Abstract
Pseudomonas aeruginosa is a ubiquitous and opportunistic bacterium that inhibits the growth of different microorganisms, including Gram-positive bacteria and fungi such as Candida spp. and Aspergillus fumigatus. In this study, we investigated the interaction between P. aeruginosa and Cryptococcus spp. We found that P. aeruginosa PA14 and, to a lesser extent, PAO1 significantly inhibited the growth of Cryptococcus spp. The inhibition of growth was observed on solid medium by the visualization of a zone of inhibition of yeast growth and in liquid culture by viable cell counting. Interestingly, such inhibition was only observed when P. aeruginosa and Cryptococcus were co-cultured. Minimal inhibition was observed when cell–cell contact was prevented using a separation membrane, suggesting that cell contact is required for inhibition. Using mutant strains of Pseudomonas quinoline signaling, we showed that P. aeruginosa inhibited the growth of Cryptococcus spp. by producing antifungal molecules pyocyanin, a redox-active phenazine, and 2-heptyl-3,4-dihydroxyquinoline (PQS), an extracellular quorum-sensing signal. Because both P. aeruginosa and Cryptococcus neoformans are commonly found in lung infections of immunocompromised patients, this study may have important implication for the interaction of these microbes in both an ecological and a clinical point of view.
Keywords: Cryptococcus spp.; Pseudomonas aeruginosa; Quorum sensing; 2-heptyl-3,4-dihydroxyquinoline; Pyocyanin
Introduction
Cryptococcus neoformans is an important, widely distributed, fungal human pathogen that causes a life-threatening meningoencephalitis in immunocompromised hosts. In recent years, it has been shown to cause serious pulmonary infection also in individuals with competent immune system [1, 2]. The cryptococcal infection develops after exposure to fungal basidiospores or encapsulated yeast found ubiquitously in the environment, such as pigeon guano and eucalyptus trees. The port of entry of Cryptococcus species is the lung. When spores or yeasts are inhaled, the infection can be restricted to the lung or can disseminate to other tissues. Following inhalation, alveolar macrophages are the first line of defense against Cryptococcus, contributing to the clearance or containment of the fungal pathogen in the granuloma [3]. This occurs normally in immunocompetent subjects. In immunocompromised subjects, the yeast cells can disseminate from the lung to the brain within host cells (intracellularly) or in the bloodstream (extracellularly), leading to the development of life-threatening disease [4–6]. Thus, the lung is an important organ for the containment of cryptococcal infection.
The respiratory tract is also a common organ infected by Pseudomonas aeruginosa (Pa), a ubiquitous Gram-negative bacterium commonly found in soil and water, following aspiration of the organism from the upper respiratory tract, especially in patients on mechanical ventilation. Alternatively, pneumonia may occur as a result of bacteremia spread to the lungs. This is observed commonly in patients following chemotherapy-induced neutropenia or in patients with AIDS [7, 8]. In Pa and in other bacteria, modulation and coordination of gene expression are influenced by population density via the production of small molecules that impact the production of virulence factors. This mechanism based on cell–cell communication is known as quorum sensing (QS).
QS molecules are capable of not only controlling gene expression in Pa but also affecting host immune response and the growth of other microorganisms [9]. Several groups have demonstrated the capacity of Pa to inhibit yeast growth in vitro, namely Candida spp., Aspergillus fumigatus and Saccharomyces cerevisiae [10, 11]. Interest in the inhibitory effect of several strains of Pa against C. neoformans in vitro started in 1954, when Fisher et al. reported that strains of Pa produced a substance that inhibits the growth of C. neoformans in vitro, but the inhibitor has not been successfully separated [12]. Twenty years later, Teoh-Chan et al. reported an initial attempt to isolate and characterize the inhibitory substances involved in 1974 [13]. However, the literature about the inhibitory effect of Pa on C. neoformans is equivocal [12, 13]. In this study, we revealed the mechanism used by Pa to inhibit the growth of Cryptococcus spp. through the production of QS molecules. We found that the Pa PQS and, more profoundly, pyocyanin do significantly inhibit cryptococcal growth. Interestingly, this inhibition was mainly detected when C. neoformans was co-cultured with Pa, suggesting that physical contact triggers the production of antifungal molecules in Pa.
Materials and Methods
Experimental Strains
Cryptococcus neoformans var. grubii serotype A strain H99, C. neoformans var. neoformans serotype D strain JEC21, C. gattii serotype B (strain MMRL 1336) and serotype C (strain MMRL 1343) [the latter two strains were a kind gift from Wiley Schell, Duke University Medical Center, Durham, NC, USA), and P. aeruginosa wild-type strains PAO1 and PA14 were used in this study. PAO1 and PA14 are the two commonly used laboratory strains of Pa. Numerous reports showed that the clinical isolate PA14 is significantly more virulent than PAO1 in a wide range of hosts [14, 15]. P. aeruginosa PAO1 mutant strains pqsAB, pqsB and pqsE and E. coli strain DH5a were used in this study. Cryptococcus species were grown at 30°C in YPD broth (2% peptone, 1% yeast extract, 2% dextrose, BD) with shaking at 250 rpm. PAO1 and PA14 were cultured in two different media LB and YPD. Pa showed similar profile of growth in both media. PAO1 mutant strains were grown in LB (Luria–Bertani) at 37°C with shaking at 250 rpm.
YNB Plate Assay
C. neoformans var. grubii serotype A, C. neoformans var. neoformans serotype D and C. gattii serotype B and serotype C were grown overnight, and the cells were washed twice with sterile water and resuspended in YNB broth (Yeast Nitrogen Broth, Sigma-Aldrich). 300 µl of 1 × 108 cells/ml were spread on YNB agar plates using a glass spreader. Then, sterile filter paper disks were placed on the plates. Both PAO1 or PA14 were grown overnight at 37°C in LB media, and 10-µl drops of an 8 × 109 CFU/ml Pa cultures were spotted on the filter disks, following by incubation at 30°C for up to 72 h. The same procedure was used for PAO1 mutant strains: pqsAB, pqsB and pqsE.
Survival Assay of C. neoformans H99 Co-cultured with Pa Strains
C. neoformans and Pa strains PAO1 or PA14 were grown at 30°C in YPD broth overnight. Different C. neoformans dilutions were performed (107, 106, 105, 104 and 103 cells/ml) and inoculated with 3 × 109 CFU/ml Pa strains onto 96-well plate. The plate was incubated at 30°C for 24 h. C. neoformans and Pa cells viability was determined by CFU counting on YNB and LB plates, respectively. YNB and LB plates were used to count C. neoformans and Pa cells in mixed culture conditions, respectively. On YNB plates, C. neoformans was growing faster than Pa, allowing us to count C. neoformans, whereas LB plates do not support the growth of C. neoformans and were chosen to count Pa.
