Abstract
AMPA-subtype ionotropic glutamate receptors (AMPARs) mediate fast excitatory neurotransmission, and contribute to high cognitive processes such as learning and memory. In the brain, AMPAR trafficking, gating, and pharmacology is tightly controlled by transmembrane AMPAR regulatory proteins (TARPs). Here, we used cryo-EM to elucidate the structural basis of AMPAR regulation by one of these auxiliary proteins, TARP γ2 or stargazin (STZ). Our structures illuminate the variable interaction stoichiometry of the AMPAR-TARP complex, with one or two TARP molecules binding one tetrameric AMPAR. Analysis of the AMPAR-STZ binding interfaces suggests that electrostatic interactions between the extracellular domains of AMPAR and STZ play an important role in modulating AMPAR function through contact surfaces that are conserved across AMPARs and TARPs. We propose a model explaining how TARPs stabilize the activated state of AMPARs and how the interactions between AMPARs and their auxiliary proteins control fast excitatory synaptic transmission.
One Sentence Summary
AMPA receptor-stargazin complexes show how auxiliary subunits control synaptic function through variable interaction stoichiometry and electrostatics.
Excitatory neurotransmission is mediated predominantly by ionotropic glutamate receptors (iGluRs) (1). iGluRs are tetrameric ligand-gated ion channels found in the postsynaptic densities of neurons, and are typically activated by glutamate released from presynaptic terminals, resulting in ion flux and postsynaptic depolarization (1, 2). The AMPAR subtype iGluRs exhibit kinetics at the millisecond timescale, and mediate fast neurotransmission in excitatory synapses, directly impacting synaptic plasticity, learning, and memory (1). Aberrations in AMPAR function are implicated in a wide range of diseases, from developmental diseases such as Fragile-X syndrome (3), to psychiatric disorders (1), acute trauma in ischemic stroke (4, 5), epileptic seizures (6, 7) and chronic neurodegenerative disorders such as Parkinson’s and Alzheimer’s diseases (1).
Structural studies have revealed the three-layer architecture of AMPARs, which includes a two-layer extracellular domain (ECD) composed of two amino-terminal domain (ATD) and ligand-binding domain (LBD) dimers, and a channel-forming transmembrane domain (TMD), and have provided insights into the gating mechanism, through models obtained by crystallography (8–11) and cryo-electron microscopy (cryo-EM) (12). However, in cells AMPARs exist as complexes with various soluble and membrane proteins that alter their function (13–15). The prototypical transmembrane AMPAR regulatory protein (TARP) γ2, or stargazin (STZ), controls AMPAR synaptic targeting, gating, and pharmacology (16–18). TARPs are linked to the pathophysiology of several neurological and psychiatric disorders (19, 20), making TARPs and TARP-AMPAR complexes targets for a variety of human diseases. However, despite negative-stain EM studies showing that STZ contributes to the TMD of native AMPAR complexes (21), detailed structural information on the AMPAR-TARP interactions, including stoichiometry (22–24), remains elusive – a key barrier in informed therapeutic design.
