia. Proc Natl Acad Sci USA. 2013;110:3555–3560. doi: 10.1073/pnas.1218510110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ince-Dunn G, Okano HJ, Jensen KB, Park WY, Zhong R, Ule J, Mele A, Fak JJ, Yang C, Zhang C, Yoo J, Herre M, Okano H, Noebels JL, Darnell RB. Neuronal Elav-like (Hu) Proteins Regulate RNA Splicing and Abundance to Control Glutamate Levels and Neuronal Excitability. Neuron. 2012;75:1067–1079. doi: 10.1016/j.neuron.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov AV, Malygin AA, Karpova GG. Eukaryotic ribosomal proteins: Interactions with their own pre-mRNAs and their involvement in splicing regulation. Mol Biol. 2006;40:570–578. [PubMed] [Google Scholar]
- Jackson RJ, Hellen CUT, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe AE, Shin J, Collado-Torres L, Leek JT, Tao R, Li C, Gao Y, Jia Y, Maher BJ, Hyde TM, Kleinman JE, Weinberger DR. Developmental regulation of human cortex transcription and its clinical relevance at single base resolution. Nat Neurosci. 2014;18:154–161. doi: 10.1038/nn.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapasi P, Chaudhuri S, Vyas K, Baus D, Komar AA, Fox PL, Merrick WC, Mazumder B. L13a blocks 48S assembly: role of a general initiation factor in mRNA-specific translational control. Mol Cell. 2007;25:113–126. doi: 10.1016/j.molcel.2006.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbstein K. Quality control mechanisms during ribosome maturation. Trends Cell Biol. 2013;23:242–250. doi: 10.1016/j.tcb.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher RJ, Bear MF. The autistic neuron: troubled translation? Cell. 2008;135:401–406. doi: 10.1016/j.cell.2008.10.017. [DOI] [PubMed] [Google Scholar]
- Kondrashov N, Pusic A, Stumpf CR, Shimizu K, Hsieh AC, Xue S, Ishijima J, Shiroishi T, Barna M. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell. 2011;145:383–397. doi: 10.1016/j.cell.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong J, Lasko P. Translational control in cellular and developmental processes. Nat Rev Genet. 2012;13:383–394. doi: 10.1038/nrg3184. [DOI] [PubMed] [Google Scholar]
- Korneev SA, Park J, O’Shea M. Neuronal Expression of Neural Nitric Oxide Synthase (nNOS) Protein Is Suppressed by an Antisense RNA Transcribed from an NOS Pseudogene. J Neurosci. 1999;19:7711–7720. doi: 10.1523/JNEUROSCI.19-18-07711.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostović I, Judaš M. Transient patterns of cortical lamination during prenatal life: Do they have implications for treatment? Neurosci Biobehav Rev. 2007;31:1157–1168. doi: 10.1016/j.neubiorev.2007.04.018. [DOI] [PubMed] [Google Scholar]
- Kraushar ML, Thompson K, Wijeratne HRS, Viljetic B, Sakers K, Marson JW, Kontoyiannis DL, Buyske S, Hart RP, Rasin M-R. Temporally defined neocortical translation and polysome assembly are determined by the RNA-binding protein Hu antigen R. Proc Natl Acad Sci USA. 2014;111:E3815–E3824. doi: 10.1073/pnas.1408305111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraushar ML, Viljetic B, Wijeratne HRS, Thompson K, Jiao X, Pike JW, Medvedeva V, Groszer M, Kiledjian M, Hart RP, Rasin M-R. Thalamic WNT3 Secretion Spatiotemporally Regulates the Neocortical Ribosome Signature and mRNA Translation to Specify Neocortical Cell Subtypes. J Neurosci. 2015;35:10911–10926. doi: 10.1523/JNEUROSCI.0601-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D, Hurt E, Baßler J. Driving ribosome assembly. Biochim Biophys Acta. 2010;1803:673–683. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- Kriegstein A, Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguesse S, Creppe C, Nedialkova DD, Prévot P-P, Borgs L, Huysseune S, Franco B, Duysens G, Krusy N, Lee G, Thelen N, Thiry M, Close P, Chariot A, Malgrange B, Leidel SA, Godin JD, Nguyen L. A Dynamic Unfolded Protein Response Contributes to the Control of Cortical Neurogenesis. Dev Cell. 2015;35:553–567. doi: 10.1016/j.devcel.2015.11.005. [DOI] [PubMed] [Google Scholar]
- Lebedeva S, Jens M, Theil K, Schwanhäusser B, Selbach M, Lanthaler M, Rajewsky N. Transcriptome-wide Analysis of Regulatory Interactions of the RNA-Binding Protein HuR. Mol Cell. 2011;43:340–352. doi: 10.1016/j.molcel.2011.06.008. [DOI] [PubMed] [Google Scholar]
