Abstract
Production of cellular reactive oxygen species (ROS) is typically associated with protein and DNA damage, toxicity, and death. However, ROS are also essential regulators of signaling and work in concert with redox-sensitive proteins to regulate cell homeostasis during stress. In this review, we focus on the redox regulation of mitophagy, a process that contributes to energetic tone as well as mitochondrial form and function. Mitophagy has been increasingly implicated in diseases including Parkinson’s, Amyotrophic Lateral Sclerosis, and cancer. Although these disease states employ different genetic mutations, they share the common factors of redox dysregulation and autophagic signaling. This review highlights key redox sensitive signaling molecules which can enhance neuronal survival by promoting temporally and spatially controlled autophagic signaling and mitophagy.
Keywords: Mitophagy, Reactive oxygen species, Energetics, FOXO, HIF, Sirtuin, Atg, p66shc, Neurodegeneration, Amyotrophic lateral sclerosis, Isocitrate dehydrogenase, Mitochondria, Parkinson’s disease
1. Introduction
Reactive oxygen species (ROS) and other free radicals are typically associated with stress, DNA and protein damage, and used as a marker of irrevocable damage and cell death. It has become increasingly clear, however, that free radicals function as important signaling molecules in response to stress [1,2] and modify protein structure and function in ways that are essential to adaptation and survival. In this chapter we will discuss ROS production as a consequence of aerobic respiration, how mitophagic containment of injured organelles promotes cell survival and how impaired redox regulation of autophagic signaling molecules leads to cell dysfunction.
Eukaryotic cells have evolved symbiotic relationships with mitochondria to enhance energetic status and cellular and organismal complexity. On average, eukaryotic cells contain one billion ATP molecules, and the turnover from ATP to ADP and back occurs approximately three times per minute [3]. The central nervous system (CNS) relies on ATP generated primarily by aerobic respiration and, as reviewed by Erecinska and Silver, the brain at rest utilizes over 20% of total oxygen consumption and 0.3–0.8 μmol of glucose per gram of weight per minute (μmol/g/min). The resulting ATP production in neurons from consumed oxygen and glucose at rest is approximately 25–32 μmol/g/min [4]. Neurons require high levels of ATP – as much as 20–25 μmol/g/min – for maintaining membrane polarity, rapid intra- and inter-cellular signaling, and synaptic transmission. For example, neurons contain 6250 ρmol of the Na+/K+ pump/g of tissue (cardiac muscle contains only 1500–2000 ρmol/g) [5] with the highest activity reported in synaptosomes [6]. Neurons need 50–60% of total oxygen delivered to the brain and 12–16 μmol ATP/g/min [4] to maintain ion homeostasis alone. While the liver and heart consume more net ATP, the only organ with comparable levels of energy expenditure for membrane polarity and ionic balance is the kidney [7].
In addition to being the primary source of energy production in the form of ATP, mitochondria produce the majority of free radicals and other ROS. In studies measuring superoxide anions (O2−▪) in rat lung slices, about 9% of total oxygen uptake resulted in O2−▪ formation, while other studies suggest that about 1–3% of oxygen used in the mitochondria alone form O2−▪. Neurons, especially those that are most active and excitatory, have higher oxidative metabolic activity and higher cristae packing density of mitochondria than glia and other cell types [8]. Mitochondria are densely packed in extended dendrites and axon terminals. The fact that neurons are largely post mitotic places these cells under intense pressure to promote appropriate mitochondrial biogenesis fusion, fission, and mitophagy. Seminal work by Chang and Reynolds in 2006 aimed to measure the turnover rate and number of mitochondria at baseline in neurons. Interestingly, they found that mitochondria morphology and trafficking is dependent on the age of neurons [9]. For instance, a mature neuron at 14 days in vitro (DIV14) will contain a similar amount of mitochondria as a young neuron at DIV5 (~0.08 mitochondria/μm); however, mitochondria in mature neurons are elongated and form reticular networks, whereas in young neurons, mitochondria are shorter and more motile [9]. This difference in mitochondrial morphology with respect to neuron maturity points to the temporal control of proteins governing mitochondrial dynamics. David Chan pioneered work a decade ago demonstrating that fusion and fission of mitochondria is coordinated by essential proteins which are exquisitely sensitive to genetic mutations and cellular stress [10]. In addition to maladaptive changes in fusion and fission molecules, functional changes in these proteins by redox modification have increasingly been appreciated as central regulators of mitochondrial function.
In the last five years, engulfment of damaged mitochondria by molecules linked to autophagy has played a more prominent role with respect to understanding the pathological cascades associated with Parkinson’s disease, Amyotrophic Lateral Sclerosis (ALS), Alzheimer’s disease and other disorders. Autophagy is a highly controlled process in which cells engulf proteins and organelles within intracellular, lipid bilayer compartments and degrade contents after fusion with lysosomes. The process was initially described in yeast and now hundreds of proteins have been associated with this highly complex pathway [11–13]. Autophagic signaling can be promoted in response to a wide variety of stressors including ionic dysfunction, protein aggregation and proteasome failure, energetic stress and oxygen deprivation. Autophagy can also be targeted toward specific organelles – notably the mitochondria – where they can undergo engulfment and digestion; a process that is important given that mitochondria are both one of the primary sources and sensitive targets of ROS. Moreover, mitophagy may be particularly sensitive to ROS given that antioxidants, such as glutathione, can block mitophagy while autophagy continues, as revealed in yeast models [14].
2. Reactive oxygen species
2.1. ROS formation
Free radicals are a class of small molecules that contain one or more unpaired electrons, rendering them highly reactive; those derived from oxygen molecules are called reactive oxygen species (ROS), and those from nitric oxide, reactive nitrogen species (RNS). ROS are generated by a number of reactions promoted by xanthine oxidase, microsomal enzymes, cyclooxygenase and lipoxygenase as well as more abundant mechanisms including ROS produced by NADPH oxidases and the electron transport chain (ETC) [15].
The activity of NADPH oxidases, membrane-associated enzymes that utilize oxygen to catalyze the production of O2−▪, is essential for engulfment of injured cells and innate immunity. NADPH oxidase deficiency is associated with X-linked chronic granulomatous disease, a devastating disorder which leaves patients with poor immune function [16]. Assembly of the NADPH complex at lysosomes produces a large increase in oxygen consumption, referred to as a “respiratory burst”. In response, superoxide dismutase (SOD) converts O2−▪ into hydrogen peroxide (H2O2), which kills off the engulfed cells or particles. In addition, high concentrations of catalase to detoxify H2O2, as in erythrocytes, are able to protect other cells by absorbing diffused H2O2 [17] Another mechanism for O2−▪ clearance is via reaction with nitric oxide to produce peroxynitrite, which can subsequently be catalyzed to form hydroxyl radicals.
