SUMMARY
The extent to which alternative splicing and long intergenic noncoding RNAs (lincRNAs) contribute to the specialized functions of cells within an organ is poorly understood. We generated a comprehensive dataset of gene expression from individual cell types of the Arabidopsis root. Comparisons across cell types revealed that alternative splicing tends to remove parts of coding regions from a longer, major isoform, providing evidence for a progressive mechanism of splicing. Cell type-specific intron retention suggested a possible origin for this common form of alternative splicing. Coordinated alternative splicing across developmental stages pointed to a role in regulating differentiation. Consistent with this hypothesis, distinct isoforms of a transcription factor were shown to control developmental transitions. LincRNAs were generally lowly expressed at the level of individual cell types, but co-expression clusters provided clues as to their function. Our results highlight insights gained from analysis of expression at the level of individual cell types.
Keywords: RNA-seq, Arabidopsis, alternative splicing, lincRNA, transcriptome. proteome
eTOC
Li et al. present a comprehensive dataset of gene expression generated using short-read sequencing from individual cell types and developmental zones of the Arabidopsis root, complemented by long-read sequencing and quantitative proteomic analyses. The data in this resource characterize cell type and developmental stage-specific alternative splicing and lincRNA expression.
Introduction
Multicellular organisms evolved transcriptional and post-transcriptional mechanisms to convert genetic information into RNA and proteins, which determine the identity of specific cell types. Alternative splicing is a post-transcriptional mechanism proposed to increase proteome diversity (Djebali et al., 2012, Brown et al., 2014). In recent years, an increasing number of alternatively spliced isoforms and thousands of long intergenic non-coding RNAs (lincRNAs) have been identified in insects, worms, mammals and plants by RNA sequencing (Liu et al., 2012, Cabili et al., 2011, Barbosa-Morais et al., 2012, Gerstein et al., 2014, Gupta et al., 2015, Loraine et al., 2013). Some tissue-specific alternative splicing variants and lincRNAs have been shown to play important roles during development and in response to stress in different species (Sun et al., 2013, Tsai et al., 2010, Irimia and Blencowe, 2012). To date, the extent to which alternative splicing produces cell type-specific isoforms in plants, and how these isoforms contribute to proteome diversity, is largely unknown. To address these questions, we performed integrated transcriptome and proteome analysis using both short- and long-read RNA sequencing and quantitative mass spectrometry to generate a comprehensive expression map of the Arabidopsis root.
Arabidopsis roots are organized in such a way as to facilitate systematic characterization. They consist of nested cylindrical layers representing the major cell types. From the outside in, these are the epidermis, cortex, endodermis and stele (consisting of the pericycle, phloem, xylem and procambium). All root cells are generated from stem cells surrounding the quiescent center (QC). The lateral and columella root cap cells are located at the root tip and provide protection to this stem cell niche (Figure 1A). Developmental stages of the major cell types are defined along the longitudinal axis of the root, with immature cells near the stem cell niche and cells of increasing maturity located in the shootward direction (Figure 1A).
Figure 1.
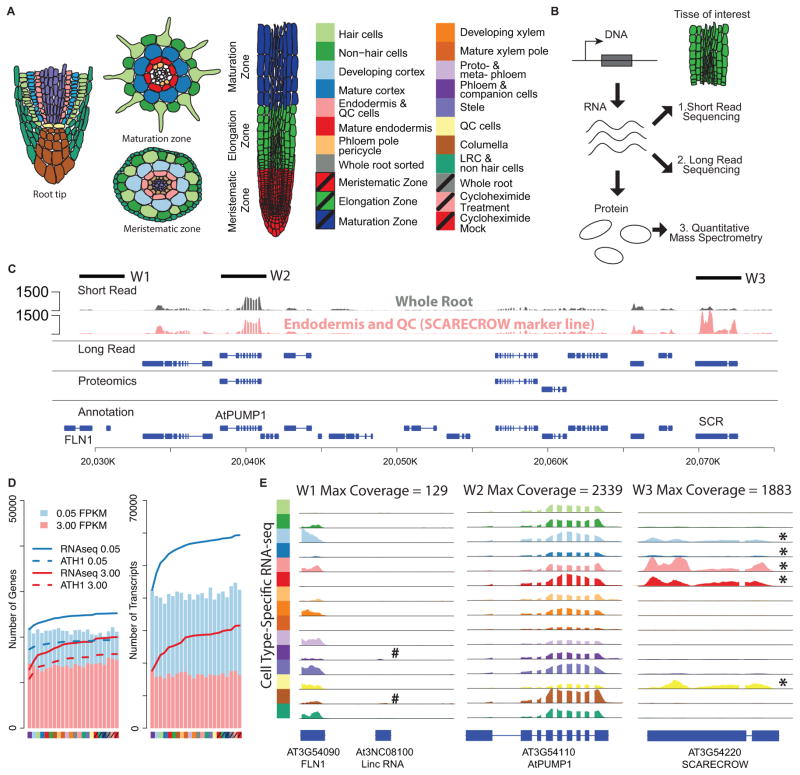
Cell-specific expression profiling using RNA-seq. (A) Cell types and (B) experimental techniques used to profile RNA and protein expression levels. (C) Example of data coverage by three different technologies. Each row represent genes identified by one technology: short read (Illumina sequencing), long read (PacBio sequencing), proteomics, and conventional genome annotation. RNA-seq read pileups in whole root and SCARECROW marker line that marks the endodermis and QC cells. Solid lines indicate four windows of 3Kb each. (D) Number of detected protein coding genes and isoforms in each cell type. Cumulative numbers of detected genes or transcripts are shown for RNA-seq (solid line) and microarray (dashed line) analyses. (E) RNA-seq read pileups for three chromosomal regions in all cell types. # An annotated lincRNA detected in specific cell types. * cell type-specific gene expression (p<0.01). Note: The expression level of this lincRNA gene (At3N08100) is very low as compared to the neighboring mRNA gene (AT4G54090) and almost invisible in this plot. See also Figure S1
Previous studies using microarrays and low-throughput mass spectrometry demonstrated that cell type-specific expression profiling can identify dominant expression patterns and signature genes with cell-type specific functions (Brady et al., 2007, Birnbaum et al., 2003, Petricka et al., 2012). However, expression profiling using microarrays is limited to the genes represented on the array, and previous proteomics analyses were limited by the available technology, with only a few hundred proteins identified for each cell type. In this study, we performed a comprehensive analysis of cell type and developmental stage-specific alternative splicing and lincRNA expression using short-read, paired-end RNA-seq. We complemented this analysis with long-read Pacific BioSciences (PacBio) sequencing and quantitative mass spectrometry (Figures 1B and C). From these three platforms, we identified > 24,000 new splice isoforms and >1000 non-coding RNAs and detected > 12,000 peptides from over >5,000 proteins.
Key insights from the analysis of these extensive data are: 1) Alternative splicing between cell types is rarely a binary process, but results in a difference in degree between major and minor isoforms; 2) Alternative splicing tends to remove parts of coding regions from a longer, major isoform, providing evidence for a progressive mechanism of splicing; 3) Intron retention of evolutionarily conserved introns suggests a possible origin for a common form of alternative splicing; 4) Coordinated alternative splicing appears to play a role in regulating differentiation in the root; 5) Distinct isoforms of a well-characterized transcription factor control different developmental processes; 6) For the majority of detected proteins, peptide abundance correlates well with major isoform abundance in different developmental stages; and 7) LincRNAs are generally lowly expressed, even when profiled at the level of individual cell types but co-expression clusters provide clues as to their function.
RESULTS
A comprehensive cell-type specific RNA expression map of the Arabidopsis root
RNA-seq reads were generated from total RNA isolated from 15 root cell types, three developmental zones and whole roots of Arabidopsis (Figure 1A, 3 biological replicates for each sample, 57 libraries total, Table S1). To enrich for specific cell types we utilized fluorescence-activated cell sorting of green fluorescent protein (GFP) marked cell populations (Birnbaum et al., 2005). For the developmental stages, we hand-dissected the meristematic, elongation and differentiation zones (Figure 1A). RNA-seq was obtained from 100 bp and 125 bp, paired-end reads on an Illumina HiSeq2000 (details of all libraries are given in Table S1). Approximately 3.3 billion reads (an average of 24.8 million read pairs per library) were uniquely mapped to the Arabidopsis genome and transcriptome (Supplemental Experimental Procedures and Figure S1A).
