Abstract
We report the first case of a child in the United States infected with an organism producing a Verona Integron-Encoded Metallo-β-Lactamase. This child succumbed to a ventilator-associated pneumonia caused by a Klebsiella pneumoniae producing this resistance mechanism.
Keywords: carbapenemase, CRE, metallo-β-lactamase, pediatrics, VIM
CASE REPORT
A full-term, 2-year-old male with congenital hydrocephalus requiring chronic invasive mechanical ventilation was transferred from a tertiary care hospital in Kuwait to a pediatric intensive care unit (PICU) in Maryland for ventriculoperitoneal shunt placement in May 2014. Upon arrival in the United States, he did not have evidence of an active infection. A rectal surveillance swab was obtained at the time of PICU admission because he received medical care abroad, per recommendations from the United States Centers for Disease Control and Prevention [1].
The rectal swab was processed in real time and inoculated into tryptic soy broth containing a 30-µg ceftriaxone disk and incubated at 37°C. After overnight incubation, 100 µL of broth sample with visible turbidity was plated on MacConkey agar with a 30-µg ceftriaxone disk and incubated at 37°C overnight. All isolates growing within 19 mm of the ceftriaxone disk (resistant zone diameter) underwent routine identification and antimicrobial susceptibility testing using the Phoenix Automated System (BD Diagnostics, Sparks, Maryland). An isolate of Klebsiella pneumoniae was recovered. The antibiotic minimum inhibitory concentrations are shown in Table 1. Because the isolate was carbapenem resistant, further phenotypic and molecular testing was performed. The organism was both modified Hodge test and metallo-β-lactamase (MβL) Etest positive (bioMérieux, Durham, North Carolina).
Table 1.
Results of Phenotypic and Antibiotic Susceptibility Testing of Klebsiella pneumoniae Isolates Recovered From Rectal Swabs and Bronchoalveolar Lavage Fluid From a Child and an Escherichia coli Isolate After Conjugation
| Source of Isolate | Modified Hodge Test | Metallo-β- lactamase Etest | Minimum Inhibitory Concentration (µg/mL)a |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aztreonam | Ceftriaxone | Cefepime | Piperacillin | Ertapenem | Imipenem | Meropenem | Doripenem | Ciprofloxacin | Gentamicin | Amikacin | Tobramycin | Tetracycline | Tigecyclineb | Colistinc | Fosfomycind | |||
| Admission rectal swab | Positive | Positive | 8 (1) | >32 (R) | 16 (R) | >64 (R) | >8 (R) | >8 (R) | >8 (R) | >2 (R) | >2 (R) | >8 (R) | <8 (S) | >8 (R) | >8 (R) | 2 (I) | 0.12 (S) | 16 (S) |
| Rectal swab 4 months into hospitalizatione | Positive | Positive | 8 (I) | >32 (R) | 16 (R) | >64 (R) | >8 (R) | >8 (R) | >8 (R) | >2 (R) | >2 (R) | >8 (R) | <8 (S) | >8 (R) | >8 (R) | 4 (R) | 0.12 (S) | 16 (S) |
| Bronchoalveolar lavage fluid sample obtained 5 months into hospitalization | Positive | Positive | 8 (I) | >32 (R) | 16 (R) | >64 (R) | >8 (R) | >8 (R) | >8 (R) | >2 (R) | >2 (R) | >8 (R) | <8 (S) | >8 (R) | >8 (R) | 4 (R) | 0.12 (S) | 16 (S) |
| E coli strain | Negative | Negative | ≤1 (S) | ≤1 (S) | ≤1 (S) | ≤1 (S) | ≤0.5 (S) | ≤1 (S) | ≤1 (S) | ≤1 (S) | ≤0.5 (S) | ≤1 (S) | ≤2 (S) | ≤1 (S) | – | ≤0.12 (S) | – | – |
| E coli J53 transconjugate strain | Positive | Positive | 8 (I) | >32 (R) | 2 (S) | >64 (R) | >8 (R) | 8 (R) | 2 (I) | 2 (I) | <0.5 (S) | 8 (I) | <8 (S) | >8 (R) | >8 (R) | <0.25 (S) | 0.06 (S) | 2 (S) |
aSusceptibility results are based on Clinical and Laboratory Standards Institute (CLSI) recommended breakpoints when available.