C. neoformans H99-PAO1 or PA14 Interaction
Both PAO1 and PA14 were grown in 100 ml of YPD at 30°C for 24 h. C. neoformans was grown in 50 ml of YPD at 30°C for 24 h. Pa and C. neoformans cultures were washed twice with sterile water and harvested at 8,000×g for 20 min and at 3,000 rpm for 10 min, respectively. Pa cultures were split into two flasks: Pa was cultured alone in one flask, and Pa was co-cultured with C. neoformans in the other flask, for 48 h at 30°C. Pa alone and the Pa–C. neoformans co-culture were pelleted by centrifugation at 8,000×g for 20 min. The supernatants were filtered with vacuum-driven disposable filtration system with 0.22-µm pore size (Millipore), frozen and lyophilized using a bench top freeze dryer. 500 mg of dried co-culture supernatants (1.26 × 1015 CFU Pa strains and 1 × 109 CFU C. neoformans) and Pa supernatants (1 × 1015 CFU Pa strains) was resuspended in 1 ml of YPD broth. Cryptococcus neoformans was grown overnight and 300 µL of 1 × 108 cells/ml culture was spread on YPD agar plates using a glass spreader. Different amounts of dried co-culture supernatants (e.g., 12.5, 25, 50, 75 and 100 mg) and dried Pa supernatants (100 mg) resuspended in 200 µl of YPD were added into 8-mm wells made on the YPD plates. The plates were incubated at 30°C for up to 72 h. We used 100 mg of dried YPD broth resuspended in 200 µl of YPD as a control.
U-Tube Assay
A sterile membrane with 0.22-µm pore size was used to separate the two sides of the U-tube. In one side of the tube, 25 ml of YPD with 1.5 × 109 CFU/ml of either Pa strains was cultured, whereas in the other side, 25 ml of YPD with 1 × 105 cells/ml of C. neoformans was inoculated. Separately, Erlenmeyer flasks were used for controls, including co-cultures of C. neoformans with PAO1 or PA14, C. neoformans with E. coli and cultures of C. neoformans alone and PAO1 or PA14 alone, maintaining the same number of C. neoformans and Pa used in the U-tube. The U-tube and the flasks were incubated for 48 h at 30°C. C. neoformans and Pa cell viability was determined by CFU counting on YNB and LB plates, respectively.
Fungal Viability Assay in Physiological Condition
Overnight cultures of C. neoformans in YPD broth at 30°C and Pa strains in LB broth at 37°C were pelleted, washed with sterile water and resuspended in DMEM buffered with 50 mM HEPES (pH 7.2). 1.5 × 109 CFU/ml of PAO1 or PA14 and 2 × 104 cells/ml of C. neoformans were co-cultured at 37°C with 5% CO2 for 24 and 48 h. C. neoformans and Pa cells viability was determined by CFU counting.
Mutant Generation
The pqs mutant strains were created in strain PAO1 by gene replacement described previously [16]. Briefly, a DNA fragment, which contained a gentamicin resistance cassette flanked by the 5′ and 3′ ends of the target gene, was cloned into pEX18ApGW [16]. The resultant plasmid was conjugated from E. coli strain SM10 into the Pa wild-type strain PAO1 with the selection of gentamicin (30 µg/ml) on Pseudomonas isolation agar plate (BD, Becton, Dickinson and Company). Merodiploids formed via a single crossover event was resolved through 5% sucrose selection in the presence of gentamicin. The gentamicin resistance cassette was subsequently removed by Flp recombinase, resulting in an unmarked deletion mutant of the target gene. The presence of the deletion in the correct region was verified by PCR and sequencing.
Pyocyanin Assay
For pyocyanin quantification, cultures of PAO1 or pqs mutants were grown overnight in LB at 37°C with shaking (250 rpm) to reach OD600 of 2.0. Pyocyanin was extracted with chloroform from cell-free supernatants, acidified by 0.2 N HCl and assayed spectrophotometrically at 520 nm as previously described [17]. Concentrations, expressed in mg/l, were determined by multiplying the optical density at 520 nm by 17.072 [17].
Microtiter Assay
C. neoformans was grown overnight, washed and resuspended in YNB buffered with 25 mM HEPES (pH 7.2). 2 × 104 cells/ml of C. neoformans were incubated in a 96-well plate containing 4-hydroxy-2-heptylquinoline (HHQ) [Qingdao Vochem], 3,4-dihydroxy-2-heptylquinoline (PQS) [Sigma-Aldrich], 2,4-dihydroxyquinoline (DHQ) [Fluka] and pyocyanin [Cayman Chemical] at different concentrations (1.5, 3.5, 7, 14, 28, 56, 112 and 225 µg/ml). The compounds were dissolved in DMSO. The final concentration of DMSO was less than 1–1.5%. The plate was incubated for 48 h at 30°C. The growth of yeast cells was assessed by monitoring optical density at 495 nm (OD495), and the surviving viable yeast cells were determined by CFU counting.
Statistics
Data from each experimental group were subjected to an analysis of normality and variance. Statistical significance between the means of different experimental data sets composed of normally distributed values was analyzed using two-tailed Student’s t test. For all statistical tests, standard deviation with P-values less than 0.05 was considered significant. All experiments were done twice, with similar results each time, or three times when statistics was applied.
Results
Pa Inhibited Growth of Cryptococcus spp
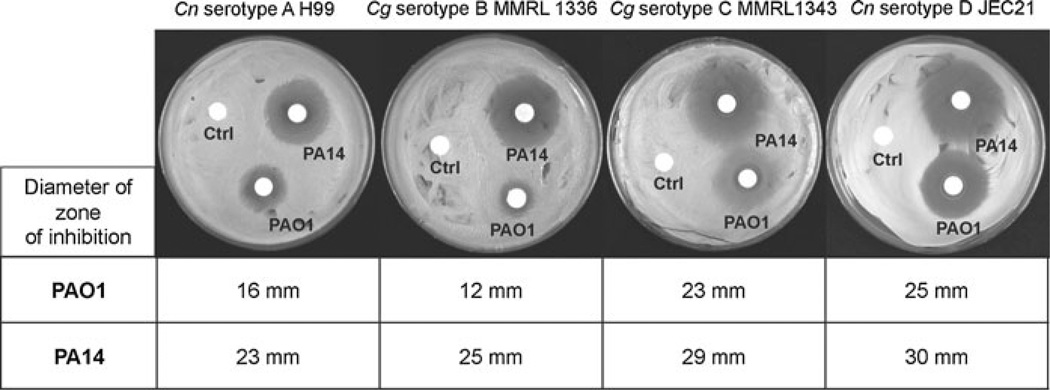
Previous reports reveal that Pa inhibits the growth of different microbes [10–13]. However, the mechanism that underlines the inhibitory effects of Pa on Cryptococcus species has not been investigated. Our results showed that when PAO1 or PA14 was inoculated on paper disks on YNB plate, which contained a lawn of Cryptococcus species, a zone of inhibition developed around the disks within 24 h. The diameters of the zone of inhibition of PA14 against serotypes, A, B, C and D of Cryptococcus were 23, 25, 29 and 30 mm, respectively. In contrast, reduced diameters of the zone of inhibition of PAO1 against Cryptococcus species were observed (16, 12, 23 and 25 mm) (Fig. 1).
Fig. 1.
