Abstract
Coronary artery disease and atherosclerosis are complex pathologies that develop over time due to genetic and environmental factors. Differential expression of miRNAs has been identified in patients with coronary artery disease and atherosclerosis, however, their association with cardiovascular disease risk factors, including hyperlipidemia, hypertension, obesity, diabetes, lack of physical activity and smoking, remains unclear. This review examines the role of miRNAs as either biomarkers or potential contributors to the pathophysiology of these aforementioned risk factors. It is intended to provide an overview of the published literature which describes alterations in miRNA levels in both human and animal studies of cardiovascular risk factors and when known, the possible mechanism by which these miRNAs may exert either beneficial or deleterious effects. The intent of this review is engage clinical, translational, and basic scientists to design future collaborative studies to further elucidate the potential role of miRNAs in cardiovascular diseases.
Keywords: miRNA, cardiovascular disease, coronary artery disease, risk factors, review
Introduction
Cardiovascular disease (CVD) is the leading cause of death for men and women in the western world. Coronary artery disease (CAD) is the most common form of cardiovascular disease and its incidence rate continues to rise across all populations independent of socio-economic status 1,2. CVD is a key cause of myocardial infarction3 and cerebrovascular accidents such as strokes and is associated with modifiable risk factors such as diabetes, obesity, hypertension, hyperlipidemia, sedentary lifestyle, and smoking.
In addition to known clinical risk factors, emerging research has established the presence of genetic predispositions to CAD. Single nucleotide polymorphisms (SNPs) are the most common form of genetic variation within the human genome, with more than 10 million SNPs having been identified 4. To date, more than 80 SNPs have been associated with acute coronary syndromes and related diseases5 suggesting that genetic susceptibility to CAD may play an important role in the development and severity of this disease. However, with the development of transcriptomics 6, a number of studies have revealed differential gene expression in cardiovascular diseases in the absence of identified SNPs7. These findings suggest that additional epigenetic factors including transcriptional and post-transcriptional regulation of gene expression may also play critical roles in the development of CAD and warrant further exploration.
A novel class of small RNAs known as microRNAs (miRNAs) were identified as important modulators of gene expression. miRNAs are evolutionarily conserved single-stranded non-coding RNAs (ncRNAs) approximately 20–22 nucleotides in length8,9. They function by binding to complementary sequences on messenger RNA (mRNA) and blocking translation of the message to protein. To date, over 1,880 unique human miRNAs have been identified and most are predicted to target and inhibit hundreds of genes10. Thus, miRNAs have the potential to modulate the expression of a large proportion of the human genome through post-transcriptional regulation. Further, recent work has demonstrated that miRNAs can be exported extracellularly either in exosomes or chaperoned by proteins and that extracellular miRNAs can be taken up by recipient cells and influence their cell functions11. Taken together, these observations suggest that miRNAs represent a crucial mechanism of epigenetic modification that can impact cellular function both locally and distantly. In this review, we provide background on miRNAs and discuss the potential associations between them and CVD with a specific focus on how known clinical risk factors may impact miRNA expression and contribute to the development of CVD.
miRNA synthesis, processing, and function
The initial discovery of the Lin-4 microRNA in Caenorhabditis elegans (C. elegans) was documented in 1993 by Ambros’ and Ruvkun’s research groups12,13. A loss of function mutation in lin-4 in these animals resulted in abnormal structure, sterility and dysregulation of developmental signaling pathways in late larval stages due to ongoing expression of the lin-14 gene targeted by lin-4. Then in 2000, Ruvkun’s laboratory reported that let-7, another non-coding RNA, targeted and inhibited the lin-41 gene14. Subsequent interest in the role of miRNAs in the gene regulation of other species prompted independent teams of investigators to identify a number of miRNAs in both the mouse and human genomes8,15. Since then, ongoing investigations have identified thousands of distinct miRNAs in a wide variety of species and have provided a better understanding of their biosynthesis, function, localization, and role in human health and disease.
miRNA genes are initially transcribed into RNA transcripts consisting of several hundred nucleotides (nt) and multiple stem-loop structures. These primary miRNA, or “pri-miRNA”, are ultimately processed into discrete individual stem-loop structures approximately 70 nt in length known as preliminary miRNA, or “pre-miRNA”, by the nuclear RNase Drosha and its binding partner DiGeorge Syndrome Critical Region 8 (DGCR8)12. Exportin 5 then couples with RanGTP to transport pre-miRNAs out of the nucleus and into the cytoplasm16–18. Once in the cytoplasm, pre-miRNAs dissociate from the exportin 5-RanGTP shuttle and are severed into shorter double-stranded RNA fragments by Dicer19. Following this final processing step, the Dicer-TRBP (the human immunodeficiency virus transactivating response RNA-binding protein) recruits argonaute 2 (Ago2) and additional proteins to bind the resultant ds-miRNA leading to formation of the RNA-induced silencing complex (RISC)20. The RISC separates the dsRNA into a guide strand and a passenger strand and facilitates binding of the guide strand to the 3’ untranslated region (UTR) region of mRNA targets. Binding of miRNA to mRNA is dependent upon sequence complementarity. Near perfect complimentary sequences between miRNA and mRNA leads to degradation by Ago2, while partial sequence matching prevents the binding of ribosomes to the mRNA21. Ultimately, loss of protein expression occurs through both mechanisms.
A growing body of evidence suggests that each miRNA has multiple targets and many mRNA are likely modulated by multiple miRNAs22. Further, miRNA expression patterns differ between healthy subjects and those with CVD and other cardiovascular diseases. It has become increasingly clearer that extracellular miRNAs reflect alterations in endogenous cells or tissues23.
Circulating miRNAs and their implications in cardiovascular disease
Cellular release of miRNAs
As early as 2008, scientists detected extracellular miRNAs in body fluids24. This observation led to the discovery that miRNA circulate in the bloodstream and, unlike most RNA, are remarkably resilient to degradation. Accumulating evidence suggests that endogenous circulating miRNA are protected from RNAse and other forms of degradation due to packaging in extracellular vesicles or association with RNA-binding protein complexessuch as Ago2 or lipoproteins including HDL and LDL 25,26. The mechanisms that govern the release of miRNA into the extracellular space are incompletely understood; however, the discovery that the contents of daughter exosomes differ from those of parent cells, suggests the existence of an active and selective transport process.
miRNA facilitates cell-to-cell communication
In vitro and in vivo studies have shed light on the potential role of extracellular miRNAs as mediators of communication between cells. Valadi et al. observed that extracellular miRNA encapsulated in exosomes can be taken up by cells and trigger changes in gene expression and function in these recipient cells11. Further, vesicular structures released from coronary artery endothelial cells were shown to restore endothelial homeostasis in mice with vascular injury through the delivery of miR-12627. As a growing body of evidence suggests that cellular stress responses lead to a robust release of extracellular microvesicles28, circulating miRNA may be abundant in the setting of a variety of diseases and may result in key alterations in any cells that internalize them. Accordingly, miRNA expression patterns differ between healthy subjects and those with CVD and other cardiovascular diseases. Whether these miRNAs play a role in the pathogenesis of the disease or only serve as biomarkers is a topic of considerable debate.
Circulating miRNAs as biomarkers
Biomarkers are used to assess both the presence and progression of disease as well as responses to treatment. miRNAs in the circulation are stable and easy to detect. However, their use as biomarkers have been challenging leading to difficulties in making scientific inferences. Effective isolation of miRNAs from plasma and tissue is contingent upon proper sample processing and handling techniques that minimize miRNA degradation and artefactual elevations. Due to variability in pre-analytic processing, which includes the method for the collection of the blood, circulating miRNA levels in diseased populations tend to vary across studies. The interpretation of the results is also confounded by the observation that the cells of interest often have multiple miRNAs so focusing on a single miNA could lead to a misinterpretation. Thus, well designed clinical trials validating the use of specific circulating miRNAs as biomarkers in cardiovascular diseases are needed. An overview of both clinical and basic science studies examining changes in microRNA profiles in response to CVD risk factors is discussed below and summarized in Table 1.
Table 1.
Association between differentially expressed microRNAs and CVD risk factors.
| Groups | miRNA | Source | Sample | Expression | Reference |
|---|---|---|---|---|---|
| Tobacco smoke exposure | miR-126-3p, 5p | Human | Plasma | Up-regulation | 49 |
| miR-126-3p | Human | Plasma | Up-regulation | 51 | |
| miR-126-5p, 101, 199, 34 | Rat | Lung | Down-regulation | 52 | |
| miR-223 | Human | Plasma | Up-regulation | 49 | |
| miR-223-5p | Human | Blood | Up-regulation | 55 | |
| miR-223 | Human | pPMVs | Down-regulation | 56 | |
| miR-223 | Rat | Lung | Down-regulation | 58 | |
| Hypertension | miR-320, 26b, 21 |
Rat | Aorta | Up-regulation | 64 |
| Rat | Aorta | Down-regulation | 64 | ||
| miR-208a | Rat | Plasma | Up-regulation | 72 | |
| Hyperlipidemia | miR-122 | Human | Plasma | Up-regulation | 80 |
| miR-30c | Bream | Liver | Up-regulation | 92 | |
| miR-33a/b | Human | Plasma | Unchanged | 80 | |
| miR-223 | Human | Serum | Up-regulation | 108 | |
| Obesity and diabetes | miR-26a | Human | Liver | Down-regulation | 112 |
| miR-26a, miR-126 | Human | cMPs | Down-regulation | 153 | |
| miR-103/107 | Mice | Liver | Up-regulation | 120 | |
| miR-143/145 | Mice | Adipose Liver | Up-regulation | 129 | |
| miR-802 | Human Mice | Liver | Up-regulation | 131 | |
| Physical activity | miR-29 | Rat | Heart | Up-regulation | 134 |
| MiR-1, 133a 133b | Human | Vastus lateralis | Down-regulation | 135 | |
| miR-222 | Human | Serum | Up-regulation | 140 | |
| miR-222,146a | Human | Serum | Up-regulation | 141 |
CVD, cardiovascular disease; miR, microRNA
Cardiovascular risk factors
Risk factors for cardiovascular disease are deemed as such due to their inherent correlation with cardiovascular events. The most prominent modifiable risk factors associated with cardiovascular disease include tobacco exposure, hypertension, hyperlipidemia, obesity, diabetes, and physical inactivity. Mechanisms whereby cardiovascular disease risk factors increase cardiovascular disease incidence and CVD mortality have been reviewed elsewhere29. Interestingly, studies examining differential regulation of microRNAs in the pathogenesis of cardiovascular disease are underway30. In this review, we discuss the known associations between miRNA expression levels and clinical risk factors of coronary artery disease. Our goal is to call attention to the possibility that miRNAs may be key determinants in gene expression and cell function in the setting of these risk factors and, therefore, may play important roles in the development of CAD. Moreover, by curating the miRNAs with known associations to CAD risk factors, we hope to facilitate future basic, translational and clinical research investigating the mechanistic role that these miRNAs may play in this highly prevalent and costly disease.