The major source of ROS in aerobic cells, however, is cellular respiration and oxidative phosphorylation within the mitochondria. In the process of transferring electrons between the protein complexes that mediate phosphorylation of ADP to ATP and reducing oxygen to water, the ETC generates free radicals. Electrons that escape the ETC primarily through mitochondrial complexes I and III transfer directly to oxygen to create O2−▪ [18–20]. Due to the high demand of ATP and increased oxidative phosphorylation to meet that demand, neurons are particularly prone to generating concentrated amounts of ROS. The intracellular environment of neurons is thus highly reducing and, as will be further discussed, contains various mechanisms to combat oxidation.
2.2. ROS modify specific amino acids
The location of amino acids within proteins is a central regulator of protein function. Post-translational modifications, such as lipidation or glycosylation, at specific amino acids alter protein behavior through inactivation or enhancing activity or binding with other proteins. Similarly, ROS serve as reversible post-translational modifiers of protein structure and function. Amino acid oxidation can alter protein binding, interaction, and translocation or confer previously unknown properties and binding partners. It is helpful to realize that much like post-translational modification by phosphorylation, the structure and availability of amino acids determines their vulnerability to ROS-mediated modifications.
Thiol groups are organosulfur compounds that contain a carbon-bonded sulfhydryl (–C–SH or R–SH) group (where R represents an alkane, alkene, or other carbon-containing group of atoms). Proteins which contain these compounds are most easily oxidized by ROS, RNS, and some metal ions. Oxidized thiol groups can proceed to form thiyl radicals or disulphides. In fact, a third of proteins synthesized by neurons contain disulfide bonds [21] that are catalyzed by protein disulfide isomerases in the endoplasmic reticulum [22].
The amino acids cysteine and methionine contain reactive thiol groups that confer redox sensitivity to proteins. Similarly, the thiol containing amino acid histidine makes it a target for singlet O2 reactions. Other less reactive amino acids include tyrosine, phenylalanine, valine, proline, arginine, and tryptophan which can be oxidized by oxygen radicals, forming peroxides and peroxyl radicals [23]. Interaction with ROS causes these residues to break down and generate highly reactive intermediate radicals.
While beyond the scope of this review, it should be noted that reactive nitrogen species, such as nitric oxide, are also able to post-translationally modify proteins to regulate their function by S-nitrosylation [24]. Using site-specific mapping of S-nitrosylation of cysteine residues in tissues of healthy mice, a recent study identified a substantial number of oxidatively modified proteins that regulate oxidative phosphorylation, glycolysis, and other pathways for metabolism [25]. The authors report that 20–25% of S-nitrosylated proteins were mitochondrial proteins in brain, liver, and kidney tissues compared to 56% in heart tissue. In addition, when comparing modified proteomes between wildtype and endothelial nitric oxide synthase (eNOS) knockout mice, the group found that mitochondrial proteomes were more than 70% dependent on eNOS activity for S-nitrosylation, suggesting an important role for RNS in normal metabolic regulation. Recent reviews [26,27] can be referenced for more information on these molecules and the link between NO formation and inhibition of autophaghosome formation in neurons [28].
2.3. ROS regulation & redox homeostasis
The tight regulation of ROS is key to energetic status, cell signaling and responses to stress. The ways in which cells regulate redox state include general mechanisms associated with increasing the activity of antioxidants, as well as utilizing redox-sensitive enzymes to decrease ROS (summarized in Table 1). Furthermore, tight spatial constraints on ROS can be enforced by organelles and protein complexes, which localize ROS for efficient removal by co-localized antioxidants, such as thioredoxin and glutathione.
Table 1.
Primary types of reactive oxygen species produced in neurons. ROS are primarily produced in aerobically dependent organisms due to inefficient electron transfer in mitochondria. Reduction of the highly reactive superoxide radical results in the formation of hydrogen peroxide, a more stable signaling molecule. Reduction of hydrogen peroxide results in formation hydroxyl radicals. Each of these ROS can be detoxified by catalytic and non-catalytic antioxidants.
| Reactive oxygen species | Structure | Reactivity | Sources | Detoxification |
|---|---|---|---|---|
| Superoxide radical |
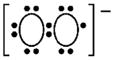
|
High | Mitochondria, ETC NOX isoforms NOS |
SOD GSH Trx |
| Hydrogen peroxide |
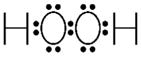
|
Low | Spontaneous reduction of O2−▪ SOD reduction of O2−▪ |
Catalase GPx |
| Hydroxyl radical |
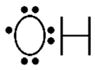
|
Very high | Breakdown of organic peroxides | GSH |
2.3.1. Endogenous non-enzymatic antioxidants reduce intracellular ROS
The tripeptide glutathione (GSH; γ–L-Glutamyl-L-cysteinylglycine) is the most abundant low molecular weight thiol in mammalian cells and constitutes a major cellular defense against ROS. In the process of reduction, GSH is oxidized into glutathione disulfide (GSSG) and can be reduced back to GSH via glutathione reductase. In most cell types, the rate-limiting step for increasing GSH synthesis is the uptake of cysteine. Interestingly, the CNS maintains an enhanced GSH pool through enriched expression of the cysteine/glutamate exchanger (xCT) in meningeal cells, as well as in both neurons and astrocytes in cerebral cortex [29] offering enhanced redox buffering in the CNS. GSH primarily acts as a ROS scavenger, directly reducing hydroxyl radicals and superoxides. In addition, GSH provides an essential function in the maintenance of other reducing enzymes, such as vitamin C and E, by reducing them to their active states.
While the redox status of neurons and other cells tends to be viewed as a homogeneous environment, quite the opposite is true. Thioredoxin (Trx) is a family of proteins that act as endogenous reducing agents with an oxidizable dithiol active site that, after activation, can be reduced back by thioredoxin reductase. Each member of the Trx family exhibits specific intracellular localization. For instance, Trx-1, the most abundant Trx protein in eukaryotes, is present in both the cytosol and nucleus, while Trx-2 localizes to mitochondria. Although they exist at a low intracellular concentration – on the order of μmol – the Trx proteins may be more specific than GSH in its rapid response to certain toxicants. In response to H2O2, hypoxia [30], ischemic-reperfusion injury [31,32], radiation [33,34], and other toxicants that increase intracellular free radicals, Trx-1 translocates into the nucleus where it can directly or indirectly associate with transcription factors, such as nuclear factor kappa-light-chain-enhancer of activated B cells (NFκB) [35] and activator protein 1 (AP-1) [36]. This compartmentalization of Trx may serve as a discreet mechanism of protection, functioning as a ROS scavenger at the nucleus and as a regulator of gene expression under stress. Also contributing to compartmentalized areas of ROS regulation, the mitochondria-specific Trx-2 interacts with components of the ETC and is thought to play a role in regulating mitochondrial membrane potential [37,38] and mitochondrial permeability transition pore activity [39]. Furthermore, Trx-2 directly reduces H2O2 as a peroxiredoxin cofactor, whereas GSH contributes minimally to H2O2 clearance [40,41]. Thus, despite minute overall concentration compared to GSH, Trx serves as an essential redox regulator through distinct antioxidant roles in mitochondria and nuclei, as well as in specific peroxide clearance.
