Abstract
Lineage development is a stepwise process, governed by stage-specific regulatory factors and associated markers. Astrocytes are one of the principle cell types in the CNS and the stages associated with their development remain very poorly defined. To identify these stages, we performed gene-expression profiling on astrocyte precursor populations in the spinal cord, identifying distinct patterns of gene induction during their development that are strongly correlated with human astrocytes. Validation studies identified a new cohort of astrocyte-associated genes during development and demonstrated their expression in reactive astrocytes in human white matter injury (WMI). Functional studies on one of these genes revealed that mice lacking Asef exhibited impaired astrocyte differentiation during development and repair after WMI, coupled with compromised blood–brain barrier integrity in the adult CNS. These studies have identified distinct stages of astrocyte lineage development associated with human WMI and, together with our functional analysis of Asef, highlight the parallels between astrocyte development and their reactive counterparts associated with injury.
SIGNIFICANCE STATEMENT Astrocytes play a central role in CNS function and associated diseases. Yet the mechanisms that control their development remain poorly defined. Using the developing mouse spinal cord as a model system, we identify molecular changes that occur in developing astrocytes. These molecular signatures are strongly correlated with human astrocyte expression profiles and validation in mouse spinal cord identifies a host of new genes associated with the astrocyte lineage. These genes are present in reactive astrocytes in human white matter injury, and functional studies reveal that one of these genes, Asef, contributes to reactive astrocyte responses after injury. These studies identify distinct stages of astrocyte lineage development and highlight the parallels between astrocyte development and their reactive counterparts associated with injury.
Keywords: astrocyte precursors, blood–brain barrier, reactive astrocytes, white matter injury
Introduction
Development of cell lineages follows a sequential series of differentiative steps, commencing with the specification of stem cells toward a given cellular identity and culminating with the differentiation of lineage-specific progenitors into mature populations that execute specific physiological functions. In the CNS, the paradigm of stepwise lineage differentiation is best exemplified during the differentiation of neuronal (Lu et al., 2015; Toma and Hanashima, 2015) and oligodendrocyte lineages (Emery, 2010; Goldman and Kuypers, 2015; Bergles and Richardson, 2016), where the expression of specific genes at distinct phases of development allows for the discrimination of progenitor and mature populations. Understanding these fundamental properties of CNS lineage development is crucial for the construction of a more comprehensive atlas of CNS development, which will ultimately lend greater insight into normal and pathological states.
The other principal cell type in the CNS is the astrocyte and, despite their relative abundance, whether they possess an analogous, stepwise developmental program in vivo remains poorly defined (Rowitch and Kriegstein, 2010; Molofsky et al., 2012; Chaboub and Deneen, 2013; Molofsky and Deneen, 2015). The developing spinal cord represents a well characterized model for examining astrocyte development, where specification is demarcated by induction of GLAST (glutamate/aspartate transporter), Aldh1l1, and NFIA (nuclear factor I-A) at embryonic day (E) 12.5 (Deneen et al., 2006; Cahoy et al., 2008) and differentiation is indicated by expression of maturation makers, including GFAP, glutamine synthase (GS), and aldolase C at ∼E18.5 (Bachoo et al., 2004; Molofsky et al., 2012; Anlauf and Derouiche, 2013; Chaboub and Deneen, 2013). Markers and regulatory factors associated with the intervening time points between E12.5 and E18.5 remain undefined, though recent profiling studies in the developing spinal cord identified specific gene-expression patterns during this interval (Molofsky et al., 2013). These findings suggest that astrocytes are endowed with stage-specific developmental programs, though rigorous analysis of temporal and spatial gene-expression patterns during this interval are necessary to prove this model in vivo.
Astrocytes play crucial roles in a wide range of CNS functions (Sofroniew and Vinters, 2010; Molofsky et al., 2012; Chaboub and Deneen, 2013; Molofsky and Deneen, 2015) and are associated with nearly every form of neurological disease, from glioma and neurodegenerative disorders to autism and injury (Molofsky et al., 2012; John Lin and Deneen, 2013; Molofsky and Deneen, 2015). Under these pathological states, astrocytes can become reactive, demonstrating increased proliferation, hypertrophic morphologies, and induction of GFAP expression (Sofroniew, 2009; Anderson et al., 2014). These properties of reactive astrocytes are also key features of astrocyte development. Therefore, many of the processes that oversee the development of astrocytes are also likely to contribute to the production of their disease-associated, reactive analogues. Despite these parallels, how developmental processes are reused during the production of reactive astrocytes and how the associated factors contribute to CNS injury or disease pathogenesis remain poorly defined.
To identify these intermediate stages of astrocyte development, we used the GLAST-dsRed reporter mouse line (Regan et al., 2007) and FACS-based approaches to access populations across the E12.5–postnatal day (P) 7 interval in the spinal cord. Gene-expression profiling studies revealed stage-specific induction patterns and identified Asef, Tom1l1, Mfge8, and Gpr37l1 as new markers of the intermediate stages of astrocyte development. We validated expression of these astrocyte genes in human white matter injury (WMI) and performed functional studies implicating Asef in astrocyte development and response after WMI. Put together, our study identifies new stages of astrocyte lineage development and their associated markers, and demonstrates that these processes are conserved and contribute to CNS injury.
Materials and Methods
Mouse lines.
The Glast-dsRed transgenic mouse line was obtained from Dr. Jeffrey Rothstein at Johns Hopkins University (Regan et al., 2007). The Asef knock-out (KO) mouse line was obtained from Dr. Tetsu Akiyama from the University of Tokyo (Kawasaki et al., 2009). We crossed the Asef KO mouse with the Aldhl1-GFP mouse (Gene Expression Nervous System Atlas) to generate Aldh1l1-GFP;Asef+/− and Aldh1l1-GFP;Asef−/− mice used to study the loss of Asef on glia development. Wild-type ICR (imprinting control region) mice were obtained from Charles River for developmental time course of spinal cord expression. E12.5 and E14.5 embryos were harvested and fixed overnight in 4% PFA then immersed in 30% sucrose overnight. For E16.5 and E18.5 embryos, mothers were perfused with 1× PBS then 4% PFA. Embryos were postfixed for 2 h, then immersed overnight in 30% sucrose. P4 and P7 pups perfused with 1× PBS then 4% PFA, followed by 2 h 4% PFA postfix and overnight 30% sucrose. All procedures were approved by the Institutional Animal Care and Use Committee at Baylor College of Medicine and conform to the US Public Health Service Policy on Human Care and Use of Laboratory Animals.
FACS and RNA profiling.
Glast-dsRed spinal cords were harvested at six different time points, and dissociated with papain (Worthington Biochemical) and mechanical force. Single cells were incubated with α-CD15/LexA antibody (Developmental Studies Hybridoma Bank) for 1–2 h at room temperature, washed three times with 1× PBS, and then incubated for 1 h at room temperature with secondary antibody anti-mouse Alexa Fluor 488 (Invitrogen). After three 1× PBS washes, the samples were sorted using BD FACSAria cell sorter (BD Biosciences). After sorting, cells were spun down and washed in cold 1× PBS twice and cells were frozen at −80°C. RNA extraction was performed using QIAshredder and RNA easy miniprep kits (Qiagen). RNA was then sent to Miltenyi Biotec for microarray using Agilent Whole Mouse Genome Oligo Microarrays (one color). The control −/− populations from each time point were isolated in duplicate. All other populations were isolated in triplicates. Data can be accessed in the Gene Expression Omnibus repository, accession number GSE80826.
Bioinformatics.
Expression array data were processed using Limma packages in R. Data were background corrected and quantile normalized before analysis. Differential expression analysis was performed to identify genes with significantly altered expression (≥4-fold change in expression with p < 0.01) at each of the time points. The set of top differential genes found for any treatment group were clustered, using a supervised approach as described previously (Creighton et al., 2008). Expression heat maps were generated using JavaTreeView (Saldanha, 2004). For visual display, expression profiles of biological triplicates within each experimental group were averaged together. Using the centered datasets, for each human astrocyte or oligodendrocyte dataset and Glast-dsRed spinal cord profile, we computed the global interprofile correlation (by Pearson's), using all genes in common between the two datasets (Zhang et al., 2014, 2016). For visual display, global correlation profiles of biological triplicates within each Glast-dsRed population group were averaged together.
In situ hybridization.
Nonradioactive in situ hybridization (ISH) was performed as previously described (Deneen et al., 2006). The mouse probes mGlast, mFGFR3, mPDGFRα, mouse myelin proteolipid protein, mouse myelin basic protein (mMBP) were previously described (Deneen et al., 2006). All other probes were made from expressed sequence tags obtained from Dharmacon for mTom1l1 (EcoRI/T3), mMfge8 (EcoRI/T3), mAsef (EcoRI/T3), and mGpr37l1 (NotI/SP6). Double ISH was performed using α-Digoxigenin-POD (Roche) for our candidate genes and α-Fluorescein-POD (Roche) for Glast, and developed with TSA Plus Cy5 Fluorescence and TSA Plus Fluorescein Fluorescence Systems (PerkinElmer). Double ISH/immunohistochemistry (IHC) was performed as previously described (Deneen et al., 2006). Briefly, ISH was performed first and developed with α-Digoxigenin-POD (Roche) and TSA Plus Cy5 Fluorescence System (PerkinElmer), then fluorescence staining was performed using α-GFAP (rabbit, DAKO) or α-GS (mouse, Millipore), with anti-mouse or anti-rabbit Alexa Fluor 488 (Invitrogen). All fluorescent slides were then counterstained with DAPI and mounted in Vectashield (Vector Laboratories).
IHC.
IHC on frozen tissue was performed as previously described (Deneen et al., 2006; Kang et al., 2012; Lee and Deneen, 2012; Lee et al., 2015). Briefly, tissue was incubated in PBS, followed by antigen retrieval when needed, blocking, and primary antibody incubation overnight at 4°C. The following antibodies were used: α-GFAP (rabbit, DAKO; mouse, Millipore), α-GS (mouse, Millipore), α-olig2 (rabbit, Millipore), α-iba1 (rabbit, Wako Pure Chemical Industries), α-AQP4 (rabbit, Millipore), α-dystroglycan (α-DAG; mouse, Leica), α-albumin (rabbit, Abcam), α-fibrinogen-FITC (Dako), α-SNTA1 (Alomone Labs), α-PDGFRβ (rabbit, Abcam), α-MBP (mouse, Covance), and α-BrDU (rat, Abcam). α-CD45-FITC was a gift from Dr. Margaret Goodell (Baylor College of Medicine). We used Alexa Fluor 488 or Alexa Fluor 568 (Invitrogen) for secondary antibodies.