Pa inhibited the growth of Cryptococcus spp. PAO1 and PA14 were inoculated on paper disks, on YNB plates with C. neoformans var. grubii serotype A (Cn serotype A H99), C. gattii serotypes B (Cg serotype B MMRL 1336), C. gattii serotypes C (Cg serotype C MMRL1343) and C. neoformans var. neoformans serotype D (Cn serotype D JEC21). Control disks containing LB medium were placed on the plates (indicated with ctrl on the plates). The diameters of the zone of inhibition were determined after 72 h at 30°C
Pa Fungicidal Effect Against C. neoformans in Co-culture
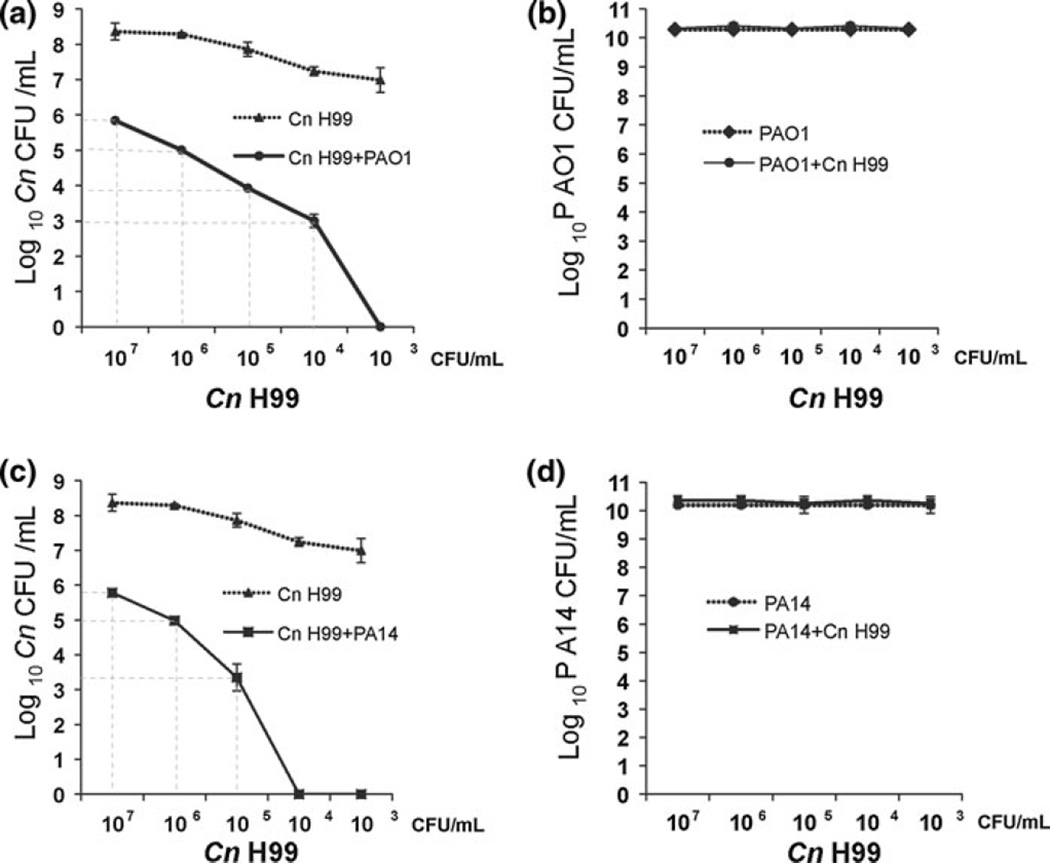
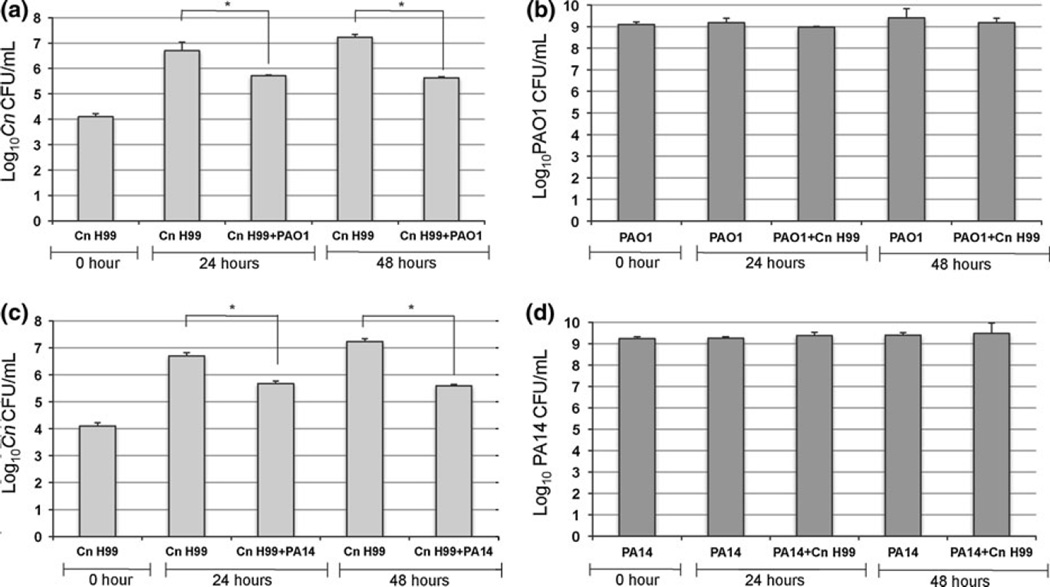
To determine whether the inhibitory effects of Pa on C. neoformans growth were fungistatic or fungicidal, we performed a viability assay of C. neoformans in co-cultures with PAO1 or PA14. Different C. neoformans dilutions were mixed with Pa strains (Fig. 2). After co-incubation of PAO1 and C. neoformans, we observed, in the first 4 dilutions, 1 log of decrease in C. neoformans cell number compared with the start culture. The last dilution showed no remaining viable cells of C. neoformans (Fig. 2a). In contrast, in the first 3 dilutions of co-cultures of PA14 and C. neoformans, we observed 1 log of decrease in C. neoformans cells number, but there was no remaining viable cells of C. neoformans in the last two dilutions (Fig. 2c). These results showed that the inhibitory effect of Pa on C. neoformans is fungicidal and the killing effect is dependent on the relative cell density. To evaluate whether Pa growth is affected in co-culture with C. neoformans, the number of Pa cells was counted. The results showed that C. neoformans does not affect Pa growth (Fig. 2b, d). Both PAO1 and PA14 grew in the condition tested, and therefore, the different inhibitory effect is not the result of different growth rate of PAO1 and PA14.
Fig. 2.
