Abstract
Purpose
The purpose of the present study is to understand Twist-related protein 1 (Twist1) spatiotemporal expression patterns and functions during early embryo development.
Methods
We performed whole-mount double immunofluorescence staining and reverse transcription (RT)-PCR analysis of the Twist1 protein and gene throughout the preimplantation development in mice.
Results
We determined that after compaction, the expression of Twist1 becomes developmentally differentiated and targeted in the inner cells of embryos. In blastocysts at E4.5, uniform staining of the inner cell mass was apparent, and it had been gradually translocated to the nucleus of hatched embryonic cells at E4.75. Furthermore, the effect of potential regulators of Twist on its expression level during blastocyst development was also sought. Accordingly, Twist1 expression appeared to be upregulated in both mRNA and protein level following culture of embryos in the presence of high glucose.
Conclusions
Our study revealed the dynamic Twist localization within the early stage of embryo. The results are discussed in terms of potential roles of Twist1 in the processes of lineage segregation, hatching, and implantation in post-compaction embryos and in blastocysts.
Keywords: Twist, Preimplantation, Mouse, Embryo
Introduction
Preimplantation development is a conserved process in mammalians and is vital for successful implantation and pregnancy. This period of development is characterized by development of the zygote through consecutive cleavage divisions, activation of embryonic transcription, and morphogenetic events, including compaction and cavitation, that result in the formation of the blastocyst [1]. During the mid-eight-cell stage, the mouse embryo undergoes a morphological restructuring process known as compaction. Specialization occurs, and distinct lineages can be distinguished within the developing blastocyst: the outer cells contribute to the trophectoderm (TE) lineage, whereas the inside cells from the inner cell mass (ICM) later segregate into the epiblast (EPI) and primitive endoderm (PE) lineages [1]. Around the fifth day, the embryo frees itself from the enveloping zona pellucida that prevents ectopic implantation during preimplantation. Through a series of expansion-contraction cycles in the embryo, thinning of ZP leads to focal rupture. This is immediately followed by extrusion of the embryo through the ruptured zona [2]. The mature blastocyst is then ready to implant; it adheres to the uterine wall, allowing the TE to invade. The peri-hatching blastocyst invariably exhibits extensive membranous extensions of the TE and exhibits undulating movements during zona escape [3]. One of the important aspects of blastocyst hatching is the involvement of regulatory molecules, primarily embryotrophic factors, which encompass second messengers, transcription factors, proteases, growth factors, and cytokines [4–7].
Members of the Twist family of basic helix-loop-helix (bHLH) factors comprise an evolutionarily conserved family of proteins that play a pivotal role in a number of essential developmental programs, including cell lineage determination and differentiation [8]. The Twist gene was originally identified in Drosophila as a recessive lethal mutation resulting in deficient mesoderm formation [9]. Twist has actually two different proteins—Twist-related protein 1 (Twist1) and Twist2—which share a high degree of homology [10]. Twist1 is the most studied partner and is also known as class A basic helix-loop-helix protein 38 (bHLHa38) encoded by the Twist1 gene. Studies previously shown that Twist1-null mouse embryos die at E11.5, displaying exencephaly, pharyngeal arch, and somitic defects, as well as defects in both cranial and cardiac neural crest cell populations [11, 12]. In addition to its role in embryogenesis, Twist1 plays important functions in cancer. Twist1 is expressed in mesenchymal cell populations, a transitional population present during development, which strikingly possesses many of the same migratory characteristics that cancer cells acquire during metastasis [8, 13]. Additionally, previous studies reported that high glucose concentration and epidermal growth factor (EGF)/EGFR signaling pathways induce Twist expression which indirectly leads to cellular invasion in cancer cells [14–16]
Although Twist expression and its essential role have been well defined in post-implantation development, very little is known about its expression pattern during the preimplantation period. The aim of the present study was to characterize more fully the expression pattern of Twist1 in mouse preimplantation embryos. Furthermore, the effect of the EGF and varying levels of excess glucose on the expression level of Twist1 was sought. In the present study, we used immunofluorescence staining analysis and reverse transcription (RT)-PCR analysis to investigate the stage-related expression and cellular localization of Twist1. Our results show that the Twist1 protein is present during preimplantation development and translocates to the nucleus with cellular movements that occur during the blastocyst’s hatching time. Our study results suggest that Twist1 has become a new player of lineage segregation, hatching, and implantation processes, with its functional expression after E4.5 confirmed in vitro and in vivo in mice.
Material and methods
Superovulation and embryo collection
Six-week-old BALB/c female and 12-week-old BALB/c male mice were maintained in the Experimental Animal Care and Production Unit of the School of Medicine at Akdeniz University. All protocols performed were approved by the Akdeniz University Institutional Animal Care and Use Committee (protocol no: 2011.09.65). Female mice were superovulated with an intraperitoneal (ip) injection of 5 IU of PMSG (Sigma-Aldrich), followed 48 h later by 5 IU of hCG (Sigma-Aldrich). Immediately after hCG injection, female mice were mated with 12-week-old BALB/c males, and the mating status was confirmed by the identification of a vaginal plug the following morning (considered as 0.5 dpc). Timing post-hCG was used to predict embryonic development, and two-cell embryos were obtained by puncturing the ampulla portion of the oviduct with a needle in HEPES-buffered media under the stereomicroscope (Zeiss). Embryos for the in vivo blastocyst study were obtained by flushing the uterine horns 96 h post-hCG.