2.3.2. Redox-sensitive enzymes catalyze antioxidant processes
In addition to non-catalytic antioxidants, several catalytic proteins also contribute to the regulation of redox homeostasis. The SOD family of enzymes, first discovered by Fridovich and McCord [42] are metal coordinating enzymes that play major roles in the enzymatic reduction of O2−▪ into oxygen and H2O2. Cytoplasmic copper/zinc superoxide dismutase (CuZnSOD) and mitochondria-localized manganese superoxide dismutase (MnSOD) are both active participants in this process. Both enzymes are highly expressed in neurons, and overexpression of either enzyme decreases tissue damage after focal ischemia in animal models [43,44]. Furthermore, mutations in these enzymes lead to deficiencies in buffering ROS and are linked to familial forms of ALS [45].
Catalases are a group of proteins that consist of four protein subunits that contain a buried heme group. Molecules small enough to travel through narrow channels leading to the heme center are then reduced. Catalases primarily reduce small molecules such as H2O2, utilizing a reaction mechanism by which one H2O2 is reduced to a water molecule and another oxidized to an oxygen molecule. Catalases are especially active within peroxisomes, which contain enzymes that create H2O2. Subcellular localization of catalases at organelles of high H2O2 production highlights the ways in which cells tightly regulate redox state. However, mitochondria contain very little catalase, thus any H2O2 generated by the mitochondria must either diffuse to peroxisomes or rely on other antioxidant defenses [46].
Another enzymatic antioxidant protein is glutathione peroxidase (GPx) that removes H2O2 by reducing it to a water molecule and simultaneously oxidizing a reduced glutathione. Like GSH, GPx is localized primarily to the cytosol but it is also found in mitochondria, which would assist in the removal of H2O2 generated from this organelle.
Localized concentrations of antioxidants may also play a significant role in mitophagic signaling. Interestingly, ethacrynic acid-induced depletion of GSH promoted mitophagy in yeast under conditions of starvation [14]. Conversely, increases in the GSH pool by addition of the antioxidant N-acetyl-L-cysteine prevented mitophagy during starvation, but did not impact autophagy [14]. The authors hypothesized that mitophagy was being impacted by the spatial and temporal and localization of GSH.
3. Redox regulation of autophagic signaling
The activation of autophagy and mitophagy is tightly regulated by molecular rearrangement, such as protein conformational changes and complex formation, and protein localization to initiate the transcription of autophagy related genes (Atgs). Increased generation of ROS promotes autophagy via oxidative modification of proteins involved in either the autophagic machinery or in signaling pathways that enhance autophagy.
3.1. Oxidatively-modified proteins change function to promote autophagy
3.1.1. ROS inhibition of PTEN increases Akt signal transduction to regulate FoxO transcription factors
Cellular redox status impacts lipids and protein signal transduction much in the same way that other forms of post-translational modifications do. As modifiers of protein activity, oxidative modifications impact neuronal mitophagy and potentially serve as essential regulators of cell growth, signaling and fate.
One example of the impact of redox status on cell fate decisions is the effect of ROS on growth factor mediated signaling. Molecules such as vascular endothelial growth factor (VEGF) and neuronal growth factor (NGF) activate signaling cascades to promote cell survival signaling by activation of the PI3K/Akt pathway [47,48]. Activation of the PI3K/Akt pathway promotes transcription of pro-survival genes, and the main regulatory member of this cascade is Phosphatase and Tensin homologue (PTEN), a tyrosine phosphatase that normally dephosphorylates PI3K, decreasing Akt signal transduction. Akt is a serine/threonine-specific protein kinase which promotes cell survival by inhibition of apoptosis and activation of transcription factors that increase expression of genes that regulate cell survival, proliferation, metabolism, and migration [49].
PTEN is oxidatively modified by H2O2, which produces a disulfide bond in the PTEN active site. Oxidation of PTEN by H2O2 leads to reversible inactivation of PTEN activity, prolonging PI3K and Akt association – an effect that promotes cell survival in the presence of high H2O2 concentrations in vitro [50] and in in vivo models of ischemia where the reperfusion stress of flooding a system with oxygen causes a burst of ROS [51]. Additionally, studies in which Akt signaling is inhibited demonstrate that mitochondrial derived ROS production increases promoting autophagy [52]. Moreover, modification of Akt by ROS can be adaptive by increasing the nuclear export of Forkhead box protein O1 (FoxO1), driving the production of antioxidant enzyme Trx. This signaling loop combats further increases in ROS [50] and enhances the activity of Atg family proteins and facilitating autophagic signaling.
In addition to the indirect effects of PTEN modification on Akt, the Forkhead family of proteins also responds to Akt signaling. The Forkhead transcriptional regulators play diverse roles in eukaryotic cell growth, differentiation, and survival. Indeed, the mammalian FoxO family was initially identified as essential tumor suppressors [53,54] and are direct substrates of Akt. When Akt phosphorylates FoxO transcription factors, both proteins are exported from the nucleus to the cytoplasm via a nuclear export sequence. Following Akt phosphorylation, FoxOs interact with 14-3-3 proteins promoting the cytoplasmic localization of FoxO and alterations in cell signaling independent of DNA binding. Direct interacting partners of FoxOs include other transcription factors, such as β-catenin and Smad, HOX proteins, and hormone receptors [55].
Engagement of FoxO transcription has been increasingly associated with the induction of autophagy-related genes, underscoring the importance of these proteins and the autophagic pathway in mutagenesis and oncogenic transformation [56]. FoxO3 upregulates genes such as Beclin 1, ATG12, ATG4, LC3, and others, with essential roles in autophagic recognition, containment, engulfment and digestion [57]. FoxO3 is a positive regulator of BNIP3 expression – a mitochondrially associated protein that promotes autophagosome formation in cell culture models of skeletal muscle atrophy [57] as well as mitophagy in animal models of hypoxia [58].
In yeast, FoxO1 stimulates autophagy via interaction with Atg7 upon acetylation by Sirtuin-2, a redox sensitive transcription factor [59]. Using mammalian cells, the same group found FoxO3 promotes FoxO1-mediated autophagy, wherein FoxO3 upregulates PI3K subunit expression, thus increasing Akt activity [60]. In consequence, FoxO1 is exported from the nucleus, where it then promotes its pro-autophagic activity [59,60]. Taken together, these data support a model in which redox status of the cell is capable of directly modifying a sentinel protein to promote autophagy via growth factor engagement, kinase activation, transcriptional regulation and subcellular relocalization of proteins and protein complexes (Fig. 1).