- Leone DP, Srinivasan K, Chen B, Alcamo E, McConnell SK. The determination of projection neuron identity in the developing cerebral cortex. Curr Opin Neurobiol. 2008;18:28–35. doi: 10.1016/j.conb.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Bendito G, Molnár Z. Thalamocortical development: how are we going to get there? Nat Rev Neurosci. 2003;4:276–289. doi: 10.1038/nrn1075. [DOI] [PubMed] [Google Scholar]
- Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malygin AA, Parakhnevitch NM, Ivanov AV, Eperon IC, Karpova GG. Human ribosomal protein S13 regulates expression of its own gene at the splicing step by a feedback mechanism. Nucleic Acids Res. 2007;35:6414–6423. doi: 10.1093/nar/gkm701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauro VP, Edelman GM. The ribosome filter redux. Cell Cycle. 2007;6:2246–2251. doi: 10.4161/cc.6.18.4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder B, Sampath P, Seshadri V. Regulated Release of L13a from the 60S Ribosomal Subunit as A Mechanism of Transcript-Specific Translational Control. Cell. 2003;115:187–198. doi: 10.1016/s0092-8674(03)00773-6. [DOI] [PubMed] [Google Scholar]
- Meyuhas O. Synthesis of the translational apparatus is regulated at the translational level. Eur J Biochem. 2000;267:6321–6330. doi: 10.1046/j.1432-1327.2000.01719.x. [DOI] [PubMed] [Google Scholar]
- Michaelson JJ, et al. Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell. 2012;151:1431–1442. doi: 10.1016/j.cell.2012.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JA, et al. Transcriptional landscape of the prenatal human brain. Nature. 2014;508:199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrovich QM, Anderson P. Unproductively spliced ribosomal protein mRNAs are natural targets of mRNA surveillance in C. elegans. Genes Dev. 2000;14:2173–2184. doi: 10.1101/gad.819900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Menezes JRL, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Goff LA, Brettler AC, Chen H-H, Brown JR, Hrvatin S, Rinn JL, Arlotta P. DeCoN: Genome-wide Analysis of In Vivo Transcriptional Dynamics during Pyramidal Neuron Fate Selection in Neocortex. Neuron. 2015;85:275–288. doi: 10.1016/j.neuron.2014.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow EM, et al. Identifying autism loci and genes by tracing recent shared ancestry. Science. 2008;321:218–223. doi: 10.1126/science.1157657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro JB, Sanbonmatsu KY, Spahn CMT, Blanchard SC. Navigating the ribosome’s metastable energy landscape. Trends Biochem Sci. 2009;34:390–400. doi: 10.1016/j.tibs.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro EM, Mah N, Andrade-Navarro MA. Functional evidence of post-transcriptional regulation by pseudogenes. Biochimie. 2011;93:1916–1921. doi: 10.1016/j.biochi.2011.07.024. [DOI] [PubMed] [Google Scholar]
- Nair A, Treiber JM, Shukla DK, Shih P, Muller RA. Impaired thalamocortical connectivity in autism spectrum disorder: A study of functional and anatomical connectivity. Brain. 2013;136:1942–1955. doi: 10.1093/brain/awt079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narla A, Ebert BL. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2011;115:3196–3205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura M, Gourse R, Baughman G. Regulation of the synthesis of ribosomes and ribosomal components. Annu Rev Biochem. 1984;53:75–117. doi: 10.1146/annurev.bi.53.070184.000451. [DOI] [PubMed] [Google Scholar]
- Numata S, Ye T, Hyde TM, Guitart-Navarro X, Tao R, Wininger M, Colantuoni C, Weinberger DR, Kleinman JE, Lipska BK. DNA methylation signatures in development and aging of the human prefrontal cortex. Am J Hum Genet. 2012;90:260–272. doi: 10.1016/j.ajhg.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano HJ, Darnell RB. A hierarchy of Hu RNA binding proteins in developing and adult neurons. J Neurosci. 1997;17:3024–3037. doi: 10.1523/JNEUROSCI.17-09-03024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikshak NN, Luo R, Zhang A, Won H, Lowe JK, Chandran V, Horvath S, Geschwind DH. