Abstract
Olfactory receptors are believed to play a central role in insects host-seeking, mating, and ovipositing. On the basis of male and female antennal transcriptome of adult Apolygus lucorum, a total of 110 candidate A. lucorum odorant receptors (AlucOR) were identified in this study including five previously annotated AlucORs. All the sequences were validated by cloning and sequencing. Tissue expression profiles analysis by RT-PCR indicated most AlucORs were antennal highly expressed genes. The qPCR measurements further revealed 40 AlucORs were significantly higher in the antennae. One AlucOR was primarily expressed in the female antennae, while nine AlucORs exhibited male-biased expression patterns. Additionally, both the RPKM value and RT-qPCR analysis showed AlucOR83 and AlucOR21 were much higher abundant in male antennae than in female antennae, suggesting their different roles in chemoreception of gender. Phylogenetic analysis of ORs from several Hemipteran species demonstrated that most AlucORs had orthologous genes, and five AlucOR-specific clades were defined. In addition, a sub-clade of potential male-based sex pheromone receptors were also identified in the phylogenetic tree of AlucORs. Our results will facilitate the functional studies of AlucORs, and thereby provide a foundation for novel pest management approaches based on these genes.
The detection and discrimination of semiochemicals in the environment by specialized sensilla plays an important role in insect survival and reproduction1,2. For insects, chemosensory sensilla distribute over the surface of chemosensory tissues including antennae, palps, mouth parts, tarsi, and many other organs3, which mediate many key behaviors, such as host-seeking, mate choice, oviposition site selection, and predator avoidance2. The antenna is a specialized organ for insect sensing, which is the most significant organs of olfaction, housing thousands of olfactory sensory neurons (OSNs) that extend their dendrites up into the sensilla and project their axons towards the brain4,5. Volatile chemicals can be transformed into electrical signals by these OSNs and then these signals were preliminarily integrated and sent to higher brain centers by antennal lobe (AL) to finally generate a behavioral response6. Diverse olfactory proteins are evolved in this olfactory sensation process, including odorant-binding proteins (OBP), chemosensory proteins (CSPs), sensory neuron membrane proteins (SNMPs), odorant-degrading enzymes (ODEs), ionotropic receptors (IRs), and odorant receptors (ORs)2,3,7,8. OBPs are thought to be the first proteins that selectively bind to liposoluble odor molecules, and acted as a carrier to transport odorants through water-soluble lymph within sensilla to the ORs in the membrane of ORN dendrites9. After activating the ORs, distinct ODEs will degrade odorants and maintain the sensitivity of ORNs2,7,10. Thus far, information on this peripheral olfactory process is very limited, especially in the odorants inactivation step.
Insects mainly rely on ORs to perform the long distance detection of volatile molecules11. ORs, which span the dendritic membrane with seven alpha helices, and present an inverted topology (intracellular N-terminus) compared with mammalian ORs, are responsible for the conversion of chemical message to an electrical signal12. A typical OR unit functions as a dimer complex with the odorant receptor co-receptor (Orco), which is highly conserved among insect species13. Orco is believed to interact with each of the divergent ORs forming ligand-gated ion channels and to enhance odorant responsiveness12,14. Since the first insect ORs discovered in the fruit fly Drosophila melanogaster5,15, multiple OR repertoires have been identified in a variety of insect species through whole-genome sequencing, including Diptera, Hymenoptera, Lepidoptera, Coleoptera, Hemiptera, and Blattodea. The number of OR genes varies significantly from 62 in Drosophila melanogaster to 259 in Tribolium castaneum and up to 350 in Camponotus floridanus15,16,17, reflecting a various evolution of insect OR genes. Silencing of the olfactory co-receptor gene in Apolygus lucorum, Lymantria dispar and Dendroctonus armandi leads to electroantennographic (EAG) response declining to major semiochemicals18,19,20. In recent years, many Lepidoptera insect pheromone receptors were explored by using the Xenopus oocyte expression system21,22,23. However, to date, the exact functions of insect OR genes are largely unknown.
The green plant bug A. lucorum is one of the most destructive agricultural insects in China, feeding on over 150 recorded host plants24, including cotton, fruits, and vegetables25,26. With successful promotion and cultivation of transgenic insect resistant cotton since 1997, the population of this non-target pest has been increasing gradually in the past two decades, causing large economic losses27. Not surprisingly, the widespread planting of Bacillus thuringiensis (Bt) cotton have effectively controlled Lepidopteran pests and reduced the use of chemical pesticides, and as a side effect causing secondary non-target insects becoming major pests in the cotton field27. It was reported that A. lucorum was a typical representative that emerged as the key pest after the wide adoption of Bt cotton. To target and exploit simple and effective coping strategies, lots of studies on A. lucorum have been performed, including its physiology, chemoecology, and insecticide resistance in order to develop novel control methods28,29. However, the molecular components and mechanisms comprising A. lucorum olfactory system that could be potential novel targets for controlling this mirid bug have not been fully elucidated. In field experiments, six electrophysiological active compounds including m-xylene, butyl acrylate, butyl propionate, butyl butyrate, (Z)-3-hexen-1-ol, and (Z)-3-hexenyl acetate from flowering Artemisia plants were considered to be the effective substance to attract A. lucorum adults30. Moreover, an antenna highly expressed olfactory receptor gene, AlucOR28, was identified to sensitively tune to (Z)-3-hexenyl acetate and several flowering compounds31. Further study on the molecular mechanisms underlying the detections of these compounds is needed.
The identification of genes encoding OR families is a key step toward understanding the characteristics of A. lucorum olfactory systems. However, till now, our understandings for the OR genes of A. lucorum at the molecular level is very limited. To elucidate the molecular mechanism of A. lucorum olfactory system and to design novel coping strategies against this green plant bug, we performed a transcriptome analysis of both female and male antennae, and a total of 110 OR genes from A. lucorum were identified successfully. The expression patterns of these candidate ORs in different tissues were also examined by using reverse transcription PCR (RT-PCR) and quantitative real-time PCR (qPCR) in this study.
Results
Analysis of A. lucorum antennae transcriptome
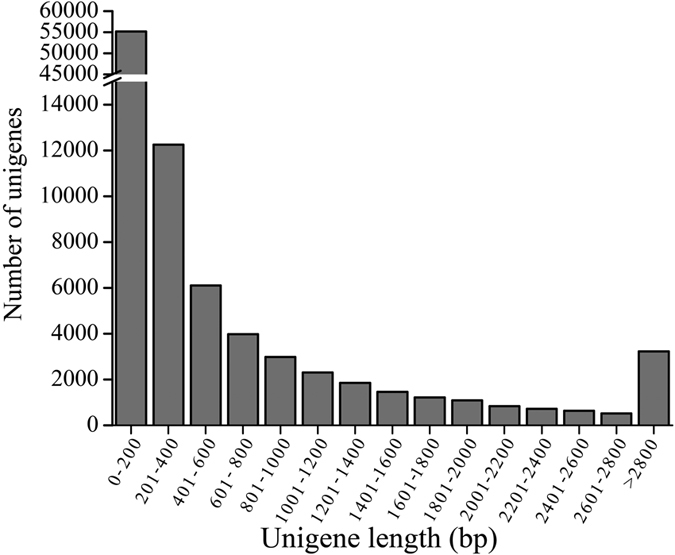
To identify candidate OR genes from A. lucorum, two transcriptomes of the male and female antennae were generated by HiSeq 2500 platform. A total of 88,020,104 (length between 150 to 200 bp) and 84,676,900 raw reads (length between 150 to 200 bp) were produced from the female and male antennae samples, and after filtering, 85,592,106 and 82,394,110 clean reads were assembled into 73,247 (mean length 829 bp) and 75,881 (mean length 808 bp) unigenes, respectively. The assembly of all clean reads together led to the generation of 133,447 transcripts with a mean length of 842 bp. After merging and clustering, 94,321 unigenes with a mean length of 698 bp and N50 of 1288 bp were acquired (Table 1), and 16,821 unigenes were larger than 1,000 bp in length, which comprised 17.80% of all unigenes (Fig. 1).