Figure 3.
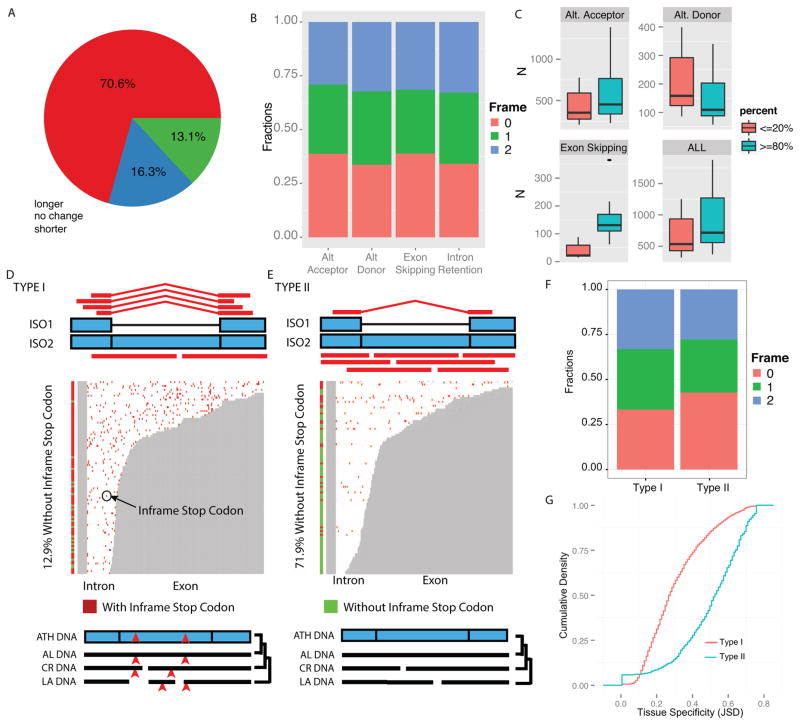
Coding potential of splice isoforms, local splicing and intron retention events. (A) Distribution of Open Reading Frame (ORF) length of DM isoforms as compared to CM isoforms. (B) Fraction of alternative splicing events that maintain or change reading frames. (C) Number of local splicing events with local PSI (LPSI) < 20% compared to those with LPS > 80%, in all cell types. (D) and (E) are pileup plots for intron retention events. Grey and white areas represent exon and intron regions respectively. In isoforms with intron retention, the white areas are kept as exons. Vertical bars with red and green color are used to indicate whether a given intron retention event contains in frame stop codon. (D) For Type I intron retention events, more reads support the spliced isoform than the intron retention isoform. The intron has in-frame stop codons (indicated by red points and carats). In multiple genome alignment, many stop codons are found in the intron regions (Text S1.2.8). (E) For Type II intron retention events, more reads support the intron retention isoform. In multiple genome alignment, the intron is free of in-frame stop codons in other related species. ATH, Arabidopsis thaliana. AL, Arabidopsis lyrata. CR, Capsella rubella. LA, Leavenworthia alabamica. (F) Distributions of reading frame changes for Type 1 and Type 2 intron retention events. (G) Comparison of the cumulative distribution of tissue specificity (based on JSD) of Type 1 and Type 2 intron retention events.
A detailed analysis of gene expression levels were performed and summarized in Figure S1 and see Experimental Procedures. Comparison with published ATH1 GeneChip data (Brady et al., 2007, Birnbaum et al., 2003) for the same cell types indicated that our RNA-seq results retain relative gene expression levels and provide high reproducibility (see Experimental Procedures, Table S1). In addition, we identified many more genes in large gene families (for example, see Figure S1B) than was the case for the microarray analysis. Overall, we detected 92.1% of the annotated protein-coding genes and 90.2% of the isoforms in at least one cell type or one developmental stage (Fig. 1D, Fragments Per Kilobasepairs per Million reads, FPKM >0.05, Supplemental Experimental Procedures and Figure S1C). Our RNA-seq reads provide an estimated 8,100-fold coverage for protein coding genes (for example, see Fig. 1C and E), and demonstrate the enhanced detection gained by sorting for specific cell types. For example, SCARECROW (AT3G54220, SCR) is lowly expressed in the whole root library but highly expressed in the endodermis and QC (Figures 1C and E). By contrast, a gene involved in the mitochondrial electron transport chain, which is 30 kb away, is highly expressed across all cell types (Figure 1E, AtPUMP1). To catalog all tissue-specific or tissue-selectively expressed genes, we first identified differentially expressed genes in any cell type (using a generalized linear model, edgeR (Robinson et al., 2010), FDR <0.001) then used an entropy based approach (Kadota et al., 2006) to assign genes to different tissue types (Supplemental Experimental Procedures). To systematically evaluate the functions of genes expressed in each cell type, we performed gene ontology (GO) enrichment analysis (Alexa et al., 2006) (Supplemental Experimental Procedures, Figure S1D). We found more than 50% of enriched GO terms to be specific to a single cell type, consistent with our previous observations from microarray data (Brady et al., 2007), allowing for clear discrimination between closely related cell types. When comparing sorted cells with unsorted whole roots, we found 4% of all detected genes (1113 genes) and 1.5% of all detected isoforms (977 isoforms) changed significantly (FDR<0.01, log2 fold change >2), indicating only a small effect of cell sorting on both gene expression and alternative splicing. These results demonstrate that our RNA-seq data substantially improve upon previous expression data with better coverage and enhanced dynamic range.
Alternative splicing events are cell-type specific
Alternative splicing has been hypothesized to play important roles in producing cell-type specific transcripts and in diversifying the proteome. To characterize alternative splicing events in different cell types we first identified short-read sequences that map across adjacent exons. With this approach we were able to identify known TAIR10 splice junctions with high sensitivity (93%) and precision (89%) (Figures S2A and S2B). Using stringent criteria (see Experimental Procedures), we identified an average of 90,108 known splice junctions (sensitivity = 89.7%) and 11,173 novel splice junctions in each of the different cell types. Across all samples, we identified 108,178 known splice junctions and 74,522 new splice junctions. The majority of known splice junctions are found across many cell types and/or developmental stages. In contrast, new splice junctions tend to be found in specific cell types or developmental stages highlighting the value of cell-type specific expression profiling.
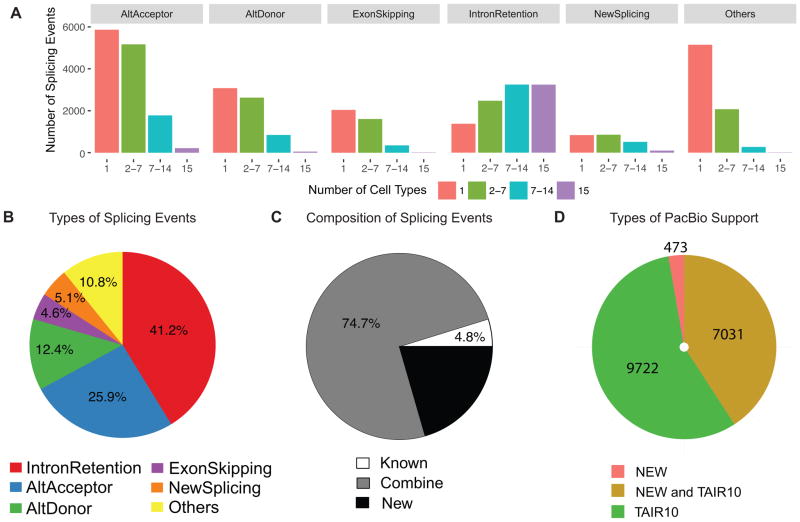
To profile the distribution of splicing events in different cell types, we grouped spliced reads into mutually exclusive groups, which were further classified into different local alternative splicing types (see Experimental Procedures). Most alternative splicing events were detected in fewer than half of the cell types (Figure 2A). We identified four major types of splicing events (Figure 2B and Figure S2C): 1) intron retention, which is, on average, the most prevalent alternative splicing type (41.2%), 2) alternative acceptor (25.9%), 3) alternative donor (12.4%) and 4) exon skipping (4.6%). We also found, in each cell type, an average of 5.1% novel splice junctions located either in known exons or known introns. We found most of these alternative splicing events (95.2%, Figure 2C) were formed by the combination of newly discovered splice junctions with known splice junctions or by newly discovered splice junctions alone. These results demonstrate that our high-resolution cell type-specific data detected new and reproducible alternative splicing events that were missed in RNA-seq data from whole organs (Marquez et al., 2012, Filichkin et al., 2010).