Pa has fungicidal effect against C. neoformans in co-culture. Different C. neoformans (Cn) dilutions were mixed with Pa strains and incubated in 96-well plates. The viability of Cn in co-culture with PAO1 (a) and PA14 (c) was determined by CFU counting on YNB plates. The viability of PAO1 (b) and PA14 (d) in co-culture with Cn was determined by CFU counting on LB plates. The inhibitory effect of Pa on Cn is fungicidal and is dependent on the relative cell density
Enhanced Inhibition of C. neoformans by Co-culture Supernatants
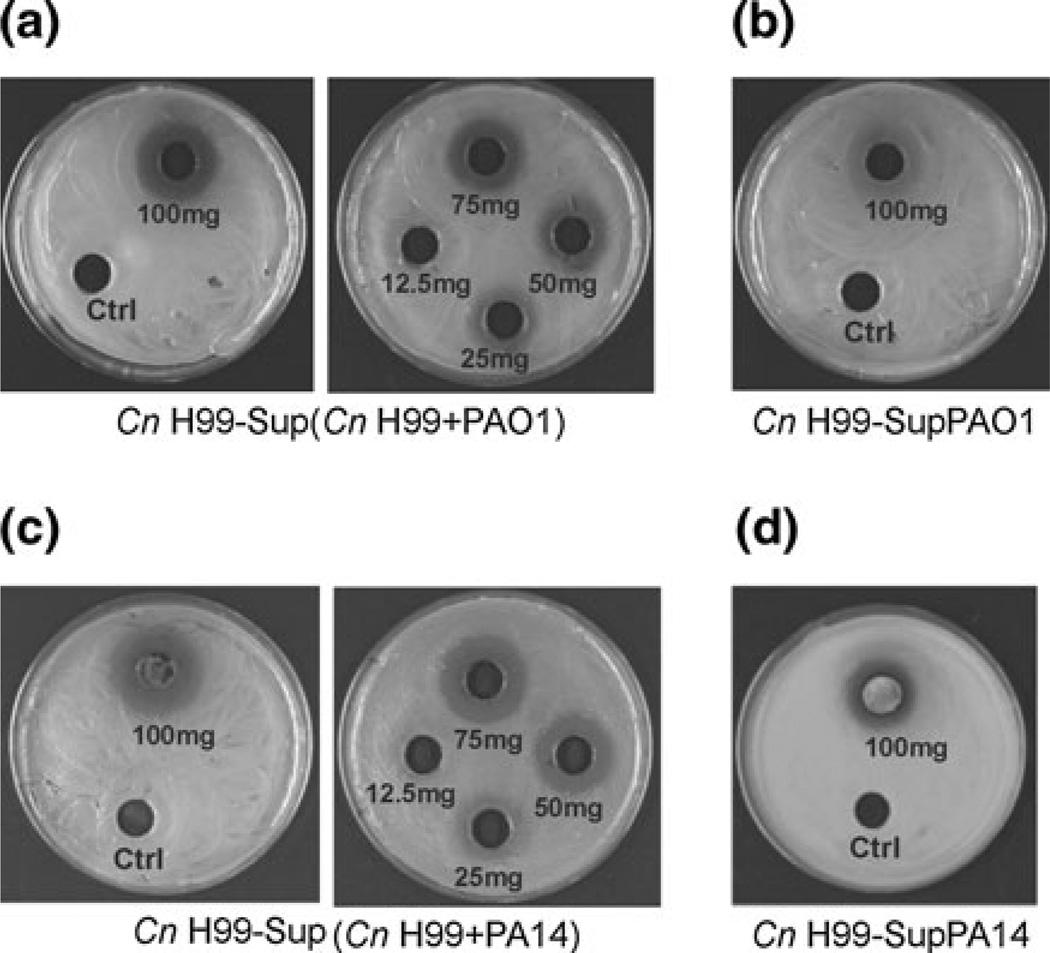
To understand the mechanism by which Pa inhibits fungal growth, we screened the effect of culture supernatants of PAO1 or PA14 and the effect of Pa–C. neoformans co-culture supernatants on C. neoformans growth. Supernatants were prepared as outlined in the Materials and Methods. We inoculated different amounts of dried co-cultured supernatants (e.g., 12.5, 25, 50, 75 and 100 mg) and dried Pa supernatants (e.g., 100 mg), on YPD plate, which contained a lawn of C. neoformans. As a control, we used dried YPD, to exclude any effects from the high concentration of salts. A diameter of zone of inhibition of 15, 16, 17 and 20 mm was observed with 25, 50, 75 and 100 mg of PAO1-C. neoformans supernatant, respectively (Fig. 3a), whereas we observed a diameter of zone of inhibition of 15, 18, 21 and 22 mm with PA14-C. neoformans supernatant (Fig. 3c). The dried PAO1 supernatant did not result in a clean zone of inhibition, whereas with dried PA14 supernatant, we estimated a zone of inhibition of 17 mm (Fig. 3b, d). These results suggest that the physical contact between Pa and C. neoformans triggers the production of antifungal molecules by Pa that inhibit C. neoformans growth. Pa supernatant alone was not able to inhibit C. neoformans growth as much as C. neoformans–Pa supernatant.
Fig. 3.
Enhanced inhibition of C. neoformans by Pa in co-culture supernatants. Dried co-culture supernatants [Sup(Cn H99 + PAO1), Sup(Cn H99 + PA14)] (a, c) and dried Pa supernatants (SupPAO1, SupPA14) (b, d), resuspended in YPD broth, were inoculated into 8-mm wells on YPD plates, which contained a lawn of C. neoformans (Cn H99). The diameters of the zone of inhibition were determined
The Cell–Cell Contact was Important for Pa to Inhibit C. neoformans
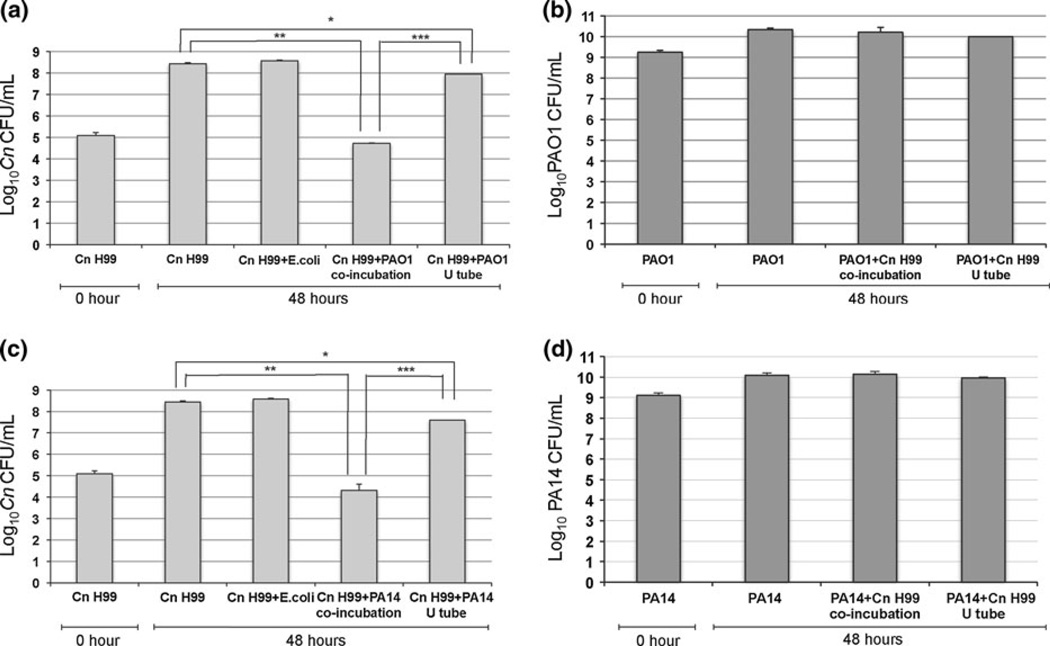
To investigate whether Pa–C. neoformans cells contact was important for inhibition, we performed an assay in YPD media using a U-tube. We cultured Pa in one side and C. neoformans in the other side of the tube; the two sides were separated with a sterile membrane, which prevented mixing of Pa and C. neoformans but allowed free exchange of nutrients and extracellular molecules. The C. neoformans growth exhibited half a log reduction when cultured in the U-tube with PAO1 (Fig. 4a), whereas 1 log reduction was observed when C. neoformans was cultured with PA14 (Fig. 4c). A 4 log inhibition was observed when C. neoformans was co-cultured with Pa strains in Erlenmeyer flasks (Fig. 4a, c). These results suggest that the cell contact is important to elicit a strong inhibition of C. neoformans by Pa. When E. coli was co-cultured with C. neoformans, no inhibitory effects were observed (Fig. 4a, c). Under these conditions, Pa growth was not affected by C. neoformans like observed before (Fig. 4b, d).