Table 1. Summary of antennal transcriptome assembley.
| Statistics proect | Transcripts | Unigenes |
|---|---|---|
| Minimum length | 201 bp | 201 bp |
| Mean length | 842 bp | 698 bp |
| Median length | 404 bp | 334 bp |
| Max length | 26,290 bp | 26,290 bp |
| N50 | 1,579 bp | 1,288 bp |
| N90 | 297 bp | 256 bp |
| Total Nucleotides | 112,413,388 | 65,824,946 |
| total number | 133,447 | 94,321 |
Figure 1. The length distribution of the assembled unigenes from A. lucorum male and female antennal transcriptome.
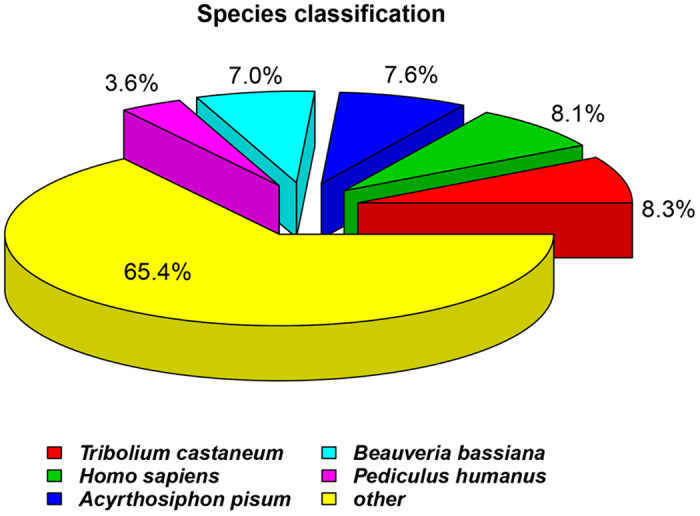
BLASTx and BLASTn homology searches of all 94,321 unigenes with an E-value <1.0E-5 showed that 33,076 unigenes (35.06%) had BLASTx hits in the Nr databases and 12,319 (13.06%) had BLASTn hits in the Nt databases. Among the annotated unigenes, the highest number of hits included 2,730 unigenes that were homologous to Tribolium castaneum sequences, and the distribution of the other best match species is shown in Fig. 2.
Figure 2. Species distribution of unigenes’ best-hit annotation term in nr database.
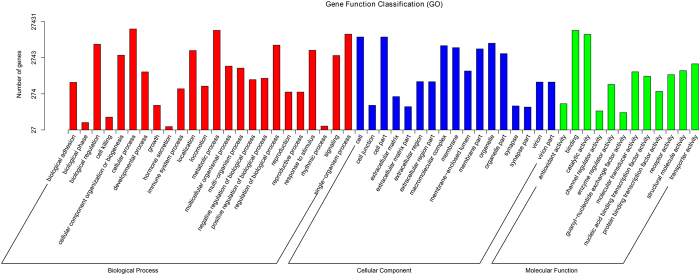
GO assignments were used to functionally classify the predicted proteins. Of all the unigenes, 27,431 (29.08%) could be classified into three functional categories: molecular function, biological process, and cellular component (Fig. 3). In molecular function category, the genes expressed in the antennae were mostly linked to binding (15,728/44.89% unigenes) and catalytic activity (12,187/34.78% unigenes). In terms of the biological process, the most represented biological processes were cellular processes (17,197/21.23% unigenes), metabolic processes (15,800/19.51% unigenes), and single-organism process (12,343/15.24% unigenes). In the cellular component terms, cell (10,304/20.06% unigenes) and cell part (10,304/20.06% unigenes) constitute the most abundant categories (Fig. 3).
Figure 3. Gene ontology classifications of the A. lucorum unigenes.
Identification of candidate ORs
A total of 110 candidate OR genes with amino acid sequences homology to known insect ORs were identified based on the antennal transcriptome data analysis of A. lucorum, among which 46 sequences encoded a full-length ORF with length ranging from 374 to 471 amino acids (Table 2). Five previously described ORs (AlucOR30, AlucOR18, AlucOR12, AlucOR28, and AlucOrco) with completed ORF were identified again in our dataset with a high level of identity (95–100%)18,31, and the remaining OR genes were named as “AlucORx” (x = 1–11, 13–17, 19–27, 29, 30–109), which was consistent with the general naming of OR genes. All the full length OR genes showed 3–8 predicted transmembrane domains and the majority of incomplete OR genes also showed multiple transmembrane domains (Table 2), which is a typical characteristic of insect OR genes, indicating that these proteins were located in the membrane of the neuron cells. Except for AlucOR25 and AlucOR88 exhibiting a high degree of similarity (67.9%), all the other candidate AlucORs were highly divergent sharing relatively low amino acid identities (19–48%) (Supplementary Table 1). In addition, all the candidate AlucORs also had relative low amino acid identities (19–63%) with the homologous ORs in other species according to the BLASTx results of NCBI (Table 2).
Table 2. List of odorant receptor genes in A. lucorum antennal transcriptome.
| Gene name | Accesion number | ORF (aa) | Completeness | TM (No.) | Accesion number | Discription | Species | Identity (%) | E-value | Score |
|---|---|---|---|---|---|---|---|---|---|---|
| AlucOR1 | KU958180 | 285 | 5′ lost | 4 | gb|AKS44360.1 | olfactory receptor 12 | Apolygus lucorum | 37 | 9.53E-53 | 189 |
| AlucOR2 | KU958181 | 403 | Complete | 6 | ref|XP_014293859.1 | odorant receptor 85b-like isoform X1 | Halyomorpha halys | 23 | 6.91E-25 | 114 |
| AlucOR3 | KU958182 | 426 | Complete | 3 | gb|AKS44362.1 | olfactory receptor 28 | Apolygus lucorum | 32 | 2.07E-64 | 224 |
| AlucOR4 | KU958183 | 440 | Complete | 6 | ref|XP_014271039.1 | odorant receptor 83a-like | Halyomorpha halys | 26 | 2.65E-23 | 110 |
| AlucOR5 | KU958184 | 429 | 5′ lost | 5 | gb|AKI29040.1 | odorant receptor 49b-2 | Bactrocera dorsalis | 29 | 2.63E-05 | 57 |
| AlucOR6 | KU958185 | 113 | 5′ and 3′ lost | 2 | gb|ALD51489.1 | odorant receptor 121 | Locusta migratoria | 33 | 1.9 | 37 |
| AlucOR7 | KU958186 | 367 | 5′ lost | 5 | ref|XP_014287492.1 | odorant receptor 4-like | Halyomorpha halys | 22 | 8.17E-10 | 71 |
| AlucOR8 | KU958187 | 99 | 5′ and 3′ lost | 0 | gb|AKS44362.1 | olfactory receptor 28 | Apolygus lucorum | 63 | 1.23E-32 | 128 |
| AlucOR9 | KU958188 | 397 | Complete | 6 | ref|XP_014274444.1 | odorant receptor 47a-like | Halyomorpha halys | 24 | 2.63E-10 | 72 |
| AlucOR10 | KU958189 | 378 | Complete | 3 | ref|XP_003689890.2| | odorant receptor Or1-like | Apis florea | 24 | 0.025 | 47 |
| AlucOR11 | KU958190 | 398 | 5′ lost | 6 | ref|XP_014250665.1 | uncharacterized protein LOC106667308 | Cimex lectularius | 26 | 1.07E-26 | 120 |
| AlucOR13 | KU958192 | 438 | Complete | 6 | gb|AKS44360.1 | olfactory receptor 12 | Apolygus lucorum | 30 | 2.67E-47 | 179 |
| AlucOR14 | KU958193 | 375 | Complete | 4 | ref|XP_014258533.1 | odorant receptor 47a-like | Cimex lectularius | 31 | 2.68E-20 | 100 |
| AlucOR15 | KU958194 | 107 | 5′ lost | 1 | ref|XP_014249919.1 | putative odorant receptor 69a, isoform A | Cimex lectularius | 43 | 1.97E-20 | 94 |
| AlucOR16 | KU958195 | 390 | 5′ lost | 5 | ref|XP_014290317.1 | odorant receptor 4-like | Halyomorpha halys | 22 | 1.09E-14 | 85 |
| AlucOR17 | KU958196 | 424 | 5′ lost | 5 | ref|XP_014287936.1 | uncharacterized protein LOC106688133 isoform X1 | Halyomorpha halys | 30 | 1.13E-23 | 110 |