Embryo culture treatment
Two-cell embryos were isolated in M2 medium containing 4 mg/mL−1 of bovine serum albumin (BSA), washed, pooled, and cultured in KSOM/0.25 % BSA (v/v) (Millipore) medium under mineral oil (Sigma) in 5 % CO2 at 37.5 °C. For studies of the Twist regulation by different microenvironments, morula-blastocyst stage embryos at E3.0 were cultured for 42 h in the presence of physiologic 5 mM d-glucose (control), 20 mM d-glucose (moderate), 52 mM d-glucose (high) (Sigma), and 20 ng/mL EGF (Endothelial Cell Systems). In vitro culture for each group was performed in 50-μL culture drops, overlaid with 3 mL of light mineral oil (Sigma-Aldrich) in a 35-mm culture dish (Becton–Dickinson).
RNA extraction and reverse transcription
RNA extraction and reverse transcription protocols were performed as described in our previous studies [17, 18]. Briefly, a pool of 30 blastocysts from each group was obtained and stored in lysis buffer at −80 °C until use. Total RNA was extracted using the RNAqueous-Micro Kit (Ambion) according to the manufacturer’s instructions. To eliminate DNA contamination, extracted RNA was treated with DNase I (Ambion). The RT reaction was performed using the RETROscript kit (Ambion) according to the manufacturer’s instructions. The samples were incubated with random decamer primers at 85 °C for 3 min to remove any secondary structures and were then incubated with an RT reaction composed of 2 μL of 10× RT buffer, 4 μL of 1.25 mM dNTP mix, 1 μL of RNase inhibitor (10 units/μL), and 1 μL of Moloney murine leukemia virus (MMLV)-RT (100 units/μL) at 44 °C for 1 h. Finally, the MMLV-RT enzyme was inactivated at 92 °C for 10 min.
PCR amplification
Amplifications were carried out with 35 cycles of PCR, during which the initial 5-min denaturation at 95 °C was followed by a “touchdown” program for 10 cycles at 92 °C for 20 s, 65 °C for 20 s (−1 °C per cycle), and 72 °C for 60 s and a final extension at 72 °C for 10 min, in a volume of 25-μL reaction mixture containing 10× PCR buffer (Qiagen, Valencia, CA, USA), 0.125 mM of each dNTP (Roche, Indianapolis, IN, USA), 0.5 μM of each primer (Keck Facility, CT, USA), and 2 units of Taq DNA polymerase (Qiagen, Valencia, CA, USA). The primer pairs were used to amplify for beta actin F: 5′-TGCGTGACATCAAAGAGAAG-3′, R: 5′-CGGATGTCAACGTCACACTT-3′, for twist F: 5′-ATG ATG CAG GAC GTG TCC AGC TCG CCA GTC TCG CCG G-3′, R:5′-GTG GGA CGC GGA CAT GGA CCA GGC CCC C-3′. All PCR products were separated on 1.5 % agarose/TBE gels and visualized by ethidium bromide staining. The relative gene expression levels were calculated using the ImageJ method. Thus, the relative Twist gene expressions in the embryos obtained from groups were reported as the fold change. Note that the RT-PCR reactions were established as triplicates and that this experiment was repeated twice.
Whole-mount indirect immunofluorescence staining
Staining protocol was performed as described in our previous study [18]. Briefly, embryos at the appropriate stage were first washed in 1 % BSA in PBS (0.1 M, pH 7.4) and were then fixed in 3 % paraformaldehyde (PFA) in PBS for 20 min at room temperature and washed three times in 1 % BSA in PBS. The fixed embryos were permeabilized and blocked by incubation for 1 h in 2 % BSA in PBS with 0.01 % Triton X-100 at room temperature. The embryos were then washed three times in 1 % BSA in PBS. The primary antibodies, rabbit anti-Twist (Santa Cruz) (1:100), mouse anti-Cdx2 (BioGenex) (1:200), and goat anti-Oct3/4 (Abcam) (1:100), were diluted in 1 % BSA in PBS with 0.01 % Tween 20 and incubated at 4 °C overnight. To detect the primary antibody, the embryos were incubated with anti-rabbit Alexa Flour 488-conjugated, anti-mouse Alexa Flour 555-conjugated, and anti-goat Alexa Flour 555-conjugated secondary antibodies (Invitrogen) diluted 1:350 in 1 % BSA in PBS with 0.01 % Tween 20 for 1 h at room temperature. To visualize DNA within nuclei, embryos were treated with 1 mg/mL (1:2000) DAPI for 10 min at room temperature. Fully processed embryos were mounted onto glass slides in 10 μL of a glycerol/PBS (1:1) mixture and analyzed with fluorescence microscopy (Olympus, Tokyo, Japan).
