Abstract
Introduction
Rapid identification of bloodstream pathogens provides crucial information that can improve the choice of antimicrobial therapy for children. Previous impact studies have primarily focused on adults. Our objective was to evaluate the impact of rapid testing in a children’s hospital on time to organism identification and antibiotic use in the setting of an established antimicrobial stewardship program.
Methods
We conducted a retrospective study over three consecutive time periods (spanning January 2013–August 2015) as our hospital sequentially introduced two rapid testing methods for positive blood cultures. An antimicrobial stewardship program was active throughout the study. In the baseline period, no rapid diagnostic methods were routinely utilized. In the second period (PNAFISH), a fluorescent in situ hybridization test was implemented for gram-positive organisms and in the third a rapid multiplex PCR (rmPCR) test was employed. For children with positive blood cultures, time to organism identification use and duration of select antimicrobial therapies were compared between periods.
Results
Positive blood cultures were analyzed. Median overall time to organism identification was 23, 11, and 0 h in the baseline, PNAFISH, and rmPCR periods, respectively (p < 0.001 for both PNAFISH and rmPCR vs. baseline). For gram-negative organisms, only rmPCR performed significantly faster than baseline (p < 0.001). The duration of vancomycin use for coagulase-negative staphylococci was shorter in both the PNAFISH and rmPCR periods (mean 31 h in the baseline period, 12 and 14 h in the PNAFISH and rmPCR periods, respectively). For MSSA bacteremia, use of vancomycin was significantly decreased only in the rmPCR period (32% of patients vs. 64 and 72% in the baseline and PNAFISH periods; mean duration of 9 h vs. 30 and 26 h). There was no difference in use or duration of broad-spectrum gram-negative therapy across the three time periods.
Conclusion
Rapid diagnostic testing for children with positive blood cultures results in faster time to identification and can influence antibiotic prescribing in the setting of active antimicrobial stewardship particularly for gram-positive pathogens.
Funding
Merck.
Keywords: Antibiotic use, Antimicrobial stewardship, Bloodstream infection, Molecular testing, Rapid diagnostic methods
Introduction
Rapid testing for identification of pathogens in bloodstream infections has the potential to provide crucial information that can improve patient care. There are a number of potential benefits to rapid pathogen identification, including decreased mortality from sepsis, more targeted diagnostic and therapeutic interventions, and more rapid optimization of antimicrobial therapy.
The Infectious Diseases Society of America has called for research on the impact of rapid diagnostic testing on the use of antimicrobials [1]. Studies in adults evaluating the clinical impact of rapid blood culture diagnostics on antimicrobial use have shown decreases in both the time to effective or optimal therapy and the reduction in the use of broad-spectrum agents [2–8]. Decreased use of broad-spectrum therapy for patients who can be treated with a more narrow-spectrum agent could reduce the incidence of adverse outcomes such as Clostridium difficile colitis or the emergence of resistant pathogens [9–11]. Studies evaluating the impact of rapid diagnostic methods for blood cultures in children are more limited. Initial studies focused primarily on the diagnostic accuracy and functionality of the tests [12–16]; however, more recent reports have shown benefits of rapid identification on overall antibiotic use in children [2, 17–22].
In adult studies, antimicrobial stewardship program (ASP) activities have proved to be a key component in effectively translating rapid diagnostics results into changes in patient care. Prior pediatrics studies evaluating the impact of rapid blood culture diagnostics have had either limited formal ASP involvement [20, 21] or implemented stewardship simultaneously with the rapid diagnostic test [22]. Our institution has had an active ASP since 2010. Our ASP provides prospective audit and feedback guidance to providers regarding antimicrobial therapy. Three years after our ASP was started our institution sequentially implemented two rapid diagnostic platforms for pathogen identification in positive blood cultures over 2 years (2013–2015). First, a fluorescent in situ hybridization (PNA FISH) test targeting gram-positive organism was implemented, followed by a rapid multiplex PCR (rmPCR) with an expanded panel targeting both gram-positive and -negative pathogens that replaced most use of the PNA FISH. Our objective in this study was to evaluate the impact of these tests on time to organism identification in our hospital and to examine the impact of rapid testing on antibiotic use for specific pathogens in the setting of an established ASP.
Methods
Statement of Ethics Compliance
Approval for this study was obtained from the Institutional Review Board of the University of Utah and Primary Children’s Hospital (PCH) with a waiver of informed consent. All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1964, as revised in 2013.
Setting
PCH is a 289-bed children’s hospital that is part of Intermountain Healthcare. PCH serves as both the pediatric community hospital for Salt Lake County, Utah, and as the tertiary pediatric referral center for five states in the Intermountain West (Utah, Idaho, Wyoming, Nevada, and Montana).
Study Design and Population
This was a retrospective study of positive blood cultures from hospitalized patients evaluated at PCH between 1 January 2013 and 15 August 2015. Positive blood cultures were identified from the Intermountain Healthcare Enterprise Data Warehouse (EDW); only the first positive blood culture from an episode of bacteremia was included in the study. Episodes of bacteremia started with the first day of positive blood cultures in a patient with no known positive blood cultures in the previous 7 days. If children had multiple episodes of bacteremia, each episode was included in the analysis. Polymicrobial bacteremias were excluded from analysis. Data abstracted from the EDW included demographics, site of care, timing of blood culture results reporting, time to organism identification, antimicrobial therapy given, and duration.
Study Timeline and Methods to Identify Organisms in Positive Blood Cultures
The study was divided into three distinct periods of 1–1.5 years based on the type of diagnostic method used. Periods were defined as: baseline (1 January–31 December 2013), PNAFISH (1 January 2013–31 January 2014), and rmPCR (1 February 2014–15 August 2015). Throughout the study, blood cultures were processed in the clinical microbiology laboratory using the BACTEC automated blood culture system (BD Diagnostic Systems, Franklin Lakes, NJ). Gram stains and rapid diagnostic tests (when available and indicated) were performed 24 h a day, 7 days a week, by trained microbiology staff. Post gram stain, organisms were inoculated onto culture media and subsequently identified using the VITEK 2® system (bioMérieux, Marcy L’Étoile, France) and other biochemical tests as appropriate.