Figure 2.
Cell type-specific splicing events. (A) The majority of local splicing events are cell type specific. Distribution of types of local alternative splicing events according to the number of cell types where each event is found. (B) Distribution of types of local splicing events. (C) Majority of local splicing events are formed by the combination of newly identified splice junctions and known splice junctions. (D) Number of assembled isoforms supported by PacBio reads. See also Figures S2 and S3
Validation of splice isoforms by PacBio sequencing
Alternatively spliced isoforms identified from short read sequences could arise from pre-spliced RNA. In particular, this could be the case for putative intron retention events. To address this issue, we first performed qRT-PCR experiments using cDNAs generated from polyA selected RNAs. We selected newly identified alternative splicing events in the four major categories and validated 12 using qRT-PCR (Figure S3A–S3D, Table S2).
To systematically validate isoforms assembled from short reads, we performed PacBio long-read sequencing. We generated libraries from polyA-selected RNAs from whole roots and performed sequencing in 10 Single Molecule Real Time sequencing (SMRT) cells for a total of ~181,000 long-read sequences. We identified 17,516 high-quality consensus isoforms, among which, 98.6% could be mapped to 6,742 genes in the TAIR10 genome. We found that most consensus isoforms support TAIR 10 isoforms or map to transcript regions that are shared by TAIR 10 and new isoforms (Figure 2D, Figures S3E–S3F). In particular, we found support for 1454 intron retention events (Figures S3G), providing confirmation for our short-read sequencing results. We found a low detection level of new isoforms in the PacBio dataset, however, this is due to dilution of cell-type specific isoforms in whole root data sets (Figures S3H).
Most genes encode a single major isoform and multiple minor isoforms
To quantify the cell-type specific expression levels of different isoforms, we assembled full-length transcripts from short reads and carried out all pairwise comparisons of isoform expression levels between different cell types (Trapnell et al., 2012). We identified 81% more putative isoforms than are present in the TAIR10 genome annotation. On average, there are 55,493 different transcripts per cell type (FPKM>0.05, Figure 1D) and 88% of multi-isoform genes encode 2 to 6 isoforms. We calculated the percentage-spliced-in value (PSI, see Experimental Procedures) as a measure of the relative expression levels of different isoforms of the same gene in each cell type. For example, the major isoform of any gene is defined as an isoform with median PSI values higher than 50% across all cell types. In other words, over 50% of the transcripts from this locus are of this isoform in at least half of the examined cell types. We also defined common minor isoforms as those expressed above 5% PSI in at least one cell type and rare minor isoforms as those expressed below 5% PSI in all cell types. Strikingly, the vast majority (85.2%) of multi-isoform genes have one major isoform that is expressed at high level in the majority of cell types (median PSI > 50%). Of all the isoforms identified in our study, there are 21.7% major isoforms, 59.4% common minor isoforms and 18.9% rare minor isoforms. These results suggest that, for most genes, the primary product of splicing is a major isoform, but the total number of common minor isoforms exceeds the number of major isoforms and rare minor isoforms. Unlike in animals where there is frequently a presence/absence difference of splice isoforms between cell types, our results indicate that, in plants, the difference is primarily one of relative levels of expression.
Major isoforms generally have longer coding regions
In metazoans, exon skipping is the most common form of alternative splicing. Therefore the prevailing view is that alternative splicing increases proteome diversity primarily through exon and domain recombination resulting in difference in coding regions (Gogos et al., 1992). By contrast, we found that the majority of the major isoforms (70.5%, Figure 3A) have longer coding regions than the common minor isoforms. A trivial explanation could have been that minor isoforms have shorter mRNAs and thus shorter coding sequences. However, more than 66% of the four major types of splicing extend the major transcripts, suggesting that transcript length does not bias the estimated coding sequence length.
Because full-length transcripts are generated by the combinatorial regulation of local splicing events (for example, alternative donor sites or acceptor sites, see Figure S2E), we analyzed the local splicing events that generate the minor isoforms. For all four types of important alternative splicing events (alternative 3′ acceptors, alternative 5′ donors, exon skipping, and intron retention), slightly more than 33% of the alternative splicing events maintain the reading frame (Figure 3B). Other local splicing events change the reading frame and disrupt the coding region by switching to a frame that has no coding potential leading to a premature termination codon, which potentially causes non-sense mediated decay (NMD) (Leviatan et al., 2013, Kalyna et al., 2012).
We analyzed splicing events in which the frame is maintained to exclude potential NMD targeted isoforms from this subset. To identify local splicing events that favor longer or shorter isoforms, we calculated the longer isoforms’ PSI (LPSI, see Experimental Procedures and Figure S2D) and determined how many local splicing events have LPSI ≥ 80% or ≤ 20% across multiple cell types. For alternative 3′ acceptors or exon-skipping events (Figure 3C) we found more have LPSI ≥ 80% than LPSI≤20%. In contrast, for 5′ alternative donor events more have LPSI ≤20% than LPSI≥80% (Figure 3C). For an alternative acceptor event, the longer isoform uses an upstream acceptor site (Figure S2F). For an alternative donor event, the shorter isoform uses an upstream donor site (Figure S2G). For both alternative acceptor and alternative donor sites, upstream acceptor or donor sites are found more often in all cell types. These results suggest a co-transcriptional splicing model in which the splicing machinery tends to choose the first available, i.e. upstream, donor or acceptor site to generate a highly expressed isoform. Even though use of an upstream donor site would result in a shorter isoform, alternative donor sites are far less prevalent than the other types of local splicing events that result in highly expressed longer isoforms (Figure 2B). For exon-skipping events, our results suggest that the splicing machinery tends to occupy all the donor and acceptor sites (Figure S2H) and favors the production of the longer isoform without exon-skipping.
Cell-type specific intron retention events suggest a mechanism for intron birth
Similar to results from entire plant organs (Filichkin et al., 2010), our data indicate that intron retention is the most prevalent alternative splicing event in individual cell types. We found 3,713 intron retention events for which the isoform that retains the intron was the common minor isoform and the spliced isoform was the major isoform. We named these Type I intron retention events (Figure 3D). We also found 1,344 intron retention events for which the isoform that retains the intron is the major isoform and the spliced isoform is the common minor isoform. We named these Type II intron retention events (Figure 3E). Strikingly, Type II events are more likely to maintain the coding frame (Figure 3F, p<1.72e-30, Chi-squared test) and have introns that are free of in frame stop codons (71.9%) as compared to Type I events (12.9%, p<2.2e-16, chi square test). We found that 87.5% of the introns in Type II events with full coding potential are conserved as coding regions in other crucifer species (Haudry et al., 2013). In contrast, only 26.6% of introns in Type I events are conserved as coding regions in other crucifer species. To quantify the tissue specificity of Type I and Type II events, we used an information theoretic metric (Jason-Shannon Divergence), which compares the expression of the spliced isoforms (ISO1 in Figures 3D and E) to the expected null distribution of non-tissue specific alternative splicing. We found Type II events are significantly enriched in tissue specific data as compared to Type I events (Figure 3G, p<1e-150, KS test). Type II events have also been called “exitrons” in an analysis of flowers and whole seedlings (Marquez et al., 2015). Our cell-type specific analysis suggests the interesting possibility that intron formation is coupled with cell-type specific partial exon excision of the coding sequences providing new functionality in specific cell types.