Fig. 4.
Cell–cell contact is important for Pa to inhibit C. neoformans. An assay in YPD media using a U-tube was performed. A sterile membrane was used to separate the two sides of the U-tube, in which Pa strains and C. neoformans (Cn) were cultured. Separately, Erlenmeyer flasks were used for controls, including co-cultures of Cn with PAO1 or PA14, Cn with E. coli and cultures of Cn alone and PAO1 or PA14 alone. The inhibitory effect of Pa on Cn, in the U-tube and in co-culture, was determined by CFU counting (a, c). When E. coli and Cn were co-cultured, no inhibitory effects of Cn were observed (a, c). The growth of PAO1 and PA14 was not affected by Cn (b, d). P values were calculated by Student’s t test, *P < 0.05; **P < 0.01; ***P < 0.001
Pa Inhibited C. neoformans in Physiological Condition
To determine whether Pa inhibits the growth of C. neoformans in physiological condition, we co-cultured PAO1 or PA14 with C. neoformans in DMEM (pH 7.2) at 37°C with 5% CO2 for 24 and 48 h. Both PAO1 and PA14 inhibited the growth of C. neoformans in DMEM; however, the degree of inhibition in DMEM (Fig. 5a, c) was significantly reduced compared to co-cultures in YPD (Fig. 4a, c). In DMEM, the growth of C. neoformans exhibited a 2 log reduction when co-cultured with PAO1 for 48 h (Fig. 5a), whereas a 4 log reduction was observed when C. neoformans was co-cultured with PAO1 in YPD (Fig. 4a). Similar results were observed with the PA14 strain (Figs. 4c, 5c). The reduced inhibition of C. neoformans growth by Pa could be the result of reduced replication of Pa in DMEM. Pa cell number counting showed that there was no significant increase in the number of Pa when co-cultured with C. neoformans in DMEM (Fig. 5b, d), suggesting that DMEM does not support the growth of Pa as well as YPD, in which Pa cell number increased by 1 log when co-cultured with C. neoformans (Fig. 4b, d). These results suggest that the inhibition of C. neoformans growth depends on the cell density of Pa and relative ratio of Pa and C. neoformans.
Fig. 5.
Pa inhibited C. neoformans in physiological condition. PAO1 and PA14 were co-cultured with C. neoformans (Cn) in DMEM for 24 and 48 h. Viable Cn and Pa were measured by CFU on YNB and LB plates, respectively. Cn inhibition was observed with both Pa strains (a, c). Pa cells number counting showed no significant increase in the number of cells in DMEM (b, d). P-values were calculated by Student’s t test, *P < 0.05
Effects of PQS Mutants of PAO1 on the Growth of C. neoformans
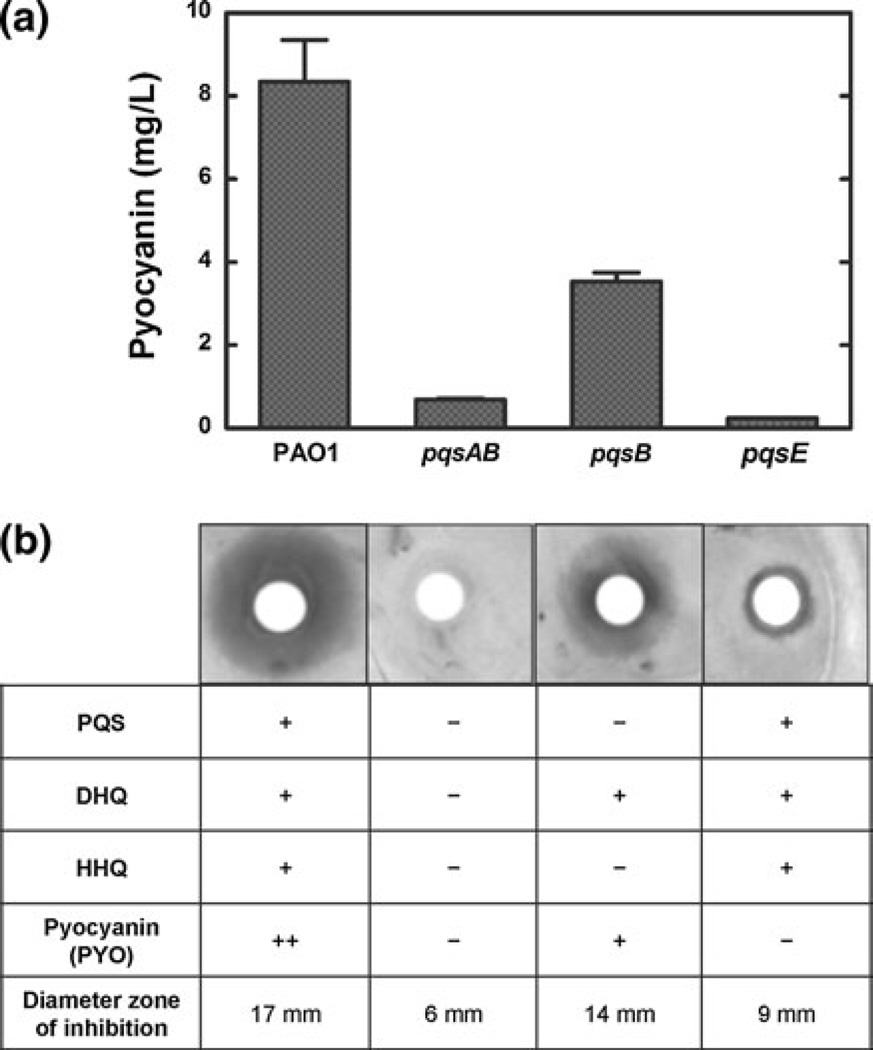
It has been shown that extracellular compounds secreted by Pa, including quorum-sensing molecules and phenazines (such as pyocyanin), affect the growth of different microbes [9, 11]. Pseudomonas quinolone signaling plays an essential role in activating the production of pyocyanin [18]. Proteins encoded by the pqsABCDE operon are required for both PQS synthesis and activation of pyocyanin production. Specifically, PqsA, PqsB, PqsC and PqsD are required for the formation of extracellular quinolones including 4-hydroxy-2-heptylquinoline (HHQ), 3,4-dihydroxy-2-heptylquinoline (PQS) and 2,4-dihydroxyquinoline (DHQ). PqsE is not required for the biosynthesis of quinolones but is essential for the activation of pyocyanin production. To determine whether the extracellular quinolones and pyocyanin are involved in the inhibition of C. neoformans, we tested the effects of pqs mutants of PAO1 on the growth of C. neoformans. Three mutant strains of PAO1 (pqsAB, pqsB and pqsE) were chosen to distinguish the effects of alkylquinolones (including HHQ and PQS), DHQ and pyocyanin. pqsAB mutant is defective in both PqsA and PqsB and does not produce any extracellular quinolones. pqsB mutant lacks alkylquinolones [19] but produces normal level of DHQ. pqsE mutant produces extracellular quinolones at the similar