Abstract
RNA transcripts fold into secondary structures via intricate patterns of base pairing. These secondary structures impart catalytic, ligand binding, and scaffolding functions to a wide array of RNAs, forming a critical node of biological regulation. Among their many functions, RNA structural elements modulate epigenetic marks, alter mRNA stability and translation, regulate alternative splicing, transduce signals, and scaffold large macromolecular complexes. Thus, the study of RNA secondary structure is critical to understanding the function and regulation of RNA transcripts.
Here, we review the origins, form, and function of RNA secondary structure, focusing on plants. We then provide an overview of methods for probing secondary structure, from physical methods such as X-ray crystallography and nuclear magnetic resonance imaging (NMR) to chemical and nuclease probing methods. Marriage with high-throughput sequencing has enabled these latter methods to scale across whole transcriptomes, yielding tremendous new insights into the form and function of RNA secondary structure.
INTRODUCTION
What is RNA secondary structure?
All RNAs have the capacity to base pair via Watson-Crick, Hoogsteen, or sugar-edge patterns of hydrogen bonds (100, 153). Intermolecular RNA base pairing underlies the coding and replicative abilities of RNA, and enables RNA to serve as a specificity factor in guiding the activity of processes like RNA-directed DNA methylation (RdDM) and microRNA-mediated gene silencing. Intramolecular RNA base pairing is the basis of RNA secondary structure, and is a critical determinant of overall macromolecular folding. In conjunction with cofactors and RNA binding proteins (RBPs), secondary structure forms higher order tertiary structures and confers catalytic, regulatory, and scaffolding functions to RNA. In turn, disrupting the secondary structure of both coding and noncoding RNAs can cause widespread physiological perturbations. For instance, improper transfer RNA (tRNA) folding disrupts its intricate set of interactions with tRNA synthetases, cofactors, and the ribosome that are required for translation, thus impeding a process fundamental to life (7, 26). Secondary structure is known to be equally necessary to the functions of ribosomal RNAs (rRNAs) (128, 144, 164, 192), small nuclear RNAs (snRNAs) (39, 114), small nucleolar RNAs (snoRNAs) (44, 81, 82, 101, 127), and microRNAs (miRNAs) (19, 23, 92, 134, 145). Additionally, recent studies are beginning to demonstrate the importance of structure in long noncoding RNAs (lncRNAs) (129, 141, 174, 180) and messenger RNAs (mRNAs) (29, 49, 103, 149). Thus, a complete understanding of the regulation and functionality of RNAs will require methods to probe and manipulate RNA secondary structure. Here, we review these methods in the context of the form, origins, and function of RNA secondary structure.
How is RNA secondary structure formed?
As with protein folding, the formation of RNA secondary structure is not a simple matter of maximizing the number of stable chemical bonds to minimize free energy. Instead, RNA secondary structure is constrained by transcription, steric crowding, RBPs, and interacting ions. For instance, RNA folding is co-transcriptional, leading to “sequential folding” that can vary with the speed of RNA polymerase (RNAP) elongation (153). Moreover, RNA folding is guided by proteins and ribozymes with RNA chaperone activity during its initial formation to avoid “kinetic folding traps” (local free energy minima) and improper conformations (71, 109, 123, 153, 172). Thus, the correct in vivo structure of RNA may differ substantially from structures that spontaneously form in vitro or the minimum free energy (MFE) structures predicted in silico.
Chaperones are a diverse group of proteins functionally defined through their ability to facilitate RNA or protein refolding. RNA refolding is sometimes facilitated by ATP-dependent DEAD-box helicase domains (109, 123), but can also occur in the absence of external energy. Since chaperones are characterized by their abundance of disordered amino acids, a passive “entropy transfer” model has been proposed in which chaperones adopt the disordered conformation of actively folding RNAs, thus stabilizing RNA folding intermediates and enabling a more complete “conformational search” (71, 153, 172). Regardless of their mechanism, chaperones generally lack sequence specificity and possess a wide array of potential targets (71, 153, 172). As a result, loss of chaperone activity usually causes widespread misfolding and pleiotropic phenotypes (153).
In plants, the best characterized of these phenotypes involve osmotic stress (20, 45, 93) and cold shock (21, 66, 77, 78, 125, 139), both of which can over-stabilize RNA secondary structure. For instance, Arabidopsis thaliana (Arabidopsis) cold shock domain protein 3 (AtCSP3) shows in vitro RNA chaperone activity and promotes freezing tolerance (78). Similar CSPs have been characterized in rice (21, 77) and wheat (125). RNA chaperones can likewise confer tolerance to salt stress (20, 93), though it has yet to be determined whether this phenotype is directly related to their RNA chaperone activity. Intriguingly, bacterial CSPs can complement plant CSP mutants in promoting stress tolerance (20), and vice versa (76), suggesting broadly conserved chaperone functions. The functions of plant RNA chaperones in stress response have been recently reviewed in depth (71, 151), and suggest that correct RNA folding is crucial for plant physiology.
RNA binding proteins and their interaction with RNA secondary structure
Chaperones are only a small subset of the numerous RBPs that constrain and actively remodel RNA secondary structures throughout the RNA lifecycle. For instance, many RBPs contain RNA binding domains (RBDs) that preferentially bind to specific structural conformations of RNA (113). For instance, the RNA recognition motif (RRM) (28, 133) and K-homology (KH) domain (4, 14) specifically recognize single-stranded RNA (ssRNA), while the double-stranded RNA binding domain (dsRBD) preferentially binds double-stranded RNA (dsRNA) (150). RBPs can also target specific structural patterns, as illustrated by the sterile alpha motif (SAM) domain that only targets stem-loops in a “shape-specific” manner (130). Tandem arrays of RBDs can likewise yield preference for more complex higher-order sequence and structural elements. Notably, both RNA binding elements and RBPs undergo structural rearrangements in response to binding, in a type of induced fit (189). In this manner, RBPs can stabilize certain patterns of secondary structure, constraining the set of possible structural conformations that a RNA molecule can adopt.
It is equally important to note that even sequence-specific RBP target recognition depends on structure. For instance, non-Watson-Crick RNA base pairing can facilitate enlargement of the major groove of RNA helices, making it easier for ligands to bind dsRNA (153). Furthermore, other sequences are only accessible when exposed at the bulged loop of a stem-loop structure, such as the elements recognized by the zinc fingers of TFIIIA (111). Conversely, excess structure can sterically hinder binding of RBPs (72). Thus, definition of both sequence and structural motifs (48) will be an important part in understanding how RBPs recognize their target transcripts and ultimately form post-transcriptional regulatory networks (170).
Beyond constraining structure, there are also classes of RBPs that actively remodel RNA base pairing. For instance, ATP-dependent RNA helicases (most notably the ribosome) actively unwind RNA, consistent with the observation that RNA secondary structure in vivo is less than observed in vitro in a partially ATP-dependent manner (149). Conversely, RNA annealers such as Hfq and dsRBD-containing proteins speed the process of folding (124, 142). RNA secondary structure can likewise be remodeled by nonprotein ligands, such as metabolite-triggered riboswitches (11) and inorganic ions (32). As a result, methods that measure RNA secondary structure outside its native context may in fact yield incorrect predictions. In particular, algorithms that utilize free energy minimization such as RNAFold (61) often yield very different predictions of secondary structure than empirical structure mapping techniques (116, 176). Likewise, probing deproteinated RNA could yield different results than probing RNA in the context of its native RBPs. Thus, when studying RNA secondary structure it is critical to understand the limitations of each technique, since RNA folding occurs among a network of chaperones, RBPs, and cofactors.
CLASSES OF KNOWN STRUCTURED RNA MOLECULES
Riboswitches
RNA secondary structure is dynamic, and thus provides a mechanism to rapidly regulate RNAs. Perhaps the most striking example of dynamic regulation of RNA structure is based upon bistable riboswitches, an ancient class of structural elements that rapidly change conformations in response to binding of a ligand. Riboswitches are comprised of a structured RNA aptamer which binds a ligand, and an expression platform that interacts with other RBPs involved in transcript expression (148). Additionally, they contain conserved single-stranded portions that suggest the importance of tertiary structure (177). Ligand binding is coupled to structural rearrangements of the expression platform, resulting in altered transcript translation and/or stability (148). Currently, there are numerous known examples of riboswitches in prokaryotes, but only the thiamine pyrophosphate (TPP) riboswitch is known to be present in eukaryotes (120). In plants, thiamine is the ligand for the TPP riboswitch, which resides in the 3’ untranslated region (UTR) of the thiamine biosynthesis gene THIC and contains an alternative splice site. Thus, thiamine binding effects changes in splicing, polyadenylation, and ultimately transcript stability (11). The TPP riboswitch is highly conserved from algae to vascular plants (10, 11), and is also present in animals, fungi, and eubacteria, suggesting it is a fundamental component of pyrimidine metabolism. Discovery of additional riboswitches remains an open area of investigation in plants as well as eukaryotes as a whole.
Structure as an osmolarity and temperature sensor
RNA secondary structure is highly sensitive to temperature and ion osmolarity (32, 33, 75, 94), which is demonstrated by the importance of RNA chaperones to abiotic stress response (20, 21, 66, 77, 78, 93, 125, 139). However, this sensitivity could also provide sessile plants with a unique opportunity to rapidly sense changes in temperature and salinity. As with riboswitches, these sensory RNA structures are best characterized in prokaryotes, which are known to possess “RNA thermometers” that transduce RNA melting into regulatory changes (68, 85, 126). One example is bacterial heat shock transcripts, which contain structured elements that inhibit translation, but “melt” in higher temperatures (126). Since it remains unclear how plants directly sense temperature (117), systematic elucidation of plant RNA structural elements could provide insights into potential RNA thermometers and osmolarity sensors.
Ribozymes
RNA has the unique property of combining both catalytic and coding potential, perhaps owing to its role as a prototypical macromolecule in life on earth (47). These catalytic RNAs are known as ribozymes. One broad function of these RNAs is to direct RNA cleavage, either in cis (self-cleavage) or in trans (cleavage of other transcripts). For instance, the ribonucleoprotein (RNP) RNase P can direct cleavage of tRNAs with only its RNA core (53, 74). Cis-acting ribozymes include small repetitive self-cleaving RNAs, such as hammerhead and hairpin satellite sequences (18, 43). Self-splicing introns are also prominent examples of cis-acting ribozymes, and are common across bacteria and endosymbionts such as chloroplasts (178). Self-splicing introns were discovered over 30 years ago (88) and have been reviewed in depth (15, 56, 155). Interestingly, self-splicing introns were likely evolved to form catalytic components of the spliceosome, as evidenced by extensive homology between the U2:U6 complex of small nuclear RNAs (snRNAs) and group II self-splicing introns (39, 114). Additionally, the bacterial glucosamine-6-phosphate (GlcN6P) riboswitch has been shown to also function as a ribozyme, which cleaves an mRNA involved in GlcN6P metabolism (190). Thus, there is overlap between ribozymes and RNAs that were originally thought to function simply as structural scaffolds or sensors, hinting that many more ribozymes remain to be discovered.
tRNAs, rRNAs, and the translational machinery
The translation machinery is composed of an intricate array of RNPs, whose RNA components must properly fold over both time and space to function. For instance, folding of rRNAs is critical not only to form scaffolds and maintain the integrity of the ribosome, it may also play a role in the catalytic reactions of translation (144, 192). More specifically, structural studies have revealed that the peptidyl transferase center of the ribosome is composed of RNA, and is most likely a ribozyme (128, 164). Consistent with this hypothesis, certain drugs that inhibit translation directly target rRNAs (121, 122, 144). tRNAs must likewise adopt specific L-shaped tertiary structures, which are based upon “cloverleaf” secondary structures, in order to become aminoacylated and facilitate codon-anticodon interactions (7, 80, 147). tRNA structure is also modified by post-transcriptional covalent chemical modifications, which are guided by another class of highly structured RNAs called small nucleolar RNAs (snoRNAs).