Baseline Study Period (1 January–31 December 2013)
During the baseline period, no rapid diagnostic methods were used for positive blood cultures. Gram stain was performed immediately when a blood culture signaled positive, and results were exported into the electronic information system. In addition, the positive blood culture gram stain result was called to the treating physician within 1 h of the positive BACTEC alert. After biochemical identification, Staphylococcus aureus was sub-cultured onto solid media and early growth tested for the presence of PBP2a using the PBP2a SA Culture Colony Test (Alere™, Waltham, MA).
PNAFISH Study Period (1 January 2013–31 January 2014)
Peptide nucleic acid (PNA) fluorescent in situ hybridization (QuickFISH®, AvanDx, Inc., Woburn,MA) was implemented in the PCH microbiology laboratory in January, 2013. QuickFISH was performed for gram stains showing gram-positive cocci in clusters (Staphylococcus QuickFISH™) starting in January 2013 and was expanded to gram-positive cocci in chains (Enterococcus QuickFISH™) in May 2013. Staphylococcus QuickFISH identifies Staphylococcus aureus and coagulase-negative staphylococci (CONS), and Enterococcus QuickFISH identifies Enterococcus faecalis and non-faecalis enterococci in about 20 min. PNA-FISH assays for gram-negative organisms and Candida spp., while available commercially, were not used in the PCH laboratory. Microbiology staff reported QuickFISH results for gram-positive organisms when available as the first notification of a positive blood culture. If QuickFISH was not indicated (i.e., for gram-negative organisms) or results were not available within an hour, the treating physician was notified of the gram stain result only. Further organism identification was the same as in the baseline period, including sub-culture of S. aureus after identification for the detection of PBP2a using the PBP2a SA Culture Colony Test.
rmPCR Study Period (1 February 2014–15 August 2015)
A multiplex PCR test [FilmArray® blood culture ID panel (BCID), BioFire Diagnostics LLC., Salt Lake City, UT] was implemented in the PCH microbiology laboratory in February 2014. This assay detects 24 pathogens and 3 antibiotic resistance genes, including Staphylococcus aureus, Streptococcus pneumoniae, Enterococcus species, Escherichia coli, Klebsiella species, Enterobacter cloacae complex as well as mecA (methicillin resistance), vanA/B (vancomycin resistance), and KPC (carbapenem resistance) in about 1 h [23]. BCID was performed on the first positive aerobic blood culture and replaced most, but not all, use of QuickFISH. QuickFISH was still performed for gram-positive organisms in anaerobic blood cultures and to distinguish E. faecalis from other enterococci. Microbiology staff called rapid diagnostic test results (BCID or QuickFISH) to the treating physician as the first report of a positive culture; if rapid diagnostic results were not available within an hour of positive BACTEC notification, the gram stain alone was reported. Further organism identification proceeded as before, although PBP2a testing was not used routinely for S. aureus, as mecA testing is part of the BCID test panel.
Time to Organism Identification
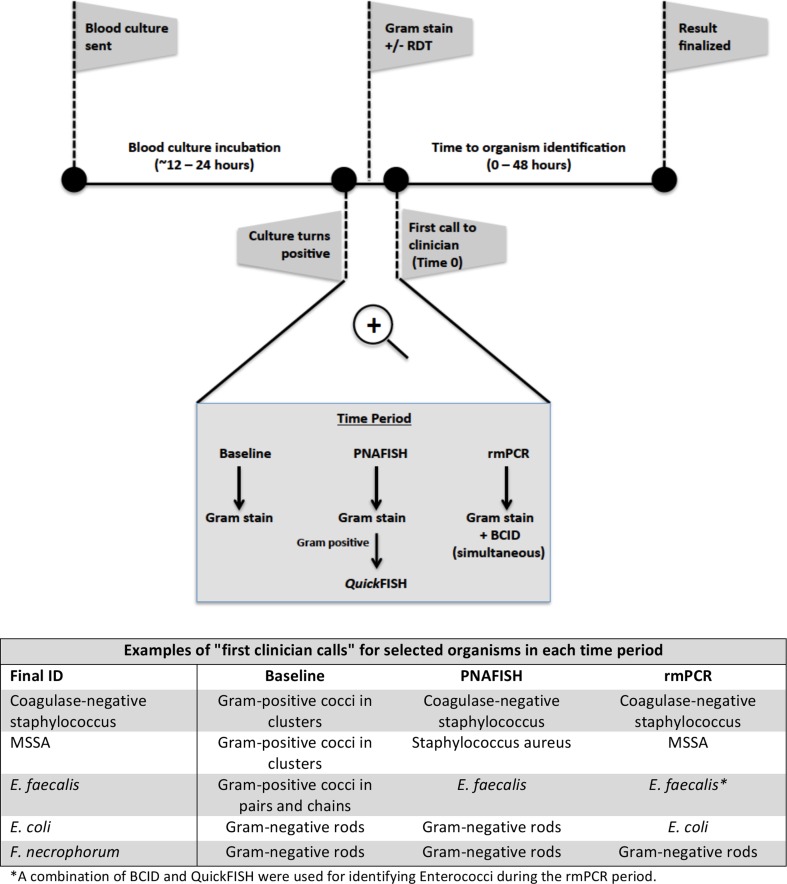
First clinician notification of a positive blood culture result, whether gram stain, QuickFISH, or BCID result, was considered “Time 0” for all analyses in this study. Organism identification was defined as the first identification of an organism to a group or genus (e.g., Enterobacteriaceae, Enterococcus spp.) or species level identification (Fig. 1). Time to organism identification was calculated as the difference in time in hours between the first clinician notification (Time 0) and the identification of the organism.
Fig. 1.
