Abstract
Objective:
To evaluate the feasibilities of controlled aliasing in parallel imaging results in higher acceleration with volumetric interpolated breath-hold examination (CAIPIRINHA-VIBE), radial acquisition of VIBE (Radial-VIBE) with k-space-weighted image contrast (KWIC) reconstruction (KWIC-Radial-VIBE) and conventional-VIBE (c-VIBE) for free-breathing dynamic contrast-enhanced (DCE)-MRI of the abdomen.
Methods:
23 prospectively enrolled patients underwent DCE-MRI of the abdomen with CAIPIRINHA-VIBE (n = 10), KWIC-Radial-VIBE (n = 6) or c-VIBE (n = 7). Qualitative image quality of the DCE-MR images and perfusion maps was independently scored by two abdominal radiologists using a 5-point scale (from 1, uninterpretable, to 5, very good). For quantitative analysis, the signal-to-noise ratio (SNR) of the liver and goodness-of-fit (GOF) of the time–intensity curve were measured.
Results:
In the three tested sequences, DCE-MRI had good temporal (5 s) and spatial resolution (1.48 × 1.48 × 4 mm/voxel). Interobserver agreement in the qualitative analysis was good (ĸ = 0.753; 95% confidence interval, 0.610–0.895). Therefore, the mean scores were used in the data analysis. Overall image quality was comparable between CAIPIRINHA-VIBE (3.52 ± 0.55) and KWIC-Radial-VIBE (3.72 ± 0.37; p = 1.000), and both were significantly better than c-VIBE (2.71 ± 0.34; p < 0.001). Perfusion map quality score was highest with KWIC-Radial-VIBE (4.33 ± 0.65), followed by CAIPIRINHA-VIBE (3.70 ± 0.73) and c-VIBE (3.14 ± 0.66), but without statistical significance between CAIPIRINHA-VIBE and KWIC-Radial-VIBE (p = 0.167). The SNR of the liver and GOF of the time–intensity curve did not significantly differ between the three sequences (p = 0.116 and 0.224, respectively).
Conclusion:
CAIPIRINHA-VIBE and KWIC-Radial-VIBE provide comparably better performance than c-VIBE. Both can be feasible sequences with acceptable good image quality for free-breathing DCE-MRI.
Advances in knowledge:
CAIPIRINHA-VIBE and KWIC-Radial-VIBE provide comparably better quality of free-breathing DCE-MRIs than c-VIBE.
INTRODUCTION
Dynamic contrast-enhanced (DCE)-MRI is a promising method for evaluating tumour vascularity. It has been used in many clinical Phase I or IIa trials of molecular targeted agents, including antiangiogenic and vascular disrupting agents, to evaluate the pharmacodynamic effects of the drugs on tumours.1 Most of these clinical trials included target tumours in the abdomen. However, respiratory motion artefact in DCE-MRI of the abdomen is a challenging issue because DCE-MRI scans continuously for several minutes while the patient breathes quietly.2
Currently, three-dimensional (3D) gradient-echo T1 weighted images are most widely used for DCE-MRI.2 However, conventional 3D T1 weighted images such as volumetric interpolated breath-hold examination (VIBE) are not motion insensitive.3 Recently, new motion-resistant sequences were proposed, including radial acquisition of VIBE (Radial-VIBE) and controlled aliasing in parallel imaging results in higher acceleration with VIBE (CAIPIRINHA-VIBE).4
Radial-VIBE with k-space-weighted image contrast (KWIC) reconstruction (KWIC-Radial-VIBE) has been proposed for free-breathing DCE-MRI of the abdomen.3,5 According to a recent study, KWIC-Radial-VIBE is insensitive to respiratory motion, enabling the acquisition of high-quality images for free-breathing examinations.3 However, this sequence is still experimental and is not yet commercially available. CAIPIRINHA-VIBE is a commercialized sequence using a new parallel acquisition technique, which can improve image quality, reduce scan time and lessen breathing motion artefacts.6
We postulated that CAIPIRINHA-VIBE or KWIC-Radial-VIBE is more suitable for free-breathing DCE-MRI of the abdomen than conventional-VIBE (c-VIBE). Therefore, we aimed to evaluate the feasibilities of CAIPIRINHA-VIBE and KWIC-Radial-VIBE for free-breathing DCE-MRI of the abdomen by comparing CAIPIRINHA-VIBE, KWIC-Radial-VIBE and c-VIBE sequences.