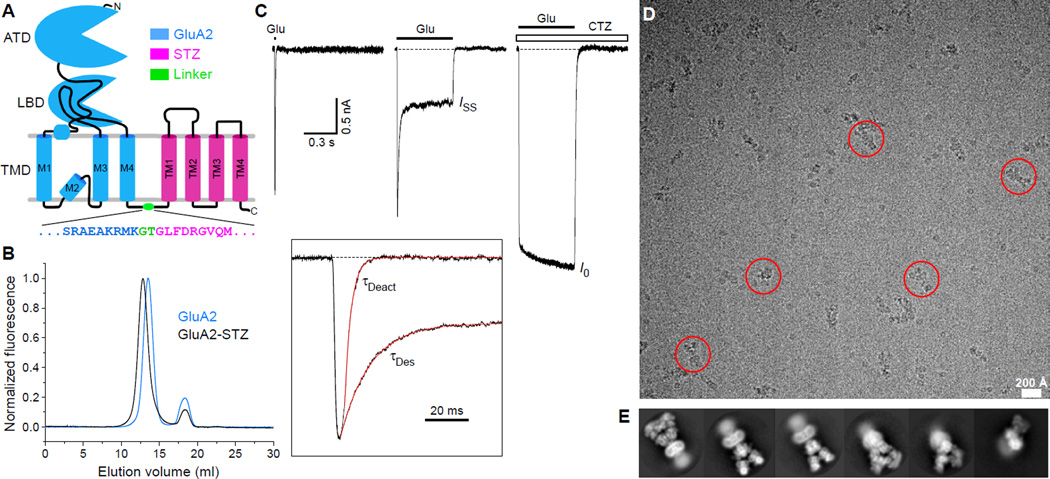
Here, we used cryo-EM to elucidate the structural basis for STZ modulation of GluA2 function. To form the complex between GluA2 and STZ, we used a tandem construct, GluA2-STZ, where the N-terminus of STZ was fused to the C-terminus of GluA2 by a glycine-threonine (GT) linker (Fig. 1A) (25). Purified GluA2-STZ eluted from the size-exclusion column as a monodisperse peak, shifted leftward compared to non-fused GluA2 (Fig. 1B), and ran as a higher molecular-weight band on SDS-PAGE (fig. S1). To assess functionality, we recorded glutamate-activated GluA2-STZ currents using patch-clamp electrophysiology. Compared to wild-type GluA2, GluA2-STZ showed reduced desensitization and slower rates of deactivation, desensitization and recovery from desensitization (Fig. 1C; fig. S2), as expected (26–30). Thus, the presence of the GT linker between GluA2 and STZ in our GluA2-STZ construct did not significantly affect modulation of GluA2 function by STZ.
Fig 1. Design, function, and cryo-EM characterization of GluA2-STZ.
(A) Topology of the GluA2-STZ construct with GluA2 (blue) and STZ (magenta) connected by a glycine-threonine linker (green). Expanded is the amino acid sequence encompassing the linker region. (B) FSEC profiles of purified GluA2-STZ (black) and GluA2 (blue) followed by tryptophan fluorescence. (C) Representative whole-cell currents recorded at −60 mV membrane potential from an HEK293 cell expressing GluA2-STZ in response to 2 ms or 500 ms applications of 3 mM glutamate (Glu) alone or application of Glu in the continuous presence of 30 µM cyclothiazide (CTZ). Inset shows normalized currents in response to 2 ms and 500 ms applications of Glu alone fitted using single exponentials with the time constants τDeact and τDes. (D) Drift-corrected cryo-EM micrograph of GluA2-STZ, with example particles highlighted by red circles. (E) Representative two-dimensional class averages.
Cryo-EM micrographs of purified GluA2-STZ bound to antagonist ZK200775 gave initial insight into the particle details (Fig. 1D), and initial views of GluA2-STZ from two-dimensional classes suggested high data quality, with visible linkers between the LBD and TMD, clear secondary structure features in both the ECD and TMD, and diverse particle orientations (Fig. 1E). However, atypical for the GluA2 three-layer topology (fig. S3A–B), GluA2-STZ showed a four-layer architecture, where below the TMD layer is a fourth layer (Fig. 1D) that appears disordered (Fig. 1E). A closer look at the GluA2-STZ particles (25) suggested that the disordered fourth layer under the TMD is likely composed of unbound STZ and is a result of the tandem construct design, which defines the protomer ratio but not interaction stoichiometry.