bNo tigecycline CLSI interpretive criteria available; European Committee on Antimicrobial Susceptibility Testing (EUCAST) defines tigecycline susceptibility as ≤1 µg/mL.
cNo colistin CLSI interpretive criteria available for Enterobacteriaceae, EUCAST defines colistin susceptibility as ≤2 µg/mL.
dNo fosfomycin CLSI interpretive criteria available for Enterobacteriaceae outside of urinary tract isolates, EUCAST defines fosfomycin susceptibility as ≤32 µg/mL.
eAdditional weekly rectal swabs are not included to simplify table but showed identical phenotypic testing and susceptibility patterns.
Upon identification of a carbapenemase-producing K pneumoniae from the admission rectal surveillance swab, the child was placed on contact precautions for the duration of his hospitalization. He remained hospitalized for 5 months, and weekly rectal surveillance swabs remained positive for this organism throughout the hospitalization. Active rectal surveillance swabs at the time of admission and weekly thereafter for all children hospitalized in the PICU were conducted [2]. No transmission of this resistant isolate to other children occurred. The patient developed a ventilator-associated pneumonia with a bronchoalveolar lavage fluid sample identifying the same carbapenem-resistant K pneumoniae isolate during his fifth month of hospitalization. Because he was known to be colonized with a carbapenemase-producing Enterobacteriaceae, he was empirically treated with extended-infusion meropenem and amikacin. Regrettably, on his fourth day of antibiotic therapy for ventilator-associated pneumonia, the patient died.
The genetic relatedness of our patient's rectal surveillance isolates and his clinical isolate was assessed by repetitive sequence-based polymerase chain reaction (rep-PCR) and multilocus sequence typing (MLST). Isolates with ≥95% similarity were considered to be of the same rep-PCR type. Sequences of 7 housekeeping genes (gapA, infB, mdh, pgi, phoE, rpoB, and tonB) were identified using the Pasteur MLST database (http://www.pasteur.fr/recherche/genopole/PF8/mlst). Rectal surveillance swabs and the clinical isolate were identical by rep-PCR and identified as sequence type 14 (ST14) by MLST.
To identify the β-lactamase genes associated with the carbapenem-resistant phenotype, the Check-MDR CT103 XL assay (CheckPoints, Wageningen, Netherlands) was performed on the surveillance and clinical isolates. We selected this platform because the Check-MDR CT103 XL assay combines PCR amplification and deoxyribonucleic acid microarray technologies for the detection of extended-spectrum β-lactamase (ESBL) genes, plasmid-mediated AmpC β-lactamase genes (pAmpC), and an extended panel of carbapenemase genes (blaKPC, blaNDM, blaVIM, blaIMP, blaOXA−48−like, blaGES, blaGIM, blaSPM, blaOXA−23−like, blaOXA−24/40−like, blaOXA−58−like). All tested isolates contained both the carbapenemase blaVIM and the pAmpC blaCMY−2−group. They were confirmed to be blaVIM−4 and blaCMY−4 by sequencing [3].