IHC on paraffin tissue was previously described (Fancy et al., 2012). Adult brain tissues from multiple sclerosis (MS) patients were obtained from the tissue bank at the Rocky Mountain MS Center at University of Colorado. Hypoxic ischemic encephalopathy (HIE) tissue was obtained through Dr. Carrie Mohila at Texas Children's Hospital. Briefly, after deparaffinization, tissue was subjected to antigen retrieval for 7 min in 1× Na citrate, pH 6.0, followed by 15 min of 3% H2O2 incubation, blocking, and finally primary antibody incubation overnight. The following antibodies were used: α-asef (rabbit, Proteintech), α-tom1l1 (rabbit, Thermo Fisher Scientific), α-mfge8 (goat, Santa Cruz Biotechnology), and α-Gpr37l1 (mouse, MaB Technologies). Following washes, slides were incubated with ImmPRESS HRP anti-mouse, anti-rabbit, or anti-goat (Vector Laboratories). Colorimetric reaction was done using ImmPACT DAB peroxidase (Vector Laboratories). Slides were counterstained with Harris Hematoxylin (Poly Scientific R&D), rehydrated, and mounted using Permount (Fisher Chemicals). For double fluorescence IHC, we labeled Asef, Tom1l1, and Gpr37l1 with ImmPRESS AP anti-mouse or rabbit followed by Vector Red Alkaline Phosphatase colorimetric reaction, which fluoresces in the red channel. Next, GFAP staining was performed and visualized with Alexa Fluor 488 (Invitrogen). Slides were counterstained with DAPI and mounted in Vectashield.
Induction of demyelination with lysolecithin in mouse spinal cord.
Lysolecithin-induced demyelinated lesions were generated as previously described (Fancy et al., 2012; Lee et al., 2015). Briefly, 1% solution of lysolecithin (Sigma-Aldrich) was injected into the ventral white matter of the spinal cord of 10–12-week-old female Asef+/− and Asef−/− mice. Mice were perfused at 10 d postlesion (dpl) with 1× PBS and 4% PFA, postfixed overnight in PFA, and immersed in 30% sucrose overnight. Spinal cords were analyzed by ISH and fluorescence IHC.
Oligodendrocyte precursor cell culture and differentiation.
Oligodendrocyte progenitor (OLP) culture and differentiation were performed as described previously (Fancy et al., 2012; Glasgow et al., 2014; Lee et al., 2015). Briefly, Asef KO and Het E14.5 cortices were dissected, dissociated, and cultured in DMEM/F12, B27 supplement (Invitrogen), and 10 ng/ml EGF (R&D Systems) and 10 ng/ml bFGF (R&D Systems). Neurospheres were allowed to form for 4–6 d. Dissociated neurospheres were then plated on poly-d-lysine-coated coverslips in oligosphere proliferation media (OPM) composed of neural precursor medium supplemented with 10 ng/ml platelet-derived growth factor (PDGF; Peprotech), but without EGF. After 2 d, OLPs were induced to differentiate by replacing OPM with basal chemically defined medium supplemented with 15 nm tri-iodothyronine (Sigma-Aldrich), 10 ng/ml ciliary neurotrophic factor (Peprotech), and 5 mg/ml N-acetyl-L-cysteine (Sigma-Aldrich) for 7 d. Some wells were treated with BrDU (Sigma-Aldrich; 10 μm) for 1 h before harvesting. Cells were then fixed and stained by fluorescence IHC.
Coimmunoprecipitation.
Human HEK293T were maintained in DMEM, in the presence of 10% FBS and penicillin/streptomycin. A 3xFlag human Asef construct was obtained from Dr. Akiyama (University of Tokyo) and subcloned into 3xFlag-pcDNA. GFP-Rac1 was obtained from Dr. Kimberley Tolias (Baylor College of Medicine). Human influenza hemagglutinin (HA) SNTA was obtained from the Dana Farber/Harvard Cancer Center DNA Resource Core, and subcloned into 3xHA-PCS2. A combination of 5 μg of each constructs (and empty construct), with a maximum of 15 μg of DNA, was transfected using Superfect (Qiagen). Lysates were quantified using Bradford assay and the same amount of protein was used for immunoprecipitation. We performed immunoprecipitation using anti-HA (rat, Roche), and analyzed by SDS-PAGE/immunoblotting using anti-flag (mouse, Sigma-Aldrich), anti-GFP (rabbit, Genetex), and anti-HA (rabbit, Santa Cruz Biotechnology).
Evans blue quantification.
Evans blue was quantified as previously described (Menezes et al., 2014). Briefly, 6-month-old mice or P20 pups were injected with a filtered 2% Evans blue solution in PBS by intraperitoneal injection (150 μl/10 g of body weight). Twenty-four hours later, mice were perfused with 1× PBS, and spinal cords were harvested, weighed, and incubated with 500 μl of formamide for 24 h at 65°C at 750 rpm. Next day, samples were centrifuged for 30 min at 13,200 rpm and absorbance was read at 600 nm. In parallel, a standard curve of Evans blue was performed to calculate the amount of Evans in ng/μl/mg of tissue.
Results
Prospective isolation of astrocyte precursors in the developing spinal cord
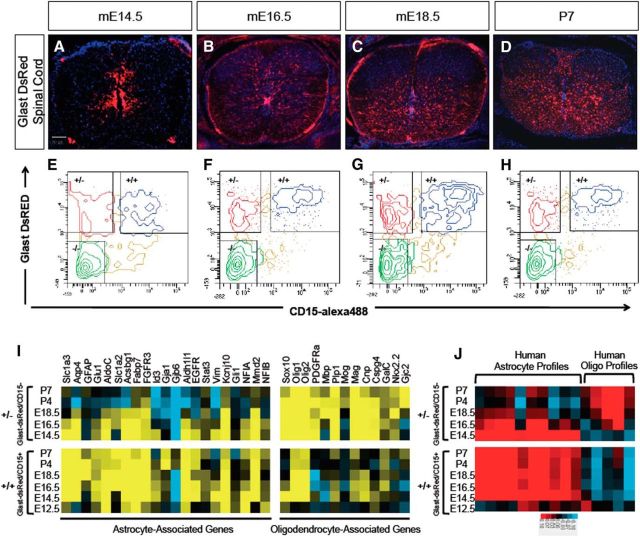
To begin dissecting the intermediate stages of astrocyte development, we performed in vivo temporal gene-expression profiling of astrocyte precursor cells from the spinal cord of Glast-dsRed mice (Regan et al., 2007). Glast is a glutamate transporter and its induction coincides with the gliogenic switch and continues into mature astrocytes in the spinal cord (Deneen et al., 2006). We chose six time points between mE12.5, the initiation of gliogenesis, and mP7, an early postnatal stage where astrocytes express mature markers. During this developmental interval, Glast-expressing cells emerge from the ventricular zone (VZ), populating both the mantle zone and the white matter (Fig. 1A–D). However, a subset of Glast-expressing cells remains in the VZ, where progenitor cells are located, marking cell populations in various lineages and differentiative states during spinal cord development.
Figure 1.
Isolation and expression profiling of Glast-dsRed populations. A–D, Glast-dsRed expression in the developing spinal cord. E–H, FACS plots showing prospective isolation of Glast-dsRed/CD15+ (+/+) and Glast-dsRed/Cd15− (+/−) populations across the E14.5–P7 interval. I, Heatmap analysis demonstrating expression of established astrocyte and oligodendrocyte lineage genes in the +/+ and +/− populations. J, Heat map showing intersample Pearson's correlations (red, positive; bright red or blue denotes r value >0.1 or <−0.1, respectively) between microarray profiles of Glast-dsRed populations and mRNA-Seq profiles from independent gene expression samples of human astrocytes and oligodendrocytes (Zhang et al., 2014).
To subfractionate prospective Glast-expressing astrocyte precursors from other potential Glast-expressing lineages, we used the cell-surface marker LexA/CD15, which has been used previously to purify astrocytes and various progenitor populations (Capela and Temple, 2002, 2006; Imura et al., 2006). Upon dissection of glast-dsRed spinal cords, we incubated dissociated cells with the CD15 antibody before FACS isolation. We isolated three populations at each time point: Glast-dsRed+/CD15− (henceforth +/−), Glast-dsRed+/CD15+ (henceforth +/+), Glast-dsRed−/CD15− (henceforth −/−; Fig. 1E–H). At E12.5, however, we only analyzed CD15+ populations (data not shown). Overall, these data demonstrate that combining CD15 with the Glast-dsRed mouse line is a suitable approach for prospectively isolating subpopulations of glial precursor populations in the developing spinal cord.
Astrocyte precursors demonstrate stage-specific gene-induction patterns
To delineate the molecular changes that occur over the E12.5–P7 interval, we FACS-isolated each population, performed microarray analysis from the prepared RNA, and subjected the resulting datasets to bioinformatics analysis (see Materials and Methods). To confirm that these populations are endowed with molecular features of glial cells, we assessed the expression of a cohort of established astrocyte and oligodendrocyte genes within each +/+ and +/− population across the time course. As indicated in Figure 1I, the +/− populations were enriched for oligodendrocyte genes, while the +/+ populations were enriched for astrocyte genes. Comparison with human astrocyte and oligodendrocyte gene-expression datasets revealed a similar association between the +/+ and +/− populations and the astrocyte and oligodendrocyte lineages (Fig. 1J; Zhang et al., 2014, 2016). Interestingly, in both mouse and human datasets, the +/− population demonstrated a gradual extinction of astrocyte-associated genes (Fig. 1I,J). The enrichment of astrocyte-associated genes in the +/+ population supports the use of CD15 as a means of enriching the bulk Glast-dsRed population for astrocytes and their precursors.
