Abstract
Objective
To conduct a comprehensive mapping of the genomic DNA methylation in CDKN2A, which codes for the p16INK4A and p14ARF proteins, and 14 of the most promising DNA methylation marker candidates previously reported to be associated with progression of low-grade cervical intraepithelial neoplasia (CIN1) to cervical cancer.
Methods
We analyzed DNA methylation in 68 HIV-seropositive and negative women with incident CIN1, CIN2, CIN3 and invasive cervical cancer, assaying 120 CpG dinucleotide sites spanning APC, CDH1, CDH13, CDKN2A, CDKN2B, DAPK1, FHIT, GSTP1, HIC1, MGMT, MLH1, RARB, RASSF1, TERT and TIMP3 using the Illumina Infinium array. Validation was performed using high resolution mapping of the target genes with HELP-tagging for 286 CpGs, followed by fine mapping of candidate genes with targeted bisulfite sequencing. We assessed for statistical differences in DNA methylation levels for each CpG loci assayed using univariate and multivariate methods correcting for multiple comparisons.
Results
In our discovery sample set, we identified dose dependent differences in DNA methylation with grade of disease in CDKN2A, APC, MGMT, MLH1 and HIC1, whereas single CpG locus differences between CIN2/3 and cancer groups were seen for CDH13, DAPK1 and TERT. Only those CpGs in the gene body of CDKN2A showed a monotonic increase in methylation between persistent CIN1, CIN2, CIN3 and cancers.
Conclusion
Our data suggests a novel link between early cervical disease progression and DNA methylation in a region downstream of the CDKN2A transcription start site that may lead to increased p16INK4A/p14ARF expression prior to development of malignant disease.
INTRODUCTION
The vast majority of low grade cervical intraepithelial neoplasia (CIN1) will regress spontaneously without treatment [1, 2]. CIN1 that contains oncogenic human papillomavirus (HPV), particularly HPV 16 or 18, the two oncogenic HPV types that are associated with over 70% of cervical cancers, are more likely to persist and progress [3–5]. CIN1 is commonly over-treated and over-managed because of the inability to distinguish the few CIN1 likely to progress. Additionally, lesion persistence and progression to high-grade cervical intraepithelial neoplasia (CIN2/3) are more common in HIV-seropositive women, which were previously thought to be driven by global immune dysfunction. Understanding the acquired molecular events that determine which few early CIN lesions will progress to cancer is a major goal with clear clinical implications.
Transcription regulation of host genes through the methylation of CpG dinucleotide sites within DNA is increasingly accepted as playing a critical role in tumorigenesis [6–11]. However, because of the cross-sectional design of prior studies, it remains unclear whether the identified epigenetic changes preceded or followed the disease. Also, prior studies focused primarily on gene promoter regions where DNA methylation would be expected to silence genes, whereas the broad profile of aberrant DNA methylation events within genes associated with cervical tumorigenesis remains unknown [6, 12]. This is a major limitation, since differential methylation of intragenic sites (i.e., within exonic and intronic regions) have been associated with differential, and often increased, expression of tumor suppressor genes predictive of clinical progression in cervical cancer [13, 14]. In particular, a number of studies have found conflicting associations between aberrant methylation and expression of known tumor suppressor genes, including CDKN2A, which codes for the p16INK4A and p14ARF proteins, with cervical cancer [15, 16] and high-grade CIN [17–19]. To address these limitations, we prospectively recruited and followed women with CIN1, CIN2, CIN3 and cancer, and collected cervical tissue for in-depth DNA methylation analysis and sequencing of CpG loci spanning across 15 candidate tumor suppressor genes, including APC, CDH1, CDH13, CDKN2A, CDKN2B, DAPK1, FHIT, GSTP1, HIC1, MGMT, MLH1, RARB, RASSF1, TERT and TIMP3, that have been previously reported to be associated with progression of CIN1 to cervical cancer [6].
METHODS
Study sample
Subjects included 68 women with histologically-confirmed CIN1 (N=27), CIN2 (N=8), CIN3 (N=10), and invasive cervical carcinoma (N=23) treated at the affiliated teaching hospitals for Albert Einstein College of Medicine – Montefiore Medical Center and North Bronx Hospital Center. HIV-seropositive and negative patients who had incident, histologically-confirmed CIN1 were enrolled and followed prospectively every 3 to 6 months for a maximum of 2 years (4 visits) with repeat biopsies done at each visit. Liquid-based cytology samples were also collected for HPV genotyping by polymerase chain reaction (PCR) protocol described below [20]. CD4+T-cell counts and HIV viral load levels in the HIV-seropositive patients assessed within six-months of their CIN/cervical cancer diagnosis were abstracted from the electronic medical records. Written informed consent was obtained from all subjects prior to participating prior in the study under IRB approved protocols. Patient characteristics are described in Table 1.
Table 1.
Distribution of study subject characteristics by disease grade
| CIN1 | CIN2 | CIN3 | Cancer | |||||
|---|---|---|---|---|---|---|---|---|
| (N=27) | (N=8) | (N=10) | (N=23) | |||||
| Age (range)* | 37.5 | (24–56) | 35 | (21–52) | 37 | (29–50) | 54.3 | (24–75) |
| Race | ||||||||
| African-American | 11 | 41% | 6 | 75% | 4 | 40.0% | 12 | 52% |
| Caucasian | 10 | 37% | 2 | 25% | 6 | 60.0% | 9 | 39% |
| Asian/Other | 0 | 0% | 0 | 0% | 0 | 0.0% | 1 | 4% |
| Unknown | 6 | 22% | 0 | 0% | 0 | 0.0% | 1 | 4% |
| Ethnicity | ||||||||
| Hispanic | 15 | 56% | 5 | 62.5% | 6 | 60.0% | 5 | 22% |
| Non-Hispanic | 12 | 44% | 3 | 37.5% | 4 | 40.0% | 18 | 78% |
| HIV serostatus | ||||||||
| HIV negative | 20 | 74% | 3 | 37.5% | 9 | 90.0% | 22 | 96% |
| HIV positive | 7 | 26% | 5 | 62.5% | 1 | 10.0% | 1 | 4% |
| HPV DNA status | ||||||||
| HPV negative | 5 | 19% | 1 | 12.5% | 2 | 20.0% | 0 | 0% |
| HPV DNA positive | 22 | 81% | 7 | 87.5% | 8 | 80.0% | 23 | 100% |
| High-risk HPV positive† | 15 | 56% | 5 | 62.5% | 8 | 80.0% | 23 | 100% |
Mean age (range).
High-risk HPV included types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58 & 59.