The majority of isoforms do not appear to be subject to nonsense mediated decay
Nonsense mediated decay (NMD) has been proposed as a post-transcriptional level of regulation in which certain sequence features such as premature termination codons serve as triggers. Because many of the splice isoforms we identified result in premature termination codons as well as other potential NMD triggers, we sought to determine the prevalence of NMD among these isoforms. One approach could have been to analyze mutants in NMD components. Unfortunately, available mutants have dramatically altered root development precluding their use for this purpose (Drechsel et al., 2013, Yoine et al., 2006). To examine expression of NMD targets without affecting root development, we used treatment with cycloheximide, as many cycloheximide-sensitive transcripts are also NMD targets (Drechsel et al., 2013, Kalyna et al., 2012). Because cycloheximide treatment affects expression of both genes and splice isoforms, we identified putative NMD targets by selecting isoforms that are differentially expressed and up-regulated more than 20% as compared to the corresponding gene expression changes. Based on these criteria, we identified 2,518 putative NMD target transcripts, which account for 4% of the isoforms identified in our study. This suggests that only a small proportion of isoforms with premature termination codons actually undergo NMD. However, this could be an underestimate as some alternative isoforms are cell type-specific and could be targets of NMD. These wouldn’t be detected by our experimental approach.
Coordinated alternative splicing is more prevalent across developmental zones than between cell types
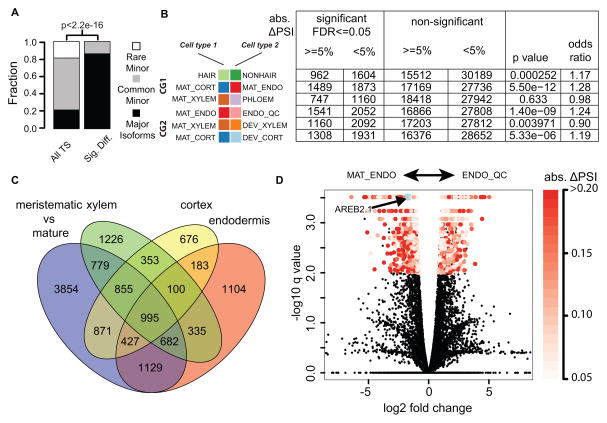
To determine the relative importance of alternative splicing versus gene expression in different cell types, we carried out pairwise comparisons between each of the different cell populations that we sorted, suggesting that alternative splicing mainly regulates major isoforms. We found that major isoforms tend to be differentially expressed between cell types (FDR<0.01, p < 2e-16, Chi-square test, Figure 4A). To obtain a finer-grained analysis of the relative importance of splicing versus expression we chose three pairwise comparisons between cell types in adjacent cell layers (Figure 4B, comparison group 1, CG1: cell type differences). We also performed three pairwise comparisons between developmental stages in the same cell lineage (Figure 4B, comparison group 2, CG2: maturation process). On average, ~5,600 isoforms were differentially expressed in each pairwise comparison (Figure 4B). An isoform can be differentially expressed between two cell types with or without substantial changes in the relative amounts of alternative splicing. To account for this, we measured the change in percentage spliced in (ΔPSI) for the isoforms. We found that, in both comparisons, differentially-expressed isoforms tend to have absolute ΔPSI higher than 5% as compared to isoforms that are not differentially expressed (Fisher exact test, Figure 4B), suggesting that alternative splicing plays an important role in modulating isoform expression between different cell types and during maturation processes. However, we found more isoforms differentially expressed in the three comparisons with developmental zone differences as compared to the comparisons across cell types. This suggests that alternative splicing may be a more common form of regulation during cellular maturation than in cell type specification. Furthermore, among the isoforms differentially expressed in all three cellular maturation comparisons, most of the differentially expressed genes are similarly regulated in the maturing cell types (Figures S4A and S4B, red arrows), suggesting there is a common set of isoforms that is regulated during the maturation process.
Figure 4.
Differentially expressed splice isoforms. (A) Comparison of the distribution of differentially expressed isoforms with genome wide distribution. Among all the transcript isoforms identified in our study, a majority are common minor isoforms. However, most of the differentially expressed isoforms are dominant major isoforms. (B) Pairwise comparisons. CG1: comparison group 1, cell type differences. CG2: comparison group 2, maturation processes. Table shows number of significantly differentially expressed isoforms. (C) Venn diagram shows overlap of isoforms that are differentially regulated through maturation processes and by cell type specific expression. (D) Volcano plot of comparison between mature endodermis and endodermis/QC marker lines. AREB2.1 (major isoforms of AREB2 gene) is significantly up-regulated in mature endodermis (highlighted by light blue star). See also Figure S4 and S5
To test this hypothesis, we identified differentially expressed isoforms using RNA-seq data generated form resected segments of the three developmental zones in the root (Figure 1A), effectively pooling all cell types present in each zone. We compared isoforms that are differentially expressed between the least mature segment (the meristematic zone) and the most mature segment (the differentiation zone) with transcripts present in all three zones (CG2, Figure 4C). We found 3311 differentially expressed isoforms (Figure 4C), which is 3.3 times the average number of cell type-specific differentially expressed isoforms. This supports the hypothesis that alternative splicing may be used to regulate maturation independent of cell type. In contrast, we found fewer splice isoforms that are co-activated or co-repressed across different cell types than among the three developmental comparisons (Figure S4A, red arrows). A possible mechanism underlying tissue-specific alternative splicing is differential expression of Serine/Arginine-rich (SR) proteins, which regulate splicing. Analysis of their mRNA levels indicates that their differential expression is greater across developmental zones than between cell types (Figure S5 and Experimental Procedures), which is consistent with the proposed mechanism. Together, these results suggest an important role for alternative splicing in regulating differentiation in the root.
Major isoforms correlate with peptide abundance across developmental zones
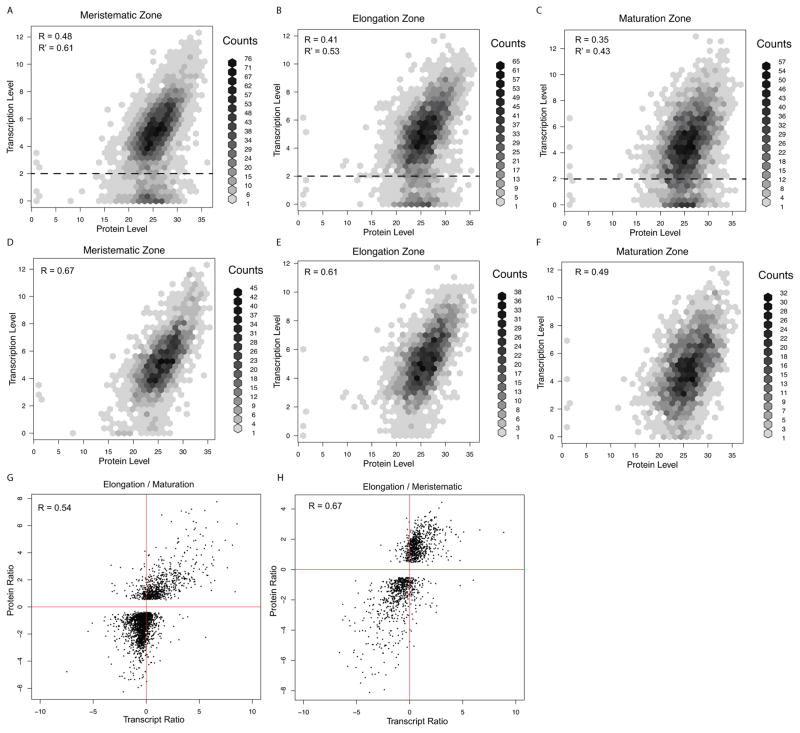
To determine if splice isoform levels correlate with protein concentrations, we used quantitative mass spectrometry (see Methods) to identify peptides expressed in the three developmental zones of the root. We mapped a total of 16,774 peptides, of which 320 uniquely mapped to individual isoforms (Table S4), the others mapped to shared regions between at least two isoforms. This is consistent with alternative splicing leading to relatively small changes in coding regions. A comparison of all transcripts and proteins inferred from peptide abundance, revealed a positive correlation in expression levels (Figures 5A–C, average Pearson Correlation Coefficients PCC = 0.41). There appeared to be two populations of transcripts: one correlated well with protein expression levels while the other showed little or no correlation. The population with low correlation appeared to be primarily those with low transcript abundance and moderate to high protein levels. Removing transcripts with very low abundance resulted in an increase in average correlation (PCC =0.52) (Figures 5A–C). Strikingly, the highest correlation was between major isoforms and protein levels (average PCC = 0.59, Figures 5D–F, Figure S6). In addition, the relative fold change of transcripts across developmental zones has an even higher correlation with relative fold change in protein levels (average PCC=0.61, Figures 5G and H). These results suggest that regulation of expression of major isoforms plays an important role in controlling the level of many proteins in the root.