Statistical analysis
All results were first subjected to a normality test, and once passed, they were analyzed by a repeated measure ANOVA test followed by a Holm-Sidak test. All error bars represent standard error of the mean (SEM) values. A P value of <0.05 was considered significant. Statistical calculations were performed using SigmaStat for Windows, version 3.5 (Jandel Scientific Corp).
Results
Twist1 expression during preimplantation development
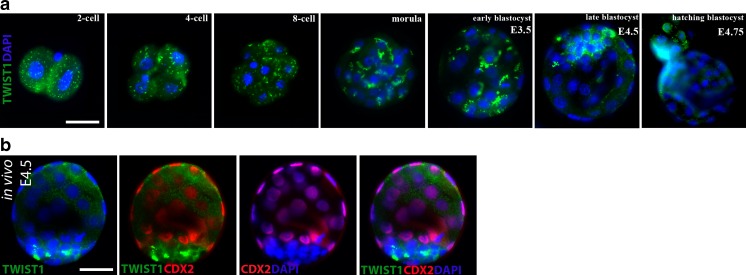
We first aimed to characterize the expression patterns of the Twist1 protein during mouse preimplantation development by immunofluorescence staining. Twist1 expression was observed similarly in the two-cell, four-cell, and eight-cell stages as dot-like reactions that were scattered in the cytoplasm (Fig. 1a). The scattered dot-like staining in two- to eight-cell stages began to clump in the cytoplasm of inner cells at both the morula and early blastocyst stage. Interestingly, the alteration of expression patterns was observed with the formation of morula as the inner cells displayed more concentrated areas of staining within their cytoplasm (Fig. 1a). During blastocyst formation, Twist1 was intensively present in the ICM (Fig. 1a). At the E4.5, Twist1 was distinctly visible in the cytoplasm and perinuclear areas of embryonic cells in the ICM layer of the fully expanded blastocysts, whereas the TE layer showed no expression (Fig. 1a). Moreover, we investigated the embryos at the blastocyst stage while hatching out of the zona pellucida (E4.75); significantly hatched embryonic cells displayed clear perinuclear and nuclear expression of Twist1 (Fig. 1a).
Fig. 1.
Cellular localization of Twist1 in mouse preimplantation embryos. a Representative images of Twist1 protein during in vitro embryo development (N = 10 embryos per group). Whereas cleavage stage (two- to eight-cell), morula, and early blastocyst stage (E3.5) embryos showed accumulated staining pattern in the cytoplasm, staining was visible in the cytoplasm and perinuclear areas of embryonic cells in ICM layer of late blastocyst at E4.5, and it had been gradually translocated to the nucleus of hatched embryonic cells at E4.75. b Representative images of double-labeled blastocysts obtained in vivo at E4.5 (N = 14 embryos). Twist1 was normally distributed in the cytoplasm and perinuclear areas of embryonic cells in the ICM layer, as shown in the control groups. The green signal indicates positive staining for Twist1, the red signal indicates positive staining for Oct43/4, and the blue signal (DAPI) indicates nuclei of the embryonic cells. Scale bar represents 25 μm
Differential expression of Twist1 in ICM layer at E4.5
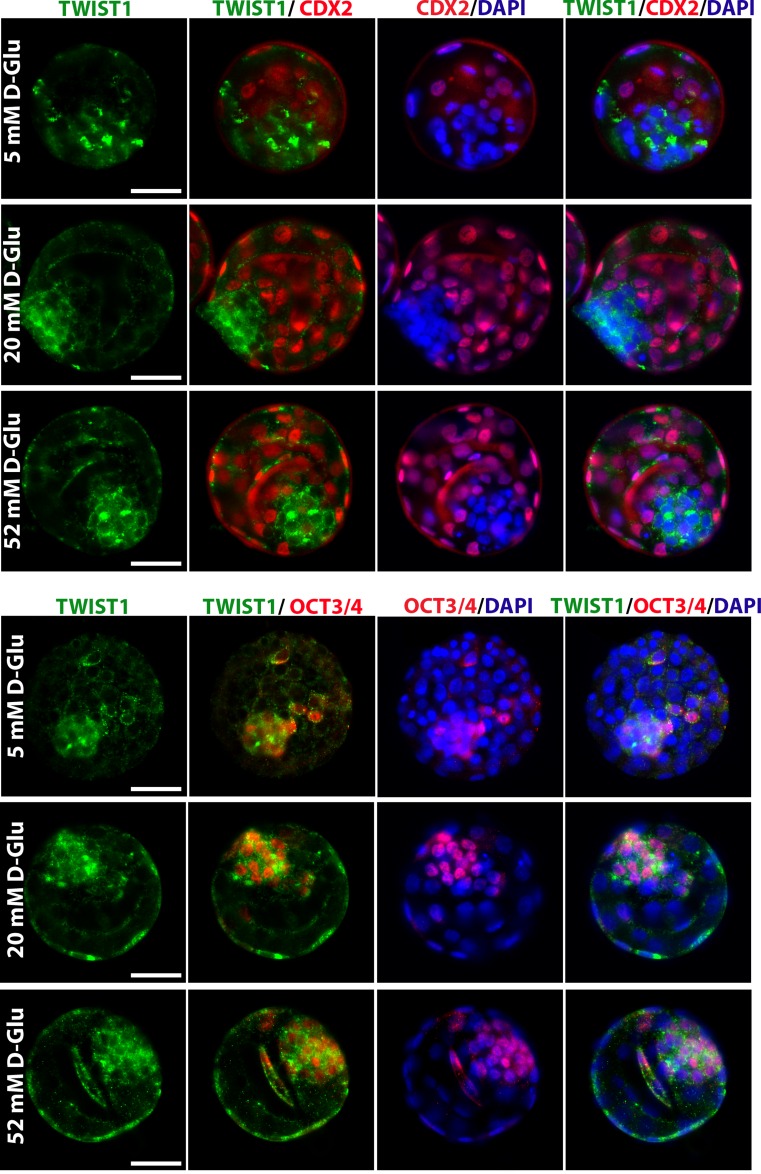