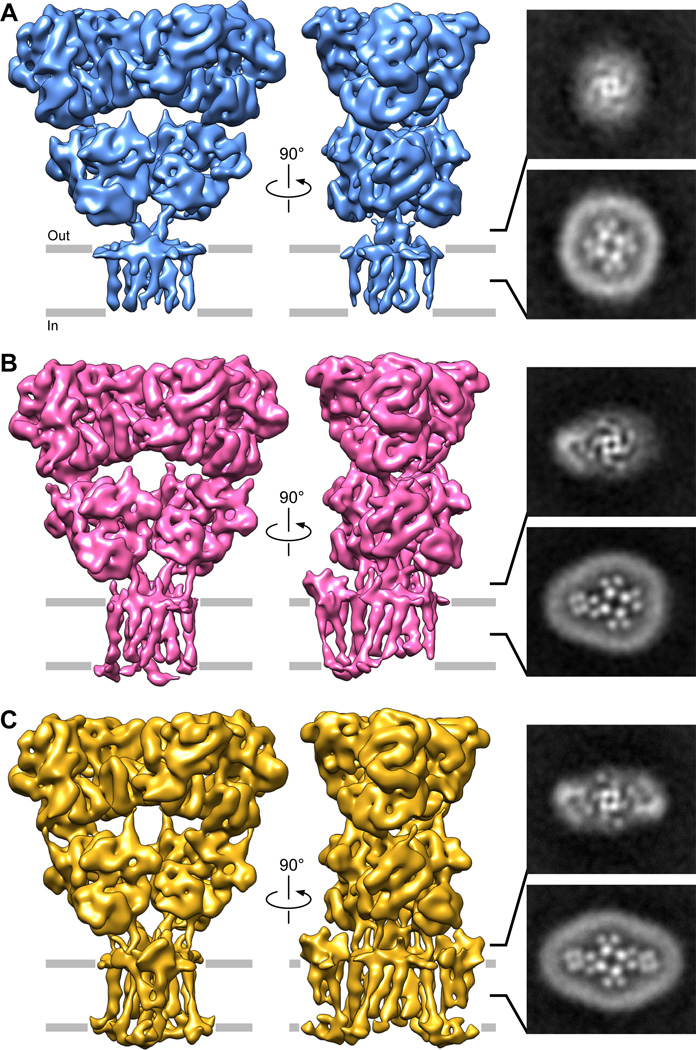
We identified multiple STZ-bound states of the GluA2-STZ complex through additional image processing (fig. S4). One state resembles the map for GluA2 alone (fig. S3C) and shows no STZ bound (GluA2-0xSTZ, Fig. 2A). A second state showed a single STZ assembled around GluA2, which we call GluA2-1xSTZ (Fig. 2B). We also identified a third state of the GluA2-STZ particles, where two STZ molecules are assembled around the GluA2 core, termed GluA2-2xSTZ (Fig. 2C). For all three states, we observed no preferred particle orientation in the refined maps (fig. S5), with most particles contributing to the single-bound STZ state (fig. S6). A closer look through 2D slices of the three-dimensional refined cryo-EM density maps in the TMD and LBD-TMD linkers highlights the differences in stoichiometry, where zero (Fig. 2A), one (Fig. 2B) or two (Fig. 2C) STZ TMDs and ECDs are visible around the GluA2 core periphery, respectively. Correspondingly, the disordered fourth layer (Fig. 1E) in these three stoichiometric states presumably has four, three and two STZ molecules from the tandem construct that are not bound around the GluA2 core (25). Based on our density maps (Fig. 2), we propose that the preferred stoichiometry of the GluA2-STZ interaction is one or two STZ protomers to one tetramer of GluA2. The existence of multiple stoichiometric states suggests that STZ expression levels could have a profound effect on AMPAR-mediated neurotransmission.
Fig. 2. Stoichiometric states of GluA2-STZ.
On the left, cryo-EM density maps of (A) STZ-unbound, (B) single STZ-bound, and (C) double STZ-bound states of GluA2-STZ filtered to 8.7 Å, 6.4 Å, and 7.8 Å resolution, respectively. For each state, two-dimensional slices made parallel to the membrane through the refined, non-filtered map are shown on the right: one through the middle of GluA2 LBD-TMD linker region and another through the middle of GluA2 TMD. Details of collection and refinement are included in fig. S6.