snoRNAs
snoRNAs are relatively short (~150 nucleotides (nt)), highly structured noncoding RNAs that generally direct pseudouridylation and methylation of target tRNAs, rRNAs, and snRNAs (40, 75, 76, 117). Additionally, snoRNAs may play a role as RNA chaperones in aiding the folding of these same targets (101). In addition to forming secondary structures, snoRNAs pair with their target RNAs to form intermolecular (duplex) structures. RNA modifying proteins associate with the secondary structure of a snoRNA to form a snoRNP, and use these structural features as guideposts in directing highly stereotyped RNA modifications (44, 81, 82, 127).
miRNAs and siRNAs
microRNAs (miRNAs) and small interfering RNAs (siRNAs) are both components of the eukaryotic RNA silencing machinery, which ultimately uses these small RNAs (smRNAs) to direct transcript silencing and translational repression. In plants, these smRNAs likewise modulate epigenetic marks via RNA-directed DNA methylation (RdDM) (95). miRNAs and siRNAs diverge in their functional roles, but both share the requirement for precursors with paired secondary structures. The Arabidopsis miRNA processing protein DICER-LIKE1 (DCL1), for instance, targets imperfect stem-loops present in dedicated primary miRNA transcripts, forming pre-miRNAs that are cleaved once more into mature 21 nt miRNAs (92, 134, 145). The siRNA machinery targets a wide array of perfectly complementary RNAs produced from viruses, transposons, antisense transcripts, and transcripts made double stranded by RNA-dependent RNA polymerases (19, 23). Given the strict requirement for secondary structure, it is interesting to note that mRNAs with a high degree of secondary structure tend to be processed into smRNAs (103), suggesting structure may be a more general signal for DCL processing of other RNAs.
lncRNAs
Long noncoding RNAs (lncRNAs) are a class of transcripts that neither code for proteins nor bear resemblance to other known classes of noncoding RNAs (e.g. rRNAs, tRNAs). Compared to mRNAs, they show little conservation at the primary sequence. However, some lncRNAs show striking conservation at the level of synteny and function (129, 174), suggesting that selection may act upon structure rather than sequence. Accordingly, one of the “archetypal” functions of lncRNAs is to bind proteins via structural aptamers, and recruit their activity to modify DNA (141, 180). One of the first characterized examples of this functionality was the polycomb-recruiting lncRNA HOTAIR (146), and in plants similar mechanisms have been uncovered for the lncRNAs COLDAIR and COOLAIR, which recruit the polycomb repressive complex to the Arabidopsis FLOWERING LOCUS C (FLC) during vernalization (60, 168). lncRNAs can also function as molecular “scaffolds” that maintain the integrity of large complexes. One such example is TERC, an RNA component of the telomerase complex with no apparent catalytic activity (24, 193).
Coding mRNAs
A growing body of evidence is indicating that secondary structure regulates nearly every step of the mRNA lifecycle, including transcription (182), 5’ capping (31), splicing (17, 67, 105, 143, 184), polyadenylation (83, 131), nuclear export (52), localization (16, 165), translation (87, 167, 186), and turnover (48).The best characterized structural elements in mRNAs include internal ribosome entry sites (IRES) to recruit the ribosome (138), histone stem loops to recruit stabilizing factors to non-polyadenylated histone mRNAs (188), and iron response elements (IRE) to recruit RBPs in an iron-dependent manner (59). mRNA can likewise contain riboswitches (120), and even produce miRNAs from their introns and less often exons. Thus, secondary structure confers even further layers of complexity to the “molecular palimpsest” of mRNA. In this review, we will focus significant attention on the new insights into mRNA secondary structure gleaned from recent, transcriptome-wide structure mapping.
ORIGINS OF RNA SECONDARY STRUCTURE
The need for RNA molecules to perform a variety of coding, catalytic, and structural functions is best framed though an understanding of its origins. According to the generally accepted RNA world hypothesis, the first biological systems predated proteins and DNA, and were centrally dependent on RNA molecules (47, 70, 98). Eventually DNA would become the primary coding molecule, and protein the primary catalytic molecule, as life evolved into more familiar and complex systems. However, the earliest RNA molecules had the burden of storing genetic information and transmitting that code into a functional form without the benefit of other cellular machinery. To this end, RNAs adopted complex patterns of folding to expand their functions beyond the coding potential of their primary sequence (6). In turn, folding enabled RNAs to function as catalysts (ribozymes), scaffolds, and sensors.
However, the need for this dual functionality led to conflicting selective pressures, in what is referred to as Eigen’s Paradox. Specifically, these early RNA molecules had to be relatively short (less than 100 nt) in order to maintain genetic fidelity when undergoing replication in the absence of any proofreading machinery (35). However, there are only a handful of functional structural conformations that a RNA molecule of this size can adopt (90). Complex structures would require longer primary sequences, at the expense of genetic fidelity. One model that addresses this paradox posits some mutational flexibility that allow for numerous nucleotide changes while still preserving functional structure(89, 90), for instance through sequence covariation.
Another pitfall of this dual functionality is that structure and coding can be mutually inhibitory. For example, templating efficiency is correlated with a lack of structure (97, 166), while catalytic activity is known to require structural complexity (171). A potential model addressing this problem is to adopt a strategy of sub-molecular specialization, whereby complementary RNA molecules provide distinct and compatible functions (65). In this scenario, the complimentary strands of a heteroduplex dsRNA molecule, which possess similar degrees of Watson-Crick base pairing, could diverge in their secondary structure through G:U wobble base pairing, which form via Hoogsteen patterns of hydrogen bonding. While wobble base pairing stabilizes a folding structure in the catalytic strand, the reverse compliment A:C is not a stable base pair. In this manner, one strand could adopt more base pairing interactions to function as a catalyst (e.g. replication enzyme), while the other maintains less structure to preserve genetic information(65). Thus, non Watson-Crick base pairs can decouple stabilizing secondary structure across complimentary strands.
There are several lines of evidence that support the hypothesis of sub-molecular specialization. First, the computational models developed to study this theory of early biological systems support this idea by showing strong selection for division of labor when assuming moderate to strong tradeoff between templating and enzymatic activity, (13) and an increase in RNA molecule fitness when wobble base pairing is permitted (65). Finally, the sub-molecular specialization model is observed in “living fossil” species such as primitive viroids, which have physically asymmetric genomes (40). However, as with much of the work aimed at proving the existence of an initial RNA world, verification in the form of biochemical validation is lacking, in large part due to a dearth in understanding the exact biochemical and physical properties of the environment found in such a primordial world.
METHODS FOR PROBING RNA SECONDARY STRUCTURE
Physical methods
The earliest studies of RNA folding were designed to characterize the three dimensional shape of both prokaryotic (79) and eukaryotic (80, 147) tRNAs via X-ray crystallography. The high degree of structure and short length of tRNAs allows them to form crystallized structures more easily than other classes of RNA (62). Although it was a powerful technique in the early studies of RNA secondary structure, X-ray crystallography is limited to transcripts that readily form crystals (Table 1). Outside of small, highly structured RNAs like tRNAs, there are few classes of RNA that can be readily studied using this approach. Additionally, this technique utilizes in vitro folded transcripts, providing only a snapshot of the most energetically stable structure that forms in the buffer tested.
Table 1.
Summary of methods used to probe RNA secondary structure.
| Method | Biases/Limitations | RNA specificity |
Mechanism of Method | Experimental Context |
Refs | |
|---|---|---|---|---|---|---|
| Adducts | DMS | A,C specific | ssRNA | Alkalates the N-1 in A and the N- 3 in C |
in vivo and in vitro |
2, 3, 55, 64, 96, 99, 135, 136, 185, 194 |
| Diethyl Pyrocarbonate |
A specific | ssRNA | Carboxylates N-7 in A | in vitro | 135, 136 | |
| Hydrazine | U specific | ssRNA | Nucleophilic attack of U, removing the base |
in vitro | 135, 136 | |
| NAI/NAI-N3 | No bias, labels ribose sugar |
ssRNA | Acylates 2’ hydroxyl of unpaired nucleotides |
in vivo and in vitro |
55, 119, 187 | |
| Nucleases | RNase A | Cleaves after purines | ssRNA | Leaves 5’OH and 3’P | in vitro | 110, 173 |
| RNase T1 | Preferrential cleavage after guanisines |
ssRNA | Leaves 5’OH and 3’P | in vitro | 110 | |
| RNase U2 | Cleaves after pyrimidines |
ssRNA | Leaves 5’OH and 3’P | in vitro | 81, 179 | |
| RNase V1 | None | dsRNA | Leaves 5’P and 3’OH | in vitro | 38, 108 | |
| Nuclease P1 | None | ssRNA | Leaves 5’P and 3’OH | in vitro | 27, 84 | |
| Nuclease S1 | None | ssRNA | Leaves 5’P and 3’OH | in vitro | 27, 84 | |
| RNase I | None | ssRNA | Leaves 5’OH and 3’P | in vitro | 27, 84 | |
| Other | NMR | None | N/A | Aligns molecules in a magnetic field |
in vitro | 9, 12, 63, 195 |
| X-Ray Crystallography |
Must be crystallizable RNA, in vitro folding only |
N/A | Scatters X-rays in an interpretable pattern around an RNA crystal |
in vitro | 62, 79, 80, 147 | |
| In silico algorithms | Difficult to predict in vivo folding |
N/A | mostly predicts based on free energy and conservation |
in silico | 50, 51, 115, 197 |
Three categories of methods are covered; chemical adducts (red), RNases (green), and other (blue).
Conversely, the dynamics of RNA folding can be characterized using solution-state nuclear magnetic resonance (NMR). As opposed to crystallography, NMR can examine the dynamics of RNA folding. Early studies have focused on identifying dynamic secondary structure rearrangement in the lead-dependent ribozyme during autolytic cleavage (63), and in the U6 snRNA (9). More recent techniques have allowed greater resolution, allowing characterization of conformational changes on the picosecond time scale (12, 195). To date, both NMR and X-ray crystallography are still considered the gold standard in RNA secondary structure probing, revealing the three-dimensional shape of the transcript. However, they are very time and labor-intensive techniques, requiring exhaustive tests in numerous buffer conditions. These limitations prevent such physical methods from being utilized on a large scale.
In silico algorithms
Most algorithms to computationally predict RNA folding patterns are based on minimizing free energy (51, 115, 197). Though widely used, many of these algorithms do not account for protein interactions, evolutionary sequence conservation, or RNA dynamics (Table 1). Additionally, the fidelity of in silico techniques is known to decrease as a function of increasing RNA sequence length, often failing to reproduce known rRNA structures (197). As opposed to earlier algorithms, the Rfam algorithm offers some improvement by prioritizing the structure of evolutionarily conserved nucleotides, leading to higher fidelity (50). However, Rfam is still limited by sequence length, and its database of secondary structure does not include models for any full mRNA molecules. Therefore, experimentally probing structure is necessary to produce reliable models for mRNA folding.
Nuclease-based methods
Early studies of ribonucleases (RNases) revealed that many of these enzymes specifically cleave ssRNA. This discovery led to nuclease-based footprinting experiments to describe secondary structure of tRNAs (22). In these experiments, the tRNAs were treated with very low concentrations of a single-stranded RNase (ssRNase) in order to induce a single cleavage event within each transcript. This resulted in a population of partially digested transcripts, with each one terminating on a single-stranded nucleotide.
These fragments were then analyzed via end labeling, primer extension, and Sanger sequencing. During an end labeling experiment, the 5’ phosphate group is removed from the tRNA via phosphatase treatment, followed by addition of a radiolabeled phosphate through a polynucleotide kinase (PNK) reaction utilizing 32P-γ-ATP. This allows the 5’ end of each tRNA fragment to be visualized on film after separation via polyacrylamide gel electrophoresis (PAGE). Alternatively, the 3’ end of a fragment can be visualized via primer extension. In this technique, a radiolabeled primer is used in a reverse transcriptase (RT) reaction. The resulting DNA is then labeled near its 3’ end, and can be visualized via PAGE. These fragments can then be extracted from the gel and undergo Sanger sequencing (34).
In addition to secondary structure biases, RNases can have nucleotide biases, preferentially cleaving after one or more nucleotides. ssRNases with such a bias include RNase A, T1, and U2. RNase T1 preferentially hydrolyzes after guanosines (110), while RNase A and U2 cleave after purines and pyrimidines, respectively (Table 1) (173, 179). In contrast, nuclease P1, nuclease S1, and RNase I are ssRNases which cleave after each single-stranded nucleotide with equal efficiency (Table 1) (27, 84). These latter enzymes are therefore the preferred ssRNases for footprinting assays.