METHODS AND MATERIALS
Patients
This prospective study was approved by our institutional review board. All participants provided written informed consent. Adult patients who visited the oncology department and met the following eligibility criteria were contacted about study enrolment: (1) aged ≥20 years with an underlying malignancy in the abdomen; (2) required MRI for evaluation of the liver; and (3) without contraindications for MRI. Patients who agreed to participate were randomly assigned to one of three DCE-MRI protocols.
Because this was an exploratory and feasibility study, the sample size was determined in consideration of study period and clinical practice and not through a mathematical calculation. Patients were randomly assigned to one of three DCE-MRI protocols by using a computerized random number generator (https://randomizer.org) to minimize any potential bias from researcher selection. Randomization was performed when the patients were enrolled in our study and provided informed consent. Neither blocking nor stratification variables was used during the randomization.
MRI acquisition
A 3.0-T MR scanner (Magnetom Skyra; Siemens Healthcare, Erlangen, Germany) with an 18-channel body matrix coil and a table-mounted 32-channel spine matrix coil was used. One of three DCE-MRI protocols (i.e. CAIPIRINHA-VIBE, KWIC-Radial-VIBE or c-VIBE) was performed for each patient. The scan coverage of these sequences was 208 mm (52 slices × 4-mm thickness), which was sufficient for covering the entire liver in all the patients. The detailed MRI parameters for the DCE-MRI protocols are summarized in Table 1.
Table 1.
MRI scan parameters
| Parameters | CAIPIRINHA-VIBE | KWIC-Radial-VIBE | c-VIBE |
|---|---|---|---|
| Repetition time (ms) | 3.8 | 3.7 | 3.2 |
| Echo time (ms) | 1.6 | 1.4 | 1.1 |
| Field of view (mm2) | 380 × 380 | 380 × 380 | 380 × 380 |
| Flip angle (°) | 25 | 25 | 25 |
| Slice thickness (mm) | 4 | 4 | 4 |
| Matrix | 256 × 256 | 256 × 256 | 256 × 256 |
| Bandwidth (kHz) | 890 | 1300 | 810 |
| Number of excitation | 1 | 1 | 1 |
| Acceleration factor | 2 × 2 | None | 2 |
| Acquisition time (s) | 305 | 305 | 305 |
CAIPIRINHA-VIBE, controlled aliasing in parallel imaging results in higher acceleration with volumetric interpolated breath-hold examination; c-VIBE, conventional-VIBE; KWIC-Radial-VIBE, radial-VIBE with k-space-weighted image contrast reconstruction.
Free-breathing DCE-MRIs with three different sequences were acquired. Before contrast agent injection, five-phase baseline acquisitions were performed. When the sixth phase acquisition was started, 0.1 mmol kg−1 of body weight of gadoteric acid (Dotarem®; Guerbet, Villepinte, France) was injected at a rate of 2 ml s−1, followed by a saline chase of 20 ml at the same injection rate. The three sequences had the same base resolution of 256, and all scan parameters were adjusted to fix the acquisition time. A free-breathing dynamic series was repeatedly obtained at 60 time points over 305 s in all the sequences. CAIPIRINHA-VIBE was performed with an acceleration factor of 4 (2 each in the phase- and partition-encoding directions) and a reordering shift of 1. Regarding c-VIBE, generalized autocalibrating partially parallel acquisition7 with an acceleration factor of 2 in the phase-encoding direction was used. An acceleration factor of 4 (2 × 2) for c-VIBE was excluded at preliminary testing because it showed relatively severe parallel imaging artefact when we set up the DCE-MRI protocol with a temporal resolution of 5 s. KWIC-Radial-VIBE was composed of 10 full-frame sets. By using the KWIC view-sharing technique, six undersampled time-resolved subframe image sets were reconstructed from one full-frame set. A full-frame set was composed of 324 radial projection views (i.e. 54 projection views per interleaved subset). Only the time-resolved subframe images were used for the DCE-MRI analysis.