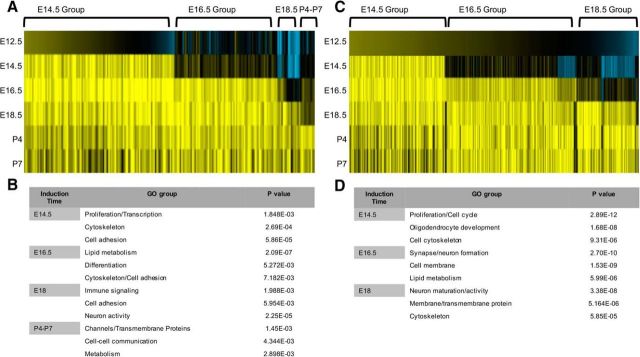
We identified dynamic and stage-specific patterns of gene induction in both the +/+ and +/− populations across the E14.5–P7 developmental interval (Fig. 2). Focusing on the +/+ population because it is enriched in established astrocyte gene signatures, we found that most changes in gene expression occurred relatively early in this population, with two broad groups of genes induced at E14.5 and E16.5 (Fig. 2A; Tables 1, 2). Gene ontology analysis of these groups revealed enrichment for genes associated with cell proliferation, cytoskeletal regulation, and cell adhesion (Fig. 2B; Tables 1, 2). Subsequent gene ontology analysis of the E18.5 and P4–P7 induction groups demonstrated enrichment for genes with metabolic and channel-associated functions (Fig. 2B; Tables 3, 4). Together, these gene induction patterns suggest two phases of astrocyte precursor differentiation across this interval: (1) proliferative/migratory and (2) metabolic/physiological.
Figure 2.
Identification of gene induction groups in astrocyte precursors. A, Heatmap analysis of the gene-expression changes across the E12.5–P7 interval in the +/+ populations. B, Gene ontology analysis of each induction group and the associated p values for gene sets associated with a given ontology in the +/+ populations. C, Heatmap analysis of the gene-expression changes across the E12.5–P7 interval in the +/− populations. D, Gene ontology analysis of each induction group and the associated p values for gene sets associated with a given ontology in the +/− populations.
Table 1.
Top genes induced at E14.5 in +/+ population
| Probe ID | Gene symbol | Relative fold change at E12.5 +/+ | Relative fold change at E14.5 +/+ |
|---|---|---|---|
| A_51_P355427 | Timp4 | 1.26 | 7.07 |
| A_51_P170463 | Gpr17 | 0.70 | 6.53 |
| A_52_P124734 | Cybrd1 | 0.87 | 5.80 |
| A_55_P2034585 | Gm6145 | 0.44 | 5.69 |
| A_51_P271311 | Slc6a11 | 1.07 | 5.66 |
| A_66_P136095 | Ctnna3 | 0.09 | 5.41 |
| A_51_P176352 | Ndrg2 | 1.10 | 5.20 |
| A_51_P268193 | Slc7a10 | 0.17 | 5.13 |
| A_51_P330044 | Cyp2j9 | 0.65 | 4.93 |
| A_51_P258690 | Scrg1 | 0.14 | 4.74 |
| A_55_P1959228 | Slc4a4 | −0.16 | 4.63 |
| A_51_P437240 | Emp2 | 0.60 | 4.63 |
| A_51_P312336 | Slc14a1 | 0.88 | 4.61 |
| A_55_P2156304 | Kcnj16 | 1.09 | 4.47 |
| A_55_P2026405 | Foxb1 | 1.29 | 4.42 |
| A_55_P1996674 | Itih3 | −0.07 | 4.37 |
| A_51_P473953 | Arhgef26 | 0.99 | 4.35 |
| A_51_P255682 | Mfge8 | 1.65 | 3.97 |
| A_55_P1959606 | Tom1l1 | 0.58 | 3.12 |
| A_51_P260504 | Arhgef4 | 0.36 | 2.84 |
Table 2.
Top genes induced at E16.5 in +/+ population
| Probe ID | Gene symbol | Relative fold change at E14.5 +/+ | Relative fold change at E16.5 +/+ |
|---|---|---|---|
| A_51_P336060 | Lcat | 1.98 | 4.17 |
| A_52_P625640 | Trim9 | 1.95 | 4.07 |
| A_51_P456465 | Cldn10 | 1.77 | 4.30 |
| A_51_P249193 | Gsg1l | 1.77 | 3.24 |
| A_55_P1954231 | Lrtm2 | 1.73 | 3.51 |
| A_52_P304720 | Crlf1 | 1.72 | 4.94 |
| A_52_P1008476 | Pou3f3 | 1.63 | 3.12 |
| A_55_P1963154 | Folh1 | 1.61 | 3.26 |
| A_52_P651248 | Zdhhc2 | 1.59 | 4.24 |
| A_66_P108276 | Gsg1l | 1.56 | 3.86 |
| A_51_P184300 | Dtna | 1.55 | 3.38 |
| A_55_P2036392 | Slc39a12 | 1.54 | 3.09 |
| A_52_P317393 | Gpr56 | 1.53 | 3.27 |
| A_52_P153291 | Ackr4 | 1.53 | 4.20 |
| A_55_P2110725 | Bai2 | 1.47 | 3.18 |
| A_55_P1991960 | Nwd1 | 1.45 | 3.17 |
| A_55_P2120060 | Dab1 | 1.43 | 3.90 |
| A_51_P513311 | Rxrg | 1.35 | 3.45 |
| A_55_P2066299 | Gpr137b | 1.34 | 3.12 |
| A_66_P110490 | Polr3h | 1.33 | 3.39 |
Table 3.
Top genes induced at E18.5 in +/+ population
| Probe ID | Gene symbol | Relative fold change at E16.5 +/+ | Relative fold change at E18.5 +/+ |
|---|---|---|---|
| A_55_P1969032 | Rgs9 | 1.05 | 3.95 |
| A_55_P1981909 | Ptprt | 0.47 | 3.61 |
| A_66_P105319 | Gm15706 | 0.86 | 3.56 |
| A_55_P1968858 | Cadps | 0.94 | 3.23 |
| A_52_P118310 | Chn1 | 0.95 | 3.05 |
| A_55_P2152248 | Cpe | −0.96 | 3.01 |
| A_51_P304125 | Slc29a2 | 0.89 | 3.00 |
| A_55_P2089076 | Msmp | 0.26 | 2.74 |
| A_55_P2044602 | Siah3 | 0.62 | 2.69 |
| A_52_P11402 | Lrrc24 | 0.92 | 2.69 |
| A_55_P2157245 | Gfap | −1.19 | 2.69 |
| A_51_P395373 | Ptprt | 0.50 | 2.66 |
| A_55_P2014229 | Sema4a | −0.50 | 2.63 |
| A_55_P2124741 | Cyp2j13 | −0.28 | 2.61 |
| A_55_P2014760 | Fam135b | 0.77 | 2.58 |
| A_55_P2120466 | Snx11 | 0.95 | 2.37 |
| A_52_P17982 | B3gat1 | 0.20 | 2.37 |
| A_52_P639402 | Kcnk3 | 0.57 | 2.34 |
| A_55_P1961720 | Kcnt1 | 0.48 | 2.23 |
| A_51_P445562 | Chst10 | −0.12 | 2.07 |
Table 4.
Top genes induced at P4–P7 in +/+ population
| Probe ID | Gene symbol | Relative fold change at E18.5 +/+ | Relative fold change at P4 +/+ |
|---|---|---|---|
| A_51_P452768 | Cyp4f14 | 0.35 | 3.74 |
| A_55_P2059864 | Igsf1 | −0.77 | 3.62 |
| A_52_P141687 | Gpr37l1 | 0.84 | 3.55 |
| A_51_P362066 | Chi3l1 | 0.79 | 3.49 |
| A_52_P655687 | Egfl6 | 1.39 | 3.46 |
| A_52_P174915 | Gja1 | 0.54 | 3.20 |
| A_55_P2199382 | Gm10635 | 1.13 | 2.85 |
| A_55_P1971025 | Paqr6 | 1.36 | 2.83 |
| A_55_P2184571 | Il1rapl1 | 1.03 | 2.76 |
| A_55_P2153743 | Slc26a6 | 1.58 | 2.39 |
| A_55_P2180934 | Gm8273 | 0.74 | 2.36 |
| A_55_P2110910 | Flrt1 | 1.04 | 2.34 |
| A_51_P257951 | Retnla | −0.36 | 2.33 |
| A_51_P476960 | Csmd3 | 0.65 | 2.30 |
| A_55_P2161375 | Galr1 | 1.11 | 2.29 |
| A_55_P2090065 | Myh15 | 0.24 | 2.25 |
| A_52_P600946 | Ccdc88c | 1.58 | 2.25 |
| A_55_P2032890 | Gm6588 | 1.29 | 2.22 |
| A_55_P2069974 | Kctd1 | 0.55 | 2.20 |
| A_66_P126110 | Pcdh11x | 1.12 | 2.19 |
Asef, Tom1l1, Mfge8, and Gpr37l1 are expressed in astrocyte precursors
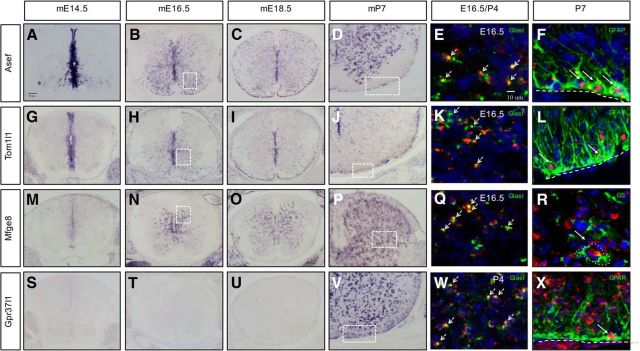
The bioinformatics analysis resulted in lists of genes from each of the +/+ induction groups (Tables 1–4), from which we extracted candidate markers and regulatory factors that contribute to astrocyte precursor development. The criteria for identifying these candidates include strong enrichment in a given +/+ induction group coupled with cross-referencing of established databases (Cahoy et al., 2008; Molofsky et al., 2013) and Allen brain atlas resources. We focused our attention on genes that had not been characterized previously in astrocyte precursors and those that demonstrated dynamic patterns of induction during this interval. To validate the expression dynamics of candidates, we used ISH to ascertain the temporal and spatial expression of the following genes: Asef/Arhgef4, Tom1l1, Mfge8, and Gpr37l1. Asef/Arhgef4 is a guanine exchange factor involved in colon cancer and cell migration via the small GTPases cdc42 and Rac1 (Kawasaki et al., 2000, 2003). Initially, Asef is induced in the VZ at the onset of gliogenesis at E12.5 (data not shown) and its expression is continuously upregulated in the mantle zone and white matter during the E14.5–P7 interval (Fig. 3A–D). Tom1l1 is a modulator of several signaling pathways, including the EGFR pathway (Li et al., 2005; Franco et al., 2006). ISH analysis indicated that Tom1l1, like Asef, is induced in the VZ during the gliogenic switch and its expression is maintained in presumptive astrocyte precursors that populate the mantle zone and white matter (Fig. 3G–J). Mfge8 is an inflammatory protein that is secreted by astrocytes and microglia (Kinugawa et al., 2013; Mills et al., 2015) and that remains uncharacterized during CNS development. We found that Mfge8-expressing cells appear at ∼E16.5, specifically in the mantle zone/gray matter, where they remain into postnatal stages (Fig. 3M–P). Gpr37l1 is a G-protein-coupled receptor recently shown to bind the neuroprotective and glioprotective factor prosaposin (Marazziti et al., 2013; Meyer et al., 2013) and to affect cerebellar development (Marazziti et al., 2013). Our analysis revealed that Gpr37l1 expression begins after birth in the early postnatal time points (P4; data not shown) in both the gray and white matter (Fig. 3S–V). Through our ISH analysis, we identified subsets of genes that are dynamically expressed during developmental gliogenesis in the spinal cord.
