Abstract
Translocator protein 18 kDa (TSPO) was previously known as the peripheral benzodiazepine receptor (PBR) in eukaryotes, where it is mainly localized to the mitochondrial outer membrane. Considerable evidence indicates that it plays regulatory roles in steroidogenesis and apoptosis and is involved in various human diseases, such as metastatic cancer, Alzheimer’s and Parkinson’s disease, inflammation, and anxiety disorders. Ligands of TSPO are widely used as diagnostic tools and treatment options, despite there being no clear understanding of the function of TSPO. An ortholog in the photosynthetic bacterium Rhodobacter was independently discovered as the tryptophan-rich sensory protein (TspO) and found to play a role in the response to changes in oxygen and light conditions that regulate photosynthesis and respiration. As part of this highly conserved protein family found in all three kingdoms, the rat TSPO is able to rescue the knockout phenotype in Rhodobacter, indicating functional as well as structural conservation. Recently, a major breakthrough in the field was achieved: the determination of atomic-resolution structures of TSPO from different species by several independent groups. This now allows us to reexamine the function of TSPO with a molecular perspective. In this review, we focus on recently determined structures of TSPO and their implications for potential functions of this ubiquitous multifaceted protein. We suggest that TSPO is an ancient bacterial receptor/stress sensor that has developed additional interactions, partners, and roles in its mitochondrial outer membrane environment in eukaryotes.
Graphical abstract
Translocator protein 18 kDa (TSPO), also widely know as the peripheral benzodiazepine receptor (PBR), has been an intense focus of research since its discovery in 1977.1 In mitochondria, it was first identified as the secondary binding site for the widely used benzodiazepine anxiolytic drugs, but with a different ligand binding profile compared to that of the central nervous system binding site, the GABAA receptor.2 TSPO is highly expressed in many tissues, especially those involved in steroidogenesis, and has been proposed to be an important player in the transport of cholesterol into mitochondria, the first and rate-limiting step for steroid hormone synthesis.3,4 A considerable and still growing body of evidence supports TSPO’s involvement in a number of other complex cellular processes, including porphyrin transport,5,6 inflammation,7,8 tumor progression,9–11 and Parkinson’s and Alzheimer’s diseases.12,13 Because of TSPO’s reproducibly high level of expression in areas of inflammation, ligands for TSPO are widely used and actively developed as imaging agents and treatment options for brain damage.14–16
Independent of its recognition in animal systems, TSPO was discovered in the carotenoid gene cluster17 in the photosynthetic bacterium Rhodobacter, a close living relative of mitochondria.18 It was named the tryptophan-rich sensory protein (TspO), for its high tryptophan content and apparent role in the regulation of the transition between photosynthesis and respiration induced by changes in levels of oxygen and light, a signaling process that may involve altered porphyrin transport.19,20 Importantly, the rat homologue of TSPO was able to rescue the TspO deletion phenotype in Rhodobacter sphaeroides, indicating a conservation of function through evolution.21 TSPO was also identified in many other evolutionarily diverse species, including the plant Arabidopsis thaliana (AtTSPO),22 moss Physcomitrella patens (PpTSPO),23 and cyanobacteria Fremyella diplosiphon (FdTSPO)24 and Synechocystis sp. PCC 6803.25 In these different species, it is a player in various stress responses, such as salt, oxidative stress, and abscisic acid.22,23,26,27 As a result of mounting evidence of its involvement in multiple cellular processes, TSPO was renamed as translocator protein 18 kDa (TSPO) in 2006 to recognize its diverse roles.28
Despite a wealth of data in the literature since the 1970s, our understanding of TSPO function is far from clear. In fact, the essential role of TSPO in embryonic development and as the cholesterol transporter in mammals was recently challenged,29–31 making this protein even more of an enigma. Induction of TSPO under various stress conditions appears to be a common theme, as seen in inflammation in animals, altered light and O2 conditions in bacteria, and exposure to high salt in plants. However, its precise role in any of these processes has not been determined. The fundamental question of whether it acts as a receptor, a transporter/translocator, or possibly an enzyme is the subject of much debate. Part of the difficulty in understanding TSPO function has related to the lack of an atomic-resolution structure and in vitro functional assay systems for biochemical characterization. Fortunately, some important breakthroughs have recently occurred. High-resolution crystal structures of TSPO from two unrelated bacteria, as well as NMR structures of the mouse TSPO (mTSPO), were determined.32–34 In addition, the ligand binding properties of purified TSPO proteins have been characterized.35,36 These new advances facilitate correlation of structure and function of TSPO in molecular detail.
This review will focus on the new TSPO structural information, areas of agreement as well as inconsistencies, and implications for TSPO function. A number of other recent reviews37–43 illustrate the strong interest in the much-debated physiological functions and drug target potential of this multifaceted protein.
CRYSTAL AND NMR STRUCTURES: SIMILARITIES AND DIFFERENCES
Crystal structures of TSPO from two unrelated bacteria32,33 and NMR structures of the mouse protein34,44 were determined in the past two years, providing long-awaited structural information and the opportunity to compare and contrast the several structures.
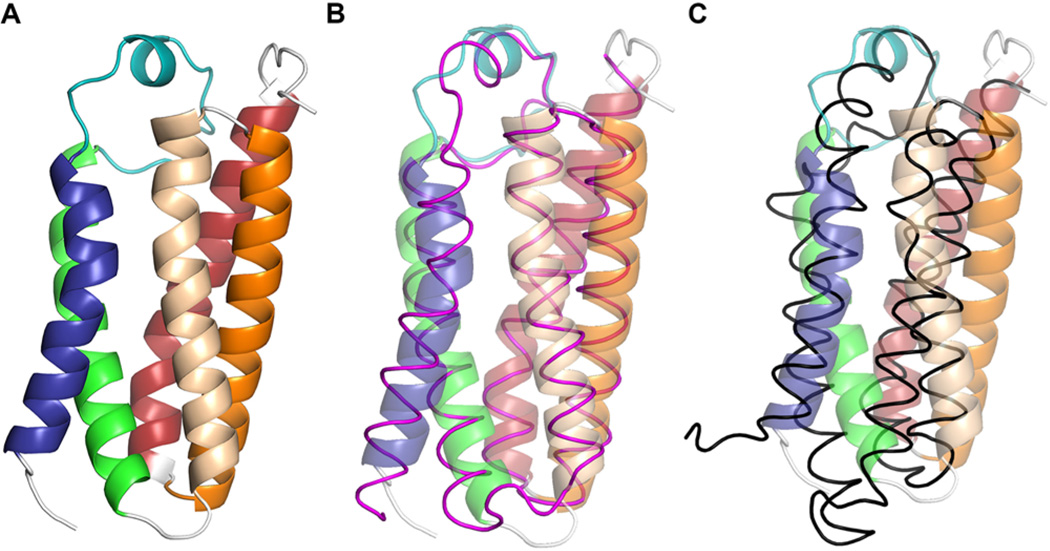
In all structures of TSPO, the monomer of the protein adopts a helical bundle structure composed of five transmembrane helices (TM) (Figure 1A) as correctly predicted by various methods for identifying TM helices. A long loop (LP1) that connects TM1 and TM2 is also observed as predicted and shows well-defined structure. Despite the fact that the crystal structures are of proteins derived from two distinctly different bacteria, one Gram-negative (R. sphaeroides) and one Gram-positive (Bacillus cereus) with only 23% sequence identity, they are remarkably similar, with or without ligand bound, suggesting a strong conservation of the structural fold during evolution. As shown in Figure 1B, the transmembrane helices of the RsTSPO and BcTSPO overlap very well and LP1 adopts a very similar structure with a signature short helix in the middle.
Figure 1.
Comparison of TSPO monomers from different species. The high-resolution crystal structure of RsTSPO A139T (PDB entry 4UC1) is colored in discrete rainbow and shown as a cartoon in panel A: TM1, blue; LP1, teal; TM2, green; TM3, wheat; TM4, orange; TM5, red. (B) Comparison of the structures of RsTSPO and BcTSPO (PDB entry 4RYQ). RsTSPO is colored the same as in panel A and shown partially transparent, while the crystal structure of BcTSPO is colored magenta and shown as a tube. (C) Comparison of the structures of RsTSPO and mTSPO (PDB entry 2MGY). RsTSPO is colored the same as in panel A and shown partially transparent, while the NMR structure of mTSPO is colored black and shown as a tube.
In contrast, the structure reported by Jaremko and coworkers, a rigorously analyzed NMR structure of the mouse homologue, mTSPO, shows distinct differences (Figure 1C). It is noteworthy that the protein used for these studies was disordered in the absence of ligand and only in the presence of excess concentrations of the ligand PK1119534 could a stable tertiary structure be obtained. This contrasts markedly with the well-defined biochemical behavior of RsTSPO and BcTSPO without ligand and their highly similar crystal structures (Figure 1B), as well as the virtually identical BcTSPO structures with and without PK11195.33 These data suggest that the disordered apo structure of mTSPO results from the conditions of purification and refolding and is not an example of a natively disordered protein as the investigators propose.45 Considering that the mTSPO was purified in sodium dodecyl sulfate (SDS) before refolding and reconstitution into dodecylphosphocholine (DPC) for the NMR experiments, there are substantive concerns about both the apo- and ligand-induced conformations.
Indeed, the NMR structure of the mTSPO determined with PK11195 bound differs from the bacterial versions in important ways. Although mTSPO shows a higher level of sequence conservation compared to RsTSPO (32% identity) and BcTSPO (27% identity) than they do to each other, the NMR structure is notably different from both (Figure 1C), with several of the transmembrane helices considerably shifted and side chains having opposite orientations.32,33 These alterations lead to substantial numbers of charged residues being exposed on the predicted membrane-embedded surface, unexpected for an integral membrane protein. It is reasonable to propose that the structural differences between mammalian and bacterial proteins, especially in the loop regions, are the result of evolutionary divergence, but at least the alignment of transmembrane regions of membrane proteins is expected to be maintained, as seen in the similarity between the structures of RsTSPO and BcTSPO. Rather than intrinsic differences, it seems more likely that conditions and detergents used in purification and NMR measurements contribute importantly to the differences in structure. Jaremko and colleagues do not report any binding studies to establish that the protein is in a native state under the conditions of their analysis, but others have shown that solubilization of RsTSPO in the ionic detergent DPC results in ~20-fold lower binding affinity for PK11195 compared to the binding affinity in the nonionic detergent decyl maltoside (DM).46 A similar detergent-induced alteration in ligand affinity was reported for the mTSPO, in which case PK11195 binding was abolished in the protein purified in SDS but recovered when the protein was reconstituted into liposomes.47 These observations indicate a substantial detergent-induced effect on binding, perhaps accounting for some of the differences between the NMR structure of the mTSPO and the crystal structures of bacterial TSPO. Jaremko and colleagues have argued that the differences arise from crystal packing effects.44,48 However, the published crystal structures were determined from at least five different crystal forms, each with unique differing packing arrangements and intermolecular contacts, yet all yield the same overall tertiary structure. Thus, it is unlikely that “crystal packing” is the cause of these major discrepancies. It is relevant to note that the recently reported NMR structure of a mutant form of the mTSPO,44 containing the human polymorphism A147T, shows less alteration compared to WT than what was observed in the crystal structure of the same mutant created in RsTSPO. The authors again suggest that the differences are due to crystal packing effects, but as discussed above, the disparity is more likely to be the result of detergent-induced effects and refolding issues. No data are provided to establish the nativeness of either the WT or the mutant mTSPO after purification or under the NMR conditions.