Generation of a perfusion map
By using the software Tissue 4D (Siemens Healthcare, Erlangen, Germany), voxel-wise perfusion maps of the initial area under the concentration curve (iAUC) in 60 s were generated because iAUC is currently most widely used in tumour perfusion analysis as recommended by Quantitative Imaging Biomarkers Alliance.5 Motion correction of the DCE-MRI scan was performed by using a non-rigid registration technique.8,9 In this study, we did not use T1 mapping for the reason that T1 values obtained from T1 mapping may be inaccurate or some errors my occur in the process of co-registration of T1 value to the dynamic data. We aimed to focus on the effect of free-breathing dynamic series on image quality. Therefore, the T1 value of the tissue (especially the liver) was assumed to be 800 ms. Then, time–intensity curves were obtained and converted to time–concentration curves. Perfusion maps were computed based on Tofts model using a population-averaged arterial input function with two exponentials.8–10
Image analysis
Qualitative analysis
Two abdominal radiologists (NS and KWK) with 7 and 13 years' experience in liver imaging independently analyzed the quality of perfusion maps by using a 5-point scale as follows: 1 = uninterpretable; 2 = poor; 3 = fair; 4 = good; and 5 = very good. The rating was based on the registration quality of the perfusion maps, the presence of misregistration artefacts and any architectural distortion of the organ.3
The readers also rated the image quality of the DCE-MRI scans (i.e. raw data of the dynamic series) in three different phases at 30 s, 1 min and 3 min after contrast agent injection, respectively, by using the same 5-point scale. Then, the sum and mean score of the ratings at those three phases were calculated. The overall image quality, lesion conspicuity, image sharpness and degree of artefacts were assessed.
Quantitative analysis
Quantitative analysis was performed for the signal-to-noise ratio (SNR) of the raw DCE-MRI scan and goodness-of-fit (GOF) of the iAUC perfusion map. First, to measure the SNR of the MRI scans obtained from new reconstruction techniques such as parallel imaging or KWIC, in which noise distribution is inhomogeneous, we used a method based on a single image voxel. In this method, SNR is defined as the ratio of the mean value to the standard deviation of the signal intensity in repeated “identical” acquisitions along the time course of the voxel.11 Accordingly, the radiologist (NS) placed regions of interest on the same spot in the liver in five sequential phases of pre-contrast MRI scans and measured the signal intensity from the regions of interest.
Second, the GOF of the time–intensity curve in the aorta and liver was evaluated because the quantitative value of the perfusion maps is greatly dependent on the curve fitting of the time–intensity curve. The GOF of the time–intensity curve describes how well the curve fits a set of observed values. The Pearson χ2 test was used to compare the GOF of the perfusion curves for each DCE-MRI protocol. The χ2 of the GOF reflects the discrepancy between the observed and expected values; thus, a small χ2 value means a better quality GOF.12
Statistical analysis
The interobserver agreement between the two reviewers for scoring image quality was analyzed with quadratic-weighted k statistics by using MedCalc® (MedCalc Software, Ostend, Belgium). The k value was interpreted as follows: poor, k < 0.2; fair, 0.2 < k ≤ 0.4; moderate, 0.2 < k ≤ 0.6; good, 0.6 < k ≤ 0.8; and very good, 0.8 < k ≤ 1.0.13 Image quality score was compared between the three sequences by using a repeated measures analysis of variance with post hoc analysis and Bonferroni correction, because each subject received two ratings by two reviewers. Quantitative values were compared between the three sequences by using one-way analysis of variance with post hoc analysis and Bonferroni correction. All statistical analyses except interobserver agreement were performed with SPSS® v. 21.0 (IBM Corp., New York, NY; formerly SPSS Inc., Chicago, IL). p-values <0.05 were considered statistically significant.
RESULTS
Patients
Among the 24 patients who initially agreed to participate in the study, 1 patient cancelled her participation at the time of MRI because of her poor general condition. Ultimately, 23 patients (14 males and 9 females; mean ± standard deviation age, 51.8 ± 13.6 years) underwent DCE-MRI. According to the random assignment process, 10 patients underwent CAIPIRINHA-VIBE, 6 patients underwent KWIC-Radial-VIBE and 7 patients underwent c-VIBE. Among these patients, 19 patients had malignant liver tumours, including cholangiocarcinomas (n = 9), metastases from pancreatic cancer (n = 5), colon cancer (n = 3) or gallbladder cancer (n = 2). The MRI findings of the remaining four patients included resolved hepatic metastases (n = 2), post-chemotherapy sinusoidal obstruction syndrome (n = 1) and focal eosinophilic infiltration (n = 1). All the patients were allowed to breathe shallowly and regularly during MRI scanning.