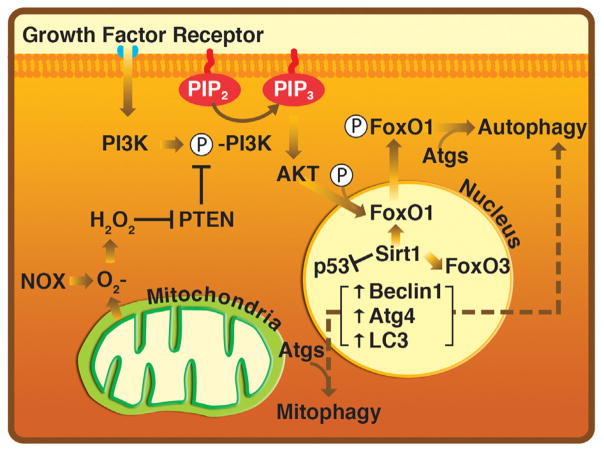
Fig. 1.
Oxidation of PTEN promotes autophagic and mitophagic signaling. ROS reversibly oxidize PTEN allowing for prolonged PI3K/Akt signaling in response to growth factor receptor stimulation. Downstream targets of Akt stimulation include pro-survival transcription factors, such as FoxO1 and FoxO3. Transport of FoxO1 out of the nucleus initiates autophagosome formation by Atg recruitment. FoxO3 is known to target pro-autophagic genes promoting autophagosome formation.
3.2. ROS regulate ATG activity
The ATG genes in yeast and respective homologues in mammals are the primary initiators and regulators of autophagy. Seminal work in yeast has shown that nutrient depletion causes vacuoles to fill with autophagosomes, a process initiated by inactivation of Tor, and that mutations of the ATG gene family disrupted that process [61–63]. Nutrient depletion has not only been associated with increased autophagic signaling, but also with changes in redox homeostasis and oxidative stress. Eisler et al. showed that prolonged amino-acid depletion induces apoptosis, as well as high concentrations of ROS. The group also showed that 40% of starving cells survived despite having high ROS, indicating that ROS is not a consequence of apoptosis, but rather a signal to initiate this process [64].
ROS is also a signaling mechanism for autophagy during acute starvation. In in vitro models of acute starvation, Scherz-Shouval et al. demonstrated that autophagosome formation depended on increases in ROS. Specifically, the activity of Atg4, a cysteine protease, was shown to be inhibited by treatment with H2O2. Under ideal growth conditions, Atg4 removes an arginine residue of Atg8, allowing Atg8 to be activated by Atg7. With H2O2 treatment Atg4 becomes oxidized at specific cysteine residues, rendering it inactive and allowing Atg8 to conjugate with autophagosomes [65].
Autophagy and mitophagy share some similar proteins, but others, such as Atg32, a newly identified mitochondrial outer membrane protein which interacts with Atg11, promotes the autophagy machinery specifically for mitophagy [66,67]. Notably, no metazoan homologues of Atg32 have been identified, but other molecules that work with Atg homologues in eukaryotic organisms such as NIX (also called BNIP3L), integrate redox sensitivity with organelle degradation mediated by Atg8 [68,69].
3.3. Oxidative modification of proteins that alter mitochondrial bioenergetic function
Some of the earliest and most well characterized molecules that have been linked to oxidative stress signaling include the serine kinase p66shc. This protein is a member of the Shc family which plays important roles in oncogenic transformation and cell signaling. Unlike other Shc family members, p66shc has evolved a glycine and proline-rich second collagen homology domain (CH2), which confers a unique ability to sense redox stress. Phosphorylation at serines 36 and 54 within the CH2 domain in response to oxidative stress results in changes in protein structure required for p66shc’s ability to serve as a redox sensing signaling molecule. In addition, cysteine 59 in the CH2 domain is the target of reversible, specific amino acid oxidation that promotes the bioactivity of p66shc allowing the formation of tetramers [70]. Polymorphisms in the protein are linked with long life [71], and mice that lack p66shc live 30% longer than wildtype animals [72] and have a significantly decreased redox burden as they age.
Phosphorylation of Ser36 of p66shc is promoted by protein kinase Cβ. Once phosphorylated, this redox sensitive molecule binds to Pin1, a prolyl hydroxylase [73]. p66shc then relocalizes to the intermembrane space of the mitochondria. p66shc activation in response to oxidative stress is rapid; our group has found Ser36 phosphorylation of p66shc occurs within 30 min after sublethal oxygen and glucose deprivation in neurons and mitochondrial localization is maximal 2–3 h after initiation of stress [2]. Once localized to mitochondria, p66shc oxidizes cytochrome c in cell free assays which is thought to promote ETC production of H2O2, and impair the ability of mitochondria to buffer calcium [74].
While these effects of p66shc activity have largely been associated with apoptosis, our group has identified p66shc as a discrete but essential mediator of neuronal metabolic tone and autophagosome formation as an adaptive mechanism to promote the removal of injured organelles following low level oxygen and glucose deprivation [2]. Indeed, interruption of p66shc activity during mild stress profoundly increased lipid and protein injury as well as the number of and autophagosomes that form. These events were all correlated with poor outcomes for neurons and a reduction in the ability to upregulate the expression of neuroprotective chaperone proteins [2]. This data supports a model in which we have come to appreciate that oxidative modification of proteins such as p66shc is not synonymous with cell death, but this modification serves as a rapid means to promote communication from the cytosol back to the mitochondria. Indeed, localization to mitochondria of activated p66shc suggests this protein may be one of several molecules that temporally and spatially mediate mitochondria homeostasis, as well as mitophagy.
3.4. Oxidatively-modified transcription factors alter activity to promote autophagy
3.4.1. SIRT1 is oxidatively modified to regulate autophagy
While cytosolic signaling molecules like Akt and p66shc promote unique redistribution in response to ROS, nuclear transcription factors also play essential roles in autophagic regulation. Notably, mammalian sirtuins function as important regulators of metabolism, cell survival, growth, and senescence. Sirtuins – mammalian homologues of yeast silent information regulator 2 (Sir2) – are Class III histone deacetylases that respond to a variety of stressors and are involved in protection against several diseases and aging. The most well-studied is Sirtuin 1 (SIRT1). Several seminal studies point to SIRT1’s capability of attenuating age-related deficiencies and protecting against oxidative stress [75].
SIRT1 mRNA is especially high in the developing CNS and heart. While most sirtuins are expressed at high levels in the brain, the most extensively studied are SIRT1[76], which is primarily located in neurons, and SIRT2, which is primarily located in oligodendrocytes and Schwann cells [75]. SIRT1-deficient mice exhibit profound developmental abnormalities, including cardiac defects, exencephaly, and perturbed retinal histology [77]. Tissues cultured from these mice contain heavily damaged mitochondria and impaired metabolic function [78]. SIRT1 deacetylase also complexes with Atg5, Atg7, and Atg8 leading to comparisons between the SIRT1−/− phenotype with ATG5−/− cells and animals [78,79]. Transgenic animals deficient in either protein displayed similar deficiencies in autophagy, lending the idea that SIRT1 deacetylase activity may be important for Atg function.
SIRT1 also acts as an indirect regulator of autophagic processes via regulation of FoxO and p53 (Fig. 1). Sirt1 forms a complex with FoxO3 in response to oxidative stress, deacetylating FoxO3 to further promote cell survival via increased DNA binding and activation of FoxO3 target genes [80].