Each subject had a biopsy of the colposcopically identified lesion for standard pathologic evaluation by two pathologists [KW, MA] and scored following lower anogenital squamous terminology (LAST) criteria [21], plus an adjacent biopsy for methylation analysis of their genomic DNA, and a biopsy of a colposcopically ‘normal’ area that was confirmed normal by H&E stain of slide during tissue processing. All biopsies were snap-frozen in liquid nitrogen within two minutes of tissue procurement. Women that had CIN1 at initial diagnosis but had no CIN at follow-up based on a pathologically normal biopsy samples collected 6 to 12 months later were defined as ‘regressed CIN1’, and those that had CIN1 on consecutive visits were defined as ‘persistent CIN1’. Our biopsy approach allowed us to assess the association between DNA methylation in a newly detected CIN1 lesion with disease regression prospectively, as well as changes in methylation in lesions over time. Women with CIN2/3 and cervical cancer were recruited with similar fresh tissue collection as for CIN1 cases, but without prospective follow-up, as these subjects required immediate treatment.
DNA methylation analyses
As is typical for genomic studies, we employed multiple assays and independent platforms to confirm our findings. Out of the subjects that had numerous time-points of tissue collection, a subset of cervical biopsies were processed and analyzed, chosen based on the amount of available tissue from the biopsy and total nucleic acid content, with two partially overlapping sample sets selected serially, first for initial analysis using the Illumina Infinium platform (discovery cohort, N=29), followed by HELP-tagging and Targeted Bisulfite Sequencing (validation cohort, N=54). Where possible, subject samples were tested in batches balanced by disease grade, HIV status and age at diagnosis (±10 years).
Illumina Infinium assay
Changes in DNA methylation associated with CIN progression to cervical cancer were assessed using the Illumina Infinium assay with the HumanMethylation27 DNA Analysis BeadChip (Illumina Inc., CA). DNA methylation levels at individual CpG loci spanning 15 candidate genes represented on the Illumina beadchip (2–25 CpG loci per gene site) were determined by measuring the fraction of methylated signal over the total signal (unmethylated + methylated fractions) in each genomic DNA sample. Bisulfite conversion of cervical tissue genomic DNA (500ng) was carried out using the EZ DNA Gold methylation kit (Zymo Research Inc., CA). Normalized M values were generated using the R package HumMeth27KQCReport function, including the X chromosome data and using an average probe p-value of 0.03 as the cutoff for sample inclusion [22]. Individual beadchip controls (DNA sample-dependent and sample-independent) confirmed efficient bisulfite conversion of DNA, hybridization specificity, base extension and target removal for all genomic DNA samples. A complete description of these controls is available from the manufacturer. Chromosome (chr) locations, RefSeq and Genebank accession numbers were based on National Center for Biotechnology Information (NCBI) build 36 (http://www.ncbi.nlm.nih.gov/mapview/stats/BuildStats.cgi?taxid=9606&build=36&ver=1).
HELP-tagging assay
To further map the CpG rich regions within candidate genes that showed differential methylation with grade of cervical disease on the Illumina array, we employed a massively-parallel sequencing protocol (HELP-tagging) developed at Einstein [23]. HELP-tagging is a more comprehensive and quantitative assay than array-based methods like the Illumina beadchip platform [24]. DNA from the cervical samples was digested with HpaII, ligated to a customized Illumina adapter that contains an EcoP15I site allowing isolation of the adjacent 27 base pair (bp) sequence, and treated with EcoP15I to generate a “tag” adjacent to the HpaII site (CCGG). We then ligated the complementary Illumina adapter to the other end of the tag, and generated a 120bp library by PCR. Because HpaII cannot cleave methylated DNA, the number of reads generated for a locus was inversely proportional to the methylation at that site, a quantitative outcome that was enhanced by using a pooled MspI representation for normalization. Results were analyzed using the Wasp cyberecosystem [25] and linked to a local mirror of the University of California, Santa Cruz (UCSC) Genome Browser for visualization.
Targeted Bisulfite Sequencing (TBS)
Validation of candidate regions identified using HumanMethylation27 and HELP-tagging was performed using an orthogonal bisulphite sequencing approach for nucleotide-resolution CpG mapping of specific regions. We designed bisulfite-conversion-based PCR primers within target regions using the University of California San Francisco (UCSF) MethPrimer tool [26]. We reduced the possibility of off-target amplicons using BiSearch to interrogate to human genome [27]. Primer sets were further optimized for PCR using random primer pools controlling for secondary structures using Life Technologies’ Multiple Primer Analyzer tool. We bisulfite converted 500ng of purified genomic DNA using a high throughput 96 well plate. After bisulfite conversion, all samples were pre-amplified using an equimolar primer mix. We then added custom dual indexed adapters and amplified the libraries using a Fluidigm Access Array (Fluidigm Inc., CA). We sequenced the resulting amplicon library using the Illuminia MiSeq platform and 150bp paired end sequencing. After spatially filtering out read clusters by removing reads that were too close to produce high confidence index reads, we aligned reads to the human genome and calculated methylation ratios using BSMAP 2.7.3. Only high confidence CpGs containing coverage of at least 50 effective reads were considered, and methylation ratios were averaged in 50bp intervals across the assay regions.
HPV DNA genotyping
DNA extracted from liquid-based cytology smears was digested with proteinase-K/Laureth-12, precipitated and purified in ethanol, and amplified by PCR with Gold-Taq using a well-described the MY09/MY11 protocol [28, 29], followed by Southern blot hybridization with generic probes for HPV and an oligonucleotide for human β-globin DNA (as a control). PCR products positive by Southern blot were analyzed using biotinylated type-specific oligonucleotide probes for >40 different HPV types, including high-risk types (16,18,31,33,35,39,45,51,52,56,58 & 59)[30]. Samples that tested positive by the generic probe mix but negative by all type-specific probes were considered to represent low-risk HPV types.
Statistical analyses
We ranked the most significant predictors of disease progression comparing average CpG methylation levels in lesion samples assessing cross-sectionally by disease grade from CIN1 to cancer, and prospectively, comparing regressing vs. persistent CIN1 lesions. We tested for statistical differences in DNA methylation levels for each CpG loci assayed using univariate parametric and non-parametric tests, where appropriate. Significant CpG sites (corrected p<0.05) were further restricted on a magnitude of difference in DNA methylation (e.g., of at least ±0.2 difference in beta for the Illumina protocol) considered sufficient to result in a ‘biologically’ significant difference on expression [31]. This approach was chosen to reduce the potential for redundant predictors and over-selection that may arise. Disease group methylation values were illustrated using box plots showing the median, interquartile range (25th and 75th percentile), and upper and lower adjacent Tukey values [32].