Tobacco exposure
Evidence suggests that tobacco exposure, including smoking or chewing, is a major risk factor for coronary heart disease and poses a threat to both the heart and blood vessels31,32. Cigarette smoke is comprised of over 4800 chemicals33 that contribute to the pathogenesis of disease. The relatively limited number of studies examining the impact of tobacco exposure on changes in miRNA expression suggest that smoking may mediate development of cardiovascular disease through promoting changes in microRNAs. Although cigarette smoke has pleiotropic effects, two smoke-induced hallmarks of CVD pathophysiology include loss of vascular tone and oxidative stress34,35. Vascular tone is maintained through proper endothelial barrier function which prevents deposition of plaque in the subendothelial space. miRNAs prevent obstruction of the endothelial barrier integrity through modulating proliferation, intercellular junction protein expression, nitric oxide production and preventing vascular permeability36. One of the most relevant microRNAs in regards to endothelial barrier function is miRNA-126.
MicroRNA-126 (miR-126) is encoded in the intron Egfl7 and is abundantly expressed in micro- and macrovascular endothelial cells37. Genetic deletion of the miR-126 gene negates vascular formation and angiogenesis during development37,38 and both its 3p and 5p strands have been identified as important determinants of endothelial homeostasis. MicroRNA-126-3p is a flow sensitive miR that enhances endothelial barrier integrity and prevents vascular permeability by modulating ERK and Akt signaling pathways37 through suppression of VCAM-1 39, SPRED-137, and PIK3R240. Whereas, microRNA-126-5p targets Dlk1 and subsequently maintains a proliferative phenotype in endothelial cells which prevents atherosclerotic plaque formation41.
Clinical investigations showed significantly elevated levels of circulating plasma miR-126 and miR-126-5p in smokers42 and patients with CAD43. XY Yu et al. found that higher circulating plasma miR-126 levels in dual antiplatelet-treated patients served as an independent risk factor for major adverse cardiovascular events44. Here, miR-126 levels significantly associated with increased time-to- a major cardiovascular event; however, the implications of miR-126 is the physiology of disease here were not studied. Overall, these studies suggest that enhanced miR-126 expression in smokers may serve as a compensatory mechanism for preventing endothelial barrier dysfunction associated with cardiovascular complications.
In vivo, Kalscheuer et al. previously demonstrated that rats exposed to 4- (methylnitrosamino)-1-(3-pyridyl)-1 butanone (NNK), a chemical found in tobacco smoke, for approximately 20 weeks, showed declines in miRNA-126-5p, miRNA-101, miRNA-199, and miRNA-34 in the lung tissue 45. The investigators additionally confirmed that NNK caused direct suppression of miRNA-126-5p levels suggesting this as a possible mechanism by which cigarette smoke impairs angiogenesis and increases the risk of atherosclerosis.
The miR-223 gene is located within the q12 locus of the X chromosome and is enriched in vascular smooth muscle cells important for maintaining the caliber of vascular tone throughout the body. Although the exact association between tobacco smoke and miR-223 expression is still under investigation, the role of miR-223 in vascular smooth muscle cell proliferation, migration, and vascular remodeling is well established46. miR-223 was previously shown to target RhoB, MLC2, and PARP-1 in order to modulate proliferation and apoptosis of vascular smooth muscle cells46. VSMC proliferation is a hallmark of atherogenesis47, a process that may be driven in smokers by increases in circulating miR-223. Whether miR-223 serves as only a biomarker of smoking and VSMC changes or participates in the disease process has not been established. Conflicting reports regarding changes in miR-223 expression in tobacco smoke exposed patients and animals exist. One group reported that circulating miR-223 is significantly increased in the plasma of smokers compared to healthy controls42. In addition, circulating miR-223-5p levels derived from maternal blood were shown to positively correlate with urine cotinine, a metabolite of nicotine48. Paradoxically, circulating miR-223 expression was decreased in platelet derived plasma microvesicles (pPMVs) in young healthy smokers49 which has previously been shown to promote platelet activation and lead to stroke development50. Similarly, Izzotti et al. reported that cigarette smoke significantly suppressed miR-223 expression in rat lung tissue51. These inconsistent reports indicate our poor understanding of how changes in circulating microRNAs coincide with changes at the tissue level. Moreover, the exact role of miR-223-3p and -5p in cigarette smoke induced changes should be resolved prior to considering these miRNAs as therapeutic agents or as biomarkers of disease.
Studies examining changes in the microRNA landscape in humans and animals exposed to cigarette smoke are fairly new thus, in vivo studies are limited. However, it is clear that cigarette smoke may induce a microRNA signature indicative of the pathophysiology of disease. Studies utilizing cigarette smoke-exposed mice with miR-223 genetically ablated may provide further insight on how miR-223 might contribute to cardiovascular abnormalities in the smoking microenvironment. The repeated discovery of a link between miR-223 and CVD suggests that tobacco modulation of miR-223 may be a mechanism by which tobacco smoke increases the risk of CVD. Further, bioinformatics analyses with experimental validation have identified FOXO1/3, MEF2C/2D, and insulin growth factor-1 receptor (IGF1R) as miR-223 targets with potential associations to cardiovascular disease52. IGF1R may have particular relevance as stretch-induced reduction in vascular smooth muscle cell (VSMC) miR-223 expression has been shown to lead to IFG1R-mediated VSMC proliferation and luminal narrowing.
Hypertension
Hypertension is a well-established risk factor for CVD. Mechanistically, it is believed that chronic hypertension leads to stress and injury of the vascular wall and subsequent vessel wall hypertrophy coupled with the deposition of atherosclerotic plaque53. Despite efforts to aggressively treat hypertension and mitigate the risk of CVD, only 52% of clinical hypertension cases are under control54 either due to non-compliance, lack of awareness, or treatment-resistant hypertension. Excessive sodium consumption is known to be associated with hypertension and has led to the recommendation of limited salt intake in patients with or at risk for hypertension. Additionally, this association has facilitated the development of animal models of salt-induced hypertension which have allowed for deeper investigations into the molecular underpinnings of hypertension including potential roles of microRNAs.
miR-320 has recently been implicated in CVD with studies showing its augmentation in the peripheral blood of patients with coronary artery disease and heart failure43,55. Environmental stressors ,such as high salt-intake, can perturb miR-320 expression leading to pathophysiologic consequences. miR-320 is an intergenic miRNA abundantly expressed in cardiomyocyte, endothelial cells and vascular smooth muscle cells56. MicroRNA analysis of Dahl Salt Sensitive (DSS) rat aortas revealed enhanced miR-320 expression and declines in miR-26b and miR-21. These changes coincided with decreased insulin growth factor-1 receptor (IGF-1R) and increased phosphatase and tensin homolog (PTEN) expression compared to controls57. Further investigation revealed that miR-320 targeted IGF-1R which led to impairments in vascular Akt/eNOS signaling. While suppression of miR-320 reduced expression of collagen in vascular smooth muscle cells, leading to prevention of hypertrophic responses to high salt57. Subsequent studies in ApoE−/− mice injected with mir-320a plasmids and fed a high-cholesterol diet demonstrated dyslipidemia and endothelial dysfunction, both of which facilitate atherosclerosis43. Similarly, knockdown of miR-320a in doxorubicin treated mice improved endothelial cell proliferation, reduced endothelial cell apoptosis, and reduced cardiac abnormalities58 suggesting that miR-320 antagonism may improve endothelial cell function that may beneficially alter the course of disease. Nonetheless, this body of work suggests that high levels of sodium chloride may lead to miR-320 upregulation, which promotes hypertension possibly through losses in insulin signaling and abnormal lipid profiles commonly observed in patients with and at risk for CVD. Thus, high salt intake leading to hypertension may also promote development of other common CVD risk. Studies examining circulating miR-320 levels in hypertensive patients are needed to determinine if miR-320 holds prognostic or diagnostic value as a biomarker of disease.
In hypertensive and coronary artery disease patients, myocardial hypertrophy serves as an independent risk factor for cardiovascular-related deaths59. The myocardium specific miR-208a/b, encoded by an intron found in the alpha myosin heavy chain (αMyHC) gene, modulates β-MyHC as well as the expression of sarcomeric contractile proteins60 and to-date has been implicated in coronary artery disease61. Genetic deletion of miR-208 proved sufficient to drive cardiac remodeling and heart failure, while subcutaneous delivery of LNA-modified anti-miR-208a circumvented cardiac remodeling, preserved cardiac function, and reduced mortality in a diastolic heart disease rat model62. Similarly, miR-208−/− mice displayed reduced cardiac hypertrophy in response to pressure overload63, while global overexpression of miR-208 induced hypertrophy in cardiac tissues60,62. Intriguingly, the role of miR-208a in salt-induced hypertension and subsequent vascular complications was substantiated using a Dahl salt-sensitive rat model in which a high salt diet increased plasma miR-208a levels in addition to cardiac remodeling and heart failure64. These data suggest that hypertension may drive cardiac abnormalities through the induction of miR-208a. These hypertrophic effects of miR-208a are mediated by their destabilizing effect on transcripts encoding hypertrophy repressors including myostatin and thyroid hormone-associated protein 1 (Thrap1/Med13)60.