Acquisition of the dynamic contrast-enhanced-MRI scans and perfusion maps
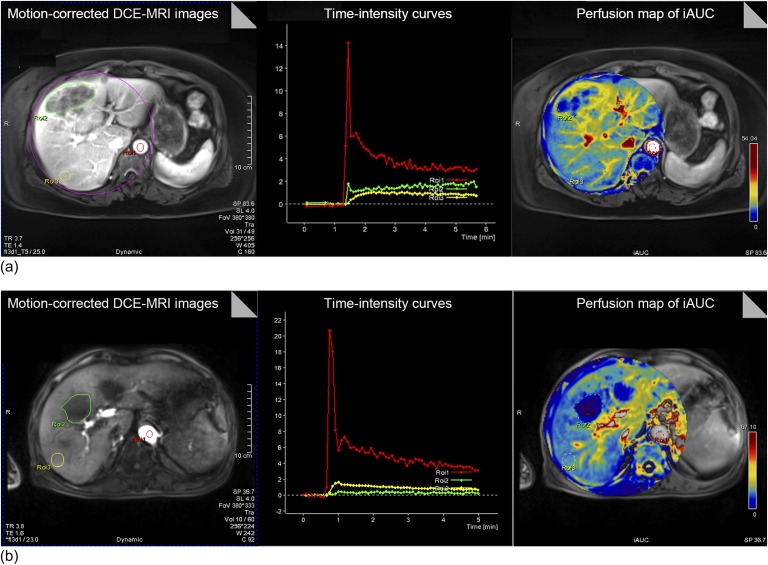
In all the three tested sequences, we obtained transverse DCE-MRI scans with a good temporal resolution of 5 s, which meets the recommendation of the Quantitative Imaging Biomarkers Alliance DCE-MRI profile and is regarded as sufficient to scan the fast initial contrast arrival. The spatial resolution was reasonably good (1.48 × 1.48 × 4 mm/voxel). The perfusion maps of the iAUC parameter were generated in all the patients (Figure 1). The perfusion map quality score was highest for KWIC-Radial-VIBE (4.33 ± 0.65), followed by CAIPIRINHA-VIBE (3.70 ± 0.73) and c-VIBE (3.14 ± 0.66), with significant differences between the three (p = 0.008). A post hoc test revealed that KWIC-Radial-VIBE significantly differed from c-VIBE (p = 0.006). However, the difference between CAIPIRINHA-VIBE and KWIC-Radial-VIBE did not reach statistical significance (p = 0.167). In general, the perfusion map in KWIC-Radial-VIBE showed less pronounced misregistration artefacts than those of the other two sequences.
Figure 1.
Generation of perfusion maps and time–intensity curve from dynamic contrast-enhanced (DCE)-MRI using radial-volumetric interpolated breath-hold examination (VIBE) with k-space-weighted image contrast reconstruction (a) and controlled aliasing in parallel imaging results in higher acceleration with VIBE (b). In each sequence, DCE-MRI scans are registered to yield motion-corrected images (left). Regions of interests (ROIs) are placed in the aorta, tumour and liver parenchyma. From motion-corrected DCE-MRI scans, time–intensity curves of the ROIs are generated (middle). Then, perfusion maps of the initial area under the concentration curve (iAUC) are created (right). FOV, field of view; TE, echo time; TR, repetition time.
Qualitative analysis of image quality
The interobserver agreement between the two reviewers for evaluating the overall image quality score of raw MRI data was good (ĸ = 0.753; 95% confidence interval, 0.610–0.895). Therefore, the mean scores were used in the data analysis.
Regarding the overall image quality of the DCE-MRI scans, CAIPIRINHA-VIBE (3.52 ± 0.55) was comparable with KWIC-Radial-VIBE (3.72 ± 0.37; p = 1.000), and both were significantly better than c-VIBE (2.71 ± 0.34; p < 0.001) (Table 2). Likewise, in the other qualitative parameters, CAIPIRINHA-VIBE and KWIC-Radial-VIBE showed comparable performances, which were both significantly better than that of c-VIBE (Figure 2).
Table 2.