| AlucOR19 | KU958198 | 69 | 5′ lost | 1 | ref|XP_014278373.1 | gustatory and pheromone receptor 32a-like | Halyomorpha halys | 58 | 6.09E-16 | 79 |
| AlucOR20 | KU958199 | 402 | 5′ lost | 6 | ref|XP_014258779.1 | putative odorant receptor 92a | Cimex lectularius | 43 | 1.4E-104 | 327 |
| AlucOR21 | KU958200 | 352 | 5′ lost | 4 | ref|XP_014294765.1 | odorant receptor 4-like | Halyomorpha halys | 26 | 1.57E-15 | 87 |
| AlucOR22 | KU958201 | 403 | Complete | 6 | ref|XP_001810505.1 | odorant receptor Or1 | Tribolium castaneum | 25 | 0.000836 | 52 |
| AlucOR23 | KU958202 | 426 | Complete | 6 | ref|XP_014287492.1 | odorant receptor 4-like | Halyomorpha halys | 26 | 8.84E-27 | 120 |
| AlucOR24 | KU958203 | 324 | 3′ lost | 5 | ref|XP_014256416.1 | odorant receptor 46a, isoform B-like | Cimex lectularius | 45 | 1.43E-84 | 272 |
| AlucOR25 | KU958204 | 430 | 5′ lost | 6 | gb|AKS44360.1 | olfactory receptor 12 | Apolygus lucorum | 30 | 1.1E-45 | 175 |
| AlucOR26 | KU958205 | 281 | 3′ lost | 4 | ref|NP_001177519.1 | odorant receptor 100 | Nasonia vitripennis | 38 | 2 | 40 |
| AlucOR27 | KU958206 | 392 | Complete | 6 | ref|XP_014287044.1 | uncharacterized protein LOC106687584 | Halyomorpha halys | 31 | 9.15E-17 | 91 |
| AlucOR29 | KU958207 | 403 | Complete | 5 | gb|ALD51384.1 | odorant receptor 61 | Locusta migratoria | 21 | 0.002 | 51 |
| AlucOR31 | KU958208 | 464 | 5′ lost | 4 | ref|XP_014270188.1 | odorant receptor 4-like isoform X1 | Halyomorpha halys | 24 | 5.4E-35 | 145 |
| AlucOR32 | KU958209 | 431 | Complete | 6 | ref|XP_014287492.1 | odorant receptor 4-like | Halyomorpha halys | 29 | 2.02E-41 | 162 |
| AlucOR33 | KU958210 | 96 | 5′ lost | 0 | ref|XP_011194820.1 | odorant receptor Or2-like | Bactrocera cucurbitae | 34 | 0.000896 | 47 |
| AlucOR34 | KU958211 | 328 | 5’ lost | 3 | ref|XP_014261896.1 | odorant receptor 85c-like | Cimex lectularius | 40 | 1.21E-73 | 244 |
| AlucOR35 | KU958212 | 374 | 5′ lost | 7 | ref|XP_014293240.1 | gustatory and odorant receptor 63a-like | Halyomorpha halys | 22 | 3.15E-11 | 75 |
| AlucOR36 | KU958213 | 387 | Complete | 6 | ref|XP_014257038.1 | odorant receptor Or2-like isoform X2 | Cimex lectularius | 31 | 2.48E-26 | 119 |
| AlucOR37 | KU958214 | 397 | 5′ lost | 6 | ref|XP_014293859.1 | odorant receptor 85b-like isoform X1 | Halyomorpha halys | 27 | 7.09E-22 | 105 |
| AlucOR38 | KU958215 | 57 | 5′ lost | 0 | ref|XP_317761.2 | AGAP007757-PA | Anopheles gambiae str. PEST | 31 | 5.6 | 35 |
| AlucOR39 | KU958216 | 394 | Complete | 5 | ref|XP_014254258.1 | uncharacterized protein LOC106669355 isoform X2 | Cimex lectularius | 45 | 6.4E-97 | 305 |
| AlucOR40 | KU958217 | 437 | Complete | 6 | ref|XP_014274444.1 | odorant receptor 47a-like | Halyomorpha halys | 23 | 2.86E-19 | 99 |
| AlucOR41 | KU958218 | 415 | Complete | 6 | gb|AKS44360.1 | olfactory receptor 12 | Apolygus lucorum | 30 | 1.84E-39 | 157 |
| AlucOR42 | KU958219 | 433 | 5′ lost | 7 | ref|XP_014289672.1 | odorant receptor 49a-like | Halyomorpha halys | 26 | 1.99E-17 | 94 |
| AlucOR43 | KU958220 | 420 | 5′ lost | 7 | ref|XP_014249551.1 | putative odorant receptor 92a | Cimex lectularius | 31 | 8.24E-55 | 199 |
| AlucOR44 | KU958221 | 393 | Complete | 8 | ref|XP_014287040.1 | odorant receptor 4-like | Halyomorpha halys | 46 | 9.11E-06 | 58 |
| AlucOR45 | KU958222 | 391 | Complete | 6 | ref|XP_014287040.1 | odorant receptor 4-like | Halyomorpha halys | 48 | 8.02E-16 | 89 |
| AlucOR46 | KU958223 | 95 | 5′ lost | 0 | ref|XP_014287040.1 | odorant receptor 4-like | Halyomorpha halys | 47 | 3.32E-19 | 91 |
| AlucOR47 | KU958224 | 337 | 5′ lost | 5 | ref|XP_014287040.1 | odorant receptor 4-like | Halyomorpha halys | 27 | 1.24E-26 | 119 |
| AlucOR48 | KU958225 | 374 | Complete | 6 | ref|XP_014278976.1 | uncharacterized protein LOC106682571 | Halyomorpha halys | 26 | 8.19E-10 | 71 |
| AlucOR49 | KU958226 | 336 | 5′ and 3′ lost | 6 | ref|XP_014258861.1 | uncharacterized protein LOC106672172 | Cimex lectularius | 25 | 9.02E-16 | 87 |
| AlucOR50 | KU958227 | 413 | Complete | 6 | ref|XP_014256564.1 | uncharacterized protein LOC106670594 isoform X1 | Cimex lectularius | 26 | 3.24E-38 | 152 |
| AlucOR51 | KU958228 | 406 | 5′ lost | 6 | ref|XP_014287492.1 | odorant receptor 4-like | Halyomorpha halys | 29 | 6.66E-42 | 162 |
| AlucOR52 | KU958229 | 437 | Complete | 6 | ref|XP_014273407.1 | uncharacterized protein LOC106679017 isoform X1 | Halyomorpha halys | 25 | 1.38E-24 | 114 |
| AlucOR53 | KU958230 | 434 | Complete | 6 | gb|AKS44361.1 | olfactory receptor 18 | Apolygus lucorum | 31 | 1.81E-49 | 185 |
| AlucOR54 | KU958231 | 411 | Complete | 5 | ref|XP_014270190.1 | odorant receptor 4-like isoform X3 | Halyomorpha halys | 23 | 2.18E-21 | 103 |
| AlucOR55 | KU958232 | 408 | 3′ lost | 4 | ref|XP_014250665.1 | uncharacterized protein LOC106667308 | Cimex lectularius | 27 | 8.02E-27 | 120 |
| AlucOR56 | KU958233 | 419 | Complete | 6 | gb|AKS44362.1 | olfactory receptor 28 | Apolygus lucorum | 57 | 6.7E-154 | 454 |
| AlucOR57 | KU958234 | 362 | 5′ lost | 5 | gb|ALR72576.1 | odorant receptor OR33 | Colaphellus bowringi | 26 | 5.39E-11 | 71 |
| AlucOR58 | KU958235 | 433 | Complete | 6 | gb|AKS44360.1 | olfactory receptor 12 | Apolygus lucorum | 40 | 2.55E-93 | 300 |
| AlucOR59 | KU958236 | 439 | Complete | 6 | ref|XP_014287936.1 | uncharacterized protein LOC106688133 isoform X1 | Halyomorpha halys | 23 | 7.58E-11 | 73 |
| AlucOR60 | KU958237 | 315 | 3′ lost | 5 | ref|XP_014249551.1 | putative odorant receptor 92a | Cimex lectularius | 33 | 1.42E-41 | 160 |