Although there are numerous ssRNases that can be used in footprinting assays, to date only one double-stranded RNase (dsRNase) has been identified and used in such experiments. Isolated from the venom of the Naja oxiana (Caspian cobra), RNase V1 preferentially cleaves dsRNA without nucleotide bias (Table 1) (38, 108). The enzyme has been shown to bind double helical RNA before cleavage, so it can also induce cleavage at single-stranded nucleotides within highly structured regions, such as loops within an RNA hairpin. Overall, when used in conjunction with ssRNases this enzyme has helped to produce a higher resolution image of secondary structure in several tRNAs and rRNAs (1, 38, 108).
Chemical-based methods
A third method of experimentally probing RNA secondary structure uses chemical adducts to modify single-stranded nucleotides. One of the first adducts used was dimethyl sulfate (DMS), which modifies unpaired adenines and cytosines. DMS was initially used in conjunction with diethyl pyrocarbonate and hydrazine, which modify adenosine and uridine respectively (Table 1), to label ssRNA in the yeast 5S rRNA and tRNAPhe (135, 136). Aniline was then used to induce strand breakage at the modified bases, allowing mapping via 5’ end labeling and PAGE. Subsequent studies revealed that RT cannot process these modified nucleotides, leading to complimentary DNA (cDNA) products terminating at the previous nucleotide, and allowing mapping via primer extension (64, 99). Although these early studies were performed in vitro, DMS has been shown to easily enter living cells (96), labeling adduct-accessible nucleotides in vivo (2, 3, 55, 185, 194).
In addition to occluding chemical adduct addition to the nucleoside, basepairing limits accessibility of the 2’ hydroxyl group on the ribose sugar (119). This limited accessibility is utilized in the selective 2’-hydroxyl acylation analyzed by primer extension sequencing (SHAPE), in which 2-methylnicotinic acid imidazolide (NAI) covalently modifies the 2’ hydroxyl of the ribose on unpaired nucleotides (Table 1) (119, 187). Unlike DMS and other nucleoside labeling based techniques, SHAPE labels the ribose sugar, and therefore has no nucleotide bias. Using a single reagent to label each accessible single-stranded nucleotide allows a higher resolution picture of the secondary structure of a transcript than DMS, diethyl pyrocarbonate, or hydrazine alone. However, dsRNA labeling chemical adducts are currently not available. While ssRNA can be directly identified by these approaches, paired bases are simply inferred by the lack of data from unlabeled nucleotides
High-throughput structure probing techniques
High-throughput sequencing techniques have revolutionized the study of RNA secondary structure. Several methods have been developed to investigate the structural landscape of eukaryotic transcriptomes. These methods utilize structure-specific nucleases or chemical adducts to identify single- or double-stranded nucleotides (Figure 1).
Figure 1. Methods for probing RNA secondary structure.
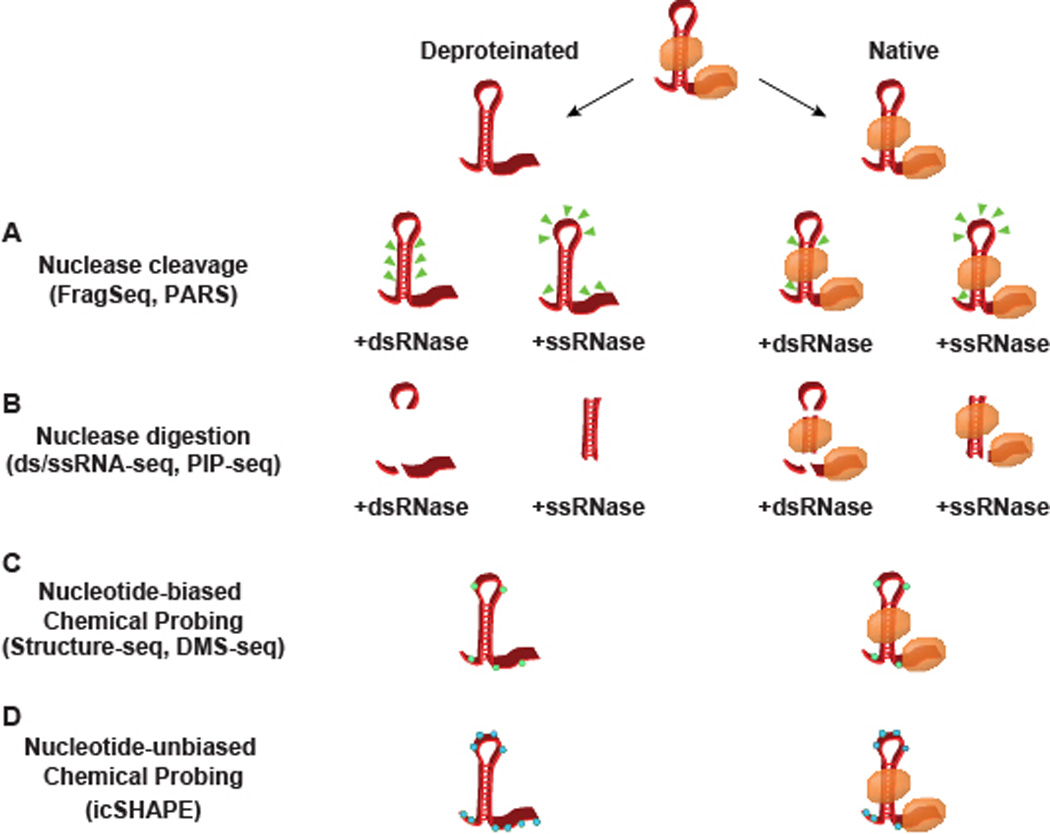
A schematic representation of the nuclease- and chemical-based probing techniques for empirically determining secondary structure. RNA can either be probed in a native state bound by RNA binding proteins (orange ovals) or deproteinated through extraction protocols or proteinase K treatement. (A) PARS assigns structure by the sites of transcript cleavage (green triangles), whereas (B) dsRNase/ssRNase-seq and PIP-seq both work by complete digestion. (C–D) Chemical probing works through reagents that preferentially adduct to nucleotides in a single-stranded confirmation, forming covalent modifications in either a (C) nucleotide-biased (green hexagons) or (D) unbiased (blue hexagons) manner. While multiple cleavage sites and covalent modifications are represented in this schematic, it is worth noting that PARS and the chemical probing techniques work with single-hit stoichiometry, with one cut/modification site interrogated per sequencing read.
Nuclease-based methods
Structure-specific RNases have been applied in a transcriptome-wide manner to reveal the global landscape of RNA secondary structure. One of the earlier genome-wide structure probing techniques was FragSeq (175), which utilized nuclease P1 to cleave ssRNA in mouse cells. This cleavage leaves a 5’ phosphate group (91), enabling their selective cloning and sequencing (Figure 1A). The 5’ most nucleotide in these reads therefore corresponds to an unpaired nucleotide, revealing ssRNA across the transcriptome with single nucleotide resolution (175). While powerful, this technique only identifies single-stranded regions, inferring dsRNA from a lack of reads. Other nuclease-based techniques have improved upon this method, identifying both single- and double-stranded regions.
A second nuclease-based approach is the parallel analysis of RNA structure (PARS) technique, which used both ss- and dsRNases to probe structure in yeast (73) and human tissue culture cells (183) (Figure 1A). To do this, the authors extracted polyadenylated RNA, which was subsequently denatured and allowed to reanneal in vitro. The renatured RNA was then treated with a dsRNase (RNase V1) or ssRNase (nuclease S1) with single-hit stoichiometry, and the resulting fragments underwent high-throughput sequencing to reveal sites of cleavage. Structure is defined as the ratio of coverage in dsRNA and ssRNA libraries, an estimate of the likelihood for a region to be single- or double-stranded. Unlike FragSeq, this technique provides a single nucleotide resolution view of both single- and double-stranded nucleotides.
The first high-throughput secondary structure analyses in plants were both nuclease-based. These studies utilized the combination of dsRNA-seq and ssRNA-seq, in which RNA is treated with either RNase I (an ssRNase) or RNase V1 (a dsRNase), respectively (Figure 1B). Unlike PARS, this technique fully digests ss- or dsRNA in a sample to allow every nucleotide of each sequencing read to be informative, offering greater sequencing depth at the expense of some resolution. In contrast, PARS and FragSeq only interrogates the structure of a single nucleotide per read. Like PARS, structure is defined by the ratio of dsRNA-seq to ssRNA-seq coverage. Although informative, each of these initial techniques required the denaturing and reannealing of RNA in vitro, thereby interrogating the folded RNA in a protein free environment. Protein interaction profile sequencing (PIP-seq) is a recently developed technique that identifies RNA secondary structure in its native state (41, 49, 158) (Figure 1B). This technique takes tissue or cells in which RNA-protein interactions have undergone crosslinking via formaldehyde or UV light, followed by ssRNA- and dsRNA-seq in both the presence and absence of proteins, allowing for simultaneous genome-wide identification of both RNA secondary structure and RNA-protein interactions.
Chemical adducts
The desire to better understand RNA secondary structure in vivo lead to the development of chemical adduct-based high-throughput approaches. These adducts can be added to tissue culture cells as well as eukaryotic organisms and modify ssRNA in vivo, revealing single-stranded protein unbound nucleotides. The first techniques were DMS-seq and Structure-seq, both of which utilized DMS to inhibit RT progression by adducting to mostly unpaired adenines and cytosines (29, 149) (Figure 1C). Like FragSeq, these data have single nucleotide resolution, with the added advantage of being in vivo assays. Although DMS only modifies single-stranded nucleotides, it is worth noting that RBP binding inhibits DMS addition (169), therefore this technique cannot differentiate between dsRNA and protein bound ssRNA sequences.
A more recently developed technique is the in vivo click selective 2’-hydroxyl acylation and profiling experiment (icSHAPE) (162). This method involves treatment of cells or tissues with 2-methylnicotinic acid imidazolide azide (NAI-N3) a cell permeable adduct that uniformly modifies the 2’-hydroxyl group of any ssRNA nucleotide (Figure 1D). The azide can then be biotinylated, allowing isolation of modified RNA with greatly reduced background, enabling SHAPE to be coupled with high-throughput sequencing. While this technique has single nucleotide resolution, it only modifies ssRNA and is subject to the same pitfalls as DMS-seq and Structure-seq.
SECONDARY STRUCTURE AND ITS FUNCTION
The advent of structure mapping, both of individual RNAs and in high-throughput, has significantly deepened our understanding of how RNA folding contributes to function. In particular, recent high-throughput methods have yielded insights into previously uncharacterized secondary structures in mRNAs and lncRNAs. Scaffolding and calatytic RNAs such as tRNAs and rRNAs have already been well-studied with physical methods and have been reviewed in depth (26, 46, 128, 164). Thus, we focus on the growing body of knowledge regarding structural elements in mRNA and lncRNAs.
Secondary structure and translation
One central role of mRNA structure is likely to regulate protein synthesis. In support of this hypothesis, high-throughput structure mapping of mRNAs consistently revealed a sharp decline in base pairing around the start and stop codons (49, 73) (Figure 2), and suggested that these regions of decreased structure are important for efficient translation (86, 87, 137). More specifically, it is thought that the single-stranded mRNA in these areas facilitates the interaction with the actively translating ribosome. Intriguingly, similar dips in secondary structure have been observed at actively translated upstream ORFs (uORFs) (181), indicating that both structure and sequence define start codons (Figure 2). Overall, stop codons also consist of an area of low structure that is highly conserved across organisms, with less well studied implications.