In addition to myocardial specific effects, miR-208 has recently been indirectly linked to the endothelial function. Studies in Dahl salt-sensitive rats show that subcutaneous administration of LNA-modified anti-miR-208a resulted in the differential expression of additional miRNAs including elevations in plasma miR-19b65. miR-19b is predominately expressed in the endothelium and prevents TNFα-induced apoptosis by targeting Apaf1 and Casp7 genes. In addition, plasma miR-19b levels are suppressed in coronary artery disease patients and inversely correlate with TNF-α, Apaf1 and Casp7 proteins66. Thus, evidence suggest that miR-208a indirectly impacts endothelial function and may mediate hypertension-induced heart disease. Additional studies utilizing antagomiRs targeting miR-208 in salt-induced hypertensive models are needed to understand the therapeutic potential of miR-208 in preventing cardiovascular disease associated with hypertension and pressure overload.
Hyperlipidemia
A nutrient-poor diet, sedentary lifestyle, and in some cases, genetic predisposition often contributes to the development of CAD by altering the lipid profile in the blood with resultant hyperlipidemia. Clinically, patients with LDL levels higher than 160mg/dL and HDL levels under 40mg/dL meet the criteria for hyperlipidemia67, an established risk factor for cardiovascular disease risk. The liver modulates lipid levels through various epigenetic mechanisms including miRNAs. Studies have shown that decreased levels of hepatic cell dicer, the endoribonuclease involved in dsRNA cleavage, resulted in elevated cholesterol ester and triglyceride levels68. In addition, dicer deficient hepatocytes displayed mild elevations in free cholesterol and fatty acid levels68. Thus, proper regulation of miRNAs is critical for lipid homeostasis and prevention of coronary artery disease induced by lipid overload . Although the role of microRNAs in lipid metabolism have been reviewed extensively elsewhere30,69,70, here we outline the importance of microRNAs in lipid metabolism related to cardiovascular disease.
LDL metabolism
Current therapeutic approaches for lowering cholesterol are contigent upon the use of statins. Statins effectively lower cholesterol in 20–40% of individuals71. Thus, novel therapeutic targets important in the development of hypercholesterolemia are needed to improve patient outcomes. miRNA-122 accounts for 70% of microRNAs in the liver72, serves as a primary regulator of lipid biosynthesis, and aberrant levels have been implicated in plasma of hyperlipidemic patients with CAD73. Studies have demonstrated that miR-122-antagomir treated mice display reduced plasma cholesterol and a reduction HMG-CoA reductase (HMGCR) expression and activity74. Subsequently, two studies using miR-122 −/− mice and pharmaceutical antagonism of miR-122 in African Green monkeys demonstrated a reduction in plasma total cholesterol levels75,76. These in vivo effects of miR-122 antagonism resulted in suppression of essential fatty acid synthesis genes Srebp1, Fasn, acetyl-coA-carboxylase (Acc) 1 and 2, and staroyl-CoA desturase (Scd)76 which are possibly indirect targets of miR-122. These observations have led to speculation that miR-122 antagonism may serve as a therapeutic tool to prevent dyslipidemia and subsequent CAD. However, recent studies demonstrated that chronic miR-122 antagonism may promote triglyceride accumulation due to increases in the lipid biosynthesis enzyme 1-acylglycerol-3-phosphate O-Acyltransferase 1(AGPAT1) and cell death-inducing DFFA-like Effector C (CIDEC) which promotes lipid droplet formation and triglyceride storage77–79. Moreover, emerging research suggests that miR-122 may possess anti-tumor properties80,81. Thus, application of such therapies to treat hyperlipidemia must be considered with caution.
Microsomal triglyceride transfer protein facilitates lipoprotein synthesis by adding lipids to apolipoprotein B (apoB) to form precursors of LDL82. Hepatic microRNA-30c targets the 3’UTR region of MTP, modulating LDL synthesis. Mice transduced with miR-30c lentiviruses and fed a Western diet displayed reduced MTP expression, hepatic lipoprotein levels and plasma cholesterol83. Subsequent investigation revealed that miR-30c degraded MTP following transcription, ultimately lowering its activity levels84. miR-30c levels were increased in the liver of blunt snout bream fed a high-fat diet 85 which lead authors to hypothesize that enhanced miR-30c may act as a compensatory mechanism to prevent hyper-production of LDL. Coinciding with this hypothesis was the finding that miR-30c transgenic mice fed a western diet displayed lower plasma cholesterol and triglyceride-rich lipoproteins. The authors further showed that miR-30c might dually impact MTP and lysophosphatidlyglycerol acyltransferase 1 (LPGAT1) to suppress plasma lipid levels 86. Modulation of MTP by miR-30c suggests that miR-30c mimetics may serve as therapeutic agents for preventing diet-induced CAD. Further studies in mice and human subjects are needed to fully understand the impact of a high fat western diet on miR-30c expression and the role of miR-30c in CVD.
HDL metabolism
Although earlier reports implicated a role for high density lipoprotein (HDL) in the removal of cholesterol from the arteries and lowering the risk of cardiovascular disease 87, recent reports have refuted these claims suggesting that clear-cut evidence supporting the “HDL hypothesis” is missing88. Nonetheless, HDL aids in the mobility of miRNAs and reciprocally, miRNAs modulate HDL synthesis.
miR-33 (miR-33a or miR-33a-5p), embedded in the sterol regulatory element binding protein 1/2 (SREBP1/2) genes, is a known regulator of cholesterol homeostasis89. miR-33 suppresses cholesterol efflux through suppression of the ABC transporter A1 (ABCA1) and rodent Abcg1 sterol transporter genes90, while preventing the expression of a host of pro-fatty acid β-oxidation genes including CRPT, CPT1a, HADHB, and AMPKa91. Multiple studies have demonstrated that genetic ablation of miR-33 enhances plasma HDL levels92,93 and promotes accumulation of atheroprotective M2 macrophages and FOXP3+ T regs in plaques leading to reduction in plaque size in an AMPK dependent manner, independent of cholesterol efflux94.
Additional studies revealed that in non-human primates, pharmaceutical antagonism of miR-33 reduced VLDL triglycerides by 50% while plasma HDL levels increased by 40% at 12 weeks. The differential effects of miR-33 on HDL and VLDL are due to active depletion of the hepatocyte cholesterol pool by ABCA1-dependent lipid efflux to apolipoprotein A-1 yielding high levels of HDL. Consequently, cholesterol depletion resulted in attenuated VLDL secretion95. Genetic ablation of miR-33a in Apoe−/− mice displayed reduction in atherosclerosis progression96 while anti-miR-33a administration promoted atherosclerotic plaque regression in Ldlr −/− mice 93.
Controversy persists regarding the therapeutic potential of miR-33 antagonism as existing animal studies focused on short-term end points. Further, one study found that prolonged miR-33 antagonism increased circulating triglyceride levels and fat accumulation in the liver of mice fed a high-fat diet97. Moreover, others have shown that anti-miR-33 therapy does not circumvent atherosclerotic plaque development in Ldlr−/− mice98. Paradoxically, recent human studies demonstrated that miR-33a/b were not elevated in the plasma of patients with CAD73. Collectively, these studies suggest that pharmaceutical antagonism of miR-33a/b may not offer long-term efficacy as a therapeutic modality for dyslipidemia despite potential beneficial effects with short term use 99. Future studies examining mechanisms involved in the short- vs long-term suppression of miR-33 and its relationship with lipid metabolism are warranted.
Cholesterol homeostasis
Several microRNAs modulate cholesterol homeostasis for the prevention of dyslipidemia including miR-223. miR-223 is abundantly expressed in myeloid, endothelial100, and hepatic cells101. Recently, Vickers et al. demonstrated that miR-223 in Huh7 cells positively correlated with intracellular cholesterol levels. Further, miR-223−/− mice exhibited elevated plasma and liver cholesterol levels. Additional studies revealed that miR-223 directly targets SR-B1, HMGCS1, and methylsterol monooxygenase 1(SC4MOL) for modulation of HDL-C uptake and cholesterol biosynthesis102. The finding that miR-223 is elevated in CAD patient’s serum103 adds a layer of complexity to the role of miR-223 in the pathogenesis of CAD. One explanation is that miR-223 has pleiotropic physiologic roles thus, its upregulation in serum may serve as a biomarker of disease or compensatory mechanism rather than a direct modulator of the pathophysiology of disease. Accordingly, further studies are needed to fully elucidate the role of miR-223 in CAD.
Obesity and diabetes
According to the World Health Organization, obesity has more than doubled throughout the world since 1980104. To-date, more than 35% of Americans are considered obese and studies suggest that 42% of the population will develop obesity by 2050104. Obesity is defined as a body mass index (BMI) of 30 or higher and results primarily from excessive feeding behaviors and a sedentary lifestyle. Obese men and women have a significantly increased propensity for developing cardiovascular disease, hyperlipidemia, hypercholesterolemia, hypertension, and type II diabetes. The crosstalk between obesity and type II diabetes pathologies is modulated on the molecular level by post-transcriptional modulators including histone deacetylases (HDACs), histone acetyltransferase (HATs), and miRNAs.
miRNA processing is required for proper neuronal function and hormone signaling that modulates feeding behaviors. Conditional neuron-specific Dicer deletion promoted the development of obesity accompanied by hyperphagia, increased food efficiency, and decreased activity. However, this study did not examine adipogenesis or changes in the hormones leptin, insulin, and TNF-alpha105. Nonetheless, a number of studies have shown the impact of miRNAs on fatty acid oxidation and lipogenesis83,106.