To confirm Twist1 expression, double immunofluorescence staining using a combination of Cdx2 as a TE layer marker or Oct3/4 as an ICM marker was performed in each blastocyst group (Figs. 1b and 2). The Twist1 signal clearly overlapped the Oct3/4 signal, whereas it did not overlap the Cdx2 signal, indicating that the expression was found in ICM cells (Figs. 1b and 2). We also performed another staining to detect Twist1 in blastocysts that were collected in vivo at E4.5 while floating in the endometrial cavity just before implantation. Twist1 expression is present in the cytoplasm and perinuclear areas of embryonic cells in ICM layer of blastocysts obtained in vivo, which is consistent with those obtained in vitro (Fig. 1b).
Fig. 2.
Twist1 protein expression in blastocysts at E4.5 which are exposed to varying levels of excess glucose. Representative images of double-labeled blastocysts after 42-h glucose treatment during morula-blastocyst transition (N = 10 embryos per group). Embryos treated with 5 mM (physiologic), 20 mM, and 52 mM glucose. The green signal indicates positive staining for Twist1, the red signal indicates positive staining for Oct43/4 or Cdx22, and the blue signal (DAPI) indicates nuclei of the embryonic cells. Scale bar represents 25 μm
The effect of the glucose and EGF supplementation on the expression level of Twist1
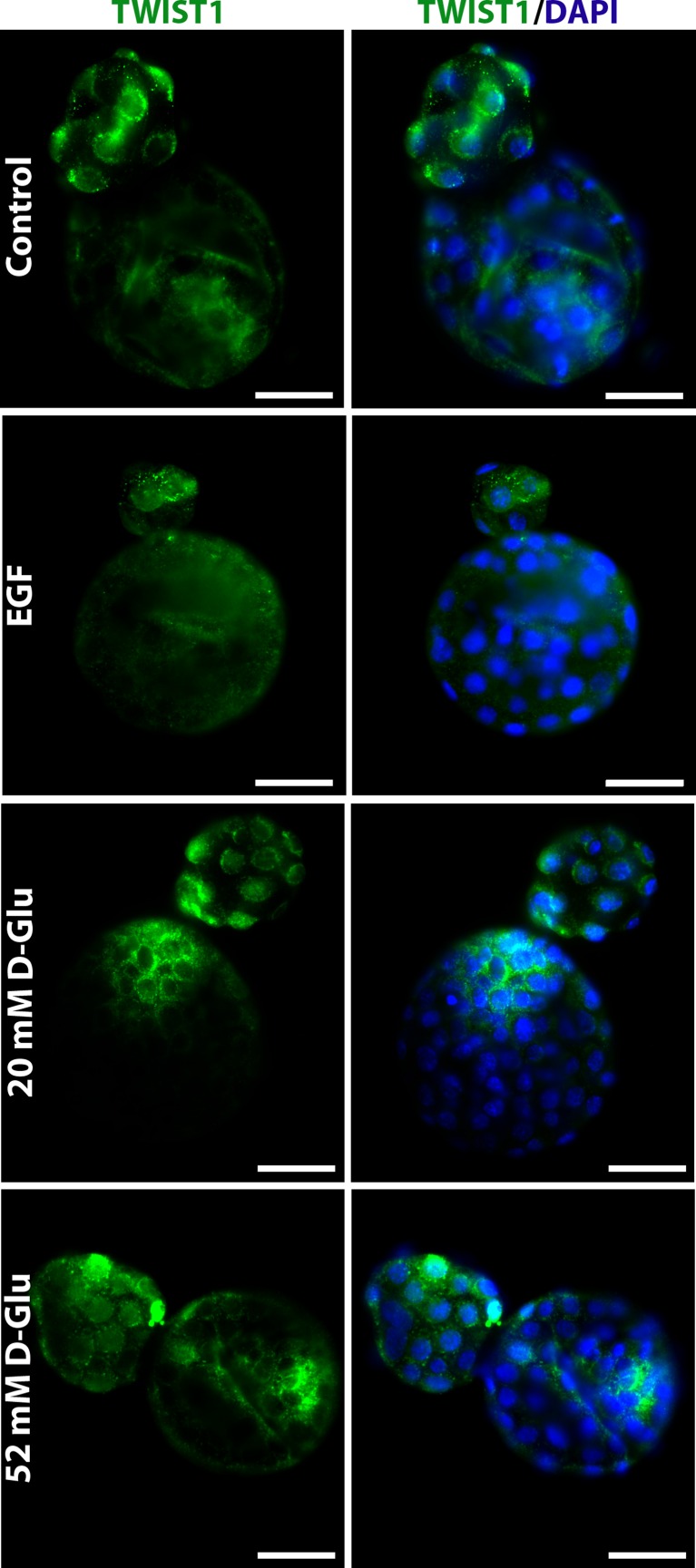
Because the protein and messenger RNA (mRNA) expression levels of Twist were found to be affected by high glucose concentration [14] and EGF stimulation [15], we next examined Twist1 protein expression in the presence of these potential regulators. That analysis revealed that the expression level of Twist1 protein was increased in blastocysts at E4.5 by the change of d-glucose concentration in the culture medium (Fig. 2). Also, increased nuclear shift of the Twist1 protein in hatched embryonic cells at E4.75 has been noticed in response to high glucose (Fig. 3). However, no expressional change of Twist1 protein was shown in blastocysts grown for 42 h in the presence of 20 ng/mL EGF and those in the control group grown in standard KSOM media up to the hatching blastocyst stage (E4.75) (Fig. 3).
Fig. 3.
Twist1 protein expression in hatching blastocysts. This analysis revealed that both the expression levels and nuclear shift of the Twist1 protein in hatched embryonic cells were increased by the change of d-glucose concentration in culture medium. However, no expressional change of Twist1 protein was shown in blastocysts after EGF supplementation of the culture media. The green signal indicates positive staining for Twist1, and the blue signal (DAPI) indicates nuclei of the embryonic cells. Scale bar represents 25 μm
Reverse transcription PCR analysis was performed to define the presence and the expression level of Twist1 mRNA in mouse blastocysts at E4.5 exposed to different microenvironments. RT-PCR analysis revealed that Twist1 mRNA was present in blastocyst stage embryos, and no significant difference was observed between the control, EGF, and 20 mM d-Glu groups. However, in the presence of high glucose concentration (52 mM d-Glu), Twist1 mRNA levels were noticeably increased in blastocysts compared with the other groups (P < 0.001) (Fig. 4).
Fig. 4.
