Abstract
SXT/R391 integrative and conjugative elements (ICEs) are self-transmissible mobile genetic elements that are found in most members of Enterobacteriaceae. Here, we determined fifteen SXT/R391 ICEs carried by Proteus isolates from food (4.2%) and diarrhoea patients (17.3%). BLASTn searches against GenBank showed that the fifteen SXT/R391 ICEs were closely related to that from different Enterobacteriaceae species, including Proteus mirabilis. Using core gene phylogenetic analysis, the fifteen SXT/R391 ICEs were grouped into six distinct clusters, including a dominant cluster and three clusters that have not been previously reported in Proteus isolates. The SXT/R391 ICEs shared a common structure with a set of conserved genes, five hotspots and two variable regions, which contained more foreign genes, including drug-resistance genes. Notably, a class A β-lactamase gene was identified in nine SXT/R391 ICEs. Collectively, the ICE-carrying isolates carried resistance genes for 20 tested drugs. Six isolates were resistant to chloramphenicol, kanamycin, streptomycin, trimethoprim-sulfamethoxazole, sulfisoxazole and tetracycline, which are drug resistances commonly encoded by ICEs. Our results demonstrate abundant genetic diversity and multidrug resistance of the SXT/R391 ICEs carried by Proteus isolates, which may have significance for public health. It is therefore necessary to continuously monitor the antimicrobial resistance and related mobile elements among Proteus isolates.
Integrative and conjugative elements (ICEs) are self-transmissible mobile genetic elements that can be excised from the chromosome of the host cell. Once excised, ICEs form a circular intermediate that can be transferred to another cell via conjugation1. Many varieties of ICEs have been found in diverse Gram-positive and Gram-negative bacteria2,3,4,5.
The SXT/R391 ICE family is one of the largest ICE families, with the most abundant diversity and members among Gram-negative bacteria6. SXT was first discovered in MO10, which is a Vibrio cholerae O139 clinical strain isolated from India in the early 1990s. SXTMO10 is an ~100 kb ICE that carries genes encoding resistance to sulfamethoxazole, trimethoprim, chloramphenicol, and streptomycin7. Since then, ICEs related to SXTMO10 have been detected in most Vibrio species in addition to V. cholerae as well as in other gammaproteobacteria8,9. R391 was first discovered in a Providencia rettgeri clinical isolate from South Africa in 196710; subsequent studies showed that it genetically and functionally belonged to the SXT family11. The R391 ICE mediates resistance to kanamycin and the heavy metal Hg11.
Comparative genomics has shown that the SXT/R391 ICEs share nearly identical sets of 52 conserved core genes that are involved in integration/excision, conjugative transfer, and regulation12. All SXT/R391 ICEs detected to date have element-specific phenotypes that are conferred by the insertion of variable DNA sequences into several sites. Variable DNA sequences are frequently found in five hotspots (HS1–HS5) and four variable regions (VRI-IV)12. The hotspots are sites within the conserved SXT/R391 ICE backbone have variable DNA present in all of the ICEs, which inserted into their intergenic regions that confer element-specific properties, these variable DNA sequences share a mosaic structure and have sizes ranging from 30–60 kb. Except for the hotspots, some SXT/R391 ICEs also contain variable regions, which encode resistance to antibiotics, heavy metals and quaternary ammonium compounds12,13.
The genus Proteus is a motile Gram-negative bacterium that survives in soil, water, and the intestinal tracts of mammals; bacteria of this genus belong to the family Enterobacteriaceae. Proteus consists of five species and three unnamed genomospecies14. Among these species, P. vulgaris and P. mirabilis are most frequently linked with food contamination and food poisoning. These species are also renowned opportunistic pathogens that cause a variety of infections in humans, including respiratory tract, wound, burn, skin, eye, ear, nose, and throat infections15.
In previous studies, a novel SXT/R391-related ICE (ICEPmiJpn1) carrying blaCMY-2 on the chromosome of a P. mirabilis clinical isolate was found and characterized. ICEPmiJpn1 was the first ICE identified to confer resistance to extended-spectrum cephalosporins16. Recently, Lei, et al. reported SXT/R391 ICEs in P. mirabilis isolates from food-producing animals in China17. However, our knowledge of the prevalence and origin of the Proteus species ICEs is limited. In this study, we screened SXT/R391-specific genes from 123 Proteus isolates collected from clinical and food samples from 2008 to 2015 in China. We detected 15 Proteus isolates that were positive for SXT/R391-related ICEs and analysed the genetic structure and evolutionary origins of each SXT/R391 ICE. Additionally, we determined their transfer capabilities and the roles of the SXT/R391 ICEs in drug resistance.
Materials and Methods
Ethics
The study was approved by the Ethics Committee of National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention, and the study was carried out in accordance with the approved guidelines. Informed consents were obtained from all the patients.
Collection of samples, bacterial isolation and identification of Proteus isolates
A total of 123 Proteus isolates were included in this study. The strains were isolated from stool samples and/or rectal swabs from diarrhoea patients (n = 75) and fresh food samples (n = 48) collected from 2008 to 2015 in three cities (Beijing, Tianjin and Ma’anshan) in China. Specimens of diarrhoea patients were collected in Cary-Blair transport media, and then each sample was incubated for 24 h at 37 °C on Salmonella-Shigella (SS) and MacConkey agar (Becton Dickinson Co., USA). Suspicious colonies were streaked on nutrient agar for incubation (37 °C, 24 h), single clone from nutrient agar was then picked for biochemical identification. The fresh food samples included meat of raw pork, beef, chicken, duck, fish and shrimp, 25 gram each food sample was put into 225 ml of Gram negative enrichment broth (Qingdao Hopebio Technology Co., Ltd, China) and enriched for 8 h at 37 °C, then streaked on SS and MacConkey agar, followed by the procedure likes that sample of diarrhoea patient. All isolates from nutrient agar were preliminarily identified as Proteus by negative for oxidase and positive for urease and phenylalanine deaminase and KIA: K/A++. The isolates were confirmed to be P. mirabilis (n = 81) and P. vulgaris (n = 42) by API20E biochemical test (BioMerieux, Lyon, France). All isolates were stored at −70 °C in LB broth containing 15% glycerol prior to use.