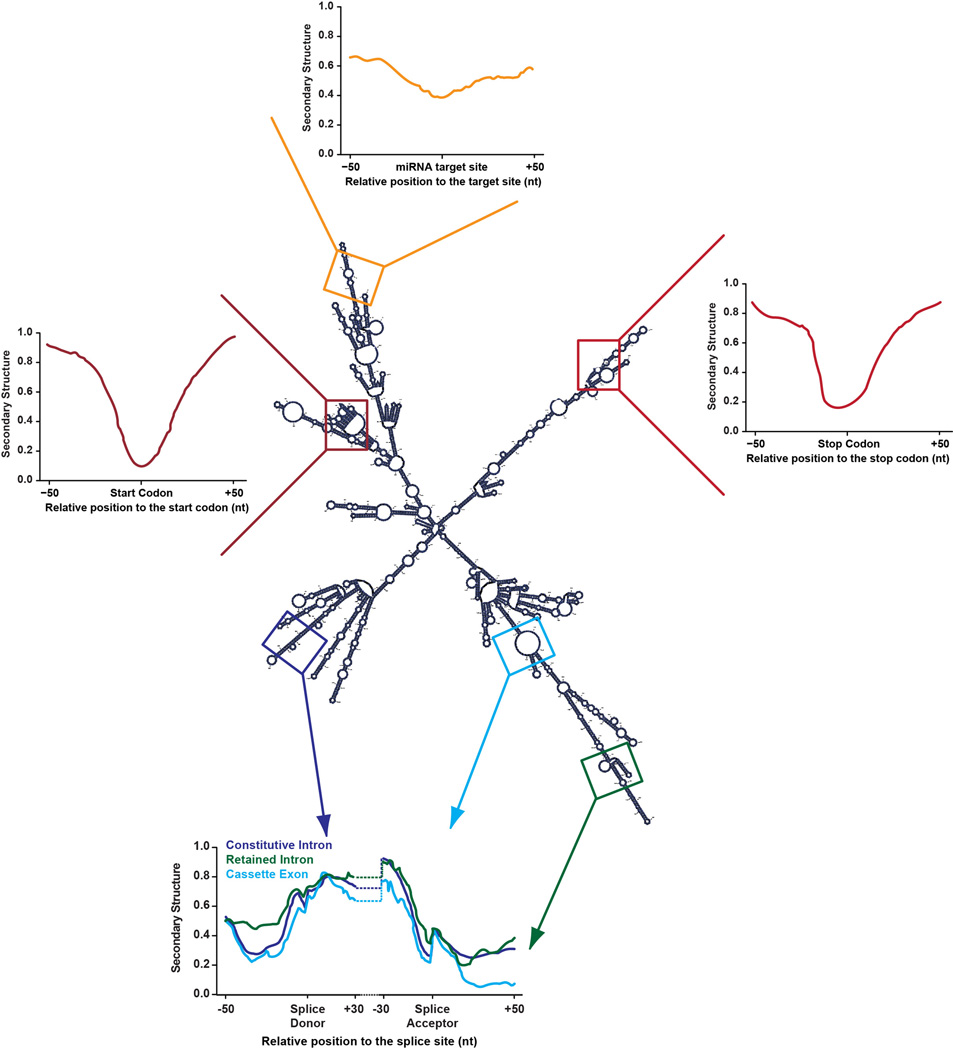
Figure 2. Structural patterns in mRNAs.
This is an example of a folded Arabidopsis thaliana mRNA. The displayed secondary structure profiles are representative of metagene patterns at start codons (dark red) miRNA target sites (orange), stop codons (red), constitutive intron (purple), retained intron (green), and cassette exon (blue) splice donor sites.
Beyond its importance in marking start and stop codons, RNA secondary structure also appears to have a functional role in defining the mRNA coding sequence (CDS). More specifically, it was recently noticed that plant CDSs but not UTRs display a three-nucleotide periodicity of secondary structure, in which every third nucleotide of each codon manifests an increased likelihood of being paired. This structural periodicity may provide a differentiating feature to allow the translational machinery to identify the protein coding from the non-coding regions of mRNAs (29). Furthermore, computational modeling has suggested that this three nucleotide periodicity is also useful for maintaining a helical structure in the mRNA CDS, potentially for the purpose of enhancing RNA stability (156). Thus, there are numerous mRNA structural features that likely have regulatory effects on translation.
Processing and stability
All canonical smRNAs (e.g. miRNAs, siRNAs) are processed from double-stranded precursors, suggesting that elements of high secondary structure in mRNAs might be similarly processed. In support of this hypothesis, nuclease-based structure mapping in Arabidopsis has revealed a positive correlation between secondary structure and smRNA processing (103). Furthermore, highly structured transcripts are in general less abundant and transcribed from more heterochromatic regions, suggesting that smRNA derived from highly structured transcripts could initiate RdDM (103). In mammals, secondary structural elements are also known to recruit RBPs that can either stabilize or destabilize mRNAs (48), so differential recruitment of RBPs might also explain the tendency of highly structured Arabidopsis RNAs to be less abundant. In support of this hypothesis, a recent study found that most regions of the Arabidopsis transcriptome that are bound by RBPs are less structured (49).
Alternative splicing
One ubiquitous form of eukaryotic post-transcriptional regulation is alternative splicing, which results in multiple mature RNA transcripts through mechanisms such as skipping of cassette exons and intron retention. RNA secondary structure has been shown to regulate alternative splicing, with highly structured regions being necessary for the production of certain splicing isoforms (160). One well-characterized example involves the human growth hormone hGH-N. This gene contains two splice acceptor sites, one located within a hairpin (acceptor site B) and a second one downstream of the hairpin. In one study, two point mutations that inhibited hairpin formation were introduced, which resulted in all transcripts using acceptor site B, while none used the downstream acceptor site. Introducing a complimentary mutations allowed the hairpin structure to refold, reducing usage of acceptor site B (37) and demonstrating this was a structure-specific phenomenon. Although there are numerous such single gene studies examining the link between splicing and RNA secondary structure (30, 36, 37, 159), global analyses have only recently been performed.
Recent global structure analyses revealed increased secondary structure at alternative splice sites. The DMS-based Structure-seq performed in whole Arabidopsis seedlings showed increased secondary structure upstream of alternative splice sites when compared to sequences of similar nucleotide composition (29). This finding was expanded upon in a PIP-seq study performed on nuclei from Arabidopsis seedlings, revealing distinct patterns of RNA secondary structure and RNA-protein interactions between alternative and constitutive splice sites (49). Specifically, retained introns were more highly structured across the upstream (donor) exon than constitutive introns, with similar structure at the downstream (acceptor) exon. Conversely, the structure at constitutive exons up- and downstream of annotated cassette exons exhibited a very different structural profile. While the upstream exon was similarly structured to constitutive introns donor splice sites, the downstream exon was significantly less structured (49) (Figure 2). These data indicated that RNA secondary structure is a global indicator of alternative splicing, and not just a feature of several specific transcripts.
microRNA targeting
Another method of post-transcriptional regulation is through mRNA stability via miRNA binding. In Arabidopsis, miRNAs are first transcribed as primary transcripts (pri-miRNAs) and are subsequently processed by DCL proteins and incorporated into an ARGONAUTE (AGO)-containing RNA induced silencing complex (RISC). These miRNAs then target mRNAs containing complementary sequences and induce RNA cleavage or inhibit translation, both of which ultimately lead to target transcript turnover (92, 134, 145). As this regulation requires formation of RNA duplexes, the miRNA target site must be accessible. In support of this hypothesis, dsRNA/ssRNA-seq analysis of unopened Arabidopsis flower buds revealed that miRNA target sites are significantly less structured than flanking regions (Figure 2), indicating that they are accessible to the miRNA bound RISC complex (103). Additionally, these results were replicated in Drosophila melanogaster and Caenorhabditis elegans via dsRNA/ssRNA-seq (102), indicating that this is a highly conserved phenomenon.
CONSERVATION OF STRUCTURE
The evolutionary conservation of secondary structure further supports its functional relevance. Perhaps the best example involves rRNAs, a fundamental component of all known organisms. Traditionally, when trying to trace evolutionary paths of organisms, it has been a common practice to tease out lineages and speciation events based on the similarities and divergences of rRNA sequences(8, 112). rRNA tends to accumulate substantial amounts of sequential changes while maintaining its structure. This covariation is due to selective pressures constraining the structure of rRNAs for their physical interactions with a multitude of protein components and biochemical activities(163).
In fact, rRNA was shown to be highly conserved in these regions of protein interaction and biochemical function, while quite variable and rapidly evolving in others (25, 132, 152). These properties made it a useful candidate for phylogenetic analysis (18), especially in bacteria that evolve rapidly and undergo horizontal gene transfer. Thus, using rRNA for phylogenetic studies has now been used in a multitude of different taxonomic groups and distinguishes taxa that would ordinarily be difficult to otherwise resolve. In fact, rRNA analyses have been used to aid our understanding in plants in a multitude of studies including bacterial symbiotes of plants (191), plant pathogens (104), and the origin of plant features themselves (154).
lncRNA conservation
lncRNAs are a class of noncoding RNAs where function is often more dependent on secondary structure than primary sequence (69, 118, 161). It is an interesting and important observation that lncRNAs display more sequence conservation than introns or random intergenic regions, but significantly less conservation than protein coding regions of mRNAs (exons) (54, 140). lncRNAs are poorly understood compared to many other RNAs, and in plants lncRNA research is particularly sparse (5). However, it is becoming clearer that plant lncRNAs often have important regulatory functions (107).
Intriguingly, despite the lack of sequence conservation in lncRNAs, there is a substantial amount of inter-species syntenic similarity, indicating a conservation of function not mediated by sequence. Even species as evolutionarily divergent as humans and zebrafish with virtually no sequence conservation at all maintaining a striking amount of synteny in their lncRNAs (174), which led to the hypothesis of conserved function. To test this idea, zebrafish embryos with a deficiency in the lncRNA cyrano were injected with the human or mouse orthologs, which only contained very small areas (~60 nt) of highly conserved primary nucleotide sequence. Remarkably, the orthologous lncRNAs were able to rescue the developmental phenotypes of the zebrafish lacking the cyrano ~60% of the time (174). This effect was not due to the conserved sequences , since introducing this 60 nt region into heterologous RNA failed to rescue the developmental defects (174). The combination of functional and syntenic conservation is highly indicative of the critical roles that structured lncRNAs play in certain cellular processes. Understanding how selective pressures affect structural maintenance of this class of RNAs needs to be a key focus moving forward.
FUTURE DIRECTIONS
Assigning Pairing Partners
Both chemical- and nuclease-based probing techniques can assign base pairing status to single nucleotides, but unlike physical methods are unable to determine the partner base of paired nucleotides. One method to overcome this problem is to leverage chemical and nuclease probing results as constraints for in silico algorithms (103, 116), for instance by assigning high-confidence paired and unpaired nucleotides (103). In fact, this approach has been shown to more accurately recapitulate known crystal structures (116). Nonetheless, constrained in silico folding is at best an educated guess, and cannot directly resolve secondary structure. Future approaches could apply new methods for mapping RNA-RNA interactions (57, 58) to map RNA secondary structural interactions at base pair resolution.
Chemical probing of double-stranded RNA
Chemical probing methods have the advantage of being directly applicable in vivo, but all have the same basic pitfall of being unable to directly probe paired regions of RNA molecules. All variants of chemical probing preferentially target relaxed RNAs, and thus provide only direct evidence from areas that lack structure. However, base pairing is inferred based on lack of evidence, which in certain cases can lead to spurious results. For instance, RBP binding will occlude chemical adduct addition, leading to the appearance of high structure regardless of actual pairing state (169). Chemical probing of dsRNA could circumvent this problem by defining base pairing based on positive rather than negative evidence. Additionally, certain methods of chemical probing rely upon RT stalling to infer adduct addition. Covalent chemical modifications likewise give rise to stalling (42), giving the appearance of low structure regardless of actual state. However, dsRNA-specific chemical probes should be equally sensitive to modification-induced stalling, and therefore bases with excessive stalls in both ssRNA- and dsRNA-probed libraries could be used to rule out covalent modifications.
Dynamic changes in secondary structure
While structural studies have examined global patterns of RNA folding in a single sample, RNA secondary structure is dynamic (195). Individual transcripts can be refolded by RBPs, or can have post-transcriptional covalent modifications resulting in drastic changes in secondary structure between samples (106). Although steady state conditions are ideal for developing novel techniques, the true biological questions need to address the dynamic nature of secondary structure, such as the effects of stress response and development on the global landscape of RNA folding. These studies can reveal additional riboswitches and other environmentally responsive structural elements within plants, allowing the identification of the complete collections of transcripts that adjust their structure in response to various stimuli. These studies of structure regulation will undoubtedly reveal new functions for RNA folding in plant biology.