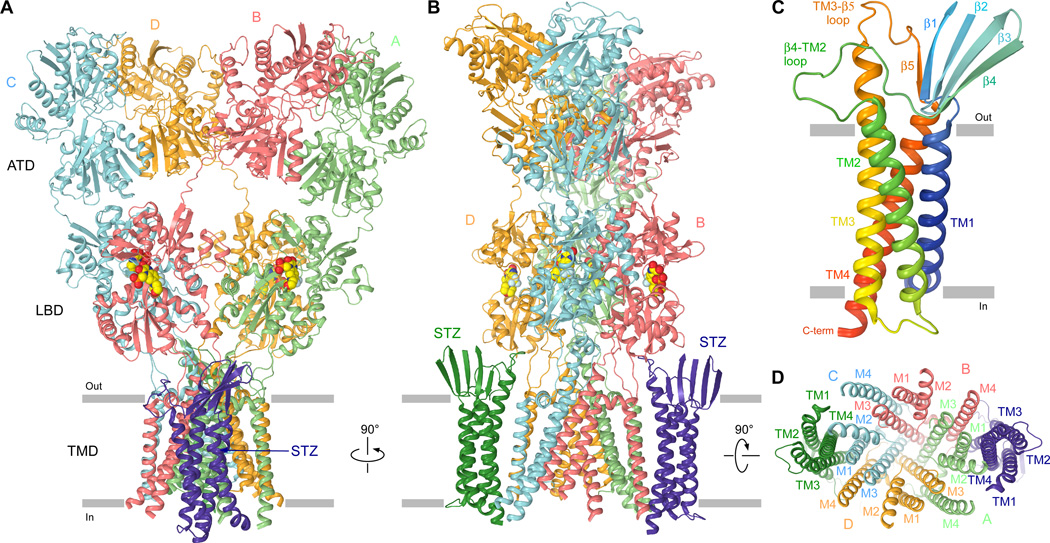
To gain further insight into the AMPAR-TARP interaction, we built a structural model of the GluA2-1xSTZ state. Guided by the two-fold symmetry of the ECD (8–12), we used the corresponding portion of the GluA2-1xSTZ map refined with C2 symmetry to 5.6 Å resolution (fig. S6) (25) to fit GluA2 ATDs and LBDs (8) and to build ATD-LBD linkers. We further used the 6.4 Å map obtained from refinement without symmetry restraints (fig. S7), to fit the GluA2 TMD region (8) and to build LBD-TMD linkers (fig. S8). We then took advantage of amino acid sequence conservation between TARP family and Claudin family proteins (fig. S9) and built a Claudin-19-based homology model of STZ. This model was fitted into the GluA2-1xSTZ density (25) (fig. S8B) confirming similar overall fold of TARPs and Claudins (31, 32) (fig. S9). The STZ and GluA2 protomers from the GluA2-1xSTZ structure were also fitted into GluA2-2xSTZ map to generate a structure of the corresponding complex (Fig. 3A–B).
Figure 3. Architecture of the GluA2-STZ complex.
A–B, D, Model of GluA2-2xSTZ viewed parallel to (A–B) or from the intracellular side of (D) the membrane. The four GluA2 subunits (A to D) are colored mint (A), coral (B), light-blue (C), and gold (D), and the STZ molecules in green and purple; TMD secondary structure elements are labeled in D. (C) Rainbow colored homology model (from blue N-terminus to red C-terminus) of STZ with the secondary structure elements labeled. In A–B, the antagonist ZK200775 is shown as a space-filling model (red/yellow).
The structure of STZ includes a TMD that represents a bundle of four transmembrane helices, TM1 to TM4, and an extracellular head domain that sits atop the TMD (Fig. 3C). The main interaction between STZ and AMPAR is mediated by a substantial interface between transmembrane helices TM3 and TM4 of STZ and M1 and M4 of GluA2 (Fig. 3D). STZ TM1 and TM2 have no direct contact to the AMPAR core, and face the lipid membrane. The STZ head domain is comprised of two extracellular polypeptide segments, between TM1 and TM2 and between TM3 and TM4 (Fig. 3C). Most of the head domain is a β-sheet that includes strands β1–4 formed by the N-terminal portion of the TM1–TM2 segment and β5 formed by the C-terminal portion of the TM3–TM4 segment. The remaining portions of the extracellular segments, the TM3-β5 loop and the β4-TM2 loop in particular, are conveniently positioned in close proximity to the LBD and LBD-TMD linkers to play a key role in regulation of AMPAR function.