Blood culture procedures and methods to identify pathogens from positive cultures in each time period are shown. “Time 0” is defined here as the first call to the clinician after the blood culture becomes positive. Table shows examples of “first clinician calls” for select organisms
Antimicrobial Stewardship Program (ASP) and Interventions
Formal antimicrobial stewardship was implemented at PCH in 2010. The PCH ASP team consists of one pediatric infectious diseases physician and a dedicated pharmacist. The PCH ASP utilizes a prospective audit and feedback model to provide antimicrobial use recommendations to clinicians. All inpatients receiving intravenous antimicrobials are reviewed by the ASP Monday through Friday, and physicians are contacted by the ASP between 0700 and 1530 regarding recommended changes to antimicrobial therapy. Throughout the study, selected antimicrobials (e.g., carbapenems, linezolid) required ASP approval between 0700 and 2200 and, if given outside of these hours, was reviewed by 1200 the next day. Vancomycin use was not restricted, but was reviewed.
Upon updating any positive sterile site culture, the PCH electronic health system generates a notification of updated results including gram stain, organism identification (if available), and susceptibility results (if available) to the ASP Pharmacist pager. For the entirety of the study period, the PCH ASP reviewed automated pages in real-time Monday through Friday from 0700 to 1530 and used this information to provide therapeutic recommendations to the treating physicians. Prior to implementation of the BCID panel, a handout describing the interpretation of results and suggestions for preferred antibiotic regimens was distributed to physicians and pharmacists throughout the hospital.
Antibiotic Use
Select antimicrobial therapy, specifically the use of broad-spectrum gram-negative agents (carbapenems, cefepime, ceftazidime, and piperacillin-tazobactam) and vancomycin, was evaluated (see below).
Vancomycin
For vancomycin, we examined overall use measured as the percentage of all patients that received at least one dose, as well as use of vancomycin specifically for children with blood cultures positive for MSSA and coagulase-negative staphylococci (CoNS). For vancomycin analyses in children with CoNS bacteremia, we excluded children with multiple positive blood cultures, infants hospitalized in the neonatal ICU, and those with central lines in order to identify a population where the positive blood culture was likely to represent contamination.
Broad-Spectrum Gram-Negative Agents
For broad-spectrum gram-negative agents, we examined overall use as well as use specifically for children with blood cultures positive for gram-negative pathogens.
Outcome Measures and Statistical Analyses
The primary outcome measures were the time to organism identification and the use of select antimicrobial therapy as described above. Descriptive statistics were used to characterize the pattern of microbiological detections and the frequency and duration of antimicrobial use. The median time to organism identification was calculated. This value is presented because for a minority of organisms time to identification is very long, resulting in a right bias. Antibiotic utilization is presented as mean hours, which is a more clinically meaningful metric. Additionally, because for many patients utilization is 0 h, leading to a large left bias in duration of use, we also present the percent of patients receiving at least one dose of each antibiotic. Categorical comparisons were performed using the chi-squared test or Fisher’s exact test, as appropriate. Continuous comparisons were performed using the Wilcoxon-Mann-Whitney U test. All statistical tests were performed two-sided with alpha equal to 0.05 using R (R Foundation for Statistical Computing, Vienna, Austria).
Results
Study Cohort
A total of 1419 bacteremic episodes were identified during the 3-year study period. Of these, 191 (13%) were polymicrobial and were excluded from analysis, leaving 1228 bacteremic episodes included in the study. The number of episodes analyzed in each time period was similar (397, 408, and 422 in the baseline, PNAFISH, and rmPCR periods, respectively). Demographic characteristics and site of treatment for included children were also similar and are shown in Table 1. We found no significant differences between periods in length of stay (8, 7, and 9 days), 30-day mortality (4.3%, 4.2%, 3.5%), and 30-da y representation to the emergency department or inpatient admission (39%, 40%, 44%) for the baseline, PNAFISH, and rmPCR periods, respectively.
Table 1.
Demographics and treatment site for included children
| Demographics | N (%) or median (IQR) | |||
|---|---|---|---|---|
| Period | ||||
| Overall (N = 1228) | Baseline (N = 397) | PNAFish (n = 408) | rmPCR (N = 422) | |
| Male | 709 (58%) | 213 (54%) | 242 (59%) | 253 (60%) |
| Age, years | 2 (0–8) | 2 (0–7) | 2 (0–8) | 2 (0–7) |
| Central line in place | 447 (36%) | 161 (41%) | 149 (37%) | 137 (32%) |
| Patient location at time 0 | ||||
| Admitted | 984 (80%) | 308 (78%) | 330 (81%) | 345 (82%) |
| General wards | 422 (34%) | 130 (33%) | 141 (35%) | 150 (36%) |
| ICU (PICU/CICU)a | 246 (20%) | 75 (19%) | 83 (20%) | 88 (21%) |
| NICUb | 131 (12%) | 42 (11%) | 44 (11%) | 45 (11%) |
| ICSc | 176 (14%) | 57 (14%) | 60 (15%) | 59 (14%) |
| Other | 9 (1%) | 4 (1%) | 2 (0%) | 3 (1%) |
| Not admitted | 244 (20%) | 89 (22%) | 78 (19%) | 77 (18%) |
| No active encounter | 216 (18%) | 78 (20%) | 68 (17%) | 70 (17%) |
| ED/RTUd | 28 (2%) | 11 (3%) | 10 (2%) | 7 (2%) |
a ICU intensive care unit; PICU pediatric ICU; CICU cardiac ICU
b NICU neonatal ICU
cImmune compromised unit: unit for patients with malignancy, organ transplantation or other immune compromising condition
d RTU rapid treatment (short stay) unit
Microbiology
Of 1228 first-positive blood cultures, 923 (75%) were positive for gram-positive organisms, 280 (23%) were positive for gram-negative organisms, and 22 (2%) were positive for Candida species. The distribution of organisms was comparable in the three periods. Specifically, the number of gram-negative pathogens as well as staphylococci, including CONS and MSSA, were not significantly different between time periods. In the PNAFISH period, 54% of organisms were identified by the rapid diagnostic test (QuickFISH), and in the rmPCR period, rapid testing (BCID or QuickFISH) identified 89% of organisms. Data are shown in Table 2.