Nuclear and cytoplasmic secondary structure
As of this writing, only a single study has examined RNA secondary structure in nuclei (49), with all previous studies having examined whole cell (mostly cytoplasmic) RNA folding (29, 73, 102, 103, 149, 162, 175, 196). While whole cell studies have consistently shown in every organism examined that the CDS of an mRNA is less structured than its UTRs, nuclear PIP-seq has revealed the opposite trend. There are many possible explanations for this trend, such as the secondary structure being rearranged by distinct cohorts of RBPs, or post-transcriptional covalent modifications leading to altered secondary structure in these two cellular locales. However, a close examination of RNA folding in both nuclear and cytoplasmic fractions must be performed to better understand these observed differences and parse apart their functional relevance.
Diversifying cell types and organisms
To date, the study of RNA secondary structure has been limited to only a few plant tissues in the Columbia (Col-0) ecotype of Arabidopsis (29, 41, 49). As RNA structure is such a dynamic moiety, it is necessary to expand these studies. This is because there is likely a substantial amount of structural variation between tissues and between individuals with mRNA polymorphisms (157). Furthermore, it is clear that RNA structure can be used as an extremely specific and subtle mechanism for fine-tuning a variety of cellular processes (29, 30, 49). However, we have yet to characterize the role of RNA secondary structure in distinguishing specific tissues and cell types.
Furthermore, there is a dearth of knowledge in our understanding of how RNA folding functions across different species of plants. Although RNA secondary structure has been investigated in a variety of animal systems, Arabidopsis is the only plant studied to date. Expanding such studies to other plants, especially those that are agriculturally important, will advance our understanding of the form and function of RNA structure in plants. This is likely to result in new insights and hypothesis generation for improvement of crop species in the future, which is an important consideration in this time of expanding world populations and global climate change.
CONCLUDING REMARKS
The advent of high-throughput structure mapping techniques has only begun to deepen our understanding of plant RNA secondary structure. Across both coding and noncoding RNAs, structure has proven to be a remarkably versatile element that enables RNAs to catalyze reactions, scaffold large macromolecular complexes, sense and transduce signals, and serve as hubs for post-transcriptional regulation. In particular, these new methods have highlighted the presence of secondary structure in mRNAs, adding an additional level of regulation to these information-dense molecules. While many challenges exist to directly measure native secondary structure across the transcriptome, the systematic study of secondary structure has already provided tremendous new insights into the form and function of RNAs, heralding a new age of structure.
Figure 3.
Acknowledgments
We thank the members of the Gregory lab for their helpful discussions. This work was funded by the NSF (Career Award MCB-1053846 and MCB-1243947 to BDG) and the National Institute of General Medical Sciences (T32GM008216-29 to SWF and T32GM007229-37 to LEV). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
REFERENCES
- 1.Andersen J, Delihas N, Hanas JS, Wu CW. 5s rna structure and interaction with transcription factor a. 1. ribonuclease probe of the structure of 5s rna from xenopus laevis oocytes. Biochemistry (Mosc.) 1984;23(24):5752–5759. doi: 10.1021/bi00319a013. [DOI] [PubMed] [Google Scholar]
- 2.Antal M, Boros E, Solymosy F, Kiss T. Analysis of the structure of human telomerase rna in vivo. Nucleic Acids Res. 2002;30(4):912–920. doi: 10.1093/nar/30.4.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ares M, Igel AH. Lethal and temperature-sensitive mutations their suppressors identify an essential structural element in u2 small nuclear rna. Genes Dev. 1990;4(12A):2132–2145. doi: 10.1101/gad.4.12a.2132. [DOI] [PubMed] [Google Scholar]
- 4.Backe PH, Messias AC, Ravelli RBG, Sattler M, Cusack S. X-ray crystallographic and nmr studies of the third kh domain of hnrnp k in complex with single-stranded nucleic acids. Structure. 2005;13(7):1055–1067. doi: 10.1016/j.str.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 5.Bai Y, Dai X, Harrison AP, Chen M. Rna regulatory networks in animals and plants: a long noncoding rna perspective. Brief. Funct. Genomics. 2015;14(2):91–101. doi: 10.1093/bfgp/elu017. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP, Unrau PJ. Constructing an rna world. Trends Cell Biol. 1999;9(12):M9–M13. [PubMed] [Google Scholar]
- 7.Bhaskaran H, Rodriguez-Hernandez A, Perona JJ. Kinetics of trna folding monitored by aminoacylation. RNA. 2012;18(3):569–580. doi: 10.1261/rna.030080.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhattacharya D, Medlin L. The phylogeny of plastids: a review based on comparisons of small-subunit ribosomal rna coding regions. J. Phycol. 1995;31(4):489–498. [Google Scholar]
- 9.Blad H, Reiter NJ, Abildgaard F, Markley JL, Butcher SE. Dynamics and metal ion binding in the u6 rna intramolecular stem-loop as analyzed by nmr. J. Mol. Biol. 2005;353(3):540–555. doi: 10.1016/j.jmb.2005.08.030. [DOI] [PubMed] [Google Scholar]
- 10.Bocobza S, Adato A, Mandel T, Shapira M, Nudler E, Aharoni A. Riboswitch-dependent gene regulation and its evolution in the plant kingdom. Genes Dev. 2007;21(22):2874–2879. doi: 10.1101/gad.443907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bocobza SE, Aharoni A. Small molecules that interact with rna: riboswitch-based gene control and its involvement in metabolic regulation in plants and algae. Plant J. 2014;79(4):693–703. doi: 10.1111/tpj.12540. [DOI] [PubMed] [Google Scholar]
- 12.Bothe JR, Nikolova EN, Eichhorn CD, Chugh J, Hansen AL, Al-Hashimi HM. Characterizing rna dynamics at atomic resolution using solution-state nmr spectroscopy. Nat. Methods. 2011;8(11):919–931. doi: 10.1038/nmeth.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boza G, Szilágyi A, Kun Á, Santos M, Szathmáry E. Evolution of the division of labor between genes and enzymes in the rna world. PLoS Comput Biol. 2014;10(12):e1003936. doi: 10.1371/journal.pcbi.1003936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Braddock DT, Louis JM, Baber JL, Levens D, Clore GM. Structure and dynamics of kh domains from fbp bound to single-stranded dna. Nature. 2002;415(6875):1051–1056. doi: 10.1038/4151051a. [DOI] [PubMed] [Google Scholar]
- 15.Brown GG, Colas des Francs-Small C, Ostersetzer-Biran O. Group ii intron splicing factors in plant mitochondria. Front. Plant Sci. 2014;5:35. doi: 10.3389/fpls.2014.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bullock SL, Ringel I, Ish-Horowicz D, Lukavsky PJ. A’-form rna helices are required for cytoplasmic mrna transport in drosophila. Nat. Struct. Mol. Biol. 2010;17(6):703–709. doi: 10.1038/nsmb.1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buratti E, Baralle FE. Influence of rna secondary structure on the pre-mrna splicing process. Mol. Cell. Biol. 2004;24(24):10505–10514. doi: 10.1128/MCB.24.24.10505-10514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buzayan JM, Gerlach WL, Bruening G. Non-enzymatic cleavage and ligation of rnas complementary to a plant virus satellite rna. Nature. 1986;323(6086):349–353. [Google Scholar]
- 19.Carthew RW, Sontheimer EJ. Origins and mechanisms of mirnas and sirnas. Cell. 2009;136(4):642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castiglioni P, Warner D, Bensen RJ, Anstrom DC, Harrison J, et al. Bacterial rna chaperones confer abiotic stress tolerance in plants and improved grain yield in maize under water-limited conditions. Plant Physiol. 2008;147(2):446–455. doi: 10.1104/pp.108.118828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaikam V, Karlson D. Functional characterization of two cold shock domain proteins from oryza sativa. Plant Cell Environ. 2008;31(7):995–1006. doi: 10.1111/j.1365-3040.2008.01811.x. [DOI] [PubMed] [Google Scholar]
- 22.Chang SH, RajBhandary UL. Studies on polynucleotides. lxxxi. yeast phenylalanine transfer ribonucleic acid: partial digestion with pancreatic ribonuclease. J. Biol. Chem. 1968;243(3):592–597. [PubMed] [Google Scholar]
- 23.Chapman EJ, Carrington JC. Specialization and evolution of endogenous small rna pathways. Nat. Rev. Genet. 2007;8(11):884–896. doi: 10.1038/nrg2179. [DOI] [PubMed] [Google Scholar]
- 24.Collins K. Physiological assembly and activity of human telomerase complexes. Mech. Ageing Dev. 2008;129(1–2):91–98. doi: 10.1016/j.mad.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Daniele Salvi GB. The analysis of rrna sequence-structure in phylogenetics: an application to the family pectinidae (mollusca: bivalvia) Mol. Phylogenet. Evol. 2010;56(3):1059–1067. doi: 10.1016/j.ympev.2010.04.025. [DOI] [PubMed] [Google Scholar]
- 26.Demeshkina N, Jenner L, Yusupova G, Yusupov M. Interactions of the ribosome with mrna and trna. Curr. Opin. Struct. Biol. 2010;20(3):325–332. doi: 10.1016/j.sbi.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 27.Desai NA, Shankar V. Single-strand-specific nucleases. FEMS Microbiol. Rev. 2003;26(5):457–491. doi: 10.1111/j.1574-6976.2003.tb00626.x. [DOI] [PubMed] [Google Scholar]
- 28.Ding J, Hayashi MK, Zhang Y, Manche L, Krainer AR, Xu R-M. Crystal structure of the two-rrm domain of hnrnp a1 (up1) complexed with single-stranded telomeric dna. Genes Dev. 1999;13(9):1102–1115. doi: 10.1101/gad.13.9.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ding Y, Tang Y, Kwok CK, Zhang Y, Bevilacqua PC, Assmann SM. In vivo genome-wide profiling of rna secondary structure reveals novel regulatory features. Nature. 2014;505:696–700. doi: 10.1038/nature12756. Structure-seq is an in vivo chemical-based structure probing method performed on Arabidopsis seedlings.