PCR screening of SXT/R391 ICEs and genomic sequencing
All 123 Proteus strains in this study were screened for the presence of SXT/R391-like ICEs using a PCR-based method targeting the intSXT gene18, which encodes a conserved SXT/R391 ICE integrase. Next-generation sequencing (NGS) was performed with the PCR-positive isolates. Genomic DNA was extracted from 5 ml of overnight cultures using a Wizard Genomic DNA Purification kit (Promega, USA) according to the manufacturer’s instructions. The extracted DNA was dissolved in Tris-EDTA buffer and stored at −20 °C prior to sequencing. The genomes were commercially sequenced using Illumina HiSeq 2000 sequencing (Illumina Inc., San Diego, CA, USA) by constructing two paired-end libraries with average insert lengths of 500 bp and 2000 bp. Then, 100× libraries were obtained with clean paired-end read data. Assembly was performed using SOAP denovo v2.0419.
Extraction, assembly and annotation of the SXT/R391 ICEs
The contigs of the fifteen SXT/R391 ICEs were extracted and assembled from whole sequenced genomes using SOAP denovo v2.0.419 against the reference SXT/R391 ICEs in the genome of P. mirabilis HI4320 (accession number: AM942759.1). The relationships between contigs were displayed using ContigScape20 with custom primer walking. Sanger sequencing was used to close the gaps in the ICE region and the results were verified by PCR. The Phred/Phrap/Consed software was used for primer design, genome assembly, editing and quality assessment (http://www.phrap.org/consed/consed.html). Regions with low quality of the genome were resequenced. Putative functions were inferred using the Basic Local Alignment Search Tool (BLAST) (http://ncbi.nlm.nih.gov/BLAST) and the ORF finder (http://www.ncbi.nlm.nih.gov/projects/gorf). The RAST (Rapid Annotation using Subsystem Technology, version 4.0)21,22,23 server pipeline was used to predict open reading frames (ORFs) and annotate the ORFs of the recovered ICEs.
Phylogenetic and structural analysis of the SXT/R391 ICEs
To investigate the evolutionary origins of the SXT/R391 ICEs in Proteus species, we performed a phylogenetic analysis. First, we used each of the fifteen SXT/R391 ICEs in a BLASTn search to obtain all high homology ICEs in the public database (last updated in Nov 2015). Then, we scanned and selected 24 ICEs as references, representative for the different evolutionary origins and species based on different scores and identifications (Supplementary Table S1). To construct the phylogenetic tree, core genes of all ICEs were identified using the OrthoMCL software. The concatenated sequences of these core genes were used for phylogenetic analysis. The bootstrap values were calculated based on 1000 replicates. The structures of the fifteen SXT/R391 ICEs were constructed by comparison with the reference ICE in the genome of P. mirabilis HI4320 (AM942759.1) using BLAST and SOAPdenovo 2.0419. The tested ICEs were compared against the reference ICE from P. mirabilis HI4320 using Pheatmap (R package version 0.7.7) to plot heatmaps of the annotated core and pan genes of each ICE.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed on P. mirabilis by broth dilution method according to the guidelines for MIC testing from the Clinical and Laboratory Standards Institute (CLSI) (2015). Twenty antimicrobials were tested as follows: ampicillin (AMP), cefazolin (CFZ), cefuroxime (CXM), ceftriaxone (CRO), cefepime (FEP), cefotetan (CTT), aztreonam (ATM), imipenem (IPM), ampicillin-sulbactam (SAM), piperacillin-tazobactam (TZP), amikacin (AMK), gentamycin (GEN), kanamycin (KAN), streptomycin (STR), sulfamethoxazole (SUL), trimethoprim-sulfamethoxazole (SXT), ciprofloxacin (CIP), chloramphenicol (CHL), tetracycline (TCY), and azithromycin (AZM). The CLSI(2015) breakpoints were used to determine susceptibility and resistance.
Conjugation experiments
A mating assay was employed to test ICE mobility. The experiment was conducted as previously described24. Transconjugants were selected on LB agar plates containing streptomycin (100 μg/ml) and kanamycin (100 μg/ml). The ICE transfer frequency was expressed as the number of transconjugants observed per recipient cell (E. coli SM10). Transconjugants were confirmed by PCR detection of intSXT and antibiotic resistance genes specific for each of the SXT/R391 ICEs. The primer information is listed in Supplementary Table S2.
Sequence data access
The reads sequences and annotated genes of the fourteen ICEs (except for ICEPmiCHN3277) in Proteus species were submitted to GenBank under accession number KX243403- KX243416.
Results
Distribution and general features of SXT/R391 ICEs among Proteus strains
Fifteen out of 123 strains (12.2%, fourteen P. mirabilis and one P. vulgaris strain) were positive for the intSXT gene, including 4.2% (2/48) of food samples (crab and pork), and 17.3% (13/75) of stool samples from the diarrhoea patients, respectively. The genomes of the 15 intSXT-positive strains were obtained and all SXT/R391 ICEs were successfully assembled with the exception of the ICE from isolate TJ3277. The general genomic features of the fifteen ICEs are summarized in Table 1. The SXT/R391 ICEs were designated according to the nomenclature proposed for this family of elements2. The lengths of the 15 ICEs ranged from 76,218 bp (ICEPmiCHN1809) to 108,335 bp (ICEPmiCHN3300), with an average length of 93,396 bp. The G + C content ranged from 44.6% (ICEPmiCHN1586) to 47.8% (ICEPmiCHN905/ICEPmiCHN3335) among the SXT/R391 ICEs, with a mean of 46.9%. The total numbers of predicted CDSs were between 53 and 99.
Table 1. Genomic features of the ICEs, antibiotic resistance patterns and transfer frequency of ICE-contained strains in this study.