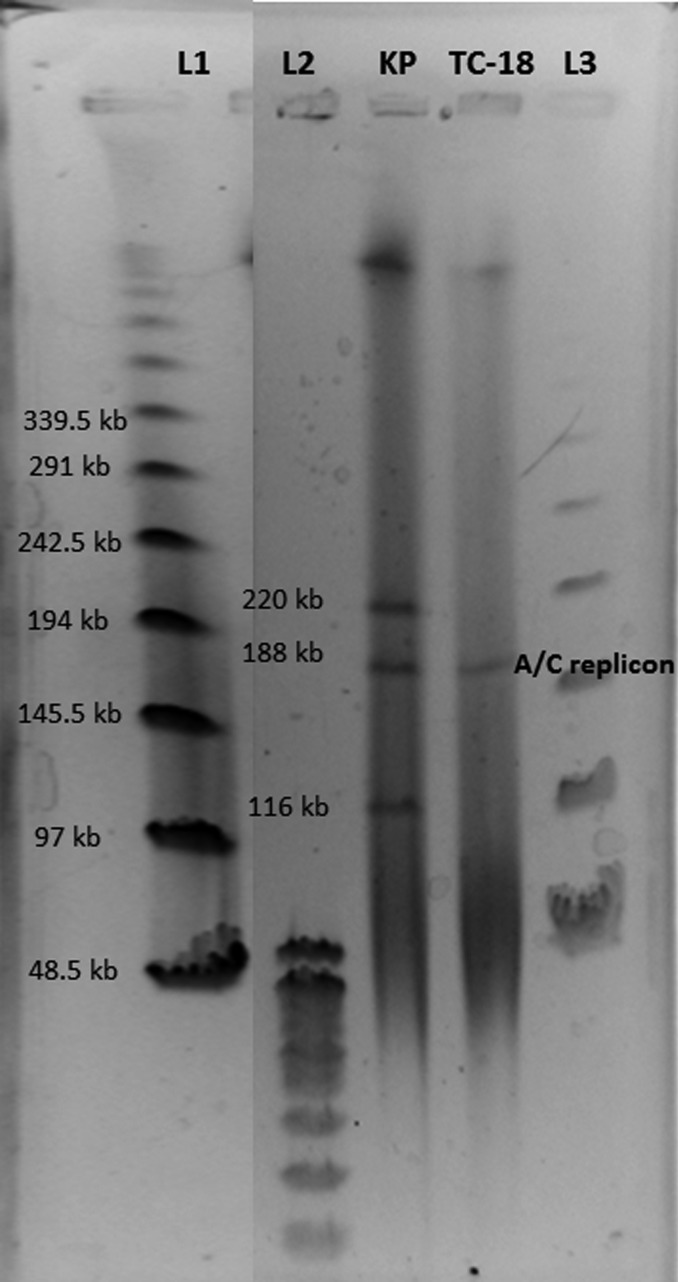
Conjugation experiments, using a variation of filter mating, succeeded in transferring the blaVIM−4-containing plasmid to the susceptible recipient strain, Escherichia coli J53 (Figure 1). The transconjugant (TC-18) was verified to be E coli and not breakthrough K pneumoniae, and antimicrobial susceptibility testing was performed (Table 1). S1 nuclease pulsed-field gel electrophoresis indicated that the parent K pneumoniae strain contained 3 plasmids and the transconjugant strain contained only 1 plasmid (Figure 1). Subsequent southern hybridization with the blaVIM probe demonstrated that the 188-kb plasmid common to both strains contained the blaVIM gene. Further PCR analysis indicated that the blaCMY−4 gene was transferred along with the blaVIM−4 gene, thereby suggesting it was also contained in the transferable 188-kb plasmid. Polymerase chain reaction-based plasmid replicon typing identified 2 of the 3 replicon types present in the K pneumoniae strain (A/C and FIIA; the third type undetermined) and that the transferable 188-kb plasmid was the A/C replicon.
Figure 1.
Southern blotting results for the metallo-β-lactamase-producing Klebsiella pneumoniae (KP) and the Escherichia coli transconjugant (TC-18). The blaVIM−4 and blaCMY−4 genes were transferable to the E coli isolate via the A/C replicon (188 kb). L1 and L3 represent the CHEF deoxyribonucleic acid (DNA) size standards, and L2 is the DNA standard size 8–48 kb (Bio-Rad, Hercules, California).
DISCUSSION
We report the first case of a Verona integron-encoded MβL (VIM) infection in a pediatric patient in the United States. Our patient remained colonized with this organism for at least 5 months and subsequently succumbed to a ventilator-associated pneumonia from this MβL-producing K pneumoniae. Other notable members of the MβL class include the “active on imipenems” (IMP) and New Delhi metallo-β-lactamases (NDM). Members of the different MβL subclasses differ not only in their high degree of sequence diversity, but also in the structure of their active sites [4]. The MβLs hydrolyze penicillins, cephalosporins, and carbapenems using zinc. In contrast to the class A and D serine-β-lactamases, notably the K pneumoniae carbapenemase and oxacillinase-types, the monobactam antibiotic aztreonam is spared from hydrolysis by the MβLs [4]. Like other carbapenemase-producers, MβLs can cause infections resulting in considerable morbidity and mortality [4].