Table 2.
Time to identification of pathogens from first clinician notification (time 0) in each period
| Period | |||
|---|---|---|---|
| Method of identification | Baseline (N = 397) | PNAFISH (N = 408) | rmPCR (N = 422) |
| Rapid diagnostic test | n/a | 222 (54%) | 376 (89%) |
| Organism type | Time to identification [median (IQR)] | p value PNAFISH vs. baseline | p value rmPCR vs. baseline | ||
|---|---|---|---|---|---|
| All | 23 (16–31) | 11 (0–29) | 0 (0–2) | p < 0.001 | p < 0.001 |
| Gram positive | 21 (15–28) [n = 301] | 0 (0–18) [n = 315] | 0 (0–1) [n = 307] | p < 0.001 | p < 0.001 |
| Gram negative | 34 (26–43) [n = 90] | 35 (29–44) [n = 84] | 1 (0–5) [n = 106] | p = 0.3 | p < 0.001 |
| Specific organisms | |||||
| Staphylococcus aureus | 20 (16–25) [n = 52] | 0 (0–0) [n = 49] | 0 (0–0) [n = 50] | p < 0.001 | p < 0.001 |
| mecA gene | 25 (24–27) [n = 7] | 0 (0–0) [n = 10] | 0 (0–0) [n = 11] | p < 0.001 | p < 0.001 |
| Coagulase negative staphylococci | 19 (15–27) [n = 133] | 0 (0–0) [n = 133] | 0 (0–1) [n = 149] | p < 0.001 | p < 0.001 |
| Enterococcus spp. | |||||
| E. faecalis | 21 (14–23) [n = 8] | 0 (0–24) [n = 13] | 0 (0–1) [n = 15] | p = 0.08 | p < 0.001 |
| E. faecium | – [n = 0] | 5 (2–7) [n = 2] | 0 (0–0) [n = 2] | n/a** | n/a** |
| Streptococcus species (all) | 22 (14–26) [n = 60] | 18 (11–23) [n = 65] | 0 (0–9) [n = 59] | p = 0.02 | p < 0.001 |
| Escherichia coli | 37 (26–42) [n = 20] | 40 (35–46) [n = 25] | 0 (0–2) [n = 37] | p = 0.1 | p < 0.001 |
| Klebsiella spp. | 37 (30–44) [n = 19] | 35 (33–40) [n = 8] | 1 (0–2) [n = 13] | p = 0.8 | p = 0.001 |
| Pseudomonas aeruginosa | 18 (13–25) [n = 7] | 24 (18–26) [n = 9] | 0 (0–0) [n = 5] | p = 0.3 | p = 0.05 |
| Enterobacter spp. | 35 (32–46) [n = 8] | 35 (32–39) [n = 5] | 0 (0) [n = 8] | p = 0.8 | p < 0.001 |
| All other gram negatives | 33 (24–44) [n = 44] | 34 (29–43) [n = 42] | 1 (0–23) [n = 51] | p = 0.5 | p = 0.05 |
| Candida spp. | 39 (44–61) [n = 5] | 30 (28–46) [n = 9] | 0 (0–0) [n = 8] | p = 0.1 | p < 0.001 |
Time to Organism Identification
In the baseline period, the median time to organism identification from time 0 across all organisms was 23 h (IQR 16–31 h; Table 2). Median time to identification for gram-positive organisms was 21 h (IQR 15–28 h) as well as median time to identification for gram-negatives was 34 h (IQR 26–43 h). Time to identification for all organisms was shorter in both the PNAFISH (median 11 h; IQR 0–29) and the rmPCR periods (median 0 h; IQR 0–2). For gram-negative organisms and Candida species, time to identification was shorter only in the rmPCR period (median 1 h; IQR 0–5) as these organisms were not included in the PNAFISH rapid testing algorithm. However, the median time to identification was shorter for gram-positive organisms (mean of 0 h) in both the PNAFISH (IQR 0–18 h) and rmPCR periods (IQR 0-1 h).
Antibiotic Utilization
Vancomycin
Overall, there was a trend toward decreased use of vancomycin for children with positive blood cultures when comparing the baseline period to the periods in which rapid diagnostics were used (Table 3). In both the PNAFISH and rmPCR periods, there was a decrease in the percentage of children with CoNS bacteremia who received any vancomycin as well as a significant trend toward shorter duration of vancomycin therapy. While there was no significant difference in the use of vancomycin for children with MSSA bacteremia in the PNAFISH period when compared to baseline (64% and 72% of children treated with vancomycin, respectively), the percentage of children with blood cultures positive for MSSA who received any doses of vancomycin declined considerably during the rmPCR period (32%; p = 0.01 for rmPCR compared to baseline). This was also reflected in an overall decreased duration of vancomycin use for children with MSSA bacteremia (mean duration of 9 vs. 30 h in the baseline period).
Table 3.