Recent studies have shown that lean individuals have higher levels of miR-26a in the liver compared to overweight subjects107. In the same study, the authors observed that two independent obese mouse models displayed attenuated miR-26a expression compared to controls. Members of the miRNA-26 family are located in the gene encoding the carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase (CTDSP) family108. CTDSP negatively regulates RNA polymerase II (RNAPII) through dephosphorylation. miR-26a is known to modulate endothelial cell-associated angiogenesis109, cardiac hypertrophy110, and VSMC differentiation111, making it a major player in the preservation of cardiovascular health. miR-26a targets genes involved in the insulin signaling pathway (Glycogen synthase kinase 3 beta and Protein Kinase C-delta ), fatty acid metabolism (Acyl-CoA Snythetase Long-Chain Family Member 3 and 4), and gluconeogenesis (Phosphoenolpyruvate carboxykinase 1 and transcription factor 7-like 2)107. Studies in diabetic mellitus patients revealed lower circulating levels of miR-26a and miR-126 in circulating microparticles (MPs) and this phenotype conferred an independently higher risk for the development of CAD112. Additional studies in mice demonstrated that overexpression of miR-26a in mice fed a high fat diet improved insulin sensitivity113. Together, these data suggest that dysregulation of miR-26a in the setting of obesity could exacerbate the development of insulin resistance, type II diabetes, and subsequent vascular abnormalities leading to cardiovascular disease.
MicroRNAs 103/107 are located on chromosome 10 and are members of the miR-107 family involved in energy metabolism. miR-103 resides in the pantothenate kinase (PANK) intron while miR-107 resides intergenically114. PANK regulates pantothenate phosphorylation important for generation of Coenzyme A (CoA) involved in metabolic pathways. Liver analysis in obese ob/ob mice revealed enhanced miR-103/107 expression 115 associated with diet-induced obesity. Pharmacological attenuation of miR-103/107 further demonstrated improvements in glucose regulation and insulin sensitivity, suggesting that miR-103/107 are negative regulators of metabolic homeostasis. Investigation by Trajkovski et al. showed that the miR-103/107 may modulate insulin resistance through targeting caveolin-1 found in the cell membrane115. Although prior studies have highlighted a role for miR-103 in promoting adipogenesis116,117 the aforementioned studies did not address the cross talk between adipogenesis and insulin sensitivity pathways. These studies did, however, set the framework for development of microRNA therapeutics that could potentially improve insulin sensitivity while simultaneously reducing lipid accumulation in vivo.
MicroRNA 143/145 are differentially regulated in patients with coronary artery disease118, unstable angina, and acute myocardial infarction119. miRNA143/145 lie in close proximity on the human chromosome 5 and murine chromosome 18120–122. These genes are co-transcribed as a bicistronic transcript122 and appear to be modulated by the serum response factor SRF and Nkx2-5121. Targets of miR-143/145 include epidermal growth factor receptor, RAS, MAP kinase kinase , and extracellular signal-regulated protein kinases 1 and 2, all of which are related to cell proliferation. In fact, miRNA-143/145 are the primary regulators of vascular smooth muscle cell contractility, preventing VSMC proliferation and subsequent accelerated lesion formation123. Mice deficient in miR-143/145 develop atherosclerotic lesions spontaneously, even in the absence of elevated lipids120. Interestingly, in dietary mouse models of obesity, it was shown that miR-143 and miR-145 were upregulated in the adult adipose tissue and liver 124. Further, mice overexpressing miR-143 suppressed oxysterol-binding protein related protein (Orp) 8125 associated with enhanced adipocyte differentiation markers including PPAR gamma, aP2, and plasma leptin125. ORP8 and leptin suppress insulin sensitivity by preventing the insulin-mediated activation of AKT and PI3K, respectively125. Whether obesity triggers miR-143/145 as an adaptive mechanism for increased adipogenesis and subsequent depletion of excessive circulating lipids is debatable; however, the net effect of miR-143/145 signaling is suppression of atherogenesis and vascular remodeling.
MicroRNA-802 is increased in the liver of two obese mouse models and obese human subjects126. Overexpression of miR-802 suppresses insulin signaling and promotes hepatic gluconeogenesis by suppressing hepatocyte nuclear factor 1 homeobox B (Hnf1b). Hnf1b is a liver specific factor that modulates insulin secretion while simultaneously inhibiting reactive oxygen species (ROS) generation via PI3K/Akt and MEK/ERK signaling pathways127. Further, investigation by Kornfeld et al. revealed that overexpression of Hnf1b in Leprdb/db mice lead to improved insulin sensitivity and glucose tolerance126. Further analysis of the role of miR-802 in insulin signaling through Hnf1b is needed. Moreover, identification of potential atherogenic targets modulated by miR-802 are missing.
Physical activity
Lack of physical activity is commonly noted as a risk factor for the development of CAD. Physical activity is defined as an activity that requires physical exertion and typically leads to increased heart rate for a transient amount of time. The current American Heart Association recommendation for physical activity is 150 minutes per week of moderate exercise or 75 minutes per week of vigorous exercise. Following these recommendations is hypothesized to aid in the prevention of coronary heart disease and stroke by lowering blood pressure and cholesterol128.
It is well established that physical activity impacts cellular and molecular pathways involved in health and disease. Skeletal muscle and circulating miRNAs are differentially impacted by aerobic exercise and may provide insight on how the pathogenesis of disease is prevented through physical activity.
The general importance of miRNAs in exercise-mediated improvements in cardiovascular physiology was demonstrated by miR-29 induction and correlative declines in miR-1, miR-133a, and miR-133b in the left ventricle (LV) of exercised trained rats. The fact that miR-29 modulated collagen proteins was well illustrated by the correlative declines in COLIAI, COLIIIAI, and total collagen proteins upon exercise training induced miR-29 expression. Further, this study showed that collagen suppression was a key element in differentiating physiological from pathophysiological hypertrophy129 presumably because miR-1 and miR-133a have been previously characterized as pro-hypertrophic.
miRNA-1-1/133a-2 and miR-1-2/133a-1 are bicistronic expressed in skeletal muscle in addition to cardiomyocytes. The myomiRNAs miR-1, 133a, 133b, and 206 levels were reduced in the vastus lateralis of healthy men following 12 weeks of exercise130. Furthermore, Ringholm et al. observed reduced miR-133a following aerobic training. Aerobic training induced changes in miR-133 expression could be explained by the muscle adaptation processes that encompass a hypertrophic response131. Thus, short-term suppression of miR-133 may invite physiological adaptation to short term environmental stressors.
Aberrant levels of miRNA-222 have been identified in patients with CAD. miR-222 is a miRNA known to induce endothelial dysfunction, which precedes atherogenesis, by suppressing PGC-1α132 and STAT5A133. In CAD patients, miR-221/222 levels were drastically higher in EPCs compared to non-CAD patients134. Treatment with atorvastatin, a lipid lowering drug, lead to reductions in miR-221/222 expression in EPCs and increased the number of circulating EPCs in this patient population134. Bye et al. detected increased serum miR-222 in healthy individuals with low VO2max (a surrogate measure of aerobic fitness) vs patients with a high VO2max 135. Conversely, in a study examining the impacts of cycling and rowing on circulating miRNAs, investigators found that miR-222 and miR-146a were increased by acute exercise before and after training 136. Although these studies provide insight into alterations in microRNA in response to aerobic exercise and physical activity, the implications of these changes remains unclear. Routine physical exertion has consistently proven to improve cardiovascular health. Changes in miRNA levels may provide insight on the therapeutic potential of exercise and how it prevents development of cardiovascular disease on a molecular level. Clinical trials examining changes in circulating and tissue miRNA levels in response to physical activity are needed to further understand the prognostic and therapeutic value of these exercise-associated miRNAs.
Current challenges and future perspectives
Use of microRNAs as Biomarkers of disease
The discovery that microRNAs were present in nucleated blood cells, plasma, platelets, and erythrocytes opened up the opportunities to study their roles in pathological processes. Following investigation of microRNA stability, investigators observed that plasma microRNAs were extremely stable even under fluctuating conditions (ie changes in temperature, pH); whereas exogenous microRNAs added to samples were quickly degraded by plasma RNAses. This phenomenon is possibly due to the finding that plasma miRNAs are chaperoned by proteins such as AGO2 and the lipoprotein HDL. This observation provided substantial evidence for use of microRNAs as diagnostic tools. Multiple studies examined the diagnostic potential of microRNAs in various diseases as evidenced in Table 1; however, investigators quickly recognized obstacles that would interfere with the acquisition of reliable and reproducible cell-free plasma microRNA analysis.
One factor influencing the quality of microRNAs included high interference from proteins and other plasma components. Plasma contains proteins and other factors that contribute to coagulation. Thus, blood must be treated with citrate or some other anticoagulant to prevent loss of microRNAs137. Low quantities of microRNA in plasma and serum may go undetected in standard analyses. Other challenges include variability in RNA isolation and PCR methodologies across studies, and inconsistencies in internal control or reference gene selection for fold change calculations in plasma and serum samples. Data normalization is a major complication that arises during RNA analysis due to changes in commonly selected housekeeping genes such as U6, likely to change under pathological conditions. Indeed, U6 should probably not be used as an internal standard. Some investigators have shown that using a spike-in normalization technique may serve as a reasonable alternative24. Accordingly, others have found that using plasma volume is the best way to account for variability in results138. Clearly, the method of collection and preparation can artifactually influence the results and must be taken into consideration when evaluating miRNAs as circulating biomarkers of pathologic processes.
microRNA therapies and targets
Since 2001, microRNAs have emerged as biomarkers and possible therapeutic targets for the diagnosis and treatment of diseases. Manipulation of RNA using miRNA mimics and antagomirs holds significant therapeutic potential for treating a variety of diseases. With recent technological advances, identification and validation of potential therapeutic miRNA targets are readily available. However, delivery of anti-miRs and microRNAs in vivo may prove to be challenging. Various delivery and targeting methods exist for miRNA therapeutics and will be discussed below.