3.3.2. ROS can increase HIF1α-mediated autophagic signaling and mitophagy
A cell’s ability to sense intracellular oxygen levels is critical for cell survival and adaptation. The primary sensors of oxygen levels within the cell are prolyl hydroxylase domain-containing enzymes (PHDs) and hypoxia-inducible factors (HIFs) [81,82]. When oxygen supply is ideal, PHDs hydroxylate proline residues on HIFα subunits [83]. Hydroxylated HIF is then ubiquitinated by the E3 ubiquitin ligase von Hippel-Lindau protein (pVHL) and is subsequently degraded. In low oxygen conditions, however, PHDs decrease activity, stabilizing HIF and inducing transcription of target genes such as vascular endothelial growth factor, cyclooxygenase-2, and erythropoietin [84].
Several studies demonstrated that mitochondrial ROS regulate HIF proteins at the level of pVHL induced degradation and expression of genes regulated by HIF such as metallothionein. Using both pharmacological and genetic techniques, Brunelle and colleagues subjected HEK293 cells to hypoxia following inactivation of mitochondrial complex III and demonstrated a rapid increase in HIF1α degradation [85]. Although ROS is not required for HIF transcription, the authors of these studies posit that ROS generated by the mitochondria left shifted the dose response curve of oxygen to prolyl hydroxylase activity [86]. In addition, a separate study found that a functional ETC and mitochondria-derived ROS are necessary for hypoxia-induced HIF1α stabilization [87].
This model, however, is limited to mild hypoxia. During periods of anoxia, HIF can be stabilized in the absence ROS. Other studies provide evidence for HIF1α as an essential cofactor for metallothionein gene transcription in response to hypoxic stress [88]. Metallothionein proteins are potent oxidant scavengers which regulate heavy metal sequestration and metabolism. Triggers such as NO promote the release of zinc from metallothioneins which can be a potent means to induce apoptotic cell death in neurons. Furthermore, elevated levels of ROS following hypoxic stress can stimulate zinc release from metallothioneines, which inhibits further HIF1α activity.
HIFs are essential transcriptional regulators of genes with essential functions in energy metabolism and vascular remodeling which optimize available oxygen and glucose. HIF stabilization also aides in autophagic signaling via transcription of BNIP3 and BNIP3L, which are members of the so-called BH3-only subfamily of Bcl-2 family proteins that heterodimerize and antagonize the activity of the largely prosurvival proteins (Bcl-2 and Bcl-XL), Semenza and colleagues presented convincing evidence that hypoxia-induced HIF1α stabilization and regulation of BNIP3 reduced the numberof mitochondria in hypoxic cells through mitophagy [89]. The mechanism proposed involves elevated BNIP3 in competition with Beclin-1 for binding with Bcl2. Bound Bcl2–BNIP3 increases levels of free Beclin-1, which then triggers formation of autophagosomes. Similar reports from other groups revealed that both BNIP3 and BNIP3L are crucial for hypoxia-induced autophagy through disruption of Bcl2-Beclin1 binding [90]. Additionally, rodent models of hypoxia tolerance demonstrate that BNIP3 expression is strongly correlated with mitochondrial clearance and mitophagic signaling in cardiac tissue depending on the degree of hypoxia [58].
Taken together, redox status of the cell is a key factor in determining HIF1α stability and thus regulation of downstream modulators of autophagy and mitophagy (Fig. 2).
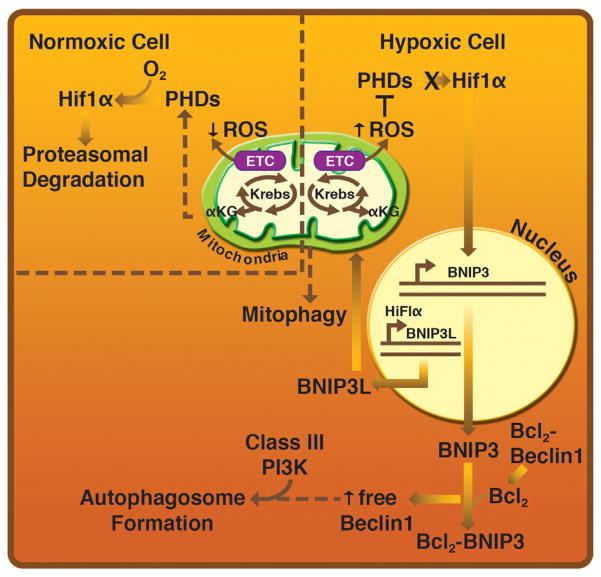
Fig. 2.
ROS inhibit PHD activity to promote autophagy and mitophagy via HIF-regulated transcription. Hypoxia and reperfusion increase production of ROS by perturbing oxidative phosphorylation. ROS inhibit the HIF1α degrading PHD complex, increasing expression of cytosolic HIF1α. HIF1α relocalizes to the nucleus when associated with HIF1β, targeting pro-survival genes, including BNIP3 and BNIP3L as well as pro-death molecules based on the extent and duration of oxygen deprivation. When BNIP3L is synthesized, it associates with mitochondria complexes with Bcl2 in competition with Beclin1. The liberation of Beclin1 promotes autophagosome formation.
Although these and other studies of hypoxia-induced signaling have been essential to understanding cellular response to oxygen levels and ROS, it is important to remember that neurons rely heavily on oxygen levels, which in turn promotes high levels of ROS within the CNS. This effect is compounded by the largely post-mitotic nature of these cells. Although hypoxic conditions disrupt neuronal metabolic and redox tone, not all neurons are equally vulnerable to hypoxia; for instance, CA1 hippocampal neurons are some of the most sensitive [91]. At sea level, atmospheric oxygen is approximately 20%, but oxygen present in vivo in tissue is lower than ambient oxygen [92]. Furthermore, levels of oxygen vary between tissue in vivo (20–40 mmHg) [93], cells in vivo (1–10 mmHg) and cells grown in culture (150 mmHg) [94], providing yet another caveat to studying the consequences of hypoxic stress.
Therefore, in attempting to capture the CNS responses to hypoxia, our group – among others – is particularly cautious in using immortalized cell lines. These mitotic cultures are primarily glycolytic, although they can be forced to rely on oxidative phosphorylation for ATP production by substituting glucose with galactose [95]. This discrepancy makes the use of primary cultures for the investigation of hypoxia-dependent signaling far more appealing from a physiological standpoint.
4. Conclusions
In summary, ROS-mediated signaling may occur independently of cell death mechanisms and may instead promote survival by activating specific regulators of mitophagy and autophagy. ROS working in concert with target redox-sensitive proteins is another facet of intracellular signaling that impacts the ways in which we study neuropathological diseases associated with high concentrations of ROS. In order to expand this area of study, better methods for differentiating between mitophagy and autophagy are needed. Klionsky and colleagues recently released guidelines for studying autophagy [96], yet the identification of methods specifically targeting mitophagy is limited. As for studying ROS-mediated signaling, many groups have taken advantage of redox sensors, such as dyes and fluorescent markers, as well as free radical spin traps to inhibit ROS [97]. Additionally, novel methods for isolating redox-sensitive proteins can be coupled with mass spectrometry to identify proteins oxidized at specific amino acids [98,99]. This method holds promise in pin-pointing modified protein targets that would affect the pathways involved following cellular stress. Furthermore, the connection between in vivo and in vitro disease models for oxidative stress and neuropathological disease must be re-evaluated and tuned more toward recapitulation of physiological conditions.