THE FIRST EXTRAMEMBRANE LOOP, LP1, IS STRUCTURALLY CONSERVED
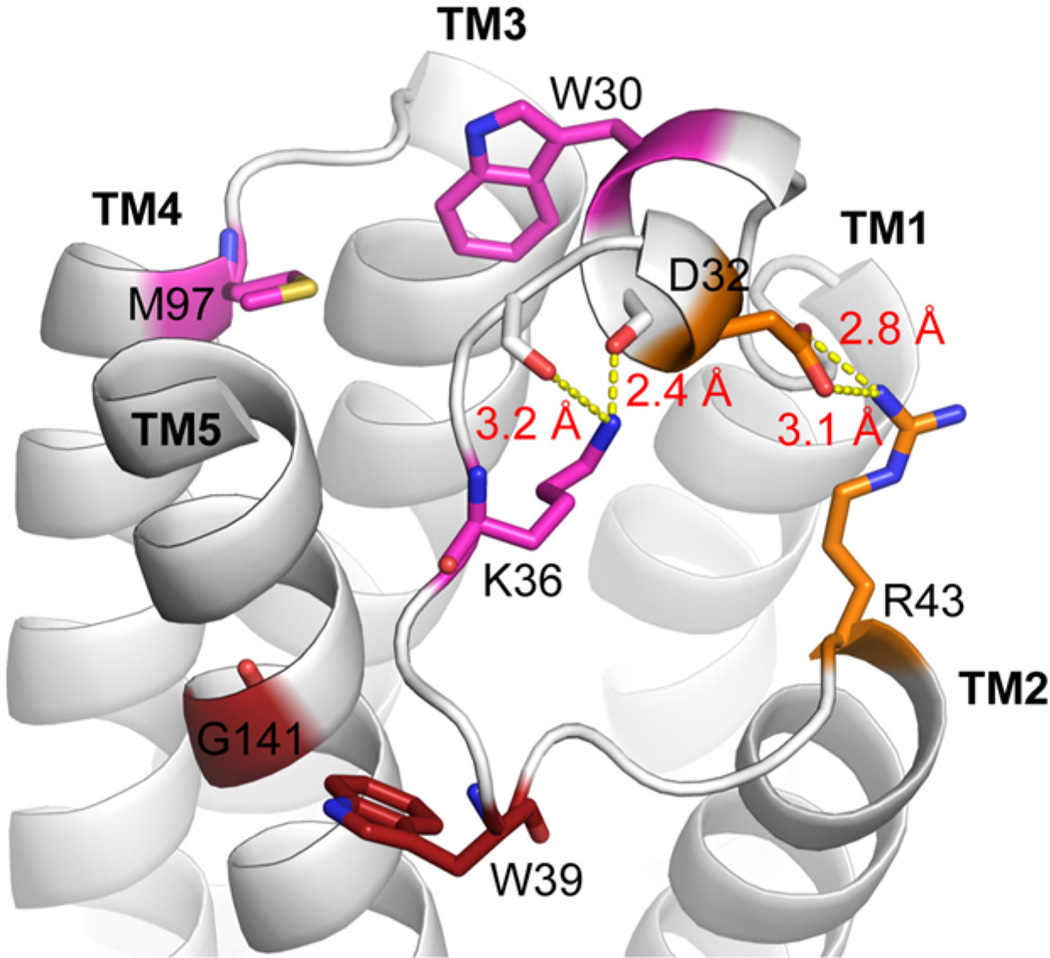
LP1 has previously been proposed to play an important regulatory role in binding of ligand to TSPO.49,50 Along with significant sequence identity, LP1 in RsTSPO and LP1 in BcTSPO are almost identical in structure, including the signature short helix in the middle of the loop. Evolutionary covariance analysis was previously used to identify important side chain interactions across evolution within TSPO and shows remarkable agreement with the crystal structures in predicting the interacting residues.35 Interestingly, the interaction between W39 on LP1 and G141 on TM5 (Figure 2), clearly seen in the crystal structure of RsTSPO, was identified by this analysis as one of the most confident predictions of close interactions. Three other interacting pairs involving residues on the short helix within LP1 are also predicted and verified, including W30 with M97, W30 with K36, and D32 with R43 in TM2. Figure 2 shows that all predicted pairs are positioned to have favorable side chain interactions in the crystal structure of RsTSPO. Four salt bridges involving predicted residue pairs are also observed, suggesting an evolutionarily stable structure of LP1. These covariance predictions and structural observations have two important implications with regard to the function of LP1. First, despite limited conservation of LP1 sequence across species, it forms a rather stable structural motif instead of a random flexible loop, especially the second half starting from the highly conserved W30, which suggests an important functional role such as acting as a lid of the central cavity. Second, conserved interactions of LP1 with TM5 could play a role in regulating a lateral entrance to the central cavity for some hydrophobic ligands. Considering that the proposed cholesterol binding site is located on TM5, as is the A139T51 mutation that has a lower cholesterol affinity and shows a significantly narrower opening between TM5 and TM2, it is possible that TM5 and TM2 form a lateral gate in TSPO. The interaction of the second half of LP1 with TM5 (W39/G141) and TM2 (D32/R43) could contribute to its regulation. None of these interactions are seen in the NMR structures of mouse TSPO, where the conformation of LP1 is modeled quite differently, but several are conserved in the BcTSPO structure.
Figure 2.
External loop, LP1, which has a defined, conserved structure across evolution and interacts with TM5 and TM2. Residues on LP1 and interacting pairs predicted by covariance analysis are shown as sticks and colored in matching colors, with red having the highest-confidence pairing across evolution, followed by magenta and orange. K36, D32, and R43 form predicted salt bridges (yellow dotted lines) with backbone and side chains maintaining a defined structure for LP1. W39 and G141 also closely interact as predicted. Conservation of interacting pairs suggests the structure of LP1 and its interaction with TM5 and TM2 is an important structural element for TSPO across different species and may play an essential functional role (figure created in Pymol from PDB entry 4UC1).
OLIGOMERIC STATES: IS THE RSTSPO DIMER SIGNIFICANT?
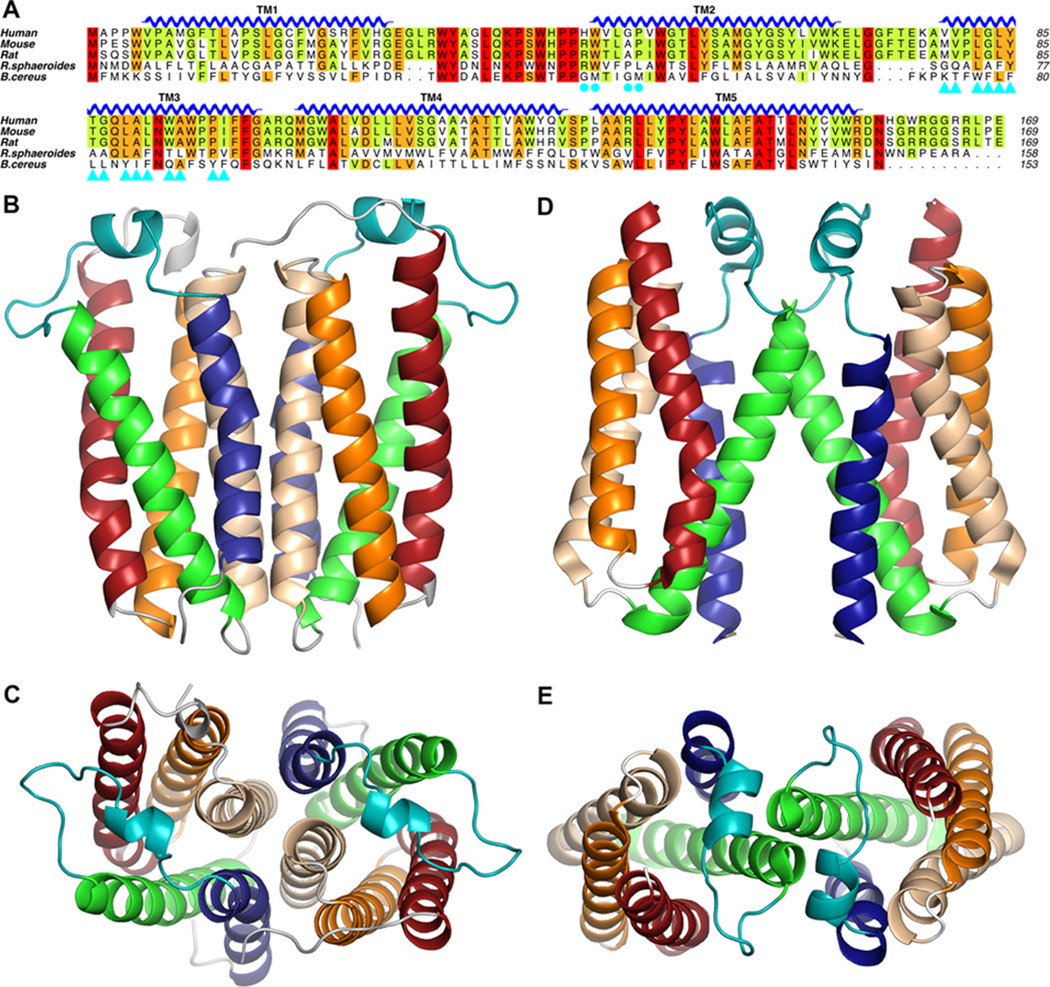
Dimerization and oligomerization have been reported for TSPO in human, mouse, and R. sphaeroides in vivo and in vitro.46,52,53 The observation of the highly stable dimeric structure of purified RsTSPO in solution46 and in several different crystal forms32,54 emphasizes the likely importance of the dimer. However, the structure of BcTSPO was determined in both a dimeric and monomeric state, while the NMR structure of mTSPO was found to be a monomer. In addition, the dimer of BcTSPO has a completely different interface compared to that of the RsTSPO dimer (Figure 3). These observations bring up important questions that need to be addressed; namely, is the dimer interface conserved, and is the dimer functionally significant? RsTSPO forms a very tight and flat dimer interface, and no monomer species is observed in solution or in the crystallized state.46 These findings lend credence to the physiological significance of the dimer form, also supported by the fact that the main interacting residues in the dimer interface (Figure 3A, cyan triangles), contributed mostly by TM3, are conserved between RsTSPO and the mammalian proteins but not in BcTSPO. In contrast, the dimer interface in the BcTSPO structure has a much smaller interface involving predominantly the top of TM2, with a sequence quite unique to BcTSPO (cyan dots) (Figure 3). Considering that a monomeric form was used to grow these crystals and the BcTSPO dimer is observed in only some of the crystal forms, this dimer may be a consequence of crystal packing. However, it could also be an alternative interface for higher-order oligomerization, as observed in both RsTSPO and the mammalian TSPO.53,54 The nature of any oligomeric state of mammalian TSPO cannot be ascertained, because the only structures currently available were obtained by NMR in a monomeric state that may be critical for obtaining high-quality NMR data. Considering the evolutionary relationship between R. sphaeroides and mitochondria,18 as well as the sequence conservation in the TM3 region (Figure 3A), the same dimer interface is plausible for the mammalian proteins, but alternative and dynamic53,55 oligomeric forms involving different partners could be required to accomplish the complex functions of TSPO in mitochondria.