Although endogenous miRNAs are incredibly stable in plasma due to their association with RNA “protective” proteins, exogenous miRNA are rapidly degraded due to ribonucleases present in the blood. Thus, chemical modifications of phosphodiester oligodeoxynucleotides (ODNs) such as locked nucleic acid (LNA)139, 2’-O-methyl-(2’O-Me) or 2’-O-methoxyethyl—oligonucleotides (2’-O-MOE)140 have been used as one method to prolong the half-life of these nucleic acids to improve therapeutic efficacy. The first in-human phase I clinical trial using LNA was published in 2013141. The authors observed minimal changes in castration-resistant prostate cancer patients administered EZN-4176, designed to inhibit the hinge region of (exon 4) of the androgen receptor141. On the contrary, a 2013 study published by Janssen et al. in the New England Journal of Medicine showed reductions in hepatitis C virus RNA levels with administration of Miravirsen, an LNA-modified DNA phosphorothiate antisense nucleotide that inhibits miR-122142. Phase I and II trials examining the efficacy of Miravirsen in Hepatitis C patients who are non-responsive to IFN-α treatment are currently ongoing143. In addition, a recent phase I clinical trial, conducted by Regulus Therapeutics, demonstrated that RG-101, a N-acetyl galactosamine (GalNAc)-conjugated-anti-miR-122 therapy, was safe and effective for treating HCV infection (www.regulusrx.com). Experimental design for a phase II clinical trial is currently in progress. Lipid-based delivery systems for RNAi and microRNA are one of the most commonly used systems in preclinical studies. MRX34, a liposomal-encapsulated miR-34 mimic replacement therapy, was proven to be safe and effective in ongoing phase I clinical trials144. Thus far, delivery of microRNAs and anti-miRs has proven challenging145. However, collaborative efforts amongst biochemist, geneticists, and bioengineers are advancing the development of microRNA-based therapies for cancer, autoimmune diseases, and atherosclerosis.
Conclusions
Patients with CAD have differentially expressed miRNAs due to a variety of signaling pathways activated during disease processes. Although there are a number of studies that outline the influence of risk factors on the pathogenesis of cardiovascular disease, epigenetic modifications initiated by these factors are becoming increasingly clearer.
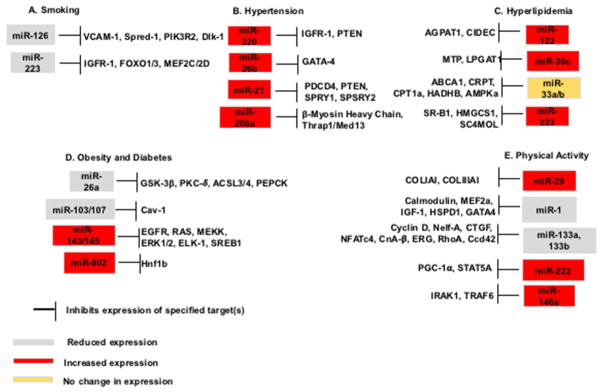
CAD risk factors are associated with differentially expressed microRNAs. However, extrapolation of these findings to understand clinically relevant pathologies is currently limited. Still, the clinical applicability of scientific discoveries in the miRNA field is promising as ongoing clinical trials using miRNAs as biomarkers of treatment efficacy are underway146. Development of disease diagnostic tools and “precise” therapies may greatly impact the epidemiological burden of CVD and CAD. Future studies are needed to determine the actual clinical value of measuring miRNAs in extracellular compartments such as plasma, serum, sputum, and urine. Further, although a number of targets of microRNAs have been identified as outlined in Figure 1, bioinformatics and basic science investigations are still needed to determine potential targets of microRNAs and their links to cardiovascular disease.
Fig. 1. The differential expression of selected microRNAs in the presence of cardiovascular risk factors and their respective targets.
(A) MicroRNAs differentially regulated in the presence of cigarette smoke or cigarette smoke by-products and their targets. (B) Increased expression of microRNAs found in hypertensive patients and mouse models and their cardiovascular disease related targets. (C) Hyperlipidemic and cholesterol metabolism associated microRNAs and their specified targets. (D) In patients with obesity and/or diabetes, miR-26a and miR-103/107 are reduced in expression, whereas miR-143/145 and miR-802 are increased in expression. These microRNAs play a role in insulin signaling and atherosclerotic plaque formation. (E) Physical inactivity and exercise impact the development of cardiovascular disease. MicroRNAs shown to response to changes in exercise include miR-29, miR-1, miR-133a, 133b, miR-222, and miR-146a. MicroRNAs in grey are down-regulated, those in red are upregulated, and microRNAs highlighted in yellow are not changed under specified conditions.
In conclusion, the impact of cardiovascular disease risk factors on miRNA expression and vice versa is novel and should be further investigated. Underpinnings of CAD modulated by risk factor mediated changes in miRNA may provide insight on CAD and CVD prevention in the future. Moreover, differential regulation of miRNAs may unveil pathways involved in the development of CAD independent of conventional risk factors.
Key points.
MicroRNAs play a potential major role in the development of cardiovascular diseases.
Cardiovascular disease risk factors differentially modulate microRNA expression in human and mouse models of disease.
Inferences made from observations of differentially expressed microRNAs in plasma and tissues must be made with caution.
MicroRNAs may serve as a potential therapeutic target for alleviation of cardiovascular diseases.
Acknowledgments
Financial support
The authors are supported by the following: JNJB by National Heart, Lung, and Blood Institute 5T32HL007260-39, AJG by National Institutes of Health/National Center for Advancing Translational Sciences grants KL2 TR000060 and UL1 TR 000062 JAC by National Institutes of Health grant GMS 5R01GM027673; PVH by National Institutes of Health/National Center for Advancing Translational Sciences grant UL1 TR 000062, HF by National Institutes of Health grant GMS 1R01GM113995.
Footnotes
Conflict of interest
The authors declared they do not have anything to disclose regarding conflict of interest with respect to this manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gouda HN, Critchley J, Powles J, Capewell S. Why choice of metric matters in public health analyses: a case study of the attribution of credit for the decline in coronary heart disease mortality in the US and other populations. BMC Public Health. 2012;12:88. doi: 10.1186/1471-2458-12-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lawlor DA, Smith GD, Leon DA, Sterne JA, Ebrahim S. Secular trends in mortality by stroke subtype in the 20th century: a retrospective analysis. Lancet. 2002;360(9348):1818–1823. doi: 10.1016/S0140-6736(02)11769-7. [DOI] [PubMed] [Google Scholar]
- 3.Mozaffarian D, Benjamin EJ, Go AS, et al. Heart disease and stroke statistics--2015 update: a report from the American Heart Association. Circulation. 2015;131(4):e29–322. doi: 10.1161/CIR.0000000000000152. [DOI] [PubMed] [Google Scholar]
- 4.Green ED, Watson JD, Collins FS. Human Genome Project: Twenty-five years of big biology. Nature. 2015;526(7571):29–31. doi: 10.1038/526029a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ozaki K. Genetic background of acute coronary syndrome. Nihon Rinsho. 2010;68(4):615–620. [PubMed] [Google Scholar]
- 6.Sui W, Liu F, Chen J, Ou M, Dai Y. Microarray technology for analysis of microRNA expression in renal biopsies of lupus nephritis patients. Methods Mol Biol. 2014;1134:211–220. doi: 10.1007/978-1-4939-0326-9_16. [DOI] [PubMed] [Google Scholar]
- 7.Abi Khalil C. The emerging role of epigenetics in cardiovascular disease. Ther Adv Chronic Dis. 2014;5(4):178–187. doi: 10.1177/2040622314529325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 9.Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5(5):396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- 10.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39(Database issue):D152–157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 12.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 13.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75(5):855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 14.Reinhart BJ, Slack FJ, Basson M, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403(6772):901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 15.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294(5543):858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 16.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21(17):4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18(24):3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303(5654):95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 19.Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15(20):2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hauptmann J, Meister G. Argonaute regulation: two roads to the same destination. Dev Cell. 2013;25(6):553–554. doi: 10.1016/j.devcel.2013.06.009. [DOI] [PubMed] [Google Scholar]
- 21.Pratt AJ, MacRae IJ. The RNA-induced silencing complex: a versatile gene-silencing machine. J Biol Chem. 2009;284(27):17897–17901. doi: 10.1074/jbc.R900012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olson EN. MicroRNAs as therapeutic targets and biomarkers of cardiovascular disease. Sci Transl Med. 2014;6(239):239ps233. doi: 10.1126/scitranslmed.3009008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee Y, El Andaloussi S, Wood MJ. Exosomes and microvesicles: extracellular vesicles for genetic information transfer and gene therapy. Hum Mol Genet. 2012;21(R1):R125–134. doi: 10.1093/hmg/dds317. [DOI] [PubMed] [Google Scholar]