Figure 3.
Expression of Asef, Tom1l1, Mfge8, and Gpr37l1 in astrocyte precursors. A–D, ISH demonstrating expression of Asef in the developing spinal cord. E, F, Coexpression of Asef with Glast and GFAP. G–J, ISH demonstrating expression of Tom1l1 in the developing spinal cord. K, L, Coexpression of Tom1l1 with Glast and GFAP. M–P, ISH demonstrating expression of Mfge8 in the developing spinal cord. Q, R, Coexpression of Mfge8 with Glast and GS. S–V, ISH demonstrating Gpr37l1 expression in the developing spinal cord. W, X, Coexpression of Gpr37l1 with Glast and GFAP. Dashed boxed in B, H, and N correspond to E, K, and Q, respectively. Dashed boxes in D, J, P, and V correspond to F, L, R, and X respectively. Note that W corresponds to P4 spinal cord. Arrows denote coexpression of a given gene with the listed marker.
We next sought to confirm that these factors colocalize in Glast-expressing populations and further establish their expression in specific glial sublineages. Toward this end, we performed double-fluorescence ISH with these genes and Glast at either E16.5 or P4 for Gpr37l1, finding that each marker was predominately coexpressed with Glast (Fig. 3E,K,Q,W). Next, we assessed their coexpression with differentiated astrocyte markers, GFAP or GS, using ISH combined with immunostaining. For Asef, Tom1l1, and Gpr37l1, we confirmed colocalization with GFAP in the white matter of P7 spinal cords (Fig. 3F,L,X). For Mfge8, which is only expressed in the gray matter, we found colocalization with GS (Fig. 3R). These ISH studies validate the temporal profiling studies on astrocyte precursors and establish a set of new astrocyte precursor markers in the developing and early postnatal spinal cord.
Astrocyte precursor genes are expressed in human WMI
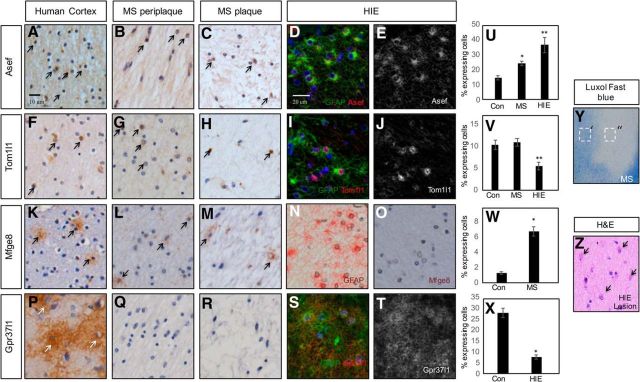
Many processes that oversee astrocyte development, including proliferation and induction of GFAP, are reused after injury. To test whether this group of astrocyte-associated genes are found in human neurological diseases that contain a reactive astrocyte component, we performed IHC on paraffin-embedded human brain tissue obtained from the Rocky Mountain MS Center at University of Colorado (MS lesion and controlateral white matter) and Texas Children's Hospital (HIE lesions). Among these genes, all are expressed in the adult human CNS, as seen by positive staining on the contralateral side (no MS lesions) of adult brains (Fig. 4A,F,K,P) further validating the link between the +/+ astrocyte population and human astrocyte expression datasets (Fig. 1J). Next, we examined expression of our candidate genes in WMI from both adult chronic inactive MS lesions and fetal HIE lesions. As indicated in Figure 4, we found that ASEF, TOM1l1, and MFGE8, but not GPR37l1, are expressed in the core of MS lesions (Fig. 4C,H,M) and periplaque regions (Fig. 4B,G,L). Staining of HIE lesions revealed that ASEF, TOM1l1, and GPR37l1 are present and colocalize with reactive astrocytes (Fig. 4D,E,I,J,S,T). These data, in conjunction with our developmental studies, reinforce the parallels between glial development and human neurological disorders, and implicate these genes in the pathogenesis of MS and HIE.
Figure 4.
ASEF, TOM1l1, MFGE8, and GPR37l1 are expressed in human WMI. A–C, IHC demonstrating expression of ASEF in adult human cortex (A) and in human MS plaques (B, C). D, E, Double immunofluorescence in neonatal HIE reveals coexpression of ASEF and GFAP. F–H, IHC demonstrating expression of TOM1l1 in adult human cortex (F) and in human MS plaques (G, H). I, J, Double immunofluorescence in neonatal HIE reveals coexpression of TOM1l1 and GFAP. K–M, IHC demonstrating expression of MFGE8 in adult human cortex (K) and in human MS plaques (L, M). N, O, IHC for GFAP (N) and MFGE8 (O) in neonatal HIE in adjacent section. No expression of Mfge8 is found in HIE with active gliosis. P–R, IHC demonstrating expression of GPR37l1 in adult human cortex (P) and no expression in human MS plaques (Q, R). S, T, Double immunofluorescence in neonatal HIE reveals coexpression of GPR37l1 and GFAP. Arrows denote IHC detection of expression of a given gene. U, Quantification of ASEF expression in MS and HIE; * and **p < 0.0001. V, Quantification of TOM1l1 expression in MS and HIE; * is not significant and **p = 0.005. W, Quantification of MFEG8 expression in MS; *p < 0.0001. X, Quantification of GPR37l1 expression in HIE; * and **p < 0.0001. Y, Luxol fast blue staining of human MS lesion. Dashed boxes denote periplaque (′) or plaque (″) regions. Z, H&E stain on neonatal HIE tissue. Arrows denote reactive gliosis.
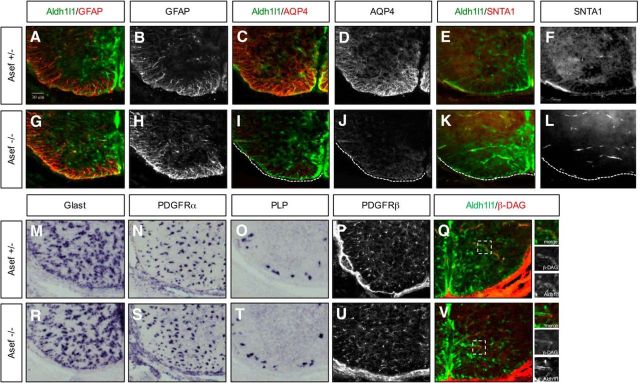
Asef is required for AQP4 and SNTA1 expression in the spinal cord
Because these candidate genes are expressed during astrocyte development and in human WMI, we examined their contributions to developmental and injury response. We focused on Asef, as the germline KO is viable, and fertile, without any gross defects (Kawasaki et al., 2000). Close examination of embryonic spinal cord development did not reveal any defects or delays in astrocyte or oligodendocyte specification, migration, or gross differentiation (Fig. 5A,B,G,H,M–O,R–T). However, further examination at E18.5 revealed a decrease in the expression of AQP4 in Aldh1l1-GFP astrocytes (Fig. 5C,D,I,J). Because AQP4 is expressed at astrocyte end feet, we examined the expression of a series of end-feet-associated proteins, finding that SNTA1, but not β-DAG, expression is also reduced in Aldh1l1-GFP astrocytes in the absence of Asef (Fig. 5E,F,K,L,Q–V). Together, these data indicate that Asef does not influence the specification or gross differentiation of oligodendrocytes or astrocytes, but is required for AQP4 and SNTA1 expression in astrocytes during spinal cord development.
Figure 5.
Asef regulates astrocyte differentiation in developing spinal cord. A–L, Immunofluorescence staining of astrocyte lineage markers at E18.5 in Aldh1l1-GFP;Asef+/− and Aldh1l1-GFP;Asef−/− mice. Aldh1l1-GFP;Asef−/− mice demonstrate reduced AQP4 (C, D vs I, J) and SNTA1 expression (E, F vs K, L) compared with Aldh1l1-GFP;Asef+/− mice. M–O, R–T, ISH of astrocyte precursor (Glast; M–R) and oligodendrocyte lineage makers (N–S, O–T) at E18.5 in Aldh1l1-GFP;Asef+/− and Aldh1l1-GFP;Asef−/− mice. P, U, Immunofluorescence staining of pericytes markers at E18.5. PDGFRβ expression is no different in Aldh1l1-GFP;Asef+/− and Aldh1l1-GFP;Asef−/− mice. Q, V, Immunofluorescence staining of β-DAG at E18.5 show no difference between Aldh1l1-GFP;Asef+/− and Aldh1l1-GFP;Asef−/− mice. Dashed boxes are blown up in adjacent panels and demonstrate colocalization of β-DAG in Aldh1l1-GFP-expressing cells.
Asef influences blood–brain barrier integrity and associates with SNTA1
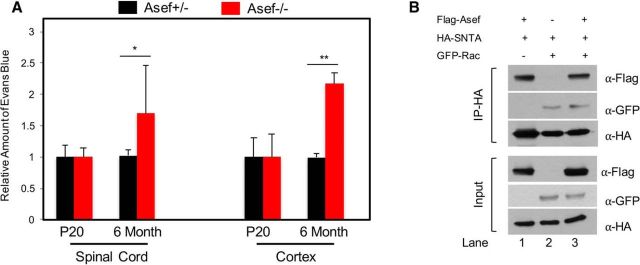
Because Asef influences the expression of key astrocyte end-feet proteins that have also been implicated in the generation and maintenance of the blood–brain barrier (BBB), we next assessed the integrity and permeability of the BBB using the Evans blue assay. Analysis of P20 mice revealed no difference in the amount of Evans blue extracted from the cortex or spinal cord of Asef+/− and Asef−/− mice (Fig. 6A), indicating that the developmental establishment of the BBB is maintained. Interestingly, mice lacking AQP4 and SNTA1 demonstrate BBB defects in adults (Neely et al., 2001; Amiry-Moghaddam et al., 2004; Bragg et al., 2006; Eilert-Olsen et al., 2012), suggesting that they play a role in maintaining the homeostatic integrity of the BBB. Therefore, we performed the Evans blue assay on 6-month-old Asef+/− and Asef−/− mice, finding an increase in the amount of Evans blue extracted from both the spinal cord and cortex of Asef−/− animals (Fig. 6A). Together these results suggest that the developmental defects in the expression of key astrocyte end-feet proteins in the Asef−/− mice manifests in the maintenance of BBB integrity in the adult.