Figure 3.
RsTSPO and BcTSPO form different dimers. A sequence alignment of TSPOs from human, mouse, rat, R. sphaeroides, and B. cereus is shown in panel A, while the two different dimer assemblies for RsTSPO (B and C; PDB entry 4UC1) and BcTSPO (D and E; PDB entry 4RYJ) are shown in side views and top views. In panel A, residues on the dimer interface of RsTSPO are labeled as cyan triangles while residues for the BcTSPO interface are labeled as cyan dots.
WHAT HAVE WE LEARNED ABOUT PK11195 AND PROTOPORPHYRIN IX BINDING SITES?
TSPO can be fairly described as an “orphan receptor” because there is still no consensus about which ligands are physiologically significant. Development of more specific ligands for TSPO continues to be an important focus, because TSPO ligands are widely used as a biomarker for brain inflammation as well as treatment for various diseases.16,56 More high-resolution structures with different ligands bound will undoubtedly facilitate the drug development effort as well as provide new opportunities for an improved understanding of TSPO function and its regulation.
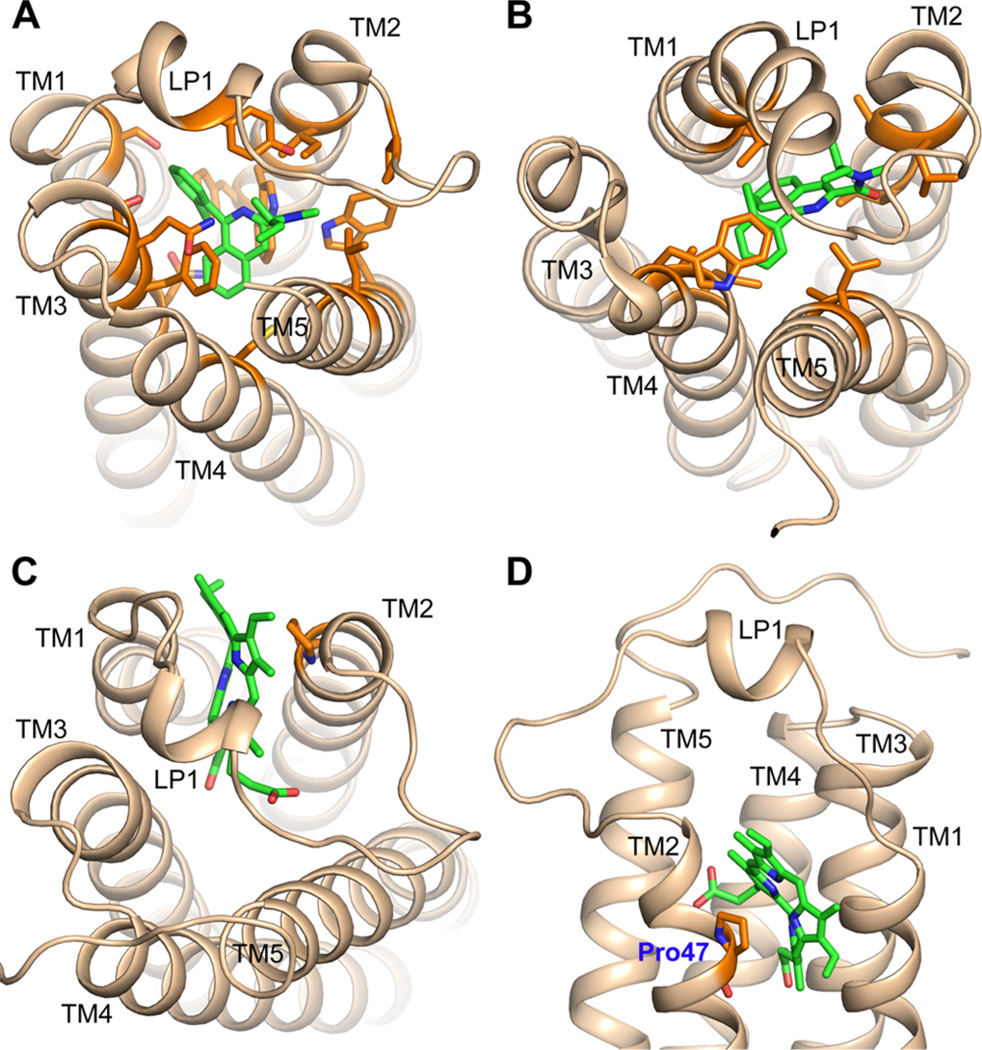
The synthetic ligand, PK11195, often used as a diagnostic of TSPO involvement, was resolved in the crystal structure of BcTSPO (Figure 4A) as well as the NMR structure of mTSPO (Figure 4B), while a potential endogenous ligand, a porphyrintype molecule, was resolved in the crystal structure of RsTSPO (Figure 4C,D). In all three structures, these ligands bind to the central cavity but in different positions. PK11195 is observed in the middle of the cavity, interacting with residues from all five transmembrane helices (Figure 4A,B), whereas the porphyrin is less deeply inserted in a region between TM1 and TM2 (Figure 4C,D). While the central cavity is no doubt playing a major role in ligand binding, the residues involved remain to be further confirmed, because PK11195 interacts in a different orientation and with a different set of residues in the crystal structure of BcTSPO (Figure 4A) compared to the NMR structure of mTSPO (Figure 4B). The altered positions of the side chains and charge due to the conditions required for the NMR structure determination are likely sources of differences, but the precise position is somewhat ambiguous in BcTSPO as well, because it is found in a relatively low-resolution crystal structure (3.5 Å) and thus is not well resolved. Unambiguous identification of the interacting residues will therefore await additional structures and comparative binding analyses.
Figure 4.
Ligand binding sites for TSPO from different species. Ligand binding sites in currently available structures of different species are positioned in the central cavity but have different interacting residues. TSPO proteins are colored wheat, while ligands (PK11195 and porphyrin) are colored green. Side chains interacting with the ligands are shown as orange sticks. (A) BcTSPO with PK11195 bound (PDB entry 4RYI). (B) mTSPO with PK11195 bound (PDB entry 2MGY). (C) RsTSPO with a representative porphyrin bound (PDB entry 4UC1). (D) As in panel C but from the top to show the position of residue P47, which is a histidine and binds heme in plant TSPO.
The other ligand found in the crystal structures was in the RsTSPO, identified on the basis of a ring-shaped density that could be best fit by a porphyrin-like molecule (Figure 4C,D). This ligand identification is supported by spectra of the purified protein that suggest the presence of a porphyrin oxidation product.32 An interesting additional clue comes from studies of the Arabidopsis homologue of TSPO in which a histidine residue (H91) has been identified to be critical for heme binding.57 In the RsTSPO structure, a proline residue (P47) takes the place of that histidine, but if a heme were bound in the same place as the observed porphyrin, the heme iron would be located correctly to be ligated by a histidine at position 47 (Figure 4D). This observation supports the identification of a porphyrin in this site.
Although the current structures give many important and testable clues regarding the liganded states of TSPO, further studies will be required to establish precisely how PK11195, porphyrin, and many other ligands are bound and how their binding may influence the conformation or aggregation state of the protein.
NEW CLUES FROM STRUCTURE REGARDING THE CHOLESTEROL BINDING SITE
TSPO was initially proposed to play an important role in cholesterol metabolism because high-level expression of the protein was observed in steroidogenic tissue and TSPO ligands were observed to regulate steroidogenesis.3,58,59 Its involvement in cholesterol regulation was further strengthened by findings that TSPO binds cholesterol with nanomolar affinity.47,60 However, the precise mechanism of TSPO–cholesterol interaction and the role of TSPO in the transport of cholesterol into mitochondria remain elusive. The embryonic lethality phenotype first observed in the TSPO knockout mouse58 has recently been challenged,29–31,61 emphasizing the need to further investigate the function of TSPO. Mounting evidence suggests that TSPO is not the transporter for cholesterol but rather is involved in at least two dynamic complexes spanning both the outer and inner mitochondrial membranes, and potentially the ER, which play a role in cholesterol transport and processing.39,55 TSPO’s unusually high affinity for cholesterol and the effects of its ligands on both transport and processing make TSPO a likely player in, and regulator of, these processes, especially under stress conditions,62 as well as a promising drug target. Nevertheless, the binding site for cholesterol remains to be precisely defined.
A cholesterol recognition/interaction amino acid consensus (CRAC) sequence was identified as the cholesterol binding site on the TM5 region of mTSPO by deletion mutational analysis.60 Confirmation of this site from crystallographic studies is not yet available, but the existing structures of TSPO from the three different species all show that the CRAC sequence is on the membrane-exposed surface of the protein, rather than forming part of a central binding site or a channel at the dimer interface of TSPO. If confirmed, this location of the CRAC site would suggest that a complex of TSPO with other binding partners may be required for cholesterol transport32,39 and is consistent with other ligands binding independently and affecting cholesterol binding allosterically.
INSIGHT INTO CHOLESTEROL BINDING FROM MUTAGENESIS OF RSTSPO
We have taken advantage of the well-characterized RsTSPO protein to further define the nature of cholesterol binding. This bacterial homologue has an intrinsically lower binding affinity for cholesterol, ~1000-fold lower than the nanomolar affinity of human TSPO.46 By comparative analysis of TSPO sequences, we identified a region one helix turn before the CRAC sequence that was distinctly less hydrophobic (ATA vs LAF) in the bacterial TSPO and much less conserved. When the mammalian version of the three-amino acid sequence was substituted into the bacterial TSPO, the binding affinity of RsTSPO for cholesterol was increased to a level similar to that of human TSPO.35 The result, defining a cholesterol binding enhancement motif,35 provides compelling evidence of the location of the cholesterol binding site. It also suggests a potential binding orientation for cholesterol in this region: the ring structure of the cholesterol could associate with the main CRAC sequence while the alkyl tail binds to the enhancement motif, consistent with the evolution of the enhancement motif in the mammalian TSPO family proteins35 toward a higher affinity for steroid-type molecules with a hydrophobic tail.
COMPUTATIONAL PREDICTION OF A CHOLESTEROL BINDING SITE
To identify potential cholesterol binding sites, we applied the CholMine algorithm63 to the RsTSPO crystal structure. This method combines SimSite3D analysis (three-dimensional surface comparison and alignment) with knowledge of conserved interactions in known crystal structures with cholesterol or cholate already bound, to search other protein structures for three-dimensional cholesterol/cholate binding sites. Compared to traditional sequence motif-based prediction, the CholMine method works across diverse protein families and shows a higher accuracy (~82%) in predicting true cholesterol or cholate binding sites for either soluble or membrane proteins.