Although MβL were first identified in the 1960s [4], they only came to prominence in the 1990s. Verona integron-encoded MβL-1 was discovered in Pseudomonas aeruginosa in 1996 [5]. Currently, more than 25 VIM allotypes belonging to 3 sublineages are recognized. blaVIM genes are predominantly found in P aeruginosa and other Gram-negative nonfermenters, but they are increasingly being reported in a number of Enterobacteriaceae [4]. Gram-negative bacteria possessing blaVIM are still mostly confined to the Mediterranean basin and Middle East. In one study in Greece, among 178 consecutive K pneumoniae bloodstream isolates, approximately 40% were blaVIM−1 positive [6]. The rapid dissemination of blaVIM in Greece has led to an endemic situation within a short period of time [6]. Imported cases involving acquired VIM carbapenemases are now described in both children and adults throughout Europe, Asia, and South America [4]. There have been a few isolated cases of infections caused by organisms producing VIMs reported in adults in the United States [7–10]. The first US case of an Enterobacteriaceae that produced a blaVIM was identified in 2010 [9]. This case involved a woman who required 12 days of hospitalization in Greece after becoming ill on a cruise, and upon return to the United States she developed septicemia from a VIM-producing K pneumoniae. In 2015, there was a report of 6 neonates in a neonatal intensive care unit in Kentucky becoming colonized with Enterobacteriaceae containing the blaVIM gene. Fortunately, no clinical infections were reported, but this cluster highlights the potential for rapid propagation of this resistance mechanism between critically ill patients [11].
In general, the dissemination of acquired MβL genes among Gram-negative organisms is mediated by mobile gene cassettes inserted into integrons [4]. The blaVIM gene is generally found to be part of a type 1 integron. These integrons are usually harbored by transferable plasmids with a high capacity for horizontal transmission. Most integrons containing gene cassettes for MβL also harbor additional gene cassettes carrying resistance determinants for a wide variety of antibiotic classes. Therefore, MβL-producing organisms frequently exhibit complex multidrug-resistance. Although aztreonam is not hydrolyzed by MβLs, its activity is often impaired by additional acquired mechanisms of resistance by organisms producing MβLs, such as ESBL or AmpC β-lactamase enzymes. This was likely the case with our patient whose K pneumoniae isolate was nonsusceptible to aztreonam and carried a pAmpC gene (blaCMY−4). Of concern, unlike the serine carbapenemases, there are very few novel agents under development with the potential to inhibit the production of MβLs [12, 13].
Metallo-β-lactamase-producing bacteria have caused a number of outbreaks throughout the world. An outbreak in an Australian ICU involved the MβL gene blaIMP−4, thought to be imported from an East Asian patient [13]. After the first detection of blaIMP−4 in a P aeruginosa isolate, the blaIMP−4 gene was subsequently identified in hospital-acquired isolates of Gram-negative pathogens of 5 different species, including several Enterobacteriaceae, over a 7-month period [14]. This outbreak was particularly concerning as it suggested that intraspecies and interspecies gene transfer plays a major role in the dissemination of MβL within healthcare settings. This case highlights the importance of screening patients for carbapenemases who have had previous healthcare exposure in endemic regions of the world. Because our patient was screened upon admission to the ICU, we believe the early recognition of colonization with an MβL-producing organism and the swift application of contact precautions likely prevented dissemination of this organism in our PICU.
CONCLUSIONS
In summary, our case reminds us that MβL-producing organisms with complex multidrug-resistant phenotypes can be a formidable clinical challenge. It is likely only a matter of time until they become endemic in pediatric healthcare settings. Identifying imported MβLs is important to prevent this resistance mechanism from becoming endemic in the United States, as it is in several other regions of the world.
Acknowledgments
Disclaimer. The contents of this study are solely the responsibility of the authors and do not necessarily represent the official view of the Fisher Center or Johns Hopkins University School of Medicine.
