Abstract
RNA-binding proteins (RBPs) acting at various steps in the post-transcriptional regulation of gene expression play crucial roles in neuronal development and synaptic plasticity. Genetic mutations affecting several RBPs and associated factors lead to diverse neurological symptoms, as characterized by neurodevelopmental and neuropsychiatric disorders, neuromuscular and neurodegenerative diseases, and can often be multisystemic diseases. We will highlight the physiological roles of a few specific proteins in molecular mechanisms of cytoplasmic mRNA regulation, and how these processes are dysregulated in genetic disease. Recent advances in computational biology and genomewide analysis, integrated with diverse experimental approaches and model systems, have provided new insights into conserved mechanisms and the shared pathobiology of mRNA dysregulation in disease. Progress has been made to understand the pathobiology of disease mechanisms for myotonic dystrophy, spinal muscular atrophy, and fragile X syndrome, with broader implications for other RBP-associated genetic neurological diseases. This gained knowledge of underlying basic mechanisms has paved the way to the development of therapeutic strategies targeting disease mechanisms.
Keywords: Fragile X Mental Retardation Protein (FMRP), Fragile X Syndrome (FXS), Muscleblind-like Splicing Regulator (MBNL), Myotonic Dystrophy (DM), RNA Binding Protein Fox-1 Homolog 1 (RBFOX1), Spinal Muscular Atrophy (SMA), Survival of Motor Neuron (SMN)
Dysregulation of mRNP assembly and localization in spinal muscular atrophy (SMA)
The assembly of mRNA-binding proteins (mRBPs) and mRNAs into messenger ribonucleoproteins (mRNPs) determines the fate of the transcripts during all steps of post-transcriptional regulation, including its localization and translation. Recent studies have shown that both hyperassembly or hypoassembly of various RNPs can lead to human neurodegenerative diseases, including spinal muscular atrophy (SMA) and amyotrophic lateral sclerosis (ALS) (Shukla and Parker, 2016). Although there has been a lot of progress to understand basic principles of RNP hyperassembly and formation of persistent stress granules, it is still not clear how the proper assembly of specific transport mRNPs is regulated (Li et al., 2013). The molecular machinery that brings together transcripts with a specific set of proteins that regulate their translocation and local translation remains unknown.
In the case of spliceosomal small nuclear ribonucleoproteins (snRNPs), it has been shown that the faithful assembly of Sm proteins into a hepatameric complex on the uridine-rich snRNAs depends on the activity of a multiprotein assemblysome consisting of SMN and associated gemin proteins (Meister et al., 2001; Pellizzoni et al., 2002). Reduced SMN protein levels cause SMA, which is characterized by axonal dying back of spinal motor neurons leading to muscular atrophy and typically death in early childhood. While widespread splicing defects occur in all SMA models characterized thus far (Gabanella et al., 2007; Zhang et al., 2008, 2013; Lotti et al., 2012), the role of these defects in the disease process may very well represent a secondary nonspecific effect of neurodegeneration, and their significance for SMA pathogenesis remains to be fully elucidated (Bäumer et al., 2009).
The interaction of SMN with diverse mRBPs and its localization to mobile RNA transport granules in axons in vitro and in vivo (Dombert et al., 2014; Hao le et al., 2015) has led to the hypothesis that SMN may have a noncanonical role in axonal mRNA metabolism that may explain the vulnerability of motor neurons to reduced SMN protein levels (Briese et al., 2005; Rossoll and Bassell, 2009; Fallini et al., 2012). Work from several laboratories has demonstrated SMN-dependent defects in the localization and local translation of axonal mRNAs and mRBPs. Since the discovery of defective axonal localization of β-actin mRNA and protein in SMA motor neurons (Rossoll et al., 2003), SMN has been found to regulate the axonal localization of transcripts encoding growth-associated protein 43 (GAP43) and neuritin/cpg15 (Akten et al., 2011; Fallini et al., 2011, 2014, 2016; Hubers et al., 2011; Sanchez et al., 2013) (Fig. 1). This mislocalization is accompanied by reduced local translation in axonal growth cones and can be rescued by overexpression of the SMN-interacting mRBPs HuD and IMP1/ZBP1 (Fallini et al., 2016). Based on these findings, a more general role for SMN in RNP complex assembly that goes beyond its well-characterized function in snRNP assembly and splicing, including a critical role in the assembly of mRNP transport granules, appears likely (Li et al., 2014; Donlin-Asp et al., 2016; So et al., 2016).
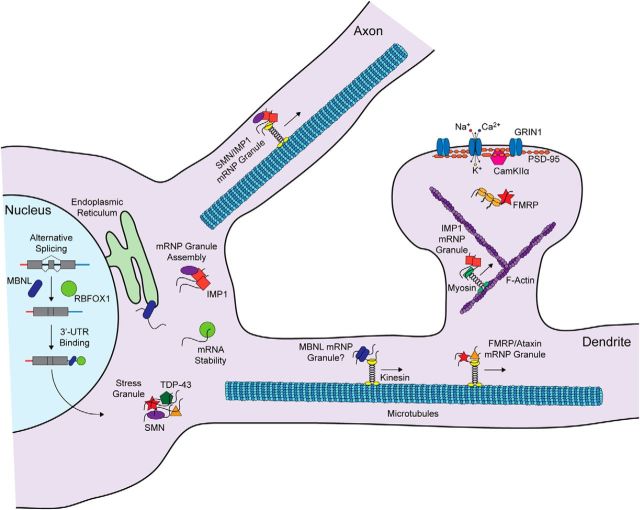
Figure 1.
Spatiotemporal regulation of RNA processing and local translation in neurons by RNA-binding proteins. In the nucleus, MBNL and RBFOX proteins regulate splicing and bind 3′-UTR sequences. Both proteins coregulate many transcripts. In the cytoplasm, MBNL binds to the rough endoplasmic reticulum (RER) where it facilitates the synthesis of membrane proteins, and also associates with the cytoskeleton to regulate mRNA localization. The cytoplasmic isoform of RBFOX1 regulates mRNA stability. Several RNA-binding proteins (e.g., FMRP, Ataxin, TDP-43, and Smn) are associated with stress granules. FMRP and Imp1 regulate mRNA granule transport by kinesin and local translation in dendritic spines. Imp1 also regulates mRNA localization in axons, and Smn plays a role in the assembly of Imp1 RNA transport granules. RNA localization and translation in spines may involve myosin motor and anchoring to F-actin. Locally synthesized proteins include glutamate receptor subunits, components of the postsynaptic density, and signaling proteins.
An open question remains what is the scope of snRNP and mRNP assembly defects in SMA in vivo, and how these defects contribute to the specific motor neuron degeneration observed in SMA. In a disease of general RNP hypoassembly, one can expect widespread downstream effects on various post-transcriptional regulation mechanisms (Donlin-Asp et al., 2016; Shukla and Parker, 2016). It will be interesting to find out whether rescuing mRNP assembly and localization defects can mitigate the SMA disease phenotype. Future work will need to show which defects are most relevant for the SMA pathogenesis and will provide insight into potential therapeutic strategies that target RNP assembly.