Twist1 mRNA expression in mouse blastocysts. Significant differences were observed in the presence of high glucose concentration (52 mM d-Glu), where the Twist1 mRNA levels were remarkably increased in blastocysts compared with the other groups (*P < 0.001). N = 30 embryos per group. MDA-MB-231 (breast cancer cell line) was used as a positive control for Twist1. Cntrl control blastocyst, NC negative control
Discussion
Twist family bHLH proteins share a partially overlapping and expansive domain of expression throughout many stages of embryonic development. Twist1 function is associated with a variety of developmental processes at early organogenesis, including head mesenchyme, branchial and pharyngeal arches, somites, and cranial and cardiac neural crest cell development [11, 19]. Twist has been considered, therefore, to be a mesoderm-determining factor and thus of great interest in the study of mesoderm determination and specification. However, little is known about Twist1 expression in preimplantation embryo development. Therefore, we evaluated the expression pattern of Twist1 protein throughout preimplantation embryo development in mice. Using potential regulators of the Twist transcription factor, including glucose and EGF, Twist1 regulation at the gene and protein levels was also analyzed during blastocyst development. The current study provides the first evidence that the Twist1 protein is present throughout preimplantation and is differentially expressed and localized after the formation of the blastocyst lineages at E4.5. Furthermore, our results also revealed that the Twist1 protein becomes functional at the time of blastocyst development when it is found in perinuclear and nuclear areas and translocates to the nucleus around the time of hatching. Differential expression of the Twist1 protein in the ICM layer indicates that this transcript might be important for lineage commitment. Moreover, translocation of Twist1 to the nucleus around the time of hatching indicates the possible role of Twist1 in the process of blastocyst hatching and/or implantation.
In the present study, we first observed that whereas cleavage stage (two- to eight-cell), morula (E2.5), and early blastocyst stage (E3.5) embryos showed an accumulated staining pattern in the cytoplasm, staining was visible in the cytoplasm and perinuclear areas of embryonic cells in ICM layer of late blastocysts at E4.5; it had gradually been translocated to the nucleus of hatched embryonic cells at E4.75. A number of studies have shown previously that the functional Twist is present in perinuclear and nuclear areas of the cells [20]. In light of the literature, we think that Twist1 is not functional before the late blastocyst stage at E4.5, and after that stage, it becomes functional by localizating and concentrating in perinuclear and nuclear areas. However, its function at this location is currently unknown. Possibly, Twist1 requires for transcriptional activation of cellular invasion-related molecules such as N-cadherin, fibronectin, and vimentin that they will be in or close to the nuclei [21, 22].