Financial support. This publication was made possible by support from the Sherrilyn and Ken Fisher Center for Environmental Infectious Diseases, Division of Infectious Diseases of the Johns Hopkins University School of Medicine.
R. A. B. was supported by funding from the VA Merit Review Board, National Institutes of Health (NIH), and Veterans Integrated Service Network 10 Geriatric Research, Education, and Clinical Center. L. K. L. was supported by funding from the NIH (grant 5K08AI112506-02).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1.Facility Guidance for Control of Carbapenem-resistant Enterobacteriaceae (CRE). Centers for Disease Control and Prevention. Available at: http://www.cdc.gov/hai/organisms/cre/cre-toolkit/index.html. Accessed 25 February 2016.
- 2.Suwantarat N, Logan LK, Carroll KC et al. . The prevalence and molecular epidemiology of multidrug-resistant Enterobacteriaceae colonization in a pediatric intensive care unit. Infect Control Hosp Epidemiol 2016; 37:535–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perez F, Hujer AM, Marshall SH et al. . Extensively drug-resistant Pseudomonas aeruginosa isolates containing blaVIM-2 and elements of Salmonella genomic island 2: a new genetic resistance determinant in Northeast Ohio. Antimicrob Agents Chemother 2014; 58:5929–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cornaglia G, Giamarellou H, Rossolini AM. Metallo-β-lactamases: a last frontier for β-lactams? Lancet Infect Dis 2011; 11:381–93. [DOI] [PubMed] [Google Scholar]
- 5.Lauretti L, Riccio ML, Mazzariol A et al. . Cloning and characterization of blaVIM, a new integron-borne metallo-beta-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob Agents Chemother 1999; 43:1584–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Psichogiou M, Tassios PT, Avlamis A et al. . Ongoing epidemic of blaVIM−1 positive Klebsiella pneumoniae in Athens, Greece: a prospective survey. J Antimicrob Chemother 2008; 61:59–63. [DOI] [PubMed] [Google Scholar]
- 7.Toleman MA, Rolston K, Jones RN et al. . blaVIM−7, an evolutionary distinct metallo-β-lactamase gene in a Pseudomonas aeruginosa isolate from the United States. Antimicrob Ag Chemother 2004; 48:329–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lolans K, Queenan AM, Bush K et al. . First nosocomial outbreak of Pseudomonas aeruginosa producing an integron-borne metallo-β-lactamase (VIM-2) in the United States. Antimicrob Agents Chemother 2005; 49:3528–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention (CDC). Update: detection of a Verona integron-encoded metallo-β-lactamase in Klebsiella pneumoniae—United States, 2010. MMWR Morb Mortal Wkly Rep 2010; 59:1212. [PubMed] [Google Scholar]
- 10.Aboufaycal H, Sader HS, Rolston K et al. . blaVIM−2 and blaVIM−7 carbapenemase-producing Pseudomonas aeruginosa isolates detected in a tertiary care medical center in the United States: report from the MYSTIC program. J Clin Microbiol 2007; 45:614–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yaffee AQ, Roser L, Daniels K et al. . Notes from the Field: Verona integron-encoded metallo-beta-lactamase–producing carbapenem-resistant Enterobacteriaceae in a neonatal and adult intensive care unit—Kentucky, 2015. MMWR Morb Mortal Wkly Rep 2016; 65:190. [DOI] [PubMed] [Google Scholar]
- 12.Biedenbach DJ, Kazmierczak K, Bouchillon SK et al. . In vitro activity of azteronam-avibactam against a global collection of Gram-negative pathogens from 2012-2013. Antimicrob Agents Chemother 2015; 59:4239–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Page MG, Dantier C, Desarbre E. In vitro properties of BAL30072, a novel siderophore sulfactam with activity against multiresistant Gram-negative bacilli. Antimicrob Agents Chemother 2010; 54:2291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peleg AY, Franklin C, Bell JM et al. . Dissemination of the metallo-β-lactamase gene blaIMP-4 among Gram-negative pathogens in a clinical setting in Australia. Clin Infect Dis 2005; 41:1549–56. [DOI] [PubMed] [Google Scholar]