levels as the wild-type PAO1. In terms of pyocyanin production, pqsAB and pqsE mutants are defective, whereas pqsB secreted pyocyanin at a level that was about 50% of the wild-type PAO1 (Fig. 6a). No inhibition on the growth of C. neoformans by the pqsAB mutant was observed (Fig. 6b). pqsB exhibited inhibition on C. neoformans growth on YNB plates (14 mm, diameter of the zone of inhibition), although the zone of inhibition of pqsB mutant was smaller than that of the wild type (17 mm) (Fig. 6b). pqsE mutant marginally inhibited C. neoformans growth with a small zone of inhibition (9 mm) outside the paper disk (Fig. 6b). These results, combined with the different compositions of extracellular compounds secreted by the mutants (Fig. 6), suggest that the inhibition of C. neoformans growth by Pa is mainly dependent on pyocyanin and, to a lesser degree, alkylquinolones such as HHQ and PQS.
Fig. 6.
Effects of pqs mutants of PAO1 on the growth of C. neoformans. The pyocyanin production was measured for all mutants (a). pqsB mutant secreted pyocyanin at a level that was about 50% of the wild-type PAO1 (a). PAO1, pqsAB, pqsB and pqsE were inoculated on paper disks, on YNB plates, which contained a lawn of C. neoformans (Cn). The diameters of the zone of inhibition were determined after 72 h at 30°C (b)
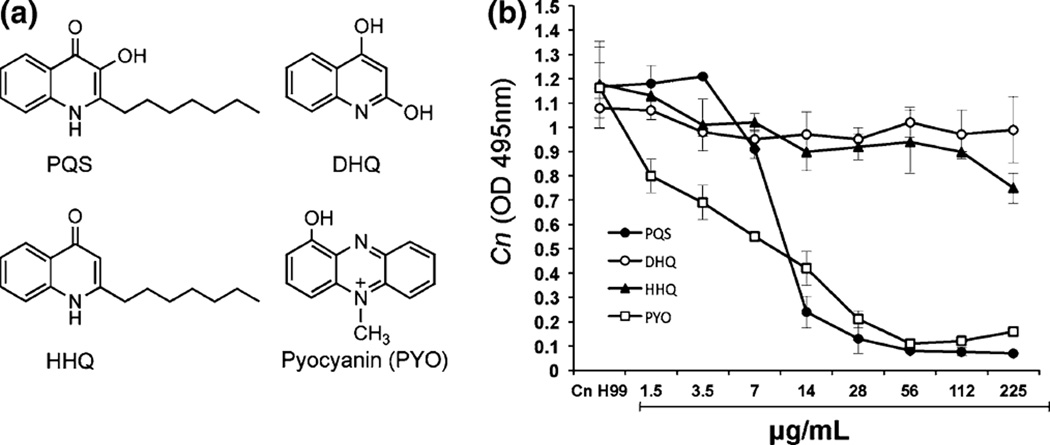
The effects of quinolones and pyocyanin on C. neoformans were further investigated using a microtiter dilution method in YNB with different concentrations of HHQ, PQS, DHQ and pyocyanin (Fig. 7a). The viable C. neoformans cells were assessed by monitoring optical density at 495 nm and CFU. Both pyocyanin and PQS completely inhibited C. neoformans growth at 28 µg/ml. However, pyocyanin exhibited more pronounced inhibition than PQS at lower concentrations (less than 14 µg/ml), demonstrating that pyocyanin is a more potent antifungal agent than PQS (Fig. 7b). HHQ partially inhibited the growth of C. neoformans only at the highest concentration tested. No inhibition on C. neoformans by DHQ was observed. These results corroborated the data obtained with the pqs mutants in that pyocyanin is a major contributing factor in the inhibition of C. neoformans.We also determined the number of viable C. neoformans cells by withdrawing aliquots from the microtiter plates. Viable cells were present in wells with pyocyanin or PQS (data not shown), demonstrating that the inhibitory effects on C. neoformans growth are static but not fungicidal.
Fig. 7.
Antifungal activity of pyocyanin and PQS against C. neoformans. Chemical structures of 3,4-dihydroxy-2-heptylquinoline (PQS), 2,4-dihydroxyquinoline (DHQ), 4-hydroxy-2-heptylquinoline (HHQ) and pyocyanin (a). C. neoformans (Cn) was incubated in microtiter plates with different concentration of PQS, DHQ, HHQ and PYO. The growth of yeast cells was assessed by monitoring optical density at 495 nm (b)
Discussion
In the present study, we investigated the ability of Pa strains PAO1 and PA14 to inhibit the growth of Cryptococcus species. Our results demonstrated that, in mixed culture conditions, Pa exhibited a fungicidal effect on C. neoformans, dependent on the relative cell density. Physical contact between Pa and C. neoformans was important for inhibition as demonstrated by experiments using U-tubes (Fig. 4), indicating that cell–cell contacts could activate the production of antifungal molecules by Pa that impeded C. neoformans growth. Our results also showed that the major antifungal molecule produced by Pa was the redoxactive metabolite, pyocyanin. In addition, alkylquinolones, such as HHQ and PQS, exhibited antifungal proprieties against C. neoformans. These findings are consistent with earlier reports that showed Pa inhibited the growth of Gram-positive bacteria and yeasts by heat-stable molecules involving quorum-sensing mechanism [20, 21].
