Abstract
Abscisic acid (ABA) is a classical phytohormone which plays an important role in plant stress resistance. Moreover, ABA is also found to regulate the activation of innate immune cells and glucose homeostasis in mammals. Therefore, this ‘stress hormone’ is of great importance to theoretical research and agricultural and medical applications. Botrytis cinerea is a well-known phytopathogenic ascomycete that synthesizes ABA via a pathway substantially different from higher plants. Identification of the functional genes involved in ABA biosynthesis in B. cinerea would be of special interest. We developed an ABA-overproducing mutant strain, B. cinerea TBC-A, previously and obtained a 41.5-Mb genome sequence of B. cinerea TBC-A. In this study, the transcriptomes of B. cinerea TBC-A and its ancestral strain TBC-6 were sequenced under identical fermentation conditions. A stringent comparative transcriptome analysis was performed to identify differentially expressed genes participating in the metabolic pathways related to ABA biosynthesis in B. cinerea. This study provides the first global view of the transcriptional changes underlying the very different ABA productivity of the B. cinerea strains and will expand our knowledge of the molecular basis for ABA biosynthesis in B. cinerea.
Abscisic acid (ABA), best known as a plant hormone, is a C15 sesquiterpene with one asymmetric carbon atom at C-1′, resulting in the S and R (or + and −, respectively) enantiomers1. The natural occurring form is S-ABA2. ABA plays important roles in enhancing plant tolerance to various kinds of stresses caused by abiotic or biotic factors3,4. The agricultural usage of ABA as a plant growth regulator has been well established2. Besides, ABA activity has also been reported in sponges5, and most recently in mammals with the function of immune modulation and glucose homeostasis regulation6,7. Thus ABA is also recognized as a candidate for medical applications8,9.
Although the application potential of ABA has drawn more attention recently, the high cost makes its wide use impractical10. Thus it is necessary to develop cost-effective approaches to synthesize ABA. It has been established by many researchers that ABA could be produced via various schemes of chemical synthesis9,11. However, chemically synthesized ABA is a mixture of approximately equal amounts of S-ABA and R-ABA9, which results in a loss of bioactivity2,12. Therefore, it is necessary to acquire the optically pure isomer of S-ABA.
It has been reported that a group of fungal species can synthesize S-ABA as a secondary metabolite13,14,15, which holds promise for S-ABA production at the industrial scale. For example, fermentative production of S-ABA using the fungus Botrytis cinerea and Cercospora rosicola were explored13,15. Labelling experiments have also been performed to elucidate the ABA biosynthetic pathway of B. cinerea and several Cercospora species, and a pathway different from plants has been postulated. In plants, ABA is biosynthesized via carotenoids derived from the 2-C-methyl-D-erythritol-4-phosphate (MEP) pathway16,17. In contrast, fungi biosynthesize ABA via a direct pathway from isopentenyl diphosphate (IDP) and farnesyl diphosphate (FDP), which are derived from the mevalonic acid (MVA) pathway18,19. After a series of reactions of cyclization, isomerization and oxidization, ABA is synthesized from FPP20,21,22. In addition, different fungal species employ different biosynthetic intermediates. It has been reported that ABA was synthesized via 1′,4′-dihydroxy-γ-ionylideneacetic acid in Cercospora cruenta22,23, while 1′,4′-trans-diol-ABA was detected as the main ABA intermediate in B. cinerea and Cercospora pini-densiflorae21,24, and 1′-deoxy-ABA was identified as a more important intermediate than 1′,4′-trans-diol-ABA in Cercospora rosicola25. Therefore, the genes responsible for fungal ABA biosynthesis should be quite distinct from plants. However, the molecular mechanism driving ABA biosynthesis in fungi is still illusive, and only a gene cluster consisting of four genes (bcaba1–4) was revealed26,27, in which at least 3 genes were presumed to be responsible for the hydroxylation of carbon atoms C-4′, C-1′ and the oxidation of C-4′ of ABA in B. cinerea.
Although little is known in the molecular mechanisms underlying fungal ABA biosynthesis, strain improvement by the traditional mutate-and-screen method has been empirically employed before these strains can be used in an industrial setting for biotechnological ABA production. In China, a wild-type strain B. cinerea TBC-6 has been genetically improved by multiple rounds of mutagenesis and screening over the past 20 years, generating mutant strains TB-31 and TB-3-H8 with substantially increased ABA yields28. Further strain improvement generated the B. cinerea TBC-A strain (Fig. 1A) with an ABA productivity of 2.0 g·L−1 at the industrial scale, which greatly reduced the cost of ABA. With the availability of the entire genome sequence of B. cinerea TBC-A and other B. cinerea strains29, as well as the advent of efficient methods for large-scale comparative transcriptome analysis, such as RNA-seq30,31,32, we are able to get important clues on the fundamental mechanisms underlying ABA overproduction in the industrial B. cinerea strains.
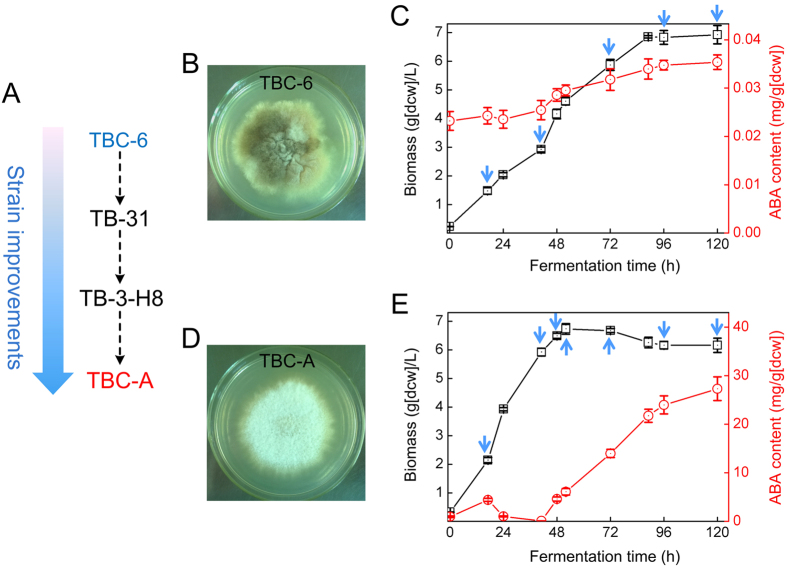
Figure 1. Phenotypes of B. cinerea TBC-A and B. cinerea TBC-6.
(A) Schematic of the B. cinerea strain genealogy during the mutagenesis and screening processes. Colonies formed by B. cinerea TBC-6 (B) and B. cinerea TBC-A (D) on CYA plates 5 d after inoculation. Time course of ABA production (red circles) and cellular growth (black squares) of B. cinerea TBC-6 (C) and TBC-A (E) in SF medium are shown. Blue arrows indicate when the culture samples were subjected to transcriptome sequencing. The error bars represent standard deviations from three independent cultures.
In this study, we performed RNA-seq analysis on the evolved mutant strain B. cinerea TBC-A and its ancestral strain TBC-6 grown under the same fermentation conditions. The gene expression profiling of 11,274 annotated genes in TBC-A and TBC-6 samples was described. A comparative transcriptome analysis was performed to identify the differentially expressed genes which may potentially contribute to the different ABA productivity of the two strains. This study presents the first exploration of the transcriptomic changes underlying the very different phenotypic outcomes of ABA production between the industrial and wild-type B. cinerea strains.
Results
Phenotypic differences between the ABA hyper-producing mutant TBC-A and the wild-type strain TBC-6
The phenotypic differences between B. cinerea TBC-A and the ancestral strain TBC-6 are shown in Fig. 1. In comparison to TBC-6, TBC-A exhibited faster growth, reduced sporulation and less pigmentation on Czapek yeast extract agar (CYA) plates. Typical time-course profiles of cell growth and ABA production for shake-flask fermentation of TBC-A and TBC-6 under the same culture conditions were also studied. We noticed that while the total biomass was approximately the same, the shapes of the growth curves and ABA production curves were markedly different between the strains (Fig. 1). Three distinct phases could be distinguished during the fermentation process of B. cinerea TBC-A: an initial period, with a moderate increase in ABA concentration lasting until 17 h, followed by a decline of ABA content until 41 h, and a third period of rapidly increasing ABA titers from 41 h to 120 h. With respect to the ancestral strain TBC-6, the period with a moderate increase in ABA titers was longer, lasting until 120 h, and resulted in a final ABA content that was approximately 1,000-fold lower than that of TBC-A.