Antibiotic utilization in the baseline, PNAFish, and rmPCR periods
| Denominatora | Antibiotic used? | Duration of use | p valuec | ||
|---|---|---|---|---|---|
| Yes | ≥24 hb | Mean (±SD) duration, h | |||
| Vancomycin | 1228 | 566 (46%) | 452 (37%) | 44 (±98) | – |
| Baseline | 397 | 197 (50%) | 161 (41%) | 53 (±119) | Ref. |
| PNAFish | 408 | 196 (48%) | 164 (40%) | 44 (±94) | 0.20 |
| rmPCR | 422 | 172 (41%) | 126 (30%) | 35 (±79) | 0.01 |
| Vancomycin + CoNSd | 248 | 66 (27%) | 54 (22%) | 19 (±57) | – |
| Baseline | 82 | 27 (33%) | 25 (30%) | 31 (±81) | Ref. |
| PNAFish | 74 | 17 (23%) | 14 (19%) | 12 (±29) | 0.05 |
| rmPCR | 91 | 21 (23%) | 14 (15%) | 14 (±45) | 0.10 |
| Vancomycin + MSSA | 124 | 69 (56%) | 49 (40%) | 22 (±33) | – |
| Baseline | 44 | 28 (64%) | 21 (48%) | 30 (±45) | Ref. |
| PNAFish | 39 | 28 (72%) | 22 (56%) | 26 (±23) | 0.55 |
| rmPCR | 41 | 13 (32%) | 6 (15%) | 9 (±19) | 0.01 |
| Broad-spectrum gram-negative antibioticse | 1228 | 265 (22%) | 212 (17%) | 28 (±105) | – |
| Baseline | 397 | 68 (17%) | 59 (15%) | 26 (±108) | Ref. |
| PNAFish | 408 | 88 (22%) | 72 (18%) | 27 (±94) | 0.91 |
| rmPCR | 422 | 108 (26%) | 80 (19%) | 31 (±111) | 0.56 |
| Broad-spectrum gram-negative antibiotics + gram-negative organismse | 280 | 104 (37%) | 87 (31%) | 47 (±118) | – |
| Baseline | 90 | 31 (34%) | 29 (32%) | 53 (±143) | Ref. |
| PNAFish | 84 | 24 (29%) | 21 (25%) | 37 (±102) | 0.40 |
| rmPCR | 106 | 49 (46%) | 37 (35%) | 51 (±105) | 0.88 |
| Carbapenems | 1228 | 130 (11%) | 118 (10%) | 20 (±100) | – |
| Baseline | 397 | 44 (11%) | 43 (11%) | 20 (±95) | Ref. |
| PNAFish | 408 | 38 (9%) | 33 (8%) | 20 (±97) | 0.99 |
| rmPCR | 422 | 47 (11%) | 41 (10%) | 20 (±107) | 0.98 |
| Carbapenems + gram-negative organisms | 280 | 70 (25%) | 64 (23%) | 40 (±123) | – |
| Baseline | 90 | 29 (32%) | 29 (32%) | 55 (±144) | Ref. |
| PNAFish | 84 | 12 (14%) | 10 (12%) | 29 (±117) | 0.19 |
| rmPCR | 106 | 29 (27%) | 25 (24%) | 37 (±107) | 0.32 |
aDenominator is the number of bacteremic episodes in each category. For example, the total number of bacteremic episodes in the baseline period is 397; the total number of bacteremic episodes in which CoNS was identified is 82 in the baseline period
bTotal number (percent) of episodes in which the antibiotic was used for >24 h. The denominator is the total number of episodes in the category
cAll p values are reported in comparison to the duration in the baseline period
dCoNS infections were included for children in whom CoNS was isolated from a blood culture that was not drawn from a central line and not drawn in the newborn intensive care unit
eBroad-spectrum gram-negative antibiotics include carbapenems, piperacillin/tazobactam, and cefepime
Broad-Spectrum Gram-Negative Antibiotics
In contrast to vancomycin, we found no difference between periods in the use of broad-spectrum gram-negative agents, including carbapenems. Overall, 22% of children with a positive blood culture received broad-spectrum gram-negative agents and 37% because their blood culture was positive for a gram-negative pathogen. Duration of therapy was also not statistically different between periods. Overall mean duration of broad-spectrum gram-negative therapy was 28, 47 h if a gram-negative pathogen was identified. Results are shown in Table 3.
Discussion
We evaluated the impact of two different rapid molecular diagnostics on the time to identification of organisms as well as select antibiotic prescribing for bloodstream pathogens in a children’s hospital with a mature ASP. We found that the use of both PNA FISH and rmPCR significantly decreased the median time to identification of gram-positive organisms, and rmPCR also decreased the time to identification for gram-negative organisms. Rapid identification of CoNS in both the PNAFISH and rmPCR periods was associated with decreased use of vancomycin, but only rmPCR was associated with a significant decrease in vancomycin exposure for children with MSSA. In contrast, we did not observe changes in the use of broad-spectrum gram-negative antibiotics associated with the use of either diagnostic test.
Prior studies have shown decreased time to identification of organisms using rapid diagnostic methods, such as QuickFISH, FilmArray BCID, and Verigene compared to standard culture-based methods [18, 20, 24–27]. While this is not unexpected, direct comparative studies between methods have not been performed in pediatrics. As the majority of rapid diagnostic tests for positive blood cultures identify only a limited set of pre-defined pathogens, the overall impact of each test may vary depending on differences in the epidemiology of bloodstream pathogens. In our population we found that both PNA FISH and rmPCR provided identification for gram-positive pathogens a full day sooner than standard techniques. Using a combined panel identifying both gram-positive and -negative organisms, rmPCR characterized 89% of all positive blood culture organisms within an hour. QuickFISH is available for gram-negative organisms as well but was not implemented at our institution.
Recent studies in adults have demonstrated that rapid identification of bloodstream pathogens can decrease the use of vancomycin [2, 6, 7, 28]. This finding was replicated for children in our study. Relative to a period with no rapid testing, we found that both PNA FISH and rmPCR decreased the duration of vancomycin therapy for all children with positive blood cultures. There were, however, specific differences in the effect of testing on different pathogens, for example, CoNS and MSSA. CoNS is a pathogen for which identification alone predicts care. When considered pathogenic, CoNS is almost exclusively treated with vancomycin; however, in many cases it is simply a blood culture contaminant, requiring no antibiotic therapy. As identification of CoNS (or, more specifically, differentiation of CoNS from S. aureus) drives therapy decisions, we observed a decrease in vancomycin therapy in both the PNAFISH and rmPCR periods for children in whom CoNS was likely a contaminant organism (excluding children with multiple positive blood cultures, infants hospitalized in the neonatal ICU, and those with central lines; see “Methods”).