- 30.Donahue CP, Muratore C, Wu JY, Kosik KS, Wolfe MS. Stabilization of the tau exon 10 stem loop alters pre-mrna splicing. J. Biol. Chem. 2006;281(33):23302–23306. doi: 10.1074/jbc.C600143200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong H, Ray D, Ren S, Zhang B, Puig-Basagoiti F, et al. Distinct rna elements confer specificity to flavivirus rna cap methylation events. J. Virol. 2007;81(9):4412–4421. doi: 10.1128/JVI.02455-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Draper DE. A guide to ions and rna structure. RNA. 2004;10(3):335–343. doi: 10.1261/rna.5205404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Draper DE. Rna folding: thermodynamic and molecular descriptions of the roles of ions. Biophys. J. 2008;95(12):5489–5495. doi: 10.1529/biophysj.108.131813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ehresmann C, Baudin F, Mougel M, Romby P, Ebel J-P, Ehresmann B. Probing the structure of rnas in solution. Nucleic Acid Res. 1987;15(22):9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eigen M. Selforganization of matter and the evolution of biological macromolecules. Naturwissenschaften. 1971;58(10):465–523. doi: 10.1007/BF00623322. [DOI] [PubMed] [Google Scholar]
- 36.Eperon LP, Graham IR, Griffiths AD, Eperon IC. Effects of rna secondary structure on alternative splicing of pre-mrna: is folding limited to a region behind the transcribing rna polymerase? Cell. 1988;54(3):393–401. doi: 10.1016/0092-8674(88)90202-4. [DOI] [PubMed] [Google Scholar]
- 37.Estes PA, Cooke NE, Liebhaber SA. A native rna secondary structure controls alternative splice-site selection and generates two human growth hormone isoforms. J. Biol. Chem. 1992;267(21):14902–14908. [PubMed] [Google Scholar]
- 38.Favorova OO, Fasiolo F, Keith G, Vassilenko SK, Ebel JP. Partial digestion of trna--aminoacyl-trna synthetase complexes with cobra venom ribonuclease. Biochemistry (Mosc.) 1981;20(4):1006–1011. doi: 10.1021/bi00507a055. [DOI] [PubMed] [Google Scholar]
- 39.Fica SM, Tuttle N, Novak T, Li N-S, Lu J, et al. Rna catalyses nuclear pre-mrna splicing. Nature. 2013;503(7475):229–234. doi: 10.1038/nature12734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Flores R, Gago-Zachert S, Serra P, Sanjuán R, Elena SF. Viroids: survivors from the rna world? Annu. Rev. Microbiol. 2014;68(1):395–414. doi: 10.1146/annurev-micro-091313-103416. [DOI] [PubMed] [Google Scholar]
- 41.Foley SW, Vandivier LE, Kuksa PP, Gregory BD. Transcriptome-wide measurement of plant rna secondary structure. Curr. Opin. Plant Biol. 2015;27:36–43. doi: 10.1016/j.pbi.2015.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Foley SW, Vandivier LE, Kuksa PP, Gregory BD. Transcriptome-wide measurement of plant rna secondary structure. Curr. Opin. Plant Biol. 2015;27:36–43. doi: 10.1016/j.pbi.2015.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Forster AC, Symons RH. Self-cleavage of plus and minus rnas of a virusoid and a structural model for the active sites. Cell. 1987;49(2):211–220. doi: 10.1016/0092-8674(87)90562-9. [DOI] [PubMed] [Google Scholar]
- 44.Ganot P, Bortolin M-L, Kiss T. Site-specific pseudouridine formation in preribosomal rna is guided by small nucleolar rnas. Cell. 1997;89(5):799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 45.Chaulk GS, Smith−Frieday MN, Arthur DC, Culham DE, Edwards RA, et al. Proq is an rna chaperone that controls prop levels in escherichia coli. Biochemistry (Mosc.) 2011;50(15):3095–3106. doi: 10.1021/bi101683a. [DOI] [PubMed] [Google Scholar]
- 46.Giegé R, Jühling F, Pütz J, Stadler P, Sauter C, Florentz C. Structure of transfer rnas: similarity and variability. Wiley Interdiscip. Rev. RNA. 2012;3(1):37–61. doi: 10.1002/wrna.103. [DOI] [PubMed] [Google Scholar]
- 47.Gilbert W. Origin of life: the rna world. Nature. 1986;319(6055):618–618. [Google Scholar]
- 48.Goodarzi H, Najafabadi HS, Oikonomou P, Greco TM, Fish L, et al. Systematic discovery of structural elements governing stability of mammalian messenger rnas. Nature. 2012;485(7397):264–268. doi: 10.1038/nature11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gosai SJ, Foley SW, Wang D, Silverman IM, Selamoglu N, et al. Global analysis of the rna-protein interaction and rna secondary structure landscapes of the arabidopsis nucleus. Mol Cell. 2015;57:376–388. doi: 10.1016/j.molcel.2014.12.004. PIP-seq was performed on nuclei extracted from Arabidopsis seedlings, probing both RNA-protein interactions and RNA secondary structure simultaneously.
- 50.Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR. Rfam: an rna family database. Nucleic Acids Res. 2003;31(1):439–441. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL. The vienna rna websuite. Nucleic Acids Res. 2008;36:W70–W74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grüter P, Tabernero C, von Kobbe C, Schmitt C, Saavedra C, et al. Tap, the human homolog of mex67p, mediates cte-dependent rna export from the nucleus. Mol. Cell. 1998;1(5):649–659. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- 53.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The rna moiety of ribonuclease p is the catalytic subunit of the enzyme. Cell. 1983;35(3, Part 2):849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 54.Guttman M, Amit I, Garber M, French C, Lin MF, et al. Chromatin signature reveals over a thousand highly conserved large non-coding rnas in mammals. Nature. 2009;458(7235):223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harris KA, Crothers DM, Ullu E. In vivo structural analysis of spliced leader rnas in trypanosoma brucei and leptomonas collosoma: a flexible structure that is independent of cap4 methylations. RNA N. Y. N. 1995;1(4):351–362. [PMC free article] [PubMed] [Google Scholar]
- 56.Haugen P, Simon DM, Bhattacharya D. The natural history of group i introns. Trends Genet. TIG. 2005;21(2):111–119. doi: 10.1016/j.tig.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 57.Helwak A, Kudla G, Dudnakova T, Tollervey D. Mapping the human mirna interactome by clash reveals frequent noncanonical binding. Cell. 2013;153(3):654–665. doi: 10.1016/j.cell.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Helwak A, Tollervey D. Mapping the mirna interactome by cross-linking ligation and sequencing of hybrids (clash) Nat. Protoc. 2014;9(3):711–728. doi: 10.1038/nprot.2014.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hentze MW, Caughman SW, Rouault TA, Barriocanal JG, Dancis A, et al. Identification of the iron-responsive element for the translational regulation of human ferritin mrna. Science. 1987;238(4833):1570–1573. doi: 10.1126/science.3685996. [DOI] [PubMed] [Google Scholar]
- 60.Heo JB, Sung S. Vernalization-mediated epigenetic silencing by a long intronic noncoding rna. Science. 2011;331(6013):76–79. doi: 10.1126/science.1197349. [DOI] [PubMed] [Google Scholar]
- 61.Hofacker IL. Vienna rna secondary structure server. Nucleic Acids Res. 2003;31(13):3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Holbrook SR, Kim SH. Rna crystallography. Biopolymers. 1997;44(1):3–21. doi: 10.1002/(SICI)1097-0282(1997)44:1<3::AID-BIP2>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 63.Hoogstraten CG, Legault P, Pardi A. Nmr solution structure of the lead-dependent ribozyme: evidence for dynamics in rna catalysis1. J. Mol. Biol. 1998;284(2):337–350. doi: 10.1006/jmbi.1998.2182. [DOI] [PubMed] [Google Scholar]
- 64.Inoue T, Cech TR. Secondary structure of the circular form of the tetrahymena rrna intervening sequence: a technique for rna structure analysis using chemical probes and reverse transcriptase. Proc Natl Acad Sci. 1985;82:648–652. doi: 10.1073/pnas.82.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ivica NA, Obermayer B, Campbell GW, Rajamani S, Gerland U, Chen IA. The paradox of dual roles in the rna world: resolving the conflict between stable folding and templating ability. J. Mol. Evol. 2013;77(3):55–63. doi: 10.1007/s00239-013-9584-x. Both experimental and computational approaches demonstrate the role of wobble base pairing in promoting a division of labor between RNA strands.
- 66.Jiang W, Hou Y, Inouye M. Cspa, the major cold-shock protein of escherichia coli, is an rna chaperone. J. Biol. Chem. 1997;272(1):196–202. doi: 10.1074/jbc.272.1.196. [DOI] [PubMed] [Google Scholar]
- 67.Jin Y, Yang Y, Zhang P. New insights into rna secondary structure in the alternative splicing of pre-mrnas. RNA Biol. 2011;8(3):450–457. doi: 10.4161/rna.8.3.15388. [DOI] [PubMed] [Google Scholar]
- 68.Johansson J, Mandin P, Renzoni A, Chiaruttini C, Springer M, Cossart P. An rna thermosensor controls expression of virulence genes in listeria monocytogenes. Cell. 2002;110(5):551–561. doi: 10.1016/s0092-8674(02)00905-4. [DOI] [PubMed] [Google Scholar]
- 69.Johnsson P, Lipovich L, Grandér D, Morris KV. Evolutionary conservation of long non-coding rnas; sequence, structure, function. Biochim. Biophys. Acta BBA - Gen. Subj. 2014;1840(3):1063–1071. doi: 10.1016/j.bbagen.2013.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Joyce GF. Rna evolution and the origins of life. Nature. 1989;338(6212):217–224. doi: 10.1038/338217a0. A review of plant RNA chaperones that function to refold their target transcripts in response to cold stress.
- 71.Kang H, Park SJ, Kwak KJ. Plant rna chaperones in stress response. Trends Plant Sci. 2013;18(2):100–106. doi: 10.1016/j.tplants.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 72. Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microrna target recognition. Nat. Genet. 2007;39(10):1278–1284. doi: 10.1038/ng2135. PARS was one of the first transcriptome-wide structure probing techniques developed in yeast.
- 73.Kertesz M, Wan Y, Mazor E, Rinn JL, Nutter RC, et al. Genome-wide measurement of rna secondary structure in yeast. Nature. 2010;467(7311):103–107. doi: 10.1038/nature09322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kikovska E, Svärd SG, Kirsebom LA. Eukaryotic rnase p rna mediates cleavage in the absence of protein. Proc. Natl. Acad. Sci. U. S. A. 2007;104(7):2062–2067. doi: 10.1073/pnas.0607326104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kilburn D, Roh JH, Guo L, Briber RM, Woodson SA. Molecular crowding stabilizes folded rna structure by the excluded volume effect. J. Am. Chem. Soc. 2010;132(25):8690–8696. doi: 10.1021/ja101500g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim JS, Park SJ, Kwak KJ, Kim YO, Kim JY, et al. Cold shock domain proteins and glycine-rich rna-binding proteins from arabidopsis thaliana can promote the cold adaptation process in escherichia coli. Nucleic Acids Res. 2007;35(2):506–516. doi: 10.1093/nar/gkl1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim JY, Kim WY, Kwak KJ, Oh SH, Han YS, Kang H. Glycine-rich rna-binding proteins are functionally conserved in arabidopsis thaliana and oryza sativa during cold adaptation process. J. Exp. Bot. 2010;61(9):2317–2325. doi: 10.1093/jxb/erq058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim M-H, Sasaki K, Imai R. Cold shock domain protein 3 regulates freezing tolerance in arabidopsis thaliana. J. Biol. Chem. 2009;284(35):23454–23460. doi: 10.1074/jbc.M109.025791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim SH, Rich A. Single crystals of transfer rna: an x-ray diffraction study. Science. 1968;162(3860):1381–1384. doi: 10.1126/science.162.3860.1381. [DOI] [PubMed] [Google Scholar]
- 80.Kim SH, Suddath FL, Quigley GJ, McPherson A, Sussman JL, et al. Three-dimensional tertiary structure of yeast phenylalanine transfer rna. Science. 1974;185(4149):435–440. doi: 10.1126/science.185.4149.435. [DOI] [PubMed] [Google Scholar]
- 81.Kiss-László Z, Henry Y, Bachellerie J-P, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal rna: a novel function for small nucleolar rnas. Cell. 1996;85(7):1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 82.Kiss T. Small nucleolar rnas: an abundant group of noncoding rnas with diverse cellular functions. Cell. 2002;109(2):145–148. doi: 10.1016/s0092-8674(02)00718-3. [DOI] [PubMed] [Google Scholar]
- 83.Klasens BI, Das AT, Berkhout B. Inhibition of polyadenylation by stable rna secondary structure. Nucleic Acids Res. 1998;26(8):1870–1876. doi: 10.1093/nar/26.8.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Knapp G. Enzymatic approaches to probing of rna secondary and tertiary structure. Methods Enzymol. 1989;180:192–212. doi: 10.1016/0076-6879(89)80102-8. [DOI] [PubMed] [Google Scholar]
- 85.Kortmann J, Narberhaus F. Bacterial rna thermometers: molecular zippers and switches. Nat. Rev. Microbiol. 2012;10(4):255–265. doi: 10.1038/nrmicro2730. [DOI] [PubMed] [Google Scholar]
- 86.Kozak M. Influences of mrna secondary structure on initiation by eukaryotic ribosomes. Proc. Natl. Acad. Sci. U. S. A. 1986;83(9):2850–2854. doi: 10.1073/pnas.83.9.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kozak M. Leader length and secondary structure modulate mrna function under conditions of stress. Mol. Cell. Biol. 1988;8(7):2737–2744. doi: 10.1128/mcb.8.7.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR. Self-splicing rna: autoexcision and autocyclization of the ribosomal rna intervening sequence of tetrahymena. Cell. 1982;31(1):147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- 89.Kun Á, Santos M, Szathmáry E. Real ribozymes suggest a relaxed error threshold. Nat. Genet. 2005;37(9):1008–1011. doi: 10.1038/ng1621. [DOI] [PubMed] [Google Scholar]
- 90.Kun Á, Szilágyi A, Könnyű B, Boza G, Zachar I, Szathmáry E. The dynamics of the rna world: insights and challenges. Ann. N. Y. Acad. Sci. 2015;1341:75–95. doi: 10.1111/nyas.12700. [DOI] [PubMed] [Google Scholar]
- 91.Kuninaka A, Kibi M, Yoshino H, Sakaguchi K. Studies on 5′-phosphodiesterases in microorganisms. Agric. Biol. Chem. 1961;25(9):693–701. [Google Scholar]
- 92.Kurihara Y, Watanabe Y. Arabidopsis micro-rna biogenesis through dicer-like 1 protein functions. Proc. Natl. Acad. Sci. U. S. A. 2004;101(34):12753–12758. doi: 10.1073/pnas.0403115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kwak KJ, Kim YO, Kang H. Characterization of transgenic arabidopsis plants overexpressing gr-rbp4 under high salinity, dehydration, or cold stress. J. Exp. Bot. 2005;56(421):3007–3016. doi: 10.1093/jxb/eri298. [DOI] [PubMed] [Google Scholar]
- 94.Lambert D, Draper DE. Effects of osmolytes on rna secondary and tertiary structure stabilities and rna-mg2+ interactions. J. Mol. Biol. 2007;370(5):993–1005. doi: 10.1016/j.jmb.2007.03.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Law JA, Jacobsen SE. Establishing, maintaining and modifying dna methylation patterns in plants and animals. Nat. Rev. Genet. 2010;11(3):204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lawley PD, Brookes P. Further studies on the alkylation of nucleic acids and their constituent nucleotides. Biochem. J. 1963;89:127–138. doi: 10.1042/bj0890127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lawrence MS, Bartel DP. Processivity of ribozyme-catalyzed rna polymerization. Biochemistry (Mosc.) 2003;42(29):8748–8755. doi: 10.1021/bi034228l. [DOI] [PubMed] [Google Scholar]
- 98.Lazcano A, Miller SL. The origin and early evolution of life: prebiotic chemistry, the pre-rna world, and time. Cell. 1996;85(6):793–798. doi: 10.1016/s0092-8674(00)81263-5. [DOI] [PubMed] [Google Scholar]
- 99.Lempereur L, Nicoloso M, Riehl N, Ehresmann C, Ehresmann B, Bachellerie JP. Conformation of yeast 18s rrna. direct chemical probing of the 5’ domain in ribosomal subunits and in deproteinized rna by reverse transcriptase mapping of dimethyl sulfate-accessible. Nucleic Acids Res. 1985;13(23):8339–8357. doi: 10.1093/nar/13.23.8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Leontis NB, Westhof E. Geometric nomenclature and classification of rna base pairs. RNA. 2001;7(4):499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lestrade L, Weber MJ. Snorna-lbme-db, a comprehensive database of human h/aca and c/d box snornas. Nucleic Acids Res. 2006;34(suppl 1):D158–D162. doi: 10.1093/nar/gkj002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li F, Zheng Q, Ryvkin P, Dragomir I, Desai Y, et al. Global analysis of rna secondary structure in two metazoans. Cell Rep. 2012;1:69–82. doi: 10.1016/j.celrep.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 103. Li F, Zheng Q, Vandivier LE, Willmann MR, Chen Y, Gregory BD. Regulatory impact of rna secondary structure across the arabidopsis transcriptome. Plant Cell Online. 2012;24(11):4346–4359. doi: 10.1105/tpc.112.104232. dsRNA/ssRNA-seq was performed in flower buds from Arabidopsis revealing many transcriptome-wide structural features.