| Strain ID | Location | Species | Sample source | Year of isolation | ICE name | ICE length (bp) | G + C (%) | No. of predicted CDSs (RAST) | Antibiotic resistance phenotype | Transfer frequency | SXT/R391 ICE gene |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dfhR | floR | strB/A | sul2 | tetA/R | |||||||||||
| 08MAS2213 | Maanshan | P. vulgaris | food | 2008 | ICEPvuCHN2213 | 94,340 | 46.8 | 92 | AMP-AZM-CHL-CFZ-SAM-STR-SXT-SUL | 1.0 × 10−5 | − | + | + | + | − |
| 08MAS1586 | Maanshan | P. mirabilis | food | 2008 | ICEPmiCHN1586 | 99,355 | 46.6 | 85 | AMP-AZM-CHL-CIP-SAM-STR-SXT-SUL-TCY | 2.5 × 10−2 | + | + | + | + | − |
| 09MAS2407 | Maanshan | P. mirabilis | stool | 2009 | ICEPmiCHN2407 | 97,078 | 47 | 95 | AMP-AZM-CHL-CFZ-CIP-KAN-STR-SXT-SUL-TCY | 9.5 × 10−4 | − | − | − | − | + |
| 09MAS2410 | Maanshan | P. mirabilis | stool | 2009 | ICEPmiCHN2410 | 93,537 | 46.5 | 86 | AMP-AZM-CHL-CFZ-CIP-KAN-STR-SXT-SUL-TCY | NT | − | − | − | − | − |
| 09MAS2416 | Maanshan | P. mirabilis | stool | 2009 | ICEPmiCHN2416 | 92,556 | 46.7 | 86 | AMP-AZM-CHL-CFZ-CIP-GEN-KAN-STR-SXT-SUL-TCY | NT | − | − | − | − | − |
| MD20140901 | Beijing | P. mirabilis | stool | 2014 | ICEPmiCHN901 | 89,493 | 47.3 | 88 | AMP-AZM-ATM-AMK-CHL-CXM-CFZ-CRO-CIP-GEN-SAM -KAN-STR-SXT-SUL-TCY | NT | + | + | + | + | − |
| MD20140902 | Beijing | P. mirabilis | stool | 2014 | ICEPmiCHN902 | 89,096 | 47.1 | 86 | AMP-AZM-ATM-AMK-CHL-CXM-CFZ-CRO-CIP-SAM-KAN-STR-SXT-SUL-TCY | NT | + | + | + | + | − |
| MD20140903 | Beijing | P. mirabilis | stool | 2014 | ICEPmiCHN903 | 89,644 | 47.3 | 86 | AMP-AZM-AMK-CHL-CXM-CFZ-CRO-CIP-SAM -KAN-STR-SXT-SUL-TCY | NT | + | + | + | + | − |
| MD20140904 | Beijing | P. mirabilis | stool | 2014 | ICEPmiCHN904 | 94,942 | 47.7 | 99 | AMP-AZM-ATM-AMK-CHL-CXM-CFZ-CRO-CIP-FEP-SAM -KAN-STR-SXT-SUL-TCY | NT | − | + | + | + | + |
| MD20140905 | Beijing | P. mirabilis | stool | 2014 | ICEPmiCHN905 | 94,956 | 47.8 | 98 | AMP-AZM-ATM-AMK-CHL-CXM-CFZ-CRO-CIP-GEN-SAM -KAN-STR-SXT-SUL-TCY | NT | − | + | + | + | + |
| TJ1809 | Tianjin | P. mirabilis | stool | 2013 | ICEPmiCHN1809 | 76,218 | 47.1 | 67 | AMP-AZM-CHL-CIP-KAN-SXT-SUL-TCY | 4.0 × 10−5 | − | − | − | − | − |
| TJ3237 | Tianjin | P. mirabilis | stool | 2013 | ICEPmiCHN3237 | 87,215 | 46.8 | 74 | AMP-AZM-CFZ-SXT-SUL-TCY | NT | − | − | − | − | − |
| TJ3277 | Tianjin | P. mirabilis | stool | 2013 | ICEPmiCHN3277 | 104,175 | 46.7 | 53 | AMP-AZM-CFZ-SXT-SUL-TCY | NT | NT | NT | NT | NT | NT |
| TJ3300 | Tianjin | P. mirabilis | stool | 2013 | ICEPmiCHN3300 | 108,335 | 46.7 | 98 | AZM-CHL-STR-SXT-SUL | NT | + | + | + | + | − |
| TJ3335 | Tianjin | P. mirabilis | stool | 2013 | ICEPmiCHN3335 | 89,996 | 47.8 | 84 | AZM-CHL-KAN-STR-SXT-SUL-TCY | 5.0 × 10−6 | − | + | + | + | − |
Abbreviations: AMP: ampicillin, AZM: azithromycin, CFZ: cefazolin, CXM: cefuroxime, CRO: ceftriaxone, FEP: cefepime, CTT: cefotetan, ATM: aztreonam, IPM: imipenem, SAM: ampicillin-sulbactam, TZP: piperacillin-tazobactam, AMK: amikacin, GEN: gentamycin, KAN: kanamycin, STR: streptomycin, SUL: sulfamethoxazole, SXT: trimethoprim-sulfamethoxazole, CIP: ciprofloxacin, CHL: chloramphenicol, TCY: tetracycline. Abbreviations in bold indicated positive resistance phenotype but negative for their gene at ICE. “+”: positive, “−”: negative, “NT”: not test.
Blast searches of closely related SXT/R391 ICEs in GenBank
As shown in Table 2, the 15 SXT/R391 ICEs were closely related to many different ICEs based on some indicators, such as the highest max score, query coverage and identity. These ICEs derived from Alteromonas macleodii, Alteromonas mediterranea, Vibrio cholerae, Vibrio alginolyticus, Proteus mirabilis and Providencia stuartii. Due to the closely related scores among the hits of each ICE, we listed the top three alignment results for each ICE; the max score ranged from 19,287 to 1.36E + 05, the total score (bits) ranged from 73,099 to 1.95E + 05, the highest query ranged from 46% to 99% and the identity ranged from 96% to100%. Notably, ICEPmiCHN3335 (host strain TJ3335, isolated from the stool of a patient from Tianjin city, see Table 1) was closely related to ICEPmiChn1 (host strain PM13C04, isolated from a chicken in Hubei, China, in 2013, see Supplementary Table S1) with a highest query and identity of 99%. However, some ICEs, such as ICEPmiCHN2407, ICEPmiCHN2410 and ICEPmiCHN2416, only exhibited query coverage from 46% to 67% and identity from 96% to 99%.
Table 2. BLASTn searches against GenBank of the 15 SXT/R391-like ICEs.