Using the HELP-tagging assay, we assessed for differences in DNA methylation at all HpaII sites proximal to the Illumina probes within candidate genes using parametric and non-parametric tests. We estimated the relative odds of methylation at each HpaII site for CIN2/3 and cervical cancer compared to persistent CIN1 using polytomous multivariate logistic regression models implemented in the nnet package in R. We used a change in point estimate selection method to assess for confounding (e.g., by detection of high-risk HPV DNA or HIV serostatus) comparing covariate adjusted OR controlling a priori for sample batch and age [33]. In addition, we performed non-parametric tests for trend using the Jonckheere-Terpstra test implemented using the clinfun R package on a covariate-adjusted data generated using the ComBat R package. A similar approach was used to test for significant differences in average DNA methylation measured across the multiple 50bp regions assessed by targeted bisulfite sequencing. We employed a strict method to control for multiple testing by setting a Bonferroni threshold of significance using an alpha=0.05 divided by either the number of assayed HpaII sites in a specific region for Help-tagging or the number of bins in each amplicon for TBS. All statistical tests were two-tailed.
RESULTS
We analyzed DNA methylation using the Illumina Infinium (HumanMethylation27) platform in a discovery sample set of HIV-negative women with incident CIN1 followed prospectively, including 6 with CIN1 that regressed and 11 with persistent CIN1, plus a convenience sample of 3 CIN2, 2 CIN3 and 7 cervical cancer patients. A total of 120 CpG loci spanning 15 candidate genes, including tumor suppressor genes APC, CDH1, CDH13, CDKN2A, CDKN2B, DAPK1, FHIT, GSTP1, HIC1, MGMT, MLH1, RARB, RASSF1, TERT and TIMP3. Nine CpG loci (8%) did not pass initial quality control (average control probe p-values <0.03 for all samples) and were excluded. We identified multiple CpG loci that showed significant dose-dependent increases in methylation with grade of disease, including several within the CDKN2A gene (Table S1). With respect to the other genes that had significant differences in methylation with grade of disease, CDKN2A, APC, MGMT, RASSF1 and HIC1 showed dose-dependent increases, whereas MLH1, CDH13, RARB, CDKN2B, TERT and DAPK1 showed larger differences either between persistent CIN1 and CIN2/3, or between CIN2/3 and cancer (Figure S1).
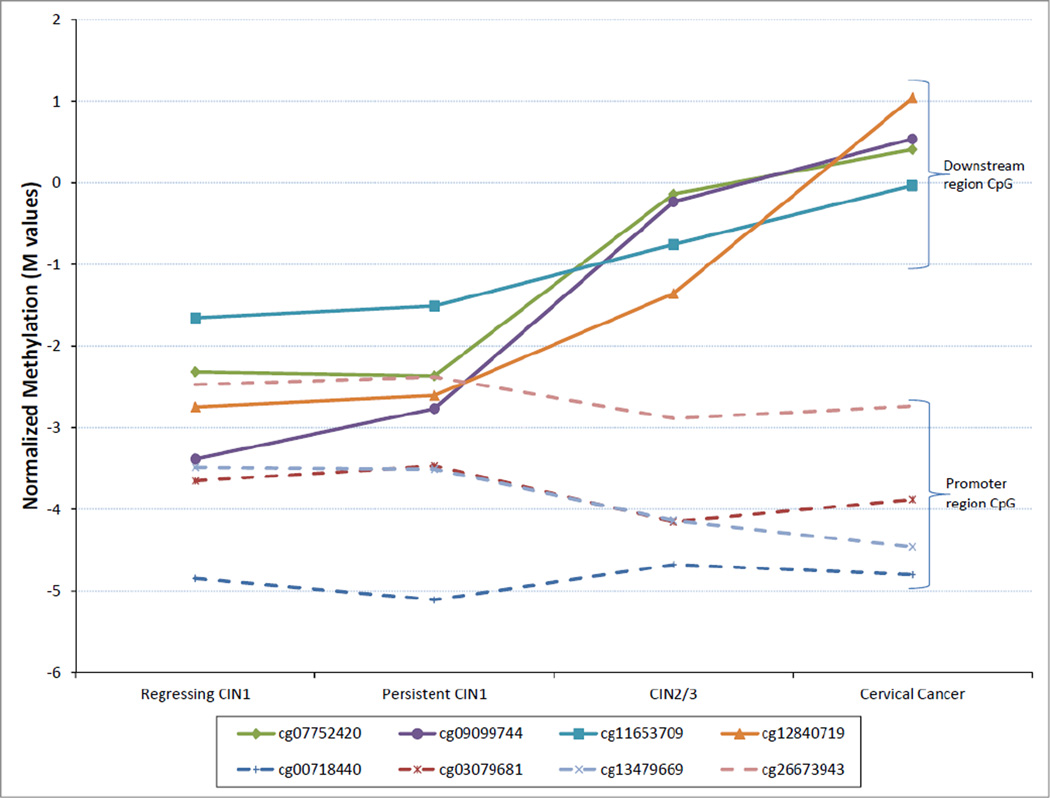
Interestingly, further assessment of the CDKN2A gene revealed monotonic increases in methylation between persistent CIN1, CIN2, CIN3 and cancers for CpG located downstream of the transcription start site (chr9:21958106–21958899 NCBI build 36), whereas no differences, or even decreases, in methylation were seen for loci within or near the p14ARF promoter region (chr9:21983444–21986348; Figure 1). Subgroup analyses excluding CIN2, and CIN1 cases that regressed, remained significant for most of the CpG loci identified in the larger sample, including within the downstream regions of CDKN2A, APC, MGMT, HIC1 and MLH1 (Table S1).
Figure 1.
Median DNA methylation (M) in CIN1 (by disease progression), CIN2/3 and cervical cancer samples for CpG loci within CDKN2A. The Illumina CG codes for each CpG tested on the Illumina Infinium (Human Methylation 27) array are listed in the figure legend with the corresponding line markers. CpGs located downstream or near the gene promoter region are indicated in the right margin.
To assess whether changes in DNA methylation predict persistence of low-grade cervical lesions, a clinically intervenable endpoint in-of-itself, CIN1 cases (n=15) with two or more follow-up visits and biopsy samples with sufficient DNA for bisulfite conversion were also assayed using the Illumina Infinium platform. We assessed for the changes in methylation comparing paired biopsies collected at diagnosis and 6–12 months later from women with persistent vs. regressing CIN1 lesions. No significant increases in CDKN2A methylation were detected for any of the measured CpG loci. Genes that showed changes (increases or decreases) in methylation associated with CIN1 persistence vs. regression included CDH1, GSTP1, MGMT, MLH1, RASSF1 and TIMP3, but these did not remain significant after Bonferroni correction.