Transcriptome sequencing of B. cinera TBC-A and TBC-6 samples
To characterize the gene expression profiles of B. cinerea, transcriptome sequencing was conducted to the strains TBC-A and TBC-6. Twelve culture samples were collected at different time points of the liquid fermentation process. Total RNA was extracted, purity and integrity of the RNA samples were checked (Supplementary Figure S1) and the transcriptomes were sequenced using an Illumina HiSeq™ 2500 platform. Based on the divergent growth curves and ABA production curves of TBC-A and TBC-6 (Fig. 1C,E), 7 samples from the TBC-A strain were collected at 17 h, 41 h, 48 h, 52 h, 72 h, 96 h and 120 h after the start of fermentation, and 5 samples from the TBC-6 strain were collected at 17 h, 41 h, 72 h, 96 h and 120 h. A total of 279.68 million (279,677,616) paired-end reads were obtained with an average of 23.31 million clean reads for each sample (Supplementary Table S1). We previously produced a 41.5 Mb draft genome sequence of B. cinerea TBC-A, and the genome annotation predicted 11,274 putative protein-coding genes (data not published). Alignment of the reads to the B. cinerea TBC-A genome sequence revealed that an average of 14.97 million reads (64.22% of the total clean reads) per sample could be mapped uniquely to the genome sequence. With a reads per kilobase of exon region per million mapped reads (RPKM) cutoff value of 1, 75.32–83.30% of genes were detected and expressed in the 12 samples of TBC-A and TBC-6 (Table 1).
Table 1. Distribution of genome-wide gene transcription levels derived from the RNA-seq data.
| Sample | TBC-A -17 h | TBC-A -41 h | TBC-A -48 h | TBC-A -52 h | TBC-A -72 h | TBC-A -96 h | TBC-A -120 h | TBC-6 -17 h | TBC-6 -41 h | TBC-6 -72 h | TBC-6 -96 h | TBC-6 -120 h |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number of highly expressed genes | 1587 | 1548 | 1425 | 1531 | 1591 | 1542 | 1528 | 1622 | 1410 | 1231 | 1379 | 1095 |
| Number of moderately expressed genes | 4543 | 4877 | 4736 | 4674 | 4799 | 4869 | 4802 | 4943 | 4264 | 4047 | 4237 | 4251 |
| Number of lowly expressed genes | 2362 | 2966 | 3022 | 2895 | 2762 | 2771 | 2756 | 2810 | 3215 | 3713 | 3282 | 3822 |
| Number of not expressed genes | 2782 | 1883 | 2091 | 2174 | 2122 | 2092 | 2188 | 1899 | 2385 | 2283 | 2376 | 2106 |
| Total number of expressed genes | 8492 | 9391 | 9183 | 9100 | 9152 | 9182 | 9086 | 9375 | 8889 | 8991 | 8898 | 9168 |
| Expressed genes % | 75.32 | 83.30 | 81.45 | 80.72 | 81.18 | 81.44 | 80.59 | 83.16 | 78.85 | 79.75 | 78.92 | 81.32 |
All genes are devided into four categories according to their RPKM values: genes not expressed (RPKM < 1), genes expressed at low levels (1 ≤ RPKM < 10), genes expressed at medium levels (10 ≤ RPKM < 100) and genes expressed at high levels (RPKM ≥ 100).
The distribution of genome-wide gene transcription levels derived from the RNA-seq data is described in Table 1. The RNA-seq results revealed that the most genes were expressed at 17 h (9,375 genes) and 41 h (9,391 genes) after the start of fermentation in TBC-6 and TBC-A, respectively. From a global view, all genes were divided into four categories according to their RPKM values, and the majority of genes were moderately expressed (10 ≤ RPKM < 100) during the fermentation time course for both strains.
To validate the accuracy and reproducibility of the RNA-seq data, quantitative real-time PCR (qRT-PCR) was performed on 19 selected genes to determine their expression levels in all 12 B. cinerea samples (Supplementary Fig. S2). The qRT-PCR results generally agreed with the RNA-seq data, suggesting that the RNA-seq data were reliable.
Identification of differentially expressed genes between the transcriptomes
In order to interpret the molecular basis of the increased ABA production in the B. cinerea TBC-A mutant at the transcription level, comparative transcriptome analysis was performed to identify the differentially expressed genes (DEGs) between the 12 transcriptomes. DEGs were identified between the samples using the threshold log2 |fold change| ≥ 1, a false discovery rate (FDR) < 0.001 and RPKM value of at least one sample larger than 10. As shown in Fig. 2, markedly more DEGs were identified between the 5 TBC-6 samples and their corresponding TBC-A samples for all time points. 1,928, 3,112, 3,855, 3,105 and 3,827 of all the 11,274 annotated genes were differentially expressed between the strains TBC-A and TBC-6 at 17 h, 41 h, 72 h, 96 h and 120 h time points during the fermentation process, respectively. Transcription levels of 728 genes were upregulated in the TBC-A-17 h sample compared with the TBC-6–17 h sample, while more than 2,000 genes were identified with upregulated transcription levels in TBC-A strain compared with TBC-6 strain at 41 h, 72 h, 96 h and 120 h time points of the fermentation process (Fig. 2).
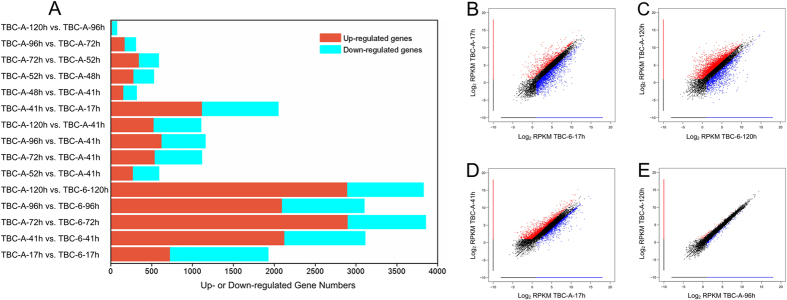
Figure 2. Genome-wide gene expression changes between B. cinerea strains TBC-A and TBC-6.
(A) Numbers of differentially expressed genes among the 12 culture samples at specific time points during the fermentation process. The graphs of upregulated genes are shown in red, and those for the downregulated genes are shown in blue. The criteria for the selection of DEGs was a log2 |fold change| ≥ 1, FDR < 0.001 and RPKM of at least one sample larger than 10. Scatter plots representing differential gene expression levels (represented as log2 RPKM) in (B) TBC-A-17 h vs. TBC-6-17 h, (C) TBC-A-120 h vs. TBC-6-120 h, (D) TBC-A-41 h vs. TBC-A-17 h and (E) TBC-A-120 h vs. TBC-A-96 h are shown. Color dots represent significantly differentially expressed genes that were overexpressed in the sample plotted on the X-axis (marked in red) or overexpressed in the sample plotted on the Y-axis (marked in blue). An additional threshold of RPKM ≥ 2 was used for the selection of DEGs in the scatterplots.
The number of DEGs between samples from adjacent time points was also calculated for the TBC-A strain. Surprisingly, 1,118 genes were upregulated and 933 genes were downregulated in the TBC-A-41 h sample compared with the TBC-A-17 h sample (Fig. 2A,D), which indicated the gene expression profiles shifted dramatically when TBC-A transitioned from its lag phase into its late-exponential phase during the fermentation process. Significantly fewer DEGs were identified in the other pairwise comparisons between the TBC-A samples. Remarkably, only 75 DEGs were identified in the TBC-A-120 h sample compared to the TBC-A-96 h sample (Fig. 2A,E). This result revealed that the expression profiles of the genes at the stationary phase of the fermentation process are nearly identical with only minor differences.