| SXT | Description of ICEs from species | Max score | Total Score (bits) | Query cover | Identity | Accession |
|---|---|---|---|---|---|---|
| ICEPvuCHN2213 | Alteromonas macleodii str. MED64’E | 1.30E + 05 | 1.71E + 05 | 86% | 99% | CP004848.1 |
| Vibrio cholerae strain AHV1003 | 1.13E + 05 | 1.74E + 05 | 92% | 99% | KT151663.1 | |
| Vibrio cholerae strain TSY216 | 1.13E + 05 | 1.74E + 05 | 92% | 99% | CP007653.1 | |
| ICEPmiCHN1586 | Vibrio cholerae strain ICDC-2605 | 57943 | 1.95E + 05 | 95% | 100% | KT151661.1 |
| Vibrio cholerae strain ICDC-1605 | 57943 | 1.95E + 05 | 95% | 100% | KT151656.1 | |
| Vibrio cholerae strain ICDC-143 | 57943 | 1.95E + 05 | 95% | 100% | KT151654.1 | |
| ICEPmiCHN2407 | Proteus mirabilis ICEPmiChn1 | 32073 | 1.08E + 05 | 64% | 98% | KT962845.1 |
| Vibrio cholerae O37 strain MZ03 | 29263 | 73099 | 46% | 96% | JQ345361.1 | |
| Vibrio alginolyticus strain HN396 | 26856 | 84281 | 53% | 96% | KT072770.1 | |
| ICEPmiCHN2410 | Proteus mirabilis ICEPmiChn1 | 32058 | 1.08E + 05 | 66% | 98% | KT962845.1 |
| Vibrio cholerae O37 strain MZ03 | 29263 | 73099 | 47% | 96% | JQ345361.1 | |
| Vibrio alginolyticus strain HN396 | 26856 | 84281 | 55% | 96% | KT072770.1 | |
| ICEPmiCHN2416 | Proteus mirabilis, ICEPmiChn1 | 37102 | 1.08E + 05 | 67% | 99% | KT962845.1 |
| Vibrio cholerae O37 strain MZ03 | 29263 | 73099 | 48% | 96% | JQ345361.1 | |
| Vibrio alginolyticus strain HN396 | 26856 | 84281 | 55% | 96% | KT072770.1 | |
| ICEPmiCHN901 | Proteus mirabilis ICEPmiChn1 | 42494 | 1.32E + 05 | 77% | 99% | KT962845.1 |
| Vibrio cholerae strain ICDC-2605 | 34481 | 1.26E + 05 | 73% | 99% | KT151661.1 | |
| Vibrio cholerae strain ICDC-1605 | 34481 | 1.26E + 05 | 73% | 99% | KT151656.1 | |
| ICEPmiCHN902 | Proteus mirabilis ICEPmiChn1 | 40945 | 1.31E + 05 | 76% | 99% | KT962845.1 |
| Vibrio cholerae strain wujiang-2 | 36490 | 1.14E + 05 | 69% | 96% | KT151664.1 | |
| Vibrio cholerae O1 str. KW3 | 36490 | 1.19E + 05 | 72% | 96% | CP006947.1 | |
| ICEPmiCHN903 | Proteus mirabilis ICEPmiChn1 | 39305 | 1.33E + 05 | 77% | 99% | KT962845.1 |
| Vibrio cholerae strain wujiang-2 | 36490 | 1.14E + 05 | 69% | 96% | KT151664.1 | |
| Vibrio cholerae O1 str. KW3 | 36490 | 1.20E + 05 | 72% | 96% | CP006947.1 | |
| ICEPmiCHN904 | Proteus mirabilis ICEPmiChn1 | 42512 | 1.35E + 05 | 74% | 99% | KT962845.1 |
| Vibrio cholerae strain wujiang-2 | 36490 | 1.12E + 05 | 64% | 96% | KT151664.1 | |
| Vibrio cholerae O1 str. KW3 | 36490 | 1.27E + 05 | 68% | 96% | CP006947.1 | |
| ICEPmiCHN905 | Proteus mirabilis ICEPmiChn1 | 42505 | 1.37E + 05 | 75% | 99% | KT962845.1 |
| Vibrio cholerae strain wujiang-2 | 36490 | 1.16E + 05 | 65% | 96% | KT151664.1 | |
| Vibrio cholerae O1 str. KW3 | 36490 | 1.22E + 05 | 68% | 96% | CP006947.1 | |
| ICEPmiCHN1809 | Providencia stuartii strain ATCC 33672 | 25080 | 1.02E + 05 | 79% | 97% | CP008920.1 |
| Vibrio alginolyticus strain HN437 | 19398 | 78001 | 62% | 98% | KT072771.1 | |
| Vibrio cholerae Ind4 ICEVchind4 | 19287 | 81235 | 64% | 97% | GQ463141.1 | |
| ICEPmiCHN3237 | Proteus mirabilis strain HI4320 | 1.36E + 05 | 1.45E + 05 | 90% | 99% | AM942759.1 |
| Vibrio cholerae strain ICDC-4210 | 1.36E + 05 | 1.65E + 05 | 96% | 99% | KT151662.1 | |
| Vibrio cholerae strain ICDC-2605 | 1.36E + 05 | 1.58E + 05 | 96% | 99% | KT151661.1 | |
| ICEPmiCHN3277 | Vibrio cholerae strain ICDC-4210 | 6584 | 1.38E + 05 | 71% | 99% | KT151662.1 |
| Vibrio cholerae strain ICDC-2605 | 6584 | 1.37E + 05 | 71% | 99% | KT151661.1 | |
| Proteus mirabilis strain HI4320 | 6584 | 1.35E + 05 | 70% | 99% | AM942759.1 | |
| ICEPmiCHN3300 | Vibrio cholerae MJ-1236 | 72417 | 1.80E + 05 | 87% | 99% | CP001485.1 |
| Alteromonas mediterranea strain U10 | 49808 | 91294 | 49% | 97% | CP013930.1 | |
| Alteromonas mediterranea strain UM8 | 49808 | 88620 | 47% | 97% | CP013928.1 | |
| ICEPmiCHN3335 | Proteus mirabilis ICEPmiChn1 | 1.24E + 05 | 1.68E + 05 | 99% | 99% | KT962845.1 |
| Vibrio alginolyticus strain A056 | 30134 | 1.28E + 05 | 81% | 96% | KR231688.1 | |
| Vibrio cholerae O37 strain MZ03 | 9198 | 77187 | 52% | 96% | JQ345361.1 |
Phylogenetic analysis of ICEs in Proteus isolates
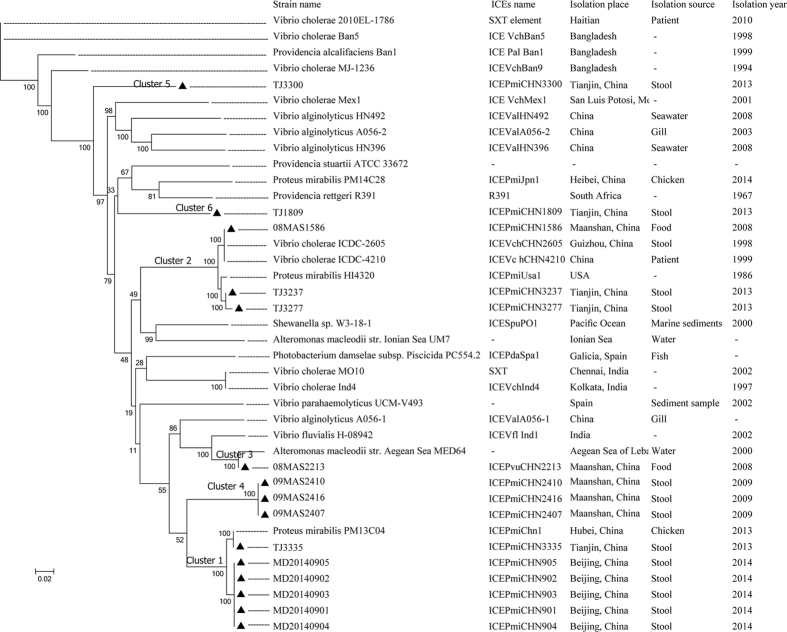
The fifteen SXT/R391 ICEs were grouped into six clusters based on the core gene phylogenetic analysis (Fig. 1). Six ICEs, their contained strains isolated from the stool of diarrhoeal patients in two cities (Table 1), belonged to cluster I, with the reference ICEPmiChn1, which was an ICE contained in a P. mirabilis strain isolated from a chicken in Hubei, China (Supplementary Table S1). Three ICEs (strains isolated from food and stool samples from diarrhoea patients in two cities in China) belonged to cluster II, with references including ICEs from V. cholerae strain ICDC-4210 (isolated from the stool of a patient in Jiangxi, China) and P. mirabilis strain HI4320. Cluster III contained ICEPvuCHN2213 from a P. vulgaris strain isolated from food that was closely related to the reference ICE of Alteromonas macleodii MED64 (from waters off the Aegean Sea)25. The remaining five ICEs were grouped into three distinct clusters (IV, V and VI) with no references; thus, these ICEs were designated as novel ICEs.