Global approaches for studying RNA dysregulation in myotonic dystrophy
The dominantly inherited neuromuscular disease myotonic dystrophy (DM) is caused by an expanded CTG (DM1) or CCTG (DM2) microsatellite repeat in the 3′-UTR of the dystrophia myotonica protein kinase gene, or the first intron of the CCHC-type zinc finger nucleic acid binding protein, respectively (Cooper, 2009; Lee and Cooper, 2009). These repeats are transcribed into RNA and sequester members of the Muscleblind-like (MBNL) family of RNA-binding proteins into nuclear foci (Fig. 1) (Miller et al., 2000; Cooper, 2009). MBNL proteins are deeply evolutionarily conserved and are required for the terminal differentiation of a number of cell types, including neurons, skeletal muscle, and cardiac muscle (Begemann et al., 1997; Artero et al., 1998). They have been most extensively studied in the context of alternative splicing regulation, as they are required for the transition from fetal-to-adult isoform expression throughout development (Pascual et al., 2006). Loss of MBNL function in DM leads to numerous missplicing events (Kanadia et al., 2003a, b), some of which have been linked to specific phenotypes in DM, such as Chloride Channel 1 (myotonia) (Charlet et al., 2002), Insulin receptor (insulin resistance) (Savkur et al., 2001), Bridging integrator 1 (muscle weakness) (Fugier et al., 2011), CaV1.1 calcium channel (muscle weakness) (Tang et al., 2012), sodium channel 5a (cardiac arrhythmia) (Wahbi et al., 2013; Freyermuth et al., 2016), and cardiac troponin T (cardiac arrhythmia) (Philips et al., 1998). These changes all essentially reflect shifts in expression of adult-to-fetal isoforms. Recent studies in human DM1 and mouse models reveal missplicing of numerous mRNAs encoding proteins that play important cytoskeletal and synaptic functions (Charizanis et al., 2012; Goodwin et al., 2015). A key open question is whether specific missplicing events are linked to specific CNS symptoms, which include profound hypersomnia and intellectual disability (Thornton, 2014).
Although a role for MBNLs in regulating alternative splicing is well established, previous studies have also indicated a role for MBNLs in regulating mRNA localization and local translation. MBNL2 was shown to interact with the 3′-UTR of Integrin α3 and carry it to the plasma membrane, where it is locally translated (Adereth et al., 2005). MBNL2 has also been suggested to enhance the stability of mRNAs encoding extracellular matrix components (Du et al., 2010), and MBNL1 has been implicated in the regulation of message stability (Wang et al., 2015; Masuda et al., 2012). Using a combination of genomics and biochemical approaches, MBNL1 was shown to be a global regulator of RNA localization and membrane-associated translation. RNAseq of subcellular compartments from mouse myoblasts showed that 3′-UTR CLIP targets of MBNL1 relocalize toward the insoluble and away from the membrane compartments, following depletion of MBNL1 and 2 (E. T. Wang et al., 2012). Many of the dysregulated mRNAs were found to encode secreted proteins, extracellular matrix components, and proteins involved in cell–cell communication and synapse function, with potential implications for neuromuscular junctions in muscle. Ribosome footprinting of myoblasts depleted of MBNL1/2 also indicated decreased translation of those mRNAs whose normal localization to the membrane compartment was relocalized toward the insoluble compartment. MBNL1 and MBNL2 CLIPseq in mouse and human brain corroborate many MBNL binding sites in the 3′-UTRs of numerous mRNAs encoding synapse and cytoskeletal proteins (E. T. Wang et al., 2012).
These observations raise a number of outstanding questions that require further investigation to answer. Whether regulation of RNA localization by MBNLs is achieved through diffusion/anchoring and/or active transport along cytoskeletal filaments is unknown. The specific membrane and insoluble compartments from which MBNL targets are relocalized are also unclear, although the rough endoplasmic reticulum likely comprises a large fraction of the membrane compartment. The role of MBNLs in regulating mRNA localization and translation in fully differentiated tissues, such as muscle and neurons, has also not been fully explored; it is likely that localization patterns are more complex in tissues than in cell culture, and possible that mislocalization of mRNAs in MBNL-depleted tissues has significant physiological consequences. The mRNAs whose splicing patterns are regulated by MBNLs also tend to exhibit 3′-UTR binding and regulation of localization by MBNLs; whether this coordination exists for functional or biophysical reasons has not been addressed. Similar questions could be asked of additional RNA-binding proteins implicated in neurological disease, and further investigation may yield general principles that apply to regulation of RNA localization and local translation across other RBPs and other diseases.
Gene-distal 3′-UTR sequences often regulate mRNA localization
Although several hundred transcripts are preferentially localized to neuronal projections (Taylor et al., 2009; Zivraj et al., 2010; Gumy et al., 2011; Cajigas et al., 2012; Minis et al., 2013), for the overwhelming majority of these transcripts, the RNA sequences within them that drive their localization are unknown. In cases where the localizing sequence is known, it is often located in the 3′-UTR of the transcript (Andreassi and Riccio, 2009). Still, 3′-UTRs are often >1 kb in length, making identification of localization elements difficult.
If multiple transcript isoforms are expressed from a single gene locus, however, then this problem becomes more manageable, especially if the isoforms display differing localization patterns. By comparing sequence elements present in localization-competent isoforms with those in localization-incompetent isoforms, the search space for relevant features can be considerably narrowed (Taliaferro et al., 2016). Given that many known RNA localization elements lie in 3′-UTRs, this effect is, perhaps not surprisingly, often most apparent in genes that express isoforms that differ in 3′-UTR composition.
For example, during Drosophila oocyte development, two isoforms of the cyclin B gene are expressed: one containing a longer 3′-UTR and another containing a shorter 3′-UTR that is a subset of the long one. Transcripts containing the longer 3′-UTR become localized to the posterior pole of the oocyte, whereas those with the shorter 3′-UTR do not, implying the existence of a localization element in the sequence specific to the long 3′-UTR (Dalby and Glover, 1992). Similarly, two Bdnf transcript isoforms that differ in polyadenylation site, and thus 3′-UTR length, are expressed in mouse brain cortex. Transcripts that contain the long 3′-UTR are efficiently trafficked to dendrites, whereas those that contain the short UTR remain in the soma (An et al., 2008).
Recently, the linkage of gene-distal polyadenylation and RNA localization to neuronal processes was shown to extend beyond these isolated examples as a more general phenomenon affecting hundreds of genes (Taliaferro et al., 2016). The analysis of splice isoform abundances in soma and neurite cell fractions revealed a strong preference for gene-distal UTR sequences in neurites. A similar profile of in vivo ribosome-associated transcripts in mouse neurons also demonstrated many examples of alternative 3′-UTR isoform use associated with differential isoform localization in axons (Shigeoka et al., 2016). Thus, the composition of a transcript's 3′-UTR and its RNA localization fate seem to be tightly linked.
The mechanisms and consequences of 3′-UTR regulation and localization are becoming better understood. The regulation of 3′-UTR composition through alternative cleavage and polyadenylation (APA) has been well correlated to broader cellular states. In general, gene-proximal polyadenylation sites are preferentially used following oncogenic transformation and cellular reprogramming (Sandberg et al., 2008; Mayr and Bartel, 2009). Conversely, gene-distal polyadenylation sites are more often used as cells progress along developmental pathways (Ji et al., 2009; Miura et al., 2013), with neurons in particular expressing long 3′-UTRs that have generally unknown function (Miura et al., 2013). The localization competency of many genes may therefore be a function of the developmental or oncogenic state of the cell.
Similarly, perturbations to factors that regulate alternative cleavage and polyadenylation or 3′-UTR-mediated transcript stability may disrupt the post-transcriptional regulation of transcripts containing particular 3′-UTRs, indirectly resulting in the mislocalization of many transcripts, thus contributing to disease phenotypes. Muscleblind-like proteins, as described above, are known RNA localization factors in their own right (Adereth et al., 2005; E. T. Wang et al., 2012; Taliaferro et al., 2016), and regulators of transcript stability through 3′-UTR binding (Wang et al., 2015). RNA-binding proteins from the CELF family also bind 3′-UTRs and regulate transcript levels (Wang et al., 2015). Functional impairments for both of these factors are associated with neurological diseases (Gallo and Spickett, 2010; Goodwin et al., 2015).
Regulation of synaptic and autism-related genes by Rbfox1 in the cytoplasm of neurons
Post-transcriptional mechanisms, including mRNA localization, stability, and regulated translation provide a means of building and controlling neural circuits with exquisite temporal and spatial specificity (D. O. Wang et al., 2010; Jung et al., 2014). RBPs play critical roles during these processes by regulating RNA metabolism to modify gene expression in neurons (Zhou et al., 2014). This idea gains further support from human genetic studies that show that mutations in the genes of several RBPs are associated with neurological diseases (Liu-Yesucevitz et al., 2011; Gao and Taylor, 2014).