- 24.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108(12):5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wagner J, Riwanto M, Besler C, et al. Characterization of levels and cellular transfer of circulating lipoprotein-bound microRNAs. Arterioscler Thromb Vasc Biol. 2013;33(6):1392–1400. doi: 10.1161/ATVBAHA.112.300741. [DOI] [PubMed] [Google Scholar]
- 27.Jansen F, Yang X, Hoelscher M, et al. Endothelial microparticle-mediated transfer of MicroRNA-126 promotes vascular endothelial cell repair via SPRED1 and is abrogated in glucose-damaged endothelial microparticles. Circulation. 2013;128(18):2026–2038. doi: 10.1161/CIRCULATIONAHA.113.001720. [DOI] [PubMed] [Google Scholar]
- 28.Emde A, Hornstein E. miRNAs at the interface of cellular stress and disease. EMBO J. 2014;33(13):1428–1437. doi: 10.15252/embj.201488142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rader DJ, Daugherty A. Translating molecular discoveries into new therapies for atherosclerosis. Nature. 2008;451(7181):904–913. doi: 10.1038/nature06796. [DOI] [PubMed] [Google Scholar]
- 30.Feinberg MW, Moore KJ. MicroRNA Regulation of Atherosclerosis. Circ Res. 2016;118(4):703–720. doi: 10.1161/CIRCRESAHA.115.306300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gupta SP, Gupta MS, Moga RL. Smoking as a coronary risk factor--comparative evaluation of cigarette paper and bidi leaf. Indian J Med Sci. 1980;34(7):163–167. [PubMed] [Google Scholar]
- 32.Huxley RR, Woodward M. Cigarette smoking as a risk factor for coronary heart disease in women compared with men: a systematic review and meta-analysis of prospective cohort studies. Lancet. 2011;378(9799):1297–1305. doi: 10.1016/S0140-6736(11)60781-2. [DOI] [PubMed] [Google Scholar]
- 33.Hoffmann D, Hoffmann I, El-Bayoumy K. The less harmful cigarette: a controversial issue. a tribute to Ernst L. Wynder. Chem Res Toxicol. 2001;14(7):767–790. doi: 10.1021/tx000260u. [DOI] [PubMed] [Google Scholar]
- 34.Tang GJ, Wang HY, Wang JY, et al. Novel role of AMP-activated protein kinase signaling in cigarette smoke induction of IL-8 in human lung epithelial cells and lung inflammation in mice. Free Radic Biol Med. 2011;50(11):1492–1502. doi: 10.1016/j.freeradbiomed.2011.02.030. [DOI] [PubMed] [Google Scholar]
- 35.Csordas A, Bernhard D. The biology behind the atherothrombotic effects of cigarette smoke. Nat Rev Cardiol. 2013;10(4):219–230. doi: 10.1038/nrcardio.2013.8. [DOI] [PubMed] [Google Scholar]
- 36.Chamorro-Jorganes A, Araldi E, Suarez Y. MicroRNAs as pharmacological targets in endothelial cell function and dysfunction. Pharmacol Res. 2013;75:15–27. doi: 10.1016/j.phrs.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang S, Aurora AB, Johnson BA, et al. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15(2):261–271. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fish JE, Santoro MM, Morton SU, et al. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15(2):272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harris TA, Yamakuchi M, Ferlito M, Mendell JT, Lowenstein CJ. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc Natl Acad Sci U S A. 2008;105(5):1516–1521. doi: 10.1073/pnas.0707493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang J, Lan H, Huang X, Liu B, Tong Y. MicroRNA-126 inhibits tumor cell growth and its expression level correlates with poor survival in non-small cell lung cancer patients. PLoS One. 2012;7(8):e42978. doi: 10.1371/journal.pone.0042978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schober A, Nazari-Jahantigh M, Wei Y, et al. MicroRNA-126-5p promotes endothelial proliferation and limits atherosclerosis by suppressing Dlk1. Nat Med. 2014;20(4):368–376. doi: 10.1038/nm.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takahashi K, Yokota S, Tatsumi N, Fukami T, Yokoi T, Nakajima M. Cigarette smoking substantially alters plasma microRNA profiles in healthy subjects. Toxicol Appl Pharmacol. 2013;272(1):154– 160. doi: 10.1016/j.taap.2013.05.018. [DOI] [PubMed] [Google Scholar]
- 43.Chen C, Wang Y, Yang S, et al. miR-320a contributes to atherogenesis by augmenting multiple risk factors and down-regulating SRF. J Cell Mol Med. 2015;19(5):970–985. doi: 10.1111/jcmm.12483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu XY, Chen JY, Zheng ZW, et al. Plasma miR-126 as a potential marker predicting major adverse cardiac events in dual antiplatelet-treated patients after percutaneous coronary intervention. EuroIntervention. 2013;9(5):546–554. doi: 10.4244/EIJV9I5A90. [DOI] [PubMed] [Google Scholar]
- 45.Kalscheuer S, Zhang X, Zeng Y, Upadhyaya P. Differential expression of microRNAs in early-stage neoplastic transformation in the lungs of F344 rats chronically treated with the tobacco carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Carcinogenesis. 2008;29(12):2394–2399. doi: 10.1093/carcin/bgn209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zeng Y, Zhang X, Kang K, et al. MicroRNA-223 Attenuates Hypoxia-induced Vascular Remodeling by Targeting RhoB/MLC2 in Pulmonary Arterial Smooth Muscle Cells. Sci Rep. 2016;6:24900. doi: 10.1038/srep24900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bennett MR, Sinha S, Owens GK. Vascular Smooth Muscle Cells in Atherosclerosis. Circ Res. 2016;118(4):692–702. doi: 10.1161/CIRCRESAHA.115.306361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Herberth G, Bauer M, Gasch M, et al. Maternal and cord blood miR-223 expression associates with prenatal tobacco smoke exposure and low regulatory T-cell numbers. J Allergy Clin Immunol. 2014;133(2):543–550. doi: 10.1016/j.jaci.2013.06.036. [DOI] [PubMed] [Google Scholar]
- 49.Badrnya S, Baumgartner R, Assinger A. Smoking alters circulating plasma microvesicle pattern and microRNA signatures. Thromb Haemost. 2014;112(1):128–136. doi: 10.1160/TH13-11-0977. [DOI] [PubMed] [Google Scholar]
- 50.Duan X, Zhan Q, Song B, et al. Detection of platelet microRNA expression in patients with diabetes mellitus with or without ischemic stroke. J Diabetes Complications. 2014;28(5):705–710. doi: 10.1016/j.jdiacomp.2014.04.012. [DOI] [PubMed] [Google Scholar]
- 51.Izzotti A, Calin GA, Arrigo P, Steele VE, Croce CM, De Flora S. Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke. FASEB J. 2009;23(3):806–812. doi: 10.1096/fj.08-121384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haneklaus M, Gerlic M, O'Neill LA, Masters SL. miR-223: infection, inflammation and cancer. J Intern Med. 2013;274(3):215–226. doi: 10.1111/joim.12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heistad DD, Baumbach GL, Faraci FM, Armstrong ML. Sick vessel syndrome: vascular changes in hypertension and atherosclerosis. J Hum Hypertens. 1995;9(6):449–453. [PubMed] [Google Scholar]
- 54.Nwankwo T, Yoon SS, Burt V, Gu Q. Hypertension among adults in the United States: National Health and Nutrition Examination Survey, 2011–2012. NCHS Data Brief. 2013;(133):1–8. [PubMed] [Google Scholar]
- 55.Goren Y, Kushnir M, Zafrir B, Tabak S, Lewis BS, Amir O. Serum levels of microRNAs in patients with heart failure. Eur J Heart Fail. 2012;14(2):147–154. doi: 10.1093/eurjhf/hfr155. [DOI] [PubMed] [Google Scholar]
- 56.Wang XH, Qian RZ, Zhang W, Chen SF, Jin HM, Hu RM. MicroRNA–320 expression in myocardial microvascular endothelial cells and its relationship with insulin-like growth factor-1 in type 2 diabetic rats. Clin Exp Pharmacol Physiol. 2009;36(2):181–188. doi: 10.1111/j.1440-1681.2008.05057.x. [DOI] [PubMed] [Google Scholar]
- 57.Ling S, Nanhwan M, Qian J, et al. Modulation of microRNAs in hypertension-induced arterial remodeling through the beta1 and beta3-adrenoreceptor pathways. J Mol Cell Cardiol. 2013;65:127–136. doi: 10.1016/j.yjmcc.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 58.Yin Z, Zhao Y, Li H, et al. miR-320a mediates doxorubicin-induced cardiotoxicity by targeting VEGF signal pathway. Aging (Albany NY) 2016;8(1):192–207. doi: 10.18632/aging.100876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vakili BA, Okin PM, Devereux RB. Prognostic implications of left ventricular hypertrophy. Am Heart J. 2001;141(3):334–341. doi: 10.1067/mhj.2001.113218. [DOI] [PubMed] [Google Scholar]
- 60.Callis TE, Pandya K, Seok HY, et al. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J Clin Invest. 2009;119(9):2772–2786. doi: 10.1172/JCI36154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang J, Pei Y, Zhong Y, Jiang S, Shao J, Gong J. Altered serum microRNAs as novel diagnostic biomarkers for atypical coronary artery disease. PLoS One. 2014;9(9):e107012. doi: 10.1371/journal.pone.0107012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Montgomery RL, Hullinger TG, Semus HM, et al. Therapeutic inhibition of miR-208a improves cardiac function and survival during heart failure. Circulation. 2011;124(14):1537–1547. doi: 10.1161/CIRCULATIONAHA.111.030932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316(5824):575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 64.Dickinson B, Zhang Y, Petrick JS, Heck G, Ivashuta S, Marshall WS. Lack of detectable oral bioavailability of plant microRNAs after feeding in mice. Nat Biotechnol. 2013;31(11):965–967. doi: 10.1038/nbt.2737. [DOI] [PubMed] [Google Scholar]
- 65.Dickinson BA, Semus HM, Montgomery RL, et al. Plasma microRNAs serve as biomarkers of therapeutic efficacy and disease progression in hypertension-induced heart failure. Eur J Heart Fail. 2013;15(6):650–659. doi: 10.1093/eurjhf/hft018. [DOI] [PubMed] [Google Scholar]
- 66.Lin L, Zheng Y, Tu Y, et al. MicroRNA-144 suppresses tumorigenesis and tumor progression of astrocytoma by targeting EZH2. Hum Pathol. 2015;46(7):971–980. doi: 10.1016/j.humpath.2015.01.023. [DOI] [PubMed] [Google Scholar]