In order to identify the principle biological functions of the DEGs, we then mapped them to terms in gene ontology (GO) database and KEGG database, looking for significantly enriched terms compared to the entire genome background. It was found that the DEGs were significantly annotated to GO terms of amino acid metabolism, catabolism, catalytic activity, cell surface, metabolism, oxidoreductase activity and transport in TBC-A samples compared with their corresponding TBC-6 samples (Supplementary Table S2). Additionally, about 15 KEGG pathways, including carbohydrate metabolism, amino acid metabolism, lipid metabolism, transport and catabolism, xenobiotics biodegradation and metabolism, metabolism of cofactors and vitamins, metabolism of other amino acids and biosynthesis of other secondary metabolites, were significantly altered in the TBC-A samples compared to their corresponding TBC-6 samples (Supplementary Table S3). The remarkable transcriptional changes in carbohydrate metabolism, sugar transport, terpenoid backbone biosynthesis and a metabolic network related to acetyl-CoA and ABA biosynthesis in B. cinerea TBC-A compared to B. cinerea TBC-6, especially during the fermentation phase with a rapid increase in ABA titers, are described in detail below.
Differentially expressed genes encoding polysaccharide hydrolases and sugar transporters
The comparative transcriptome analysis showed that a considerable number of genes encoding sugar transporters and glycoside hydrolases (GHs) were significantly upregulated in TBC-A compared with TBC-6, especially during the late exponential and stationary phases of the fermentation process. As shown in Fig. 3 and Supplementary Table S4, the differentially expressed sugar transporter and saccharide-degrading genes were categorized by their functions in carbon source utilization.
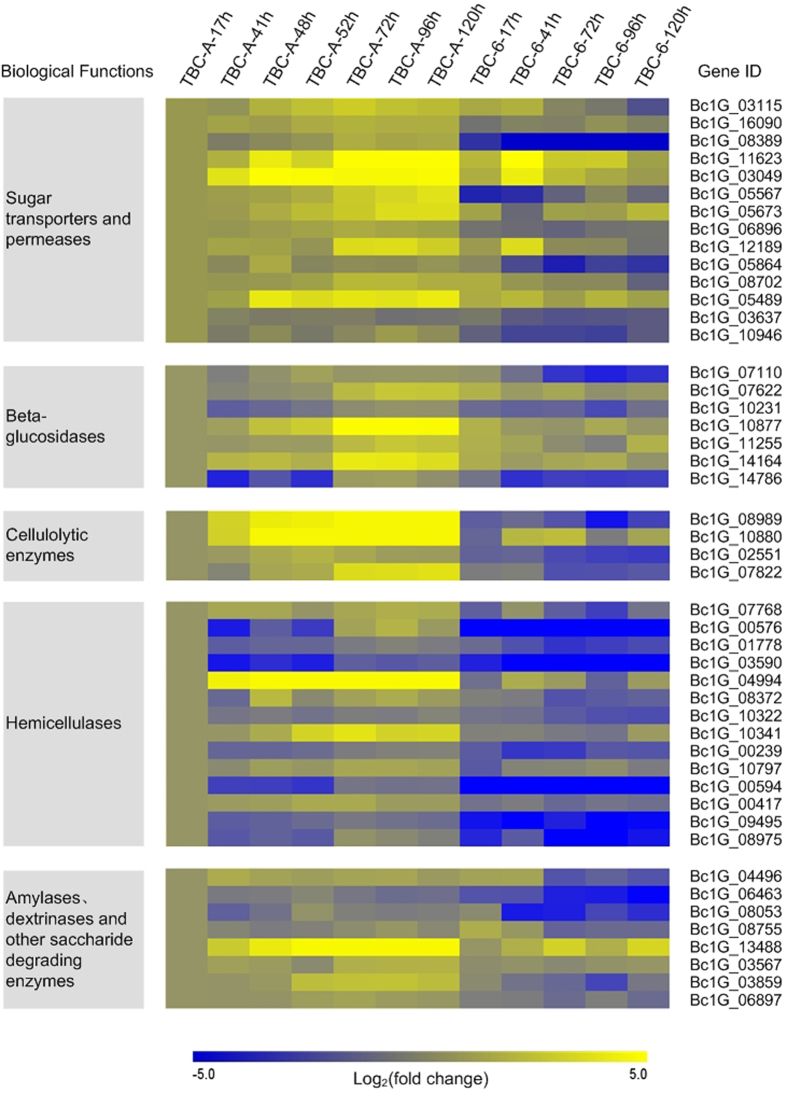
Figure 3. Selected carbon source utilization-related genes differentially expressed (p value < 0.05) between TBC-A and TBC-6 during the fermentation process.
Normalized RPKM values of the genes in the 12 samples were used, and the log2 fold change was calculated relative to the TBC-A-17 h sample for each gene in each sample to generate the heat map. Yellow indicates enhanced gene expression and blue for repressed gene expression. The color bar below indicates the expression scales [represented as log2 (fold change)].
Monosaccharides and disaccharides, such as glucose, galactose, xylose, sucrose, lactose and maltose, are nutrients that can be transported by sugar transporters or permeases into fungal cells. The gene transcription levels of several monosaccharide transporters and disaccharide permeases—including two glucose transporters rco-3; two high-affinity glucose transporters33,34 (HGT1); a glucose/galactose transporter (gluP); four lactose permeases (LAC12), which also transport galactose with low affinity35; three high-affinity maltose/proton symporters (MAL61 and MAL11) and two proton-coupled glucoside transporters (sut1), with a higher substrate affinity to maltose than sucrose36—were all upregulated dramatically in TBC-A compared with TBC-6 (p value < 0.05), especially during the stationary phase of the fermentation process. For example, the transcript abundance of Bc1G_08389, which encodes a HGT1, was more than 236.68-fold higher in the TBC-A-120 h sample than that of the TBC-6-120 h sample (RPKM values 357.38 vs. 1.51).
Expression levels of genes encoding plant saccharide degrading enzymes, such as cellulases, beta-glucosidases and hemicellulases, were also upregulated in TBC-A compared to TBC-6 during the fermentation process (Fig. 3 and Supplementary Table S4). Three groups of cellulolytic enzymes are required to digest cellulose efficiently37: endo-β-1,4-glucanases, cellobiohydrolases and β-glucosidases. The transcriptome analysis revealed that the transcription levels of genes encoding two exoglucanases or cellobiohydrolases (CBH), two endoglucanases and seven putative β-glucosidases (BGL) were upregulated significantly in TBC-A compared to TBC-6 (p value < 0.05). Of them, two genes encoding CBH (Bc1G_08989 and Bc1G_10880) were almost not expressed (RPKM < 1) in TBC-6 samples but showed significantly induced expression in TBC-A samples. For example, The RPKM value of Bc1G_08989 and Bc1G_10880 were upregulated 391.86-fold (RPKM values 76.804 vs. 0.196) and 54.37-fold (RPKM values 16.312 vs. 0.300) in TBC-A-120 h sample than that of the TBC-6-120 h sample respectively. The transcriptome analysis also showed that the transcription levels of many genes encoding hemicelluloses-degrading enzymes were upregulated in TBC-A compared to TBC-6. The gene transcription levels of three β-1, 4-endoxylanase (XYN), two β-1,4-xylosidase (XYL), a xyloglucan-specific β-1,4-endoglucanase (XEG) and a β-1,4-endomannanase (MAN) were induced significantly in TBC-A compared to TBC-6 (p value < 0.05). Among them, one gene encoding XEG (Bc1G_00594) was almost not expressed (RPKM < 0.5) in TBC-6 but was strongly induced in TBC-A. It was reported that these enzyme sets are required to hydrolyze the hemicellulose backbones of xylans, xyloglucan and mannan polysaccharides38,39, respectively. In addition, the transcription levels of several genes encoding other hemicellulose-degrading enzymes, which can release branching residues from xylan chains, such as N-arabinose, L-fucose, alpha-linked D-galactose, ferulic acid and acetyl residues, were also upregulated in TBC-A compared to TBC-6 (p value < 0.05), especially during the stationary phase of the fermentation process.
The analysis of significantly differentially expressed genes between the 12 samples also revealed that the transcription levels of genes encoding several amylases, dextrinases and other saccharide-degrading enzymes, such as β-galactosidases, which hydrolyze lactose into galactose and glucose, were induced in TBC-A compared to TBC-6 (p value < 0.05). In conclusion, the increased transcription levels of genes encoding polysaccharide hydrolases and sugar transporters in TBC-A suggested that the supply of saccharides as the nutrients was enhanced in this B. cinerea mutant strain. Glucose that is transported into fungal cells can enter the glycolysis and pentose phosphate pathways for subsequent primary and secondary metabolism (Fig. 4).