The RBP, RBFOX1, is the vertebrate homolog of the Caenorhabditis elegans Feminizing Gene 1 on X gene product (also known as A2BP1). Each of RBFOX1's paralogs, RBFOX2 (RBM9) and RBFOX3 (NeuN), has distinct but overlapping expression patterns (Kuroyanagi, 2009). Chromosomal translocations and copy number variations in Rbfox1 have been associated with intellectual disability, epilepsy, and autism (Martin et al., 2007; Sebat et al., 2007). Furthermore, transcriptome analysis of autistic and normal brain tissue identified RBFOX1 as a master regulator of autism-related genes (Voineagu et al., 2011). The CNS-specific conditional Rbfox1 knock-out mice exhibit spontaneous seizures, indicating that RBFOX1 regulates neuronal excitability (Gehman et al., 2011). All three RBFOX paralogs bind the RNA motif (U)GCAUG and have a well-characterized function in regulating alternative splicing in the nucleus of cells. Rbfox1 itself is alternatively spliced into nuclear and cytoplasmic isoforms, and the function of the cytoplasmic RBFOX1 has only recently been investigated (Hamada et al., 2015; Carreira-Rosario et al., 2016; Lee et al., 2016). In neurons, RBFOX1 binds to the 3′-UTR of its target mRNAs in the cytoplasm and promotes the stability of these transcripts. These cytoplasmic targets are enriched for synaptic and autism-related genes, supporting the hypothesis that RBFOX1 promotes mRNA stability of autism-related genes in the brain (Ray et al., 2013). One potential mechanism for this regulation is to antagonize microRNA-binding and the repression activity of microRNA machinery. Future work is needed to know whether Rbfox proteins may play a role in mRNA localization.
Several hundreds of transcripts have now been identified as targets of RBFOX proteins by different mechanisms. For example, the cytoplasmic targets of RBFOX1 are enriched in genes involved in calcium signaling pathway. In neurons, the splicing of Camk2d and Camk2 g is regulated by RBFOX1 in the nucleus, and the mRNA concentration of Camk2a, Camk2b, Camk4, and Ppp3r1 is affected by RBFOX1 in the cytoplasm (Lee et al., 2016), making it challenging to understand the contributions of individual RBFOX1 targets to any neural circuit phenotype. In C. elegans nervous system, RBFOX, CELF, and PTB family proteins regulate the splicing of insulin receptor daf-2 in a single neuron, and misregulation of this splicing leads to behavioral changes (Tomioka et al., 2016), underscoring the need for cell-type-specific approaches in charactering the physiological function of RBPs in neurons.
A growing literature has revealed the multifunctionality of RBPs (Heraud-Farlow and Kiebler, 2014; Vanharanta et al., 2014), and this is the case for RBFOX family proteins as well. Drosophila cytoplasmic RBFOX1 represses translation of one of its target mRNAs, the translational regulator pumilio, to regulate germ cell differentiation (Carreira-Rosario et al., 2016), indicating that RBFOX1 may either enhance or repress gene expression in the cytoplasm of cells. Recent studies showed that RBFOX2 and RBFOX3 can regulate the biogenesis of microRNAs, and RBFOX2 can bind nascent RNAs to regulate polycomb complex 2 targeting (Kim et al., 2014; Chen et al., 2016; Wei et al., 2016). These data suggest that RBPs might have diverse functions in neurons and an increased understanding of RNA regulation will lead to novel therapies for a range of brain disorders.
Role of FMRP and Ataxin-2 in RNA granules, synapse function, and behavior
Individual mRNPs formed by interaction of mRNAs with cis-element-interacting RNA-binding proteins as well as other regulatory factors assemble into larger and heterogeneous RNA granules through trans-mRNP interactions. In such RNA granule assemblies, sequestered and translationally repressed mRNAs are transported on cytoskeletal filaments to specific intracellular locations (Kiebler and Bassell, 2006; Zeitelhofer et al., 2008). In neurons, activity-induced disassembly of RNP granules plays an important physiological role to enable local translation of dendritically localized mRNAs (Krichevsky and Kosik, 2001; H. Wang and Tiedge, 2004; Zeitelhofer et al., 2008). Prion-like domains with low amino acid complexity (LC domain) found predominantly on RNA-binding proteins are considered to mediate the assembly and disassembly of mRNP granules (Malinovska et al., 2013). Aberrant translation arising from abnormal mRNP assembly is causally related to developmental and degenerative disorders of the nervous system (Liu-Yesucevitz et al., 2011; Tolino et al., 2012). Consistently, mutations in the LC domains of these proteins are predominantly linked to heritable forms of these disorders (Ramaswami et al., 2013; Toretsky and Wright, 2014). Genetic mutations affecting LC domains of RBPs affect stress granule dynamics and biophysical properties, which have been linked to neurodegenerative diseases (Kim et al., 2013; Molliex et al., 2015; Protter and Parker, 2016).
In neurons, mRNA localization and local translation ensure spatiotemporal regulation of synaptic plasticity required for formation of stable memories. Notable among memory-associated RNA regulatory proteins are Staufen/Pumilio pathway components, as well as Atx2 and FMRP, which are linked to trinucleotide expansion disorders (Dubnau et al., 2003; Orr and Zoghbi, 2007; Bolduc et al., 2008; McCann et al., 2011; Sudhakaran et al., 2014). CGG repeats in the 5′UTR of the FMR1 gene and CAG expansion in the reading frame of the Atx2 gene lead to genetic disorders; fragile X syndrome (FXS), and spinocerebellar ataxia type 2 (SCA2) and ALS, respectively (Verkerk et al., 1991; O'Donnell and Warren, 2002; Al-Ramahi et al., 2007; Elden et al., 2010). Recent studies demonstrate that the affected proteins are required for mRNP assembly and/or translational regulation underlying memory formation, in a manner that exhibits shared and distinct mechanisms. The KH domain in FMRP and the Lsm domain in Atx2, respectively, mediates their interaction with RNA (Neuwald and Koonin, 1998; Lewis et al., 2000). FMRP and Atx2 are both required in the same subset of central olfactory neurons for formation of long-term but not short-term olfactory habituation in Drosophila. Whereas loss-of-function mutations in the genes have no dominant effects, dfmr1 and atx2 mutations show strong transdominant genetic interactions. Several lines of evidence indicate that FMRP and Atx2 proteins interact biochemically, and jointly bind and regulate the expression of dendritic CaMKIIα mRNA that is required for long-term memory (Sudhakaran et al., 2014). Excess protein synthesis is thought to underlie Fmr1 loss-of-function phenotypes, including defective long-term memory formation (Bolduc et al., 2008). Although both dFMR1 and Atx2 possess prion-like LC domains and are mRNA granule components, in vivo, FMRP is dispensable and Atx2 is crucial for assembly of the majority of RNA granules visible in neurons that encode habituation associated memory (Sudhakaran et al., 2014). One potential explanation for this observation is that dFMR1 is only required for assembly of a subset of mRNP granules. Alternatively, the Q/N domain of FMRP, which does not exhibit typical prion-like behavior in yeast assays, may facilitate protein–protein interactions involved in its function as a translational repressor (Banerjee et al., 2010).
Although the loss-of-function effects of dfmr1 and atx2 mutations are similar, FXS-related pathologies are generally not associated with inclusion bodies, whereas ALS or SCA2 patients show formation of inclusion bodies in affected neurons (Orr, 2012). This is consistent with the observation that fragile X is an X-linked disorder (caused by loss of FMRP function) and SCA2 is autosomal dominant, caused by a polyglutamine expansion that confers enhanced aggregation efficiency to Atx2.