- 67.Expert Panel on Detection E, Treatment of High Blood Cholesterol in A. Executive Summary of The Third Report of The National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, And Treatment of High Blood Cholesterol In Adults (Adult Treatment Panel III) JAMA. 2001;285(19):2486–2497. doi: 10.1001/jama.285.19.2486. [DOI] [PubMed] [Google Scholar]
- 68.Sekine S, Ogawa R, Ito R, et al. Disruption of Dicer1 induces dysregulated fetal gene expression and promotes hepatocarcinogenesis. Gastroenterology. 2009;136(7):2304–2315. e2301–2304. doi: 10.1053/j.gastro.2009.02.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rotllan N, Price N, Pati P, Goedeke L, Fernandez-Hernando C. microRNAs in lipoprotein metabolism and cardiometabolic disorders. Atherosclerosis. 2016;246:352–360. doi: 10.1016/j.atherosclerosis.2016.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Willeit P, Skroblin P, Kiechl S, Fernandez-Hernando C, Mayr M. Liver microRNAs: potential mediators and biomarkers for metabolic and cardiovascular disease? Eur Heart J. 2016 doi: 10.1093/eurheartj/ehw146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cheng AY, Leiter LA. Implications of recent clinical trials for the National Cholesterol Education Program Adult Treatment Panel III guidelines. Curr Opin Cardiol. 2006;21(4):400–404. doi: 10.1097/01.hco.0000231412.15049.fb. [DOI] [PubMed] [Google Scholar]
- 72.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissuespecific microRNAs from mouse. Curr Biol. 2002;12(9):735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 73.Gao W, He HW, Wang ZM, et al. Plasma levels of lipometabolism-related miR-122 and miR-370 are increased in patients with hyperlipidemia and associated with coronary artery disease. Lipids Health Dis. 2012;11:55. doi: 10.1186/1476-511X-11-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Krutzfeldt J, Rajewsky N, Braich R, et al. Silencing of microRNAs In vivo with 'antagomirs'. Nature. 2005;438(7068):685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 75.Elmen J, Lindow M, Silahtaroglu A, et al. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008;36(4):1153–1162. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Esau C, Davis S, Murray SF, et al. miR-122 regulation of lipid metabolism revealed by In vivo antisense targeting. Cell Metab. 2006;3(2):87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 77.Hsu SH, Wang B, Kota J, et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest. 2012;122(8):2871–2883. doi: 10.1172/JCI63539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Takeuchi K, Reue K. Biochemistry, physiology, and genetics of GPAT, AGPAT, and lipin enzymes in triglyceride synthesis. Am J Physiol Endocrinol Metab. 2009;296(6):E1195–1209. doi: 10.1152/ajpendo.90958.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Keller P, Petrie JT, De Rose P, et al. Fat-specific protein 27 regulates storage of triacylglycerol. J Biol Chem. 2008;283(21):14355–14365. doi: 10.1074/jbc.M708323200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang B, Hsu SH, Wang X, et al. Reciprocal regulation of microRNA-122 and c-Myc in hepatocellular cancer: role of E2F1 and transcription factor dimerization partner 2. Hepatology. 2014;59(2):555–566. doi: 10.1002/hep.26712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xu Y, Xia F, Ma L, et al. MicroRNA-122 sensitizes HCC cancer cells to adriamycin and vincristine through modulating expression of MDR and inducing cell cycle arrest. Cancer Lett. 2011;310(2):160–169. doi: 10.1016/j.canlet.2011.06.027. [DOI] [PubMed] [Google Scholar]
- 82.Hussain MM, Rava P, Pan X, et al. Microsomal triglyceride transfer protein in plasma and cellular lipid metabolism. Curr Opin Lipidol. 2008;19(3):277–284. doi: 10.1097/MOL.0b013e3282feea85. [DOI] [PubMed] [Google Scholar]
- 83.Soh J, Iqbal J, Queiroz J, Fernandez-Hernando C, Hussain MM. MicroRNA-30c reduces hyperlipidemia and atherosclerosis in mice by decreasing lipid synthesis and lipoprotein secretion. Nat Med. 2013;19(7):892–900. doi: 10.1038/nm.3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Soh J, Hussain MM. Supplementary site interactions are critical for the regulation of microsomal triglyceride transfer protein by microRNA-30c. Nutr Metab (Lond) 2013;10(1):56. doi: 10.1186/1743-7075-10-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang D, Lu K, Dong Z, Jiang G, Xu W, Liu W. The effect of exposure to a high-fat diet on microRNA expression in the liver of blunt snout bream (Megalobrama amblycephala) PLoS One. 2014;9(5):e96132. doi: 10.1371/journal.pone.0096132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vickers KC, Moore KJ. Small RNA overcomes the challenges of therapeutic targeting of microsomal triglyceride transfer protein. Circ Res. 2013;113(11):1189–1191. doi: 10.1161/CIRCRESAHA.113.302732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chapman MJ. The potential role of HDL- and LDL-cholesterol modulation in atheromatous plaque development. Curr Med Res Opin. 2005;21(Suppl 6):S17–22. doi: 10.1185/030079905X59111. [DOI] [PubMed] [Google Scholar]
- 88.Vergeer M, Holleboom AG, Kastelein JJ, Kuivenhoven JA. The HDL hypothesis: does high-density lipoprotein protect from atherosclerosis? J Lipid Res. 2010;51(8):2058–2073. doi: 10.1194/jlr.R001610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Horie T, Ono K, Horiguchi M, et al. MicroRNA-33 encoded by an intron of sterol regulatory element-binding protein 2 (Srebp2) regulates HDL In vivo. Proc Natl Acad Sci U S A. 2010;107(40):17321–17326. doi: 10.1073/pnas.1008499107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Marquart TJ, Allen RM, Ory DS, Baldan A. miR-33 links SREBP-2 induction to repression of sterol transporters. Proc Natl Acad Sci U S A. 2010;107(27):12228–12232. doi: 10.1073/pnas.1005191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gerin I, Clerbaux LA, Haumont O, et al. Expression of miR-33 from an SREBP2 intron inhibits cholesterol export and fatty acid oxidation. J Biol Chem. 2010;285(44):33652–33661. doi: 10.1074/jbc.M110.152090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rayner KJ, Esau CC, Hussain FN, et al. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature. 2011;478(7369):404–407. doi: 10.1038/nature10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rayner KJ, Sheedy FJ, Esau CC, et al. Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J Clin Invest. 2011;121(7):2921–2931. doi: 10.1172/JCI57275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ouimet M, Ediriweera HN, Gundra UM, et al. MicroRNA-33-dependent regulation of macrophage metabolism directs immune cell polarization in atherosclerosis. J Clin Invest. 2015;2015 doi: 10.1172/JCI81676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sahoo D, Trischuk TC, Chan T, et al. ABCA1-dependent lipid efflux to apolipoprotein A-I mediates HDL particle formation and decreases VLDL secretion from murine hepatocytes. J Lipid Res. 2004;45(6):1122–1131. doi: 10.1194/jlr.M300529-JLR200. [DOI] [PubMed] [Google Scholar]
- 96.Horie T, Baba O, Kuwabara Y, et al. MicroRNA-33 deficiency reduces the progression of atherosclerotic plaque in ApoE−/− mice. J Am Heart Assoc. 2012;1(6):e003376. doi: 10.1161/JAHA.112.003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Goedeke L, Salerno A, Ramirez CM, et al. Long-term therapeutic silencing of miR-33 increases circulating triglyceride levels and hepatic lipid accumulation in mice. EMBO Mol Med. 2014;6(9):1133–1141. doi: 10.15252/emmm.201404046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Marquart TJ, Wu J, Lusis AJ, Baldan A. Anti-miR-33 therapy does not alter the progression of atherosclerosis in low-density lipoprotein receptor-deficient mice. Arterioscler Thromb Vasc Biol. 2013;33(3):455–458. doi: 10.1161/ATVBAHA.112.300639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dai Y, Wang D, Tian X, et al. Insights into the application of let-7 family as promising biomarker in cancer screening. Tumour Biol. 2015;36(7):5233–5239. doi: 10.1007/s13277-015-3180-1. [DOI] [PubMed] [Google Scholar]
- 100.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303(5654):83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 101.Vickers KC, Landstreet SR, Levin MG, et al. MicroRNA-223 coordinates cholesterol homeostasis. Proc Natl Acad Sci U S A. 2014;111(40):14518–14523. doi: 10.1073/pnas.1215767111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vickers KC, Remaley AT. HDL and cholesterol: life after the divorce? J Lipid Res. 2014;55(1):4–12. doi: 10.1194/jlr.R035964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schulte C, Molz S, Appelbaum S, et al. miRNA-197 and miRNA-223 Predict Cardiovascular Death in a Cohort of Patients with Symptomatic Coronary Artery Disease. PLoS One. 2015;10(12):e0145930. doi: 10.1371/journal.pone.0145930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Finucane MM, Stevens GA, Cowan MJ, et al. National, regional, and global trends in body-mass index since 1980: systematic analysis of health examination surveys and epidemiological studies with 960 country-years and 9.1 million participants. Lancet. 2011;377(9765):557–567. doi: 10.1016/S0140-6736(10)62037-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mang GM, Pradervand S, Du NH, et al. A neuron-specific deletion of the microRNA-processing enzyme DICER induces severe but transient obesity in mice. PLoS One. 2015;10(1):e0116760. doi: 10.1371/journal.pone.0116760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang H, Luo J, Zhang T, et al. MicroRNA-26a/b and their host genes synergistically regulate triacylglycerol synthesis by targeting the INSIG1 gene. RNA Biol. 2016:1–11. doi: 10.1080/15476286.2016.1164365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fu X, Dong B, Tian Y, et al. MicroRNA-26a regulates insulin sensitivity and metabolism of glucose and lipids. J Clin Invest. 2015;125(6):2497–2509. doi: 10.1172/JCI75438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gao J, Liu QG. The role of miR-26 in tumors and normal tissues (Review) Oncol Lett. 2011;2(6):1019–1023. doi: 10.3892/ol.2011.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Icli B, Wara AK, Moslehi J, et al. MicroRNA-26a regulates pathological and physiological angiogenesis by targeting BMP/SMAD1 signaling. Circ Res. 2013;113(11):1231–1241. doi: 10.1161/CIRCRESAHA.113.301780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wei C, Kim IK, Kumar S, et al. NF-kappaB mediated miR-26a regulation in cardiac fibrosis. J Cell Physiol. 2013;228(7):1433–1442. doi: 10.1002/jcp.24296. [DOI] [PubMed] [Google Scholar]