Figure 5.
Expression correlation between quantitative proteomic data and RNAseq data. RNAseq data are transformd by log2(FPKM+1). Proteomic data are log2 transformed. (A), (B) and (C) show correlation between transcripts and their corresponding protein levels. R is Pearson Correlation Coefficient (PCC), R′ is PCC after removing isoforms with expression levels below dashed line (log2(FPKM+1) < 2). (D), (E) and (F) show correlation between all major transcripts and their corresponding protein levels. (G) and (H) show the correlation between ratios of transcripts and ratios of protein levels. See also Figure S6
A minor isoform of a transcription factor regulates root differentiation
To determine if an alternatively spliced isoform can perform specific biological functions, we focused on candidate genes from transcription factors that are differentially expressed across developmental zones and are alternatively spliced (Figure 4D, red points, FDR < 0.05 and absolute Δ PSI > 5%). We selected ABSCISIC ACID RESPONSIVE ELEMENT (ABRE) BINDING PROTEIN 2 (AREB2) because it is a well-characterized transcription factor and its alternative isoforms have different amino acid sequences in the conserved protein domains (Figure S7). ABA treatment inhibits root growth and promotes premature differentiation of the root meristem. AREB1, AREB2, and ABRE BINDING FACTOR3 (ABF3) are Basic Leucine Zipper (b-zip) transcription factors that function downstream of the ABA signal. The roots of the areb1, areb2, abf3 triple mutant show higher resistance to ABA treatment than single and double mutants of these genes, indicating that AREB1, AREB2, and ABF3 have redundant functions (Yoshida et al., 2010).
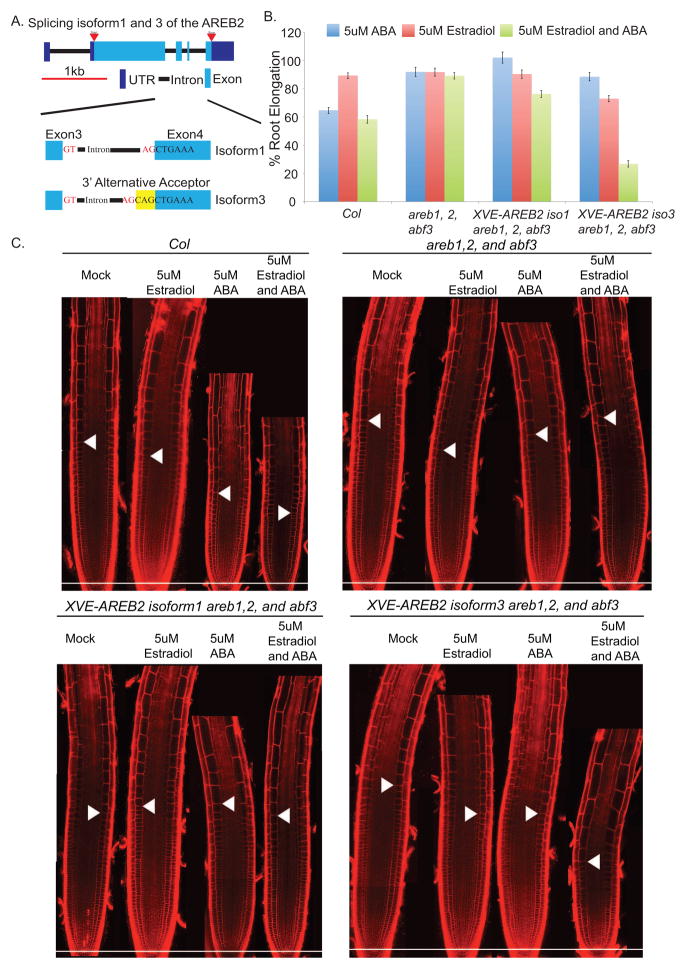
AREB2 encodes a dominant major isoform, AREB2-iso1 (which we will refer to as iso1), and a common minor isoform AREB2-iso3 (iso3) (Figure 6A). The isoforms differ in their splice acceptor sites at the fourth exon (Figure 6A), which results in the insertion of a single glutamine residue within a conserved alpha helical domain of iso3 (Figures S7A and S7B). Computational structure modeling (Kallberg et al., 2012) suggests that the additional amino acid could significantly alter the three-dimensional structure and affect dimerization (Figures S7C–F). The absolute values of ΔPSI are > 5% for iso3 in comparisons between mature and developing endodermis and between mature and developing cortex. Furthermore, iso3 is highly expressed in the maturation zone as compared to the other two developmental zones (Figures S7J). These results suggest that this isoform may play a role in maturation of the root. To test this hypothesis, the cDNA of each isoform was ectopically expressed in the areb1,areb2,abf3 triple mutant using an estradiol inducible promoter. Induction permits selective activation in a well-controlled fashion such that off-target effects can be assessed and minimized. After treatment with estradiol, induction of both isoforms was confirmed by qRT-PCR (Figure S7I) and the PCR products were separated by capillary electrophoresis to distinguish the 3 base pair difference (Figure S7I). The ABA resistant phenotypes were examined after induction of each isoform in the triple mutant background. Ectopic over-expression of iso1 had no detectable effects on root elongation, root meristem development, or root hair development (Figures 6B, 6C, and Figure S7). By contrast, ectopic over-expression of iso3 inhibited root elongation (Figure 6B) and resulted in premature differentiation of the root meristem as compared with the triple mutant (Fig. 6C and Figure S7). Ectopic expression of iso3 also caused root hairs to form at an earlier development stage and generated a smaller root meristem (Figure 6C and Figure S7). However, ectopic expression of iso3 did not appear to affect root patterning (Figure 6C). These results indicate that the minor splice variant, AREB2-iso3 plays a role in regulating differentiation of the root.
Figure 6.
Ectopic expression of AREB2 isoforms. (A) Schematic of AREB2 isoforms (B) Average root elongation of Col-0, areb1,2, abf3 triple mutant, XVE-AREB2-Iso1, and XVE-AREB2–Iso3 in areb1,2, abf3 on MS media with Mock, 5uM Estradiol, 5uM ABA, and 5uM Estradiol and ABA. 9-day-old seedlings were grown on MS medium for 5 days before transfer. Average root lengths in each condition were divided by the average root length under Mock treatment. (N>16, ±SD, *p<0.001) (C) Optical sections of root meristem of Col-0, areb1,2, abf3, XVE-AREB2-Iso1 in areb1,2, abf3, XVE-AREB2-Iso3 in areb1,2, abf3 under the same conditions as (B). White arrowheads indicate the first elongated cell in the cortex. Scale bar, 100um. See also Figure S7
We hypothesized that the additional amino acid present in iso3 could inhibit homo-dimerization of iso1 preventing its nuclear localization (Figure S7). To test this hypothesis, both isoforms were ectopically expressed in a line that contained a GFP tagged version of iso1 driven by its native promoter (pAREB2-GFP-AREB2-iso1) (Yoshida et al., 2010). In the absence of stimulation, the GFP signal of iso1 was detected in the nuclei of most cell types of the root (Figure S7). It was previously shown that over-expression of iso1 does not alter the phenotype, but can still be activated by ABA (Kang et al., 2002, Umezawa et al., 2013). Similarly, we observed only subtle changes in the nuclear localization of GFP- iso1 after ectopic expression of iso1 and ABA treatment (Figure S7). By contrast, addition of ABA to a line ectopically expressing iso3 significantly inhibited the nuclear localization of GFP-iso1 (Figure S7). Interestingly, ectopic expression of iso3 without ABA treatment did not strongly inhibit root elongation (Figure 6B and C). These experiments suggest that AREB2-iso3 prevents AREB2-iso1 from localizing to the nucleus in an ABA-dependent manner.