The work in Drosophila argues that LC-domain containing RNA regulatory proteins may be generally involved in the activity-dependent translational regulation of memory-associated mRNAs, such as CaMKII. Consistent with this, Q/N domains of FMRP are required for the formation of long-term memory (Banerjee et al., 2010). A prediction of this model is that similar domains of Atx2 will also be required for long-term memory and that loss of function mutations in human Atx2 or its closely related paralog Atx2L could cause symptoms that overlap with FXS.
The PI3K/mammalian target of rapamycin (mTOR) pathway is important for local protein synthesis and regulated by FMRP: implications for FXS and other autism spectrum disorders
A prerequisite for synaptic translation of localized mRNAs is that components of the protein synthesis machinery are present and active in dendrites. The PI3K/mTOR pathway is an essential regulator of mRNA translation in neurons and localized to dendrites and synapses (Banko et al., 2006; Ohno et al., 2014). A few studies show that activation of the PI3K/mTOR pathway is important for stimulus-dependent local mRNA translation in dendrites. A PI3K inhibitor prevents metabotropic glutamate receptor 1/5 (mGlu1/5)-mediated protein synthesis in synaptic fractions (Gross et al., 2010), and mTOR inhibition by rapamycin or siRNA-mediated silencing blocks BDNF-induced synaptic protein synthesis in isolated dendrites and axons (Takei et al., 2004). Apart from the effect on general dendritic protein synthesis, mTOR also regulates the local translation of specific mRNAs, such as CaMKIIα and Kv1.1 (Sosanya et al., 2013, 2015).
Recent studies suggest that the expression of the PI3K complex is controlled through activity-dependent protein synthesis and degradation (Gross et al., 2010; Briz et al., 2013). Large-scale screens for mRNAs associated with FMRP, the protein lost in FXS, have identified several PI3K/mTOR pathway components (Brown et al., 2001; Darnell et al., 2011; Ascano et al., 2012). Two of these, the PI3K catalytic subunit p110β and the regulatory subunit PI3K enhancer (PIKE), have been confirmed independently to bind to FMRP leading to upregulated p110β and PIKE protein levels in cells from patients with FXS and/or in FXS mouse models (Gross et al., 2010; Sharma et al., 2010; Gross and Bassell, 2012; Kumari et al., 2014). FXS is characterized by increased and stimulus-insensitive synaptic protein synthesis, which may underlie defects in synaptic plasticity and neuronal function (Darnell and Klann, 2013). The fact that FMRP directly controls central regulators of general and dendritic mRNA translation makes them attractive candidates for therapeutic intervention to correct dysregulated local protein synthesis, which may restore synaptic plasticity in FXS. In line with this hypothesis, genetic reduction of p110β or PIKE restored stimulus-induced synaptic protein synthesis in Fmr1 knock-out mice and reversed deficits on the cellular, behavioral, and cognitive level in both mouse and fly models of FXS (Gross et al., 2015a, b; Monyak et al., 2016). As a next step, it will be important to assess whether selective inhibitors of p110β, which are available from cancer research, likewise rescue phenotypes in animal and human cell models. Inhibitors of PI3K/mTOR signaling components could have a broader applicability in neurodevelopmental disorders, as defects in expression or activity of the PI3K/mTOR complex leading to dysregulated protein synthesis have been associated with different forms of autism spectrum disorders (Kelleher and Bear, 2008; Cuscó et al., 2009; Gross, 2016).
FMRP is transported into dendrites and synapses (Antar et al., 2004) and regulates the local translation of its targets at synapses (Ifrim et al., 2015; Liu and Cline, 2016). An open question is whether the components of the PI3K/mTOR pathway, which are regulated by FMRP, are translated locally. There is evidence for local translation of p110β as the mRNA is localized into dendrites in vivo and associates with actively translating polysomes in synaptic fractions (Gross et al., 2010), but so far, it is unknown whether mRNAs for PIKE or any of the other potential FMRP targets within the PI3K/mTOR pathway are present in dendrites and may be locally translated. It will be interesting to investigate whether the local synthesis of the PI3K/mTOR complex, an important regulator of protein synthesis (Kye et al., 2014), adds another layer of complexity to the regulation of synaptic mRNA translation.
In conclusion, here we have highlighted recent progress in our understanding of some proteins that are linked to local regulation of mRNA and neurogenetic diseases. The field of RNA-based neurological disease has grown rapidly (Gao and Taylor, 2014). We present a model that integrates the proposed roles of several RBPs and associated factors in splicing, RNA granule assembly (both stress granules and RNA transport granules), cytoskeletal-based transport, association with RER for synthesis and localization of membrane proteins, and local regulation of mRNA translation in axonal and/or dendritic compartments.
Footnotes
E.T.W. is a consultant for 5AM Ventures. The remaining authors declare no competing financial interests.
References
- Adereth Y, Dammai V, Kose N, Li R, Hsu T. RNA-dependent integrin alpha3 protein localization regulated by the Muscleblind-like protein MLP1. Nat Cell Biol. 2005;7:1240–1247. doi: 10.1038/ncb1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akten B, Kye MJ, Hao le T, Wertz MH, Singh S, Nie D, Huang J, Merianda TT, Twiss JL, Beattie CE, Steen JA, Sahin M. Interaction of survival of motor neuron (SMN) and HuD proteins with mRNA cpg15 rescues motor neuron axonal deficits. Proc Natl Acad Sci U S A. 2011;108:10337–10342. doi: 10.1073/pnas.1104928108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Ramahi I, Pérez AM, Lim J, Zhang M, Sorensen R, de Haro M, Branco J, Pulst SM, Zoghbi HY, Botas J. dAtaxin-2 mediates expanded Ataxin-1-induced neurodegeneration in a Drosophila model of SCA1. PLoS Genet. 2007;3:e234. doi: 10.1371/journal.pgen.0030234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An JJ, Gharami K, Liao G-Y, Woo NH, Lau AG, Vanevski F, Torre ER, Jones KR, Feng Y, Lu B, Xu B. Distinct role of long 3′ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell. 2008;134:175–187. doi: 10.1016/j.cell.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreassi C, Riccio A. To localize or not to localize: mRNA fate is in 3′UTR ends. Trends Cell Biol. 2009;19:465–474. doi: 10.1016/j.tcb.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Antar LN, Afroz R, Dictenberg JB, Carroll RC, Bassell GJ. Metabotropic glutamate receptor activation regulates fragile X mental retardation protein and FMR1 mRNA localization differentially in dendrites and at synapses. J Neurosci. 2004;24:2648–2655. doi: 10.1523/JNEUROSCI.0099-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artero R, Prokop A, Paricio N, Begemann G, Pueyo I, Mlodzik M, Perez-Alonso M, Baylies MK. The muscleblind gene participates in the organization of Z-bands and epidermal attachments of Drosophila muscles and is regulated by Dmef2. Dev Biol. 1998;195:131–143. doi: 10.1006/dbio.1997.8833. [DOI] [PubMed] [Google Scholar]