In contrast, optimizing care for children with S. aureus bacteremia requires identification of both the pathogen and a resistance marker. Thus, we observed a significant reduction in duration of vancomycin use for MSSA bacteremia during the rmPCR but not the PNAFISH period. In both adult and pediatric studies, treatment of MSSA bacteremia with nafcillin or cefazolin decreases mortality when compared to treatment with vancomycin [29–32]. The reduction in vancomycin use observed in our study is likely related to the fact that the rmPCR used both identified S. aureus and evaluates for the presence or absence of mecA simultaneously, whereas PNAFISH identified only the pathogen. Nguyen et al. showed a decrease in the duration of vancomycin use when using PCR to detect the mecA gene in positive blood cultures [6]. Beal et al., using a multiplex DNA hybridization assay, found that the rapid detection of MSSA (vs. MRSA) allowed for a reduction in time to optimal antibiotics of 21 h for adult patients [33]. Our findings for both CoNS and MSSA are similar to those of Pardo et al. who looked at the impact of the BCID panel on the management of patients with blood cultures positive for gram-positive organisms [34].
We did not find a reduction in the use of broad-spectrum gram-negative agents with the introduction of either PNAFISH or rmPCR. Explanations of these findings include our low, but non-trivial institutional rate of gram-negative resistance for pathogens in bacteremia (10% ceftriaxone resistance among gram-negative pathogens) as well as ASP activities resulting in well-controlled use of broad-spectrum agents prior to the start of this study. In a previous study, Buss et al. failed to improve outcomes in high-risk children with gram-negative bacteremia utilizing MALDI-TOF combined with a robust ASP in a large tertiary care hospital with low institutional resistance rates similar to ours [35]. Additionally, while the rmPCR test used in our study reported resistance to carbapenems mediated by the KPC gene, it did not provide information regarding susceptibility or resistance to third-generation cephalosporins. Without data on resistance, the ability and desire of clinicians to rapidly de-escalate therapy for bloodstream infection is limited. Finally, many gram-negative bloodstream infections are part of a polymicrobial or complex process such as an intra-abdominal infection, necessitating broad-spectrum therapy even when the identified pathogen could be more narrowly treated. However, with low resistance rates among gram-negative organisms in children’s hospitals in general [36, 37] as well as lower mortality from sepsis [38, 39], the opportunity for early de-escalation and reduction in selection pressure is great. Expansion of rapid testing to include gram-negative resistance markers is critical.
This study has several limitations. First, it was performed at a single freestanding children’s hospital and our results may not be generalizable to other settings with different bacterial epidemiology and resistance patterns. Second, this was a retrospective study comparing trends over time and did not use randomization or concurrent controls. Finally, our focus was on utilization of select agents overall and in specific clinical circumstances; we did not examine time to effective or optimal therapy for patients individually. It is possible that we may have under-estimated the beneficial impact of rapid diagnostics for positive blood cultures as a result. There are other potential benefits to more rapid identification of bloodstream pathogens that remain unmeasured in our study as well as others. These include the ability to more rapidly direct other aspects of care, including diagnostic evaluation and source control. Future studies should attempt to incorporate endpoints that relate to these aspects of clinical care to more fully understand the impact of rapid testing. In addition, formal economic analysis should be performed to determine the economic value and cost-effectiveness of using these platforms. Despite these limitations, this is the largest study to our knowledge to evaluate the impact of rapid diagnostic testing on antibiotic prescribing for children with positive blood cultures and the first to our knowledge to evaluate two different rapid molecular methods with conventional culture-based methods over consecutive periods in the context of a mature ASP.
Conclusions
In conclusion, the introduction of rapid diagnostic testing for children with positive blood cultures results in faster time to organism identification. More importantly, such tests can be effective in decreasing unnecessary antimicrobial use and can influence prescribing of certain antibiotics for select organisms, particularly vancomycin use for MSSA and CoNS in certain clinical settings. Further expansion of these panels to include more comprehensive detection of gram-negative resistance mechanisms is critical to further improvements in antimicrobial use. Implementation of rapid diagnostic tests can have a powerful positive impact on care, even in the setting of a mature ASP, and the benefits are likely augmented by such a program.
Acknowledgements
The authors dedicate this paper to the memory of Chris Stockmann, who passed away after it was submitted, for his important contributions to this and many other studies.
This work was supported by an antimicrobial stewardship grant from Merck to Angela Fimbres Veesenmeyer, Adam L. Hersh, and Anne J. Blaschke. All authors had full access to all of the data in this study and take complete responsibility for the integrity of the data and accuracy of the data analysis.
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this manuscript, take responsibility for the integrity of the work as a whole, and have given final approval to the version to be published.
Disclosures
Adam L. Hersh received research funding from Agency for Healthcare Research and Quality, Centers for Disease Control and Prevention and Merck. Chris Stockmann has collaborated with BioFire Diagnostics on NIH and CDC funded studies. He also received grant support from the Thrasher Research Fund, the Primary Children’s Hospital Foundation, and Merck for work unrelated to this project. Andrew T. Pavia has collaborated with BioFire Diagnostics on NIH and CDC funded studies. Anne J. Blaschke has collaborated with BioFire Diagnostics on NIH and CDC funded studies. Anne J. Blaschke also has intellectual property in BioFire Diagnostics through the University of Utah, has received research funding from BioFire Diagnostics and bioMerieux, Inc., for investigator-initiated research, and has acted as a paid advisor to BioFire Diagnostics and BioFire Defense regarding risk assessment for FDA-cleared products. Anne J. Blaschke has received research funding from Gilead Sciences and Merck for investigator-initiated research. Angela Fimbres Veesenmeyer, Jared A. Olson, Kent Korgenski, and Emily A. Thorell declare no conflicts of interest.
Compliance with Ethics Guidelines
Approval for this study was obtained from the Institutional Review Board of the University of Utah and Primary Children’s Hospital (PCH) with a waiver of informed consent. All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1964, as revised in 2013.
Data Availability
The data sets used for the current study are available from the corresponding author on reasonable request.
Open Access
This article is distributed under the terms of the Creative Commons Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/), which permits any noncommercial use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Footnotes
Enhanced content
To view enhanced content for this article go to http://www.medengine.com/Redeem/8517F0600FFF30F6.