Comparison of the three dynamic contrast-enhanced-MRI sequences through qualitative analysis
| Parameters | CAIPIRINHA-VIBE | KWIC-Radial-VIBE | c-VIBE | p-value of RMANOVA |
p-values of Bonferroni post hoc testa |
||
|---|---|---|---|---|---|---|---|
| CAIPIRINHA-VIBE vs KWIC | CAIPIRINHA-VIBE vs c-VIBE | KWIC vs c-VIBE | |||||
| Overall image quality | 3.52 (3.25–3.79) | 3.72 (3.38–4.07) | 2.71 (2.39–3.04) | <0.001 | 1.000 | 0.002 | 0.001 |
| Lesion conspicuity | 3.60 (3.30–3.90) | 3.61 (3.22–4.00) | 2.81 (2.45–3.17) | 0.004 | 1.000 | 0.007 | 0.015 |
| Image sharpness | 3.43 (3.17–3.70) | 3.42 (3.07–3.76) | 2.71 (2.40–3.02) | 0.003 | 1.000 | 0.004 | 0.013 |
| Artefacts | 3.45 (3.12–3.78) | 3.42 (3.00–3.84) | 2.69 (2.30–3.01) | 0.012 | 1.000 | 0.016 | 0.047 |
CAIPIRINHA-VIBE, controlled aliasing in parallel imaging results in higher acceleration with volumetric interpolated breath-hold examination; c-VIBE, conventional-VIBE; KWIC-Radial-VIBE, radial-VIBE with k-space-weighted image contrast reconstruction; RMANOVA, repeated measures analysis of variance.
The numbers in parenthesis represent the 95% confidence interval of the average score of image quality.
Adjusted p-values using Bonferroni correction.
Figure 2.
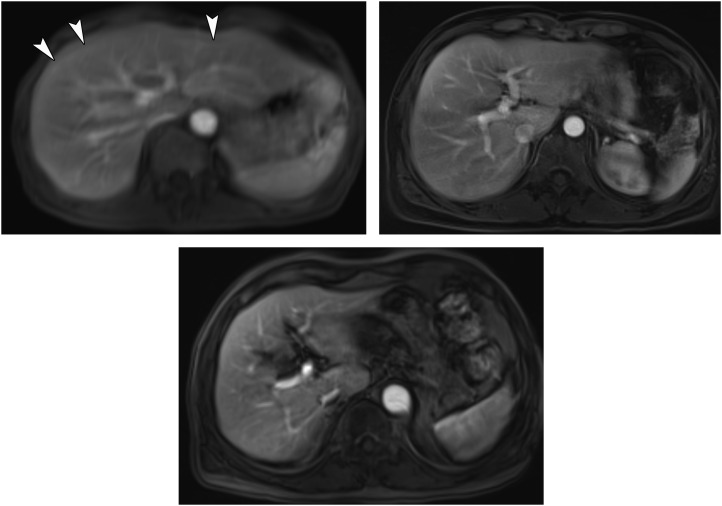
Raw dynamic contrast-enhanced-MR images using conventional-volumetric interpolated breath-hold examination (c-VIBE), radial-VIBE with k-space-weighted image contrast reconstruction (KWIC-Radial-VIBE) and controlled aliasing in parallel imaging results in higher acceleration with VIBE (CAIPIRINHA-VIBE): (a) an MR image obtained using c-VIBE reveals an overall blurred image with poor conspicuity of hepatic vessels. The prominent motion artefacts due to breathing motion can be noted (arrowheads). (b) MR images obtained using KWIC-Radial-VIBE and (c) CAIPIRINHA-VIBE show superior image quality with less motion artefacts compared with (a).
Quantitative analysis
The SNR of the liver was highest with KWIC-Radial-VIBE (24.39 ± 1.92), followed by c-VIBE (20.24 ± 5.80) and CAIPIRINHA-VIBE (17.62 ± 7.41) (Table 3). However, these differences did not reach statistical significance (p = 0.116).
Table 3.
Comparison of the three dynamic contrast-enhanced-MRI sequences through quantitative analysis
| Parameters | CAIPIRINHA-VIBE | KWIC-Radial-VIBE | c-VIBE | p-value of ANOVA |
p-values of Bonferroni post hoc testa |
||
|---|---|---|---|---|---|---|---|
| CAIPIRINHA-VIBE vs KWIC | CAIPIRINHA-VIBE vs c-VIBE | KWIC vs c-VIBE | |||||
| SNR | |||||||
| SNR of the liver | 17.62 (12.32–22.92) | 24.39 (22.38–26.40) | 20.24 (14.88–25.60) | 0.116 | 0.121 | 1.000 | 0.680 |
| SI of the liver | 76.76 (67.83–85.70) | 108.53 (100.99–116.06) | 93.88 (79.03–108.73) | <0.001 | <0.001 | 0.042 | 0.074 |
| SD of the liver | 4.81 (3.72–5.91) | 4.59 (4.08–5.90) | 5.02 (3.57–6.47) | 0.850 | 1.000 | 1.000 | 1.000 |
| GOF | |||||||
| Aorta | 2.05 (0.98–3.12) | 6.31 (0–14.60) | 4.65 (0.54–8.75) | 0.224 | 0.293 | 0.841 | 1.000 |
| Liver | 0.17 (0.10–0.24) | 0.10 (0.08–0.12) | 0.15 (0.09–0.22) | 0.216 | 0.264 | 1.000 | 0.641 |
ANOVA, analysis of variance; CAIPIRINHA-VIBE, controlled aliasing in parallel imaging results in higher acceleration with volumetric interpolated breath-hold examination; c-VIBE, conventional-VIBE; GOF, goodness-of-fit; KWIC-Radial-VIBE, radial-VIBE with k-space-weighted image contrast reconstruction; SD, standard deviation; SI, signal intensity; SNR, signal-to-noise ratio.