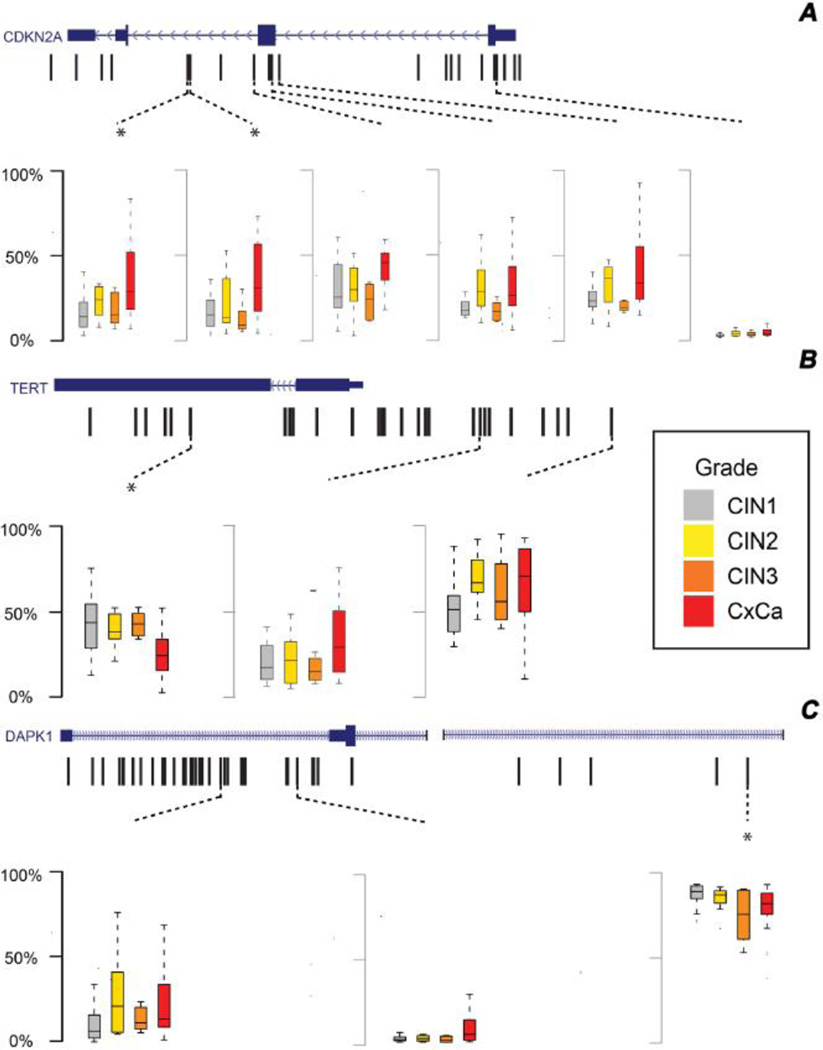
To further explore the apparent associations between DNA methylation and grade of disease, and to broaden coverage to include gene promoter regions not assayed by the Illumina Infinium platform, we assessed the methylation levels across 286 HpaII sites proximal to the HumanMethylation27 probes using HELP-tagging on a partially overlapping sample of 59 biopsies from HIV-seropositive and negative persistent CIN1 (n=20), CIN2 (n=8), CIN3 (n=8) and (n=23) cervical cancer cases. Figure 2a illustrates the distribution of percent DNA methylation (shown as box plots) across the four lesion grade groups at CpG loci within CDKN2A. A significant trend in increasing methylation with grade of disease was detected at 5 HpaII sites in the CpG island and shore approximately 1kb to 2.5kb upstream of the HumanMethlation27 probes. The trends were similar to those seen for neighboring CpG sites probed by the Illumina array, and after Bonferroni correction, the two closest loci within the CpG island shore remained significant. Although there was one CpG significantly differentially methylated in the CDKN2A p16INK4a promoter (not assayed by the HumanMethylation27 beadchip), this result did not remain significant after Bonferroni correction. The DAPK1 gene showed significant increases in DNA methylation at HpaII sites within the gene promoter, and decreasing methylation within the gene body, although only the gene body differences remained significant after Bonferroni correction (Figure 2b). In contrast, the TERT gene only showed a significant decrease in methylation with disease grade within its promoter after Bonferroni correction (Figure 2c). Other genes that showed significant trends (increases or decreases) in methylation with grade of disease after correction for multiple comparisons were HIC1 and RARB, as well as for the CDKN2B gene region proximal to the CDKN2A promoter (Figure S2). A few CpG loci differences in methylation between persistent CIN1, CIN2, CIN3 and cancer lesions were seen in promoter regions of other genes like APC, CDH13, CDH1, FHIT, MGMT, GSTP1 and MLH1, and in the gene body of MGMT, although these were not significant after Bonferroni correction.
Figure 2.
Gene maps showing CpG loci with significant methylation differences between CIN1, CIN2, CIN3 and cervical cancer samples for (A) CDKN2A, (B) TERT, and (C) DAPK1. Refseq maps for each gene are shown on top, with the CpGs assayed by HELP-tagging indicated by black hash marks. Box plots summarizing median, interquartile range and adjacent percent (%) methylation values are shown below each map for CpGs with significant statistical differences in methylation across lesion groups (uncorrected p<0.05), with comparisons that remained significant after Bonferroni correction marked by an asterisk.
We used multivariable polytomous logistic regression to assess the strength of association between increasing DNA methylation percentage and disease grade for the CDKN2A, TERT, RARB, HIC1, DAPK1 and CDKN2B gene regions identified above, while accounting for potential confounding by sequencing batch and patient age. To optimize power, adjusted odds ratios (ORs) and 95% confidence intervals (CI) were generated comparing CIN1 to CIN2/3 and cervical cancer, respectively. Significant differences in DNA methylation between CIN1 and cancers persisted after adjustment for two of the CpG sites implicated in the gene body regions of CDKN2A (OR=1.09, 95%CI:1.02–1.18 and OR=1.08 (95%CI:1.01–1.15), while the ORs for the CIN2/3 and CIN1 comparison were somewhat attenuated and not significant (OR=1.05, 95%CI:0.98–1.12 and OR=1.01, 95%CI:0.95–1.07). Other gene regions that showed consistent results after covariate adjustment included the promoter regions of TERT, RARB and HIC1, and the gene body regions of DAPK1 and CDKN2B.
Further adjustment for detection of high-risk and low-risk HPV types in the cervical samples and for HIV seropositivity yielded similar results, but generated unstable estimates due to over-stratification. To address this, we restricted the analyses to samples positive for high-risk HPV DNA (n=39), and saw consistent associations between DNA methylation within the downstream region of CDKN2A and disease grade for the cancer vs. CIN1 (OR=1.09, 95%CI:0.98–1.22 and OR=1.13, 95%CI:1.00–1.27) and CIN2/3 vs. CIN1 comparisons (OR=1.07, 95%CI:0.93–1.24 and OR=0.97, 95%CI:0.83–1.14). Similarly, for the HIV-negative group (n=44), the ORs for DNA methylation at the two CDKN2A CpG sites were similar for the cancer vs. CIN1 (OR=1.08, 95%CI:1.00–1.17 and OR=1.15, 95%CI:1.00–1.34) and CIN2/3 vs. CIN1 comparisons (OR=1.01, 95%CI:0.94–1.09 and OR=0.91, 95%CI:0.79–1.04), and even increased somewhat for HIV-seropositive subjects (n=15), for whom we could compare the CIN1 and the pooled CIN2/3 and cancer groups (OR=1.21, 95%CI:0.98–1.51 and OR=1.08, 95%CI:0.99–1.19), although the differences were not significant.