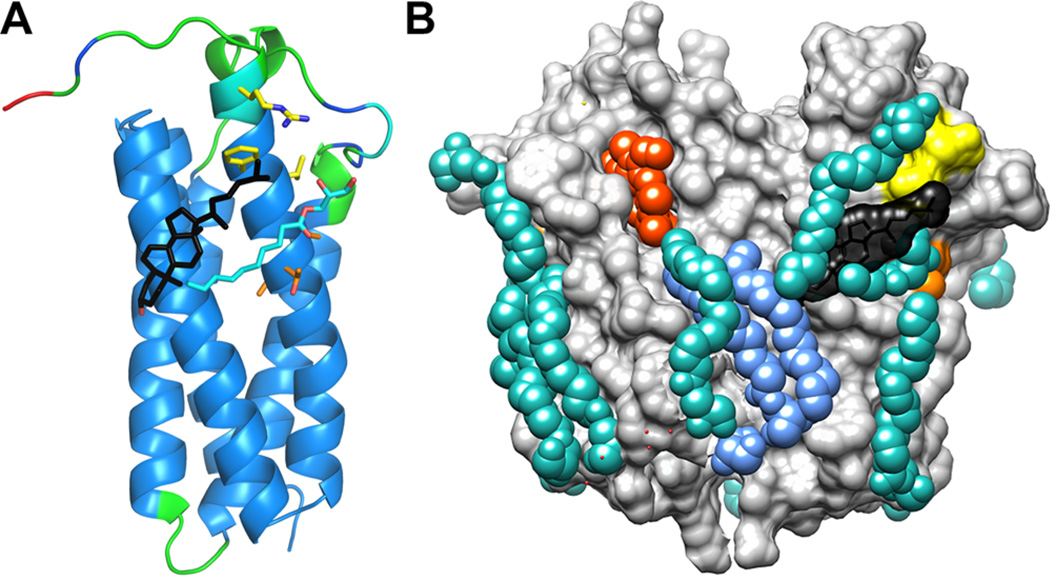
The CholMine analysis predicted a cholesterol binding site on RsTSPO close to the expected position involving TM5 (Figure 5). The most favorable position for cholesterol binding (black) traverses helices TM5 and TM4 (Figure 5A) and matches 8 of 10 characteristic interactions for cholesterol determined from a series of unrelated cholesterol-bound PDB structures. This cholesterol site parallels the binding site for a monoolein lipid (cyan) found in the same position just below the cholesterol site in all our different crystal structures of TSPO (e.g., PDB entries 4UC1, 4UC2, and 5DUO). An additional conserved lipid site is resolved in some structures just above the predicted cholesterol site, as seen in Figure 5B, which shows the location of the cholesterol site in a surface rendering of the TSPO dimer and includes more of the resolved lipids. The stacking arrangement of cholesterol with other lipids has been observed in structures of other membrane proteins, such as the β2-adrenergic receptor (PDB entry 2RH1),64 in which a palmitate (residue 415) is associated with cholesterol molecules (residues 412–414). Interestingly, the predicted cholesterol binding site in TSPO is located in the vicinity of the previously defined CRAC site (yellow) and the enhancement motif (orange), shown as surface rendering in Figure 5B, but in the reverse orientation with respect to what we predicted on the basis of the mutagenesis analysis.35 The tail is oriented toward the outside of the membrane plane and closer to the CRAC rather than the enhancement motif. As to why a bound cholesterol is not resolved in the crystal structure, it is noteworthy that monoolein is used at a high concentration under the lipidic cubic phase crystallization condition and is very likely to outcompete other lipidic ligands on the protein surface, especially those with relatively low affinity such as cholesterol for RsTSPO.
Figure 5.
Favored cholesterol binding site predicted in TSPO. (A) A CholMine-predicted cholesterol binding position (black sticks) is shown on the crystal structure of RsTSPO, where the A chain of RsTSPO A139T (PDB entry 4UC1) is represented with main chain ribbon and side chain sticks for the LAF and CRAC motif residues (A136, T137, and A138 colored yellow and L142, F144, and R148 colored orange). The main chain of TSPO is colored according to crystal temperature factor values, with blue indicating low-mobility, green moderate-mobility, and red high-mobility regions. (B) The AB dimer of RsTSPO A139T is shown in surface representation. The most favorable position for cholesterol binding is shown in black space-filling representation, located in the vicinity of the CRAC site (yellow) and LAF site (orange). Monoolein lipids are colored cyan; a phospholipid is colored blue, and a porphyrin-type ligand is colored red. In panel A, one partial monoolein that sits parallel with the predicted cholesterol binding site is shown as sticks, while in panel B, all crystallographically observed lipids are shown in space-filling representation.
BINDING OF BILAYER LIPIDS
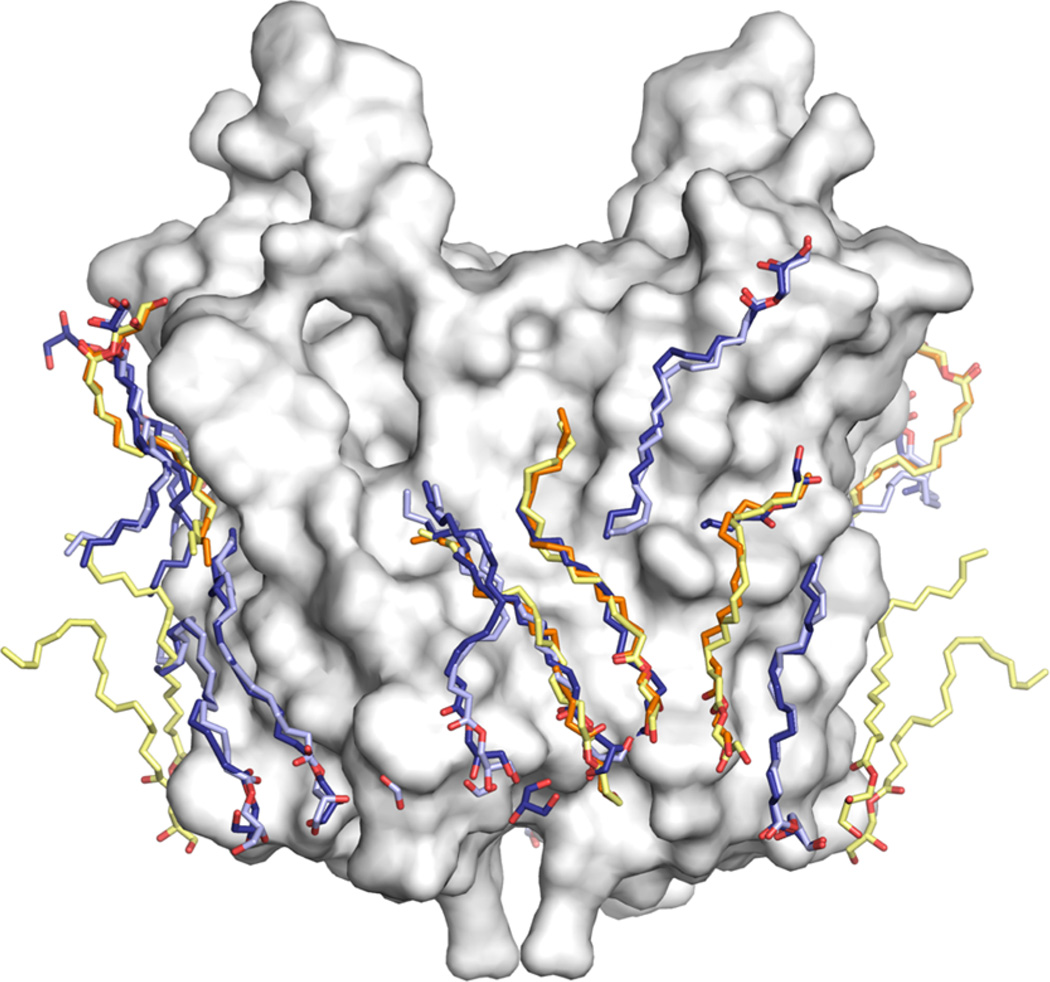
Determination of native lipid binding sites crystallographically is challenging, given the much higher concentration of nonnative lipids or detergents required for the crystallization. Nevertheless, the location of an artificial lipid or detergent in a crystal structure is frequently the site where a native lipid is bound.65–67 Notably, a number of alkyl chains identified in the crystal structures of RsTSPO are seen consistently occupying the same positions in crystals grown under very different conditions and crystal packing, suggesting that the occupied locations may represent true lipid binding sites (Figure 6). The lipids that are the most consistently well resolved in all four dimeric structures are seen on the surface spanning the dimer interface (overlay of blue, light blue, orange, and yellow), supporting the idea that the dimer is an important organizational unit providing the structural basis for strong lipid interactions of potential functional significance.
Figure 6.
Observed lipid binding sites. Lipids are consistently observed in similar positions in different crystal structures of RsTSPO grown under different conditions (shown as sticks). Lipids observed in the 1.8 Å structure (PDB entry 4UC1) are colored blue (AB dimer) and cyan (CC′ dimer), while lipids observed in the 2.4 Å structure (PDB entry 5DUO) are colored orange (AB dimer) and yellow (CC′ dimer).
FUNCTIONAL PUZZLE OF TSPO
The functions of TSPO remain elusive despite many studies in different systems and organisms. A variety of roles have been identified that often seem to be unrelated and sometimes conflicting. A general problem is recognized where conclusions are based on effects of TSPO ligands whose off-target interactions can be a concern. For instance, the benzodiazepine derivative Bz-423 was found to bind specifically to the F1F0 ATPase, but not TSPO, and induce the mitochondrial permeability transition,68,69 raising questions about observations that other benzodiazepine-related ligands considered to be specific for TSPO, such as PK11195, may also induce off-target effects.70–72 The seemingly definitive TSPO knockout experiments have also led to conflicting results.31,61,62 However, one consistent finding from knockout experiments appears to be altered mitochondrial energy metabolism, including lower oxygen consumption, membrane potential, and ATP levels.40,41,61,73 This observation fits with the additional finding of an increased level of fatty acid oxidation in the absence of TSPO,74 but how these observations relate to another emerging common theme, sensing or responding to stress, and how they may be associated with oxygen radical production and regulatory phenomena, remains to be determined.
WHAT IS THE ROLE OF TSPO IN CHOLESTEROL TRANSPORT OR TRANSLOCATION?
The evidence that TSPO is a bona fide transporter is not strong. Binding of both cholesterol and porphyrin to TSPO has been demonstrated in vitro and in vivo, but evidence of the direct involvement in transport is still lacking, partially because of the difficulties of assaying the movement of neutral hydrophobic molecules across a membrane. Disappointingly, in none of the new structures of TSPO is cholesterol resolved, nor are there any obvious channels visible within the protein monomer or at the dimer interface.32,33 The only stable consistent dimer observed, the RsTSPO dimer, has a remarkably tight interface, suggesting that this is not a transport pathway. In addition, the dimer interface does not involve any of the residues defined in the CRAC site or the enhancement motif.