Our results showed that PA14 exhibited more potent inhibition of Cryptococcus growth than PAO1. The p-value of paired t test is 0.02, demonstrating a significant difference in the diameters of the zone of inhibition of PA14 and PAO1 against Cryptococcus species (Fig. 1). PAO1 and PA14 are two commonly used laboratory strains of Pa. Strain PAO1 was first isolated in 1954 from a wound [22]. Numerous reports showed that the clinical isolate PA14 is significantly more virulent than PAO1 in multiple hosts, including mice, the nematode Caenorhabditis elegans, the insect Galleria mellonella and the plant Arabidopsis thaliana [14, 15, 23], suggesting that the greater antifungal activity of PA14 may be due to its increased virulence. Although the genomes of the two Pa strains are very similar (92% of the PA14 genome is present in PAO1, and 96% of PAO1 genome is present in PA14 [24]), unique genes of PA14 have been extensively studied for their roles in virulence. However, Lee et al. showed that the presence of these genes was not correlated with PA14 virulence, and the virulence of Pa is both combinatorial and multifactorial [24]. Therefore, future studies to identify PA14 genes that are essential for the increased inhibition of C. neoformans growth may help decipher the roles of different virulence factor on the ability of Pa to cause diseases.
Previous reports have shown an interspecies competition between Pa and yeasts, and in this study, we have clearly demonstrated that Pa inhibits the growth of C. neoformans, whereas Pa growth is not affected by Cryptococcus strains. Hogan et al. [25] demonstrated that Pa formed a biofilmon Candida albicans filaments and killed the fungus; where as C. albicans produced farnesol, a major fungal QS molecule, which reduced the production of PQS and pyocyanin by Pa [18, 26]. Our data showed that the interaction of Pa and C. neoformans is not reciprocal; production of extracellular virulence molecules by Pa inhibited the growth of Cryptococcus strains, while Pa was not affected by the presence of C. neoformans. Our results also demonstrated that Pa exhibited a fungicidal effect when co-cultured with C. neoformans (Fig. 2), whereas the effects of pyocyanin and PQS were static, suggesting that other unidentified exoproducts, such as proteases, hemolysin and rhamnolipids, may be involved in fungicidal activity of Pa. Moreover, cell–cell contact with C. neoformans may be important in mounting a fungicidal response of Pa on C. neoformans.
The lung of patients with cystic fibrosis is infected with a large spectrum of microbial pathogens. Over time, both the types of bacterial and their individual characteristics change [27, 28]. For instance, Staphylococcus aureus and Haemophilus influenzae are common bacteria found in CF patients at an early age. Eventually, more than 80% of CF patients harbor Pa. Filamentous fungal pathogens such as A. fumigatus and C. albicans are also frequently isolated from CF patients [29]. Fungal colonization could be the result of prolonged therapy with antibiotics and steroid, in addition to the defective mucus clearance. Hughes et al. [30] showed that in CF patients with Pa infection, only 10% had positive C. albicans skin tests, compared with 30% positivity in those free of Pa, suggesting the antifungal substance produced by Pa could prevent Candida infections. To date, there has been no report on the isolation of Cryptococcus species from the CF patients. Considering that both Cryptococcus and Pa are common lung pathogens, the lack of co-colonization could be the result of the antifungal effect of Pa on the growth of C. neoformans. Both pyocyanin and PQS accumulate intensively in the lung mucus of CF patients [31, 32], and these antifungal molecules may be important in the prevention of pulmonary cryptococcosis in CF patients.
Acknowledgments
We thank all members of Del Poeta, Zhang and Luberto laboratories for discussion. This work was supported in part by Grants AI56168, AI71142, AI78493 and AI87541 (to M.D.P) from the National Institute of Health, in part by RR17677 (to M. D. P. and Y-M. Z) from the Centers of Biomedical Research Excellence Program of the National Center for Research Resources, and in part by NIH C06 RR015455 from the Extramural Research Facilities Program of the National Center for Research Resources. Dr. Maurizio Del Poeta is a Burroughs Wellcome New Investigator in the Pathogenesis of Infectious Diseases.
Contributor Information
Antonella Rella, Department of Biochemistry and Molecular Biology, Medical University of South Carolina, 173 Ashley Avenue, BSB 535E, Charleston, SC 29425, USA; Department of Biomedical Science and Human Oncology, Hygiene Section, University of Bari, Bari, Italy.
Mo Wei Yang, Department of Biochemistry and Molecular Biology, Medical University of South Carolina, 173 Ashley Avenue, BSB 535E, Charleston, SC 29425, USA.
Jordon Gruber, Department of Biochemistry and Molecular Biology, Medical University of South Carolina, 173 Ashley Avenue, BSB 535E, Charleston, SC 29425, USA.
Maria Teresa Montagna, Department of Biomedical Science and Human Oncology, Hygiene Section, University of Bari, Bari, Italy.
Chiara Luberto, Department of Biochemistry and Molecular Biology, Medical University of South Carolina, 173 Ashley Avenue, BSB 535E, Charleston, SC 29425, USA.
Yong-Mei Zhang, Email: zhangym@musc.edu, Department of Biochemistry and Molecular Biology, Medical University of South Carolina, 173 Ashley Avenue, BSB 535E, Charleston, SC 29425, USA; Department of Microbiology and Immunology, Medical University of South Carolina, Charleston, SC, USA.
Maurizio Del Poeta, Email: delpoeta@musc.edu, Department of Biochemistry and Molecular Biology, Medical University of South Carolina, 173 Ashley Avenue, BSB 535E, Charleston, SC 29425, USA; Department of Microbiology and Immunology, Medical University of South Carolina, Charleston, SC, USA; Department of Craniofacial Biology, Medical University of South Carolina, Charleston, SC, USA; Division of Infectious Diseases, Medical University of South Carolina, Charleston, SC, USA.