The TE, the first differentiated cell lineage in the embryo, forms the outer layer of a blastocyst as an epithelial sheet enclosing the ICM. Whereas the ICM retains pluripotency, the TE is restricted: it undergoes epithelialization and plays essential roles in implantation. It interacts with the decidualized maternal uterus and later contributes to the fetal portions of the placenta [1]. In the mouse embryo, epithelium formation starts as compaction at the late 8-cell stage and continues for the next 24 h to completion at around the 32-cell stage. During compaction, intercellular adhesion complexes are gradually assembled, mainly through the post-translational activation of E-cadherin [23]. Whereas synthesis, accumulation, and membrane assembly of E-cadherin occur throughout cleavage, during compaction, functional E-cadherin expression is the first essential step of the TE formation. E-cadherin is required for the formation of stable adherin junctions and thus for the maintenance of the epithelial phenotype. On the other hand, Li has previously shown that Twist inhibits E-cadherin expression and increases the expression of matrix metalloproteinase 9 (MMP9) in peritoneal mesothelial cells [14]. This suggests two possibilities that will require further research. First, Twist1 downregulation in outer cells may be related to strong E-cadherin expression, polarity complexes, and the epithelialization process of the TE layer. Second, at hatching time, Twist1 translocates to the nucleus where it becomes functional and could have direct action on the MMP9, allowing cellular invasion during implantation.
Many previous studies have defined Twist as a mesoderm-determining factor. Twist1 has been located as follows: at E7.5 in the head mesenchyme, somites, and lateral plate mesoderm [24]; at E7.5 in the head mesenchyme and first pharyngeal arch [24]; at E9.5 in the cranial mesenchyme, sclerotomal mesoderm, somatopleuric mesoderm [19, 24, 25]; and between E16 and E18 in the cranial mesenchyme in the primordial of the tooth mesenchyme [8, 24]. It is known that embryonic derivatives of the EPI form the mesoderm. Thus, we believe that the differential staining of the Twist1 protein in the ICM layer that appeared in our results is not surprising and is consistent with previous studies.
Formation of the ICM in mouse embryos is dependent upon FGF signaling [26]. The level of FGF signal in the ICM directly or indirectly influences levels of Gata6 and Nanog expression, the classic PE and EPI specifiers, respectively [26, 27]. A number of studies have shown previously that the Twist function is essential for maintaining FGF signaling activity in mice [28, 29]. Twist and FGF signaling act in the same morphogenetic networks during organogenesis of post-implantation embryos [29]. Strikingly, we have determined that functional Twist1 expression is also coincident with the ICM layer specification after E3.5. That result may suggest the possible role of Twist in the ICM lineage differentiation and commitment process for regulation of the FGF signaling.
EGF is one of the key molecules that play different roles in the regulation of embryo development. EGF contributes to blastocyst formation by increasing rates of ICM and TE cells at all stages of development and also regulates the differentiation of embryo cells after the morula stage [30–32]. As known, EGF and its signaling significantly increase mRNA and protein levels of Twist1 in human cancer cells and cause tumor progression and metastasis [15]. Therefore, we aimed to investigate the possible effects of Twist1 on preimplantation embryo development by EGF stimulation. Our observation of no statistically significant expressional change of Twist1 both in mRNA and protein levels after EGF supplementation suggests that EGF activity has no influence on the expression of Twist during the earliest stage of development. However, to precisely define whether there is any functional effect of EGF and its signaling network on the Twist1 protein, further studies are needed.
Twist expression is known to be induced by glucose; however, it is unclear whether it is by glucose directly or a by secondary mediator. Li demonstrated that Twist expression is increased by high glucose concentration in mesothelial cell cultures [14]. Although glucose is not used as an energy source in the early preimplantation embryos in mice, it becomes the major energy substrate at the time of compaction: blastocysts have an absolute requirement for glucose to maintain normal proliferative, differentiative, and metabolic development [33–35]. In the present study, the observation of functional Twist expression during blastocyst development was consistent with increased glucose uptake at this stage. Our findings, which displayed developmentally differentiated, targeted expression of Twist1, may be a result of increasing intraembryonic glucose concentrations in post-compaction embryos. Additionally, due to the importance of glucose in the regulation of Twist expression, we investigated its mRNA and protein level in the blastocysts exposed to different glucose levels in vitro. Following the glucose exposure, increased Twist expression at both the mRNA and protein level suggests that the amount of glucose in the environment is critical to baseline Twist levels in mouse blastocysts. Also, results show that glucose might be involved in the regulation of Twist expression throughout early embryogenesis and that glucose-mediating Twist primarily acts on blastocyst development—possibly through regulating gene expression.
In conclusion, developmentally differentiated, targeted expression of Twist1 has been demonstrated in mouse preimplantation embryos. This study is the first report of the expression pattern of the Twist1 gene and protein in early mouse embryos. We propose that an increase in glucose uptake after compaction is directly related to functional Twist1 expression and that subsequent localization of Twist1 in the ICM layer is associated with lineage differentiation in the blastocyst, which might then induce mesoderm differentiation in the later stages of development. Future studies will reveal further roles for Twist during preimplantation development.