- 104.Lim PO, Sears BB. 16s rrna sequence indicates that plant-pathogenic mycoplasmalike organisms are evolutionarily distinct from animal mycoplasmas. J. Bacteriol. 1989;171(11):5901–5906. doi: 10.1128/jb.171.11.5901-5906.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liu HX, Goodall GJ, Kole R, Filipowicz W. Effects of secondary structure on pre-mrna splicing: hairpins sequestering the 5’ but not the 3’ splice site inhibit intron processing in nicotiana plumbaginifolia. EMBO J. 1995;14(2):377–388. doi: 10.1002/j.1460-2075.1995.tb07012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N6-methyladenosine-dependent rna structural switches regulate rna-protein interactions. Nature. 2015;518:560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liu X, Hao L, Li D, Zhu L, Hu S. Long non-coding rnas and their biological roles in plants. Genomics Proteomics Bioinformatics. 2015 doi: 10.1016/j.gpb.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lockard RE, Kumar A. Mapping trna structure in solution using double-strand-specific ribonuclease v1 from cobra venom. Nucleic Acids Res. 1981;9(19):5125–5140. doi: 10.1093/nar/9.19.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lorsch JR. Rna chaperones exist and dead box proteins get a life. Cell. 2002;109(7):797–800. doi: 10.1016/s0092-8674(02)00804-8. [DOI] [PubMed] [Google Scholar]
- 110.Loverix S, Steyaert J. Deciphering the mechanism of rnase t1. Methods Enzymol. 2001;341:305–323. doi: 10.1016/s0076-6879(01)41160-8. [DOI] [PubMed] [Google Scholar]
- 111.Lu D, Alexandra Searles M, Klug A. Crystal structure of a zinc-finger-rna complex reveals two modes of molecular recognition. Nature. 2003;426(6962):96–100. doi: 10.1038/nature02088. [DOI] [PubMed] [Google Scholar]
- 112.Ludwig W, Strunk O, Klugbauer S, Klugbauer N, Weizenegger M, et al. Bacterial phylogeny based on comparative sequence analysis (review) ELECTROPHORESIS. 1998;19(4):554–568. doi: 10.1002/elps.1150190416. [DOI] [PubMed] [Google Scholar]
- 113.Lunde BM, Moore C, Varani G. Rna-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell Biol. 2007;8(6):479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Madhani HD. Snrna catalysts in the spliceosome’s ancient core. Cell. 2013;155(6):1213–1215. doi: 10.1016/j.cell.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mathews DH. Rna secondary structure analysis using rnastructure. Curr. Protoc. Bioinforma. Ed. Board Andreas Baxevanis Al. 2014;46 doi: 10.1002/0471250953.bi1206s46. 12.6.1–12.6.25. [DOI] [PubMed] [Google Scholar]
- 116.Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of rna secondary structure. Proc. Natl. Acad. Sci. U. S. A. 2004;101(19):7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.McClung CR, Davis SJ. Ambient thermometers in plants: from physiological outputs towards mechanisms of thermal sensing. Curr. Biol. 2010;20(24):R1086–R1092. doi: 10.1016/j.cub.2010.10.035. [DOI] [PubMed] [Google Scholar]
- 118.Mercer TR, Mattick JS. Structure and function of long noncoding rnas in epigenetic regulation. Nat. Struct. Mol. Biol. 2013;20(3):300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 119.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. Rna structure analysis at single nucleotide resolution by selective 2’-hydroxyl acylation and primer extension (shape) J. Am. Chem. Soc. 2005;127(12):4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 120.Miranda-Ríos J. The thi-box riboswitch, or how rna binds thiamin pyrophosphate. Structure. 2007;15(3):259–265. doi: 10.1016/j.str.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 121.Moazed D, Noller HF. Interaction of antibiotics with functional sites in 16s ribosomal rna. Nature. 1987;327(6121):389–394. doi: 10.1038/327389a0. [DOI] [PubMed] [Google Scholar]
- 122.Moazed D, Noller HF. Chloramphenicol, erythromycin, carbomycin and vernamycin b protect overlapping sites in the peptidyl transferase region of 23s ribosomal rna. Biochimie. 1987;69(8):879–884. doi: 10.1016/0300-9084(87)90215-x. [DOI] [PubMed] [Google Scholar]
- 123.Mohr S, Stryker JM, Lambowitz AM. A dead-box protein functions as an atp-dependent rna chaperone in group i intron splicing. Cell. 2002;109(6):769–779. doi: 10.1016/s0092-8674(02)00771-7. [DOI] [PubMed] [Google Scholar]
- 124.Møller T, Franch T, Højrup P, Keene DR, Bächinger HP, et al. Hfq: a bacterial sm-like protein that mediates rna-rna interaction. Mol. Cell. 2002;9(1):23–30. doi: 10.1016/s1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- 125.Nakaminami K, Karlson DT, Imai R. Functional conservation of cold shock domains in bacteria and higher plants. Proc. Natl. Acad. Sci. 2006;103(26):10122–10127. doi: 10.1073/pnas.0603168103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Narberhaus F. Translational control of bacterial heat shock and virulence genes by temperature-sensing mrnas. RNA Biol. 2010;7(1):84–89. doi: 10.4161/rna.7.1.10501. [DOI] [PubMed] [Google Scholar]
- 127.Ni J, Tien AL, Fournier MJ. Small nucleolar rnas direct site-specific synthesis of pseudouridine in ribosomal rna. Cell. 1997;89(4):565–573. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- 128.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289(5481):920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 129.Novikova IV, Hennelly SP, Sanbonmatsu KY. Sizing up long non-coding rnas: do lncrnas have secondary and tertiary structure? BioArchitecture. 2012;2(6):189–199. doi: 10.4161/bioa.22592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Oberstrass FC, Lee A, Stefl R, Janis M, Chanfreau G, Allain FH-T. Shape-specific recognition in the structure of the vts1p sam domain with rna. Nat. Struct. Mol. Biol. 2006;13(2):160–167. doi: 10.1038/nsmb1038. [DOI] [PubMed] [Google Scholar]
- 131.Oikawa D, Tokuda M, Hosoda A, Iwawaki T. Identification of a consensus element recognized and cleaved by ire1 alpha. Nucleic Acids Res. 2010;38(18):6265–6273. doi: 10.1093/nar/gkq452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Oliverio M, Cervelli M, Mariottini P. Its2 rrna evolution and its congruence with the phylogeny of muricid neogastropods (caenogastropoda, muricoidea) Mol. Phylogenet. Evol. 2002;25(1):63–69. doi: 10.1016/s1055-7903(02)00227-0. [DOI] [PubMed] [Google Scholar]
- 133.Oubridge C, Ito N, Evans PR, Teo C-H, Nagai K. Crystal structure at 1.92 å resolution of the rna-binding domain of the u1a spliceosomal protein complexed with an rna hairpin. Nature. 1994;372(6505):432–438. doi: 10.1038/372432a0. [DOI] [PubMed] [Google Scholar]
- 134.Park W, Li J, Song R, Messing J, Chen X. Carpel factory, a dicer homolog, and hen1, a novel protein, act in microrna metabolism in arabidopsis thaliana. Curr. Biol. 2002;12(17):1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Peattie DA. Direct chemical method for sequencing rna. Proc Natl Acad Sci. 1979;76(4):1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Peattie DA, Gilbert W. Chemical probes for higher-order structure in rna. Proc Natl Acad Sci. 1980;77(6):4679–4682. doi: 10.1073/pnas.77.8.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pelletier J, Sonenberg N. Insertion mutagenesis to increase secondary structure within the 5′ noncoding region of a eukaryotic mrna reduces translational efficiency. Cell. 1985;40(3):515–526. doi: 10.1016/0092-8674(85)90200-4. [DOI] [PubMed] [Google Scholar]
- 138.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mrna directed by a sequence derived from poliovirus rna. Nature. 1988;334(6180):320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 139.Phadtare S, Inouye M, Severinov K. The nucleic acid melting activity of escherichia colicspe is critical for transcription antitermination and cold acclimation of cells. J. Biol. Chem. 2002;277(9):7239–7245. doi: 10.1074/jbc.M111496200. [DOI] [PubMed] [Google Scholar]
- 140.Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? evidence for selection within long noncoding rnas. Genome Res. 2007;17(5):556–565. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding rnas. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 142.Rajkowitsch L, Chen D, Stampfl S, Semrad K, Waldsich C, et al. Rna chaperones, rna annealers and rna helicases. RNA Biol. 2007;4(3):118–130. doi: 10.4161/rna.4.3.5445. [DOI] [PubMed] [Google Scholar]
- 143.Raker VA, Mironov AA, Gelfand MS, Pervouchine DD. Modulation of alternative splicing by long-range rna structures in drosophila. Nucleic Acids Res. 2009;37(14):4533–4544. doi: 10.1093/nar/gkp407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ramakrishnan V. The ribosome emerges from a black box. Cell. 2014;159(5):979–984. doi: 10.1016/j.cell.2014.10.052. [DOI] [PubMed] [Google Scholar]
- 145.Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. Micrornas in plants. Genes Dev. 2002;16(13):1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, et al. Functional demarcation of active and silent chromatin domains in human hox loci by noncoding rnas. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Robertus JD, Ladner JE, Finch JT, Rhodes D, Brown RS, et al. Structure of yeast phenylalanine trna at 3 a resolution. Nature. 1974;250(467):546–551. doi: 10.1038/250546a0. [DOI] [PubMed] [Google Scholar]
- 148.Roth A, Breaker RR. The structural and functional diversity of metabolite-binding riboswitches. Annu. Rev. Biochem. 2009;78:305–334. doi: 10.1146/annurev.biochem.78.070507.135656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. Genome-wide probing of rna structure reveals active unfolding of mrna structures in vivo. Nature. 2014;505(7485):701–705. doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ryter JM. Molecular basis of double-stranded rna-protein interactions: structure of a dsrna-binding domain complexed with dsrna. EMBO J. 1998;17(24):7505–7513. doi: 10.1093/emboj/17.24.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sasaki K, Imai R. Pleiotropic roles of cold shock domain proteins in plants. Front. Plant Sci. 2012;2 doi: 10.3389/fpls.2011.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Schlötterer C, Hauser MT, Haeseler von A, Tautz D. Comparative evolutionary analysis of rdna its regions in drosophila. Mol. Biol. Evol. 1994;11(3):513–522. doi: 10.1093/oxfordjournals.molbev.a040131. [DOI] [PubMed] [Google Scholar]