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell. 2013;155:1008–1021. doi: 10.1016/j.cell.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Park JM, Kim S, Kim J-A, Shepherd JD, Smith-Hicks CL, Chowdhury S, Kaufmann W, Kuhl D, Ryazanov AG, Huganir RL, Linden DJ, Worley PF. Elongation factor 2 and fragile X mental retardation protein control the dynamic translation of Arc/Arg3.1 essential for mGluR-LTD. Neuron. 2008;59:70–83. doi: 10.1016/j.neuron.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petanjek Z, Kostović I. Epigenetic regulation of fetal brain development and neurocognitive outcome. Proc Natl Acad Sci USA. 2012;109:11062–11063. doi: 10.1073/pnas.1208085109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petreanu L, Mao T, Sternson SM, Svoboda K. The subcellular organization of neocortical excitatory. Nature. 2009;457:1142–1145. doi: 10.1038/nature07709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroulakis E, Wang E. Nerve growth factor specifically stimulates translation of eukaryotic elongation factor 1A-1 (eEF1A-1) mRNA by recruitment to polyribosomes in PC12 cells. J Biol Chem. 2002;277:18718–18727. doi: 10.1074/jbc.M111782200. [DOI] [PubMed] [Google Scholar]
- Pilaz L-J, Silver DL. Post-transcriptional regulation in corticogenesis: how RNA-binding proteins help build the brain. WIREs RNA. 2015;6:501–515. doi: 10.1002/wrna.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletikos M, Sousa AMM, Sedmak G, Meyer KA, Zhu Y, Cheng F, Li M, Kawasawa YI, Sestan N. Temporal Specification and Bilaterality of Human Neocortical Topographic Gene Expression. Neuron. 2013:1–12. doi: 10.1016/j.neuron.2013.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulk A, Liiv A, Peil L, Maiväli Ü, Nierhaus K, Remme J. Ribosome reactivation by replacement of damaged proteins. Mol Microbiol. 2010;75:801–814. doi: 10.1111/j.1365-2958.2009.07002.x. [DOI] [PubMed] [Google Scholar]
- Rakic P. Evolution of the neocortex: a perspective from developmental biology. Nat Rev Neurosci. 2009;10:724–735. doi: 10.1038/nrn2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly SK, Yin J, Ayoub AE, Emera D, Leng J, Cotney J, Sarro R, Rakic P, Noonan JP. Evolutionary changes in promoter and enhancer activity during human corticogenesis. Science. 2015;347:1155–1159. doi: 10.1126/science.1260943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reschke M, Clohessy JG, Seitzer N, Goldstein DP, Breitkopf SB, Schmolze DB, Ala U, Asara JM, Beck AH, Pandolfi P. Characterization and Analysis of the Composition and Dynamics of the Mammalian Riboproteome. Cell Rep. 2013;4:1276–1287. doi: 10.1016/j.celrep.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts E, Sethi A, Montoya J, Woese CR, Luthey-Schulten Z. Molecular signatures of ribosomal evolution. Proc Natl Acad Sci USA. 2008;105:13953–13958. doi: 10.1073/pnas.0804861105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez CGG, Teixeira FKK, Czech B, Preall JBB, Zamparini ALL, Seifert JRKRK, Malone CDD, Hannon GJJ, Lehmann R. Regulation of Ribosome Biogenesis and Protein Synthesis Controls Germline Stem Cell Differentiation. Cell Stem Cell. 2016;18:276–290. doi: 10.1016/j.stem.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, et al. Multiple Recurrent De Novo CNVs, Including Duplications of the 7q11.23 Williams Syndrome Region, Are Strongly Associated with Autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santini E, Huynh TN, MacAskill AF, Carter AG, Pierre P, Ruggero D, Kaphzan H, Klann E. Exaggerated translation causes synaptic and behavioural aberrations associated with autism. Nature. 2013;493:411–415. doi: 10.1038/nature11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt GM. BDNF Regulates the Translation of a Select Group of mRNAs by a Mammalian Target of Rapamycin-Phosphatidylinositol 3-Kinase-Dependent Pathway during Neuronal Development. J Neurosci. 2004;24:7366–7377. doi: 10.1523/JNEUROSCI.1739-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavov N, Semrau S, Budnik B, Slavov N, Semrau S, Airoldi E, Budnik B, Van Oudenaarden A. Differential Stoichiometry among Core Ribosomal Proteins. Cell Rep. 2015;13:865–873. doi: 10.1016/j.celrep.2015.