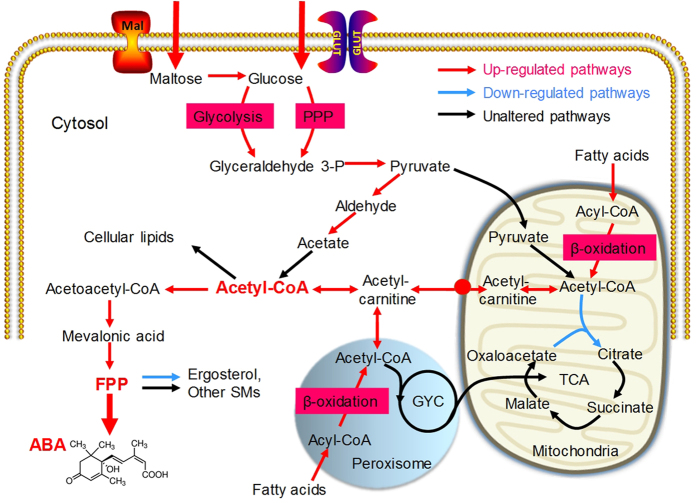
Figure 4. Schematic representation of metabolic pathways related to ABA production that were differentially regulated (p value < 0.05) between TBC-A and TBC-6 during the fermentation process.
The red arrows indicate upregulated pathways, blue arrows indicate downregulated pathways, and black arrows indicate pathways that were not significantly regulated. The most differentially regulated pathways are indicated by thick arrows. The biochemical pathways depicted are glycolysis, pentose phosphate pathway (PPP) and β-oxidation of fatty acids. Key metabolites such as cytosol acetyl-CoA, FPP and ABA are shown in bold red font. •Represents the putative mitochondrial carnitine carrier.
Differentially expressed genes involved in metabolic pathways related to the precursor acetyl-CoA
The central metabolite acetyl coenzyme A (acetyl-CoA) is a common link between primary and secondary metabolic pathways and ABA is ultimately derived from cytosolic acetyl-CoA in B. cinerea. Considering the ABA overproduction phenotype of the mutant TBC-A strain, we suspected that an enhanced generation of acetyl-CoA might occur in the cytoplasm of the B. cinerea TBC-A strain. The genes involved in acetyl-CoA biosynthesis and transportation were analyzed and the transcriptome data revealed that most of them were differentially expressed between the two strains. As illustrated in Fig. 4, transcription levels of genes involved in several metabolic pathways related to the production of cytosolic acetyl-CoA were upregulated in TBC-A compared to TBC-6, which suggested an increased supply of acetyl-CoA in the TBC-A strain.
The comparative transcriptome analysis revealed that the transcription levels of several genes involved in catabolic glycolysis and pentose phosphate pathway (PPP), including one gene (Bc1G_11968) encoding glyceraldehyde-3-phosphate dehydrogenase (GAPDH), two genes (Bc1G_03837 and Bc1G_11392) encoding phosphoglycerate mutase (PGAM), one gene (Bc1G_00350) encoding enolase (ENO), one gene (Bc1G_03095) encoding ribose-5-phosphate isomerase (RPI), one gene (Bc1G_02529) encoding Ribulose-phosphate 3-epimerase (RPE) and one gene (Bc1G_00585) encoding transketolase (TKT), were upregulated in TBC-A samples compared to those in TBC-6 samples (p value < 0.05) during the stationary phase of the fermentation process (Fig. 4 and Supplementary Table S5), which may allow the increase of carbon flux from glucose to pyruvate.
Cytosolic pyruvate can be converted into acetyl-CoA through two different pathways: the pyruvate dehydrogenase bypass (PDH-bypass), which converts pyruvate into acetyl-CoA in the cytosol40,41, and the mitochondrion-associated pyruvate dehydrogenase complex (PDH), which catalyzes the pyruvate decarboxylation reaction releasing acetyl-CoA into the mitochondrial matrix42. The gene transcription levels of two of the three enzymatic steps involved in the PDH-bypass were upregulated in TBC-A compared to TBC-6 (p value < 0.05). The transcript abundances of a pyruvate decarboxylase (PDC)-encoding gene (Bc1G_11347) and a cytosolic aldehyde dehydrogenase (ALDH)-encoding gene (Bc1G_06362) were much higher in the TBC-A-120 h sample relative to the TBC-6-120 h sample (RPKM values 731.89 vs. 131.39 and 565.31 vs. 84.03, respectively), which suggested an enhanced cytosolic acetyl-CoA production through the PDH-bypass (Fig. 4 and Supplementary Table S5). In contrast, there was very little difference in the transcription levels of the genes encoding different components of PDH between TBC-A and TBC-6. In addition, as shown in Fig. 4 and Supplementary Table S5, the transcription levels of the genes involved in the tricarboxylic acid (TCA) cycle, a primary metabolic pathway consuming acetyl-CoA in the mitochondria, were unchanged or downregulated in TBC-A compared to TBC-6.
In many fungal species, acetyl-CoA can also be generated via the β-oxidation of fatty acids in both mitochondria and peroxisomes43,44,45. In the present study, the transcriptome data revealed that the transcription levels of 18 of 21 genes participating in mitochondrial and peroxisomal β-oxidation, including the genes encoding acyl-CoA oxidase, acyl-CoA dehydrogenase, enoyl-CoA hydratase, 3-hydroxyacyl-CoA dehydrogenase and 3-ketoacyl-CoA thiolase, were upregulated in TBC-A compared to TBC-6 (p value < 0.05) (Supplementary Table S5). Enhanced transcriptional activities of the genes participating in β-oxidation could lead to increased acetyl-CoA production in the mitochondria and peroxisomes of the TBC-A strain.
In fungi, the carnitine/acetyl-carnitine shuttle is essential for the intracellular transport of acetyl-CoA among the mitochondria, peroxisomes and the cytosol46,47,48,49. In the present study, the transcription levels of Bc1G_14516 and Bc1G_14666, which encode the B. cinerea ortholog of the mitochondrial/peroxisomal carnitine acetyltransferase Cat2 and the mitochondrial carnitine carrier50,51,52,53, respectively, were upregulated in TBC-A compared to TBC-6 (p value < 0.05). These results showed that the transcription activities of the genes participating in the transportion of acetyl-CoA across the mitochondrial and peroxisomal membranes were enhanced in TBC-A compared to TBC-6 during the fermentation process.
The primary fate of cytosolic acetyl-CoA is to serve as a precursor for cellular lipids, such as fatty acids. As shown in Supplementary Table S5, the transcription levels of the genes encoding enzymes involved in fatty acid biosynthesis, such as acetyl-CoA carboxylase (ACAC), fungi type fatty acid synthase (FASN) and [acyl-carrier-protein] S-malonyltransferase, were unaltered (or downregulated slightly) in TBC-A compared to TBC-6, which indicated that the fatty acid biosynthetic activities were similar between the two strains.
In summary, compared to TBC-6, the elevated gene transcription levels of the abovementioned metabolic pathways suggested an enhanced supply of cytosolic acetyl-CoA during the fermentation process of TBC-A and we suspect that an increased supply of the precursor, acetyl-CoA, might contribute to the ABA hyper-production phenotype of the mutant TBC-A strain.
Differentially expressed genes involved in the ABA biosynthetic pathway
As a sesquiterpene, ABA is synthesized using intermediates of the terpenoid backbone biosynthetic pathway. Biosynthesis of the terpenoid backbone for ABA biosynthesis in B. cinerea can be divided into two steps: Step 1 includes all 7 enzymatic steps involved in the biosynthesis of isopentenyl pyrophosphate (IPP) via the mevalonate (MVA) pathway18,19 (Fig. 5); Step 2 includes the biosynthesis of farnesyl pyrophosphate (FPP), the immediate precursor essential for the biosynthesis of ABA from IPP54.