References
- 1.Chayakulkeeree M, Perfect JR. Cryptococcosis. Infect Dis Clin North Am. 2006;20(3):507–544. doi: 10.1016/j.idc.2006.07.001. v–vi. [DOI] [PubMed] [Google Scholar]
- 2.Fraser JA, Giles SS, Wenink EC, Geunes-Boyer SG, Wright JR, Diezmann S, et al. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature. 2005;437(7063):1360–1364. doi: 10.1038/nature04220. [DOI] [PubMed] [Google Scholar]
- 3.Mansour MK, Levitz SM. Interactions of fungi with phagocytes. Curr Opin Microbiol. 2002;5(4):359–365. doi: 10.1016/s1369-5274(02)00342-9. [DOI] [PubMed] [Google Scholar]
- 4.Feldmesser M, Tucker S, Casadevall A. Intracellular parasitism of macrophages by Cryptococcus neoformans. Trends Microbiol. 2001;9(6):273–278. doi: 10.1016/s0966-842x(01)02035-2. [DOI] [PubMed] [Google Scholar]
- 5.Levitz SM, Nong SH, Seetoo KF, Harrison TS, Speizer RA, Simons ER. Cryptococcus neoformans resides in an acidic phagolysosome of human macrophages. Infect Immun. 1999;67(2):885–890. doi: 10.1128/iai.67.2.885-890.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chretien F, Lortholary O, Kansau I, Neuville S, Gray F, Dromer F. Pathogenesis of cerebral Cryptococcus neoformans infection after fungemia. J Infect Dis. 2002;186(4):522–530. doi: 10.1086/341564. [DOI] [PubMed] [Google Scholar]
- 7.Maschmeyer G, Braveny I. Review of the incidence and prognosis of Pseudomonas aeruginosa infections in cancer patients in the 1990s. Eur J Clin Microbiol Infect Dis. 2000;19(12):915–925. doi: 10.1007/s100960000410. [DOI] [PubMed] [Google Scholar]
- 8.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48(1):1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 9.Gibson J, Sood A, Hogan DA. Pseudomonas aeruginosa-Candida albicans interactions: localization and fungal toxicity of a phenazine derivative. Appl Environ Microbiol. 2009;75(2):504–513. doi: 10.1128/AEM.01037-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kerr J. Inhibition of fungal growth by Pseudomonas aeruginosa and Pseudomonas cepacia isolated from patients with cystic fibrosis. J Infect. 1994;28(3):305–310. doi: 10.1016/s0163-4453(94)91943-7. [DOI] [PubMed] [Google Scholar]
- 11.Kerr JR, Taylor GW, Rutman A, Hoiby N, Cole PJ, Wilson R. Pseudomonas aeruginosa pyocyanin and 1-hydroxyphenazine inhibit fungal growth. J Clin Pathol. 1999;52(5):385–387. doi: 10.1136/jcp.52.5.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fisher AM. Inhibition of growth of Cryptococcus neoformans by cultures of Pseudomonas aeruginosa. Bull Johns Hopkins Hosp. 1954;95(4):157–161. [PubMed] [Google Scholar]
- 13.Teoh-Chan H, Chau PY, Ng MH, Wong PC. Inhibition of Cryptococcus neoformans by Pseudomonas aeruginosa. J Med Microbiol. 1975;8(1):77–81. doi: 10.1099/00222615-8-1-77. [DOI] [PubMed] [Google Scholar]
- 14.Choi JY, Sifri CD, Goumnerov BC, Rahme LG, Ausubel FM, Calderwood SB. Identification of virulence genes in a pathogenic strain of Pseudomonas aeruginosa by representational difference analysis. J Bacteriol. 2002;184(4):952–961. doi: 10.1128/jb.184.4.952-961.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan MW, Mahajan-Miklos S, Ausubel FM. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc Natl Acad Sci USA. 1999;96(2):715–720. doi: 10.1073/pnas.96.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi KH, Schweizer HP. An improved method for rapid generation of unmarked Pseudomonas aeruginosa deletion mutants. BMC Microbiol. 2005;5:30. doi: 10.1186/1471-2180-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tully JG, Jr, Latteri A. Paraplegia, syringomyelia tarda and neuropathic arthrosis of the shoulder: a triad. Clin Orthop Relat Res. 1978;134:244–248. [PubMed] [Google Scholar]
- 18.Heeb S, Fletcher MP, Chhabra SR, Diggle SP, Williams P, Camara M. Quinolones: from antibiotics to autoinducers. FEMS Microbiol Rev. 2011;35(2):247–274. doi: 10.1111/j.1574-6976.2010.00247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang YM, Frank MW, Zhu K, Mayasundari A, Rock CO. PqsD is responsible for the synthesis of 2, 4-dihydroxyquinoline, an extracellular metabolite produced by Pseudomonas aeruginosa. J Biol Chem. 2008;283(43):28788–28794. doi: 10.1074/jbc.M804555200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Machan ZA, Taylor GW, Pitt TL, Cole PJ, Wilson R. 2-Heptyl-4-hydroxyquinoline N-oxide, an antistaphylococcal agent produced by Pseudomonas aeruginosa. J Antimicrob Chemother. 1992;30(5):615–623. doi: 10.1093/jac/30.5.615. [DOI] [PubMed] [Google Scholar]
- 21.Mowat E, Rajendran R, Williams C, McCulloch E, Jones B, Lang S, et al. Pseudomonas aeruginosa and their small diffusible extracellular molecules inhibit Aspergillus fumigatus biofilm formation. FEMS Microbiol Lett. 2010;313(2):96–102. doi: 10.1111/j.1574-6968.2010.02130.x. [DOI] [PubMed] [Google Scholar]
- 22.Holloway BW. Genetic recombination in Pseudomonas aeruginosa. J Gen Microbiol. 1955;13(3):572–581. doi: 10.1099/00221287-13-3-572. [DOI] [PubMed] [Google Scholar]
- 23.Rahme LG, Stevens EJ, Wolfort SF, Shao J, Tompkins RG, Ausubel FM. Common virulence factors for bacterial pathogenicity in plants and animals. Science. 1995;268(5219):1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 24.Lee DG, Urbach JM, Wu G, Liberati NT, Feinbaum RL, Miyata S, et al. Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol. 2006;7(10):R90. doi: 10.1186/gb-2006-7-10-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hogan DA, Kolter R. Pseudomonas-Candida interactions: an ecological role for virulence factors. Science. 2002;296(5576):2229–2232. doi: 10.1126/science.1070784. [DOI] [PubMed] [Google Scholar]
- 26.Singh A, Del Poeta M. Lipid signalling in pathogenic fungi. Cell Microbiol. 2011;13(2):177–185. doi: 10.1111/j.1462-5822.2010.01550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oliver A. Mutators in cystic fibrosis chronic lung infection: prevalence, mechanisms, and consequences for antimicrobial therapy. Int J Med Microbiol. 2010;300(8):563–572. doi: 10.1016/j.ijmm.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 28.Hoboth C, Hoffmann R, Eichner A, Henke C, Schmoldt S, Imhof A, et al. Dynamics of adaptive microevolution of hypermutable Pseudomonas aeruginosa during chronic pulmonary infection in patients with cystic fibrosis. J Infect Dis. 2009;200(1):118–130. doi: 10.1086/599360. [DOI] [PubMed] [Google Scholar]
- 29.Bakare N, Rickerts V, Bargon J, Just-Nubling G. Prevalence of Aspergillus fumigatus and other fungal species in the sputum of adult patients with cystic fibrosis. Mycoses. 2003;46(1–2):19–23. doi: 10.1046/j.1439-0507.2003.00830.x. [DOI] [PubMed] [Google Scholar]
- 30.Hughes WT, Kim HK. Mycoflora in cystic fibrosis: some ecologic aspects of Pseudomonas aeruginosa and Candida albicans. Mycopathol Mycol Appl. 1973;50(3):261–269. doi: 10.1007/BF02053377. [DOI] [PubMed] [Google Scholar]
- 31.Taylor GW, Machan ZA, Mehmet S, Cole PJ, Wilson R. Rapid identification of 4-hydroxy-2-alkylquinolines produced by Pseudomonas aeruginosa using gas chromatography-electron-capture mass spectrometry. J Chromatogr B Biomed Appl. 1995;664(2):458–462. doi: 10.1016/0378-4347(94)00494-p. [DOI] [PubMed] [Google Scholar]
- 32.Caldwell CC, Chen Y, Goetzmann HS, Hao Y, Borchers MT, Hassett DJ, et al. Pseudomonas aeruginosa exotoxin pyocyanin causes cystic fibrosis airway pathogenesis. Am J Pathol. 2009;175(6):2473–2488. doi: 10.2353/ajpath.2009.090166. [DOI] [PMC free article] [PubMed] [Google Scholar]