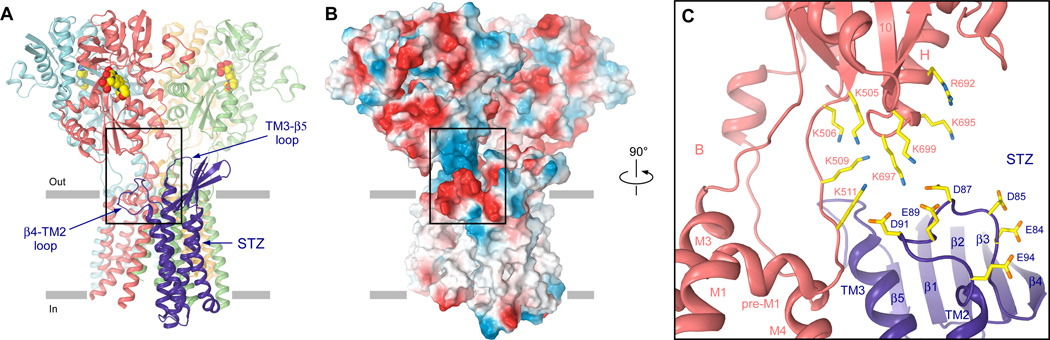
The GluA2-STZ structures indicate that the TM3-β5 and β4-TM2 loops of STZ can only interact with GluA2 subunits B and D but not A and C (Fig. 3B). At the level of the LBD, B and D represent the distal subunits (fig. S10A) that a play more important role in iGluR gating than the proximal subunits A and C (9, 33, 34). STZ molecules are thus optimally positioned in the GluA2-STZ complex to maximally affect GluA2 gating. The most likely regions of GluA2 subunits B and D to interact with STZ loops TM3-β5 and β4-TM2 are the adjacent S1-M1 linker and the LBD loop between helix H and the β stand 10 (Fig. 4A). The S1-M1 linker contains four positively-charged residues (K505, K506, K509 and K511), in addition to four positively-charged residues in the helix H-β strand 10 loop (R692, K695, K697 and K699), that form an electropositive patch on the surface of GluA2 facing STZ (Fig. 4B–C). In contrast, six negatively charged residues in the β4-TM2 loop of STZ (E84, D85, D87, E89, D91 and E94) form an electronegative patch on the surface of the STZ head domain facing GluA2.
Fig. 4. Electrostatic interactions between the STZ head domain and GluA2.
(A) Side view of GluA2-1xSTZ with GluA2 ATDs omitted and STZ β4-TM2 and TM3-β5 loops labeled. (B) Electrostatic surface of the molecule in A with highly electronegative regions shown in red and highly electropositive regions shown in blue. (C) Close-up orthogonal view of the region boxed in A and B. Charged residues in the S1-M1 linker and LBD lower lobe of GluA2, and the β4-TM2 loop of STZ are shown as sticks.
Confirming a key role of the electrostatic interactions in regulation of AMPAR function by STZ, recent mutagenesis experiments on a similar GluA2-STZ tandem construct showed that the aspartate substitution of a KGK motif, which is highly conserved in AMPARs and includes K697 and K699 residues in the helix H-β strand 10 loop, almost completely abolished the effects of STZ on GluA2 receptor function (29). In addition, the electronegative motif in the STZ β4-TM2 loop is highly conserved across type I TARPs (19), including γ2 (STZ), γ3, γ4, and γ8 (fig. S9). Similar to STZ, γ3, γ4, and γ8 slow AMPAR deactivation and desensitization kinetics (19). In contrast, type II TARPs γ5 and γ7 do not have the conserved electronegative motif in the β4-TM2 loop (fig. S9), and likely use different mechanisms to alter AMPAR gating and pharmacology (19, 35). Indeed, substitution of the TM1–TM2 extracellular stretch in STZ with that of γ5 dramatically reduced the effect of STZ on AMPAR gating and agonist efficacy to near wild-type levels (27).
We hypothesize that the overall compression of AMPARs upon activation (9–11, 36) brings the negative patch on the STZ surface closer to the positive patch on the surface of GluA2 (fig. S10B), thus enhancing their electrostatic attraction. This additional force would account for STZ-stabilization of the open conformation of AMPARs (37), and correspondingly make deactivated and desensitized states less favorable.
Supplementary Material
Acknowledgments
We thank Z. H. Yu, C. Hong, and R. Huang for assistance with data collection at the HHMI Janelia Research Campus, M. Fislage (Frank Lab) for assistance with the initial cryo-EM preparation and critical reading of the manuscript, and H. Kao (Frank Lab) for computational support. We are grateful for manuscript edits and comments provided by K. Saotome (Sobolevsky Lab) and A. K. Singh (Sobolevsky Lab), and advice on image processing provided by I.S. Fernandez (Columbia), Z. Liu (Frank Lab), O.B. Clarke (Hendrickson Lab, Columbia), A. des Georges (CUNY), and members of the Frank Lab. E.C.T. was supported NIH training grants (T32 GM008224 and GM008281). A.I.S. was supported by NIH (R01 NS083660), the Pew Scholar Award in Biomedical Sciences, the Schaefer Research Scholar Award, the Klingenstein Fellowship Award in the Neurosciences and the Irma T. Hirschl Career Scientist Award. J.F. was supported by the Howard Hughes Medical Institute and NIH (R01 GM029169). Cryo-EM density maps have been deposited in the Electron Microscopy Data Bank (EMDB) under accession numbers EMD-8229 (GluA2-0xSTZ), EMD-8230 (GluA2-1xSTZ), EMD-8231 (GluA2-2xSTZ) and EMD-8232 (GluA2). Model coordinates have been deposited in the Protein Data Bank (PDB) under accession numbers 5KBS (GluA2-0xSTZ), 5KBT (GluA2-1xSTZ), 5KBU (GluA2-2xSTZ) and 5KBV (GluA2).