References
- 1.Caliendo AM, Gilbert DN, Ginocchio CC, et al. Better tests, better care: improved diagnostics for infectious diseases. Clin Infect Dis. 2013;57(Suppl 3):S139–S170. doi: 10.1093/cid/cit578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauer KA, West JE, Balada-Llasat JM, Pancholi P, Stevenson KB, Goff DA. An antimicrobial stewardship program’s impact with rapid polymerase chain reaction methicillin-resistant Staphylococcus aureus/S. aureus blood culture test in patients with S. aureus bacteremia. Clin Infect Dis. 2010;51(9):1074–1080. doi: 10.1086/656623. [DOI] [PubMed] [Google Scholar]
- 3.Carver PL, Lin SW, DePestel DD, Newton DW. Impact of mecA gene testing and intervention by infectious disease clinical pharmacists on time to optimal antimicrobial therapy for Staphylococcus aureus bacteremia at a University Hospital. J Clin Microbiol. 2008;46(7):2381–2383. doi: 10.1128/JCM.00801-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forrest GN, Mehta S, Weekes E, Lincalis DP, Johnson JK, Venezia RA. Impact of rapid in situ hybridization testing on coagulase-negative staphylococci positive blood cultures. J Antimicrob Chemother. 2006;58(1):154–158. doi: 10.1093/jac/dkl146. [DOI] [PubMed] [Google Scholar]
- 5.Galar A, Leiva J, Espinosa M, Guillen-Grima F, Hernaez S, Yuste JR. Clinical and economic evaluation of the impact of rapid microbiological diagnostic testing. J Infect. 2012;65(4):302–309. doi: 10.1016/j.jinf.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 6.Nguyen DT, Yeh E, Perry S, et al. Real-time PCR testing for mecA reduces vancomycin usage and length of hospitalization for patients infected with methicillin-sensitive staphylococci. J Clin Microbiol. 2010;48(3):785–790. doi: 10.1128/JCM.02150-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aitken SL, Hemmige VS, Koo HL, Vuong NN, Lasco TM, Garey KW. Real-world performance of a microarray-based rapid diagnostic for Gram-positive bloodstream infections and potential utility for antimicrobial stewardship. Diagn Microbiol Infect Dis. 2015;81(1):4–8. doi: 10.1016/j.diagmicrobio.2014.09.025. [DOI] [PubMed] [Google Scholar]
- 8.Beal SG, Ciurca J, Smith G, et al. Evaluation of the nanosphere verigene gram-positive blood culture assay with the VersaTREK blood culture system and assessment of possible impact on selected patients. J Clin Microbiol. 2013;51(12):3988–3992. doi: 10.1128/JCM.01889-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ibrahim EH, Sherman G, Ward S, Fraser VJ, Kollef MH. The influence of inadequate antimicrobial treatment of bloodstream infections on patient outcomes in the ICU setting. Chest. 2000;118(1):146–155. doi: 10.1378/chest.118.1.146. [DOI] [PubMed] [Google Scholar]
- 10.Dellit TH, Owens RC, McGowan JE, Jr, et al. Infectious Diseases Society of America and the Society for Healthcare Epidemiology of America guidelines for developing an institutional program to enhance antimicrobial stewardship. Clin Infect Dis. 2007;44(2):159–177. doi: 10.1086/510393. [DOI] [PubMed] [Google Scholar]
- 11.Fridkin S, Baggs J, Fagan R, et al. Vital signs: improving antibiotic use among hospitalized patients. MMWR Morb Mortal Wkly Rep. 2014;63(9):194–200. [PMC free article] [PubMed] [Google Scholar]
- 12.Blaschke AJ, Heyrend C, Byington CL, et al. Rapid identification of pathogens from positive blood cultures by multiplex polymerase chain reaction using the FilmArray system. Diagn Microbiol Infect Dis. 2012;74(4):349–355. doi: 10.1016/j.diagmicrobio.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quiles MG, Menezes LC, Bauab Kde C, et al. Diagnosis of bacteremia in pediatric oncologic patients by in-house real-time PCR. BMC Infect Dis. 2015;15:283. doi: 10.1186/s12879-015-1033-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taira CL, Okay TS, Delgado AF, Ceccon ME, de Almeida MT, Del Negro GM. A multiplex nested PCR for the detection and identification of Candida species in blood samples of critically ill paediatric patients. BMC Infect Dis. 2014;14:406. doi: 10.1186/1471-2334-14-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sullivan KV, Turner NN, Roundtree SS, et al. Rapid detection of Gram-positive organisms by use of the Verigene Gram-positive blood culture nucleic acid test and the BacT/Alert Pediatric FAN system in a multicenter pediatric evaluation. J Clin Microbiol. 2013;51(11):3579–3584. doi: 10.1128/JCM.01224-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng X, Polanco W, Carter D, Shulman S. Rapid identification of pathogens from pediatric blood cultures by use of the FilmArray blood culture identification panel. J Clin Microbiol. 2014;52(12):4368–4371. doi: 10.1128/JCM.02133-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagel JL, Huang AM, Kunapuli A, et al. Impact of antimicrobial stewardship intervention on coagulase-negative Staphylococcus blood cultures in conjunction with rapid diagnostic testing. J Clin Microbiol. 2014;52(8):2849–2854. doi: 10.1128/JCM.00682-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perez KK, Olsen RJ, Musick WL, et al. Integrating rapid diagnostics and antimicrobial stewardship improves outcomes in patients with antibiotic-resistant Gram-negative bacteremia. J Infect. 2014;69(3):216–225. doi: 10.1016/j.jinf.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 19.Perez KK, Olsen RJ, Musick WL, et al. Integrating rapid pathogen identification and antimicrobial stewardship significantly decreases hospital costs. Arch Pathol Lab Med. 2013;137(9):1247–1254. doi: 10.5858/arpa.2012-0651-OA. [DOI] [PubMed] [Google Scholar]
- 20.Felsenstein S, Bender JM, Sposto R, Gentry M, Takemoto C, Bard JD. Impact of a rapid blood culture assay for gram-positive identification and detection of resistance markers in a pediatric hospital. Arch Pathol Lab Med. 2016;140(3):267–275. doi: 10.5858/arpa.2015-0119-OA. [DOI] [PubMed] [Google Scholar]