Figure 1. Phylogenetic tree from the maximum-likelihood analysis of the core genome alignments of the fifteen ICEs (black triangle).
The scale bar indicates substitutions per site. Bootstraps are indicated at each node.
Sequence structure and variable region characteristics of the SXT/R391 ICEs
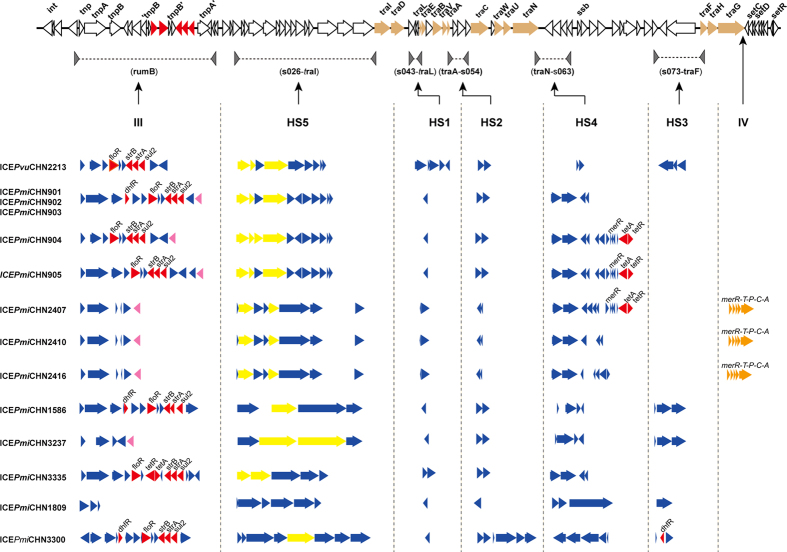
Except for SXT/R391 ICE from TJ3277, which was partially assembled, the remaining fourteen SXT/R391 ICEs shared a common structure that was identical to most SXT/R391 ICEs. In addition to the common structure, all fourteen ICEs contained five hotspots (HS1–5) and two variable (III and IV) regions (Fig. 2). VRIII, inserted into rumB gene, was found in all 14 SXT/R391 ICEs. Five ICEs (ICEPmiCHN901, ICEPmiCHN902, ICEPmiCHN903 ICEPmiCHN1586 and ICEPmiCHN3300), contained the dhfR, floR, strB/A and sul2 genes in this region, which conferred resistance to SXT, CHL, STR and SUL, respectively. The tetA and tetR tetracycline resistance genes were present in VRIII of ICEPmiCHN3335 and HS4 of ICEPmiCHN904 and ICEPmiCHN904. Notably, nine ICEs contained a β-lactamase gene in VRIII (Fig. 2 and Supplementary Table S3), which showed 100% identity to the class A β-lactamase gene bla (HMS-1) contained in plasmid R997 from P. mirabilis (GenBank: KX228735) and V. parahaemolyticus strain UCM-V493 (CP007004.1). HS1, HS2, HS4 and HS5 were detected in all ICEs. However, HS3 was detected in only five ICEs with few inserted genes; a dhfR gene was located in HS3 of ICEPmiCHN3300. Abundant foreign genes were inserted into HS4 and HS5, including genes encoding the restriction-modification system (RM) of which the Type I RM was common, serine protease, ATPase, helicase, and exonuclease. Additionally, genes encoding hypothetical proteins with unknown functions were frequently inserted into these two regions. In VRIV, a mer operon was found in ICEPmiCHN2407, ICEPmiCHN2410 and ICEPmiCHN2416, this mer operon was also reported at ICEs in bacterial strains from aquaculture environments, previously described for R391 ICEs and mediated resistance to mercury13. The annotated genes from all fifteen ICEs and the six closely related ICEs (Supplementary Table S1) were used to construct a heatmap using P. mirabilis HI4320 (AM942759.1) as the reference; ICEPmiCHN3277 was excluded because it was incompletely assembled. The remaining fourteen ICEs shared minor numbers of common genes (Supplementary Fig. S1) between each of their ICEs and the closely related ICEs (e.g., ICEPmiCHN1809 vs the ICE of Providencia stuartii strain ATCC33672, ICEPmiCHN1586 vs ICEVchCHN2605, and ICEPmiCHN3237 and ICEPmiCHN3277 vs ICEVchCHN4210 had extensive diversity in their annotated genes).
Figure 2. Genetic organization of the ICEs in this study.
The upper indicate backbone, represents the 52 conserved core genes of the SXT/R391 family ICEs. Below the common structure, indicate five hotspots (HS1–5) and two variable (III and IV) regions of the fourteen ICEs (except for ICEPmiCHN3277, which was incompletely assembled). Drug-resistance gene (red triangle); the restriction-modification system (RM) (yellow triangle); β-lactamase encoding gene (pink triangle); mercury resistance gene (orange triangle); other genes (blue triangle). Detailed annotated genes from all fifteen ICEs are listed in Supplementary Table S3.