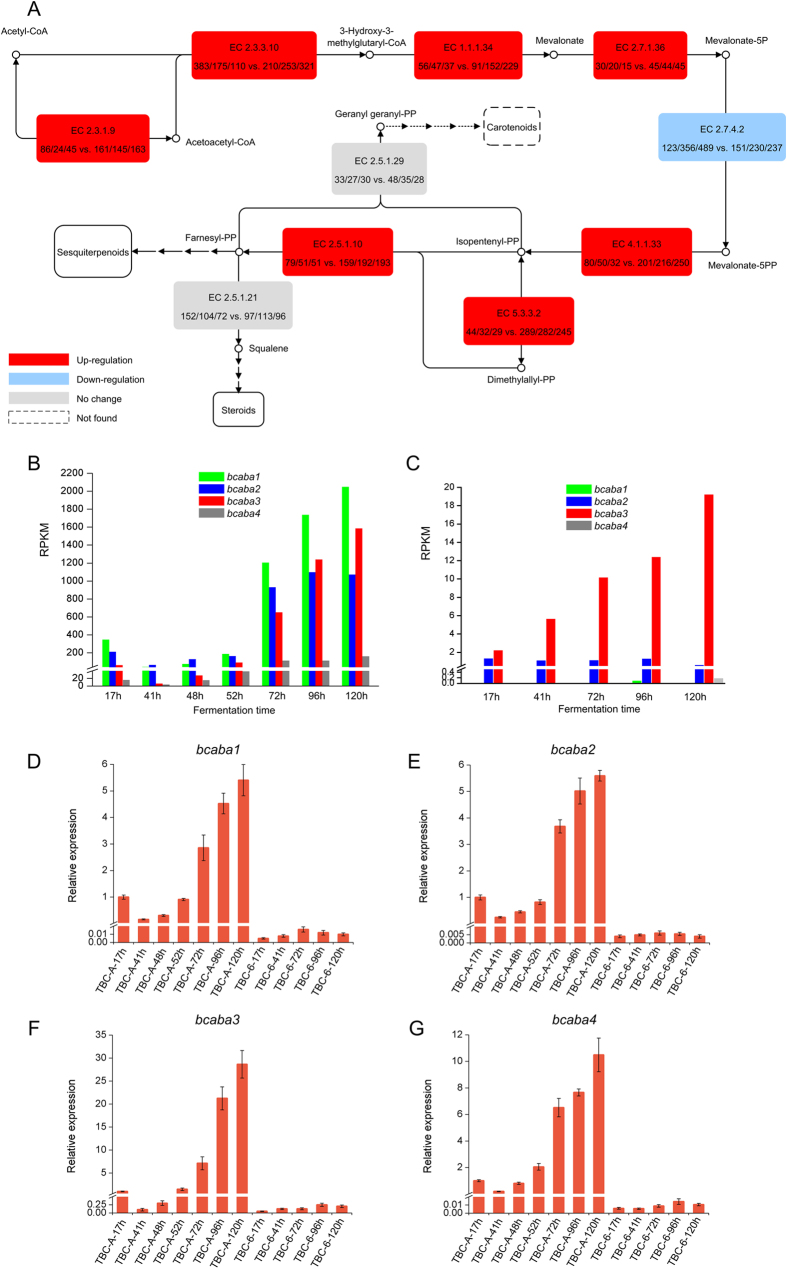
Figure 5. Divergent expression profiles of the genes involved in terpenoid backbone biosynthesis and the ABA biosynthetic gene cluster in TBC-A and TBC-6 under identical fermentation conditions.
(A) Schematic representation of the transcript abundance of terpenoid backbone biosynthetic genes in TBC-A and TBC-6 samples. Red boxes indicate increased gene expression, blue indicates repressed gene expression and gray indicates no significant differences were observed in TBC-A samples compared to TBC-6 samples. The numbers in the boxes correspond to the EC (Enzyme Commission) numbers, and the expression levels of the genes encoding these enzymes were calculated as RPKM values in the samples TBC-6-41 h, TBC-6-72 h, TBC-6-120 h,TBC-A-41 h, TBC-A-72 h and TBC-A-120 h. The dotted arrows and boxes indicate enzymatic steps or compounds that were not detected. The expression patterns of the 4 genes located in the ABA biosynthetic gene cluster were evaluated for TBC-A (B) and TBC-6 (C) during the fermentation process. Relative expression levels of bcaba1 (D), bcaba2 (E), bcaba3 (F) and bcaba4 (G) transcripts in the 12 culture samples of TBC-A and TBC-6 are shown. EC 2.3.1.9: acetyl-CoA acetyltransferase; EC 2.3.3.10: hydroxymethylglutaryl-CoA synthase; EC 1.1.1.34: 3-hydroxy-3-methylglutaryl-CoA reductase; EC 2.7.1.36: mevalonate kinase; EC 2.7.4.2: phosphomevalonate kinase; EC 4.1.1.33: diphosphomevalonate decarboxylase; EC 5.3.3.2: isopentenyl-diphosphate delta-isomerase; EC 2.5.1.10: farnesyl pyrophosphate synthase; EC 2.5.1.29: geranylgeranyl diphosphate synthase; EC 2.5.1.21: squalene synthase.
The comparative transcriptome analysis revealed that most genes involved in the MVA pathway as well as those involved in Step 2 were differentially expressed between the two strains. As shown in Fig. 5 and Supplementary Table S6, the transcription levels of most of the key MVA pathway genes were upregulated (p value < 0.05), with the exception of genes encoding hydroxymethylglutaryl-CoA synthase (HMGCS, encoded by Bc1G_09652 whose transcription level was also upregulated in the TBC-A-120 h sample compared with the TBC-6-120 h sample) and phosphomevalonate kinase (PMVK, encoded by Bc1G_07491). Additionally, two genes encoding acetoacetyl-CoA synthase (AACS, encoded by Bc1G_08841 and Bc1G_02728), which catalyzes the condensation reaction of acetyl-CoA and malonyl-CoA to produce acetoacetyl-CoA55, were detected in this study. The expression level of Bc1G_08841 and Bc1G_02728 did not differ obviously in TBC-A compered to TBC-6. For the enzymatic steps in Step 2 of the FPP backbone biosynthesis, the gene transcription levels of isopentenyl-diphosphate delta-isomerase (IDI, encoded by Bc1G_11480) and farnesyl pyrophosphate synthase (FPPS, encoded by Bc1G_02940), which catalyze the final steps of FPP biosynthesis, were also upregulated dramatically in TBC-A compared to TBC-6 (p value < 0.05).
Siewers et al. reported an ABA biosynthetic gene cluster in B. cinerea that consisted of the four genes bcaba1–426. bcaba1 and bcaba2 encode two putative P450 monooxygenases thought to be responsible for the hydroxylation of carbon atoms C-4′ and C-1′; bcaba4 encodes a putative short-chain dehydrogenase/reductase that may be responsible for the oxidation of the C-4′ carbon atom of ABA, while the function of bcaba3 is unknown. The gene expression of bcaba1–4 was identified in the transcriptomes of TBC-A and TBC-6. As shown in Fig. 5B-5C, the transcription levels of bcaba1–4 were very different between TBC-A and TBC-6 (p value < 0.05). Of them, bcaba1 and bcaba4 were almost not expressed (RPKM < 0.5) in TBC-6 samples. But they showed significantly induced expression in TBC-A samples (log2|fold change| > 5). In addition, the transcriptome data indicated a dramatic increase in bcaba1–4 gene expression when TBC-A transitioned from its late logarithmic phase into the stationary phase of the fermentation process. QRT-PCR was performed on the bcaba1–4 genes to validate their transcription levels in all 12 B. cinerea samples (Fig. 5D–G), and the qRT-PCR results agreed with the RNA-seq data, confirming the hyper-transcription levels of bcaba1–4 during the stationary phase of the fermentation process of TBC-A strain. It is suggested that the high expression levels of the bcaba1–4 genes in the ABA biosynthetic gene cluster play a key role in the ABA overproduction phenotype of the TBC-A strain.
Discussion
Our laboratory has developed an ABA-overproducing mutant strain, B. cinerea TBC-A, specifically for fermentative ABA production at the industrial scale. Although whole-genome sequencing has been performed on this mutant strain, the molecular basis beneath the ABA overproduction phenotype was unclear. Identification of the genes that may contribute to the overproduction of ABA in TBC-A has profound importance, especially for understanding the molecular mechanism underlying ABA biosynthesis and for rational strain improvement for better ABA productivity of B. cinerea strains.