- Ascano M, Jr, Mukherjee N, Bandaru P, Miller JB, Nusbaum JD, Corcoran DL, Langlois C, Munschauer M, Dewell S, Hafner M, Williams Z, Ohler U, Tuschl T. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature. 2012;492:382–386. doi: 10.1038/nature11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee P, Schoenfeld BP, Bell AJ, Choi CH, Bradley MP, Hinchey P, Kollaros M, Park JH, McBride SM, Dockendorff TC. Short- and long-term memory are modulated by multiple isoforms of the fragile X mental retardation protein. J Neurosci. 2010;30:6782–6792. doi: 10.1523/JNEUROSCI.6369-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banko JL, Hou L, Poulin F, Sonenberg N, Klann E. Regulation of eukaryotic initiation factor 4E by converging signaling pathways during metabotropic glutamate receptor-dependent long-term depression. J Neurosci. 2006;26:2167–2173. doi: 10.1523/JNEUROSCI.5196-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäumer D, Lee S, Nicholson G, Davies JL, Parkinson NJ, Murray LM, Gillingwater TH, Ansorge O, Davies KE, Talbot K. Alternative splicing events are a late feature of pathology in a mouse model of spinal muscular atrophy. PLoS Genet. 2009;5:e1000773. doi: 10.1371/journal.pgen.1000773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begemann G, Paricio N, Artero R, Kiss I, Pérez-Alonso M, Mlodzik M. muscleblind, a gene required for photoreceptor differentiation in Drosophila, encodes novel nuclear Cys3His-type zinc-finger-containing proteins. Development. 1997;124:4321–4331. doi: 10.1242/dev.124.21.4321. [DOI] [PubMed] [Google Scholar]
- Bolduc FV, Bell K, Cox H, Broadie KS, Tully T. Excess protein synthesis in Drosophila Fragile X mutants impairs long-term memory. Nat Neurosci. 2008;11:1143–1145. doi: 10.1038/nn.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briese M, Esmaeili B, Sattelle DB. Is spinal muscular atrophy the result of defects in motor neuron processes? Bioessays. 2005;27:946–957. doi: 10.1002/bies.20283. [DOI] [PubMed] [Google Scholar]
- Briz V, Hsu YT, Li Y, Lee E, Bi X, Baudry M. Calpain-2-mediated PTEN degradation contributes to BDNF-induced stimulation of dendritic protein synthesis. J Neurosci. 2013;33:4317–4328. doi: 10.1523/JNEUROSCI.4907-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown V, Jin P, Ceman S, Darnell JC, O'Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, Darnell RB, Warren ST. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/S0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- Cajigas IJ, Tushev G, Will TJ, tom Dieck S, Fuerst N, Schuman EM. The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron. 2012;74:453–466. doi: 10.1016/j.neuron.2012.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreira-Rosario A, Bhargava V, Hillebrand J, Kollipara RK, Ramaswami M, Buszczak M. Repression of Pumilio protein expression by Rbfox1 promotes germ cell differentiation. Dev Cell. 2016;36:562–571. doi: 10.1016/j.devcel.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charizanis K, Lee KY, Batra R, Goodwin M, Zhang C, Yuan Y, Shiue L, Cline M, Scotti MM, Xia G, Kumar A, Ashizawa T, Clark HB, Kimura T, Takahashi MP, Fujimura H, Jinnai K, Yoshikawa H, Gomes-Pereira M, Gourdon G, et al. Muscleblind-like 2-mediated alternative splicing in the developing brain and dysregulation in myotonic dystrophy. Neuron. 2012;75:437–450. doi: 10.1016/j.neuron.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlet BN, Savkur RS, Singh G, Philips AV, Grice EA, Cooper TA. Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol Cell. 2002;10:45–53. doi: 10.1016/S1097-2765(02)00572-5. [DOI] [PubMed] [Google Scholar]
- Chen Y, Zubovic L, Yang F, Godin K, Pavelitz T, Castellanos J, Macchi P, Varani G. Rbfox proteins regulate microRNA biogenesis by sequence-specific binding to their precursors and target downstream Dicer. Nucleic Acids Res. 2016;44:4381–4395. doi: 10.1093/nar/gkw177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TA. Chemical reversal of the RNA gain of function in myotonic dystrophy. Proc Natl Acad Sci U S A. 2009;106:18433–18434. doi: 10.1073/pnas.0910643106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuscó I, Medrano A, Gener B, Vilardell M, Gallastegui F, Villa O, González E, Rodríguez-Santiago B, Vilella E, Del Campo M, Pérez-Jurado LA. Autism-specific copy number variants further implicate the phosphatidylinositol signaling pathway and the glutamatergic synapse in the etiology of the disorder. Hum Mol Genet. 2009;18:1795–1804. doi: 10.1093/hmg/ddp092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalby B, Glover DM. 3′ non-translated sequences in Drosophila cyclin B transcripts direct posterior pole accumulation late in oogenesis and peri-nuclear association in syncytial embryos. Development. 1992;115:989–997. doi: 10.1242/dev.115.4.989. [DOI] [PubMed] [Google Scholar]
- Darnell JC, Klann E. The translation of translational control by FMRP: therapeutic targets for FXS. Nat Neurosci. 2013;16:1530–1536. doi: 10.1038/nn.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, Licatalosi DD, Richter JD, Darnell RB. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombert B, Sivadasan R, Simon CM, Jablonka S, Sendtner M. Presynaptic localization of Smn and hnRNP R in axon terminals of embryonic and postnatal mouse motoneurons. PLoS One. 2014;9:e110846. doi: 10.1371/journal.pone.0110846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlin-Asp PG, Bassell GJ, Rossoll W. A role for the survival of motor neuron protein in mRNP assembly and transport. Curr Opin Neurobiol. 2016;39:53–61. doi: 10.1016/j.conb.2016.04.004. [DOI] [PubMed] [Google Scholar]
- Du H, Cline MS, Osborne RJ, Tuttle DL, Clark TA, Donohue JP, Hall MP, Shiue L, Swanson MS, Thornton CA, Ares M., Jr Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat Struct Mol Biol. 2010;17:187–193. doi: 10.1038/nsmb.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau J, Chiang AS, Grady L, Barditch J, Gossweiler S, McNeil J, Smith P, Buldoc F, Scott R, Certa U, Broger C, Tully T. The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr Biol. 2003;13:286–296. doi: 10.1016/S0960-9822(03)00064-2. [DOI] [PubMed] [Google Scholar]
- Elden AC, Kim HJ, Hart MP, Chen-Plotkin AS, Johnson BS, Fang X, Armakola M, Geser F, Greene R, Lu MM, Padmanabhan A, Clay-Falcone D, McCluskey L, Elman L, Juhr D, Gruber PJ, Rüb U, Auburger G, Trojanowski JQ, Lee VM, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466:1069–1075. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallini C, Zhang H, Su Y, Silani V, Singer RH, Rossoll W, Bassell GJ. The Survival of Motor Neuron (SMN) protein interacts with the mRNA-binding protein HuD and regulates localization of poly(A) mRNA in primary motor neuron axons. J Neurosci. 2011;31:3914–3925. doi: 10.1523/JNEUROSCI.3631-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallini C, Bassell GJ, Rossoll W. Spinal muscular atrophy: the role of SMN in axonal mRNA regulation. Brain Res. 2012;1462:81–92. doi: 10.1016/j.brainres.2012.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallini C, Rouanet JP, Donlin-Asp PG, Guo P, Zhang H, Singer RH, Rossoll W, Bassell GJ. Dynamics of survival of motor neuron (SMN) protein interaction with the mRNA-binding protein IMP1 facilitates its trafficking into motor neuron axons. Dev Neurobiol. 2014;74:319–332. doi: 10.1002/dneu.22111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallini C, Donlin-Asp PG, Rouanet JP, Bassell GJ, Rossoll W. Deficiency of the survival of motor neuron protein impairs mRNA localization and local translation in the growth cone of motor neurons. J Neurosci. 2016;36:3811–3820. doi: 10.1523/JNEUROSCI.2396-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyermuth F, Rau F, Kokunai Y, Linke T, Sellier C, Nakamori M, Kino Y, Arandel L, Jollet A, Thibault C, Philipps M, Vicaire S, Jost B, Udd B, Day JW, Duboc D, Wahbi K, Matsumura T, Fujimura H, Mochizuki H, et al. Splicing misregulation of SCN5A contributes to cardiac-conduction delay and heart arrhythmia in myotonic dystrophy. Nat Commun. 2016;7:11067. doi: 10.1038/ncomms11067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugier C, Klein AF, Hammer C, Vassilopoulos S, Ivarsson Y, Toussaint A, Tosch V, Vignaud A, Ferry A, Messaddeq N, Kokunai Y, Tsuburaya R, de la Grange P, Dembele D, Francois V, Precigout G, Boulade-Ladame C, Hummel MC, Lopez de Munain A, Sergeant N, et al. Misregulated alternative splicing of BIN1 is associated with T tubule alterations and muscle weakness in myotonic dystrophy. Nat Med. 2011;17:720–725. doi: 10.1038/nm.2374. [DOI] [PubMed] [Google Scholar]