References and Notes
- 1.Traynelis SF, et al. Pharmacol Rev. 2010 Sep;62:405. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar J, Mayer ML. Annu Rev Physiol. 2013;75:313. doi: 10.1146/annurev-physiol-030212-183711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uzunova G, Hollander E, Shepherd J. Curr Neuropharmacol. 2014 Jan;12:71. doi: 10.2174/1570159X113116660046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morrell CN, et al. J Exp Med. 2008 Mar 17;205:575. doi: 10.1084/jem.20071474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang F, Guo A, Liu C, Comb M, Hu B. Stroke. 2013 Jan;44:170. doi: 10.1161/STROKEAHA.112.667253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chapman AG. J Nutr. 2000 Apr;130:1043S. doi: 10.1093/jn/130.4.1043S. [DOI] [PubMed] [Google Scholar]
- 7.Rogawski MA. Acta Neurol Scand Suppl. 2013:9. doi: 10.1111/ane.12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sobolevsky AI, Rosconi MP, Gouaux E. Nature. 2009 Dec 10;462:745. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen L, Durr KL, Gouaux E. Science. 2014 Aug 29;345:1021. doi: 10.1126/science.1258409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Durr KL, et al. Cell. 2014 Aug 14;158:778. doi: 10.1016/j.cell.2014.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yelshanskaya MV, Li M, Sobolevsky AI. Science. 2014 Aug 29;345:1070. doi: 10.1126/science.1256508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyerson JR, et al. Nature. 2014 Aug 3; [Google Scholar]
- 13.Bredt DS, Nicoll RA. Neuron. 2003 Oct 9;40:361. doi: 10.1016/s0896-6273(03)00640-8. [DOI] [PubMed] [Google Scholar]
- 14.Howe JR. J Physiol. 2015 Jan 1;593:61. doi: 10.1113/jphysiol.2014.273904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwenk J, et al. Neuron. 2012 May 24;74:621. doi: 10.1016/j.neuron.2012.03.034. [DOI] [PubMed] [Google Scholar]
- 16.Chen L, et al. Nature. 2000 Dec 21–28;408:936. [Google Scholar]
- 17.Jackson AC, Nicoll RA. J Neurosci. 2011 Mar 16;31:3939. doi: 10.1523/JNEUROSCI.5134-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vandenberghe W, Nicoll RA, Bredt DS. Proc Natl Acad Sci U S A. 2005 Jan 11;102:485. doi: 10.1073/pnas.0408269102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato AS, Gill MB, Yu H, Nisenbaum ES, Bredt DS. Trends Neurosci. 2010 May;33:241. doi: 10.1016/j.tins.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 20.Jackson AC, Nicoll RA. Neuron. 2011 Apr 28;70:178. doi: 10.1016/j.neuron.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakagawa T, Cheng Y, Ramm E, Sheng M, Walz T. Nature. 2005 Feb 3;433:545. doi: 10.1038/nature03328. [DOI] [PubMed] [Google Scholar]
- 22.Hastie P, et al. Proc Natl Acad Sci U S A. 2013 Mar 26;110:5163. doi: 10.1073/pnas.1218765110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim KS, Yan D, Tomita S. J Neurosci. 2010 Jan 20;30:1064. doi: 10.1523/JNEUROSCI.3909-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi Y, Lu W, Milstein AD, Nicoll RA. Neuron. 2009 Jun 11;62:633. doi: 10.1016/j.neuron.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.See Supplementary Materials and Methods.