It was previously assumed that the ABA biosynthetic genes in phytopathogenic fungi might be transferred from plants in a process of coevolution between host plants and parasites56. However, labeling experiments elucidated that fungal species biosynthesize ABA via a direct route from FPP21,25,57. ABA biosynthesis in B. cinerea can be divided into four steps: (i) Production of the C5 monomer IPP and dimethylallyl diphosphate (DMAPP). Feeding experiments using [1-13C]-D-glucose demonstrated that IPP and DMAPP are synthesized from the MVA pathway, in contrast to the MEP pathway in higher plants18,19. (ii) Head-to-tail condensations of IPP with a DMAPP starter unit to generate the linear prenyl diphosphate precursor FPP54. (iii) Cyclization and/or rearrangement of FPP to form the initial carbon backbone of terpenes. Although isomers of ionylideneacetate and/or ionylideneethanol have been identified from several fungal species21,22,24,58, the experimental evidence for FPP cyclization in the ABA biosynthetic pathway is still missing. (iv) Oxidation of the sesquiterpene skeleton to produce ABA. 18O labeling experiments demonstrated that the oxygen atoms at C-1, C-1′ and C-4′ of ABA were derived from atmospheric oxygen in B. cinerea21. In addition, 1′,4′-trans-diol ABA is likely the predominant precursor whose endogenous level is correlated with ABA production59. Siewers et al. reported an ABA biosynthetic gene cluster consisting of four genes bcaba1–426,27, which were presumed to be responsible for the oxidation and/or hydroxylation at carbon atoms C-1′ and C-4′ of ABA in B. cinerea. It is obviously that the biosynthetic mechanisms of ABA, especially the regulatory genes and the genes responsible for other enzymatic steps of ABA biosynthesis, such as the FPP cyclization/rearrangement, were not well explained in B. cinerea.
The comparative transcriptome analysis between B. cinerea TBC-A and its ancestral strain TBC-6 in the present study facilitates the understanding of the crucial genes and metabolic pathways related to ABA biosynthesis. Significantly increased transcription levels of genes involved in the MVA pathway and the biosynthesis of FPP were found in the ABA-overproducing mutant TBC-A. Furthermore, the genes involved in the MEP pathway and the biosynthesis of carotenoids (with the exception of phytoene) were not found. So we conclude that the ABA biosynthetic pathway of B. cinerea is indeed different from that of higher plants and B. cinerea did not inherited the ABA biosynthetic genes via horizontal gene transfer from plants. Furthermore, the transcriptome data revealed that changes in the expression levels of bcaba1–4 genes in the ABA biosynthetic gene cluster were consistent with the pattern of ABA production during the fermentation process of TBC-A (Figs 1 and 5), confirming the key roles this gene cluster playing in ABA biosynthesis.
We also paid particular attention to the candidate genes encoding terpene synthases (TSs) in TBC-A29,60, including 6 sesquiterpene cyclases (STCs)-encoding genes (Bc1G_00729, Bc1G_06357, Bc1G_09560, Bc1G_10537, Bc1G_14308 and Bc1G_16381) and 3 diterpene cyclases (DTCs)-encoding genes (Bc1G_06148, Bc1G_06751 and Bc1G_13295). The transcriptome data showed that the expression levels of these TS genes, which contribute to the cyclization and/or rearrangement of FPP derivatives, were unchanged or suppressed in TBC-A compared to TBC-6. These results indicate that the competitive biosynthesis of other terpenes is not induced in TBC-A.
Identification of functional genes involved in the ABA biosynthetic pathway in B. cinerea has profound importance. The comparative transcriptome analysis identified dozens of genes with similar expression patterns to those of bcaba1–4 during the fermentation process of TBC-A and TBC-6 (Supplementary Table S7). Most of these genes encode hypothetical proteins or predicted proteins with unknown functions, while several putative oxidoreductase and cytochrome P450 encoding genes were also identified. We propose that these genes may also be related to ABA biosynthesis in B. cinerea. Targeted inactivation of these genes will be performed to evaluate their impact on ABA biosynthesis.
The transcriptome data generated in this study provide a significant contribution to elucidating the differentially expressed genes involved in ABA biosynthesis, terpenoid biosynthesis, primary metabolism and other secondary metabolic pathways between B. cinerea TBC-A with TBC-6. Based on the comparative transcriptome analysis, we provide the first comprehensive view of whole-genome-scale gene expression differences which may contribute to the ABA overproduction in the TBC-A strain. This study should have its profound theoretical significance for elucidating the mechanism basis for ABA biosynthesis and facilitating further rational improvement of B. cinerea strains to promote the biotechnological production of ABA and other valuable secondary metabolites.
Methods
Strains and culture conditions
The B. cinerea wild-type strain TBC-6, first isolated in southwestern China more than 20 years ago and the ABA hyper-producing strain B. cinerea TBC-A were used in this study. B. cinerea strains TBC-A and TBC-6 were revived by growing on potato dextrose agar (PDA) slants at 26 °C for 7 d. Conidial suspensions with a final concentration of 1 × 106 conidia·mL−1 were inoculated into the seed culture (SC) medium containing (g·L−1) 30.0 of glucose, 10.0 of yeast extract, 10.0 of wheat bran, 10.0 of sucrose, 3.0 of maltose, 15.0 of NH4NO3 and 2.0 of MgSO4·7H2O. Seed cultivation was performed at 26 °C at 180 rpm on an orbital shaker. After 50 h of cultivation (late-exponential phase), 2.5 mL of the growth phase uniformed seed culture was inoculated into 50 mL shake-flask fermentation (SF) medium which contained (g·L−1): 5.0 of dextrin, 20.0 of glucose, 10.0 of yeast extract, 20.0 of soluble starch, 20.0 of soybean meal, 10.0 of wheat bran, 5.0 of maltose, 10.0 of fatty acids, 15.0 of NH4NO3, 2.0 of MgSO4·7H2O and 1.0 of K2HPO4. Shake-flask cultivation of TBC-A and TBC-6 was performed at 26 °C at 180 rpm on an orbital shaker.
Biomass and extracellular ABA quantification
The mycelia were collected from 10 mL culture samples of three randomly selected shake flasks, vacuum-filtered, washed with demineralized water, dried in a heat oven for 120 min at 100 °C and immediately weighed. The culture filtrates of each shake flask were collected for extracellular ABA quantification. ABA content was determined with a plant hormone abscisic acid ELISA kit (CUSABIO, Cat # CSB-E09159PI) following the manufacturer’s instructions. The ABA content was also confirmed by HPLC with the Agilent 1200 Pure Liquid Chromatography system (an Agilent 1260 Infinity Quaternary Pump VL with an Agilent 1260 Infinity Standard Autosampler and an Agilent 1260 Infinity Variable Wavelength Detector) and a Luna® 5 μm C18(2) LC Column (Phenomenex, Cat # 00G-4252-E0). ABA was quantified by the absorption at 254 nm using a standard curve generated using a commercial S-(+)-ABA (98% w/w, Lomon Bio Technology Co., Ltd., Sichuan, China). The quantity of ABA was calculated from the ratio between the integrals of the standard sample and that of the corresponding curve of the experimental samples. All these measurements were performed independently in triplicate, and the values are shown as the means ± standard error in the figures.
RNA extraction and library preparation
Ten milliliter culture samples from three randomly selected shake flasks were mixed and vacuum filtered. The mycelium pellets were washed with demineralized water, dried with filter paper, immediately frozen in liquid nitrogen and ground into powder. Total RNA extraction was performed with an E.Z.N.A.TM Fungal RNA Miniprep Kit (OMEGA, Cat # R6840-01) following the manufacturers’ instructions, and on-membrane DNaseI digestion was performed with an E.Z.N.A. RNase-Free DNase I Set (OMEGA, Cat # E1091) to remove residual DNA. RNA quality and concentration were measured using a NanoDrop spectro-photometer (Thermo Fisher Scientific, Waltham, MA, USA) and an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). RNA samples were purified and used for library construction with a NEBNext® UltraTM RNA Library Prep Kit (NEB, USA) following the manufacturers’ instructions.