- Gabanella F, Butchbach ME, Saieva L, Carissimi C, Burghes AH, Pellizzoni L. Ribonucleoprotein assembly defects correlate with spinal muscular atrophy severity and preferentially affect a subset of spliceosomal snRNPs. PLoS One. 2007;2:e921. doi: 10.1371/journal.pone.0000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo J-M, Spickett C. The role of CELF proteins in neurological disorders. RNA Biol. 2010;7:474–479. doi: 10.4161/rna.7.4.12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao FB, Taylor JP. RNA metabolism in neurological disease. Brain Res. 2014;1584:1–2. doi: 10.1016/j.brainres.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehman LT, Stoilov P, Maguire J, Damianov A, Lin CH, Shiue L, Ares M, Jr, Mody I, Black DL. The splicing regulator Rbfox1 (A2BP1) controls neuronal excitation in the mammalian brain. Nat Genet. 2011;43:706–711. doi: 10.1038/ng.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin M, Mohan A, Batra R, Lee KY, Charizanis K, Fernández Gómez FJ, Eddarkaoui S, Sergeant N, Buée L, Kimura T, Clark HB, Dalton J, Takamura K, Weyn-Vanhentenryck SM, Zhang C, Reid T, Ranum LP, Day JW, Swanson MS. MBNL sequestration by toxic RNAs and RNA misprocessing in the myotonic dystrophy brain. Cell Rep. 2015;12:1159–1168. doi: 10.1016/j.celrep.2015.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross C. Defective phosphoinositide metabolism in autism. J Neurosci Res. 2016 doi: 10.1002/jnr.23797. doi: 10.1002/jnr.23797. Advance online publication. Retrieved Jul. 4, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross C, Bassell GJ. Excess protein synthesis in FXS patient lymphoblastoid cells can be rescued with a p110beta-selective inhibitor. Mol Med. 2012;18:336–345. doi: 10.2119/molmed.2011.00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross C, Nakamoto M, Yao X, Chan CB, Yim SY, Ye K, Warren ST, Bassell GJ. Excess phosphoinositide 3-kinase subunit synthesis and activity as a novel therapeutic target in fragile X syndrome. J Neurosci. 2010;30:10624–10638. doi: 10.1523/JNEUROSCI.0402-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross C, Chang CW, Kelly SM, Bhattacharya A, McBride SM, Danielson SW, Jiang MQ, Chan CB, Ye K, Gibson JR, Klann E, Jongens TA, Moberg KH, Huber KM, Bassell GJ. Increased expression of the PI3K enhancer PIKE mediates deficits in synaptic plasticity and behavior in fragile X syndrome. Cell Rep. 2015a;11:727–736. doi: 10.1016/j.celrep.2015.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross C, Raj N, Molinaro G, Allen AG, Whyte AJ, Gibson JR, Huber KM, Gourley SL, Bassell GJ. Selective role of the catalytic PI3K subunit p110beta in impaired higher order cognition in fragile X syndrome. Cell Rep. 2015b;11:681–688. doi: 10.1016/j.celrep.2015.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumy LF, Yeo GSH, Tung Y-CL, Zivraj KH, Willis D, Coppola G, Lam BYH, Twiss JL, Holt CE, Fawcett JW. Transcriptome analysis of embryonic and adult sensory axons reveals changes in mRNA repertoire localization. Rna. 2011;17:85–98. doi: 10.1261/rna.2386111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada N, Ito H, Iwamoto I, Morishita R, Tabata H, Nagata K. Role of the cytoplasmic isoform of RBFOX1/A2BP1 in establishing the architecture of the developing cerebral cortex. Mol Autism. 2015;6:56. doi: 10.1186/s13229-015-0049-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao le T, Duy PQ, Jontes JD, Beattie CE. Motoneuron development influences dorsal root ganglia survival and Schwann cell development in a vertebrate model of spinal muscular atrophy. Hum Mol Genet. 2015;24:346–360. doi: 10.1093/hmg/ddu447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heraud-Farlow JE, Kiebler MA. The multifunctional Staufen proteins: conserved roles from neurogenesis to synaptic plasticity. Trends Neurosci. 2014;37:470–479. doi: 10.1016/j.tins.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubers L, Valderrama-Carvajal H, Laframboise J, Timbers J, Sanchez G, Côté J. HuD interacts with survival motor neuron protein and can rescue spinal muscular atrophy-like neuronal defects. Hum Mol Genet. 2011;20:553–579. doi: 10.1093/hmg/ddq500. [DOI] [PubMed] [Google Scholar]
- Ifrim MF, Williams KR, Bassell GJ. Single molecule imaging of PSD-95 mRNA translation in dendrites and its dysregulation in a mouse model of fragile X syndrome. J Neurosci. 2015;35:7116–7130. doi: 10.1523/JNEUROSCI.2802-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Lee JY, Pan Z, Jiang B, Tian B. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci U S A. 2009;106:7028–7033. doi: 10.1073/pnas.0900028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H, Gkogkas CG, Sonenberg N, Holt CE. Remote control of gene function by local translation. Cell. 2014;157:26–40. doi: 10.1016/j.cell.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanadia RN, Johnstone KA, Mankodi A, Lungu C, Thornton CA, Esson D, Timmers AM, Hauswirth WW, Swanson MS. A muscleblind knockout model for myotonic dystrophy. Science. 2003a;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- Kanadia RN, Urbinati CR, Crusselle VJ, Luo D, Lee YJ, Harrison JK, Oh SP, Swanson MS. Developmental expression of mouse muscleblind genes Mbnl1, Mbnl2 and Mbnl3. Gene Expr Patterns. 2003b;3:459–462. doi: 10.1016/S1567-133X(03)00064-4. [DOI] [PubMed] [Google Scholar]
- Kelleher RJ, 3rd, Bear MF. The autistic neuron: troubled translation? Cell. 2008;135:401–406. doi: 10.1016/j.cell.2008.10.017. [DOI] [PubMed] [Google Scholar]
- Kiebler MA, Bassell GJ. Neuronal RNA granules: movers and makers. Neuron. 2006;51:685–690. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- Kim KK, Yang Y, Zhu J, Adelstein RS, Kawamoto S. Rbfox3 controls the biogenesis of a subset of microRNAs. Nat Struct Mol Biol. 2014;21:901–910. doi: 10.1038/nsmb.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A, Kanagaraj AP, Carter R, Boylan KB, Wojtas AM, Rademakers R, Pinkus JL, Greenberg SA, Trojanowski JQ, Traynor BJ, Smith BN, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495:467–473. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky AM, Kosik KS. Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron. 2001;32:683–696. doi: 10.1016/S0896-6273(01)00508-6. [DOI] [PubMed] [Google Scholar]