In light of the structural information, we proposed an external pathway for cholesterol that would require a dimer of dimers or another binding partner for TSPO to facilitate movement.32 In fact, experiments in mammalian cell culture systems provide evidence55 of a 800 kDa protein complex being required to demonstrate the cholesterol side chain cleavage activity that occurs in the mitochondrial matrix. In this complex, TSPO is proposed to be the cholesterol binding/sequestering site, along with StAR, and the voltage-dependent anion channel (VDAC) is proposed to be the direct binding partner of TSPO that facilitates transport. However, deleting TSPO in at least one mouse model causes no obvious change in development or steroidogenic behavior, suggesting a regulatory role or possible functional redundancy, as well as the requirement for association with partner proteins.29–31 Several other TSPO−/− mouse constructs have yielded conflicting reports regarding lethality and other characteristics.31,41,42,58,61 These differences have been discussed in detail,39,62,75 clarifying some of the complex issues involved in the methodology that may explain the discrepancies. Recent results from conditional, cell-targeted TSPO deletion show altered development and hormone-driven steroid synthesis,62 in keeping with the role of TSPO as a stress response player in steroid metabolism.
TSPO and VDAC are consistently observed in the cholesterol transduceosome and metabolon complexes that bridge the mitochondrial inner and outer membrane and are responsible for hormone-induced steroidogenesis.39,55,76,77 Direct interaction of TSPO with VDAC has also been demonstrated with various methods, including blue native PAGE,55 immunoprecipitation and confocal microscopy,77 and copurification.78 However, the mode of interaction and the role in cholesterol transport by this complex at a molecular level are still unknown. With the availability of high-resolution crystal structures of both TSPO32,33 and VDAC,79 these questions can now be investigated in more detail for the first time. Current knowledge of TSPO and VDAC suggests that a possible transport pathway could be through the interface of a VDAC and TSPO complex. This is supported by the fact that cholesterol binding sites on both TSPO and VDAC are predicted to be on the outside surface of the proteins.32,35,80 Identifying the binding interface of TSPO and VDAC will be critical to understanding a transport mechanism. Computational docking (protein–protein and ligand–protein) and evolutionary covariance analysis could provide some useful clues. Docking predictions for cholesterol on VDAC are already available.81 However, the presence of TSPO is likely to change the energy landscape significantly. Other components of the transduceosome and metabolon complexes need also be considered because they would be expected to have a significant impact on the overall cholesterol transport activity.
High-affinity cholesterol binding by TSPO appears to be a relatively new development during evolution, given the lack of cholesterol in bacteria and the increasing importance of cholesterol and steroid hormones in the physiology of mammals.35,62,76 However, structural and biochemical analyses suggest that a binding site that can recognize cholesterol-like molecules already exists in bacterial TSPO, perhaps optimized for other related ligands such as hopanoids.35,76 The unusually high affinity of mammalian TSPO for cholesterol and the requirement for a dedicated complex for transport into mitochondria for steroidogenesis suggest that the main function of TSPO may be to sequester cholesterol for further processing. The new structural evidence implies that cholesterol and porphyrin are bound at two distinct binding sites, but given the small size of the protein, they may nevertheless influence each other. The cholesterol binding function is expected to have evolved as a result of pressures different from those exerted on the porphyrin binding ability, yet it should be noted that cholesterol synthesis and further conversion to steroid hormones depend on heme-containing enzymes, suggesting a possible interplay between the development of cholesterol binding and porphyrin binding capacities in TSPO.
AN ORPHAN RECEPTOR, A PROTOTYPE TRANSPORTER, A TRANSLOCATOR?
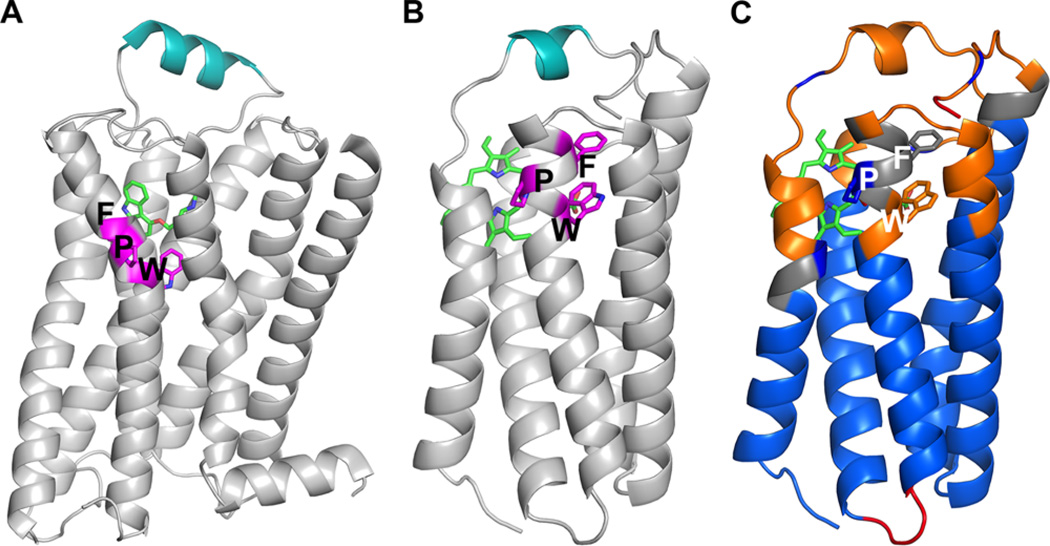
Originally identified as the peripheral benzodiazepine receptor, TSPO binding of various synthetic and drug ligands and their effects on various pathological conditions have been extensively investigated.8,82 However, the endogenous ligand(s) and physiological functions of TSPO are still not established. Both cholesterol and porphyrins are proposed to be endogenous ligands for TSPO, and given their apparent binding at different sites on the protein, different modes of action32,35,46 are expected. Recent studies of the structure and function of G-protein-coupled receptors (GPCRs) and various membrane transporters invite comparison with TSPO. Panels A and B of Figure 7 show that TSPO has a shape very similar to that of the inner helical bundle of the GPCR with the ligand binding site centrally located and facing out of the membrane toward the cytoplasm. LP1 of TSPO is also similar in helical conformation to an extracellular loop of the GPCR β2-adrenergic receptor, in which it acts as a lid of the ligand binding site, suggesting a similar regulatory role for LP1. Interestingly, the highly conserved WxPxF motif in the β1- and β2-adrenergic receptors,83–85 at the hinge of helix 6 where a large conformational change occurs during G-protein activation, is also found in TSPO as a highly conserved WtPvF motif on TM3.
Figure 7.
TSPO structure that resembles GPCR structure with a similar “toggle switch”. TSPO resembles GPCRs in the overall architecture with the extracellular loop (teal) on top of the central ligand binding site. A highly conserved WxPxF motif was also found within a transmembrane helix in RsTSPO (B), similar to that identified in the structure of β2-adrenergic receptor (A). ProFlex analysis of RsTSPO A139T indicates that the WxPxF motif in RsTSPO creates a flexible hinge, centered on the tryptophan, between two segments of TM3 (C). WxPxF motifs are colored magenta in panels A and B, while panel C is colored by main chain flexibility based on ProFlex analysis.87,88 ProFlex analysis was performed on a ligand-free version of the A139T crystal structure (chain B of PDB entry 4UC1). It allows identification of regions of the TSPO structure with different degrees of stability: blue regions are highly constrained and mutually rigid, due to the presence of a dense network of hydrophobic interactions and hydrogen bonds; gray regions have borderline stability due to a weak network of hydrophobic and hydrogen bond interactions; orange regions are more flexible; and red regions are highly flexible, with few stabilizing noncovalent interactions. Note that the relative flexibility of the helix 1–2 and 3–4 loops and the free C-terminal region of RsTSPO A139T (loops and short helix at the top of the figure) predicted by ProFlex are very similar to the flexibility of these regions indicated by crystallographic temperature factors in Figure 5A.
To examine the possibility of conformational change in TSPO, we applied the ProFlex analysis to crystal structures of RsTSPO. ProFlex is software designed to identify rigid versus less stable regions in protein structures86 and therefore can identify likely regions for conformational change. This method of analysis of a crystal structure is able to correctly identify flexible main chain regions of soluble as well as membrane proteins with functional significance.87,88 ProFlex analysis of the structure of the A139T mutant of RsTSPO (PDB entry 4UC1) indicates that the analogous WtPvF region in RsTSPO (side chains shown as sticks in Figure 7C), like that in GPCRs (Figure 7A), also forms a helical hinge that can be independently flexible. In RsTSPO, this hinge is adjacent to the proposed binding site for a porphyrin (green), suggesting that binding of ligand to TSPO could influence a conformational change in the helices, promoting a signaling event. However, given that the bottom (intermembrane) half of TSPO appears to be quite rigid in current structures (based on conserved conformation and low temperature factors), the signaling mechanism is likely to be very different than in GPCRs. Considering the location of the WxPxF motif at the dimer interface of RsTSPO, one possibility is that ligand binding would alter the oligomeric state of TSPO, with resultant modification of interactions with itself or other partners.
On the other hand, TSPO can also be considered as half of a prototype transporter, in particular those with 10 transmembrane helices, for example, the LeuT family neurotransmitter sodium symporters (NSS). Interestingly, a “5+5 internal repeat” often exists89 in these larger transporters with an inverted repeat topology. The equivalent in TSPO would be an antiparallel dimer, as seen in EmrE.90,91 Inconsistent with this arrangement in TSPO, both the dimers seen in the crystal structures of RsTSPO and BcTSPO are parallel dimers.
As distinct from a transporter, a translocator may be considered a more general term for the activity of moving a substance from A to B, as opposed to specifically across a membrane. As discussed above, a translocator role for TSPO has been proposed,55,92,93 involving the sequestration of cholesterol (or porphyrin) and transfer to a protein partner. This appears to be consistent with current evidence and with the structural characteristics of the dimer of RsTSPO, in which the predicted cholesterol binding site is found on the membrane-exposed surface where a protein partner could associate (Figure 5).