| AlucOR61 | KU958238 | 266 | 3′ lost | 3 | ref|XP_014273330.1 | odorant receptor 85b-like | Halyomorpha halys | 33 | 1.35E-31 | 131 |
| AlucOR62 | KU958239 | 224 | 5′ lost | 4 | gb|AKS44361.1 | olfactory receptor 18 | Apolygus lucorum | 25 | 0.000493 | 51 |
| AlucOR63 | KU958240 | 380 | Complete | 5 | ref|XP_014249919.1 | putative odorant receptor 69a, isoform A | Cimex lectularius | 31 | 7.33E-38 | 151 |
| AlucOR64 | KU958241 | 316 | 3′ lost | 4 | gb|AKS44361.1 | olfactory receptor 18 | Apolygus lucorum | 23 | 1.12E-17 | 93 |
| AlucOR65 | KU958242 | 449 | 5′ lost | 7 | ref|XP_014273407.1 | uncharacterized protein LOC106679017 isoform X1 | Halyomorpha halys | 28 | 4.03E-28 | 125 |
| AlucOR66 | KU958243 | 88 | 5′ lost | 0 | gb|AKS44360.1 | olfactory receptor 12 | Apolygus lucorum | 41 | 5.46E-14 | 76 |
| AlucOR67 | KU958244 | 417 | Complete | 5 | ref|XP_014254257.1 | uncharacterized protein LOC106669355 isoform X1 | Cimex lectularius | 42 | 1.07E-96 | 307 |
| AlucOR68 | KU958245 | 394 | Complete | 6 | ref|XP_014287040.1 | odorant receptor 4-like | Halyomorpha halys | 46 | 4.98E-16 | 89 |
| AlucOR69 | KU958246 | 436 | 5′ and 3′ lost | 6 | gb|AKS44360.1 | olfactory receptor 12 | Apolygus lucorum | 31 | 2.99E-44 | 171 |
| AlucOR70 | KU958247 | 384 | Complete | 7 | ref|XP_014287040.1 | odorant receptor 4-like | Halyomorpha halys | 28 | 1.84E-42 | 164 |
| AlucOR71 | KU958248 | 376 | Complete | 6 | ref|XP_014258590.1 | odorant receptor 67a isoform X1 | Cimex lectularius | 25 | 1.11E-18 | 96 |
| AlucOR72 | KU958249 | 235 | 5′ lost | 3 | gb|AKS44360.1 | olfactory receptor 12 | Apolygus lucorum | 30 | 2.77E-21 | 101 |
| AlucOR73 | KU958250 | 423 | Complete | 6 | ref|XP_014292012.1 | odorant receptor 22c-like | Halyomorpha halys | 26 | 7.75E-43 | 166 |
| AlucOR74 | KU958251 | 267 | 3′ lost | 4 | ref|XP_014249919.1 | putative odorant receptor 69a, isoform A | Cimex lectularius | 26 | 5.22E-07 | 61 |
| AlucOR75 | KU958252 | 383 | Complete | 3 | ref|XP_014249919.1 | putative odorant receptor 69a, isoform A | Cimex lectularius | 29 | 2.55E-44 | 169 |
| AlucOR76 | KU958253 | 182 | 5′ and 3′ lost | 3 | ref|XP_014250664.1 | odorant receptor 43a-like | Cimex lectularius | 32 | 3.66E-12 | 73 |
| AlucOR77 | KU958254 | 98 | 5′ lost | 0 | gb|ALR72557.1 | odorant receptor OR12 | Colaphellus bowringi | 38 | 4.54E-06 | 52 |
| AlucOR78 | KU958255 | 403 | 5′ lost | 6 | gb|AJO62227.1 | olfactory receptor OR8 | Tenebriomolitor | 25 | 1.63E-06 | 61 |
| AlucOR79 | KU958256 | 382 | Complete | 6 | ref|XP_014258533.1 | odorant receptor 47a-like | Cimex lectularius | 19 | 2.71E-07 | 63 |
| AlucOR80 | KU958257 | 411 | 5′ lost | 6 | ref|XP_014289200.1 | odorant receptor 82a-like | Halyomorpha halys | 55 | 1.5E-133 | 401 |
| AlucOR81 | KU958258 | 426 | 5′ lost | 4 | ref|XP_014287492.1 | odorant receptor 4-like | Halyomorpha halys | 23 | 1.07E-24 | 114 |
| AlucOR82 | KU958259 | 105 | 5′ and 3′ lost | 2 | ref|XP_014290637.1 | uncharacterized protein LOC106689927 isoform X1 | Halyomorpha halys | 33 | 1.38E-05 | 53 |
| AlucOR83 | KU958260 | 372 | 5′ and 3′ lost | 6 | ref|XP_014289672.1 | odorant receptor 49a-like | Halyomorpha halys | 28 | 0.018 | 47 |
| AlucOR84 | KU958261 | 437 | 5′ lost | 6 | gb|AKS44361.1 | olfactory receptor 18 | Apolygus lucorum | 35 | 2.13E-72 | 246 |
| AlucOR85 | KU958262 | 434 | Complete | 4 | ref|XP_014270188.1 | odorant receptor 4-like isoform X1 | Halyomorpha halys | 29 | 8.44E-38 | 152 |
| AlucOR86 | KU958263 | 409 | 5′ lost | 6 | ref|XP_014250665.1 | uncharacterized protein LOC106667308 | Cimex lectularius | 30 | 5.03E-42 | 163 |
| AlucOR87 | KU958264 | 407 | 5′ lost | 4 | ref|XP_014274899.1 | uncharacterized protein LOC106679982 | Halyomorpha halys | 32 | 3.29E-50 | 186 |
| AlucOR88 | KU958265 | 430 | 5′ lost | 6 | gb|AKS44360.1 | olfactory receptor 12 | Apolygus lucorum | 32 | 1.29E-49 | 186 |
| AlucOR89 | KU958266 | 406 | Complete | 6 | ref|XP_014256564.1 | uncharacterized protein LOC106670594 isoform X1 | Cimex lectularius | 26 | 6.79E-30 | 129 |
| AlucOR90 | KU958267 | 444 | 5′ lost | 5 | ref|XP_014289672.1 | odorant receptor 49a-like | Halyomorpha halys | 26 | 7.75E-19 | 98 |
| AlucOR91 | KU958268 | 74 | 5′ and 3′ lost | 1 | ref|NP_001177702.1 | odorant receptor 156 | Nasonia vitripennis | 37 | 0.007 | 44 |
| AlucOR92 | KU958269 | 136 | 5′ and 3′ lost | 2 | gb|AKS44362.1 | olfactory receptor 28 | Apolygus lucorum | 58 | 1.25E-35 | 137 |
| AlucOR93 | KU958270 | 393 | 5′ lost | 5 | ref|XP_014256808.1 | odorant receptor 46a, isoform B-like | Cimex lectularius | 28 | 2.63E-08 | 66 |
| AlucOR94 | KU958271 | 420 | 5′ lost | 3 | ref|XP_014275211.1 | odorant receptor 24a-like | Halyomorpha halys | 26 | 0.032 | 47 |
| AlucOR95 | KU958272 | 115 | 5′ lost | 0 | gb|AKS44361.1 | olfactory receptor 18 | Apolygus lucorum | 40 | 6.45E-19 | 91 |
| AlucOR96 | KU958273 | 386 | Complete | 6 | ref|XP_014294765.1 | odorant receptor 4-like | Halyomorpha halys | 24 | 0.071 | 46 |
| AlucOR97 | KU958274 | 432 | 5′ lost | 7 | gb|AKS44360.1 | olfactory receptor 12 | Apolygus lucorum | 29 | 1.04E-47 | 180 |
| AlucOR98 | KU958275 | 122 | 5′ lost | 2 | gb|AKS44360.1 | olfactory receptor 12 | Apolygus lucorum | 31 | 1.67E-09 | 65 |
| AlucOR99 | KU958276 | 390 | 5′ lost | 6 | ref|XP_014256808.1 | odorant receptor 46a, isoform B-like | Cimex lectularius | 28 | 1.09E-22 | 107 |
| AlucOR100 | KU958277 | 402 | Complete | 3 | gb|ALD51419.1 | odorant receptor 136 | Locusta migratoria | 26 | 0.13 | 44 |
| AlucOR101 | KU958278 | 378 | 5′ lost | 4 | ref|XP_014294439.1 | odorant receptor Or1-like isoform X1 | Halyomorpha halys | 36 | 2.77E-70 | 237 |
| AlucOR102 | KU958279 | 58 | 5′ and 3′ lost | 1 | ref|XP_015594072.1 | odorant receptor 13a-like isoform X3 | Cephus cinctus | 32 | 0.049 | 41 |