LincRNAs are expressed at low levels in individual cell types
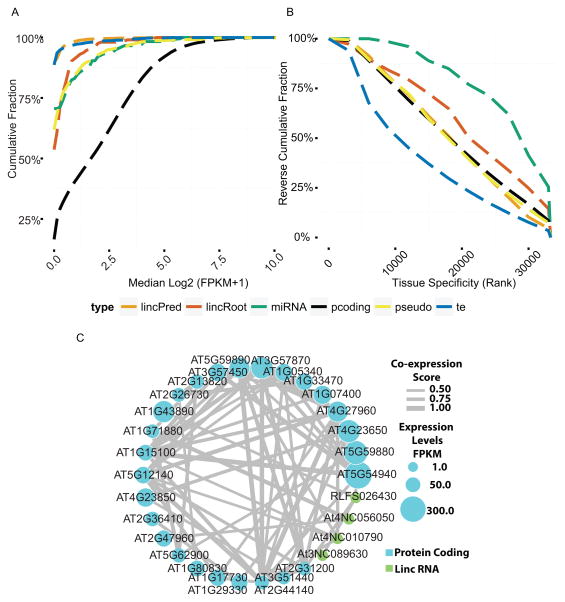
A large number of lincRNAs have been annotated, the majority of which are expressed at low levels in plant organ and seedling samples (Liu et al., 2012). It has been suggested that lincRNAs are likely to be highly cell-type specific and the signal is diluted when measured in whole organs. To test this hypothesis, we characterized both the median expression levels and tissue specificities of known lincRNAs and compared them to well-annotated components such as mRNA, pre-miRNA and transposable element transcripts (Experimental Procedures). Our cell-type specific data show that lincRNA expression is much lower than most mRNAs even in specialized cell types (e.g. At3NC081060, maximum coverage is 10 reads as shown in Figure 1D, and Figure 7A). This suggests that lincRNAs must perform whatever function they do at relatively low concentrations.
Figure 7.
Expression pattern of lincRNAs: (A) Median expression levels and (B) cell type specificity of differentially expressed RNA in the cell type-specific transcriptome. lincPred: lincRNA found in lincRNA database. lincRoot: lincRNA found in our data. (C) Co-expression network of mRNA and lincRNAs.
We identified 430 new lincRNAs not found in the lincRNA database (Liu et al., 2012). These newly identified lincRNAs have higher median expression levels and are slightly more cell type-specific than other lincRNAs (Figures 7A and 7B, Table S3). Furthermore, because the resolution of the previously used tiling array is limited by the location of the probes, we found 203 lincRNAs that overlap with previously annotated lincRNAs but extend either in the 5′ and/or 3′ directions. Overall, we found 1267 lincRNAs from the lincRNA database and in our dataset that are expressed consistently (>0.05 FPKM in all replicates per cell type) in at least one cell type. Among these genes, only 313 (25%) were found in more than half of the cell types.
Although thousands of lincRNAs have been predicted in the Arabidopsis genome, their functions remain mostly unknown. Our cell-type specific transcriptome data provides the first opportunity to identify lincRNAs and mRNAs that co-vary across multiple cell types. As a proof of concept, we constructed 26 co-expression networks of protein coding genes and lincRNAs that are differentially expressed across all cell types. For example, seven lincRNAs were found to be co-expressed with 26 protein-coding genes (Figure 7C). These co-expression clusters provide entry points to determine the function of the associated lincRNAs.
DISCUSSION
In metazoans, it is commonly believed that alternative splicing increases proteome diversity by mediating exon and domain recombination (Gogos et al., 1992). Our analysis indicates that major isoforms tend to have longer coding regions, suggesting that in plants, alternative splicing increases proteome diversity by shortening the coding sequences of major splice isoforms. This is the result of using upstream acceptor and donor sites for production of major isoforms and is favored by the kinetics of co-transcriptional splicing (Djebali et al., 2012, Ameur et al., 2011), which has not been directly observed in plants. Our results suggest a hierarchical model in which the major isoform is the primary product of the splicing machinery, which uses upstream donor and acceptor sites to produce a full-length coding sequence (Figure S2). Our analysis also shows that more than 80% of multi-isoform genes encode a single major isoform. These observations are in agreement with recent findings in human tissues and mammalian cell lines (Gonzalez-Porta et al., 2013). We found major isoforms tend to have better correlation with protein expression levels than minor isoforms, indicating that major isoforms are likely to play an important role in regulating protein expression levels.
We found that expression of alternatively spliced isoforms is more consistent across developmental zones than between cell types suggesting that alternative splicing may serve to regulate the process of differentiation. Consistent with this hypothesis, ectopic expression of a minor isoform of the transcription factor, AREB2 led to a change in the timing of differentiation. Splice isoforms of SR genes have been shown to regulate alternative splicing (Xing et al., 2015). Our data indicate that SR splice isoforms vary in expression across developmental zones and these expression differences are larger than differences previously detected among organs and developmental ages (Experimental Procedures and Figure S5). Thus, differential SR expression across developmental zones could be responsible for the observed coordinated regulation of alternative splicing
In Arabidopsis, intron retention has been reported to be the most prevalent splicing event (Ner-Gaon et al., 2004). A number of mechanisms such as intron translocation and transposable element-mediated intron formation have been proposed to explain the distribution and abundance of introns in eukaryotic genomes (Roy and Gilbert, 2006, Catania and Lynch, 2008). Our analysis provides evidence that, at least, some intron formation is coupled with cell type-specific partial exon excision, which is consistent with the observation that introns are more abundant in most multicellular organisms than in single cell organisms (Ast, 2004). Our results raise the possibility that these isoforms could emerge as cell type-specific splicing events that delete part of the complete coding region (Marquez et al., 2015). The short isoform is likely to perform a subset of the functions of the full-length protein and contribute to cell type-specific functions in a multicellular organism.
The low level of expression of numerous lincRNAs in whole plant organs (Liu et al., 2012) raised the possibility that they are primarily expressed in a cell-type specific manner. Our data indicates that even at the level of individual cell types, lincRNA are expressed at low levels. At the same time, our data set allows for co-expression clustering of lincRNAs and protein-coding RNAs providing numerous hypotheses as to functions for these RNA species.
In summary, this work provides a comprehensive map of cell type and developmental stage-specific expression of multiple RNA species within a developing organ, which can be used to guide functional characterization of both coding and non-coding RNAs. All expression data and gene annotations for newly assembled isoforms and non-coding RNAs are provided as supplemental information (Tables S3 and S5, Files S6 and File S7)
Experimental Procedures
Read mapping and analysis
The paired-end (2 × 100 bases or 2 × 125 bases) libraries were mapped to the Arabidopsis genome (TAIR10) and transcriptome using STAR version 2.4.2 (Dobin et al., 2013) and a GTF annotation file from the TAIR website (http://arabidopsis.org). Raw reads and alignment files will be uploaded to NCBI-SRA. See Supplemental Experimental Procedures for details about the alignment parameters. In the alignment files, only properly paired reads that mapped to unique genomic locations were kept. On average, 24 million reads mapped to one unique location in the genome, 17% of reads were spliced reads (Table S1).
Gene expression analysis
A simulation (Ramskold et al., 2009) was performed to determine the detection threshold in FPKM (fragments per kilobase per million reads) for protein coding genes in our data. For known coding regions, the number of read pairs in each annotated gene region was counted using featureCount (Liao et al., 2014) and gene expression levels were summarized as FPKM. We choose 0.05 as our threshold to determine the number of expressed genes in each sample (see Supplemental Experimental Procedures). For the gene family coverage analysis (Figure S1B), gene family annotations were downloaded from TAIR10.
Splice junction detection and alternative splicing discovery
We identified unannotated splice junctions using spliced reads by requiring a minimum of 8 nt mapped to the neighboring exon (Kim et al., 2013). We further required that a splice junction has to be supported by at least one read in two biological replicates for each sample to ensure that the putatively novel splice junctions are supported by biological replicates.
To identify local splicing events, the spliced reads from all libraries were transformed into a list of potential splice junctions. To discover novel alternative splicing events, splice junctions were grouped based on their spliced-out and spliced-in locations according to the following rules: 1) one splice junction can only belong to one group, 2) for each group, all splice junctions were within the 3′ and 5′ boundaries of the group and 3) in each group, any splice junction has to overlap with at least one other splice junction in the same group. Newly identified junctions, as well as known junctions, supported by RNA-seq reads from our data were exhaustively searched for these junction groups. For simple alternative donor and alternative acceptor events (Figure S2C), we first identified junction groups with only two splice junctions, and then required that the two splice junctions are spliced-out (or spliced-in) at the same genomic location and spliced-in (or spliced-out) at different genomic locations. For an exon-skipping event, there are three exons involved: a left exon, a right exon, and a middle exon. The left and right exons were included in both transcripts and the middle exon was not included in one of the alternative isoforms (Figure S2C). To identify exon skipping events, we required a combination of three splice junctions: a long splice junction (n1) that connects the left exon and the right exon, and two shorter splice junctions (s1 and s2) that connect the left exon to the middle exon and the middle exon to the right exon. Complex splicing patterns, such as tri-acceptors and tri-donors (Figure S2C), where three splice junctions spliced-out (or spliced-in) at the same location, were found to be rare (Figure 2B).