- 111.Leeper NJ, Cooke JP. MicroRNA and mechanisms of impaired angiogenesis in diabetes mellitus. Circulation. 2011;123(3):236–238. doi: 10.1161/CIRCULATIONAHA.110.003855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jansen F, Wang H, Przybilla D, et al. Vascular endothelial microparticles-incorporated microRNAs are altered in patients with diabetes mellitus. Cardiovasc Diabetol. 2016;15(1):49. doi: 10.1186/s12933-016-0367-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Guay C, Menoud V, Rome S, Regazzi R. Horizontal transfer of exosomal microRNAs transduce apoptotic signals between pancreatic beta-cells. Cell Commun Signal. 2015;13:17. doi: 10.1186/s12964-015-0097-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Finnerty JR, Wang WX, Hebert SS, Wilfred BR, Mao G, Nelson PT. The miR-15/107 group of microRNA genes: evolutionary biology, cellular functions, and roles in human diseases. J Mol Biol. 2010;402(3):491–509. doi: 10.1016/j.jmb.2010.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Trajkovski M, Hausser J, Soutschek J, et al. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474(7353):649–653. doi: 10.1038/nature10112. [DOI] [PubMed] [Google Scholar]
- 116.Xie H, Lim B, Lodish HF. MicroRNAs induced during adipogenesis that accelerate fat cell development are downregulated in obesity. Diabetes. 2009;58(5):1050–1057. doi: 10.2337/db08-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Xie H, Sun L, Lodish HF. Targeting microRNAs in obesity. Expert Opin Ther Targets. 2009;13(10):1227–1238. doi: 10.1517/14728220903190707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fichtlscherer S, De Rosa S, Fox H, et al. Circulating microRNAs in patients with coronary artery disease. Circ Res. 2010;107(5):677–684. doi: 10.1161/CIRCRESAHA.109.215566. [DOI] [PubMed] [Google Scholar]
- 119.Rayner KJ, Moore KJ. MicroRNA control of high-density lipoprotein metabolism and function. Circ Res. 2014;114(1):183–192. doi: 10.1161/CIRCRESAHA.114.300645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Boettger T, Beetz N, Kostin S, et al. Acquisition of the contractile phenotype by murine arterial smooth muscle cells depends on the Mir143/145 gene cluster. J Clin Invest. 2009;119(9):2634–2647. doi: 10.1172/JCI38864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cordes KR, Sheehy NT, White MP, et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460(7256):705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Xin M, Small EM, Sutherland LB, et al. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 2009;23(18):2166–2178. doi: 10.1101/gad.1842409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wei Y, Nazari-Jahantigh M, Neth P, Weber C, Schober A. MicroRNA-126, -145, and -155: a therapeutic triad in atherosclerosis? Arterioscler Thromb Vasc Biol. 2013;33(3):449–454. doi: 10.1161/ATVBAHA.112.300279. [DOI] [PubMed] [Google Scholar]
- 124.Takanabe R, Ono K, Abe Y, et al. Up-regulated expression of microRNA-143 in association with obesity in adipose tissue of mice fed high-fat diet. Biochem Biophys Res Commun. 2008;376(4):728–732. doi: 10.1016/j.bbrc.2008.09.050. [DOI] [PubMed] [Google Scholar]
- 125.Jordan SD, Kruger M, Willmes DM, et al. Obesity-induced overexpression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism. Nat Cell Biol. 2011;13(4):434–446. doi: 10.1038/ncb2211. [DOI] [PubMed] [Google Scholar]
- 126.Kornfeld JW, Baitzel C, Konner AC, et al. Obesity-induced overexpression of miR-802 impairs glucose metabolism through silencing of Hnf1b. Nature. 2013;494(7435):111–115. doi: 10.1038/nature11793. [DOI] [PubMed] [Google Scholar]
- 127.Quan X, Zhang L, Li Y, Liang C. TCF2 attenuates FFA-induced damage in islet beta-cells by regulating production of insulin and ROS. Int J Mol Sci. 2014;15(8):13317–13332. doi: 10.3390/ijms150813317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kraus WE, Bittner V, Appel L, et al. The National Physical Activity Plan: a call to action from the American Heart Association: a science advisory from the American Heart Association. Circulation. 2015;131(21):1932–1940. doi: 10.1161/CIR.0000000000000203. [DOI] [PubMed] [Google Scholar]
- 129.Melo SF, Fernandes T, Barauna VG, et al. Expression of MicroRNA-29 and Collagen in Cardiac Muscle after Swimming Training in Myocardial-Infarcted Rats. Cell Physiol Biochem. 2014;33(3):657–669. doi: 10.1159/000358642. [DOI] [PubMed] [Google Scholar]
- 130.Nielsen S, Akerstrom T, Rinnov A, et al. The miRNA plasma signature in response to acute aerobic exercise and endurance training. PLoS One. 2014;9(2):e87308. doi: 10.1371/journal.pone.0087308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ringholm S, Bienso RS, Kiilerich K, et al. Bed rest reduces metabolic protein content and abolishes exercise-induced mRNA responses in human skeletal muscle. Am J Physiol Endocrinol Metab. 2011;301(4):E649–658. doi: 10.1152/ajpendo.00230.2011. [DOI] [PubMed] [Google Scholar]
- 132.Xue Y, Wei Z, Ding H, et al. MicroRNA-19b/221/222 induces endothelial cell dysfunction via suppression of PGC-1alpha in the progression of atherosclerosis. Atherosclerosis. 2015;241(2):671–681. doi: 10.1016/j.atherosclerosis.2015.06.031. [DOI] [PubMed] [Google Scholar]
- 133.Dentelli P, Rosso A, Orso F, Olgasi C, Taverna D, Brizzi MF. microRNA-222 controls neovascularization by regulating signal transducer and activator of transcription 5A expression. Arterioscler Thromb Vasc Biol. 2010;30(8):1562–1568. doi: 10.1161/ATVBAHA.110.206201. [DOI] [PubMed] [Google Scholar]
- 134.Minami Y, Satoh M, Maesawa C, et al. Effect of atorvastatin on microRNA 221 /222 expression in endothelial progenitor cells obtained from patients with coronary artery disease. Eur J Clin Invest. 2009;39(5):359–367. doi: 10.1111/j.1365-2362.2009.02110.x. [DOI] [PubMed] [Google Scholar]
- 135.Bye A, Rosjo H, Aspenes ST, Condorelli G, Omland T, Wisloff U. Circulating microRNAs and aerobic fitness--the HUNT-Study. PLoS One. 2013;8(2):e57496. doi: 10.1371/journal.pone.0057496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Baggish AL, Hale A, Weiner RB, et al. Dynamic regulation of circulating microRNA during acute exhaustive exercise and sustained aerobic exercise training. J Physiol. 2011;589(Pt 16):3983–3994. doi: 10.1113/jphysiol.2011.213363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Costa MC, Leitao AL, Enguita FJ. MicroRNA profiling in plasma or serum using quantitative RTPCR. Methods Mol Biol. 2014;1182:121–129. doi: 10.1007/978-1-4939-1062-5_11. [DOI] [PubMed] [Google Scholar]
- 138.Cheng Y, Tan N, Yang J, et al. A translational study of circulating cell-free microRNA-1 in acute myocardial infarction. Clin Sci (Lond) 2010;119(2):87–95. doi: 10.1042/CS20090645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wahlestedt C, Salmi P, Good L, et al. Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. Proc Natl Acad Sci U S A. 2000;97(10):5633–5638. doi: 10.1073/pnas.97.10.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yoo BH, Bochkareva E, Bochkarev A, Mou TC, Gray DM. 2'-O-methyl-modified phosphorothioate antisense oligonucleotides have reduced non-specific effects in vitro. Nucleic Acids Res. 2004;32(6):2008–2016. doi: 10.1093/nar/gkh516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bianchini D, Omlin A, Pezaro C, et al. First-in-human Phase I study of EZN-4176, a locked nucleic acid antisense oligonucleotide to exon 4 of the androgen receptor mRNA in patients with castration-resistant prostate cancer. Br J Cancer. 2013;109(10):2579–2586. doi: 10.1038/bjc.2013.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Janssen HL, Reesink HW, Lawitz EJ, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368(18):1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 143.Lindow M, Kauppinen S. Discovering the first microRNA-targeted drug. J Cell Biol. 2012;199(3):407–412. doi: 10.1083/jcb.201208082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bouchie A. First microRNA mimic enters clinic. Nat Biotechnol. 2013;31(7):577. doi: 10.1038/nbt0713-577. [DOI] [PubMed] [Google Scholar]
- 145.Tutar L, Tutar E, Ozgur A, Tutar Y. Therapeutic Targeting of microRNAs in Cancer: Future Perspectives. Drug Dev Res. 2015;76(7):382–388. doi: 10.1002/ddr.21273. [DOI] [PubMed] [Google Scholar]
- 146.Jung EJ, Santarpia L, Kim J, et al. Plasma microRNA 210 levels correlate with sensitivity to trastuzumab and tumor presence in breast cancer patients. Cancer. 2012;118(10):2603–2614. doi: 10.1002/cncr.26565. [DOI] [PMC free article] [PubMed] [Google Scholar]