TSPO INDUCTION UNDER STRESS CONDITIONS ACROSS EVOLUTION
A diverse range of environmental stress conditions in different species and tissues affect TSPO expression levels, including salt and osmotic stress in cyanobacteria24 and Arabidopsis.22,26 Oxidative stress appears to be a common theme, and porphyrins have often been implicated as having a role in regulating it. Porphyrins bind to TSPO with micromolar affinity in all species so far characterized and are proposed to be endogenous ligands for TSPO and to play an important role in multiple processes in which TSPO is involved.5,6,46,57,73,94 In R. sphaeroides, TSPO is proposed to promote export of porphyrintype molecules from the cell to regulate photosynthetic gene expression.19 Interestingly, TSPO is reported to play a similar role in regulating the accumulation of porphyrin in mammalian cell lines95 when challenged with protoporphyrin IX, although recent studies in the TSPO knockout mouse do not confirm this role.61 In plants,37,57,96 a role for AtTSPO in scavenging of heme and porphyrins is reported, involving degradation of TSPO via an autophagy mechanism. Recent data in mouse also indicate that TSPO is involved in regulating mitochondrial ROS levels and mitophagy, dependent on the ratio of TSPO to VDAC1.73
TSPO may also respond to, or augment, oxidative stress by directly facilitating the breakdown of porphyrin. This idea that TSPO is a porphyrin-degrading enzyme was first suggested on the basis of the observation of the color change of porphyrin molecules when mixed with purified TSPO from the Chlorobium tepidum, an anaerobic phototrophic green sulfur bacteria,36 and further followed up with the TSPO isolated from Gram-positive bacterium B. cereus and other sources.33 However, the results show that this activity is relatively slow as well as light-dependent, suggesting a limited applicability to most tissues in eukaryotic organisms. Nevertheless, it is possible that binding of porphyrin to TSPO leads to distortion of the ring structure, as suggested by crystals of RsTSPO32 and observed in several heme oxygenases,97,98 which in turn could promote oxidation even in the absence of light. Considering the diverse physiology across kingdoms and the highly diverse forms of tetrapyrroles and other ligands, a variety of roles of TSPO in stress sensing or response in different organisms and tissues may have evolved.
CONCLUSION AND OUTLOOK
The tryptophan-rich sensory protein or translocator protein, TSPO, is a member of a highly conserved and ancient protein family that exhibits apparently diverse functions across species in all kingdoms. Despite the fact that the mammalian ortholog was discovered more than 30 years ago as a secondary binding site for benzodiazepine drugs, and a bacterial version has also been studied extensively, the precise functions of TSPO are still not understood. New structures of TSPO proteins from three different species provide some new clues regarding structure–function relationships. Studies of TSPO proteins from bacteria, plants, and mammals suggest that a common theme of stress sensing and response may underlie the apparent diversity of function and involve ligand-induced conformational change and interaction with protein partners. However, many challenges to developing a fuller understanding of TSPO function remain. One is the identification of the physiologically relevant protein partners of TSPO. Another is more rigorous and comprehensive in vitro characterization of function, ligand binding, and conformational effects in prokaryotic and eukaryotic proteins. The growing appreciation for the role(s) TSPO plays in environmental stress and in human disease, as well as a new phase of structure-aided mutagenic studies, should augment efforts to understand the functions of this intriguing protein and its drug target potential.
Acknowledgments
Funding
This work was supported by Michigan State University Foundation Strategic Partnership Grant “Mitochondrial Science & Medicine” (S.F.-M.) and National Institutes of Health Grant GM26916 (S.F.-M.).
We thank Dr. Carrie Hiser for careful critical reading of the manuscript.
ABBREVIATIONS
- TSPO
translocator protein 18 kDa
- PBR
peripheral-type benzodiazepine receptor
- RsTSPO
TSPO from R. sphaeroides
- mTSPO
TSPO from mouse
- BcTSPO
TSPO from B. cereus
- AtTSPO
TSPO from A. thaliana
- MPTP
mitochondrial permeability transition pore
- VDAC
voltage-dependent anion channel
- CRAC
cholesterol recognition/interaction amino acid consensus
- DM
decyl maltoside
- SDS
sodium dodecyl sulfate
- PpIX
protoporphyrin IX
- DPC
n-dodecylphosphocholine
- WT
wild-type
- PDB
Protein Data Bank
- NMR
nuclear magnetic resonance
Footnotes
The authors declare no competing financial interest.
REFERENCES
- 1.Braestrup C, Squires RF. Specific benzodiazepine receptors in rat brain characterized by high-affinity (3H)diazepam binding. Proc. Natl. Acad. Sci. U. S. A. 1977;74:3805–3809. doi: 10.1073/pnas.74.9.3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Verma A, Snyder SH. Peripheral type benzodiazepine receptors. Annu. Rev. Pharmacol. Toxicol. 1989;29:307–322. doi: 10.1146/annurev.pa.29.040189.001515. [DOI] [PubMed] [Google Scholar]
- 3.Mukhin AG, Papadopoulos V, Costa E, Krueger KE. Mitochondrial benzodiazepine receptors regulate steroid biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 1989;86:9813–9816. doi: 10.1073/pnas.86.24.9813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krueger KE, Papadopoulos V. Mitochondrial benzodiazepine receptors and the regulation of steroid biosynthesis. Annu. Rev. Pharmacol. Toxicol. 1992;32:211–237. doi: 10.1146/annurev.pa.32.040192.001235. [DOI] [PubMed] [Google Scholar]
- 5.Wendler G, Lindemann P, Lacapère JJ, Papadopoulos V. Protoporphyrin IX binding and transport by recombinant mouse PBR. Biochem. Biophys. Res. Commun. 2003;311:847–852. doi: 10.1016/j.bbrc.2003.10.070. [DOI] [PubMed] [Google Scholar]
- 6.Verma A, Nye JS, Snyder SH. Porphyrins are endogenous ligands for the mitochondrial (peripheral-type) benzodiazepine receptor. Proc. Natl. Acad. Sci. U. S. A. 1987;84:2256–2260. doi: 10.1073/pnas.84.8.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maeda J, Higuchi M, Inaji M, Ji B, Haneda E, Okauchi T, Zhang MR, Suzuki K, Suhara T. Phase-dependent roles of reactive microglia and astrocytes in nervous system injury as delineated by imaging of peripheral benzodiazepine receptor. Brain Res. 2007;1157:100–111. doi: 10.1016/j.brainres.2007.04.054. [DOI] [PubMed] [Google Scholar]
- 8.Rupprecht R, Papadopoulos V, Rammes G, Baghai TC, Fan J, Akula N, Groyer G, Adams D, Schumacher M. Translocator protein (18 kDa) (TSPO) as a therapeutic target for neurological and psychiatric disorders. Nat. Rev. Drug Discovery. 2010;9:971–988. doi: 10.1038/nrd3295. [DOI] [PubMed] [Google Scholar]
- 9.Katz Y, Ben-Baruch G, Kloog Y, Menczer J, Gavish M. Increased density of peripheral benzodiazepine-binding sites in ovarian carcinomas as compared with benign ovarian tumours and normal ovaries. Clin. Sci. 1990;78:155–158. doi: 10.1042/cs0780155. [DOI] [PubMed] [Google Scholar]
- 10.Hardwick M, Fertikh D, Culty M, Li H, Vidic B, Papadopoulos V. Peripheral-type benzodiazepine receptor (PBR) in human breast cancer: correlation of breast cancer cell aggressive phenotype with PBR expression, nuclear localization, and PBR-mediated cell proliferation and nuclear transport of cholesterol. Cancer Res. 1999;59:831–842. [PubMed] [Google Scholar]
- 11.Wu X, Gallo KA. The 18-kDa translocator protein (TSPO) disrupts mammary epithelial morphogenesis and promotes breast cancer cell migration. PLoS One. 2013;8:e71258. doi: 10.1371/journal.pone.0071258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yasuno F, Ota M, Kosaka J, Ito H, Higuchi M, Doronbekov TK, Nozaki S, Fujimura Y, Koeda M, Asada T, Suhara T. Increased binding of peripheral benzodiazepine receptor in Alzheimer’s disease measured by positron emission tomography with [11C]DAA1106. Biol. Psychiatry. 2008;64:835–841. doi: 10.1016/j.biopsych.2008.04.021. [DOI] [PubMed] [Google Scholar]
- 13.Cosenza-Nashat M, Zhao ML, Suh HS, Morgan J, Natividad R, Morgello S, Lee SC. Expression of the translocator protein of 18 kDa by microglia, macrophages and astrocytes based on immunohistochemical localization in abnormal human brain. Neuropathol. Appl. Neurobiol. 2009;35:306–328. doi: 10.1111/j.1365-2990.2008.01006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ji B, Maeda J, Sawada M, Ono M, Okauchi T, Inaji M, Zhang MR, Suzuki K, Ando K, Staufenbiel M, Trojanowski JQ, Lee VM, Higuchi M, Suhara T. Imaging of peripheral benzodiazepine receptor expression as biomarkers of detrimental versus beneficial glial responses in mouse models of Alzheimer’s and other CNS pathologies. J. Neurosci. 2008;28:12255–12267. doi: 10.1523/JNEUROSCI.2312-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Papadopoulos V, Lecanu L. Translocator protein (18 kDa) TSPO: an emerging therapeutic target in neurotrauma. Exp. Neurol. 2009;219:53–57. doi: 10.1016/j.expneurol.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen MK, Guilarte TR. Translocator protein 18 kDa (TSPO): molecular sensor of brain injury and repair. Pharmacol. Ther. 2008;118:1–17. doi: 10.1016/j.pharmthera.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Armstrong GA, Alberti M, Leach F, Hearst JE. Nucleotide sequence, organization, and nature of the protein products of the carotenoid biosynthesis gene cluster of Rhodobacter capsulatus. Mol. Gen. Genet. 1989;216:254–268. doi: 10.1007/BF00334364. [DOI] [PubMed] [Google Scholar]
- 18.Bui ET, Bradley PJ, Johnson PJ. A common evolutionary origin for mitochondria and hydrogenosomes. Proc. Natl. Acad. Sci. U. S. A. 1996;93:9651–9656. doi: 10.1073/pnas.93.18.9651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeliseev AA, Kaplan S. A novel mechanism for the regulation of photosynthesis gene expression by the TspO outer membrane protein of Rhodobacter sphaeroides 2.4.1. J. Biol. Chem. 1999;274:21234–21243. doi: 10.1074/jbc.274.30.21234. [DOI] [PubMed] [Google Scholar]
- 20.Yeliseev AA, Kaplan S. A sensory transducer homologous to the mammalian peripheral-type benzodiazepine receptor regulates photosynthetic membrane complex formation in Rhodobacter sphaeroides 2.4.1. J. Biol. Chem. 1995;270:21167–21175. doi: 10.1074/jbc.270.36.21167. [DOI] [PubMed] [Google Scholar]