Transcriptome sequencing and data analysis
The library preparations were sequenced on an Illumina HiSeq™ 2500 platform (Illumina, San Diego, CA, USA) by the Chinese National Human Genome Center (CHGC)-Shanghai. Illumina sequencing with 60× coverage of the B. cinerea genome was performed on all 12 RNA samples, and 24 million 100-bp paired-end raw reads were generated for each sample. Adaptors, ambiguous bases in the raw reads and reads of low quality were filtered out. After quality trimming, all clean reads were mapped to the reference genome of B. cinerea TBC-A using the bowtie2 (version 2.1.0) program61. Statistical analysis was conducted for the uniquely mapped reads, and RPKM values were calculated for each gene with Cufflinks 2.1.1. Genes differentially expressed between two samples were identified using the DEGseq package62 with the MARS (MA-plot-based method with Random Sampling) model. The thresholds used for the selection of DEGs were a log2|fold change| ≥ 1, FDR < 0.001 and RPKM value of at least one sample is larger than 10. A similarity search was performed against the UniProt Knowledgebase (http://www.uniprot.org/) for functional annotation of genes using a threshold of E-value < 1e−5. The heat map was generated with the Multi Experiment Viewer (MeV, v4.9) software63. RNA sequencing data were deposited in the GeneBank Sequence Read Archive (SRA) database under bioproject identifier PRJNA281796. Gene enrichment analysis was carried out with the DEGs identified. The functional classification of the DEGs was performed using the gene ontology (GO) analysis by GoPipe. Pathway-based analysis further informed the biological functions of the DEGs, indicating the significantly enriched metabolic or signal transduction pathways compared to the whole genome background. The statistical significance of the terms analyzed was calculated with hypergeometric test. A p value cut-off of 0.05 was used to identify the significantly differentially expressed genes in B. cinerea TBC-A compared to TBC-6 during the fermentation phases with rapidly increasing ABA titers. Statistical analysis was performed to TBC-A and TBC-6 samples from 72 h, 96 h and 120 h time points using the paired sample t-test. DEGs with significant changes in transcript abundance (p value < 0.05) were adopted in this study.
Quantitative RT-PCR
RNA-seq data were validated by examining the expression levels of 20 B. cinerea genes using qRT-PCR. The gDNA-free cDNA was synthesized from 1 μg of the total RNA samples used for RNA sequencing with the ReverTra Ace-α-® kit (Cat # FSK-101, TOYOBO, Japan). Quantitative PCR analysis was performed on a CFX96 Real-Time PCR Detection System (BioRad, USA) using TransStart Green qPCR SuperMix UDG (Transgen, China) with gene-specific primers (Supplementary Table S8). The relative abundances of selected transcripts were calculated using the 2−ΔΔCt method64. The B. cinerea tubulin gene (Bc1G_05600) was used as the reference gene. All qRT-PCR results represent the mean of three independent experiments.
Additional Information
How to cite this article: Ding, Z. et al. Comparative transcriptome analysis between an evolved abscisic acid-overproducing mutant Botrytis cinerea TBC-A and its ancestral strain Botrytis cinerea TBC-6. Sci. Rep. 6, 37487; doi: 10.1038/srep37487 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work received financial support from the National Natural Science Foundation of China (31501005). We acknowledge funding from the Open Research Fund of Key Laboratory of Environmental and Applied Microbiology, Chengdu Institute of Biology, Chinese Academy of Sciences (KLCAS-2016-07, KLEAMCAS201502). This work was also supported by funding from Science and Technology Service Network Initiative, Chinese Academy of Sciences (KFJ-SW-STS-143-1).
Footnotes
Author Contributions Z.D. contributed to the laboratory work, data analyses and manuscript writing; Z.Z., J.Z., D.L. and J.Z. conceived of the study and helped with the data analysis; J.Z. helped with the shake-flask fermentations; Z.Z., J.Y. and L.X. commented on the manuscript; D.S. helped with the draft of the manuscript; D.S. and H.T. conceived of and designed the study. All authors read and approved the final manuscript.
References
- Srivastava L. M. Abscisic acid In Plant Growth and Development, Hormones and Environment (ed. Srivastava L. M.) 217–232 (Elsevier, 2002). [Google Scholar]
- Yamaguchi I. et al. 4.02–plant hormones In Comprehensive Natural Products II, Chemistry and Biology (ed. Liu H. W. & Mander L.) 9–125 (Elsevier, 2010). [Google Scholar]
- Hey S. J., Byrne E. & Halford N. G. The interface between metabolic and stress signalling. Ann. Bot. 105, 197–203 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaart J. A. D. Abscisic acid metabolism and its regulation In New Comprehensive Biochemistry Volume 33, Biochemistry and Molecular Biology of Plant Hormones (ed. Hooykaas P. J. J., Hall M. A. & Libbenga K. R.) 189–207 (Elsevier, 1999). [Google Scholar]
- Zocchi E. et al. The temperature-signaling cascade in sponges involves a heat-gated cation channel, abscisic acid, and cyclic ADP-ribose. Proc. Nati. Acad. Sci. USA 98, 14859–14864 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruzzone S. et al. Abscisic acid is an endogenous cytokine in human granulocytes with cyclic ADP-ribose as second messenger. Proc. Nati. Acad. Sci. USA 104, 5759–5764 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameri P. et al. Impaired increase of plasma abscisic acid in response to oral glucose load in type 2 diabetes and in gestational diabetes. PLoS One 10, e0115992 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakthivel P., Sharma N., Klahn P., Gereke M. & Bruder D. Abscisic acid: a phytohormone and mammalian cytokine as novel pharmacon with potential for future development into clinical applications. Curr. Med. Chem. 23, 1549–1570 (2016). [DOI] [PubMed] [Google Scholar]
- Bassaganya-Riera J. et al. Mechanisms of action and medicinal applications of abscisic acid. Curr. Med. Chem. 17, 467–478 (2010). [DOI] [PubMed] [Google Scholar]
- Han X. et al. Synthesis, resolution and biological evaluation of cyclopropyl analogs of abscisic acid. Bioorg. Med. Chem. 23, 6210–6217 (2015). [DOI] [PubMed] [Google Scholar]
- Zaharia L. I., Galka M. M., Ambrose S. J. & Abrams S. R. Preparation of deuterated abscisic acid metabolites for use in mass spectrometry and feeding studies. J. Labelled Comp. Radiopharm. 48, 435–445 (2005). [Google Scholar]
- Balsevich J. J. et al. Response of cultured maize cells to (+)-abscisic acid, (−)-abscisic acid, and their metabolites. Plant Physiol. 106, 135–142 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marumo S. et al. Microbial production of abscisic acid by Botrytis cinerea. Agric. Biol. Chem. 46, 1967–1968 (1982). [Google Scholar]
- Dörffling K., Petersen W., Sprecher E., Urbasch I. & Hanssen H. P. Abscisic acid in phytopathogenic fungi of the genera Botrytis, Ceratocystis, Fusarium, and Rhizoctonia. Zeitschrift für Naturforschung C. 39, 683–684 (2014). [Google Scholar]
- Takayama T., Yoshida H., Araki K. & Nakayama K. Microbial production of abscisic acid with Cercospora rosicola. 1. Stimulation of abscisic acid accumulation by plant extracts. Biotechnol. Lett. 5, 55–58 (1983). [Google Scholar]
- Nambara E. & Marion-Poll A. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 56, 165–185 (2005). [DOI] [PubMed] [Google Scholar]
- Schwartz S. H., Qin X. & Zeevaart J. A. Elucidation of the indirect pathway of abscisic acid biosynthesis by mutants, genes, and enzymes. Plant Physiol. 131, 1591–1601 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai N., Yoshida R., Todoroki Y. & Ohigashi H. Biosynthesis of abscisic acid by the non-mevalonate pathway in plants, and by the mevalonate pathway in fungi. Biosci. Biotechnol. Biochem. 64, 1448–1458 (2000). [DOI] [PubMed] [Google Scholar]
- Oritani T. & Kiyota H. Biosynthesis and metabolism of abscisic acid and related compounds. Nat. Prod. Rep. 20, 414–425 (2003). [DOI] [PubMed] [Google Scholar]
- Tudzynski B. & Sharon A. Biosynthesis, biological role and application of fungal phytohormones In The Mycota X, Industrial Applications (ed. Osiewacz H. D.) 183–211 (Springer, 2002). [Google Scholar]
- Inomata M., Hirai N., Yoshida R. & Ohigashi H. The biosynthetic pathway to abscisic acid via ionylideneethane in the fungus Botrytis cinerea. Phytochemistry 65, 2667–2678 (2004). [DOI] [PubMed] [Google Scholar]