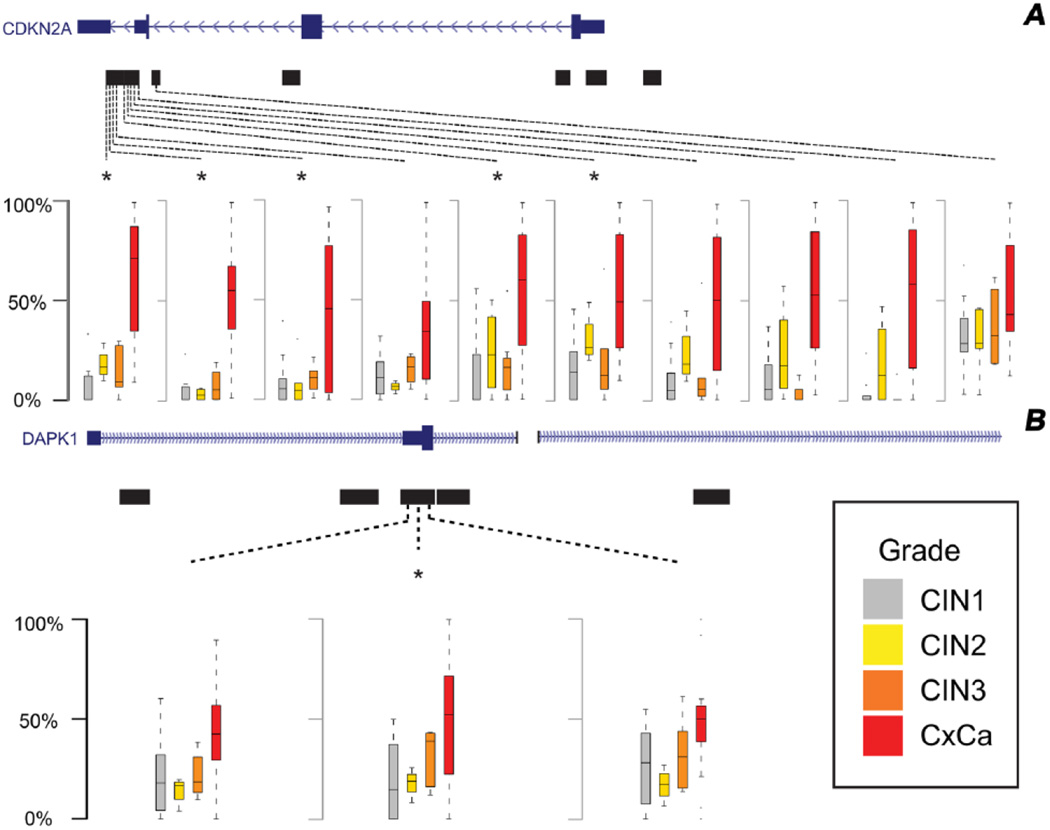
As the Illumina Infinium HumanMethylation27 array and the HELP-tagging assay implicated similar regions within the target genes tested, but did not necessarily assay overlapping CpGs, we further verified these findings through TBS. We conducted fine epigenetic mapping of the all CpG loci within specific regions of the candidate genes, including CDKN2A, CDKN2B, DAPK1, MGMT, RASSF1 and TERT in a subset of 53 biopsies (persistent CIN1 (n=20), CIN2, (n=7), CIN3 (n=7) and cervical cancer (n=19) cases). TBS confirmed there were significant increases in DNA methylation with grade of disease for a large number of CpG loci within the downstream and distal gene body regions of CDKN2A that overlapped with the HumanMethylation27 probes, whereas no significant differences were detected after Bonferroni correction for CpG loci located within the p16INK4a and p14ARF promoter regions, or in the CpG island shore identified upstream by the HELP-tagging assay (Figure 3a). The one other gene region that showed consistent increases in DNA methylation by TBS included the promoter of DAPK1 (Figure 3b), although only the differences observed for CDKN2A remained significant after adjustment for patient age when modeled using multivariable polytomous logistic regression. Further adjustment for HPV DNA type or restriction on HIV serostatus did not substantively change the observed associations between DNA methylation and disease grade.
Figure 3.
Gene maps showing CpG regions with significant differences in percent (%) methylation between CIN1, CIN2, CIN3 and cervical cancer samples for (A) CDKN2A and (B) DAPK1 genes as measured by bisulfite sequencing. The mapped regions are shown by black bars. Box plots are shown for CpGs with significant statistical differences in methylation across lesion groups (uncorrected p<0.05), with comparisons that remained significant after Bonferroni correction marked by an asterisk.
DISCUSSION
In this prospective study of epigenetic profiles in HIV-seropositive and negative women with different grades of CIN, including regressed and persistent CIN1, there were clear differences in the quantity and profile of methylated genes across grades of CIN and cervical cancer. Other cross-sectional studies in CIN have revealed patterns of aberrant DNA methylation in specific genes, including for tumor suppressor genes CDKN2A [34–37], MGMT [19, 38], HIC1, APC, CADM1, MAL, and RARβ [6, 9, 12]. However, as these prior studies were cross-sectional, the predictive nature and timing of the gene methylation has not yet been elucidated. One of the strengths of our study is the prospective collection at different time points and using subjects as their own controls, allowing us to more precisely investigate the predictive value of methylation of these specific genes. By studying these previously identified targets and many other genes along similar pathways as well as novel pathways, we had the ability to confirm previous results for validity, as well as identify other patterns of methylation yet unstudied.
Among the candidate genes identified, MGMT (represented by two CpG sites on the array) has been suggested as an intermediate to late event in cervical cancer [19, 38]. However, no differences were observed in MGMT for our patient samples of CIN after accounting for multiple testing. Moreover, when we conducted TBS of a separate but defined region of MGMT, we observed a markedly lower level of DNA methylation. In contrast, we now show that methylation of CDKN2A, which expresses the p16INK4A and p14ARF proteins, clearly begins to occur between low-grade and high-grade CIN. Immunohistochemistry staining for the p16INK4A protein has been shown to be important in defining clinically-relevant, but histologically equivocal CIN [39, 40]. In advanced cervical cancers, the majority of cells had methylated CDKN2A, lack the p16INK4A protein, but no longer express the HPV E7 oncoprotein. Thus p16INK4A inactivation may be a mechanism of blocking the cyclin D-Rb pathway in invasive cervical cancer [41], whereas p14ARF expression may be increased in HPV-positive precursor lesions [42–45].