- 26.Priel A, et al. J Neurosci. 2005 Mar 9;25:2682. doi: 10.1523/JNEUROSCI.4834-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomita S, et al. Nature. 2005 Jun 23;435:1052. doi: 10.1038/nature03624. [DOI] [PubMed] [Google Scholar]
- 28.Carbone AL, Plested AJ. Nat Commun. 2016;7:10178. doi: 10.1038/ncomms10178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dawe GB, et al. Neuron. 2016 Mar 16;89:1264. doi: 10.1016/j.neuron.2016.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang W, Devi SP, Tomita S, Howe JR. Eur J Neurosci. 2014 Apr;39:1138. doi: 10.1111/ejn.12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki H, et al. Science. 2014 Apr 18;344:304. [Google Scholar]
- 32.Saitoh Y, et al. Science. 2015 Feb 13;347:775. [Google Scholar]
- 33.Dong H, Zhou HX. Nat Commun. 2011;2:354. doi: 10.1038/ncomms1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kazi R, Dai J, Sweeney C, Zhou HX, Wollmuth LP. Nat Neurosci. 2014 Jul;17:914. doi: 10.1038/nn.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kato AS, Siuda ER, Nisenbaum ES, Bredt DS. Neuron. 2008 Sep 25;59:986. doi: 10.1016/j.neuron.2008.07.034. [DOI] [PubMed] [Google Scholar]
- 36.Sobolevsky AI. J Physiol. 2015 Jan 1;593:29. doi: 10.1113/jphysiol.2013.264911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.MacLean DM, Ramaswamy SS, Du M, Howe JR, Jayaraman V. J Gen Physiol. 2014 Dec;144:503. doi: 10.1085/jgp.201411287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morimoto-Tomita M, et al. Neuron. 2009 Jan 15;61:101. doi: 10.1016/j.neuron.2008.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomita S, et al. Nature. 2005 Jun 23;435:1052. doi: 10.1038/nature03624. [DOI] [PubMed] [Google Scholar]
- 40.Chen L, et al. Nature. 2000;408:936. doi: 10.1038/35050030. [DOI] [PubMed] [Google Scholar]
- 41.Kawate T, Gouaux E. Structure. 2006 Apr;14:673. doi: 10.1016/j.str.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 42.Dukkipati A, Park HH, Waghray D, Fischer S, Garcia KC. Protein Expr Purif. 2008 Dec;62:160. doi: 10.1016/j.pep.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robert A, Howe JR. J Neurosci. 2003 Feb 1;23:847. doi: 10.1523/JNEUROSCI.23-03-00847.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goehring A, et al. Nat Protoc. 2014 Nov;9:2574. doi: 10.1038/nprot.2014.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Russo CJ, Passmore LA. Science. 2014 Dec 12;346:1377. doi: 10.1126/science.1259530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mastronarde DN. J Struct Biol. 2005 Oct;152:36. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 47.Suloway C, et al. J Struct Biol. 2005 Jul;151:41. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 48.Grant T, Grigorieff N. Elife. 2015;4:e06980. doi: 10.7554/eLife.06980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rohou A, Grigorieff N. J Struct Biol. 2015 Nov;192:216. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scheres SH. J Struct Biol. 2012 Dec;180:519. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scheres SH. J Struct Biol. 2015 Feb;189:114. doi: 10.1016/j.jsb.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bai XC, Rajendra E, Yang G, Shi Y, Scheres SH. Elife. 2015;4 doi: 10.7554/eLife.11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pettersen EF, et al. J Comput Chem. 2004 Oct;25:1605. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 54.Kucukelbir A, Sigworth FJ, Tagare HD. Nat Methods. 2014 Jan;11:63. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li X, et al. Nat Methods. 2013 Jun;10:584. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scheres SH, Chen S. Nat Methods. 2012 Sep;9:853. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Biasini M, et al. Nucleic Acids Res. 2014 Jul;42:W252. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Emsley P, Lohkamp B, Scott WG, Cowtan K. Acta Crystallogr D Biol Crystallogr. 2010 Apr;66:486. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yifrach O, MacKinnon R. Cell. 2002 Oct 18;111:231. doi: 10.1016/s0092-8674(02)01013-9. [DOI] [PubMed] [Google Scholar]
- 60.Cais O, et al. Cell Rep. 2014 Oct 23;9:728. doi: 10.1016/j.celrep.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meyerson JR, et al. Nature. 2014 Oct 16;514:328. doi: 10.1038/nature13603. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.