| AlucOR103 | KU958280 | 385 | 5′ lost | 6 | ref|XP_014258596.1 | odorant receptor 67a isoform X4 | Cimex lectularius | 29 | 5.76E-16 | 85 |
| AlucOR104 | KU958281 | 178 | 5′ lost | 1 | ref|XP_014290637.1 | uncharacterized protein LOC106689927 isoform X1 | Halyomorpha halys | 28 | 1.58E-10 | 69 |
| AlucOR105 | KU958282 | 421 | 5′ lost | 6 | ref|XP_014249551.1 | putative odorant receptor 92a | Cimex lectularius | 33 | 4.86E-66 | 228 |
| AlucOR106 | KU958283 | 396 | Complete | 5 | ref|XP_014294765.1 | odorant receptor 4-like | Halyomorpha halys | 26 | 1.68E-12 | 79 |
| AlucOR107 | KU958284 | 435 | Complete | 6 | ref|XP_014273407.1 | uncharacterized protein LOC106679017 isoform X1 | Halyomorpha halys | 27 | 4.99E-26 | 119 |
| AlucOR108 | KU958191 | 144 | 5′ lost | 2 | ref|XP_014293240.1 | gustatory and odorant receptor 63a-like | Halyomorpha halys | 38 | 1.46E-09 | 66 |
| AlucOR109 | KU958197 | 418 | Complete | 7 | ref|XP_014244354.1 | gustatory and odorant receptor 22-like | Cimex lectularius | 50 | 6.5E-126 | 382 |
Phylogenetic analysis
In phylogenetic tree, Orco from seven Hemipteran species were easily assigned to one branch because of sharing high similarity (Fig. 4). By contrast, the other ORs are relatively divergent and formed several monophyletic clades (Fig. 4). Several species-specific subgroups were formed such as AlucOR-clade 1 to AlucOR-clade 5, indicating their closely orthologous relationship and specie-specific functions. In addition, several other AlucORs did not cluster in species-specific clades, like AlucOR30 clustered with SfurORs, AlucOR101 and AlucOR3 clustered with HhalORs, and AlucOR109 clustered with ClecORs, suggesting that some Hemipteran ORs may have common basic functions.
Figure 4. Neighbor-joining tree of candidate OR proteins (>200 aa) from Hemiptera species.
The evolutionary distances were computed using the p-distance method. Afas: Adelphocoris fasciaticollis; Alin: Adelphocoris lineolatus; Asut: Adelphocoris suturalis; Aluc: Apolygus lucorum; Hhal: Halyomorpha halys; Clec: Cimex lectularius; Sfur: Sogatella furcifera.
Transcript expressions of AlucORs
Based on RPKM value analysis of the 110 AlucORs, we found that AlucOrco was the most abundant expressed gene (RPKM >170) in antennae, followed by AlucOR104 (RPKM >53), AlucOR18 (RPKM >37), and AlucOR41 (RPKM >35) (Supplementary Table 2).
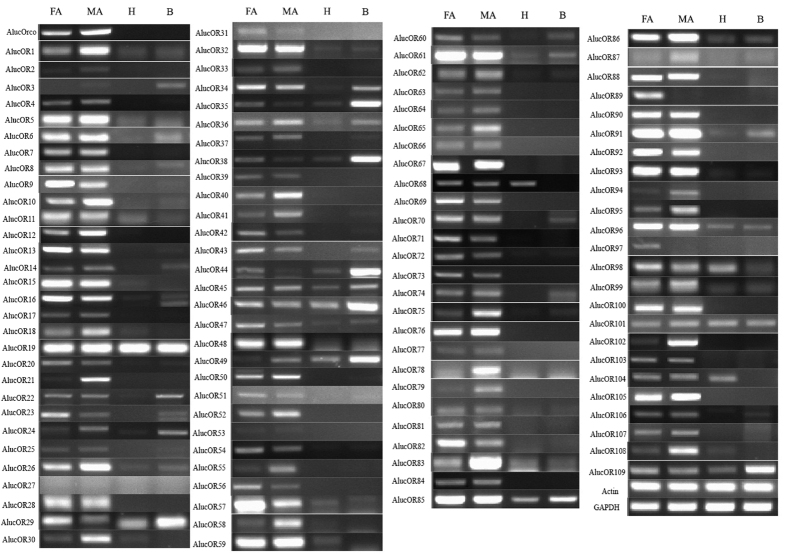
The expression profiles of AlucORs in four different tissues (female antennae, male antennae, head without antennae, and body parts without heads) were evaluated by using RT-PCR. The results revealed that AlucORs had distinct expression profiles. Four (AlucOR2, AlucOR3, AlucOR27, and AlucOR53) of the 109 AlucOR genes showed very weak or undetectable expression levels in both male and female antennae (Fig. 5). AlucOR89 and AlucOR97 were uniquely expressed in the female antennae, while AlucOR21 and AlucOR78 were uniquely expressed in the male antennae (Fig. 5). Ten AlucORs (AlucOR23, AlucOR29, AlucOR31, AlucOR35, AlucOR38, AlucOR42, AlucOR44, AlucOR60, AlucOR82, and AlucOR95) were higher expressed in the female antennae than in male antennae. Eighteen AlucORs (AlucOR1, AlucOR12, AlucOR18, AlucOR24, AlucOR30, AlucOR40, AlucOR41, AlucOR49, AlucOR55, AlucOR58, AlucOR65, AlucOR75, AlucOR79, AlucOR83, AlucOR87, AlucOR94, AlucOR102, and AlucOR108) were higher expressed in the male antennae than in female antennae. Thirteen AlucORs (AlucOR19, AlucOR29, AlucOR44, AlucOR45, AlucOR46, AlucOR49, AlucOR68, AlucOR85, AlucOR96, AlucOR98, AlucOR101, AlucOR104, and AlucOR109) could be detected highly expressed in the head and 12 AlucORs (AlucOR19, AlucOR22, AlucOR24, AlucOR29, AlucOR34, AlucOR35, AlucOR38, AlucOR44, AlucOR45, AlucOR46, AlucOR85, and AlucOR101) were abundant in the tissue of body parts. However, the remaining AlucORs appeared to be predominantly expressed in both the female and male antennae with similar expression levels (Fig. 5).
Figure 5. Tissues expression profiles of candidate AlucORs evaluated by RT-PCR.
FA: female antennae; MA: male antennae; H: heads; B: body parts without heads including body parts including thoraxes, abdomens, legs, and wings.
In order to further investigate the AlucORs transcript profile in detail, qPCR analysis was performed to measure relative expression levels of the 110 AlucOR genes in seven different tissue samples (female antennae, male antennae, heads without antennae, thoraxes, abdomens, legs and wings). The results revealed the expression levels of 40 AlucOR genes were significantly higher abundant (more than five times) in the antennae than in other body parts (Fig. S3), among which five OR genes (AlucOR2, AlucOR53, AlucOR96, AlucOR24, AlucOR100) showed similar expressions between the sexes. The expression level of AlucOR91 in female antennae was 7.4 times that of in other tissues, and nine AlucORs (AlucOR4, AlucOR13, AlucOR14, AlucOR21, AlucOR65, AlucOR71, AlucOR81, AlucOR83, and AlucOR102) in the male antennae were 5.5 to 38.1 fold higher expressed than in other tissues (Fig. S3). Comparative analysis of expression level of AlucOR genes between male and female antennae revealed that the expression level of 25 AlucOR genes in male antennae were 3 times higher than that in female antennae and seven AlucOR genes in female antennae were 3 times higher than that in male antennae. In addition to these antennal highly expressed AlucOR genes, we also identified some other tissues highly expressed OR genes, including two head highly expressed, two abdomen highly expressed and four wing highly expressed OR genes (Supplementary Table 3).