- Kumari D, Bhattacharya A, Nadel J, Moulton K, Zeak NM, Glicksman A, Dobkin C, Brick DJ, Schwartz PH, Smith CB, Klann E, Usdin K. Identification of fragile X syndrome-specific molecular markers in human fibroblasts: a useful model to test the efficacy of therapeutic drugs. Hum Mutat. 2014;35:1485–1494. doi: 10.1002/humu.22699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroyanagi H. Fox-1 family of RNA-binding proteins. Cell Mol Life Sci. 2009;66:3895–3907. doi: 10.1007/s00018-009-0120-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kye MJ, Niederst ED, Wertz MH, Goncalves Ido C, Akten B, Dover KZ, Peters M, Riessland M, Neveu P, Wirth B, Kosik KS, Sardi SP, Monani UR, Passini MA, Sahin M. SMN regulates axonal local translation via miR-183/mTOR pathway. Hum Mol Genet. 2014;23:6318–6331. doi: 10.1093/hmg/ddu350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Damianov A, Lin CH, Fontes M, Parikshak NN, Anderson ES, Geschwind DH, Black DL, Martin KC. Cytoplasmic Rbfox1 regulates the expression of synaptic and autism-related genes. Neuron. 2016;89:113–128. doi: 10.1016/j.neuron.2015.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Cooper TA. Pathogenic mechanisms of myotonic dystrophy. Biochem Soc Trans. 2009;37:1281–1286. doi: 10.1042/BST0371281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis HA, Musunuru K, Jensen KB, Edo C, Chen H, Darnell RB, Burley SK. Sequence-specific RNA-binding by a Nova KH domain: implications for paraneoplastic disease and the fragile X syndrome. Cell. 2000;100:323–332. doi: 10.1016/S0092-8674(00)80668-6. [DOI] [PubMed] [Google Scholar]
- Li YR, King OD, Shorter J, Gitler AD. Stress granules as crucibles of ALS pathogenesis. J Cell Biol. 2013;201:361–372. doi: 10.1083/jcb.201302044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Yesucevitz L, Bassell GJ, Gitler AD, Hart AC, Klann E, Richter JD, Warren ST, Wolozin B. Local RNA translation at the synapse and in disease. J Neurosci. 2011;31:16086–16093. doi: 10.1523/JNEUROSCI.4105-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HH, Cline HT. Fragile X mental retardation protein is required to maintain visual conditioning-induced behavioral plasticity by limiting local protein synthesis. J Neurosci. 2016;36:7325–7339. doi: 10.1523/JNEUROSCI.4282-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li DK, Tisdale S, Lotti F, Pellizzoni L. SMN control of RNP assembly: from post-transcriptional gene regulation to motor neuron disease. Semin Cell Dev Biol. 2014;32:22–29. doi: 10.1016/j.semcdb.2014.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotti F, Imlach WL, Saieva L, Beck ES, Hao le T, Li DK, Jiao W, Mentis GZ, Beattie CE, McCabe BD, Pellizzoni L. An SMN-dependent U12 splicing event essential for motor circuit function. Cell. 2012;151:440–454. doi: 10.1016/j.cell.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinovska L, Kroschwald S, Alberti S. Protein disorder, prion propensities, and self-organizing macromolecular collectives. Biochim Biophys Acta. 2013;1834:918–931. doi: 10.1016/j.bbapap.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Martin CL, Duvall JA, Ilkin Y, Simon JS, Arreaza MG, Wilkes K, Alvarez-Retuerto A, Whichello A, Powell CM, Rao K, Cook E, Geschwind DH. Cytogenetic and molecular characterization of A2BP1/FOX1 as a candidate gene for autism. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:869–876. doi: 10.1002/ajmg.b.30530. [DOI] [PubMed] [Google Scholar]
- Masuda A, Andersen HS, Doktor TK, Okamoto T, Ito M, Andresen BS, Ohno K. CUGBP1 and MBNL1 preferentially bind to 3′ UTRs and facilitate mRNA decay. Scientific Rep. 2012;2:209. doi: 10.1038/srep00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann C, Holohan EE, Das S, Dervan A, Larkin A, Lee JA, Rodrigues V, Parker R, Ramaswami M. The Ataxin-2 protein is required for microRNA function and synapse-specific long-term olfactory habituation. Proc Natl Acad Sci U S A. 2011;108:E655–E662. doi: 10.1073/pnas.1107198108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Bühler D, Pillai R, Lottspeich F, Fischer U. A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nat Cell Biol. 2001;3:945–949. doi: 10.1038/ncb1101-945. [DOI] [PubMed] [Google Scholar]
- Miller JW, Urbinati CR, Teng-Umnuay P, Stenberg MG, Byrne BJ, Thornton CA, Swanson MS. Recruitment of human muscleblind proteins to (CUG) (n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minis A, Dahary D, Manor O, Leshkowitz D, Pilpel Y, Yaron A. Subcellular transcriptomics-Dissection of the mRNA composition in the axonal compartment of sensory neurons. Devel Neurobio. 2013;74:365–381. doi: 10.1002/dneu.22140. [DOI] [PubMed] [Google Scholar]
- Miura P, Shenker S, Andreu-Agullo C, Westholm JO, Lai EC. Widespread and extensive lengthening of 3′ UTRs in the mammalian brain. Genome Res. 2013;23:812–825. doi: 10.1101/gr.146886.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monyak RE, Emerson D, Schoenfeld BP, Zheng X, Chambers DB, Rosenfelt C, Langer S, Hinchey P, Choi CH, McDonald TV, et al. Insulin signaling misregulation underlies circadian and cognitive deficits in a Drosophila fragile X model. Mol Psychiatry. 2016 doi: 10.1038/mp.2016.51. doi: 10.1038/mp.2016.51. Advance online publication. Retrieved Apr. 19, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald AF, Koonin EV. Ataxin-2, global regulators of bacterial gene expression, and spliceosomal snRNP proteins share a conserved domain. J Mol Med. 1998;76:3–5. doi: 10.1007/s109-1998-8098-0. [DOI] [PubMed] [Google Scholar]
- O'Donnell WT, Warren ST. A decade of molecular studies of fragile X syndrome. Annu Rev Neurosci. 2002;25:315–338. doi: 10.1146/annurev.neuro.25.112701.142909. [DOI] [PubMed] [Google Scholar]
- Ohno H, Kato S, Naito Y, Kunitomo H, Tomioka M, Iino Y. Role of synaptic phosphatidylinositol 3-kinase in a behavioral learning response in C. elegans. Science. 2014;345:313–317. doi: 10.1126/science.1250709. [DOI] [PubMed] [Google Scholar]
- Orr HT. Cell biology of spinocerebellar ataxia. J Cell Biol. 2012;197:167–177. doi: 10.1083/jcb.201105092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- Pascual M, Vicente M, Monferrer L, Artero R. The Muscleblind family of proteins: an emerging class of regulators of developmentally programmed alternative splicing. Differentiation. 2006;74:65–80. doi: 10.1111/j.1432-0436.2006.00060.x. [DOI] [PubMed] [Google Scholar]
- Pellizzoni L, Yong J, Dreyfuss G. Essential role for the SMN complex in the specificity of snRNP assembly. Science. 2002;298:1775–1779. doi: 10.1126/science.1074962. [DOI] [PubMed] [Google Scholar]
- Philips AV, Timchenko LT, Cooper TA. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- Protter DS, Parker R. Principles and properties of stress granules. Trends Cell Biol. 2016;26:668–679. doi: 10.1016/j.tcb.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswami M, Taylor JP, Parker R. Altered ribostasis: RNA-protein granules in degenerative disorders. Cell. 2013;154:727–736. doi: 10.1016/j.cell.2013.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, Na H, Irimia M, Matzat LH, Dale RK, Smith SA, Yarosh CA, Kelly SM, Nabet B, Mecenas D, Li W, et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013;499:172–177. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossoll W, Jablonka S, Andreassi C, Kröning AK, Karle K, Monani UR, Sendtner M. Smn, the spinal muscular atrophy-determining gene product, modulates axon growth and localization of beta-actin mRNA in growth cones of motoneurons. J Cell Biol. 2003;163:801–812. doi: 10.1083/jcb.200304128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossoll W, Bassell GJ. Spinal muscular atrophy and a model for survival of motor neuron protein function in axonal ribonucleoprotein complexes. Results Probl Cell Differ. 2009;48:289–326. doi: 10.1007/400_2009_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez G, Dury AY, Murray LM, Biondi O, Tadesse H, El Fatimy R, Kothary R, Charbonnier F, Khandjian EW, Côté J. A novel function for the survival motoneuron protein as a translational regulator. Hum Mol Genet. 2013;22:668–684. doi: 10.1093/hmg/dds474. [DOI] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savkur RS, Philips AV, Cooper TA. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat Genet. 