References
- 1.Adler V, et al. Role of redox potential and reactive oxygen species in stress signaling. Oncogene. 1999;18(45):6104–6111. doi: 10.1038/sj.onc.1203128. [DOI] [PubMed] [Google Scholar]
- 2.Brown JE, et al. Essential role of the redox-sensitive kinase p66shc in determining energetic and oxidative status and cell fate in neuronal preconditioning. J Neurosci. 2010;30(15):5242–5252. doi: 10.1523/JNEUROSCI.6366-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kornberg A. For the Love of Enzymes: the Odyssey of a Biochemist. Vol. 9. Harvard University Press; Cambridge, Mass: 1989. p. 336. [Google Scholar]
- 4.Erecinska M, Silver IA. ATP and brain function. J Cereb Blood Flow Metab. 1989;9(1):2–19. doi: 10.1038/jcbfm.1989.2. [DOI] [PubMed] [Google Scholar]
- 5.Hansen O, Clausen T. Quantitative determination of Na+-K+-ATPase and other sarcolemmal components in muscle cells. Am J Physiol. 1988;254(1 Pt 1):C1–C7. doi: 10.1152/ajpcell.1988.254.1.C1. [DOI] [PubMed] [Google Scholar]
- 6.Atterwill CK, Cunningham VJ, Balazs R. Characterization of Na+, K+-ATPase in cultured and separated neuronal and glial cells from rat cerebellum. J Neurochem. 1984;43(1):8–18. doi: 10.1111/j.1471-4159.1984.tb06672.x. [DOI] [PubMed] [Google Scholar]
- 7.Soltoff SP. ATP and the regulation of renal cell function. Annu Rev Physiol. 1986;48:9–31. doi: 10.1146/annurev.ph.48.030186.000301. [DOI] [PubMed] [Google Scholar]
- 8.Pysh JJ, Khan T. Variations in mitochondrial structure and content of neurons and neuroglia in rat brain: an electron microscopic study. Brain Res. 1972;36(1):1–18. doi: 10.1016/0006-8993(72)90762-7. [DOI] [PubMed] [Google Scholar]
- 9.Chang DT, Reynolds IJ. Differences in mitochondrial movement and morphology in young and mature primary cortical neurons in culture. Neuroscience. 2006;141(2):727–736. doi: 10.1016/j.neuroscience.2006.01.034. [DOI] [PubMed] [Google Scholar]
- 10.Chan DC. Fusion and fission: interlinked processes critical for mitochondrial health. Annu Rev Genet. 2012;46:265–287. doi: 10.1146/annurev-genet-110410-132529. [DOI] [PubMed] [Google Scholar]
- 11.Abeliovich H. Mitophagy: the life-or-death dichotomy includes yeast. Autophagy. 2007;3(3):275–277. doi: 10.4161/auto.3915. [DOI] [PubMed] [Google Scholar]
- 12.Lemasters JJ. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res. 2005;8(1):3–5. doi: 10.1089/rej.2005.8.3. [DOI] [PubMed] [Google Scholar]
- 13.Homma K, Suzuki K, Sugawara H. The autophagy database: an all-inclusive information resource on autophagy that provides nourishment for research. Nucleic Acids Res. 2011;39:D986–D990. doi: 10.1093/nar/gkq995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deffieu M, et al. Glutathione participates in the regulation of mitophagy in yeast. J Biol Chem. 2009;284(22):14828–14837. doi: 10.1074/jbc.M109.005181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai H, Harrison DG. Endothelial dysfunction in cardiovascular diseases: the role of oxidant stress. Circ Res. 2000;87(10):840–844. doi: 10.1161/01.res.87.10.840. [DOI] [PubMed] [Google Scholar]
- 16.Hohn DC, Lehrer RI. NADPH oxidase deficiency in X-linked chronic granulomatous disease. J Clin Invest. 1975;55(4):707–713. doi: 10.1172/JCI107980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winterbourn CC, Stern A. Human red cells scavenge extracellular hydrogen peroxide and inhibit formation of hypochlorous acid and hydroxyl radical. J Clin Invest. 1987;80(5):1486–1491. doi: 10.1172/JCI113230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turrens JF, Boveris A. Generation of superoxide anion by the NADH dehydrogenase of bovine heart mitochondria. Biochem J. 1980;191(2):421–427. doi: 10.1042/bj1910421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boveris A, Cadenas E, Stoppani AO. Role of ubiquinone in the mitochondrial generation of hydrogen peroxide. Biochem J. 1976;156(2):435–444. doi: 10.1042/bj1560435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cadenas E, et al. Production of superoxide radicals and hydrogen peroxide by NADH-ubiquinone reductase and ubiquinol-cytochrome c reductase from beef-heart mitochondria. Arch Biochem Biophys. 1977;180(2):248–257. doi: 10.1016/0003-9861(77)90035-2. [DOI] [PubMed] [Google Scholar]
- 21.Kosuri P, et al. Protein folding drives disulfide formation. Cell. 2012;151(4):794–806. doi: 10.1016/j.cell.2012.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 23.Dean RT, et al. Biochemistry and pathology of radical-mediated protein oxidation. Biochem J. 1997;324(Pt 1):1–18. doi: 10.1042/bj3240001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stamler JS, Lamas S, Fang FC. Nitrosylation: the prototypic redox-based signaling mechanism. Cell. 2001;106(6):675–683. doi: 10.1016/s0092-8674(01)00495-0. [DOI] [PubMed] [Google Scholar]
- 25.Doulias PT, et al. Nitric oxide regulates mitochondrial fatty acid metabolism through reversible protein s-nitrosylation. Sci Signal. 2013;6(256):rs1. doi: 10.1126/scisignal.2003252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gow AJ, et al. Biological significance of nitric oxide-mediated protein modifications. Am J Physiol Lung Cell Mol Physiol. 2004;287(2):L262–L268. doi: 10.1152/ajplung.00295.2003. [DOI] [PubMed] [Google Scholar]
- 27.Haldar SM, Stamler JS. S-Nitrosylation at the interface of autophagy and disease. Mol Cell. 2011;43(1):1–3. doi: 10.1016/j.molcel.2011.06.014. [DOI] [PubMed] [Google Scholar]
- 28.Sarkar S, et al. Complex inhibitory effects of nitric oxide on autophagy. Mol Cell. 2011;43(1):19–32. doi: 10.1016/j.molcel.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shih AY, et al. Cystine/glutamate exchange modulates glutathione supply for neuroprotection from oxidative stress and cell proliferation. J Neurosci. 2006;26(41):10514–10523. doi: 10.1523/JNEUROSCI.3178-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ema M, et al. Molecular mechanisms of transcription activation by HLF and HIF1alpha in response to hypoxia: their stabilization and redox signal-induced interaction with CBP/p300. EMBO J. 1999;18(7):1905–1914. doi: 10.1093/emboj/18.7.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takagi Y, et al. Expression and distribution of redox regulatory protein, thioredoxin during transient focal brain ischemia in the rat. Neurosci Lett. 1998;251(1):25–28. doi: 10.1016/s0304-3940(98)00492-3. [DOI] [PubMed] [Google Scholar]
- 32.Takagi Y, et al. Redox control of neuronal damage during brain ischemia after middle cerebral artery occlusion in the rat: immunohistochemical and hybridization studies of thioredoxin. J Cereb Blood Flow Metab. 1998;18(2):206–214. doi: 10.1097/00004647-199802000-00012. [DOI] [PubMed] [Google Scholar]