Discussion
In this work, the repertoire of ORs in A. lucorum was determined by using RNA-Seq method. After extensive sequencing, assembly, and bioinformatic analysis, a total of candidate 110 OR genes were identified, including five previously annotated OR genes (AlucOR12, AlucOR18, AlucOR28, AlucOR30, and AlucOrco). Subsequent cloning and sequencing of these OR sequences with specific primers showed that our transcriptome data was highly credible. Compared to other Hemipteran transcriptomes of Sogatella furcifera with 63ORs32, Aphis gossypii with 45ORs33, and Acyrthosiphon pisum with 73ORs34, our OR dataset of 110 sequences showed an expansion of AlucOR family, which could provide the diversity of odorant receptors that allowed A. lucorum to recognize diverse odors. Indeed, A. lucorum feed on a wide range of plants that emit complex and species specific volatiles26. Nevertheless, because the ORF of some ORs is incomplete, we cannot exclude the possibility that some of these ORs might be pseudogenes. The sequence number is much lower compared with species including Apis mellifera (163 ORs)35, Tribolium castaneum (341ORs)36, and Locusta migratoria (142 ORs)37. This may be caused by adaptation of distinct species to their hosts during evolution38.
For a better understanding the function of these AlucOR genes, tissue-specific expressions were evaluated by using RT-PCR and qPCR methods. Results showed that AlucOR genes exhibited diverse expression patterns, which could be briefly classified into five types: antennal highly expressed ORs, head highly expressed ORs, abdomen highly expressed ORs, wing highly expressed ORs, and broadly expressed ORs (Supplementary Table 2). It was also reported that some ORs could be expressed in a variety of tissues apart from the olfactory organs37,39. Antennae are important sensory organs for insect, so the majority of AlucOR genes displayed high expressions in antennae, including female and male antennal highly expressed genes. Five AlucORs (AlucOR2, AlucOR5, AlucOR24, AlucOR96, and AlucOR100) were predominantly expressed in the antennae of males and females with similar expression levels, which suggested that these ORs could play important roles in the detection of general odorants, such as host plant volatiles. In particular, we found that AlucOR91 were highly expressed in the female antennae (7.4 times higher than in other tissues) and AlucOR21 in the male antennae (38.1 times higher than in other tissues), indicating sex-specific functions of AlucOR91 and AlucOR21. According to previous studies of the insect OR functions in moths40,41,42,43,44, the male-dominant expression of ORs might be involved in the detection and discrimination of the sex pheromone or in other male-specific behaviors, while female-dominant expression of ORs might have the preferential function that is critical to female olfactory behavior, such as oviposition sites selection or male-produced courtship pheromones detection. The sex-specific functions of these ORs need to be further investigated in the future. In addition, we found eight AlucOR genes were highly expressed in heads, legs, or wings rather than antennae. The expression of broadly expressed OR genes in non-olfactory tissues suggested that they might have diverse physiological functions in other organs. The co-expression of LmigOR95 and LmigOrco were also observed in the fat body of migratory locust37. In locusts and mosquitoes, the testis-enhanced OR genes supposed to participate in the sperm chemotaxis, fertilization, or the activation of spermatozoa37,45. Further research on the broadly expressed OR genes is worthwhile to elucidate their roles in the non-olfactory tissues.
The phylogenetic analysis of 207 ORs (>200 aa) from seven Hemiptera species demonstrated that these OR genes had undergone functional differentiation due to their scattered distribution. Except for Orco, the sequences of other OR genes were differentiated into several different clades even within the conspecifics (Fig. 4), which is consistent with the previous research11,32,33. In particular, despite the diversity of OR genes, many species-specific sub-clades were clustered, such as AlucOR-clade 1 to AlucOR-clade 5 (Fig. 4), suggesting a relatively conservative of these ORs within the conspecifics. The phylogenetic tree of AlucORs (>300 aa) were also constructed, and three large lineage-specific clades were generated, including clade 1 (33 ORs), clade 2 (32 ORs), and clade 3 (21 ORs) (Fig. S1). In particular, four male antennae highly expressed ORs (AlucOR4, AlucOR21, AlucOR65, and AlucOR83) were clustered into sub-clade 1 (Fig. S1). Based on previous study on sex pheromone receptor of Lepidoptera insects42,43,44,45,46, we speculated that the sub-clade 1 could be a cluster of potential sex pheromone receptors of A. lucorum.
In conclusion, based on the transcriptome analysis of male and female antennae from A. lucorum, an extensive set of 110 candidate AlucOR genes that may be related to odorant perception were identified in our laboratory. As a crucial first step toward understanding their functions, a comprehensive examination of the expression patterns of these AlucOR genes in different tissue samples were prefromed by using RT-PCR and qPCR. Forty ORs were found to be significantly higher abundant in antenna. One female antennae specific and nine male antennae specific AlucOR genes were identified successfully. The phylogenetic relationships between AlucORs and other Hemipteran ORs were also evaluated. The results of this study will provide a valuable foundation for further elucidating the mechanisms of olfaction in A. lucorum, which also could help us use odorant receptors as targets to regulate insect olfactory behavior and broaden the applications of available tools for effective control of insect pests.
Materials and Methods
Insect rearing
A. lucorum nymphs and adults were originally collected from a cotton fields at the Langfang Experimental Station of Chinese Academy of Agricultural Sciences, Hebei Province (Latitude 39.53°N, Longitude 116.70°E), China. A laboratory colony feeding on green bean pods (Phaseolus Vulgaris L.) was cultivated in climatic chambers under a condition of 29 ± 1 °C, relative humidity (RH) 60 ± 5% and 14:10 light: dark (L:D) photoperiod47.
Sample collection
For transcriptome analysis, approximately 500 pairs of adult antennae from each sex were individually dissected and immediately immersed in liquid nitrogen, then stored at −80 °C till to the RNA isolation. For RT-PCR and qPCR analysis, different tissue samples including 500 pairs of female antennae, 500 pairs of male antennae, 200 heads without antennae, 100 thoraxes, 50 abdomens, 500 legs, 500 wings, and 50 body parts without heads were collected separately and immediately frozen in liquid nitrogen, then stored at −80 °C. Unless stated, 3–4 d old adult bugs were used in this work. All the tissue samples used for RT-PCR and qPCR were prepared in triplicate.
RAN isolation, cDNA library construction and Illumina sequencing
Total RNA was isolated from male and female antennae by using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. The concentration, quality, and quantity of RNA samples were determined with NanoDrop ND-2000 Spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA) and Qubit® RNA Assay Kit in Qubit® 2.0 Flurometer (Life Technologies, CA, USA). RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA). The cDNA library construction and Illumina sequencing of the RNA samples were performed by Novogene Bioinformatics Technology Co. Ltd, Beijing, China. Briefly, poly-A RNA was purified from 3 μg of total RNA using oligo (dT) magnetic beads and fragmented into short sequences in the fragmentation buffer. Then, random hexamer primer and M-MuLV Reverse Transcriptase (RNaseH) was used for first-strand cDNA generation, followed by synthesis of the second-strand cDNA using RNaseH and DNA polymerase I. After end repair and adaptor ligation, the library fragments were amplified by PCR and purified using the AMPure XP system (Beckman Coulter, Beverly, USA) to obtain a cDNA library. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using a TruSeq PE Cluster Kit v3-cBot-HS (Illumina) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina Hiseq 2500 platform and paired-end reads were generated.
De novo assembly and functional annotation
After removing short or low quality and adaptor sequence, each clean-read dataset of male and female antennae was assembled using the short read assembling program Trinity (r20140413p1) with min_kmer_cov set to 2 and all other parameters set default48. The resulting unigenes were further clustered by TGICL to remove redundant fragments and to acquire non-redundant unigenes as long as possible49. To annotate these unigenes, BLASTx search was performed against protein database of Nr, SwissProt, GO and COG (e - value < 10−5). The blast results were then imported into Blast2GO pipeline for Go annotations50.