Whole gene model construction and expression analysis of major isoforms
Cufflinks version 2.1.1 (Trapnell et al., 2013) and StringTie 1.0.4 (Pertea et al., 2015) were used to assemble novel and known transcripts in each of the biological samples. The transcripts from each biological sample were combined into a unified set of transcripts using Cuffmerge. Cufflinks and StringTie were then used on each library to quantify the expression of individual isoforms and gene expression levels. Isoform expression levels are highly correlated between two different quantification methods: Cufflinks and StringTie (Table S1, average Pearson correlation 0.78). The Cuffnorm estimation of gene expression is highly consistent with the FPKM estimated by counting reads mapped to each gene (R2 > 0.97 for all comparisons). The assembled gene models that cover more than one known protein coding gene locus (from TAIR10) were excluded. A premature termination codon (PTC) is defined as the presence of a splice junction (SJ) greater than 50 nucleotides downstream of the stop codon, exactly as defined in (Drechsel et al., 2013). PSI (percentage spliced in) was calculated using FPKM values estimated by cufflinks. For each multi-isoform gene, the PSI is calculated as:
| (8) |
where yij is the expression of isoform j in cell type i.
For alternative acceptor sites and alternative donor sites, the number of reads supporting the shorter junction was divided by the total number of reads supporting both splice junctions (Figure S2D). Because a shorter splice junction implies a longer isoform, the number of reads supporting the shorter splice junction was used as numerator to calculate the longer isoform PSI (LPSI) for each alternative acceptor or donor event (Figure 3C, Figure S2D). For an exon skipping event, there are three types of splice junctions: one long junction across the skipped exon, and two shorter junctions define the boundary of the middle exon (Figure S2D). The PSI value is calculated using the average number of reads mapped to the two shorter junctions ((s1+s2)/2) divided by the sum of the total number of reads (n1 + (s1+s2)/2), as suggested (Barbosa-Morais et al., 2012).
Intron retention and evolutionary analysis
Intron retention events were extracted from the cufflinks assembled whole transcripts by comparing common minor isoforms to the corresponding dominant isoform. Type I intron retention events were found when the intron was retained in the common minor isoform, whereas the Type II intron retention events were found when intron retention occurs in the dominant isoform. Multiple sequence alignment of the 9 crucifier species (Yeo et al., 2012) was downloaded from http://mustang.biol.mcgill.ca:8885. Only intron retention events with a length in a multiple of three were used for the analysis. To determine the protein coding sequences for the intron retention events, the phases of codons for the dominant isoforms were determined from the TAIR annotation, and the phases of the codons in the intron retention isoform were determined according to the phase of the dominant isoform. For the intron sequences in other species, we used Arabidopsis as a template to translate the aligned nucleotide sequences into protein sequences.
LincRNA identification and expression
Known Arabidopsis lincRNAs (RepTAS predicted lincRNAs, RNAseq predicted lincRNAs and TAIR10 annotated lincRNAs) were downloaded from a lincRNA database (Jin et al., 2013). To identify intergenic transcribed regions from our RNAseq data, we performed the following analyses. We first used Cufflinks and Stringtie to construct full-length putative transcripts for each biological sample. We then used Cuffmerge to combine putative transcripts constructed in the first step into a unified set of transcripts. In this set of transcripts, those that overlap (as short as 1 base pair) with known protein coding genes were removed. The remaining transcripts were compared to the three types of lincRNAs found in the lincRNA database. The predicted lincRNAs from our data were merged with the known lincRNAs by extending the boundaries of overlapping lincRNAs to the furthest genomic location covered by either the lincRNA database predictions or by our RNAseq data. We then filtered the resulting lincRNAs using similar criteria as the lincRNA database: 1) transcribed regions longer than 200bp; 2) the longest open reading frame does not encode more than 100 amino acids; 3) transcribed regions are 500 bp away from any protein coding gene and 4) these transcribed regions do not overlap with any transposable elements. LincRNA expression was summarized in FPKM and differentially expressed LincRNAs were identified using edgeR (FDR<0.001). Differentially expressed LincRNAs with expression higher than 0.05 FPKM in more than half of our samples were used to calculate co-expression networks between mRNA (n=5000, genes with highest variation between samples) and lincRNA (n= 877) using WGCNA package with default parameters.
Supplementary Material
Research Highlights.
Single cell expression analyses characterize alt splicing and lincRNA expression
Splicing tends to remove parts of coding regions from a longer, major isoform
Coordinated alternative splicing appears to regulate differentiation in the root
For the majority of proteins, peptide and major isoform abundance correlate well
Acknowledgments
UO acknowledges support as a long-term visitor of the Spring 2016 Algorithmic Challenges in Genomics Program at the Simons Institute for the Theory of Computing at UC Berkeley. We thank Louisa Liberman for help with RNA sample preparation and library construction; Heather Belcher for help with cell sorting; Jingyuan Zhang for help with generating over-expression lines; George Yuan at Pacific Biosciences for help with data production; Polly Hsu, Erin Sparks, Eric Rogers, Cara Winter, Jaimie Van Newman and Xinnan Dong for comments on the manuscript; the Duke Genome Sequencing Shared Resource and the New York Genome Center for sequencing the Illumina libraries; the High-Throughput Sequencing Facility at University of North Carolina for sequencing the PacBio libraries; Will Thompson at the Duke Proteomics Core Facility for proteomics analysis; Kazuko Shinozaki at University of Tokyo for the seeds of the areb1, 2 and abf3 triple mutants and pAREB2-GFP-AREB2; Scott Langdon at Duke University DNA Analysis Facility for fragment analysis and Peyton Vaughn and Alan Cowles for help in setting up the computational environments. This work was funded by a grant to P.N.B. and U.O. from the NSF Arabidopsis 2010 program (IOS-1021619), and by the Howard Hughes Medical Institute and the Gordon and Betty Moore Foundation (through Grant GBMF3405) and from the NIH (R01-GM043778) to PNB.