Acknowledgments
The authors would like to acknowledge Prof. Dr. Nidai Ozes (PhD) for his kind support to this work.
Footnotes
Capsule Overall, our results provide important insight into the expression pattern of Twist1, which displayed significant temporal and spatial differences during the earliest stages of development.
Berna Sozen and Suray Pehlivanoglu contributed equally to this work.
References
- 1.Sozen B, Can A, Demir N. Cell fate regulation during preimplantation development: a view of adhesion-linked molecular interactions. Dev Biol. 2014;395:73–83. doi: 10.1016/j.ydbio.2014.08.028. [DOI] [PubMed] [Google Scholar]
- 2.Lin SP, Lee RK, Tsai YJ. In vivo hatching phenomenon of mouse blastocysts during implantation. J Assist Reprod Genet. 2001;18:341–345. doi: 10.1023/A:1016640923269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seshagiri PB, Sen Roy S, Sireesha G, Rao RP. Cellular and molecular regulation of mammalian blastocyst hatching. J Reprod Immunol. 2009;83:79–84. doi: 10.1016/j.jri.2009.06.264. [DOI] [PubMed] [Google Scholar]
- 4.Kane MT, Morgan PM, Coonan C. Peptide growth factors and preimplantation development. Hum Reprod Update. 1997;3:137–157. doi: 10.1093/humupd/3.2.137. [DOI] [PubMed] [Google Scholar]
- 5.Sargent IL, Martin KL, Barlow DH. The use of recombinant growth factors to promote human embryo development in serum-free medium. Hum Reprod. 1998;13(Suppl 4):239–248. doi: 10.1093/humrep/13.suppl_4.239. [DOI] [PubMed] [Google Scholar]
- 6.Seshagiri PB, Mishra A, Ramesh G, Rao RP. Regulation of peri-attachment embryo development in the golden hamster: role of growth factors. J Reprod Immunol. 2002;53:203–213. doi: 10.1016/S0165-0378(01)00086-9. [DOI] [PubMed] [Google Scholar]
- 7.Simon C, Gimeno MJ, Mercader A, Frances A, Garcia Velasco J, Remohi J, et al. Cytokines-adhesion molecules-invasive proteinases. The missing paracrine/autocrine link in embryonic implantation? Mol Hum Reprod. 1996;2:405–424. doi: 10.1093/molehr/2.6.405. [DOI] [PubMed] [Google Scholar]
- 8.Barnes RM, Firulli AB. A twist of insight—the role of Twist-family bHLH factors in development. Int J Dev Biol. 2009;53:909–924. doi: 10.1387/ijdb.082747rb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simpson P. Maternal-zygotic gene interactions during formation of the dorsoventral pattern in Drosophila embryos. Genetics. 1983;105:615–632. doi: 10.1093/genetics/105.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, Wang W, Yang R, Wang T, Su T, Weng D, Tao T, Li W, Ma D, Wang S. Correlation of TWIST2 up-regulation and epithelial-mesenchymal transition during tumorigenesis and progression of cervical carcinoma. Gynecol Oncol. 2012b 124; 112-118. [DOI] [PubMed]
- 11.Soo K, O’Rourke MP, Khoo PL, Steiner KA, Wong N, Behringer RR, et al. Twist function is required for the morphogenesis of the cephalic neural tube and the differentiation of the cranial neural crest cells in the mouse embryo. Dev Biol. 2002;247:251–270. doi: 10.1006/dbio.2002.0699. [DOI] [PubMed] [Google Scholar]
- 12.Vincentz JW, Barnes RM, Rodgers R, Firulli BA, Conway SJ, Firulli AB. An absence of Twist1 results in aberrant cardiac neural crest morphogenesis. Dev Biol. 2008;320:131–139. doi: 10.1016/j.ydbio.2008.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Acloque H, Thiery JP, Nieto MA. The physiology and pathology of the EMT. Meeting on the epithelial-mesenchymal transition. EMBO Rep. 2008;9:322–326. doi: 10.1038/embor.2008.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li C, Ren Y, et al. Twist overexpression promoted epithelial-to-mesenchymal transition of human peritoneal mesothelial cells under high glucose. Nephrol Dial Transplant. 2012;27(11):4119–4124. doi: 10.1093/ndt/gfs049. [DOI] [PubMed] [Google Scholar]
- 15.Lo HW, Hsu SC, et al. Epidermal growth factor receptor cooperates with signal transducer and activator of transcription 3 to induce epithelial-mesenchymal transition in cancer cells via up-regulation of TWIST gene expression. Cancer Res. 2007;67(19):9066–9076. doi: 10.1158/0008-5472.CAN-07-0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Margetts PJ. Twist: a new player in the epithelial-mesenchymal transition of the peritoneal mesothelial cells. Nephrol Dial Transplant. 2012;27(11):3978–3981. doi: 10.1093/ndt/gfs172. [DOI] [PubMed] [Google Scholar]