- 21.Ray ST, Drew RJ, Hardiman F, Pizer B, Riordan A. Rapid identification of microorganisms by FilmArray blood culture identification panel improves clinical management in children. Pediatr Infect Dis J. 2016;35(5):e134–e138. doi: 10.1097/INF.0000000000001065. [DOI] [PubMed] [Google Scholar]
- 22.Messacar K, Hurst AL, Child J, et al. Clinical impact and provider acceptability of real-time antimicrobial stewardship decision support for rapid diagnostics in children with positive blood culture results. J Pediatr Infect Dis Soc. 2016 doi: 10.1093/jpids/piw047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salimnia H, Fairfax MR, Lephart PR, et al. Evaluation of the FilmArray blood culture identification panel: results of a multicenter controlled trial. J Clin Microbiol. 2016;54(3):687–698. doi: 10.1128/JCM.01679-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang AM, Newton D, Kunapuli A, et al. Impact of rapid organism identification via matrix-assisted laser desorption/ionization time-of-flight combined with antimicrobial stewardship team intervention in adult patients with bacteremia and candidemia. Clin Infect Dis. 2013;57(9):1237–1245. doi: 10.1093/cid/cit498. [DOI] [PubMed] [Google Scholar]
- 25.Forrest GN, Roghmann MC, Toombs LS, et al. Peptide nucleic acid fluorescent in situ hybridization for hospital-acquired enterococcal bacteremia: delivering earlier effective antimicrobial therapy. Antimicrob Agents Chemother. 2008;52(10):3558–3563. doi: 10.1128/AAC.00283-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lagace-Wiens PR, Adam HJ, Karlowsky JA, et al. Identification of blood culture isolates directly from positive blood cultures by use of matrix-assisted laser desorption ionization-time of flight mass spectrometry and a commercial extraction system: analysis of performance, cost, and turnaround time. J Clin Microbiol. 2012;50(10):3324–3328. doi: 10.1128/JCM.01479-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peters RP, Savelkoul PH, Simoons-Smit AM, Danner SA, Vandenbroucke-Grauls CM, van Agtmael MA. Faster identification of pathogens in positive blood cultures by fluorescence in situ hybridization in routine practice. J Clin Microbiol. 2006;44(1):119–123. doi: 10.1128/JCM.44.1.119-123.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Banerjee R, Teng CB, Cunningham SA, et al. Randomized trial of rapid multiplex polymerase chain reaction-based blood culture identification and susceptibility testing. J Clin Microbiol. 2015;61(7):1071–1080. doi: 10.1093/cid/civ447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim SH, Kim KH, Kim HB, et al. Outcome of vancomycin treatment in patients with methicillin-susceptible Staphylococcus aureus bacteremia. Antimicrob Agents Chemother. 2008;52(1):192–197. doi: 10.1128/AAC.00700-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schweizer ML, Furuno JP, Harris AD, et al. Comparative effectiveness of nafcillin or cefazolin versus vancomycin in methicillin-susceptible Staphylococcus aureus bacteremia. BMC Infect Dis. 2011;11:279. doi: 10.1186/1471-2334-11-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McMullan BJ, Bowen A, Blyth CC, et al. Epidemiology and mortality of Staphylococcus aureus bacteremia in Australian and New Zealand children. JAMA Pediatr. 2016;170(10):979–986. doi: 10.1001/jamapediatrics.2016.1477. [DOI] [PubMed] [Google Scholar]
- 32.Liu C, Bayer A, Cosgrove SE, et al. Clinical practice guidelines by the infectious diseases society of america for the treatment of methicillin-resistant Staphylococcus aureus infections in adults and children. Clin Infect Dis. 2011;52(3):e18–e55. doi: 10.1093/cid/ciq146. [DOI] [PubMed] [Google Scholar]
- 33.Beal SG, Thomas C, Dhiman N, et al. Antibiotic utilization improvement with the Nanosphere Verigene Gram-Positive blood culture assay. Proceedings 2015; 28(2):139–43. [DOI] [PMC free article] [PubMed]
- 34.Pardo J, Klinker KP, Borgert SJ, Butler BM, Giglio PG, Rand KH. Clinical and economic impact of antimicrobial stewardship interventions with the FilmArray blood culture identification panel. Diagn Microbiol Infect Dis. 2016;84(2):159–164. doi: 10.1016/j.diagmicrobio.2015.10.023. [DOI] [PubMed] [Google Scholar]
- 35.Buss BA, Schulz LT, Reed KD, Fox BC. MALDI-TOF Utility in a region with low antibacterial resistance rates. Clin Infect Dis. 2016;62(5):666–7. doi: 10.1093/cid/civ962. [DOI] [PubMed] [Google Scholar]
- 36.Logan LK, Braykov NP, Weinstein RA, Laxminarayan R, Program CDCEP Extended-spectrum beta-lactamase-producing and third-generation cephalosporin-resistant Enterobacteriaceae in Children: Trends in the United States, 1999–2011. J Pediatr Infect Dis Soc. 2014;3(4):320–328. doi: 10.1093/jpids/piu010. [DOI] [PubMed] [Google Scholar]
- 37.Larru B, Gong W, Vendetti N, et al. Bloodstream infections in hospitalized children: epidemiology and antimicrobial susceptibilities. Pediatr Infect Dis J. 2016;35(5):507–10. doi: 10.1097/INF.0000000000001057. [DOI] [PubMed] [Google Scholar]
- 38.Sievert DM, Ricks P, Edwards JR, et al. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009–2010. Infect Control Hosp Epidemiol. 2013;34(1):1–14. doi: 10.1086/668770. [DOI] [PubMed] [Google Scholar]
- 39.Klevens RM, Edwards JR, Gaynes RP, National Nosocomial Infections Surveillance S The impact of antimicrobial-resistant, health care-associated infections on mortality in the United States. Clin Infect Dis. 2008;47(7):927–930. doi: 10.1086/591698. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets used for the current study are available from the corresponding author on reasonable request.