- 21.Yeliseev AA, Krueger KE, Kaplan S. A mammalian mitochondrial drug receptor functions as a bacterial ″oxygen″ sensor. Proc. Natl. Acad. Sci. U. S. A. 1997;94:5101–5106. doi: 10.1073/pnas.94.10.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guillaumot D, Guillon S, Déplanque T, Vanhee C, Gumy C, Masquelier D, Morsomme P, Batoko H. The Arabidopsis TSPO-related protein is a stress and abscisic acid-regulated, endoplasmic reticulum-Golgi-localized membrane protein. Plant J. 2009;60:242–256. doi: 10.1111/j.1365-313X.2009.03950.x. [DOI] [PubMed] [Google Scholar]
- 23.Frank W, Baar KM, Qudeimat E, Woriedh M, Alawady A, Ratnadewi D, Gremillon L, Grimm B, Reski R. A mitochondrial protein homologous to the mammalian peripheral-type benzodiazepine receptor is essential for stress adaptation in plants. Plant J. 2007;51:1004–1018. doi: 10.1111/j.1365-313X.2007.03198.x. [DOI] [PubMed] [Google Scholar]
- 24.Busch AW, Montgomery BL. The Tryptophan-Rich Sensory Protein (TSPO) is Involved in Stress-Related and Light-Dependent Processes in the Cyanobacterium Fremyella diplosiphon. Front. Microbiol. 2015;6:1393. doi: 10.3389/fmicb.2015.01393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marin K, Kanesaki Y, Los DA, Murata N, Suzuki I, Hagemann M. Gene expression profiling reflects physiological processes in salt acclimation of Synechocystis sp. strain PCC 6803. Plant Physiol. 2004;136:3290–3300. doi: 10.1104/pp.104.045047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balsemão-Pires E, Jaillais Y, Olson BJ, Andrade LR, Umen JG, Chory J, Sachetto-Martins G. The Arabidopsis translocator protein (AtTSPO) is regulated at multiple levels in response to salt stress and perturbations in tetrapyrrole metabolism. BMC Plant Biol. 2011;11:108. doi: 10.1186/1471-2229-11-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hachez C, Veljanovski V, Reinhardt H, Guillaumot D, Vanhee C, Chaumont F, Batoko H. The Arabidopsis abiotic stress-induced TSPO-related protein reduces cell-surface expression of the aquaporin PIP2;7 through protein-protein interactions and autophagic degradation. Plant Cell. 2014;26:4974–4990. doi: 10.1105/tpc.114.134080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Papadopoulos V, Baraldi M, Guilarte TR, Knudsen TB, Lacapère JJ, Lindemann P, Norenberg MD, Nutt D, Weizman A, Zhang MR, Gavish M. Translocator protein (18 kDa): new nomenclature for the peripheral-type benzodiazepine receptor based on its structure and molecular function. Trends Pharmacol. Sci. 2006;27:402–409. doi: 10.1016/j.tips.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 29.Morohaku K, Pelton SH, Daugherty DJ, Butler WR, Deng W, Selvaraj V. Translocator protein/peripheral benzodiazepine receptor is not required for steroid hormone biosynthesis. Endocrinology. 2014;155:89–97. doi: 10.1210/en.2013-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Banati RB, Middleton RJ, Chan R, Hatty CR, Wai-Ying Kam W, Quin C, Graeber MB, Parmar A, Zahra D, Callaghan P, Fok S, Howell NR, Gregoire M, Szabo A, Pham T, Davis E, Liu GJ. Positron emission tomography and functional characterization of a complete PBR/TSPO knockout. Nat. Commun. 2014;5:5452. doi: 10.1038/ncomms6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tu LN, Morohaku K, Manna PR, Pelton SH, Butler WR, Stocco DM, Selvaraj V. Peripheral benzodiazepine receptor/translocator protein global knock-out mice are viable with no effects on steroid hormone biosynthesis. J. Biol. Chem. 2014;289:27444–27454. doi: 10.1074/jbc.M114.578286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li F, Liu J, Zheng Y, Garavito RM, Ferguson-Miller S. Protein structure. Crystal structures of translocator protein (TSPO) and mutant mimic of a human polymorphism. Science. 2015;347:555–558. doi: 10.1126/science.1260590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo Y, Kalathur RC, Liu Q, Kloss B, Bruni R, Ginter C, Kloppmann E, Rost B, Hendrickson WA. Protein structure. Structure and activity of tryptophan-rich TSPO proteins. Science. 2015;347:551–555. doi: 10.1126/science.aaa1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jaremko L, Jaremko M, Giller K, Becker S, Zweckstetter M. Structure of the mitochondrial translocator protein in complex with a diagnostic ligand. Science. 2014;343:1363–1366. doi: 10.1126/science.1248725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li F, Liu J, Valls L, Hiser C, Ferguson-Miller S. Identification of a Key Cholesterol Binding Enhancement Motif in Translocator Protein 18 kDa. Biochemistry. 2015;54:1441–1443. doi: 10.1021/bi5015453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ginter C, Kiburu I, Boudker O. Chemical catalysis by the translocator protein (18 kDa) Biochemistry. 2013;52:3609–3611. doi: 10.1021/bi400364z. [DOI] [PubMed] [Google Scholar]
- 37.Batoko H, Jurkiewicz P, Veljanovski V. Translocator proteins, porphyrins and abiotic stress: new light? Trends Plant Sci. 2015;20:261–263. doi: 10.1016/j.tplants.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 38.Batoko H, Veljanovski V, Jurkiewicz P. Enigmatic Translocator protein (TSPO) and cellular stress regulation. Trends Biochem. Sci. 2015;40:497–503. doi: 10.1016/j.tibs.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 39.Papadopoulos V, Aghazadeh Y, Fan J, Campioli E, Zirkin B, Midzak A. Translocator protein-mediated pharmacology of cholesterol transport and steroidogenesis. Mol. Cell. Endocrinol. 2015;408:90–98. doi: 10.1016/j.mce.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Campanella M, Turkheimer FE. TSPO: functions and applications of a mitochondrial stress response pathway. Biochem. Soc. Trans. 2015;43:593–594. doi: 10.1042/BST20150068. [DOI] [PubMed] [Google Scholar]
- 41.Gatliff J, Campanella M. TSPO: kaleidoscopic 18-kDa amid biochemical pharmacology, control and targeting of mitochondria. Biochem. J. 2016;473:107–121. doi: 10.1042/BJ20150899. [DOI] [PubMed] [Google Scholar]
- 42.Selvaraj V, Stocco DM, Tu LN. Minireview: translocator protein (TSPO) and steroidogenesis: a reappraisal. Mol. Endocrinol. 2015;29:490–501. doi: 10.1210/me.2015-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guilarte T, Loth M, Guariglia S. TSPO finds NOX2 in microglia for redox homeostasis. Trends Pharmacol. Sci. 2016 doi: 10.1016/j.tips.2016.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jaremko M, Jaremko Ł, Giller K, Becker S, Zweckstetter M. Structural Integrity of the A147T Polymorph of Mammalian TSPO. ChemBioChem. 2015;16:1483–1489. doi: 10.1002/cbic.201500217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jaremko Ł, Jaremko M, Giller K, Becker S, Zweckstetter M. Conformational Flexibility in the Transmembrane Protein TSPO. Chem. - Eur. J. 2015;21:16555–16563. doi: 10.1002/chem.201502314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li F, Xia Y, Meiler J, Ferguson-Miller S. Characterization and Modeling of the Oligomeric State and Ligand Binding Behavior of Purified Translocator Protein 18 kDa from Rhodobacter sphaeroides. Biochemistry. 2013;52:5884–5899. doi: 10.1021/bi400431t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lacapère JJ, Delavoie F, Li H, Péranzi G, Maccario J, Papadopoulos V, Vidic B. Structural and functional study of reconstituted peripheral benzodiazepine receptor. Biochem. Biophys. Res. Commun. 2001;284:536–541. doi: 10.1006/bbrc.2001.4975. [DOI] [PubMed] [Google Scholar]
- 48.Gut P, Zweckstetter M, Banati RB. Lost in translocation: the functions of the 18-kD translocator protein. Trends Endocrinol. Metab. 2015;26:349–356. doi: 10.1016/j.tem.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Farges R, Joseph-Liauzun E, Shire D, Caput D, Le Fur G, Ferrara P. Site-directed mutagenesis of the peripheral benzodiazepine receptor: identification of amino acids implicated in the binding site of Ro5–4864. Mol. Pharmacol. 1994;46:1160–1167. [PubMed] [Google Scholar]
- 50.Farges R, Joseph-Liauzun E, Shire D, Caput D, Le Fur G, Loison G, Ferrara P. Molecular basis for the different binding properties of benzodiazepines to human and bovine peripheral-type benzodiazepine receptors. FEBS Lett. 1993;335:305–308. doi: 10.1016/0014-5793(93)80407-l. [DOI] [PubMed] [Google Scholar]
- 51.Costa B, Pini S, Gabelloni P, Da Pozzo E, Abelli M, Lari L, Preve M, Lucacchini A, Cassano GB, Martini C. The spontaneous Ala147Thr amino acid substitution within the translocator protein influences pregnenolone production in lymphomonocytes of healthy individuals. Endocrinology. 2009;150:5438–5445. doi: 10.1210/en.2009-0752. [DOI] [PubMed] [Google Scholar]
- 52.Yeliseev AA, Kaplan S. TspO of Rhodobacter sphaeroides. A structural and functional model for the mammalian peripheral benzodiazepine receptor. J. Biol. Chem. 2000;275:5657–5667. doi: 10.1074/jbc.275.8.5657. [DOI] [PubMed] [Google Scholar]
- 53.Delavoie F, Li H, Hardwick M, Robert JC, Giatzakis C, Péranzi G, Yao ZX, Maccario J, Lacapère JJ, Papadopoulos V. In vivo and in vitro peripheral-type benzodiazepine receptor polymerization: functional significance in drug ligand and cholesterol binding. Biochemistry. 2003;42:4506–4519. doi: 10.1021/bi0267487. [DOI] [PubMed] [Google Scholar]
- 54.Korkhov VM, Sachse C, Short JM, Tate CG. Three-dimensional structure of TspO by electron cryomicroscopy of helical crystals. Structure. 2010;18:677–687. doi: 10.1016/j.str.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rone MB, Midzak AS, Issop L, Rammouz G, Jagannathan S, Fan J, Ye X, Blonder J, Veenstra T, Papadopoulos V. Identification of a dynamic mitochondrial protein complex driving cholesterol import, trafficking, and metabolism to steroid hormones. Mol. Endocrinol. 2012;26:1868–1882. doi: 10.1210/me.2012-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Campanella M. Editorial: The physiology and pharmacology of the mitochondrial 18 kDa translocator protein (TSPO): an emerging molecular target for diagnosis and therapy. Curr. Mol. Med. 2012;12:355. [PubMed] [Google Scholar]
- 57.Vanhee C, Zapotoczny G, Masquelier D, Ghislain M, Batoko H. The Arabidopsis multistress regulator TSPO is a heme binding membrane protein and a potential scavenger of porphyrins via an autophagy-dependent degradation mechanism. Plant Cell. 2011;23:785–805. doi: 10.1105/tpc.110.081570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Papadopoulos V, Amri H, Boujrad N, Cascio C, Culty M, Garnier M, Hardwick M, Li H, Vidic B, Brown AS, Reversa JL, Bernassau JM, Drieu K. Peripheral benzodiazepine receptor in cholesterol transport and steroidogenesis. Steroids. 1997;62:21–28. doi: 10.1016/s0039-128x(96)00154-7. [DOI] [PubMed] [Google Scholar]