- 17.Ozturk S, Yaba-Ucar A, Sozen B, Mutlu D, Demir N. Superovulation alters embryonic poly(A)-binding protein (Epab) and poly(A)-binding protein, cytoplasmic 1 (Pabpc1) gene expression in mouse oocytes and early embryos. Reprod Fertil Dev. 2014;28(3):375–83. doi: 10.1071/RD14106. [DOI] [PubMed] [Google Scholar]
- 18.Sozen B, Ozturk S, Yaba A, Demir N. The p38 MAPK signalling pathway is required for glucose metabolism, lineage specification and embryo survival during mouse preimplantation development. Mech Dev. 2015;138 Pt(3):375–98. doi: 10.1016/j.mod.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 19.Gitelman I. Twist protein in mouse embryogenesis. Dev Biol. 1997;189:205–214. doi: 10.1006/dbio.1997.8614. [DOI] [PubMed] [Google Scholar]
- 20.Brunet T, Bouclet A, Ahmadi P, Mitrossilis D, Driquez B, Brunet AC, et al. Evolutionary conservation of early mesoderm specification by mechanotransduction in Bilateria. Nat Commun. 2013;4:2821. doi: 10.1038/ncomms3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang Y, Massague J. Epithelial-mesenchymal transitions: twist in development and metastasis. Cell. 2004;118:277–279. doi: 10.1016/j.cell.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 22.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 23.Vestweber D, Gossler A, Boller K, Kemler R. Expression and distribution of cell adhesion molecule uvomorulin in mouse preimplantation embryos. Dev Biol. 1987;124:451–456. doi: 10.1016/0012-1606(87)90498-2. [DOI] [PubMed] [Google Scholar]
- 24.Fuchtbauer EM. Expression of M-twist during postimplantation development of the mouse. Dev Dyn. 1995;204:316–322. doi: 10.1002/aja.1002040309. [DOI] [PubMed] [Google Scholar]
- 25.Wolf C, Thisse C, Stoetzel C, Thisse B, Gerlinger P, Perrin-Schmitt F. The M-twist gene of Mus is expressed in subsets of mesodermal cells and is closely related to the Xenopus X-twi and the Drosophila twist genes. Dev Biol. 1991;143:363–373. doi: 10.1016/0012-1606(91)90086-I. [DOI] [PubMed] [Google Scholar]
- 26.Yamanaka Y, Lanner F, Rossant J. FGF signal-dependent segregation of primitive endoderm and epiblast in the mouse blastocyst. Development. 2010;137:715–724. doi: 10.1242/dev.043471. [DOI] [PubMed] [Google Scholar]
- 27.Gasperowicz M, Natale DR. Establishing three blastocyst lineages—then what? Biol Reprod. 2011;84:621–630. doi: 10.1095/biolreprod.110.085209. [DOI] [PubMed] [Google Scholar]
- 28.O’Rourke MP, Soo K, Behringer RR, Hui CC, Tam PP. Twist plays an essential role in FGF and SHH signal transduction during mouse limb development. Dev Biol. 2002;248:143–156. doi: 10.1006/dbio.2002.0730. [DOI] [PubMed] [Google Scholar]
- 29.Zuniga A, Quillet R, Perrin-Schmitt F, Zeller R. Mouse Twist is required for fibroblast growth factor-mediated epithelial-mesenchymal signalling and cell survival during limb morphogenesis. Mech Dev. 2002;114:51–59. doi: 10.1016/S0925-4773(02)00048-5. [DOI] [PubMed] [Google Scholar]
- 30.Dadi TD, Li MW, Lloyd KC. Decreased growth factor expression through RNA interference inhibits development of mouse preimplantation embryos. Comp Med. 2009;59:331–338. [PMC free article] [PubMed] [Google Scholar]
- 31.Martin KL, Barlow DH, Sargent IL. Heparin-binding epidermal growth factor significantly improves human blastocyst development and hatching in serum-free medium. Hum Reprod. 1998;13:1645–1652. doi: 10.1093/humrep/13.6.1645. [DOI] [PubMed] [Google Scholar]
- 32.Terada A, Minoura H, Toyoda N. Effects of epidermal growth factor on preimplantation mouse embryos. J Assist Reprod Genet. 1997;14:404–411. doi: 10.1007/BF02766149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gardner DK, Leese HJ. The role of glucose and pyruvate transport in regulating nutrient utilization by preimplantation mouse embryos. Development. 1988;104:423–429. doi: 10.1242/dev.104.3.423. [DOI] [PubMed] [Google Scholar]
- 34.Gardner DK, Pool TB, Lane M. Embryo nutrition and energy metabolism and its relationship to embryo growth, differentiation, and viability. Semin Reprod Med. 2000;18:205–218. doi: 10.1055/s-2000-12559. [DOI] [PubMed] [Google Scholar]
- 35.Pantaleon M, Scott J, Kaye PL. Nutrient sensing by the early mouse embryo: hexosamine biosynthesis and glucose signaling during preimplantation development. Biol Reprod. 2008;78:595–600. doi: 10.1095/biolreprod.107.062877. [DOI] [PubMed] [Google Scholar]