The numbers in parenthesis represent the 95% confidence interval.
Adjusted p-values using Bonferroni correction.
The GOF of the time–intensity curve for the aorta was lowest for CAIPIRINHA-VIBE (2.05 ± 1.50), followed by c-VIBE (4.65 ± 4.44) and KWIC-Radial-VIBE (6.31 ± 7.9), but this difference was also not statistically significant (p = 0.224).
DISCUSSION
Our present study demonstrated that CAIPIRINHA-VIBE and KWIC-Radial-VIBE are quite feasible for free-breathing DCE-MRI of the abdomen. Both CAIPIRINHA-VIBE and KWIC-Radial-VIBE are essentially comparable in terms of image quality and perfusion map generation. Both sequences are expected to be promising for DCE-MRI, although they have different advantages and drawbacks. In contrast, c-VIBE may not be a good choice for free-breathing DCE-MRI because of image degradation by respiratory motion artefacts. Indeed, when we fixed the temporal (5 s) and spatial resolutions (1.48 × 1.48 × 4 mm/voxel) for the three tested sequences, CAIPIRINHA-VIBE and KWIC-Radial-VIBE showed better image quality of the perfusion maps and DCE-MRI scans than c-VIBE.
It is worth noting the characteristics of CAIPIRINHA-VIBE and KWIC-Radial-VIBE with regard to using these motion-resistant sequences for free-breathing DCE-MRI. CAIPIRINHA-VIBE is a 3D T1 weighted gradient-echo sequence using a new parallel imaging scheme that modifies the acquisition pattern by shifting the sampling positions in the partition-encoding direction. The motion-resistant characteristics of CAIPIRINHA-VIBE are based on (1) reduction of the severity of aliased pixels by a delta shift in the partition-encoding direction and (2) reduction of the acquisition time.14 KWIC-Radial-VIBE acquires radial sampling in-plane and Cartesian sampling along the slice-encoding direction using a “stack-of-stars” scheme.15–17 KWIC-Radial-VIBE shows less motion sensitivity and less pronounced aliasing artefacts and provides a higher temporal resolution than c-VIBE.16,18 The motion robustness of KWIC-Radial-VIBE fundamentally results from the radial sampling pattern of k-space.
Regarding the quality of perfusion maps and DCE-MRI scans, we found no significant difference between CAIPIRINHA-VIBE and KWIC-Radial-VIBE, although the perfusion map quality score was higher for KWIC-Radial-VIBE. In general, KWIC-Radial-VIBE provided less prominent misregistration artefacts. This difference could be explained by the excellent motion insensitivity of radial k-space sampling and the time-averaging effect of radial spokes at the centre of k-space.3,17 KWIC-Radial-VIBE demonstrated a higher SNR than CAIPIRINHA-VIBE, even though this difference was not statistically significant. With KWIC reconstruction, the peripheral region of k-space consists of additional data from adjacent respiratory cycles, and the use of more data increases the SNR.15 On the other hand, in a scan accelerated by parallel imaging, such as 3D CAIPIRINHA, loss of SNR is a well-known limitation of undersampling.19,20
On the other hand, the lack of fidelity of the time–intensity curve obtained from KWIC-Radial-VIBE is a known drawback in the quantification of perfusion analysis.3 In our study, six individual subframe images were generated every 5 s with 54 radial views for the central k-space filling. Therefore, the temporal resolution of the central k-space region was 5 s. However, each subframe image also used the data from the adjacent interleaved data set of 30 s. Thus, the larger temporal window in the outer k-space results in averaging of high-frequency data.3 In particular, during the initial contrast arrival time when the signal intensity is rapidly changing, the temporal profile near the periphery of the lesions may be less accurate.15
Our study had several limitations. First, a small number of patients were included in this pilot study. Nevertheless, this pilot work provides evidence for the effectiveness of CAIPIRINHA-VIBE and KWIC-Radial-VIBE for free-breathing DCE-MRI in the abdomen and can act as a basis for further studies. Second, we did not compare the three sequences in the same patient, which may raise the issue of selection bias. It is not practical to scan the same subject twice with contrast within a single imaging session. Repeat scans on the same subject at separate times would be subject to changes in breathing pattern. Instead, we randomly assigned the MRI sequences per patient to minimize the effect of patient health status on the quality of the DCE-MRI scans. Finally, we did not compare the image quality between DCE-MRI and conventional MRI. The spatial resolution of DCE-MRI (1.48 × 1.48 × 4 mm/voxel) may have limitation for detecting and characterizing lesions compared with conventional MRI.