Figure 6.
BBB is compromised in the adult CNS in the absence of Asef. A, Analysis of the relative amount of Evans blue extracted from the spinal cord or cortex from P20 or 6-month-old mice. In each case, for each genetic condition and time point, Evans blue was extracted and quantified as μg/μl/mg of tissue. The values presented are a relative comparison between Asef+/− and Asef−/−. N > 4 for each genetic condition, at each time point. B, Immunoblot demonstrating coimmunoprecipitation of SNTA and ASEF from 293 cells transfected with tagged versions of these proteins, with and without RAC transfection. Error bars in A are SEM and *p = 0.04 and **p = 0.007.
Interestingly, both SNTA and ASEF have been shown to associate with Rho-GTPases, suggesting that they may also associate with each other (Bhat et al., 2014). Therefore, to further investigate this potential mechanism, we performed a series of coimmunoprecipitation assays finding that SNTA and Asef coimmunoprecipitate, indicating a biochemical association (Fig. 6B, lane 1). Furthermore, both proteins also associate with Rac (Fig. 6B; lanes 2, 3), suggesting that modulation of Rho-GTPase function may be linked to the BBB-associated functions of these proteins in adult mice.
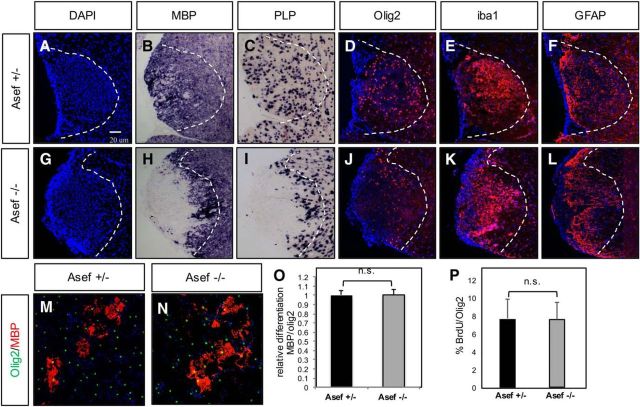
Asef is required for repair after WMI
As Asef is expressed in human WMI and regulates key astrocyte features implicated in disease pathogenesis, we examined whether it similarly contributes to repair in an associated animal-injury model. We used lysolecithin lesioning of the adult spinal cord, finding hypercellularity at the lesion site (Fig. 7A), which by 10 dpl was filled with oligodendrocyte precursor cells (OPCs) and differentiated oligodendrocytes in Asef+/− mice (Fig. 7B–D). In contrast, lesions in Asef−/− animals showed a significant delay in oligodendrocyte differentiation at 10 dpl (Fig. 7H,I), with Olig2-expressing OPCs residing at the periphery of the lesion (Fig. 7J). These observations suggest that OPCs from KO animals are able to proliferate and migrate to lesion sites and demonstrate an impaired capacity to differentiate.
Figure 7.
Loss of Asef results in delayed repair after WMI. A–L, Analysis of cellular responses after lysolecithin injury at 10 dpl in the adult spinal cord of Asef+/− (A–F) and Asef−/− (G–L) mice. Asef−/− mice demonstrate reduced expression of mature oligodendrocyte markers MBP and proteolipid protein compared with Asef+/− controls (B, C vs H, I), coupled with a redistribution of Olig2-expressing OPCs within the lesion (D vs J). Expression of reactive astrocyte (F vs L) and microglia (E vs K) markers within the lesion is unaffected. The images are representative of eight mice for each genotype, of which 2–3 sections for each marker were analyzed from each of these mice. M, N, Immunostaining of differentiated OPCs derived from Asef+/− and Asef−/− mice, with markers of mature oligodendrocytes (MBP) and precursors (Olig2). O, Quantification of the number of MBP-expressing cells across three experiments, performed in triplicate, indicates there is no difference in OPC differentiation between these genetic conditions. P, Quantification of the number of Olig2-expressing OPCs labeled with BrdU across three experiments, performed in triplicate, indicates that there is no difference in OPC proliferation between these genetic conditions.
We therefore asked whether loss of Asef influences OPC differentiation; however, in vitro culture of OPCs derived from Asef−/− OPCs did not demonstrate impaired proliferation or differentiation (Fig. 7M–P) and Asef−/− mice did not demonstrate overt defects in oligodendrocyte differentiation during development (Fig. 5N,O,Q,R). These observations suggest that impaired differentiation after injury is secondary to other cellular events in the primary lesion. While OPC differentiation and remyelination is the direct readout for WMI repair, other cell types, including microglia and reactive astrocytes, influence the repair process. Staining for markers of activated microglia or gross reactive astrocytes did not reveal any significant differences between Asef+/− or Asef−/− animals (Fig. 7E,F,K,L), suggesting the delay in oligodendrocyte differentiation after WMI in the Asef−/− mouse is likely secondary to other cellular events.
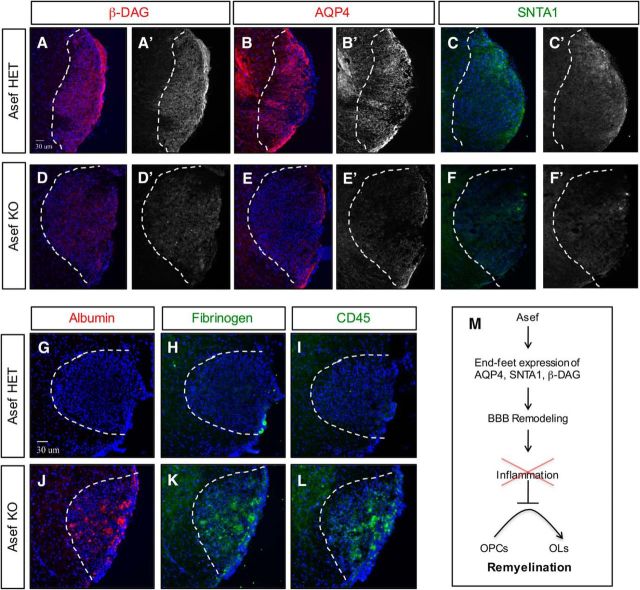
Given our observations in the Asef−/− mouse during spinal cord development, we assessed the expression of AQP4 and SNTA1, finding that both are substantially reduced in Asef−/− mice during repair (Fig. 8, compare B,C, E,F). These properties of Asef−/− reactive astrocytes parallel their developmental counterparts (Fig. 5C–F,I–L), leading us to further assess the expression of proteins associated with astrocyte end feet. Analysis of β-DAG, a key protein localized to end feet and linked to injury responses, revealed a significant decrease in the absence of Asef (Fig. 8, compare A, D). Importantly, both SNTA1 and β-DAG play a key role in the establishment and maintenance of the BBB after injury (Bragg et al., 2006; 2010; Puwarawuttipanit et al., 2006), suggesting that reactive astrocytes may not properly remodel the BBB after WMI in the absence of Asef. Therefore, these changes in the expression of key astrocyte end-feet proteins, coupled with our observations that Asef−/− mice demonstrate impaired BBB integrity in the adult CNS (Fig. 6A), led us to hypothesize that the BBB was also compromised after WMI. To test this possibility, we stained for proteins associated with BBB leakage, blood proteins (albumin and fibrinogen) and inflammatory markers (CD45), finding that both were present at very high levels in lesions from Asef−/− mice at 10 dpl (Fig. 8G–L). The increased blood and inflammatory cells in lesions from the Asef−/− mice suggest impaired BBB remodeling after WMI. Given the deleterious effects of blood proteins on OPC differentiation (Kremer et al., 2011), our observations further suggest impaired OPC differentiation after injury is the result of the presence of these proteins in the lesion (Fig. 8M).
Figure 8.
Impaired reactive astrocyte response in the absence of Asef. A–F, Immunofluorescence demonstrating expression of β-DAG, AQP4, and SNTA1 in Asef+/− (A–C) and Asef−/− (D–F) mice; sections are from adult spinal cord lysolecithin lesions at 10 dpl. Asef−/− mice exhibit decreased expression of β-DAG (A vs D), AQP4 (B vs E), and SNTA1 (C vs F) within lesions. G–L, Immunofluorescence demonstrating expression of albumin, fibrinogen, and CD45 in Asef+/− (G–I) and Asef−/− (J, K) mice at 10 dpl. Asef−/− mice exhibit increased expression of serum/blood proteins albumin and fibrinogen (G, H vs J, K), as well as increased presence of immune cells marked by CD45 (I vs L). The images are representative of four mice for each genotype, of which 2–3 sections for each marker were analyzed from each of these mice. M, Summary of Asef function in reactive astrocytes and BBB remodeling after WMI.
Discussion
In this study we performed gene-expression profiling on astrocyte precursor populations in the developing spinal cord, identifying distinct patterns of gene induction across the E12.5–P7 developmental interval and validated the expression of a subset of these genes in the astrocyte lineage. Applying these findings to human neurological disease, we found that these developmental genes are also expressed in reactive astrocyte populations in human WMI. Focusing our functional studies on Asef, we found that it plays a critical role in the expression of AQP4 and SNTA1 in astrocytes, during development and after WMI. Collectively, our studies identified distinct stages of astrocyte development in the embryonic spinal cord that are relevant to human WMI and revealed that Asef regulates astrocyte development and reactive astrocyte responses after injury.