Previous studies have also identified the intriguing result that CDKN2A, which is normally repressed in cycling cells by EZH2 via H3K27me3, and frequently undergoes DNA hypermethylation in cancer, is often overexpressed in HPV-positive carcinoma [14]. Our data suggests a novel potential link between early cervical disease progression and DNA methylation of CpGs located within the 700bp downstream region of the transcriptional start site of CDKN2A that may lead to increased p16INK4A/p14ARF expression prior to development of malignant disease. Consistent with this epigenetically driven association, we have previously shown a positive correlation between downstream CDKN2A methylation and p16INK4A/p14ARF expression in HPV-associated head and neck cancer [46].
The mechanism by which hypermethylation occurs at the downstream CpG island region from the CDKN2A locus is not clear. Differential methylation events, including some associated with CDKN2A, have also been previously observed in cancer cell lines [47]. Cervical cell line data using methylation-specific PCR suggests DNA methylation of CDKN2A occurs only occasionally within early cervical cancer cells and is not always correlated with HPV E7 expression [41]. However, our study is unique in that it used biopsied human cervical tissue, thereby overcoming the limitation of high levels of DNA methylation observed in many cell lines when compared to corresponding patient samples [48].
Current treatment strategies for the management of CIN1 are not uniform. There is always a concern amongst women’s health providers when a patient has persistent CIN1, which will regress spontaneously in the majority of cases but will progress in a significant minority. Other sensitive molecular markers such as HPV DNA testing has limited utility for identifying the lesions that might progress over time,[5, 49, 50] so providers are forced to closely observe these patients until they have a regressing lesion or they progress. This often adds stress to the patient, as well as the provider, and may lead to more aggressive management (e.g. cryotherapy, loop electrosurgical procedure, or cone biopsy) and over-treatment on the part of the provider [51], with associated long-term risks to future pregnancies [52]. New predictive point-of-care testing markers, such as establishing specific DNA methylation panels that can specifically identify clinically-relevant CIN that have a high risk of progressing have clear clinical implications.
Supplementary Material
RESEARCH HIGHLIGHTS.
There is inconsistent evidence for host gene DNA methylation in early cervical neoplasia.
We conducted comprehensive mapping of DNA methylation loci in 15 candidate genes.
Increased methylation of the downstream region of CDKN2A was associated with progression to cervical cancer.
Acknowledgments
The study was supported in part by an American Cancer Society Research Scholar Grant (RSG 08-002-01-CCE, MHE), an Einstein Cancer Center Supplement (NIH CA013330, NFS and MHE), the MSTP program at Albert Einstein College of Medicine (NIH T32 GM007288, NAW), and Einstein’s Center for Epigenomics.
Dr. Schlecht has served in the past as an advisory board member for Merck, Glaxo-Smith Klein and PDS Biotechnology. Dr. Einstein has advised, but does not receive an honorarium from any companies. In specific cases in the past 12 months, his employer during the conduct of this study, Montefiore Medical Center has received payment for time spent for advice related to clinical trials from Roche, Photocure, Papivax, Natera, Inovio, and PDS Biotechnology. If travel is required for meetings with any industry, the company pays for Dr. Einstein’s travel-related expenses. Also, Montefiore has received grant funding for research related costs of clinical trials that Dr. Einstein has been the overall PI or Montefiore PI from Roche, Photocure, Fujiboro, Eli Lilly, PDS Biotechnology, and Becton-Dickinson.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest disclosures: The other authors declare that there are no conflicts of interest.
DATA AVAILABILITY
All sequencing data generated in this study are deposited at the Gene Expression Omnibus and are available in Series GSE76986. Interim link for reviewers: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=izyrcmsafpoljgh&acc=GSE7698
REFERENCES
- 1.Clifford GM, Smith JS, Plummer M, Munoz N, Franceschi S. Human papillomavirus types in invasive cervical cancer worldwide: a meta-analysis. British journal of cancer. 2003;88:63–73. doi: 10.1038/sj.bjc.6600688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bosch FX, de Sanjose S. Chapter 1: Human papillomavirus and cervical cancer--burden and assessment of causality. Journal of the National Cancer Institute Monographs. 2003:3–13. doi: 10.1093/oxfordjournals.jncimonographs.a003479. [DOI] [PubMed] [Google Scholar]
- 3.Khan MJ, Castle PE, Lorincz AT, Wacholder S, Sherman M, Scott DR, et al. The elevated 10-year risk of cervical precancer and cancer in women with human papillomavirus (HPV) type 16 or 18 and the possible utility of type-specific HPV testing in clinical practice. Journal of the National Cancer Institute. 2005;97:1072–1079. doi: 10.1093/jnci/dji187. [DOI] [PubMed] [Google Scholar]
- 4.Castle PE, Solomon D, Schiffman M, Wheeler CM. Human papillomavirus type 16 infections and 2-year absolute risk of cervical precancer in women with equivocal or mild cytologic abnormalities. Journal of the National Cancer Institute. 2005;97:1066–1071. doi: 10.1093/jnci/dji186. [DOI] [PubMed] [Google Scholar]
- 5.Cuzick J, Thomas Cox J, Zhang G, Einstein MH, Stoler M, Trupin S, et al. Human papillomavirus testing for triage of women with low-grade squamous intraepithelial lesions. International journal of cancer Journal international du cancer. 2013;132:959–966. doi: 10.1002/ijc.27723. [DOI] [PubMed] [Google Scholar]
- 6.Wentzensen N, Sherman ME, Schiffman M, Wang SS. Utility of methylation markers in cervical cancer early detection: appraisal of the state-of-the-science. Gynecologic oncology. 2009;112:293–299. doi: 10.1016/j.ygyno.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banzai C, Nishino K, Quan J, Yoshihara K, Sekine M, Yahata T, et al. Promoter methylation of DAPK1, FHIT, MGMT, and CDKN2A genes in cervical carcinoma. International journal of clinical oncology. 2014;19:127–132. doi: 10.1007/s10147-013-0530-0. [DOI] [PubMed] [Google Scholar]