- Inomata M., Hirai N., Yoshida R. & Ohigashi H. Biosynthesis of abscisic acid by the direct pathway via ionylideneethane in a fungus, Cercospora cruenta. Biosci. Biotechnol. Biochem. 68, 2571–2580 (2004). [DOI] [PubMed] [Google Scholar]
- Yamamoto H., Inomata M., Tsuchiya S., Nakamura M. & Oritani T. Metabolism of chiral ionylideneacetic acids on the abscisic acid biosynthetic pathway in Cercospora. Biosci. Biotechnol. Biochem. 64, 2644–2650 (2000). [DOI] [PubMed] [Google Scholar]
- Okamoto M., Hirai N. & Koshimizu K. Biosynthesis of abscisic acid from α-ionylideneethanol in Cercospora pini-densiflorae. Phytochemistry 27, 3465–3469 (1988). [Google Scholar]
- Bennett R. D., Norman S. M. & Maier V. P. Intermediate steps in the biosynthesis of abscisic acid from farnesyl pyrophosphate in Cercospora rosicola. Phytochemistry 29, 3473–3477 (1990). [Google Scholar]
- Siewers V., Kokkelink L., Smedsgaard J. & Tudzynski P. Identification of an abscisic acid gene cluster in the grey mold Botrytis cinerea. Appl. Environ. Microbiol. 72, 4619–4626 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siewers V., Smedsgaard J. & Tudzynski P. The p450 monooxygenase BcABA1 is essential for abscisic acid biosynthesis in Botrytis cinerea. Appl. Environ. Microbiol. 70, 3868–3876 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong T. et al. Isolation of genes related to abscisic acid production in Botrytis cinerea TB-3-H8 by cDNA-AFLP. J. Basic Microbiol. 54, 204–214 (2014). [DOI] [PubMed] [Google Scholar]
- Amselem J. et al. Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet. 7, e1002230 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M. Y. et al. Genomic and transcriptomic analyses of the medicinal fungus Antrodia cinnamomea for its metabolite biosynthesis and sexual development. Proc. Nati. Acad. Sci. USA 111, E4743–E4752 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadista J. et al. Global genomic and transcriptomic analysis of human pancreatic islets reveals novel genes influencing glucose metabolism. Proc. Nati. Acad. Sci. USA 111, 13924–13929 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Keeffe G. et al. RNA-seq reveals the pan-transcriptomic impact of attenuating the gliotoxin self-protection mechanism in Aspergillus fumigatus. BMC Genomics 15, 894 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baruffini E., Goffrini P., Donnini C. & Lodi T. Galactose transport in Kluyveromyces lactis: major role of the glucose permease Hgt1. FEMS Yeast Res. 6, 1235–1242 (2006). [DOI] [PubMed] [Google Scholar]
- Billard P. et al. Glucose uptake in Kluyveromyces lactis: Role of the HGT1 gene in glucose transport. J. Bacteriol. 178, 5860–5866 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigamonte T. A. et al. Restricted sugar uptake by sugar-induced internalization of the yeast lactose/galactose permease Lac12. FEMS Yeast Res. 11, 243–251 (2011). [DOI] [PubMed] [Google Scholar]
- Reinders A. & Ward J. M. Functional characterization of the alpha-glucoside transporter Sut1p from Schizosaccharomyces pombe, the first fungal homologue of plant sucrose transporters. Mol. Microbiol. 39, 445–454 (2001). [DOI] [PubMed] [Google Scholar]
- Lynd L. R., Weimer P. J., van Zyl W. H. & Pretorius I. S. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 66, 506–577 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Brink J. & de Vries R. P. Fungal enzyme sets for plant polysaccharide degradation. Appl. Microbiol. Biotechnol. 91, 1477–1492 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries R. P. & Visser J. Aspergillus enzymes involved in degradation of plant cell wall polysaccharides. Microbiol. Mol. Biol. Rev. 65, 497–522 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starai V. J. & Escalante-Semerena J. C. Acetyl-coenzyme A synthetase (AMP forming). Cell. Mol. Life Sci. 61, 2020–2030 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba Y., Paradise E. M., Kirby J., Ro D. K. & Keasing J. D. Engineering of the pyruvate dehydrogenase bypass in Saccharomyces cerevisiae for high-level production of isoprenoids. Metab. Eng. 9, 160–168 (2007). [DOI] [PubMed] [Google Scholar]
- Guest J. R., Angier S. J. & Russell G. C. Structure, expression, and protein engineering of the pyruvate dehydrogenase complex of Escherichia coli. Ann. N. Y. Acad. Sci. 573, 76–99 (1989). [DOI] [PubMed] [Google Scholar]
- Galagan J. E. et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438, 1105–1115 (2005). [DOI] [PubMed] [Google Scholar]
- Maggio-Hall L. A. & Keller N. P. Mitochondrial beta-oxidation in Aspergillus nidulans. Mol. Microbiol. 54, 1173–1185 (2004). [DOI] [PubMed] [Google Scholar]
- Camões F. et al. New insights into the peroxisomal protein inventory: Acyl-CoA oxidases and -dehydrogenases are an ancient feature of peroxisomes. Biochim. Biophys. Acta. 1853, 111–125 (2015). [DOI] [PubMed] [Google Scholar]
- Elgersma Y., van Roermund C. W., Wanders R. J. & Tabak H. F. Peroxisomal and mitochondrial carnitine acetyltransferases of Saccharomyces cerevisiae are encoded by a single gene. EMBO J. 14, 3472–3479 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strijbis K. et al. Contributions of carnitine acetyltransferases to intracellular acetyl unit transport in Candida albicans. J. Biol. Chem. 285, 24335–24346 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strijbis K. et al. Carnitine-dependent transport of acetyl coenzyme A in Candida albicans is essential for growth on nonfermentable carbon sources and contributes to biofilm formation. Eukaryot. Cell 7, 610–618 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son H. et al. Mitochondrial carnitine-dependent acetyl coenzyme A transport is required for normal sexual and asexual development of the ascomycete Gibberella zeae. Eukaryot. Cell 11, 1143–1153 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez P. et al. Functional analysis of mutations in the human carnitine/acylcarnitine translocase in Aspergillus nidulans. Fungal Genet. Biol. 39, 211–220 (2003). [DOI] [PubMed] [Google Scholar]
- De Lucas J. R. et al. The Aspergillus nidulans carnitine carrier encoded by the acuH gene is exclusively located in the mitochondria. FEMS Microbiol. Lett. 201, 193–198 (2001). [DOI] [PubMed] [Google Scholar]
- Palmieri L. et al. Identification of the mitochondrial carnitine carrier in Saccharomyces cerevisiae. FEBS Lett. 462, 472–476 (1999). [DOI] [PubMed] [Google Scholar]
- van Roermund C. W., Hettema E. H., van den Berg M., Tabak H. F. & Wanders R. J. Molecular characterization of carnitine-dependent transport of acetyl-CoA from peroxisomes to mitochondria in Saccharomyces cerevisiae and identification of a plasma membrane carnitine transporter, Agp2p. EMBO J. 18, 5843–5852 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewick P. M. The biosynthesis of C5-C25 terpenoid compounds. Nat. Prod. Rep. 19, 181–222 (2002). [DOI] [PubMed] [Google Scholar]
- Okamura E., Tomita T., Sawa R., Nishiyama M. & Kuzuyama T. Unprecedented acetoacetyl-coenzyme A synthesizing enzyme of the thiolase superfamily involved in the mevalonate pathway. Proc. Nati. Acad. Sci. USA 107, 11265–11270 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman D. J. & Ragan M. A. Evolution of biochemical pathways: evidence from comparative biochemistry. Annu. Rev. Plant Physiol. 31, 639–678 (1980). [Google Scholar]
- Bennett R. D., Norman S. M. & Maier V. P. Biosynthesis of abscisic acid from farnesol derivatives in Cercospora rosicola. Phytochemistry 23, 1913–1915 (1984). [Google Scholar]
- Kato T., Oritani T. & Yamashita K. Metabolism of (2Z,4E)-γ-ionylideneethanol and (2Z,4E)-γ-ionylideneacetic acid in Cercospora cruenta. Agric. Biol. Chem. 51, 2695–2699 (1987). [Google Scholar]
- Hirai N., Okamoto M. & Koshimizu K. The 1′,4′-trans-diol of abscisic acid, a possible precursor of abscisic acid in Botrytis cinerea. Phytochemistry 25, 1865–1868 (1986). [Google Scholar]
- Christianson D. W. Structural biology and chemistry of the terpenoid cyclases. Chem. Rev. 106, 3412–3442 (2006). [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M. & Salzberg S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. K., Feng Z. X., Wang X., Wang X. W. & Zhang X. G. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26, 136–138 (2010). [DOI] [PubMed] [Google Scholar]
- Saeed A. I. et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34, 374–378 (2003). [DOI] [PubMed] [Google Scholar]
- Schmittgen T. D. & Livak K. J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.