- 33.Didier C, et al. Induction of thioredoxin by ultraviolet-A radiation prevents oxidative-mediated cell death in human skin fibroblasts. Free Radic Biol Med. 2001;31(5):585–598. doi: 10.1016/s0891-5849(01)00617-7. [DOI] [PubMed] [Google Scholar]
- 34.Wei SJ, et al. Thioredoxin nuclear translocation and interaction with redox factor-1 activates the activator protein-1 transcription factor in response to ionizing radiation. Cancer Res. 2000;60(23):6688–6695. [PubMed] [Google Scholar]
- 35.Matthews JR, et al. Thioredoxin regulates the DNA binding activity of NF-kappa B by reduction of a disulphide bond involving cysteine 62. Nucleic Acids Res. 1992;20(15):3821–3830. doi: 10.1093/nar/20.15.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hirota K, et al. AP-1 transcriptional activity is regulated by a direct association between thioredoxin and Ref-1. Proc Natl Acad Sci USA. 1997;94(8):3633–3638. doi: 10.1073/pnas.94.8.3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Damdimopoulos AE, et al. Human mitochondrial thioredoxin. Involvement in mitochondrial membrane potential and cell death. J Biol Chem. 2002;277(36):33249–33257. doi: 10.1074/jbc.M203036200. [DOI] [PubMed] [Google Scholar]
- 38.Ly JD, Grubb DR, Lawen A. The mitochondrial membrane potential (deltapsi(m)) in apoptosis; an update. Apoptosis. 2003;8(2):115–128. doi: 10.1023/a:1022945107762. [DOI] [PubMed] [Google Scholar]
- 39.He M, et al. Identification of thioredoxin-2 as a regulator of the mitochondrial permeability transition. Toxicol Sci. 2008;105(1):44–50. doi: 10.1093/toxsci/kfn116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chae JH, Lee YH, Kim CG. Transcription factor CP2 is crucial in hemoglobin synthesis during erythroid terminal differentiation in vitro. Biochem Biophys Res Commun. 1999;263(2):580–583. doi: 10.1006/bbrc.1999.1408. [DOI] [PubMed] [Google Scholar]
- 41.Drechsel DA, Patel M. Respiration-dependent H2O2 removal in brain mitochondria via the thioredoxin/peroxiredoxin system. J Biol Chem. 2010;285(36):27850–27858. doi: 10.1074/jbc.M110.101196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCord JM, Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein) J Biol Chem. 1969;244(22):6049–6055. [PubMed] [Google Scholar]
- 43.Keller JN, et al. Mitochondrial manganese superoxide dismutase prevents neural apoptosis and reduces ischemic brain injury: suppression of peroxynitrite production, lipid peroxidation, and mitochondrial dysfunction. J Neurosci. 1998;18(2):687–697. doi: 10.1523/JNEUROSCI.18-02-00687.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang G, et al. Human copper–zinc superoxide dismutase transgenic mice are highly resistant to reperfusion injury after focal cerebral ischemia. Stroke. 1994;25(1):165–170. doi: 10.1161/01.str.25.1.165. [DOI] [PubMed] [Google Scholar]
- 45.Barber SC, Mead RJ, Shaw PJ. Oxidative stress in ALS: a mechanism of neurodegeneration and a therapeutic target. Biochim Biophys Acta. 2006;1762(11–12):1051–1067. doi: 10.1016/j.bbadis.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 46.Halliwell B, Gutteridge JMC. Free Radicals in Biology and Medicine. 3. Oxford University Press; USA: 1999. [Google Scholar]
- 47.Gerber HP, et al. Vascular endothelial growth factor regulates endothelial cell survival through the phosphatidylinositol 3′-kinase/Akt signal transduction pathway. Requirement for Flk-1/KDR activation. J Biol Chem. 1998;273(46):30336–30343. doi: 10.1074/jbc.273.46.30336. [DOI] [PubMed] [Google Scholar]
- 48.Yao R, Cooper GM. Requirement for phosphatidylinositol-3 kinase in the prevention of apoptosis by nerve growth factor. Science. 1995;267(5206):2003–2006. doi: 10.1126/science.7701324. [DOI] [PubMed] [Google Scholar]
- 49.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129(7):1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee SR, et al. Reversible inactivation of the tumor suppressor PTEN by H2O2. J Biol Chem. 2002;277(23):20336–20342. doi: 10.1074/jbc.M111899200. [DOI] [PubMed] [Google Scholar]
- 51.Numajiri N, et al. On-off system for PI3-kinase-Akt signaling through S-nitrosylation of phosphatase with sequence homology to tensin (PTEN) Proc Natl Acad Sci USA. 2011;108(25):10349–10354. doi: 10.1073/pnas.1103503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Degtyarev M, et al. Akt inhibition promotes autophagy and sensitizes PTEN-null tumors to lysosomotropic agents. J Cell Biol. 2008;183(1):101–116. doi: 10.1083/jcb.200801099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salih DA, Brunet A. FoxO transcription factors in the maintenance of cellular homeostasis during aging. Curr Opin Cell Biol. 2008;20(2):126–136. doi: 10.1016/j.ceb.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Greer EL, Brunet A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene. 2005;24(50):7410–7425. doi: 10.1038/sj.onc.1209086. [DOI] [PubMed] [Google Scholar]
- 55.Tzivion G, Dobson M, Ramakrishnan G. FoxO transcription factors; Regulation by AKT and 14-3-3 proteins. Biochim Biophys Acta. 2011;1813(11):1938–1945. doi: 10.1016/j.bbamcr.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 56.Azad MB, Chen Y, Gibson SB. Regulation of autophagy by reactive oxygen species (ROS): implications for cancer progression and treatment. Antioxid Redox Signal. 2009;11(4):777–790. doi: 10.1089/ars.2008.2270. [DOI] [PubMed] [Google Scholar]
- 57.Mammucari C, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6(6):458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 58.Band M, et al. Hypoxia-induced BNIP3 expression and mitophagy: in vivo comparison of the rat and the hypoxia-tolerant mole rat Spalax ehrenbergi. FASEB J. 2009;23(7):2327–2335. doi: 10.1096/fj.08-122978. [DOI] [PubMed] [Google Scholar]
- 59.Zhao Y, et al. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat Cell Biol. 2010;12(7):665–675. doi: 10.1038/ncb2069. [DOI] [PubMed] [Google Scholar]
- 60.Zhou J, et al. FOXO3 induces FOXO1-dependent autophagy by activating the AKT1 signaling pathway. Autophagy. 2012;8(12) doi: 10.4161/auto.21830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333(1–2):169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- 62.Thumm M, et al. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994;349(2):275–280. doi: 10.1016/0014-5793(94)00672-5. [DOI] [PubMed] [Google Scholar]
- 63.Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12(Suppl 2):1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eisler H, Frohlich KU, Heidenreich E. Starvation for an essential amino acid induces apoptosis and oxidative stress in yeast. Exp Cell Res. 2004;300(2):345–353. doi: 10.1016/j.yexcr.2004.07.025. [DOI] [PubMed] [Google Scholar]