Transcript abundance analysis of unigenes
The transcript abundance of these unigenes were calculated based on the reads per kilobase per million mapped reads (RPKM) method51, using the formula: RPKM (A) = (10,00,000 × C × 1000)/(N × L), where RPKM (A) is the abundance of gene A, C is the number of reads that uniquely aligned to gene A, N is the total number of reads that uniquely aligned to all genes, and L is the number of bases in gene A. The RPKM method was able to eliminate the influence of different gene lengths and sequencing discrepancies in the calculation of expression abundance.
Verification of OR sequences by cloning and sequencing
To obtain a more reliable sequence, all the A. lucorum OR sequences from transcriptome were further confirmed by gene cloning and sequencing. Gene-specific primers amplifying the intact ORF or partial sequences of each OR gene were designed by using Primer Premier 5 software (PREMIER Biosoft International, CA, USA) based on the transcriptome sequences (Supplementary Table 4). PCR reactions were carried out in a total reaction volume of 30 μl with template cDNA of 200 ng and Takara LA Taq Polymerase (TaKaRa, Dalian, China) of 0.3 μl. The PCR cycling profile was: 95 °C for 1 min, followed by 40 cycles of 95 °C for 20 sec, 57 °C for 20 sec, 72 °C for 1 min, and a final extension at 72 °C for 10 min. The PCR products were subsequently gel-purified and cloned into pCloneEZ vector (CloneSmarter Technologies Inc., USA) and then sequenced with standard M13 primers.
Sequence and phylogenetic analysis of candidate OR genes
Candidate OR gene fragments were determined by searching for homology using Blastx and Blastn tool in NCBI. The longest open reading frame (ORF) of each unigene was identified by ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Transmembrane domains of OR genes were predicted with the TMHMM Server Version 2.0 (http://www.cbs.dtu.dk/services/TMHMM).
Alignments of amino acid sequences were performed using the program ClustalW and further edited using Jalview 2.752. A neighbor-joining tree was constructed by MEGA 6.06 based on p-distance model53. The bootstrap support of tree branches was assessed by re-sampling amino acid positions 1000 times. Phylogenetic analysis was performed with 207 ORs (>200 aa) from seven Hemipteran species. The dataset contains 92 ORs from Apolygus lucorum, 36 ORs from Halyomorpha halys, 27 ORs from Cimex lectularius, 49 ORs from Sogatella furcifera32, and three Orco genes from Adelphocoris fasciaticollis, Adelphocoris lineolatus, and Adelphocoris suturalis. The protein sequences of the 207 ORs used in this analysis are listed in Supplementary Table 5. In addition, the phylogenetic tree was constructed with 86 selected AlucORs (>300 aa).
RT-PCR analysis
The expression of AlucOR transcripts in different tissues (female antennae, male antennae, heads without antennae, and body parts without heads) were evaluated by RT-PCR. Total RNA was extracted from tissue samples by using Trizol reagent following the manufacturer’s instructions. RNA concentration and quality were checked by Nanodrop ND-2000 spectrophotometer and 1.1% agarose gel electrophoresis. For each sample, 2 μg of total RNA was used for cDNA synthesis in a total reaction volume of 20 μl by using FastQuant RT Kit (with gDNase, Tiangen Biotech, Beijing Co., Ltd.) according to the manufacturer’s protocol. The β-actin (GenBank accession number: JN616391.1) and GAPDH (GenBank accession number: JX987672.1) of A. lucorum were selected as the control genes to assess the cDNA integrity. The specific primers of target and control genes in RT-PCR were designed by using Primer Premier 5 software (Supplementary Table 6). An equal amount of cDNA (200 ng) was added to each reaction mixture (50 μl) under the following cycling conditions: 94 °C for 4 min, followed by 35 cycles of 3-step amplification of 94 °C for 30 s, 55~60 °C for 30 s, 72 °C for 50 s, and a final extension for 10 min at 72 °C. PCR products were checked on a 1% agarose gel and verified by DNA sequencing. Three repeats with three biological samples of each gene were performed in this experiment.
qPCR measurment
The relative expression levels of target AlucOR genes in seven different tissues (female antennae, male antennae, heads without antennae, thoraxes, abdomens, legs, and wings) were further examined by qPCR on an ABI Prism 7500 system (Applied Biosystems, Carlsbad, CA, USA) using a mixture of 10 μl 2 × SuperReal PreMix Plus (Tiangen Biotech, Beijing Co., Ltd.), 0.8 μl of each primer (10 μM), 200 ng of sample cDNA, 0.4 μl of 50 × ROX Reference Dye and 6 μl of sterilized ultrapure water. The reaction program was composed of 95 °C for 15 min, 40 cycles of 95 °C for 10 s and 60 °C for 32 s. All the primers used in qPCR were designed with Beacon Designer 7.9 software (PREMIER Biosoft International, CA, SA) and listed in Supplementary Table 7. A discrete amplification peak and a subsequent melting curve was checked to ensure the primer specificity. A high amplification efficiency of each primer pair was calculated by a five-fold cDNA dilution series. Negative controls without template were run in parallel for each primer pair. Each reaction was performed with three biological replicates, and each biological replicate was assessed three times. Prior to qPCR, we performed semi-quantitative RT-PCR and confirmed that β-actin is expressed at similar level in different tissues (Fig. S2), and the expression level of each AlucOR gene relative to β-actin were calculated by using the comparative 2−ΔΔCT method54. All data were statistically analyzed by using SAS 9.2® Software (SAS Institute Inc., Carey, North Carolina, USA). One-way analysis of variance (ANOVA) followed by a Tukey’s honestly significant difference (HSD) test (P < 0.05) were used to compare expression of each target gene among various tissues.
Additional Information
How to cite this article: An, X.-K. et al. Identification and expression analysis of an olfactory receptor gene family in green plant bug Apolygus lucorum (Meyer-Dür). Sci. Rep. 6, 37870; doi: 10.1038/srep37870 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported by the China National Basic Research Program (2012CB114104), the National Natural Science Foundation of China (31171858, 31321004, 31501652 and 31471778) and Research Foundation of State Key Laboratory for Biology of Plant Diseases and Insect Pests (SKLOF201514). This manuscript has been edited by the native English-speaking experts of Elsevier Language Editing Services.
Footnotes
Author Contributions Conceived and designed the experiments: X.K.A. and Y.J.Z. Sample collection: L.S., D.F.L., and Y.X.D. Performed the experiments: X.K.A., L.M.L. Data analysis: D.F.L., Y.Y.G., H.W.L. and J.J.Z. Wrote the paper: X.K.A. Manuscript revision: Y.Y.G. and J.J.Z.