Using astrocyte lineage reporter mice, in conjunction with FACS-based approaches, we prospectively isolated astrocyte precursor populations and identified distinct patterns of gene induction across embryonic and early postnatal development. Previous studies have set out to molecularly characterize astrocyte precursor populations in the developing spinal cord using the Aldh1l1-GFP reporter mouse (Molofsky et al., 2013). Here, we used an intersectional approach that combined the Glast-dsRed reporter (Regan et al., 2007) with the antibody CD15, which allowed us to interrogate two distinct populations: Glast+/CD15+ (+/+) and Glast+/CD15− (+/−). CD15 has been widely used for neural precursor cell isolation and is also expressed by astrocytes (Capela and Temple, 2002, 2006; Imura et al., 2006). Consistent with this we found that the +/+ populations show strong correlation with established astrocyte genes from both mouse and human studies, validating this approach and further distinguishing it from the other studies (Molofsky et al., 2013; Zhang et al., 2014). Importantly, subfractionation of these Glast-dsRed populations revealed that the +/− group is enriched in oligodendrocyte genes (Fig. 2C,D), highlighting the fact that many of these broad astrocyte precursor markers (like Glast and Aldh1l1) are also expressed in OPCs during early development. Consistent with this notion, at early time points the +/− population is enriched in both oligodendrocyte and astrocyte genes and eventually loses its antecedent astrocyte gene-expression pattern over the course of lineage development across both mouse and human systems, suggesting this is a conserved mechanism of lineage diversification (Fig. 1I,J; Tables 5–7). Indeed, key astrocyte fate determinants, NFIA and NFIB, which are initially expressed in OPCs and eventually downregulated, are dynamically expressed in the +/− population (Fig. 1I; Fancy et al., 2012; Glasgow et al., 2014). These observations suggest that many glial genes are initially expressed in both lineages and are later compartmentalized into the different sublineages.
Table 5.
Top genes induced at E14.5 in +/− population
| Probe ID | Gene symbol | Relative fold change at E14.5 +/+ |
|---|---|---|
| A_52_P603038 | Olig1 | 8.58 |
| A_51_P170463 | Gpr17 | 8.35 |
| A_55_P1971599 | Bcan | 7.34 |
| A_51_P258690 | Scrg1 | 6.72 |
| A_51_P355427 | Timp4 | 6.51 |
| A_55_P2105472 | Omg | 5.82 |
| A_55_P2011286 | HopX | 5.78 |
| A_51_P431543 | Prss56 | 5.71 |
| A_51_P468260 | S100b | 5.68 |
| A_51_P451588 | Plekhb1 | 5.58 |
| A_55_P2027083 | Kcnj10 | 5.34 |
| A_52_P584279 | Pnlip | 5.04 |
| A_51_P389834 | Zpld1 | 5.01 |
| A_51_P352452 | Dbx2 | 5.01 |
| A_51_P298107 | Vit | 4.99 |
| A_52_P579531 | Pdlim3 | 4.95 |
| A_52_P631547 | Cytl1 | 4.88 |
| A_52_P393314 | P2rx7 | 4.88 |
| A_55_P2156304 | Kcnj16 | 4.79 |
| A_51_P468456 | S100a3 | 4.76 |
Table 6.
Top genes induced at E16.5 in +/− population
| Probe ID | Gene symbol | Relative fold change at E14.5 +/+ | Relative fold change at E16.5 +/+ |
|---|---|---|---|
| A_55_P2043033 | Pla2g3 | 0.26 | 3.67 |
| A_55_P2227335 | Ptpre | −0.20 | 3.60 |
| A_52_P420665 | Uts2b | 1.57 | 3.54 |
| A_66_P126110 | Pcdh11x | 0.65 | 3.53 |
| A_55_P1979873 | Phactr3 | −0.72 | 3.52 |
| A_52_P179309 | AMPA3 | 0.51 | 3.50 |
| A_55_P2040260 | Acot5 | 0.21 | 3.47 |
| A_51_P506733 | P2rx7 | 1.19 | 3.47 |
| A_52_P439263 | Ugt8a | 0.66 | 3.39 |
| A_52_P707503 | Tmem212 | 0.74 | 3.33 |
| A_55_P2009206 | Sumo2 | 1.82 | 3.32 |
| A_55_P2111416 | Rcc2 | 0.78 | 3.30 |
| A_55_P1958532 | Hr | 1.59 | 3.27 |
| A_65_P09031 | Cdc42se2 | 0.40 | 3.25 |
| A_55_P2136847 | Slc1a2 | −1.41 | 3.24 |
| A_55_P1968683 | Anks1b | 0.82 | 3.23 |
| A_51_P245789 | Pcolce2 | 0.66 | 3.23 |
| A_55_P2107398 | Ncam2 | 1.03 | 3.22 |
| A_52_P184368 | Ppp1r16b | 0.94 | 3.22 |
| A_51_P306017 | Dll1 | 1.48 | 3.17 |
Table 7.
Top genes induced at E18.5 in +/− population
| Probe ID | Gene symbol | Relative fold change at E16.5 +/+ | Relative fold change at E18.5 +/+ |
|---|---|---|---|
| A_55_P2039876 | Gp1bb | 0.73 | 3.64 |
| A_52_P17982 | B3gat1 | 1.02 | 3.51 |
| A_51_P211260 | Dscaml1 | 1.28 | 3.48 |
| A_52_P604195 | Mbp | 0.68 | 3.34 |
| A_51_P102459 | Jph3 | 0.45 | 3.17 |
| A_52_P312204 | Nxph1 | 1.27 | 3.14 |
| A_52_P262201 | Atcay | 1.17 | 3.08 |
| A_52_P468343 | Bdh1 | 0.84 | 3.07 |
| A_52_P663757 | Tnik | 1.39 | 3.05 |
| A_51_P442642 | Cadm2 | 1.24 | 3.00 |
| A_52_P480266 | Atat1 | 0.88 | 2.92 |
| A_51_P220934 | Vstm5 | −0.01 | 2.91 |
| A_52_P202142 | Sv2a | −0.60 | 2.86 |
| A_55_P2085333 | Fbxo44 | 1.05 | 2.81 |
| A_55_P2066593 | Oprl1 | 0.70 | 2.81 |
| A_52_P531731 | Grin3a | 1.27 | 2.80 |
| A_51_P403799 | Sptbn2 | −0.11 | 2.78 |
| A_55_P1971139 | Fbll1 | 0.02 | 2.75 |
| A_51_P259029 | Dusp26 | 0.77 | 2.75 |
| A_66_P115072 | Prrt4 | 1.25 | 2.73 |
Focusing on the +/+ population, we identified four distinct induction groups over the E12.5–P7 interval. These induction groups can be broadly categorized based on their GO properties into two functional stages of lineage development: proliferation/migratory and metabolic/physiological (Fig. 2). Early induction groups (E14.5/E16.5) demonstrate enrichment in cell cycle/proliferation and cytoskeletonal genes, which follows the logic of astrocyte differentiation in vivo, where astrocyte precursors migrate and continue to proliferate outside germinal centers (Tien et al., 2012). In contrast, the later induction groups (E18.5/P4–P7) are enriched in immune signaling, metabolic, and ion channel activity genes, properties necessary to execute their physiological and metabolic roles subserving neurons. This biphasic wave of functional gene expression in the developing mouse spinal cord is consistent with recent in vitro studies suggesting that human astrocytes also contains two stages of lineage development (Zhang et al., 2016).
Our validation studies revealed that Asef, Gpr37l1, Mfge8, and Tom1l1 are dynamically expressed in developing astrocytes, mark intermediate stages of lineage development, and are expressed in broad subsets of astrocytes (i.e., gray vs white matter). The identification of markers associated with these intermediate stages of astrocyte development has been elusive, as the field has relied on markers associated with early specification (Glast, Aldh1l1, NFIA) or differentiation (GFAP, aldolase C, GS). This has, in turn, limited our basic understanding of lineage development and how regulatory factors operate during this process. Moreover, each of the principal cell types in the CNS has a well defined series of developmental markers associated with lineage development (Emery, 2010; Goldman and Kuypers, 2015; Lu et al., 2015; Toma and Hanashima, 2015; Bergles and Richardson, 2016) and it will be important to further expand this cohort for astrocytes by validating additional genes from these induction groups and interrogating expression in developing astrocytes in other regions of the CNS. Also, because many of the key properties of reactive astrocytes have foundations in development, including proliferation and induction of GFAP, expanding the scope of these markers and regulatory factors has critical implications for understanding the nature of reactive astrocytes and their contributions to neurological disease. The potential importance of these parallel states is underscored by our observations that each of the astrocyte precursor genes that we validated is also expressed in reactive astrocytes in human WMI, either both MS and HIE, or either lesion (Gpr37l1 vs Mfge8; Fig. 4). Therefore, by extension, it's possible that these different induction groups demarcate varying states of astrocyte reactivity across different neurological disorders, with the functions of their associated genes potentially revealing new insights into disease pathogenesis.
Toward this we focused our functional studies on Asef, which is a guanine exchange factor (GEF) for the Rho GTPase family that was originally characterized in colon cancer and shown to interact with adenomatous polyposis coli (Kawasaki et al., 2000). At the cellular level, Asef influences cytoskeleton dynamics and cell migration, through regulation of Rac or Cdc42 activity (Kawasaki et al., 2003; Mitin et al., 2007; Murayama et al., 2007). While recent studies have also implicated Asef in the regulation of the lung–blood barrier (Meng et al., 2015; Tian et al., 2015), its role in the CNS remains undefined. Here we show for the first time that Asef is expressed in the astrocyte lineage and contributes to astrocyte maturation during development and reactive astrocyte responses after WMI.
Analysis of astrocyte development in the spinal cord of the Asef KO mouse revealed specific developmental phenotypes. While proliferation, migration, and induction of maturation marker GFAP are unaffected, we detected a substantial decrease in the expression of AQP4 and SNTA1. Importantly, SNTA1 forms a complex with α-DAG and β-DAG, dystrophin, and dystrobrevin (Bragg et al., 2006, 2010; Boor et al., 2007; Bhat et al., 2013) and contributes to membrane clustering of AQP4, Kir4.1, or MLC-1 at astrocyte end feet (Nagelhus et al., 2004; Puwarawuttipanit et al., 2006; Boor et al., 2007; Masaki et al., 2010; Lien et al., 2012), suggesting that decreased expression of AQP4 could be the result of Asef regulation of SNTA1. Indeed, SNTA1 has been shown to bind Rac (Bhat et al., 2014) and the association between SNTA1 and Asef (Fig. 6B) supports a potential regulatory relationship between these proteins that contributes to BBB homeostasis in the adult and after injury.