- 153.Schroeder R, Barta A, Semrad K. Strategies for rna folding and assembly. Nat. Rev. Mol. Cell Biol. 2004;5(11):908–919. doi: 10.1038/nrm1497. [DOI] [PubMed] [Google Scholar]
- 154.Schwarz Z, Kössel H. The primary structure of 16s rdna from zea mays chloroplast is homologous to e. coli 16s rrna. Nature. 1980;283(5749):739–742. [Google Scholar]
- 155.Serganov A, Patel DJ. Ribozymes, riboswitches and beyond: regulation of gene expression without proteins. Nat. Rev. Genet. 2007;8(10):776–790. doi: 10.1038/nrg2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Shabalina SA, Ogurtsov AY, Spiridonov NA. A periodic pattern of mrna secondary structure created by the genetic code. Nucleic Acids Res. 2006;34(8):2428–2437. doi: 10.1093/nar/gkl287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Shen LX, Basilion JP, Stanton VP. Single-nucleotide polymorphisms can cause different structural folds of mrna. Proc. Natl. Acad. Sci. 1999;96(14):7871–7876. doi: 10.1073/pnas.96.14.7871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Silverman IM, Li F, Alexander A, Goff L, Trapnell C, et al. Rnase-mediated protein footprint sequencing reveals protein-binding sites throughout the human transcriptome. Genome Biol. 2014;15:R3. doi: 10.1186/gb-2014-15-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Sirand-Pugnet P, Durosay P, Clouet d’Orval BC, Brody E, Marie J. Beta-tropomyosin pre-mrna folding around a muscle-specific exon interferes with several steps of spliceosome assembly. J. Mol. Biol. 1995;251(5):591–602. doi: 10.1006/jmbi.1995.0458. [DOI] [PubMed] [Google Scholar]
- 160.Solnick D. Alternative splicing caused by rna secondary structure. Cell. 1985;43(3 Pt 2):667–676. doi: 10.1016/0092-8674(85)90239-9. [DOI] [PubMed] [Google Scholar]
- 161.Somarowthu S, Legiewicz M, Chillón I, Marcia M, Liu F, Pyle AM. Hotair forms an intricate and modular secondary structure. Mol. Cell. 2015;58(2):353–361. doi: 10.1016/j.molcel.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Spitale RC, Flynn RA, Zhang QC, Crisalli P, Lee B, et al. Structural imprints in vivo decode rna regulatory mechanisms. Nature. 2015;519(7544):486–490. doi: 10.1038/nature14263. icSHAPE is the first high-throughput chemical-based structure probing technique that probes ssRNA without nucleotide bias.
- 163.Springer MS, Douzery E. Secondary structure and patterns of evolution among mammalian mitochondrial 12s rrna molecules. J. Mol. Evol. 1996;43(4):357–373. doi: 10.1007/BF02339010. [DOI] [PubMed] [Google Scholar]
- 164.Steitz TA, Moore PB. Rna, the first macromolecular catalyst: the ribosome is a ribozyme. Trends Biochem. Sci. 2003;28(8):411–418. doi: 10.1016/S0968-0004(03)00169-5. [DOI] [PubMed] [Google Scholar]
- 165.Subramanian M, Rage F, Tabet R, Flatter E, Mandel J-L, Moine H. G-quadruplex rna structure as a signal for neurite mrna targeting. EMBO Rep. 2011;12(7):697–704. doi: 10.1038/embor.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Suo Z, Johnson KA. Effect of rna secondary structure on the kinetics of dna synthesis catalyzed by hiv-1 reverse transcriptase. Biochemistry (Mosc.) 1997;36(41):12459–12467. doi: 10.1021/bi971217h. [DOI] [PubMed] [Google Scholar]
- 167.Svitkin YV, Pause A, Haghighat A, Pyronnet S, Witherell G, et al. The requirement for eukaryotic initiation factor 4a (elf4a) in translation is in direct proportion to the degree of mrna 5’ secondary structure. RNA N. Y. N. 2001;7(3):382–394. doi: 10.1017/s135583820100108x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Swiezewski S, Liu F, Magusin A, Dean C. Cold-induced silencing by long antisense transcripts of an arabidopsis polycomb target. Nature. 2009;462(7274):799–802. doi: 10.1038/nature08618. [DOI] [PubMed] [Google Scholar]
- 169.Talkish J, May G, Lin Y, Woolford JL, Jr, McManus CJ. Mod-seq: high-throughput sequencing for chemical probing of rna structure. RNA. 2014;20:1–8. doi: 10.1261/rna.042218.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Tenenbaum SA, Christiansen J, Nielsen H. The post-transcriptional operon. Methods Mol. Biol. Clifton NJ. 2011;703:237–245. doi: 10.1007/978-1-59745-248-9_16. [DOI] [PubMed] [Google Scholar]
- 171.Thirumalai D, Hyeon C. Theory of rna folding: from hairpins to ribozymes. In: Walter DNG, Woodson DSA, Batey DRT, editors. Non-Protein Coding RNAs. Springer Berlin Heidelberg; 2009. pp. 27–47. [Google Scholar]
- 172.Tompa P, Csermely P. The role of structural disorder in the function of rna and protein chaperones. FASEB J. 2004;18(11):1169–1175. doi: 10.1096/fj.04-1584rev. [DOI] [PubMed] [Google Scholar]
- 173.Uchida T, Arima T, Egami F. Specificity of rnase u2. J. Biochem. (Tokyo) 1970;67(1):91–102. doi: 10.1093/oxfordjournals.jbchem.a129239. [DOI] [PubMed] [Google Scholar]
- 174.Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincrnas in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147(7):1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Underwood JG, Uzilov AV, Katzman S, Onodera CS, Mainzer JE, et al. Fragseq: transcriptome-wide rna structure probing using high-throughput sequencing. Nat. Methods. 2010;7(12):995–1001. doi: 10.1038/nmeth.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Vandivier L, Li F, Zheng Q, Willmann M, Chen Y, Gregory B. Arabidopsis mrna secondary structure correlates with protein function and domains. Plant Signal. Behav. 2013;8(6):e24301. doi: 10.4161/psb.24301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Vitreschak AG, Rodionov DA, Mironov AA, Gelfand MS. Riboswitches: the oldest mechanism for the regulation of gene expression? Trends Genet. 2004;20(1):44–50. doi: 10.1016/j.tig.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 178.Vogel J. Lariat formation and a hydrolytic pathway in plant chloroplast group ii intron splicing. EMBO J. 2002;21(14):3794–3803. doi: 10.1093/emboj/cdf359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Volkin E, Cohn WE. On the structure of ribonucleic acids. ii. the products of ribonuclease action. J. Biol. Chem. 1953;205(2):767–782. [PubMed] [Google Scholar]
- 180.Wang KC, Chang HY. Molecular mechanisms of long noncoding rnas. Mol. Cell. 2011;43(6):904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Wang L, Wessler SR. Role of mrna secondary structure in translational repression of the maize transcriptional activatorlc. Plant Physiol. 2001;125(3):1380–1387. doi: 10.1104/pp.125.3.1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Wanrooij PH, Uhler JP, Simonsson T, Falkenberg M, Gustafsson CM. G-quadruplex structures in rna stimulate mitochondrial transcription termination and primer formation. Proc. Natl. Acad. Sci. 2010;107(37):16072–16077. doi: 10.1073/pnas.1006026107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Wan Y, Qu K, Zhang QC, Flynn RA, Manor O, et al. Landscape and variation of rna secondary structure across the human transcriptome. Nature. 2014;505(7485):706–709. doi: 10.1038/nature12946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Warf MB, Berglund JA. Role of rna structure in regulating pre-mrna splicing. Trends Biochem. Sci. 2010;35(3):169–178. doi: 10.1016/j.tibs.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wells SE, Hughes JM, Igel AH, Ares M. Use of dimethyl sulfate to probe rna structure in vivo. Methods Enzymol. 2000;318:479–493. doi: 10.1016/s0076-6879(00)18071-1. [DOI] [PubMed] [Google Scholar]
- 186.Wen J-D, Lancaster L, Hodges C, Zeri A-C, Yoshimura SH, et al. Following translation by single ribosomes one codon at a time. Nature. 2008;452(7187):598–603. doi: 10.1038/nature06716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Wilkinson KA, Merino EJ, Weeks KM. Selective 2’-hydroxyl acylation analyzed by primer extension (shape): quantitative rna structure analysis at single nucleotide resolution. Nat. Protoc. 2006;1(3):1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 188.Williams AS, Marzluff WF. The sequence of the stem and flanking sequences at the 3′ end of histone mrna are critical determinants for the binding of the stem-loop binding protein. Nucleic Acids Res. 1995;23(4):654–662. doi: 10.1093/nar/23.4.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Williamson JR. Induced fit in rna-protein recognition. Nat. Struct. Mol. Biol. 2000;7(10):834–837. doi: 10.1038/79575. [DOI] [PubMed] [Google Scholar]
- 190.Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428(6980):281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- 191.Yang C-H, Crowley DE. Rhizosphere microbial community structure in relation to root location and plant iron nutritional status. Appl. Environ. Microbiol. 2000;66(1):345–351. doi: 10.1128/aem.66.1.345-351.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Yusupova G, Yusupov M. High-resolution structure of the eukaryotic 80s ribosome. Annu. Rev. Biochem. 2014;83(1):467–486. doi: 10.1146/annurev-biochem-060713-035445. [DOI] [PubMed] [Google Scholar]
- 193.Zappulla DC, Cech TR. Yeast telomerase rna: a flexible scaffold for protein subunits. Proc. Natl. Acad. Sci. U. S. A. 2004;101(27):10024–10229. doi: 10.1073/pnas.0403641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Zaug AJ, Cech TR. Analysis of the structure of tetrahymena nuclear rnas in vivo: telomerase rna, the self-splicing rrna intron, and u2 snrna. RNA N. Y. N. 1995;1(4):363–374. [PMC free article] [PubMed] [Google Scholar]
- 195. Zhao B, Zhang Q. Characterizing excited conformational states of rna by nmr spectroscopy. Curr. Opin. Struct. Biol. 2015;30:134–146. doi: 10.1016/j.sbi.2015.02.011. This review describes the utility of NMR and its ability to study RNA folding dynamics.
- 196. Zheng Q, Ryvkin P, Li F, Dragomir I, Valladares O, et al. Genome-wide double-stranded rna sequencing reveals the functional significance of base-paired rnas in arabidopsis. PLoS Genet. 2010;6(9):e1001141. doi: 10.1371/journal.pgen.1001141. dsRNA-seq was the first transcriptome-wide structure probing technique performed in plants, identifying dsRNA in Arabidopsis flower buds.
- 197.Zuker M, Stiegler P. Optimal computer folding of large rna sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]