CONCLUSION
In conclusion, we suggest that both CAIPIRINHA-VIBE and KWIC-Radial-VIBE are feasible sequences with acceptable high image quality for free-breathing DCE-MRI of the abdomen.
FUNDING
This study was supported by a grant (No. 2014-0351) from the Asan Institute for Life Sciences of Asan Medical Center and a grant (No. 2014R1A1A1006823) from the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and Future Planning.
Contributor Information
Nieun Seo, Email: sldmsdl@hanmail.net.
Seong J Park, Email: joon77@gmail.com.
Bohyun Kim, Email: baboojum@naver.com.
Chang K Lee, Email: leeck0326@naver.com.
Jimi Huh, Email: jimihuh.rad@gmail.com.
Jeong K Kim, Email: rialto@amc.seoul.kr.
Seung S Lee, Email: Seungsoolee@amc.seoul.kr.
In S Kim, Email: inseong.kim@siemens.com.
Dominik Nickel, Email: marcel.nickel@siemens.com.
Kyung W Kim, Email: medimash@gmail.com.
REFERENCES
- 1.O'Connor JP, Jackson A, Parker GJ, Roberts C, Jayson GC. Dynamic contrast-enhanced MRI in clinical trials of antivascular therapies. Nat Rev Clin Oncol 2012; 9: 167–77. doi: 10.1038/nrclinonc.2012.2 [DOI] [PubMed] [Google Scholar]
- 2.Thng CH, Koh TS, Collins DJ, Koh DM. Perfusion magnetic resonance imaging of the liver. World J Gastroenterol 2010; 16: 1598–609. doi: 10.3748/wjg.v16.i13.1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim KW, Lee JM, Jeon YS, Kang SE, Baek JH, Han JK, et al. Free-breathing dynamic contrast-enhanced MRI of the abdomen and chest using a radial gradient echo sequence with K-space weighted image contrast (KWIC). Eur Radiol 2013; 23: 1352–60. doi: 10.1007/s00330-012-2699-4 [DOI] [PubMed] [Google Scholar]
- 4.Kim BS, Lee KR, Goh MJ. New imaging strategies using a motion-resistant liver sequence in uncooperative patients. Biomed Res Int 2014; 2014: 142658. doi: 10.1155/2014/142658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DMT Committee. DCE MRI quantification profile. Quantitative imaging biomarkers alliance. Version 1.0 reviewed draft 2012.