Because Asef is expressed in reactive astrocytes in human WMI, we examined its role in the lysolecithin demyelinating model, finding impaired OPC differentiation in its absence. This delay in repair is likely the result of defects in BBB remodeling after injury as the lesions in the Asef mutant contained enhanced serum and blood proteins, which can suppress OPC differentiation and survival (Kremer et al., 2011). At the molecular level, the Asef mutants demonstrated decreased expression of SNTA1 and β-DAG, which play key roles in BBB formation and function. Moreover, we also witnessed decreased expression of AQP4. Together, these data suggest that loss of Asef results in reduced expression of key components of astrocyte end feet, which leads to defective BBB remodeling and increased leakage of blood proteins into the parenchyma, culminating in impaired regeneration. Importantly, these results after adult WMI parallel our observations in the adult spinal cord and cortex, where BBB integrity is also impaired (Fig. 6B), suggesting parallel regulatory role in the normal and injured CNS.
However, our observations in the adult (normal or WMI) do not completely recapitulate the developmental phenotypes of the Asef mutants, where we did not witness defects in β-DAG expression or compromised BBB integrity at perinatal stages (Figs. 5, 7). This highlights the fact that injury, pathology, or aging creates a specialized sensitized cellular state that differs from development and can directly influence gene expression or function. In the context of Asef function and BBB, it's conceivable that Asef regulation of β-DAG reflects an injury-specific relationship that affects re-establishment of the BBB and highlights the likelihood that distinct astrocyte-associated factors oversee BBB development and its re-establishment after injury. Importantly several genetic mouse models have found a correlation between altered astrocyte end feet and BBB disruption (Bragg et al., 2006, 2010; Lien et al., 2012; Alvarez et al., 2013; Menezes et al., 2014; Giocanti-Auregan et al., 2016). This, coupled with the recent observation of anti-Kir4.1 antibodies present in blood samples from human MS patients (Srivastava et al., 2012), suggests that alterations in astrocyte end feet can compromise BBB integrity, and potentially participate in MS pathogenesis. Moving forward, it will be important to discern how Asef influences the organization of astrocyte end feet after injury and how this role is linked to its established biochemical function.
Footnotes
This work was supported by grants from the Sontag Foundation (to B.D.), the National Multiple Sclerosis Society (RG-1501-02756 to B.D.), the Cancer Prevention Research Institute of Texas (RP510334 and RP160192 to B.D. and C.J.C.; and predoctoral Fellowship RP101499 to L.S.C.), and the National Institutes of Health (NS071153 to B.D.). This project was also supported in part by the Cytometry and Cell Sorting Core at Baylor College of Medicine with funding from the National Institutes of Health (P30 AI036211, P30 CA125123, and S10 RR024574) and the expert assistance of Joel M. Sederstrom; and by an Intellectual and Developmental Disabilities Research Center grant (1U54 HD083092) from the Eunice Kennedy Shriver National Institute of Child Health & Human Development. Human multiple sclerosis tissue was provided by the tissue bank at the Rocky Mountain MS Center at University of Colorado.
The authors declare no competing financial interests.
References
- Alvarez JI, Katayama T, Prat A. Glial influence on the blood brain barrier. Glia. 2013;61:1939–1958. doi: 10.1002/glia.22575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiry-Moghaddam M, Xue R, Haug FM, Neely JD, Bhardwaj A, Agre P, Adams ME, Froehner SC, Mori S, Ottersen OP. Alpha-syntrophin deletion removes the perivascular but not endothelial pool of aquaporin-4 at the blood–brain barrier and delays the development of brain edema in an experimental model of acute hyponatremia. FASEB J. 2004;18:542–544. doi: 10.1096/fj.03-0869fje. [DOI] [PubMed] [Google Scholar]
- Anderson MA, Ao Y, Sofroniew MV. Heterogeneity of reactive astrocytes. Neurosci Lett. 2014;565:23–29. doi: 10.1016/j.neulet.2013.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anlauf E, Derouiche A. Glutamine synthetase as an astrocytic marker: its cell type and vesicle localization. Front Endocrinol (Lausanne) 2013;4:144. doi: 10.3389/fendo.2013.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachoo RM, Kim RS, Ligon KL, Maher EA, Brennan C, Billings N, Chan S, Li C, Rowitch DH, Wong WH, DePinho RA. Molecular diversity of astrocytes with implications for neurological disorders. Proc Natl Acad Sci U S A. 2004;101:8384–8389. doi: 10.1073/pnas.0402140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergles DE, Richardson WD. Oligodendrocyte development and plasticity. Cold Spring Harb Perspect Biol. 2016;8:a020453. doi: 10.1101/cshperspect.a020453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat HF, Adams ME, Khanday FA. Syntrophin proteins as Santa Claus: role(s) in cell signal transduction. Cell Mol Life Sci. 2013;70:2533–2554. doi: 10.1007/s00018-012-1233-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat HF, Baba RA, Adams ME, Khanday FA. Role of SNTA1 in Rac1 activation, modulation of ROS generation, and migratory potential of human breast cancer cells. Br J Cancer. 2014;110:706–714. doi: 10.1038/bjc.2013.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boor I, Nagtegaal M, Kamphorst W, van der Valk P, Pronk JC, van Horssen J, Dinopoulos A, Bove KE, Pascual-Castroviejo I, Muntoni F, Estévez R, Scheper GC, van der Knaap MS. MLC1 is associated with the dystrophin-glycoprotein complex at astrocytic endfeet. Acta Neuropathol. 2007;114:403–410. doi: 10.1007/s00401-007-0247-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragg AD, Amiry-Moghaddam M, Ottersen OP, Adams ME, Froehner SC. Assembly of a perivascular astrocyte protein scaffold at the mammalian blood-brain barrier is dependent on alpha-syntrophin. Glia. 2006;53:879–890. doi: 10.1002/glia.20347. [DOI] [PubMed] [Google Scholar]
- Bragg AD, Das SS, Froehner SC. Dystrophin-associated protein scaffolding in brain requires alpha-dystrobrevin. Neuroreport. 2010;21:695–699. doi: 10.1097/WNR.0b013e32833b0a3b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, Thompson WJ, Barres BA. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capela A, Temple S. LeX/ssea-1 is expressed by adult mouse CNS stem cells, identifying them as nonependymal. Neuron. 2002;35:865–875. doi: 10.1016/S0896-6273(02)00835-8. [DOI] [PubMed] [Google Scholar]
- Capela A, Temple S. LeX is expressed by principle progenitor cells in the embryonic nervous system, is secreted into their environment and binds Wnt-1. Dev Biol. 2006;291:300–313. doi: 10.1016/j.ydbio.2005.12.030. [DOI] [PubMed] [Google Scholar]
- Chaboub LS, Deneen B. Astrocyte form and function in the developing central nervous system. Semin Pediatr Neurol. 2013;20:230–235. doi: 10.1016/j.spen.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton CJ, Casa A, Lazard Z, Huang S, Tsimelzon A, Hilsenbeck SG, Osborne CK, Lee AV. Insulin-like growth factor-I activates gene transcription programs strongly associated with poor breast cancer prognosis. J Clin Oncol. 2008;26:4078–4085. doi: 10.1200/JCO.2007.13.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deneen B, Ho R, Lukaszewicz A, Hochstim CJ, Gronostajski RM, Anderson DJ. The transcription factor NFIA controls the onset of gliogenesis in the developing spinal cord. Neuron. 2006;52:953–968. doi: 10.1016/j.neuron.2006.11.019. [DOI] [PubMed] [Google Scholar]
- Eilert-Olsen M, Haj-Yasein NN, Vindedal GF, Enger R, Gundersen GA, Hoddevik EH, Petersen PH, Haug FM, Skare Ø, Adams ME, Froehner SC, Burkhardt JM, Thoren AE, Nagelhus EA. Deletion of aquaporin-4 changes the perivascular glial protein scaffold without disrupting the brain endothelial barrier. Glia. 2012;60:432–440. doi: 10.1002/glia.22277. [DOI] [PubMed] [Google Scholar]
- Emery B. Regulation of oligodendrocyte differentiation and myelination. Science. 2010;330:779–782. doi: 10.1126/science.1190927. [DOI] [PubMed] [Google Scholar]
- Fancy SP, Glasgow SM, Finley M, Rowitch DH, Deneen B. Evidence that nuclear factor IA inhibits repair after white matter injury. Ann Neurol. 2012;72:224–233. doi: 10.1002/ana.23590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco M, Furstoss O, Simon V, Benistant C, Hong WJ, Roche S. The adaptor protein Tom1L1 is a negative regulator of Src mitogenic signaling induced by growth factors. Mol Cell Biol. 2006;26:1932–1947. doi: 10.1128/MCB.26.5.1932-1947.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giocanti-Auregan A, Vacca O, Bénard R, Cao S, Siqueiros L, Montañez C, Paques M, Sahel JA, Sennlaub F, Guillonneau X, Rendon A, Tadayoni R. Altered astrocyte morphology and vascular development in dystrophin-Dp71-null mice. Glia. 2016;64:716–729. doi: 10.1002/glia.22956. [DOI] [PubMed] [Google Scholar]
- Glasgow SM, Zhu W, Stolt CC, Huang TW, Chen F, LoTurco JJ, Neul JL, Wegner M, Mohila C, Deneen B. Mutual antagonism between Sox10 and NFIA regulates diversification of glial lineages and glioma subtypes. Nat Neurosci. 2014;17:1322–1329. doi: 10.1038/nn.3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman SA, Kuypers NJ. How to make an oligodendrocyte. Development. 2015;142:3983–3995. doi: 10.1242/dev.126409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imura T, Nakano I, Kornblum HI, Sofroniew MV. Phenotypic and functional heterogeneity of GFAP-expressing cells in vitro: differential expression of LeX/CD15 by GFAP-expressing multipotent neural stem cells and non-neurogenic astrocytes. Glia. 2006;53:277–293. doi: 10.1002/glia.20281. [DOI] [PubMed] [Google Scholar]