Antibiotic resistance of the Proteus isolates and their relationship with ICEs
Phenotypically, all fifteen SXT/R392 ICE-harbouring isolates presented multi-drug resistance to the 20 tested drugs (Table 1). Isolates MD20140901, MD20140904 and MD20140905 with the most antibiotic phenotypes were resistant to 16 out of 20 drugs. Even the least resistant isolate (TJ3300) was resistant to five drugs. 09MAS2407 and 09MAS2410, MD20140901 and MD20140905, and TJ3237 and TJ3277 shared same multidrug resistance patterns, respectively. Notably, all isolates were resistant to AMP and 12 isolates (in addition to TJ1809, TJ3237 and TJ3277) were resistant to CHL, STR, SXT and SUL, which are commonly encoded by SXT/R391 ICEs. MD20140901 to MD20140905 were resistant to the first, second and third-generation cephalosporins; MD20140904 was also resistant to the fourth generation cephalosporin.
All isolates positive for resistant related genes (floR, STR, SUL and SXT) at ICEs, were phenotypic resistant to their drugs (chloramphenicol, streptomycin, sulfisoxazole and trimethoprim-sulfamethoxazole), by contrast, even if isoates were phenotypic resistant to those four drugs, their ICEs not always carried those resistant related genes (Table 1 and Fig. 2).
Transfer ability
To test the transfer ability of the Proteus isolate ICEs, we selected five ICE-carrying isolates (08MAS2213, 08MAS1586, 09MAS2407, TJ1809, and TJ3335) for the mobility test. Transconjugants were obtained with a transfer frequency of 5.0 × 10−6 (TJ3335) to 2.5 × 10−2 (08MAS1586) per recipient cell (Table 1). The transconjugants were confirmed by PCR detection of the intSXT and antibiotic resistance genes (strA/B, sul2, floR, and dfrA) in the recipient cells.
Discussion
In this study, the ICEs contained in Proteus isolates showed high diversity compared to those carried in a variety of other bacterial species. Our results indicated that SXT/R391 ICEs presented a strong ability to transmit among different bacterial species as a type of self-transmissible mobile genetic element. This study revealed the epidemiology of the spatio-temporal prevalence of ICEs in Proteus. The occurrence and dispersion of Proteus isolates from different regions in China revealed the occurrence and widespread distribution of ICEs among Proteus. Furthermore, the various ICEs conferred phenotypes such as multidrug and heavy metal resistance to their host strains. To date, at least 89 SXT/R391-family ICEs have been identified (http://db-mml.sjtu.edu.cn/ICEberg/)26. Most ICEs have been investigated in V. cholerae4,27,28,29,30,31, which is the aetiological agent of the diarrhoeal disease cholera. The ICEs reported in Proteus strains including R997 (India)32, ICEPmiUSA1 (America)33, ICEPmiJpn1(Japan)16, ICEPmiSpn1(Spain)24 and ICEPmiChn1(China)17. In this study, our fifteen ICEs revealed six distinct clusters and were positioned in different branches of the phylogenetic tree. The Blastn analysis in Genbank showed that closely related ICEs might be different. Additionally, five of our ICEs grouped into three distinctive clusters representing novel ICEs (Fig. 1), which increases the number of the SXT/R391 family members. Consequently, our results indicated that abundant genetic diversities and variable types of ICEs were ubiquitous in Proteus strains, compared with the ICEs in V. cholerae and other Enterobacteriaceae. The ICE types in the Proteus strains in our study were different from a recent investigation of the SXT/R391 ICEs in P. mirabilis isolates from food-producing animals in China17, which included two types of ICEs (ICEPmiJpn1 and ICEPmiChn1). We suggest that the use of a more extensive source of isolates (food and clinical samples from different cities) might contribute to the varieties of ICEs in Proteus isolates. Regardless of the diversity of the hosts and locations, the ICEs of the SXT/R391 family share a common structure and contain 52 conserved core genes that mediate integration, recombination, DNA repair and conjugative transfer12. In this study, the ICE-harbouring Proteus strains shared a common structure even though they were isolated from different cities in China and were at different evolutionary stages. However, the 15 ICEs were closely related to many different ICEs derived from Alteromonas macleodii, A. mediterranea, V. cholerae, V. alginolyticus, P. mirabilis and Providencia stuartii. They share much different query score and identity at nucleotide level (Table 1), consequently, these ICEs formed unique variable regions. Antibiotic resistance genes were typically present within VRIII in ICEs reported in previous studies. Our results showed that most of the multidrug resistance genes were present in this region, including the genes encoding resistance for sulfamethoxazole, trimethoprim, chloramphenicol, and streptomycin that were first described in the SXT of V. cholerae O139 MO107. A class A β-lactamase gene was found, which has been reported in the plasmid from P. mirabilis (GenBank: KX228735), and the ICE from V. parahaemolyticus34. However, further research will be necessary to confirm the relationship of this gene with resistance to β-lactams. Five hotspots were located in s043-traL (HS1), traA-s054 (HS2), s073-traF (HS3), traN-s063 (HS4), and s026-traI (HS5). Variable genes were identified and predicted to encode restriction-modification systems, endonucleases, which may provide protection from invasion by foreign DNA12. ICEs without any antibiotic resistances were described12 and determinants for antibiotic resistance were not found in ICEPmiCHN1809. However, whether the successful existence of the SXT/R391 ICEs is related exclusively to the appearance of resistance determinants is inconclusive. Other genes found in the variable regions, including genes encoding unknown functions, may be related to the enhancement of the adaptability of ICEs12.
In this study, the fifteen ICEs were grouped into six clusters by the phylogenetic analysis and therefore may be representative of six different evolutionary origins. Six ICEs were placed in cluster I, these ICEs were identical to ICEPmiChn1, which is a recently reported ICE contained in P. mirabilis that was isolated from a faecal sample from chicken in Hubei, China, in 201317. ICEPmiChn1 was a predominant SXT/R391 family member in clinical Proteus sources in China and probably experienced horizontal transmission from food to humans. Because the six ICEs and the ICEPmiChn1-containing strains were isolated during and after 2013 (Supplementary Tables S1 and S2), these ICEs may have appeared during the latest period of their evolution and then presented an extensive distribution in China. Three ICEs (cluster II in Fig. 1) with references including ICEs from V. cholerae strain ICDC-4210 (isolated from Jiangxi, China, in 1999)31 and P. mirabilis strain HI4320 (USA, 1986)33, indicated that the ICEs of this cluster may have appeared as early as 1986 and have been transmitted among different bacterial species and countries. Similarly, ICEPvuCHN2213 in cluster III contained in a P. vulgaris strain isolated from food was closely related to the ICE MED6425 (from the Aegean Sea near Lebanon, 2000), which was between clusters I and II. Interestingly, five novel ICEs in this study were grouped into three distinctive clusters (IV, V and VI in Fig. 1) with no references in the three clusters, which might indicate that they were independently acquired by Proteus strains. Generally, the phylogenetic analysis suggested that ICEs could be transmitted among Proteus, V. cholerae and other Enterobacteriaceae and might share a common ancestor although they evolved independently. The acquired ICEs in Proteus were not species-specific under certain conditions; for example, to adapt to the environment and to facilitate survival under selective pressure, Proteus strains are able to acquire ICEs from different sources and evolutionary stages35.