References
- Hansson Bill S. & Stensmyr Marcus C. Evolution of Insect Olfaction. Neuron 72, 698–711 (2011). [DOI] [PubMed] [Google Scholar]
- Leal W. S. Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 58, 373–391 (2013). [DOI] [PubMed] [Google Scholar]
- Touhara K. & Vosshall L. B. Sensing odorants and pheromones with chemosensory receptors. Annu. Rev. Physiol. 71, 307–332 (2009). [DOI] [PubMed] [Google Scholar]
- Zhao X. C. et al. Fine structure and primary sensory projections of sensilla located in the labial-palp pit organ of Helicoverpa armigera (Insecta). Cell Tissue Res 353, 399–408 (2013). [DOI] [PubMed] [Google Scholar]
- Gao Q. & Chess A. Identification of candidate Drosophila olfactory receptors from genomic DNA sequence. Genomics 60, 31–39 (1999). [DOI] [PubMed] [Google Scholar]
- Martin J. P. et al. The neurobiology of insect olfaction: Sensory processing in a comparative context. Prog. Neurobiol. 95, 427–447 (2011). [DOI] [PubMed] [Google Scholar]
- Vogt R. G. et al. The insect SNMP gene family. Insect. Biochem. Molec. 39, 448–456 (2009). [DOI] [PubMed] [Google Scholar]
- Benton R., Vannice K. S., Gomez Diaz C. & Vosshall L. B. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 136, 149–162 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelosi P., Zhou J. J., Ban L. P. & Calvello M. Soluble proteins in insect chemical communication. Cell. Mol. Life Sci. 63, 1658–1676 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelosi P., Iovinella I., Felicioli A. & Dani F. R. Soluble proteins of chemical communication: an overview across arthropods. Front Physiol 5, 320 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missbach C. et al. Evolution of insect olfactory receptors. eLife 3, e02115 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K. et al. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature 452, 1002–1006 (2008). [DOI] [PubMed] [Google Scholar]
- Wicher D. et al. Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature 452, 1007–1011 (2008). [DOI] [PubMed] [Google Scholar]
- Benton R., Sachse S., Michnick S. W. & Vosshall L. B. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol 4, e20 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyne P. J. et al. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron 22, 327–338 (1999). [DOI] [PubMed] [Google Scholar]
- Zhou X. et al. Phylogenetic and transcriptomic analysis of chemosensory receptors in a pair of divergent ant species reveals sex-specific signatures of odor coding. PLoS Genet. 8, e1002930 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engsontia P. et al. The red flour beetle’s large nose: An expanded odorant receptor gene family in Tribolium castaneum. Insect. Biochem. Mol. Biol. 38, 387–397 (2008). [DOI] [PubMed] [Google Scholar]
- Zhou Y. L. et al. Silencing in Apolygus lucorum of the olfactory coreceptor Orco gene by RNA interference induces EAG response declining to two putative semiochemicals. J. Insect Physiol. 60, 31–9 (2014). [DOI] [PubMed] [Google Scholar]
- Zhang R., Gao G. & Chen H. Silencing of the olfactory co-receptor gene in Dendroctonus armandi leads to EAG response declining to major host volatiles. Sci. Rep. 6, 23136 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W. et al. Identification and knockdown of the olfactory receptor (orco) in gypsy moth, Lymantria Dispar. Int. J. Biol. Sci. 11, 772–80 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai T. et al. Identification and functional characterization of a sex pheromone receptor in the silkmoth Bombyx mori. Proc. Natl. Acad. Sci. USA 101, 16653–8 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Sakurai T., Nishioka T. & Touhara K. Insect sex-pheromone signals mediated by specific combinations of olfactory receptors. Science 307, 1638–42 (2005). [DOI] [PubMed] [Google Scholar]
- Miura N., Nakagawa T., Touhara K. & Ishikawa Y. Broadly and narrowly tuned odorant receptors are involved in female sex pheromone reception in Ostrinia moths. Insect Biochem. Mol. Biol. 40, 64–73 (2010). [DOI] [PubMed] [Google Scholar]
- Lu Y. H. et al. Species composition and seasonal abundance of pestiferous plant bugs (Hemiptera: Miridae) on Bt Cotton in China. Crop. Prot. 27, 465–472 (2008). [Google Scholar]
- Lu Y., Wu K., Wyckhuys K. A. G. & Guo Y. Overwintering hosts of Apolygus lucorum (Hemiptera: Miridae) in northern China. Crop. Prot. 29, 1026–1033 (2010). [Google Scholar]
- Lu Y. & Wu K. Biology and Control of Cotton Mirids. (Golden Shield Press, 2008). [Google Scholar]
- Lu Y. H. et al. Mirid bug outbreaks in multiple crops correlated with wide-scale adoption of Bt cotton in China. Science 328, 1151–1154 (2010). [DOI] [PubMed] [Google Scholar]
- Liu Y., Lu Y., Wu K., Wyckhuys K. A. & Xue F. Lethal and sublethal effects of endosulfan on Apolygus lucorum (Hemiptera: Miridae). J. Econ. Entomol. 101, 1805–1810 (2008). [DOI] [PubMed] [Google Scholar]
- Lu Y., Jiao Z. & Wu K. Early season host plants of Apolygus lucorum (Heteroptera: Miridae) in northern China. J. Econ. Entomol. 105, 1603–1611 (2012). [DOI] [PubMed] [Google Scholar]
- Pan H., Lu Y., Wyckhuys K. A. & Wu K. Preference of a polyphagous mirid bug, Apolygus lucorum (Meyer-Dur) for flowering host plants. PLoS One 8, e68980 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S. W., Zhang J., Liu Y., Li G. Q. & Wang G. R. An olfactory receptor from Apolygus lucorum (Meyer-Dur) mainly tuned to volatiles from flowering host plants. J. Insect Physiol. 79, 36–41 (2015). [DOI] [PubMed] [Google Scholar]
- He M., Zhang Y. N. & He P. Molecular characterization and differential expression of an olfactory receptor gene family in the white-backed planthopper sogatella furcifera based on transcriptome analysis. PLoS One 10, e0140605 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D., Liu Y., Walker W. B., Li J. & Wang G. Molecular characterization of the Aphis gossypii olfactory receptor gene families. PLoS One 9, e101187 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smadja C. M. et al. Large-scale candidate gene scan reveals the role of chemoreceptor genes in host plant specialization and speciation in the pea aphid. Evolution 66, 2723–38 (2012). [DOI] [PubMed] [Google Scholar]
- Robertson H. M. & Wanner K. W. The chemoreceptor superfamily in the honey bee, Apis mellifera: expansion of the odorant, but not gustatory, receptor family. Genome Res 16, 1395–403 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engsontia P. et al. The red flour beetle’s large nose: an expanded odorant receptor gene family in Tribolium castaneum. Insect. Biochem. Mol. Biol. 38, 387–397 (2008). [DOI] [PubMed] [Google Scholar]
- Wang Z. et al. Identification and functional analysis of olfactory receptor family reveal unusual characteristics of the olfactory system in the migratory locust. Cell. Mol. Life Sci. 72, 4429–43 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Gracia A., Vieira F. G. & Rozas J. Molecular evolution of the major chemosensory gene families in insects. Heredity 103, 208–216 (2009). [DOI] [PubMed] [Google Scholar]
- Gu S. H. et al. Molecular characterization and differential expression of olfactory genes in the antennae of the black cutworm moth Agrotis ipsilon. PloS One 9, e103420 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. N. et al. Functional characterization of sex pheromone receptors in the purple stem borer, Sesamia inferens (Walker). Insect. Mol. Boil. 23, 611–620 (2014). [DOI] [PubMed] [Google Scholar]
- Anderson A. R. et al. Molecular basis of female-specific odorant responses in Bombyx mori. Insect. Biochem. Mol. Biol. 39, 189–197 (2009). [DOI] [PubMed] [Google Scholar]
- Zhang J. et al. Identification and functional characterization of sex pheromone receptors in the common cutworm (Spodoptera litura). Chem. Senses. 40, 7–16 (2015). [DOI] [PubMed] [Google Scholar]
- Krieger J. et al. Genes encoding candidate pheromone receptors in a moth (Heliothis virescens). Proc. Natl. Acad. Sci. USA 101, 11845–11850 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger J., Große‐Wilde E., Gohl T. & Breer H. Candidate pheromone receptors of the silkmoth Bombyx mori. Eur. J. Neurosci. 21, 2167–2176 (2005). [DOI] [PubMed] [Google Scholar]
- Pitts R. J., Liu C., Zhou X., Malpartida J. C. & Zwiebel L. J. Odorant receptor-mediated sperm activation in disease vector mosquitoes. Proc. Natl. Acad. Sci. USA 111, 2566–2571 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D. D. & Löfstedt C. Moth pheromone receptors: gene sequences, function, and evolution. Front Ecol Evol 3 (2015). [Google Scholar]
- Yuan H. B. et al. Molecular characterization and expression profiling of odorant-binding proteins in Apolygus lucorum. PLoS One 10, e0140562 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea G. et al. TIGR Gene Indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics 19, 651–652 (2003). [DOI] [PubMed] [Google Scholar]
- Conesa A. et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–6 (2005). [DOI] [PubMed] [Google Scholar]
- Mortazavi A., Williams B. A., McCue K., Schaeffer L. & Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5, 621–628 (2008). [DOI] [PubMed] [Google Scholar]
- Waterhouse A. M., Procter J. B., Martin D. M., Clamp M. & Barton G. J. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A. & Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. mst197 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.