- John Lin CC, Deneen B. Astrocytes: the missing link in neurologic disease? Semin Pediatr Neurol. 2013;20:236–241. doi: 10.1016/j.spen.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang P, Lee HK, Glasgow SM, Finley M, Donti T, Gaber ZB, Graham BH, Foster AE, Novitch BG, Gronostajski RM, Deneen B. Sox9 and NFIA coordinate a transcriptional regulatory cascade during the initiation of gliogenesis. Neuron. 2012;74:79–94. doi: 10.1016/j.neuron.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki Y, Sato R, Akiyama T. Mutated APC and Asef are involved in the migration of colorectal tumour cells. Nat Cell Biol. 2003;5:211–215. doi: 10.1038/ncb937. [DOI] [PubMed] [Google Scholar]
- Kawasaki Y, Senda T, Ishidate T, Koyama R, Morishita T, Iwayama Y, Higuchi O, Akiyama T. Asef, a link between the tumor suppressor APC and G-protein signaling. Science. 2000;289:1194–1197. doi: 10.1126/science.289.5482.1194. [DOI] [PubMed] [Google Scholar]
- Kawasaki Y, Tsuji S, Muroya K, Furukawa S, Shibata Y, Okuno M, Ohwada S, Akiyama T. The adenomatous polyposis coli-associated exchange factors Asef and Asef2 are required for adenoma formation in Apc(Min/+)mice. EMBO Rep. 2009;10:1355–1362. doi: 10.1038/embor.2009.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinugawa K, Monnet Y, Lu L, Bekaert AJ, Théry C, Mallat Z, Hirsch EC, Hunot S. MFGE8 does not orchestrate clearance of apoptotic neurons in a mouse model of Parkinson's disease. Neurobiol Dis. 2013;51:192–201. doi: 10.1016/j.nbd.2012.11.010. [DOI] [PubMed] [Google Scholar]
- Kremer D, Aktas O, Hartung HP, Küry P. The complex world of oligodendroglial differentiation inhibitors. Ann Neurol. 2011;69:602–618. doi: 10.1002/ana.22415. [DOI] [PubMed] [Google Scholar]
- Lee HK, Chaboub LS, Zhu W, Zollinger D, Rasband MN, Fancy SP, Deneen B. Daam2-PIP5K is a regulatory pathway for Wnt signaling and therapeutic target for remyelination in the CNS. Neuron. 2015;85:1227–1243. doi: 10.1016/j.neuron.2015.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HK, Deneen B. Daam2 is required for dorsal patterning via modulation of canonical Wnt signaling in the developing spinal cord. Dev Cell. 2012;22:183–196. doi: 10.1016/j.devcel.2011.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Marshall C, Mei L, Dzubow L, Schmults C, Dans M, Seykora J. Srcasm modulates EGF and Src-kinase signaling in keratinocytes. J Biol Chem. 2005;280:6036–6046. doi: 10.1074/jbc.M406546200. [DOI] [PubMed] [Google Scholar]
- Lien CF, Mohanta SK, Frontczak-Baniewicz M, Swinny JD, Zablocka B, Górecki DC. Absence of glial alpha-dystrobrevin causes abnormalities of the blood–brain barrier and progressive brain edema. J Biol Chem. 2012;287:41374–41385. doi: 10.1074/jbc.M112.400044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu DC, Niu T, Alaynick WA. Molecular and cellular development of spinal cord locomotor circuitry. Front Mol Neurosci. 2015;8:25. doi: 10.3389/fnmol.2015.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marazziti D, Di Pietro C, Golini E, Mandillo S, La Sala G, Matteoni R, Tocchini-Valentini GP. Precocious cerebellum development and improved motor functions in mice lacking the astrocyte cilium-, patched 1-associated Gpr37l1 receptor. Proc Natl Acad Sci U S A. 2013;110:16486–16491. doi: 10.1073/pnas.1314819110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masaki H, Wakayama Y, Hara H, Jimi T, Unaki A, Iijima S, Oniki H, Nakano K, Kishimoto K, Hirayama Y. Immunocytochemical studies of aquaporin 4, Kir4.1, and alpha1-syntrophin in the astrocyte endfeet of mouse brain capillaries. Acta Histochem Cytochem. 2010;43:99–105. doi: 10.1267/ahc.10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menezes MJ, McClenahan FK, Leiton CV, Aranmolate A, Shan X, Colognato H. The extracellular matrix protein laminin α2 regulates the maturation and function of the blood–brain barrier. J Neurosci. 2014;34:15260–15280. doi: 10.1523/JNEUROSCI.3678-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F, Meliton A, Moldobaeva N, Mutlu G, Kawasaki Y, Akiyama T, Birukova AA. Asef mediates HGF protective effects against LPS-induced lung injury and endothelial barrier dysfunction. Am J Physiol Lung Cell Mol Physiol. 2015;308:L452–L463. doi: 10.1152/ajplung.00170.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer RC, Giddens MM, Schaefer SA, Hall RA. GPR37 and GPR37L1 are receptors for the neuroprotective and glioprotective factors prosaptide and prosaposin. Proc Natl Acad Sci U S A. 2013;110:9529–9534. doi: 10.1073/pnas.1219004110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills EA, Davis CH, Bushong EA, Boassa D, Kim KY, Ellisman MH, Marsh-Armstrong N. Astrocytes phagocytose focal dystrophies from shortening myelin segments in the optic nerve of Xenopus laevis at metamorphosis. Proc Natl Acad Sci U S A. 2015;112:10509–10514. doi: 10.1073/pnas.1506486112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitin N, Betts L, Yohe ME, Der CJ, Sondek J, Rossman KL. Release of autoinhibition of ASEF by APC leads to CDC42 activation and tumor suppression. Nat Struct Mol Biol. 2007;14:814–823. doi: 10.1038/nsmb1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AV, Deneen B. Astrocyte development: a guide for the perplexed. Glia. 2015;63:1320–1329. doi: 10.1002/glia.22836. [DOI] [PubMed] [Google Scholar]
- Molofsky AV, Krencik R, Krenick R, Ullian EM, Ullian E, Tsai HH, Deneen B, Richardson WD, Barres BA, Rowitch DH. Astrocytes and disease: a neurodevelopmental perspective. Genes Dev. 2012;26:891–907. doi: 10.1101/gad.188326.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AV, Glasgow SM, Chaboub LS, Tsai HH, Murnen AT, Kelley KW, Fancy SP, Yuen TJ, Madireddy L, Baranzini S, Deneen B, Rowitch DH, Oldham MC. Expression profiling of Aldh1l1-precursors in the developing spinal cord reveals glial lineage-specific genes and direct Sox9-Nfe2l1 interactions. Glia. 2013;61:1518–1532. doi: 10.1002/glia.22538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama K, Shirouzu M, Kawasaki Y, Kato-Murayama M, Hanawa-Suetsugu K, Sakamoto A, Katsura Y, Suenaga A, Toyama M, Terada T, Taiji M, Akiyama T, Yokoyama S. Crystal structure of the rac activator, Asef, reveals its autoinhibitory mechanism. J Biol Chem. 2007;282:4238–4242. doi: 10.1074/jbc.C600234200. [DOI] [PubMed] [Google Scholar]
- Nagelhus EA, Mathiisen TM, Ottersen OP. Aquaporin-4 in the central nervous system: cellular and subcellular distribution and coexpression with KIR4.1. Neuroscience. 2004;129:905–913. doi: 10.1016/j.neuroscience.2004.08.053. [DOI] [PubMed] [Google Scholar]
- Neely JD, Amiry-Moghaddam M, Ottersen OP, Froehner SC, Agre P, Adams ME. Syntrophin-dependent expression and localization of Aquaporin-4 water channel protein. Proc Natl Acad Sci U S A. 2001;98:14108–14113. doi: 10.1073/pnas.241508198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puwarawuttipanit W, Bragg AD, Frydenlund DS, Mylonakou MN, Nagelhus EA, Peters MF, Kotchabhakdi N, Adams ME, Froehner SC, Haug FM, Ottersen OP, Amiry-Moghaddam M. Differential effect of alpha-syntrophin knock-out on aquaporin-4 and Kir4.1 expression in retinal macroglial cells in mice. Neuroscience. 2006;137:165–175. doi: 10.1016/j.neuroscience.2005.08.051. [DOI] [PubMed] [Google Scholar]
- Regan MR, Huang YH, Kim YS, Dykes-Hoberg MI, Jin L, Watkins AM, Bergles DE, Rothstein JD. Variations in promoter activity reveal a differential expression and physiology of glutamate transporters by glia in the developing and mature CNS. J Neurosci. 2007;27:6607–6619. doi: 10.1523/JNEUROSCI.0790-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowitch DH, Kriegstein AR. Developmental genetics of vertebrate glial-cell specification. Nature. 2010;468:214–222. doi: 10.1038/nature09611. [DOI] [PubMed] [Google Scholar]
- Saldanha AJ. Java Treeview—extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Sofroniew MV. Molecular dissection of reactive astrogliosis and glial scar formation. Trends Neurosci. 2009;32:638–647. doi: 10.1016/j.tins.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofroniew MV, Vinters HV. Astrocytes: biology and pathology. Acta Neuropathol. 2010;119:7–35. doi: 10.1007/s00401-009-0619-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava R, Aslam M, Kalluri SR, Schirmer L, Buck D, Tackenberg B, Rothhammer V, Chan A, Gold R, Berthele A, Bennett JL, Korn T, Hemmer B. Potassium channel KIR4.1 as an immune target in multiple sclerosis. N Engl J Med. 2012;367:115–123. doi: 10.1056/NEJMoa1110740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, Tian Y, Gawlak G, Meng F, Kawasaki Y, Akiyama T, Birukova AA. Asef controls vascular endothelial permeability and barrier recovery in the lung. Mol Biol Cell. 2015;26:636–650. doi: 10.1091/mbc.E14-02-0725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien AC, Tsai HH, Molofsky AV, McMahon M, Foo LC, Kaul A, Dougherty JD, Heintz N, Gutmann DH, Barres BA, Rowitch DH. Regulated temporal-spatial astrocyte precursor cell proliferation involves BRAF signalling in mammalian spinal cord. Development. 2012;139:2477–2487. doi: 10.1242/dev.077214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toma K, Hanashima C. Switching modes in corticogenesis: mechanisms of neuronal subtype transitions and integration in the cerebral cortex. Front Neurosci. 2015;9:274. doi: 10.3389/fnins.2015.00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O'Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, Deng S, Liddelow SA, Zhang C, Daneman R, Maniatis T, Barres BA, Wu JQ. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Sloan SA, Clarke LE, Caneda C, Plaza CA, Blumenthal PD, Vogel H, Steinberg GK, Edwards MS, Li G, Duncan JA, 3rd, Cheshier SH, Shuer LM, Chang EF, Grant GA, Gephart MG, Barres BA. Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron. 2016;89:37–53. doi: 10.1016/j.neuron.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]