The majority of known SXT/R391 ICEs contain four types of antibiotic genes (strA/B, sul2, floR and dfrA) that confer resistance to streptomycin, sulfamethoxazole, chloramphenicol and trimethoprim, respectively. R391 contains a kanamycin-encoding gene11 and other new antibiotic resistance genes of known SXT/R391 ICEs carried have also been found (e.g. the cephalosporin resistance gene blaCMY-216 and rifampicin resistance gene)13. In this study, isolates positive for resistant related genes at SXT/R391 ICEs were not always consistent with their phenotypic resistance (Table 1), this result indicated the phenotypic resistance may not be associated with genes encoded by ICEs, determinants of other mobile genetic elements, like plasmid36, transposon37 and genomic island38 within Enterobacteriaceae, are also mediated drug resistance. However, the role of ICEs in the acquisiton and transmission of antibiotic resistance should not be neglected, our study have displayed high transfer ability of ICEs from Proteus isolates to recipient cells. In addition, the 15 SXT/R391 ICEs carried not only the four common resistance genes mentioned above, nine SXT/R391 ICEs also carried a class A β-lactamase gene. In contrast, no dominant β-lactamase genes are carried by ICEPmiChn117, suggesting that the class A β-lactamase gene might have been obtained via horizontal gene transfer or recombination. In summary, our study reported multi-drug resistance, including the increasing prevalence of class A β-lactamase-producing P. mirabilis, which is consistent with the trends in Japan16, Taiwan39 and other reports24,40. In conclusion, our results present abundant genetic diversity and multidrug resistance of ICEs carried by Proteus strains from both food sources and diarrhoeal patients. The SXT/R391 ICEs could be transferred between Proteus and other Enterobacteriaceae, thereby conferring resistance to the host and facilitating bacterial survival in the environment. Therefore, we need to strengthen the continuous monitoring of antimicrobial resistance and related mobile elements among Proteus isolates.
Additional Information
How to cite this article: Li, X. et al. SXT/R391 integrative and conjugative elements in Proteus species reveal abundant genetic diversity and multidrug resistance. Sci. Rep. 6, 37372; doi: 10.1038/srep37372 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported by the NSFC of China (30872260 and 31570134), State Key Laboratory of Infectious Disease Prevention and Control (2011SKLID201) and the Priority Project on Infectious Disease Control and Prevention (2012ZX10004215) from the Ministry of Health, China.
Footnotes
Author Contributions Designed the work: X.L. and D.W.; Performed the experiments: X.L., H.D. and Y.F.; Performed the bioinformatic studies: Y.D., P.D., Z.L., N.L. and B.Z.; Analyzed the data: X.L., D.W. and B.K.; Wrote the paper: X.L. and D.W.
References
- Burrus V. & Waldor M. K. Shaping bacterial genomes with integrative and conjugative elements. Research in microbiology 155, 376–386, doi: 10.1016/j.resmic.2004.01.012 (2004). [DOI] [PubMed] [Google Scholar]
- Burrus V., Marrero J. & Waldor M. K. The current ICE age: biology and evolution of SXT-related integrating conjugative elements. Plasmid 55, 173–183, doi: 10.1016/j.plasmid.2006.01.001 (2006). [DOI] [PubMed] [Google Scholar]
- Pembroke J. T. & Piterina A. V. A novel ICE in the genome of Shewanella putrefaciens W3-18-1: comparison with the SXT/R391 ICE-like elements. FEMS microbiology letters 264, 80–88, doi: 10.1111/j.1574-6968.2006.00452.x (2006). [DOI] [PubMed] [Google Scholar]
- Spagnoletti M. et al. Acquisition and evolution of SXT-R391 integrative conjugative elements in the seventh-pandemic Vibrio cholerae lineage. mBio 5, doi: 10.1128/mBio.01356-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eidam C. et al. Analysis and comparative genomics of ICEMh1, a novel integrative and conjugative element (ICE) of Mannheimia haemolytica. The Journal of antimicrobial chemotherapy 70, 93–97, doi: 10.1093/jac/dku361 (2015). [DOI] [PubMed] [Google Scholar]
- Daccord A., Ceccarelli D. & Burrus V. Integrating conjugative elements of the SXT/R391 family trigger the excision and drive the mobilization of a new class of Vibrio genomic islands. Molecular microbiology 78, 576–588, doi: 10.1111/j.1365-2958.2010.07364.x (2010). [DOI] [PubMed] [Google Scholar]
- Waldor M. K., Tschape H. & Mekalanos J. J. A new type of conjugative transposon encodes resistance to sulfamethoxazole, trimethoprim, and streptomycin in Vibrio cholerae O139. Journal of bacteriology 178, 4157–4165 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed A. M., Shinoda S. & Shimamoto T. A variant type of Vibrio cholerae SXT element in a multidrug-resistant strain of Vibrio fluvialis. FEMS microbiology letters 242, 241–247, doi: 10.1016/j.femsle.2004.11.012 (2005). [DOI] [PubMed] [Google Scholar]
- Osorio C. R. et al. Genomic and functional analysis of ICEPdaSpa1, a fish-pathogen-derived SXT-related integrating conjugative element that can mobilize a virulence plasmid. Journal of bacteriology 190, 3353–3361, doi: 10.1128/JB.00109-08 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coetzee J. N., Datta N. & Hedges R. W. R factors from Proteus rettgeri. Journal of general microbiology 72, 543–552, doi: 10.1099/00221287-72-3-543 (1972). [DOI] [PubMed] [Google Scholar]
- Boltner D., MacMahon C., Pembroke J. T., Strike P. & Osborn A. M. R391: a conjugative integrating mosaic comprised of phage, plasmid, and transposon elements. Journal of bacteriology 184, 5158–5169 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak R. A. et al. Comparative ICE genomics: insights into the evolution of the SXT/R391 family of ICEs. Plos genetics 5, e1000786, doi: 10.1371/journal.pgen.1000786 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Blanco A., Lemos M. L. & Osorio C. R. Integrating conjugative elements as vectors of antibiotic, mercury, and quaternary ammonium compound resistance in marine aquaculture environments. Antimicrobial agents and chemotherapy 56, 2619–2626, doi: 10.1128/AAC.05997-11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hara C. M., Brenner F. W. & Miller J. M. Classification, identification, and clinical significance of Proteus, Providencia, and Morganella. Clinical microbiology reviews 13, 534–546 