- 8.Lai HC, Lin YW, Huang TH, Yan P, Huang RL, Wang HC, et al. Identification of novel DNA methylation markers in cervical cancer. International journal of cancer Journal international du cancer. 2008;123:161–167. doi: 10.1002/ijc.23519. [DOI] [PubMed] [Google Scholar]
- 9.Wang SS, Smiraglia DJ, Wu YZ, Ghosh S, Rader JS, Cho KR, et al. Identification of novel methylation markers in cervical cancer using restriction landmark genomic scanning. Cancer research. 2008;68:2489–2497. doi: 10.1158/0008-5472.CAN-07-3194. [DOI] [PubMed] [Google Scholar]
- 10.Shivapurkar N, Sherman ME, Stastny V, Echebiri C, Rader JS, Nayar R, et al. Evaluation of candidate methylation markers to detect cervical neoplasia. Gynecologic oncology. 2007;107:549–553. doi: 10.1016/j.ygyno.2007.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Louvanto K, Franco EL, Ramanakumar AV, Vasiljevic N, Scibior-Bentkowska D, Koushik A, et al. Methylation of viral and host genes and severity of cervical lesions associated with human papillomavirus type 16. International journal of cancer Journal international du cancer. 2015;136:E638–E645. doi: 10.1002/ijc.29196. [DOI] [PubMed] [Google Scholar]
- 12.Litjens RJ, Hopman AH, van de Vijver KK, Ramaekers FC, Kruitwagen RF, Kruse AJ. Molecular biomarkers in cervical cancer diagnosis: a critical appraisal. Expert opinion on medical diagnostics. 2013;7:365–377. doi: 10.1517/17530059.2013.808621. [DOI] [PubMed] [Google Scholar]
- 13.Gil J, Peters G. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nature reviews Molecular cell biology. 2006;7:667–677. doi: 10.1038/nrm1987. [DOI] [PubMed] [Google Scholar]
- 14.Paschos K, Allday MJ. Epigenetic reprogramming of host genes in viral and microbial pathogenesis. Trends in microbiology. 2010;18:439–447. doi: 10.1016/j.tim.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jha AK, Nikbakht M, Jain V, Capalash N, Kaur J. p16(INK4a) and p15(INK4b) gene promoter methylation in cervical cancer patients. Oncology letters. 2012;3:1331–1335. doi: 10.3892/ol.2012.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lof-Ohlin ZM, Levanat S, Sabol M, Sorbe B, Nilsson TK. Promoter methylation in the PTCH gene in cervical epithelial cancer and ovarian cancer tissue as studied by eight novel Pyrosequencing(R) assays. International journal of oncology. 2011;38:685–692. doi: 10.3892/ijo.2011.895. [DOI] [PubMed] [Google Scholar]
- 17.Huang LW, Pan HS, Lin YH, Seow KM, Chen HJ, Hwang JL. P16 methylation is an early event in cervical carcinogenesis. International journal of gynecological cancer : official journal of the International Gynecological Cancer Society. 2011;21:452–456. doi: 10.1097/IGC.0b013e31821091ea. [DOI] [PubMed] [Google Scholar]
- 18.Furtado YL, Almeida G, Lattario F, Silva KS, Maldonado P, Silveira FA, et al. The presence of methylation of the p16INK4A gene and human papillomavirus in high-grade cervical squamous intraepithelial lesions. Diagnostic molecular pathology : the American journal of surgical pathology, part B. 2010;19:15–19. doi: 10.1097/PDM.0b013e3181aa8f64. [DOI] [PubMed] [Google Scholar]
- 19.Kim JH, Choi YD, Lee JS, Lee JH, Nam JH, Choi C. Assessment of DNA methylation for the detection of cervical neoplasia in liquid-based cytology specimens. Gynecologic oncology. 2010;116:99–104. doi: 10.1016/j.ygyno.2009.09.032. [DOI] [PubMed] [Google Scholar]
- 20.Burk RD, Kadish AS, Calderin S, Romney SL. Human papillomavirus infection of the cervix detected by cervicovaginal lavage and molecular hybridization: correlation with biopsy results and Papanicolaou smear. American journal of obstetrics and gynecology. 1986;154:982–989. doi: 10.1016/0002-9378(86)90733-7. [DOI] [PubMed] [Google Scholar]
- 21.Darragh TM, Colgan TJ, Thomas Cox J, Heller DS, Henry MR, Luff RD, et al. The Lower Anogenital Squamous Terminology Standardization project for HPV-associated lesions: background and consensus recommendations from the College of American Pathologists and the American Society for Colposcopy and Cervical Pathology. International journal of gynecological pathology : official journal of the International Society of Gynecological Pathologists. 2013;32:76–115. doi: 10.1097/PGP.0b013e31826916c7. [DOI] [PubMed] [Google Scholar]
- 22.Mancuso FM, Montfort M, Carreras A, Alibes A, Roma G. HumMeth27QCReport: an R package for quality control and primary analysis of Illumina Infinium methylation data. BMC research notes. 2011;4:546. doi: 10.1186/1756-0500-4-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki M, Jing Q, Lia D, Pascual M, McLellan A, Greally JM. Optimized design and data analysis of tag-based cytosine methylation assays. Genome biology. 2010;11:R36. doi: 10.1186/gb-2010-11-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ulahannan N, Greally JM. Genome-wide assays that identify and quantify modified cytosines in human disease studies. Epigenetics & chromatin. 2015;8:5. doi: 10.1186/1756-8935-8-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLellan AS, Dubin RA, Jing Q, Broin PO, Moskowitz D, Suzuki M, et al. The Wasp System: an open source environment for managing and analyzing genomic data. Genomics. 2012;100:345–351. doi: 10.1016/j.ygeno.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 26.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 27.Tusnady GE, Simon I, Varadi A, Aranyi T. BiSearch: primer-design and search tool for PCR on bisulfite-treated genomes. Nucleic acids research. 2005;33:e9. doi: 10.1093/nar/gni012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qu W, Jiang G, Cruz Y, Chang CJ, Ho GY, Klein RS, et al. PCR detection of human papillomavirus: comparison between MY09/MY11 and GP5+/GP6+ primer systems. J Clin Microbiol. 1997;35:1304–1310. doi: 10.1128/jcm.35.6.1304-1310.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Castle PE, Schiffman M, Gravitt PE, Kendall H, Fishman S, Dong H, et al. Comparisons of HPV DNA detection by MY09/11 PCR methods. Journal of medical virology. 2002;68:417–423. doi: 10.1002/jmv.10220. [DOI] [PubMed] [Google Scholar]
- 30.Clifford GM, Gallus S, Herrero R, Munoz N, Snijders PJ, Vaccarella S, et al. Worldwide distribution of human papillomavirus types in cytologically normal women in the International Agency for Research on Cancer HPV prevalence surveys: a pooled analysis. Lancet. 2005;366:991–998. doi: 10.1016/S0140-6736(05)67069-9. [DOI] [PubMed] [Google Scholar]