2001;29:40–47. doi: 10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, Leotta A, Pai D, Zhang R, Lee YH, Hicks J, Spence SJ, Lee AT, Puura K, Lehtimäki T, Ledbetter D, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Hoeffer CA, Takayasu Y, Miyawaki T, McBride SM, Klann E, Zukin RS. Dysregulation of mTOR signaling in fragile X syndrome. J Neurosci. 2010;30:694–702. doi: 10.1523/JNEUROSCI.3696-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeoka T, Jung H, Jung J, Turner-Bridger B, Ohk J, Lin JQ, Amieux PS, Holt CE. Dynamic axonal translation in developing and mature visual circuits. Cell. 2016;166:181–192. doi: 10.1016/j.cell.2016.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S, Parker R. Hypo- and hyper-assembly diseases of RNA-protein complexes. Trends Mol Med. 2016;22:615–628. doi: 10.1016/j.molmed.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So BR, Wan L, Zhang Z, Li P, Babiash E, Duan J, Younis I, Dreyfuss G. A U1 snRNP-specific assembly pathway reveals the SMN complex as a versatile hub for RNP exchange. Nat Struct Mol Biol. 2016;23:225–230. doi: 10.1038/nsmb.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosanya NM, Huang PP, Cacheaux LP, Chen CJ, Nguyen K, Perrone-Bizzozero NI, Raab-Graham KF. Degradation of high affinity HuD targets releases Kv1.1 mRNA from miR-129 repression by mTORC1. J Cell Biol. 2013;202:53–69. doi: 10.1083/jcb.201212089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosanya NM, Cacheaux LP, Workman ER, Niere F, Perrone-Bizzozero NI, Raab-Graham KF. Mammalian target of rapamycin (mTOR) tagging promotes dendritic branch variability through the capture of Ca2+/calmodulin-dependent protein kinase II α (CaMKIIα) mRNAs by the RNA-binding protein HuD. J Biol Chem. 2015;290:16357–16371. doi: 10.1074/jbc.M114.599399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhakaran IP, Hillebrand J, Dervan A, Das S, Holohan EE, Hülsmeier J, Sarov M, Parker R, VijayRaghavan K, Ramaswami M. FMRP and Ataxin-2 function together in long-term olfactory habituation and neuronal translational control. Proc Natl Acad Sci U S A. 2014;111:E99–E108. doi: 10.1073/pnas.1309543111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei N, Inamura N, Kawamura M, Namba H, Hara K, Yonezawa K, Nawa H. Brain-derived neurotrophic factor induces mammalian target of rapamycin-dependent local activation of translation machinery and protein synthesis in neuronal dendrites. J Neurosci. 2004;24:9760–9769. doi: 10.1523/JNEUROSCI.1427-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taliaferro JM, Vidaki M, Oliveira R, Olson S, Zhan L, Saxena T, Wang ET, Graveley BR, Gertler FB, Swanson MS, Burge CB. Distal alternative last exons localize mRNAs to neural projections. Mol Cell. 2016;61:821–833. doi: 10.1016/j.molcel.2016.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang ZZ, Yarotskyy V, Wei L, Sobczak K, Nakamori M, Eichinger K, Moxley RT, Dirksen RT, Thornton CA. Muscle weakness in myotonic dystrophy associated with misregulated splicing and altered gating of Ca (V)1.1 calcium channel. Hum Mol Genet. 2012;21:1312–1324. doi: 10.1093/hmg/ddr568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AM, Berchtold NC, Perreau VM, Tu CH, Jeon NL, Cotman CW. Axonal mRNA in uninjured and regenerating cortical mammalian axons. J Neurosci. 2009;29:4697–4707. doi: 10.1523/JNEUROSCI.6130-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton CA. Myotonic dystrophy. Neurol Clin. 2014;32:705–719. doi: 10.1016/j.ncl.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolino M, Köhrmann M, Kiebler MA. RNA-binding proteins involved in RNA localization and their implications in neuronal diseases. Eur J Neurosci. 2012;35:1818–1836. doi: 10.1111/j.1460-9568.2012.08160.x. [DOI] [PubMed] [Google Scholar]
- Tomioka M, Naito Y, Kuroyanagi H, Iino Y. Splicing factors control C. elegans behavioural learning in a single neuron by producing DAF-2c receptor. Nat Commun. 2016;7:11645. doi: 10.1038/ncomms11645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toretsky JA, Wright PE. Assemblages: functional units formed by cellular phase separation. J Cell Biol. 2014;206:579–588. doi: 10.1083/jcb.201404124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanharanta S, Marney CB, Shu W, Valiente M, Zou Y, Mele A, Darnell RB, Massague J. Loss of the multifunctional RNA-binding protein RBM47 as a source of selectable metastatic traits in breast cancer. eLife. 2014:3. doi: 10.7554/eLife.02734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DP, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang FP. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-H. [DOI] [PubMed] [Google Scholar]
- Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, Mill J, Cantor RM, Blencowe BJ, Geschwind DH. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahbi K, Algalarrondo V, Bécane HM, Fressart V, Beldjord C, Azibi K, Lazarus A, Berber N, Radvanyi-Hoffman H, Stojkovic T, Béhin A, Laforêt P, Eymard B, Hatem S, Duboc D. Brugada syndrome and abnormal splicing of SCN5A in myotonic dystrophy type 1. Arch Cardiovasc Dis. 2013;106:635–643. doi: 10.1016/j.acvd.2013.08.003. [DOI] [PubMed] [Google Scholar]
- Wang DO, Martin KC, Zukin RS. Spatially restricting gene expression by local translation at synapses. Trends Neurosci. 2010;33:173–182. doi: 10.1016/j.tins.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Cody NA, Jog S, Biancolella M, Wang TT, Treacy DJ, Luo S, Schroth GP, Housman DE, Reddy S, Lécuyer E, Burge CB. Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell. 2012;150:710–724. doi: 10.1016/j.cell.2012.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Ward AJ, Cherone JM, Giudice J, Wang TT, Treacy DJ, Lambert NJ, Freese P, Saxena T, Cooper TA, Burge CB. Antagonistic regulation of mRNA expression and splicing by CELF and MBNL proteins. Genome Res. 2015;25:858–871. doi: 10.1101/gr.184390.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Tiedge H. Translational control at the synapse. Neuroscientist. 2004;10:456–466. doi: 10.1177/1073858404265866. [DOI] [PubMed] [Google Scholar]
- Wei C, Xiao R, Chen L, Cui H, Zhou Y, Xue Y, Hu J, Zhou B, Tsutsui T, Qiu J, Li H, Tang L, Fu XD. RBFox2 binds nascent RNA to globally regulate polycomb complex 2 targeting in mammalian genomes. Mol Cell. 2016;62:875–889. doi: 10.1016/j.molcel.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitelhofer M, Karra D, Macchi P, Tolino M, Thomas S, Schwarz M, Kiebler M, Dahm R. Dynamic interaction between P-bodies and transport ribonucleoprotein particles in dendrites of mature hippocampal neurons. J Neurosci. 2008;28:7555–7562. doi: 10.1523/JNEUROSCI.0104-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Lotti F, Dittmar K, Younis I, Wan L, Kasim M, Dreyfuss G. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008;133:585–600. doi: 10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Pinto AM, Wan L, Wang W, Berg MG, Oliva I, Singh LN, Dengler C, Wei Z, Dreyfuss G. Dysregulation of synaptogenesis genes antecedes motor neuron pathology in spinal muscular atrophy. Proc Natl Acad Sci U S A. 2013;110:19348–19353. doi: 10.1073/pnas.1319280110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Mangelsdorf M, Liu J, Zhu L, Wu JY. RNA-binding proteins in neurological diseases. Sci China Life Sci. 2014;57:432–444. doi: 10.1007/s11427-014-4647-9. [DOI] [PubMed] [Google Scholar]
- Zivraj KH, Tung YCL, Piper M, Gumy L, Fawcett JW, Yeo GSH, Holt CE. Subcellular profiling reveals distinct and developmentally regulated repertoire of growth cone mRNAs. J Neurosci. 2010;30:15464–15478. doi: 10.1523/JNEUROSCI.1800-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]