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozalski A., Sidorczyk Z. & Kotelko K. Potential virulence factors of Proteus bacilli. Microbiology and molecular biology reviews: MMBR 61, 65–89 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada S., Ishii Y., Saga T., Tateda K. & Yamaguchi K. Chromosomally encoded blaCMY-2 located on a novel SXT/R391-related integrating conjugative element in a Proteus mirabilis clinical isolate. Antimicrobial agents and chemotherapy 54, 3545–3550, doi: 10.1128/AAC.00111-10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei C. W. et al. Characterization of SXT/R391 Integrative and Conjugative Elements in Proteus mirabilis Isolates from Food-Producing Animals in China. Antimicrobial agents and chemotherapy 60, 1935–1938, doi: 10.1128/AAC.02852-15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath B. M., O’Halloran J. A., Piterina A. V. & Pembroke J. T. Molecular tools to detect the IncJ elements: a family of integrating, antibiotic resistant mobile genetic elements. Journal of microbiological methods 66, 32–42, doi: 10.1016/j.mimet.2005.10.004 (2006). [DOI] [PubMed] [Google Scholar]
- Li R. et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome research 20, 265–272, doi: 10.1101/gr.097261.109 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang B. et al. ContigScape: a Cytoscape plugin facilitating microbial genome gap closing. BMC genomics 14, 289, doi: 10.1186/1471-2164-14-289 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz R. K. et al. The RAST Server: rapid annotations using subsystems technology. BMC genomics 9, 75, doi: 10.1186/1471-2164-9-75 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R. et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic acids research 42, D206–214, doi: 10.1093/nar/gkt1226 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brettin T. et al. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Scientific reports 5, 8365, doi: 10.1038/srep08365 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata C. et al. Prevalence of SXT/R391-like integrative and conjugative elements carrying blaCMY-2 in Proteus mirabilis. The Journal of antimicrobial chemotherapy 66, 2266–2270, doi: 10.1093/jac/dkr286 (2011). [DOI] [PubMed] [Google Scholar]
- Lopez-Perez M., Gonzaga A. & Rodriguez-Valera F. Genomic diversity of “deep ecotype” Alteromonas macleodii isolates: evidence for Pan-Mediterranean clonal frames. Genome biology and evolution 5, 1220–1232, doi: 10.1093/gbe/evt089 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi D. et al. ICEberg: a web-based resource for integrative and conjugative elements found in Bacteria. Nucleic acids research 40, D621–626, doi: 10.1093/nar/gkr846 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taviani E., Grim C. J., Chun J., Huq A. & Colwell R. R. Genomic analysis of a novel integrative conjugative element in Vibrio cholerae. FEBS letters 583, 3630–3636, doi: 10.1016/j.febslet.2009.10.041 (2009). [DOI] [PubMed] [Google Scholar]
- Bordeleau E., Brouillette E., Robichaud N. & Burrus V. Beyond antibiotic resistance: integrating conjugative elements of the SXT/R391 family that encode novel diguanylate cyclases participate to c-di-GMP signalling in Vibrio cholerae. Environmental microbiology 12, 510–523, doi: 10.1111/j.1462-2920.2009.02094.x (2010). [DOI] [PubMed] [Google Scholar]
- Taviani E. et al. Genomic analysis of ICEVchBan8: An atypical genetic element in Vibrio cholerae. FEBS letters 586, 1617–1621, doi: 10.1016/j.febslet.2012.03.064 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccarelli D. et al. A new integrative conjugative element detected in Haitian isolates of Vibrio cholerae non-O1/non-O139. Research in microbiology 164, 891–893, doi: 10.1016/j.resmic.2013.08.004 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Yu D., Yue J. & Kan B. Variations in SXT elements in epidemic Vibrio cholerae O1 El Tor strains in China. Scientific reports 6, 22733, doi: 10.1038/srep22733 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy D. B. & Pembroke J. T. Monitoring of chromosomal insertions of the IncJ elements R391 and R997 in Escherichia coli K-12. FEMS microbiology letters 174, 355–361 (1999). [DOI] [PubMed] [Google Scholar]
- Pearson M. M. et al. Complete genome sequence of uropathogenic Proteus mirabilis, a master of both adherence and motility. Journal of bacteriology 190, 4027–4037, doi: 10.1128/JB.01981-07 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalburge S. S. et al. Complete Genome Sequence of Vibrio parahaemolyticus Environmental Strain UCM-V493. Genome announcements 2, doi: 10.1128/genomeA.00159-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak R. A. & Waldor M. K. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nature reviews. Microbiology 8, 552–563, doi: 10.1038/nrmicro2382 (2010). [DOI] [PubMed] [Google Scholar]
- Pugliese N., Maimone F., Scrascia M., Materu S. F. & Pazzani C. SXT-related integrating conjugative element and IncC plasmids in Vibrio cholerae O1 strains in Eastern Africa. The Journal of antimicrobial chemotherapy 63, 438–442, doi: 10.1093/jac/dkn542 (2009). [DOI] [PubMed] [Google Scholar]
- Shimizu W. et al. Persistence and epidemic propagation of a Pseudomonas aeruginosa sequence type 235 clone harboring an IS26 composite transposon carrying the blaIMP-1 integron in Hiroshima, Japan, 2005 to 2012. Antimicrobial agents and chemotherapy 59, 2678–2687, doi: 10.1128/AAC.04207-14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girlich D., Dortet L., Poirel L. & Nordmann P. Integration of the blaNDM-1 carbapenemase gene into Proteus genomic island 1 (PGI1-PmPEL) in a Proteus mirabilis clinical isolate. The Journal of antimicrobial chemotherapy 70, 98–102, doi: 10.1093/jac/dku371 (2015). [DOI] [PubMed] [Google Scholar]
- Wang J. T. et al. Antimicrobial susceptibilities of Proteus mirabilis: a longitudinal nationwide study from the Taiwan surveillance of antimicrobial resistance (TSAR) program. BMC infectious diseases 14, 486, doi: 10.1186/1471-2334-14-486 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aberkane S. et al. High Prevalence of SXT/R391-Related Integrative and Conjugative Elements Carrying blaCMY-2 in Proteus mirabilis Isolates from Gulls in the South of France. Antimicrobial agents and chemotherapy 60, 1148–1152, doi: 10.1128/AAC.01654-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.