- 31.del Refugio Gonzalez-Losa M, Laviada Miery TM, Puerto-Solis M, Garcia-Carranca A. Molecular variants of HPV type 16 E6 among Mexican women with LSIL and invasive cancer. J Clin Virol. 2004;29:95–98. doi: 10.1016/s1386-6532(03)00094-5. [DOI] [PubMed] [Google Scholar]
- 32.Tukey JW. Exploratory Data Analysis. Reading, MA: Addison–Wesley; 1977. [Google Scholar]
- 33.Maldonado G, Greenland S. Estimating causal effects. Int J Epidemiol. 2002;31:422–438. [PubMed] [Google Scholar]
- 34.Virmani AK, Muller C, Rathi A, Zoechbauer-Mueller S, Mathis M, Gazdar AF. Aberrant methylation during cervical carcinogenesis. Clinical cancer research : an official journal of the American Association for Cancer Research. 2001;7:584–589. [PubMed] [Google Scholar]
- 35.Dong SM, Kim HS, Rha SH, Sidransky D. Promoter hypermethylation of multiple genes in carcinoma of the uterine cervix. Clinical cancer research : an official journal of the American Association for Cancer Research. 2001;7:1982–1986. [PubMed] [Google Scholar]
- 36.Narayan G, Arias-Pulido H, Koul S, Vargas H, Zhang FF, Villella J, et al. Frequent promoter methylation of CDH1, DAPK, RARB, and HIC1 genes in carcinoma of cervix uteri: its relationship to clinical outcome. Molecular cancer. 2003;2:24. doi: 10.1186/1476-4598-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin CM, Astbury K, O'Leary JJ. Molecular profiling of cervical neoplasia. Expert review of molecular diagnostics. 2006;6:217–229. doi: 10.1586/14737159.6.2.217. [DOI] [PubMed] [Google Scholar]
- 38.Terra AP, Murta EF, Maluf PJ, Caballero OL, Brait M, Adad SJ. Aberrant promoter methylation can be useful as a marker of recurrent disease in patients with cervical intraepithelial neoplasia grade III. Tumori. 2007;93:572–579. doi: 10.1177/030089160709300610. [DOI] [PubMed] [Google Scholar]
- 39.Waxman AG, Chelmow D, Darragh TM, Lawson H, Moscicki AB. Revised terminology for cervical histopathology and its implications for management of high-grade squamous intraepithelial lesions of the cervix. Obstetrics and gynecology. 2012;120:1465–1471. doi: 10.1097/aog.0b013e31827001d5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reuschenbach M, Clad A, von Knebel Doeberitz C, Wentzensen N, Rahmsdorf J, Schaffrath F, et al. Performance of p16(INK4a)-cytology, HPV mRNA, and HPV DNA testing to identify high grade cervical dysplasia in women with abnormal screening results. Gynecologic oncology. 2010 doi: 10.1016/j.ygyno.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 41.Nuovo GJ, Plaia TW, Belinsky SA, Baylin SB, Herman JG. In situ detection of the hypermethylation-induced inactivation of the p16 gene as an early event in oncogenesis. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:12754–12759. doi: 10.1073/pnas.96.22.12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.von Keyserling H, Kuhn W, Schneider A, Bergmann T, Kaufmann AM. p16INK(4)a and p14ARF mRNA expression in Pap smears is age-related. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2012;25:465–470. doi: 10.1038/modpathol.2011.179. [DOI] [PubMed] [Google Scholar]
- 43.Vazquez-Vega S, Sanchez-Suarez LP, Contreras-Paredes A, Castellanos-Juarez E, Penarroja-Flores R, Lizano-Soberon M, et al. Nuclear co-expression of p14ARF and p16INK4A in uterine cervical cancer-derived cell lines containing HPV. Cancer biomarkers : section A of Disease markers. 2010;8:341–350. doi: 10.3233/CBM-2011-0223. [DOI] [PubMed] [Google Scholar]
- 44.Bulten J, van der Avoort IA, Melchers WJ, Massuger LF, Grefte JM, Hanselaar AG, et al. p14ARF and p16INK4A, two products of the same gene, are differently expressed in cervical intraepithelial neoplasia. Gynecologic oncology. 2006;101:487–494. doi: 10.1016/j.ygyno.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 45.Wang JL, Zheng BY, Li XD, Angstrom T, Lindstrom MS, Wallin KL. Predictive significance of the alterations of p16INK4A, p14ARF, p53, and proliferating cell nuclear antigen expression in the progression of cervical cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2004;10:2407–2414. doi: 10.1158/1078-0432.ccr-03-0242. [DOI] [PubMed] [Google Scholar]
- 46.Schlecht NF, Ben-Dayan M, Anayannis N, Lleras RA, Thomas C, Wang Y, et al. Epigenetic changes in the CDKN2A locus are associated with differential expression of P16INK4A and P14ARF in HPV-positive oropharyngeal squamous cell carcinoma. Cancer medicine. 2015;4:342–353. doi: 10.1002/cam4.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sartor MA, Dolinoy DC, Jones TR, Colacino JA, Prince ME, Carey TE, et al. Genome-wide methylation and expression differences in HPV(+) and HPV(−) squamous cell carcinoma cell lines are consistent with divergent mechanisms of carcinogenesis. Epigenetics : official journal of the DNA Methylation Society. 2011;6:777–787. doi: 10.4161/epi.6.6.16216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paz MF, Fraga MF, Avila S, Guo M, Pollan M, Herman JG, et al. A systematic profile of DNA methylation in human cancer cell lines. Cancer research. 2003;63:1114–1121. [PubMed] [Google Scholar]
- 49.Group A. Human papillomavirus testing for triage of women with cytologic evidence of low-grade squamous intraepithelial lesions: baseline data from a randomized trial. The Atypical Squamous Cells of Undetermined Significance/Low-Grade Squamous Intraepithelial Lesions Triage Study (ALTS) Group. Journal of the National Cancer Institute. 2000;92:397–402. doi: 10.1093/jnci/92.5.397. [DOI] [PubMed] [Google Scholar]
- 50.Einstein MH, Garcia FA, Mitchell AL, Day SP. Age-stratified performance of the Cervista HPV 16/18 genotyping test in women with ASC-US cytology. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2011;20:1185–1189. doi: 10.1158/1055-9965.EPI-11-0116. [DOI] [PubMed] [Google Scholar]
- 51.Elit LM. Pitfalls in the diagnosis of cervical intraepithelial neoplasia 1. Journal of lower genital tract disease. 2004;8:181–187. doi: 10.1097/00128360-200407000-00004. [DOI] [PubMed] [Google Scholar]
- 52.Kyrgiou M, Koliopoulos G, Martin-Hirsch P, Arbyn M, Prendiville W, Paraskevaidis E. Obstetric outcomes after conservative treatment for intraepithelial or early invasive cervical lesions: systematic review and meta-analysis. Lancet. 2006;367:489–498. doi: 10.1016/S0140-6736(06)68181-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.