- 65.Scherz-Shouval R, et al. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26(7):1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kanki T, Klionsky DJ. Atg32 is a tag for mitochondria degradation in yeast. Autophagy. 2009;5(8):1201–1202. doi: 10.4161/auto.5.8.9747. [DOI] [PubMed] [Google Scholar]
- 67.Kanki T, et al. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17(1):98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Novak I, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11(1):45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12(1):9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gertz M, et al. Activation of the lifespan regulator p66Shc through reversible disulfide bond formation. Proc Natl Acad Sci USA. 2008;105(15):5705–5709. doi: 10.1073/pnas.0800691105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pandolfi S, et al. P66(shc) is highly expressed in fibroblasts from centenarians. Mech Ageing Dev. 2005;126(8):839–844. doi: 10.1016/j.mad.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 72.Napoli C, et al. Deletion of the p66Shc longevity gene reduces systemic and tissue oxidative stress, vascular cell apoptosis, and early atherogenesis in mice fed a high-fat diet. Proc Natl Acad Sci USA. 2003;100(4):2112–2116. doi: 10.1073/pnas.0336359100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pinton P, et al. Protein kinase C beta and prolyl isomerase 1 regulate mitochondrial effects of the life-span determinant p66Shc. Science. 2007;315(5812):659–663. doi: 10.1126/science.1135380. [DOI] [PubMed] [Google Scholar]
- 74.Giorgio M, et al. Electron transfer between cytochrome c and p66Shc generates reactive oxygen species that trigger mitochondrial apoptosis. Cell. 2005;122(2):221–233. doi: 10.1016/j.cell.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 75.Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J. 2007;404(1):1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Afshar G, Murnane JP. Characterization of a human gene with sequence homology to Saccharomyces cerevisiae SIR2. Gene. 1999;234(1):161–168. doi: 10.1016/s0378-1119(99)00162-6. [DOI] [PubMed] [Google Scholar]
- 77.Cheng HL, et al. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci USA. 2003;100(19):10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee IH, et al. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci USA. 2008;105(9):3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kuma A, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432(7020):1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 80.Tothova Z, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128(2):325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 81.Semenza GL. Oxygen sensing, homeostasis, and disease. N Engl J Med. 2011;365(6):537–547. doi: 10.1056/NEJMra1011165. [DOI] [PubMed] [Google Scholar]
- 82.Miyata T, Takizawa S, van Ypersele de Strihou C. Hypoxia. 1 Intracellular sensors for oxygen and oxidative stress: novel therapeutic targets. Am J Physiol Cell Physiol. 2011;300(2):226–231. doi: 10.1152/ajpcell.00430.2010. [DOI] [PubMed] [Google Scholar]
- 83.Kaelin WG, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30(4):393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 84.Manalo DJ, et al. Transcriptional regulation of vascular endothelial cell responses to hypoxia by HIF-1. Blood. 2005;105(2):659–669. doi: 10.1182/blood-2004-07-2958. [DOI] [PubMed] [Google Scholar]
- 85.Brunelle JK, et al. Oxygen sensing requires mitochondrial ROS but not oxidative phosphorylation. Cell Metab. 2005;1(6):409–414. doi: 10.1016/j.cmet.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 86.Kaelin WG., Jr ROS: really involved in oxygen sensing. Cell Metab. 2005;1(6):357–358. doi: 10.1016/j.cmet.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 87.Guzy RD, et al. Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metab. 2005;1(6):401–408. doi: 10.1016/j.cmet.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 88.Murphy BJ, et al. Metallothionein induction by hypoxia involves cooperative interactions between metal-responsive transcription factor-1 and hypoxia-inducible transcription factor-1alpha. Mol Cancer Res. 2008;6(3):483–490. doi: 10.1158/1541-7786.MCR-07-0341. [DOI] [PubMed] [Google Scholar]
- 89.Zhang H, et al. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem. 2008;283(16):10892–10903. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 90.Bellot G, et al. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol. 2009;29(10):2570–2581. doi: 10.1128/MCB.00166-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kirino T, Sano K. Selective vulnerability in the gerbil hippocampus following transient ischemia. Acta Neuropathol. 1984;62(3):201–208. doi: 10.1007/BF00691853. [DOI] [PubMed] [Google Scholar]
- 92.Iranon NN, Miller DL. Interactions between oxygen homeostasis, food availability, and hydrogen sulfide signaling. Front Genet. 2012;3:257. doi: 10.3389/fgene.2012.00257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Montgomery H. Oxygen tension of tissues in vivo. Circulation. 1957;15(5):646–660. doi: 10.1161/01.cir.15.5.646. [DOI] [PubMed] [Google Scholar]
- 94.Halliwell B. Oxidative stress in cell culture: an under-appreciated problem? FEBS Lett. 2003;540(1–3):3–6. doi: 10.1016/s0014-5793(03)00235-7. [DOI] [PubMed] [Google Scholar]
- 95.Stankowski JN, et al. C-terminus of heat shock cognate 70 interacting protein increases following stroke and impairs survival against acute oxidative stress. Antioxid Redox Signal. 2011;14(10):1787–1801. doi: 10.1089/ars.2010.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8(4):445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Halliwell B, Whiteman M. Measuring reactive species and oxidative damage in vivo and in cell culture: how should you do it and what do the results mean? Br J Pharmacol. 2004;142(2):231–255. doi: 10.1038/sj.bjp.0705776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Codreanu SG, et al. Biotinylated probes for the analysis of protein modification by electrophiles. Methods Mol Biol. 2012;803:77–95. doi: 10.1007/978-1-61779-364-6_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vila A, et al. Identification of protein targets of 4-hydroxynonenal using click chemistry for ex vivo biotinylation of azido and alkynyl derivatives. Chem Res Toxicol. 2008;21(2):432–444. doi: 10.1021/tx700347w. [DOI] [PMC free article] [PubMed] [Google Scholar]