- 6.AlObaidy M, Ramalho M, Busireddy KK, Liu B, Burke LM, Altun E, et al. High-resolution 3D-GRE imaging of the abdomen using controlled aliasing acceleration technique—a feasibility study. Eur Radiol 2015; 25: 3596–605. doi: 10.1007/s00330-015-3780-6 [DOI] [PubMed] [Google Scholar]
- 7.Griswold MA, Jakob PM, Heidemann RM, Nittka M, Jellus V, Wang J, et al. Generalized autocalibrating partially parallel acquisitions (GRAPPA). Magn Reson Med 2002; 47: 1202–10. doi: 10.1002/mrm.10171 [DOI] [PubMed] [Google Scholar]
- 8.Chefd'hotel C, Hermosillo G, Faugeras O. Flows of diffeomorphisms for multimodal image registration. Proceedings of the IEEE International Symposium on Biomedical Imaging. Washington, USA 2002: 753–6. doi: 10.1109/ISBI.2002.1029367 [DOI] [Google Scholar]
- 9.Chefd'hotel C, Hermosillo G, Faugeras O. A variational approach to multimodal image matching. Proceedings of the IEEE Workshop on Variational and Level Set Methods in Computer Vision, Vancouver, BC, Canada 2001: 21–8. doi: 10.1109/VLSM.2001.938877 [DOI] [Google Scholar]
- 10.Tofts PS, Brix G, Buckley DL, Evelhoch JL, Henderson E, Knopp MV, et al. Estimating kinetic parameters from dynamic contrast-enhanced T(1)-weighted MRI of a diffusable tracer: standardized quantities and symbols. J Magn Reson Imaging 1999; 10: 223–32. doi: [DOI] [PubMed] [Google Scholar]
- 11.Dietrich O, Raya JG, Reeder SB, Reiser MF, Schoenberg SO. Measurement of signal-to-noise ratios in MR images: influence of multichannel coils, parallel imaging, and reconstruction filters. J Magn Reson Imaging 2007; 26: 375–85. doi: 10.1002/jmri.20969 [DOI] [PubMed] [Google Scholar]
- 12.Knopp MV, Brix G, Junkermann HJ, Sinn HP. MR mammography with pharmacokinetic mapping for monitoring of breast cancer treatment during neoadjuvant therapy. Magn Reson Imaging Clin N Am 1994; 2: 633–58. [PubMed] [Google Scholar]
- 13.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics 1977; 33: 159–74. doi: 10.2307/2529310 [DOI] [PubMed] [Google Scholar]
- 14.Park YS, Lee CH, Kim IS, Kiefer B, Woo ST, Kim KA, et al. Usefulness of controlled aliasing in parallel imaging results in higher acceleration in gadoxetic acid-enhanced liver magnetic resonance imaging to clarify the hepatic arterial phase. Invest Radiol 2014; 49: 183–8. doi: 10.1097/RLI.0000000000000011 [DOI] [PubMed] [Google Scholar]
- 15.Lin W, Guo J, Rosen MA, Song HK. Respiratory motion-compensated radial dynamic contrast-enhanced (DCE)-MRI of chest and abdominal lesions. Magn Reson Med 2008; 60: 1135–46. doi: 10.1002/mrm.21740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song HK, Dougherty L. Dynamic MRI with projection reconstruction and KWIC processing for simultaneous high spatial and temporal resolution. Magn Reson Med 2004; 52: 815–24. doi: 10.1002/mrm.20237 [DOI] [PubMed] [Google Scholar]
- 17.Chandarana H, Block TK, Rosenkrantz AB, Lim RP, Kim D, Mossa DJ, et al. Free-breathing radial 3D fat-suppressed T1-weighted gradient echo sequence: a viable alternative for contrast-enhanced liver imaging in patients unable to suspend respiration. Invest Radiol 2011; 46: 648–53. doi: 10.1097/RLI.0b013e31821eea45 [DOI] [PubMed] [Google Scholar]
- 18.Fujinaga Y, Ohya A, Tokoro H, Yamada A, Ueda K, Ueda H, et al. Radial volumetric imaging breath-hold examination (VIBE) with k-space weighted image contrast (KWIC) for dynamic gadoxetic acid (Gd-EOB-DTPA)-enhanced MRI of the liver: advantages over Cartesian VIBE in the arterial phase. Eur Radiol 2014; 24: 1290–9. doi: 10.1007/s00330-014-3122-0 [DOI] [PubMed] [Google Scholar]
- 19.Riffel P, Attenberger UI, Kannengiesser S, Nickel MD, Arndt C, Meyer M, et al. Highly accelerated T1-weighted abdominal imaging using 2-dimensional controlled aliasing in parallel imaging results in higher acceleration: a comparison with generalized autocalibrating partially parallel acquisitions parallel imaging. Invest Radiol 2013; 48: 554–61. doi: 10.1097/RLI.0b013e31828654ff [DOI] [PubMed] [Google Scholar]
- 20.Robson PM, Grant AK, Madhuranthakam AJ, Lattanzi R, Sodickson DK, McKenzie CA. Comprehensive quantification of signal-to-noise ratio and g-factor for image-based and k-space-based parallel imaging reconstructions. Magn Reson Med 2008; 60: 895–907. doi: 10.1002/mrm.21728 [DOI] [PMC free article] [PubMed] [Google Scholar]