- 59.Krueger KE, Papadopoulos V. Peripheral-type benzodiazepine receptors mediate translocation of cholesterol from outer to inner mitochondrial membranes in adrenocortical cells. J. Biol. Chem. 1990;265:15015–15022. [PubMed] [Google Scholar]
- 60.Li H, Papadopoulos V. Peripheral-type benzodiazepine receptor function in cholesterol transport. Identification of a putative cholesterol recognition/interaction amino acid sequence and consensus pattern. Endocrinology. 1998;139:4991–4997. doi: 10.1210/endo.139.12.6390. [DOI] [PubMed] [Google Scholar]
- 61.Zhao AH, Tu LN, Mukai C, Sirivelu MP, Pillai VV, Morohaku K, Cohen R, Selvaraj V. Mitochondrial Translocator Protein (TSPO) Function Is Not Essential for Heme Biosynthesis. J. Biol. Chem. 2016;291:1591–1603. doi: 10.1074/jbc.M115.686360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fan J, Campioli E, Midzak A, Culty M, Papadopoulos V. Conditional steroidogenic cell-targeted deletion of TSPO unveils a crucial role in viability and hormone-dependent steroid formation. Proc. Natl. Acad. Sci. U. S. A. 2015;112:7261–7266. doi: 10.1073/pnas.1502670112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu N, Van Voorst JR, Johnston JB, Kuhn LA. CholMine: Determinants and Prediction of Cholesterol and Cholate Binding Across Nonhomologous Protein Structures. J. Chem. Inf. Model. 2015;55:747–759. doi: 10.1021/ci5006542. [DOI] [PubMed] [Google Scholar]
- 64.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Qin L, Hiser C, Mulichak A, Garavito RM, Ferguson-Miller S. Identification of conserved lipid/detergent-binding sites in a high-resolution structure of the membrane protein cytochrome c oxidase. Proc. Natl. Acad. Sci. U. S. A. 2006;103:16117–16122. doi: 10.1073/pnas.0606149103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Qin L, Mills DA, Buhrow L, Hiser C, Ferguson-Miller S. A conserved steroid binding site in cytochrome c oxidase. Biochemistry. 2008;47:9931–9933. doi: 10.1021/bi8013483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hiser C, Buhrow L, Liu J, Kuhn L, Ferguson-Miller S. A conserved amphipathic ligand binding region influences k-path-dependent activity of cytochrome c oxidase. Biochemistry. 2013;52:1385–1396. doi: 10.1021/bi3014505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Johnson KM, Chen X, Boitano A, Swenson L, Opipari AW, Glick GD. Identification and validation of the mitochondrial F1F0-ATPase as the molecular target of the immunomodulatory benzodiazepine Bz-423. Chem. Biol. 2005;12:485–496. doi: 10.1016/j.chembiol.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 69.Giorgio V, von Stockum S, Antoniel M, Fabbro A, Fogolari F, Forte M, Glick GD, Petronilli V, Zoratti M, Szabó I, Lippe G, Bernardi P. Dimers of mitochondrial ATP synthase form the permeability transition pore. Proc. Natl. Acad. Sci. U. S. A. 2013;110:5887–5892. doi: 10.1073/pnas.1217823110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xiao J, Liang D, Zhang H, Liu Y, Li F, Chen YH. 4′-Chlorodiazepam, a translocator protein (18 kDa) antagonist, improves cardiac functional recovery during postischemia reperfusion in rats. Exp. Biol. Med. (London, U. K.) 2010;235:478–486. doi: 10.1258/ebm.2009.009291. [DOI] [PubMed] [Google Scholar]
- 71.Schaller S, Paradis S, Ngoh GA, Assaly R, Buisson B, Drouot C, Ostuni MA, Lacapere JJ, Bassissi F, Bordet T, Berdeaux A, Jones SP, Morin D, Pruss RM. TRO40303, a new cardioprotective compound, inhibits mitochondrial permeability transition. J. Pharmacol. Exp. Ther. 2010;333:696–706. doi: 10.1124/jpet.110.167486. [DOI] [PubMed] [Google Scholar]
- 72.Seneviratne MS, Faccenda D, De Biase V, Campanella M. PK11195 inhibits mitophagy targeting the F1Fo-ATPsynthase in Bcl-2 knock-down cells. Curr. Mol. Med. 2012;12:476–482. doi: 10.2174/156652412800163406. [DOI] [PubMed] [Google Scholar]
- 73.Gatliff J, East D, Crosby J, Abeti R, Harvey R, Craigen W, Parker P, Campanella M. TSPO interacts with VDAC1 and triggers a ROS-mediated inhibition of mitochondrial quality control. Autophagy. 2014;10:2279–2296. doi: 10.4161/15548627.2014.991665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tu LN, Zhao AH, Hussein M, Stocco DM, Selvaraj V. Translocator Protein (TSPO) Affects Mitochondrial Fatty Acid Oxidation in Steroidogenic Cells. Endocrinology. 2016;157:1110–1121. doi: 10.1210/en.2015-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Papadopoulos V. On the role of the translocator protein (18-kDa) TSPO in steroid hormone biosynthesis. Endocrinology. 2014;155:15–20. doi: 10.1210/en.2013-2033. [DOI] [PubMed] [Google Scholar]
- 76.Fan J, Lindemann P, Feuilloley MG, Papadopoulos V. Structural and functional evolution of the translocator protein (18 kDa) Curr. Mol. Med. 2012;12:369–386. doi: 10.2174/1566524011207040369. [DOI] [PubMed] [Google Scholar]
- 77.Rone MB, Fan J, Papadopoulos V. Cholesterol transport in steroid biosynthesis: role of protein-protein interactions and implications in disease states. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids. 2009;1791:646–658. doi: 10.1016/j.bbalip.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Colombini M. Purification of VDAC (voltage-dependent anion-selective channel) from rat liver mitochondria. J. Membr. Biol. 1983;74:115–121. doi: 10.1007/BF01870500. [DOI] [PubMed] [Google Scholar]
- 79.Ujwal R, Cascio D, Colletier JP, Faham S, Zhang J, Toro L, Ping P, Abramson J. The crystal structure of mouse VDAC1 at 2.3 A resolution reveals mechanistic insights into metabolite gating. Proc. Natl. Acad. Sci. U. S. A. 2008;105:17742–17747. doi: 10.1073/pnas.0809634105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hiller S, Garces RG, Malia TJ, Orekhov VY, Colombini M, Wagner G. Solution structure of the integral human membrane protein VDAC-1 in detergent micelles. Science. 2008;321:1206–1210. doi: 10.1126/science.1161302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Weiser BP, Salari R, Eckenhoff RG, Brannigan G. Computational investigation of cholesterol binding sites on mitochondrial VDAC. J. Phys. Chem. B. 2014;118:9852–9860. doi: 10.1021/jp504516a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Scarf AM, Auman KM, Kassiou M. Is there any correlation between binding and functional effects at the translocator protein (TSPO) (18 kDa)? Curr. Mol. Med. 2012;12:387–397. doi: 10.2174/1566524011207040387. [DOI] [PubMed] [Google Scholar]
- 83.Ballesteros JA, Shi L, Javitch JA. Structural mimicry in G protein-coupled receptors: implications of the high-resolution structure of rhodopsin for structure-function analysis of rhodopsin-like receptors. Mol. Pharmacol. 2001;60:1–19. [PubMed] [Google Scholar]
- 84.Kobilka BK, Deupi X. Conformational complexity of G-protein-coupled receptors. Trends Pharmacol. Sci. 2007;28:397–406. doi: 10.1016/j.tips.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 85.Deupi X, Kobilka B. Activation of G protein-coupled receptors. Adv. Protein Chem. 2007;74:137–166. doi: 10.1016/S0065-3233(07)74004-4. [DOI] [PubMed] [Google Scholar]
- 86.Jacobs DJ, Rader AJ, Kuhn LA, Thorpe MF. Protein flexibility predictions using graph theory. Proteins: Struct., Funct., Genet. 2001;44:150–165. doi: 10.1002/prot.1081. [DOI] [PubMed] [Google Scholar]
- 87.Gohlke H, Kuhn LA, Case DA. Change in protein flexibility upon complex formation: analysis of Ras-Raf using molecular dynamics and a molecular framework approach. Proteins: Struct., Funct., Genet. 2004;56:322–337. doi: 10.1002/prot.20116. [DOI] [PubMed] [Google Scholar]
- 88.Buhrow L, Ferguson-Miller S, Kuhn LA. From static structure to living protein: computational analysis of cytochrome c oxidase main-chain flexibility. Biophys. J. 2012;102:2158–2166. doi: 10.1016/j.bpj.2012.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Khafizov K, Staritzbichler R, Stamm M, Forrest LR. A study of the evolution of inverted-topology repeats from LeuT-fold transporters using AlignMe. Biochemistry. 2010;49:10702–10713. doi: 10.1021/bi101256x. [DOI] [PubMed] [Google Scholar]
- 90.Nasie I, Steiner-Mordoch S, Gold A, Schuldiner S. Topologically random insertion of EmrE supports a pathway for evolution of inverted repeats in ion-coupled transporters. J. Biol. Chem. 2010;285:15234–15244. doi: 10.1074/jbc.M110.108746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lloris-Garcerá P, Bianchi F, Slusky JS, Seppälä S, Daley DO, von Heijne G. Antiparallel dimers of the small multidrug resistance protein EmrE are more stable than parallel dimers. J. Biol. Chem. 2012;287:26052–26059. doi: 10.1074/jbc.M112.357590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fan J, Papadopoulos V. Evolutionary origin of the mitochondrial cholesterol transport machinery reveals a universal mechanism of steroid hormone biosynthesis in animals. PLoS One. 2013;8:e76701. doi: 10.1371/journal.pone.0076701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lacapère JJ, Papadopoulos V. Peripheral-type benzodiazepine receptor: structure and function of a cholesterol-binding protein in steroid and bile acid biosynthesis. Steroids. 2003;68:569–585. doi: 10.1016/s0039-128x(03)00101-6. [DOI] [PubMed] [Google Scholar]
- 94.Rampon C, Bouzaffour M, Ostuni MA, Dufourcq P, Girard C, Freyssinet JM, Lacapere JJ, Schweizer-Groyer G, Vriz S. Translocator protein (18 kDa) is involved in primitive erythropoiesis in zebrafish. FASEB J. 2009;23:4181–4192. doi: 10.1096/fj.09-129262. [DOI] [PubMed] [Google Scholar]
- 95.Zeno S, Veenman L, Katz Y, Bode J, Gavish M, Zaaroor M. The 18 kDa mitochondrial translocator protein (TSPO) prevents accumulation of protoporphyrin IX. Involvement of reactive oxygen species (ROS) Curr. Mol. Med. 2012;12:494–501. doi: 10.2174/1566524011207040494. [DOI] [PubMed] [Google Scholar]
- 96.Vanhee C, Batoko H. Arabidopsis TSPO and porphyrins metabolism: a transient signaling connection? Plant Signaling Behav. 2011;6:1383–1385. doi: 10.4161/psb.6.9.16477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Takayama SJ, Ukpabi G, Murphy ME, Mauk AG. Electronic properties of the highly ruffled heme bound to the heme degrading enzyme IsdI. Proc. Natl. Acad. Sci. U. S. A. 2011;108:13071–13076. doi: 10.1073/pnas.1101459108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Unno M, Ardèvol A, Rovira C, Ikeda-Saito M. Structures of the substrate-free and product-bound forms of HmuO, a heme oxygenase from corynebacterium diphtheriae: x-ray crystallography and molecular dynamics investigation. J. Biol. Chem. 2013;288:34443–34458. doi: 10.1074/jbc.M113.486936. [DOI] [PMC free article] [PubMed] [Google Scholar]