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- State MW, Šestan N. Neuroscience. The emerging biology of autism spectrum disorders. Science. 2012;337:1301–1303. doi: 10.1126/science.1224989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunk BS, Loucks CR, Su M, Vashisth H, Cheng S, Schilling J, Brooks CL, Karbstein K, Skiniotis G. Ribosome assembly factors prevent premature translation initiation by 40S assembly intermediates. Science. 2011;333:1449–1453. doi: 10.1126/science.1208245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunk BS, Novak MN, Young CL, Karbstein K. A translation-like cycle is a quality control checkpoint for maturing 40S ribosome subunits. Cell. 2012;150:111–121. doi: 10.1016/j.cell.2012.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subtelny AO, Eichhorn SW, Chen GR, Sive H, Bartel DP. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature. 2014;508:66–71. doi: 10.1038/nature13007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafforeau L, Zorbas C, Langhendries JL, Mullineux ST, Stamatopoulou V, Mullier R, Wacheul L, Lafontaine DJ. The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of pre-rRNA processing factors. Mol Cell. 2013;51:539–551. doi: 10.1016/j.molcel.2013.08.011. [DOI] [PubMed] [Google Scholar]
- Teng T, Thomas G, Mercer CA. Growth control and ribosomopathies. Curr Opin Genet Dev. 2013;23:63–71. doi: 10.1016/j.gde.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Thomson AM, Lamy C. Functional maps of neocortical local circuitry. Front Neurosci. 2007;1:19–42. doi: 10.3389/neuro.01.1.1.002.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulmin H, Beckmann CF, O’Muircheartaigh J, Ball G, Nongena P, Makropoulos A, Ederies A, Counsell SJ, Kennea N, Arichi T, Tusor N, Rutherford MA, Azzopardi D, Gonzalez-Cinca N, Hajnal JV, Edwards AD. Specialization and integration of functional thalamocortical connectivity in the human infant. Proc Natl Acad Sci USA. 2015;112 doi: 10.1073/pnas.1422638112. 201422638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschochner H, Hurt E. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 2003;13:255–263. doi: 10.1016/s0962-8924(03)00054-0. [DOI] [PubMed] [Google Scholar]
- Vesper O, Amitai S, Belitsky M, Byrgazov K, Kaberdina AC, Engelberg-Kulka H, Moll I. Selective Translation of Leaderless mRNAs by Specialized Ribosomes Generated by MazF in Escherichia coli. Cell. 2011;147:147–157. doi: 10.1016/j.cell.2011.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, Mill J, Cantor RM, Blencowe BJ, Geschwind DH. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss LA, Arking DE, Daly MJ, Chakravarti A. A genome-wide linkage and association scan reveals novel loci for autism. Nature. 2009;461:802–808. doi: 10.1038/nature08490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward ND, Heckers S. Mapping Thalamocortical Functional Connectivity in Chronic and Early Stages of Psychotic Disorders. Biol Psychiatry. 2014 doi: 10.1016/j.biopsych.2015.06.026. S0006-3223(15)00534-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward ND, Karbasforoushan H, Heckers S. Thalamocortical dysconnectivity in schizophrenia. Am J Psychiatry. 2012;169:1092–1099. doi: 10.1176/appi.ajp.2012.12010056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S, Barna M. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat Rev Mol Cell Biol. 2012;13:355–369. doi: 10.1038/nrm3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S, Tian S, Fujii K, Kladwang W, Das R, Barna M. RNA regulons in Hox 5′ UTRs confer ribosome specificity to gene regulation. Nature. 2015;517:33–38. doi: 10.1038/nature14010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Smibert CA, Kaplan DR, Miller FD. An eIF4E1/4E-T Complex Determines the Genesis of Neurons from Precursors by Translationally Repressing a Proneurogenic Transcription Program. Neuron. 2014;84:1–17. doi: 10.1016/j.neuron.2014.10.022. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Harrison P, Gerstein M. Identification and Analysis of Over 2000 Ribosomal Protein Pseudogenes in the Human Genome. Genome Res. 2002;12:1466–1482. doi: 10.1101/gr.331902. [DOI] [PMC free article] [PubMed] [Google Scholar]