Footnotes
Author Contributions
PNB and UO conceptualized the study; SL and UO designed the computational approaches; SL and XH performed the computational analyses, MY and XH performed bench experiments; All authors wrote and revised the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexa A, Rahnenfuhrer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22:1600–7. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- Ameur A, Zaghlool A, Halvardson J, Wetterbom A, Gyllensten U, Cavelier L, Feuk L. Total RNA sequencing reveals nascent transcription and widespread co-transcriptional splicing in the human brain. Nat Struct Mol Biol. 2011;18:1435–40. doi: 10.1038/nsmb.2143. [DOI] [PubMed] [Google Scholar]
- Ast G. How did alternative splicing evolve? Nat Rev Genet. 2004;5:773–82. doi: 10.1038/nrg1451. [DOI] [PubMed] [Google Scholar]
- Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ, Slobodeniuc V, Kutter C, Watt S, Colak R, et al. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012;338:1587–93. doi: 10.1126/science.1230612. [DOI] [PubMed] [Google Scholar]
- Barta A, Kalyna M, Reddy AS. Implementing a rational and consistent nomenclature for serine/arginine-rich protein splicing factors (SR proteins) in plants. Plant Cell. 2010;22:2926–9. doi: 10.1105/tpc.110.078352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum K, Jung JW, Wang JY, Lambert GM, Hirst JA, Galbraith DW, Benfey PN. Cell type-specific expression profiling in plants via cell sorting of protoplasts from fluorescent reporter lines. Nat Methods. 2005;2:615–9. doi: 10.1038/nmeth0805-615. [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN. A gene expression map of the Arabidopsis root. Science. 2003;302:1956–60. doi: 10.1126/science.1090022. [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318:801–6. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- Brown JB, Boley N, Eisman R, May GE, Stoiber MH, Duff MO, Booth BW, Wen J, Park S, Suzuki AM, et al. Diversity and dynamics of the Drosophila transcriptome. Nature. 2014;512:393–9. doi: 10.1038/nature12962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–27. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catania F, Lynch M. Where do introns come from? PLoS Biol. 2008;6:e283. doi: 10.1371/journal.pbio.0060283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–8. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drechsel G, Kahles A, Kesarwani AK, Stauffer E, Behr J, Drewe P, Ratsch G, Wachter A. Nonsense-mediated decay of alternative precursor mRNA splicing variants is a major determinant of the Arabidopsis steady state transcriptome. Plant Cell. 2013;25:3726–42. doi: 10.1105/tpc.113.115485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filichkin SA, Priest HD, Givan SA, Shen R, Bryant DW, Fox SE, Wong WK, Mockler TC. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 2010;20:45–58. doi: 10.1101/gr.093302.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–15. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- Gerstein MB, Rozowsky J, Yan KK, Wang D, Cheng C, Brown JB, Davis CA, Hillier L, Sisu C, Li JJ, et al. Comparative analysis of the transcriptome across distant species. Nature. 2014;512:445–8. doi: 10.1038/nature13424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogos JA, Hsu T, Bolton J, Kafatos FC. Sequence discrimination by alternatively spliced isoforms of a DNA binding zinc finger domain. Science. 1992;257:1951–5. doi: 10.1126/science.1290524. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Porta M, Frankish A, Rung J, Harrow J, Brazma A. Transcriptome analysis of human tissues and cell lines reveals one dominant transcript per gene. Genome Biol. 2013;14:R70. doi: 10.1186/gb-2013-14-7-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V, Estrada AD, Blakley I, Reid R, Patel K, Meyer MD, Andersen SU, Brown AF, Lila MA, Loraine AE. RNA-Seq analysis and annotation of a draft blueberry genome assembly identifies candidate genes involved in fruit ripening, biosynthesis of bioactive compounds, and stage-specific alternative splicing. Gigascience. 2015;4:5. doi: 10.1186/s13742-015-0046-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haudry A, Platts AE, Vello E, Hoen DR, Leclercq M, Williamson RJ, Forczek E, Joly-Lopez Z, Steffen JG, Hazzouri KM, et al. An atlas of over 90,000 conserved noncoding sequences provides insight into crucifer regulatory regions. Nature Genetics. 2013;45:891–U228. doi: 10.1038/ng.2684. [DOI] [PubMed] [Google Scholar]
- Irimia M, Blencowe BJ. Alternative splicing: decoding an expansive regulatory layer. Curr Opin Cell Biol. 2012;24:323–32. doi: 10.1016/j.ceb.2012.03.005. [DOI] [PubMed] [Google Scholar]
- Jin J, Liu J, Wang H, Wong L, Chua NH. PLncDB: plant long non-coding RNA database. Bioinformatics. 2013;29:1068–71. doi: 10.1093/bioinformatics/btt107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota K, Ye J, Nakai Y, Terada T, Shimizu K. ROKU: a novel method for identification of tissue-specific genes. BMC Bioinformatics. 2006;7:294. doi: 10.1186/1471-2105-7-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallberg M, Wang H, Wang S, Peng J, Wang Z, Lu H, Xu J. Template-based protein structure modeling using the RaptorX web server. Nat Protoc. 2012;7:1511–22. doi: 10.1038/nprot.2012.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyna M, Lopato S, Barta A. Ectopic expression of atRSZ33 reveals its function in splicing and causes pleiotropic changes in development. Mol Biol Cell. 2003;14:3565–77. doi: 10.1091/mbc.E03-02-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyna M, Simpson CG, Syed NH, Lewandowska D, Marquez Y, Kusenda B, Marshall J, Fuller J, Cardle L, Mcnicol J, et al. Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res. 2012;40:2454–69. doi: 10.1093/nar/gkr932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JY, Choi HI, Im MY, Kim SY. Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell. 2002;14:343–57. doi: 10.1105/tpc.010362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leviatan N, Alkan N, Leshkowitz D, Fluhr R. Genome-wide survey of cold stress regulated alternative splicing in Arabidopsis thaliana with tiling microarray. PLoS One. 2013;8:e66511. doi: 10.1371/journal.pone.0066511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–30. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Liu J, Jung C, Xu J, Wang H, Deng S, Bernad L, Arenas-Huertero C, Chua NH. Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell. 2012;24:4333–45. doi: 10.1105/tpc.112.102855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopato S, Kalyna M, Dorner S, Kobayashi R, Krainer AR, Barta A. atSRp30, one of two SF2/ASF-like proteins from Arabidopsis thaliana, regulates splicing of specific plant genes. Genes Dev. 1999;13:987–1001. doi: 10.1101/gad.13.8.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loraine AE, Mccormick S, Estrada A, Patel K, Qin P. RNA-seq of Arabidopsis pollen uncovers novel transcription and alternative splicing. Plant Physiol. 2013;162:1092–109. doi: 10.1104/pp.112.211441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez Y, Brown JW, Simpson C, Barta A, Kalyna M. Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Research. 2012;22:1184–95. doi: 10.1101/gr.134106.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez Y, Hopfler M, Ayatollahi Z, Barta A, Kalyna M. Unmasking alternative splicing inside protein-coding exons defines exitrons and their role in proteome plasticity. Genome Res. 2015;25:995–1007. doi: 10.1101/gr.186585.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ner-Gaon H, Halachmi R, Savaldi-Goldstein S, Rubin E, Ophir R, Fluhr R. Intron retention is a major phenomenon in alternative splicing in Arabidopsis. Plant J. 2004;39:877–85. doi: 10.1111/j.1365-313X.2004.02172.x. [DOI] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33:290–5. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petricka JJ, Schauer MA, Megraw M, Breakfield NW, Thompson JW, Georgiev S, Soderblom EJ, Ohler U, Moseley MA, Grossniklaus U, et al. The protein expression landscape of the Arabidopsis root. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:6811–8. doi: 10.1073/pnas.1202546109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramskold D, Wang ET, Burge CB, Sandberg R. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput Biol. 2009;5:e1000598. doi: 10.1371/journal.pcbi.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AS, Shad Ali G. Plant serine/arginine-rich proteins: roles in precursor messenger RNA splicing, plant development, and stress responses. Wiley Interdiscip Rev RNA. 2011;2:875–89. doi: 10.1002/wrna.98. [DOI] [PubMed] [Google Scholar]
- Robinson MD, Mccarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy SW, Gilbert W. The evolution of spliceosomal introns: patterns, puzzles and progress. Nat Rev Genet. 2006;7:211–21. doi: 10.1038/nrg1807. [DOI] [PubMed] [Google Scholar]
- Sun Q, Csorba T, Skourti-Stathaki K, Proudfoot NJ, Dean C. R-loop stabilization represses antisense transcription at the Arabidopsis FLC locus. Science. 2013;340:619–21. doi: 10.1126/science.1234848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–78. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–93. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T, Sugiyama N, Takahashi F, Anderson JC, Ishihama Y, Peck SC, Shinozaki K. Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci Signal. 2013;6:rs8. doi: 10.1126/scisignal.2003509. [DOI] [PubMed] [Google Scholar]
- Xing D, Wang Y, Hamilton M, Ben-Hur A, Reddy AS. Transcriptome-Wide Identification of RNA Targets of Arabidopsis SERINE/ARGININE-RICH45 Uncovers the Unexpected Roles of This RNA Binding Protein in RNA Processing. Plant Cell. 2015;27:3294–308. doi: 10.1105/tpc.15.00641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo ZX, Chan M, Yap YS, Ang P, Rozen S, Lee AS. Improving indel detection specificity of the Ion Torrent PGM benchtop sequencer. PLoS One. 2012;7:e45798. doi: 10.1371/journal.pone.0045798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoine M, Ohto MA, Onai K, Mita S, Nakamura K. The lba1 mutation of UPF1 RNA helicase involved in nonsense-mediated mRNA decay causes pleiotropic phenotypic changes and altered sugar signalling in Arabidopsis. Plant J. 2006;47:49–62. doi: 10.1111/j.1365-313X.2006.02771.x. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Fujita Y, Sayama H, Kidokoro S, Maruyama K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 2010;61:672–85. doi: 10.1111/j.1365-313X.2009.04092.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.