Abstract
The nucleotide sequence of the RNA genome of the human hepatitis C virus (HCV) has been determined from overlapping cDNA clones. The sequence (9379 nucleotides) has a single large open reading frame that could encode a viral polyprotein precursor of 3011 amino acids. While there as little overall amino acid and nucleotide sequence homology with other viruses, the 5' HCV nucleotide sequence upstream of this large open reading frame has substantial similarity to the 5' termini of pestiviral genomes. The polyprotein also has significant sequence similarity to helicases encoded by animal pestiviruses, plant potyviruses, and human flaviviruses, and it contains sequence motifs widely conserved among viral replicases and trypsin-like proteases. A basic, presumed nucleocapsid domain is located at the N terminus upstream of a region containing numerous potential N-linked glycosylation sites. These HCV domains are located in the same relative position as observed in the pestiviruses and flaviviruses and the hydrophobic profiles of all three viral polyproteins are similar. These combined data indicate that HCV is an unusual virus that is most related to the pestiviruses. Significant genome diversity is apparent within the putative 5' structural gene region of different HCV isolates, suggesting the presence of closely related but distinct viral genotypes.
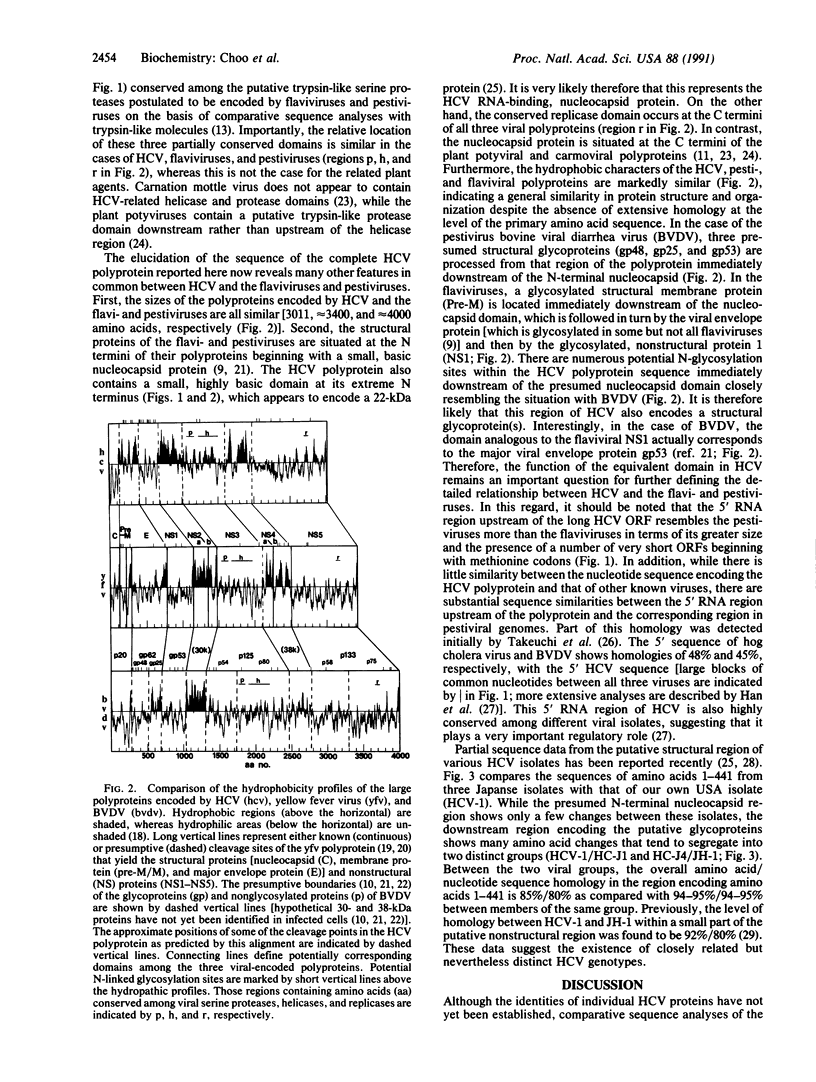
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bradley D. W., Cook E. H., Maynard J. E., McCaustland K. A., Ebert J. W., Dolana G. H., Petzel R. A., Kantor R. J., Heilbrunn A., Fields H. A. Experimental infection of chimpanzees with antihemophilic (factor VIII) materials: recovery of virus-like particles associated with non-A, non-B hepatitis. J Med Virol. 1979;3(4):253–269. doi: 10.1002/jmv.1890030403. [DOI] [PubMed] [Google Scholar]
- Bradley D. W. The agents of non-A, non-B viral hepatitis. J Virol Methods. 1985 Apr;10(4):307–319. doi: 10.1016/0166-0934(85)90047-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers T. J., McCourt D. W., Rice C. M. Yellow fever virus proteins NS2A, NS2B, and NS4B: identification and partial N-terminal amino acid sequence analysis. Virology. 1989 Mar;169(1):100–109. doi: 10.1016/0042-6822(89)90045-7. [DOI] [PubMed] [Google Scholar]
- Choo Q. L., Kuo G., Weiner A. J., Overby L. R., Bradley D. W., Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989 Apr 21;244(4902):359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- Choo Q. L., Weiner A. J., Overby L. R., Kuo G., Houghton M., Bradley D. W. Hepatitis C virus: the major causative agent of viral non-A, non-B hepatitis. Br Med Bull. 1990 Apr;46(2):423–441. doi: 10.1093/oxfordjournals.bmb.a072408. [DOI] [PubMed] [Google Scholar]
- Colett M. S., Larson R., Gold C., Strick D., Anderson D. K., Purchio A. F. Molecular cloning and nucleotide sequence of the pestivirus bovine viral diarrhea virus. Virology. 1988 Jul;165(1):191–199. doi: 10.1016/0042-6822(88)90672-1. [DOI] [PubMed] [Google Scholar]
- Collett M. S., Anderson D. K., Retzel E. Comparisons of the pestivirus bovine viral diarrhoea virus with members of the flaviviridae. J Gen Virol. 1988 Oct;69(Pt 10):2637–2643. doi: 10.1099/0022-1317-69-10-2637. [DOI] [PubMed] [Google Scholar]
- Collett M. S., Moennig V., Horzinek M. C. Recent advances in pestivirus research. J Gen Virol. 1989 Feb;70(Pt 2):253–266. doi: 10.1099/0022-1317-70-2-253. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A. E., Donchenko A. P., Koonin E. V., Blinov V. M. N-terminal domains of putative helicases of flavi- and pestiviruses may be serine proteases. Nucleic Acids Res. 1989 May 25;17(10):3889–3897. doi: 10.1093/nar/17.10.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A. E., Koonin E. V., Donchenko A. P., Blinov V. M. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 1989 Jun 26;17(12):4713–4730. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilley H., Carrington J. C., Balàzs E., Jonard G., Richards K., Morris T. J. Nucleotide sequence and genome organization of carnation mottle virus RNA. Nucleic Acids Res. 1985 Sep 25;13(18):6663–6677. doi: 10.1093/nar/13.18.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn Y. S., Galler R., Hunkapiller T., Dalrymple J. M., Strauss J. H., Strauss E. G. Nucleotide sequence of dengue 2 RNA and comparison of the encoded proteins with those of other flaviviruses. Virology. 1988 Jan;162(1):167–180. doi: 10.1016/0042-6822(88)90406-0. [DOI] [PubMed] [Google Scholar]
- Han J. H., Shyamala V., Richman K. H., Brauer M. J., Irvine B., Urdea M. S., Tekamp-Olson P., Kuo G., Choo Q. L., Houghton M. Characterization of the terminal regions of hepatitis C viral RNA: identification of conserved sequences in the 5' untranslated region and poly(A) tails at the 3' end. Proc Natl Acad Sci U S A. 1991 Mar 1;88(5):1711–1715. doi: 10.1073/pnas.88.5.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo Y., Takeuchi K., Boonmar S., Katayama T., Choo Q. L., Kuo G., Weiner A. J., Bradley D. W., Houghton M., Saito I. A cDNA fragment of hepatitis C virus isolated from an implicated donor of post-transfusion non-A, non-B hepatitis in Japan. Nucleic Acids Res. 1989 Dec 25;17(24):10367–10372. doi: 10.1093/nar/17.24.10367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo G., Choo Q. L., Alter H. J., Gitnick G. L., Redeker A. G., Purcell R. H., Miyamura T., Dienstag J. L., Alter M. J., Stevens C. E. An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science. 1989 Apr 21;244(4902):362–364. doi: 10.1126/science.2496467. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Meyers G., Rümenapf T., Thiel H. J. Molecular cloning and nucleotide sequence of the genome of hog cholera virus. Virology. 1989 Aug;171(2):555–567. doi: 10.1016/0042-6822(89)90625-9. [DOI] [PubMed] [Google Scholar]
- Miller R. H., Purcell R. H. Hepatitis C virus shares amino acid sequence similarity with pestiviruses and flaviviruses as well as members of two plant virus supergroups. Proc Natl Acad Sci U S A. 1990 Mar;87(6):2057–2061. doi: 10.1073/pnas.87.6.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto H., Okada S., Sugiyama Y., Yotsumoto S., Tanaka T., Yoshizawa H., Tsuda F., Miyakawa Y., Mayumi M. The 5'-terminal sequence of the hepatitis C virus genome. Jpn J Exp Med. 1990 Jun;60(3):167–177. [PubMed] [Google Scholar]
- Saiki R. K., Scharf S., Faloona F., Mullis K. B., Horn G. T., Erlich H. A., Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985 Dec 20;230(4732):1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- Strauss J. H., Strauss E. G. Evolution of RNA viruses. Annu Rev Microbiol. 1988;42:657–683. doi: 10.1146/annurev.mi.42.100188.003301. [DOI] [PubMed] [Google Scholar]
- Takeuchi K., Kubo Y., Boonmar S., Watanabe Y., Katayama T., Choo Q. L., Kuo G., Houghton M., Saito I., Miyamura T. Nucleotide sequence of core and envelope genes of the hepatitis C virus genome derived directly from human healthy carriers. Nucleic Acids Res. 1990 Aug 11;18(15):4626–4626. doi: 10.1093/nar/18.15.4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi K., Kubo Y., Boonmar S., Watanabe Y., Katayama T., Choo Q. L., Kuo G., Houghton M., Saito I., Miyamura T. The putative nucleocapsid and envelope protein genes of hepatitis C virus determined by comparison of the nucleotide sequences of two isolates derived from an experimentally infected chimpanzee and healthy human carriers. J Gen Virol. 1990 Dec;71(Pt 12):3027–3033. doi: 10.1099/0022-1317-71-12-3027. [DOI] [PubMed] [Google Scholar]
- Westaway E. G., Brinton M. A., Gaidamovich SYa, Horzinek M. C., Igarashi A., Käriäinen L., Lvov D. K., Porterfield J. S., Russell P. K., Trent D. W. Flaviviridae. Intervirology. 1985;24(4):183–192. doi: 10.1159/000149642. [DOI] [PubMed] [Google Scholar]



