Abstract
Recent sequencing efforts in melanoma have elucidated many previously unknown molecular pathways and biological mechanisms involved in melanoma development and progression and have yielded a number of promising targets for molecular therapy. As sequencing technologies have become more sophisticated and have revealed an ever-increasing complexity of the genetic landscape of melanoma, it has become clear that sequencing methods applied to clinical specimens have to reliably capture not only recurrent “hotspot” mutations like BRAFV600 and NRASQ61 or “mini-hotspot” mutations like exon 11 and 13 c-KIT but also heterogeneous somatic mutations dispersed across multiple functionally conserved regions of genes or entire genes. One such example in melanoma is the ERBB4 receptor, or HER4, a member of the Erb receptor family, which has recently been shown to be a major oncogenic “driver” in melanoma. Mutated ERBB4 signaling activates both aberrant ERBB4 and PI3K-AKT signal transduction, mediates sensitivity to small-molecule inhibition with the dual-tyrosine kinase inhibitor lapatinib, and has recently also been implied in oncogenic glutamatergic signaling in melanoma. Mutations involving the ERBB4 gene act as “gain-of-function” mutations and predominantly involve the extracellular domains of the receptor. Additional sequencing efforts have recently identified recurrent mutations (“mini-hotspots”) or mutation clusters which affect the regulation of, e.g., ligand binding, arrangement of extracellular domain alignment, or intramolecular tether formation.
In this chapter, we describe the methods used to determine the mutation status of all exons of the ERBB4 gene in clinical specimens obtained from patients afflicted by metastatic melanoma. Upon slight modifications, this protocol can also be used for mutational analysis of other oncogenes affected by “non- hotspot” mutations dispersed across multiple exons. This sequencing technique has successfully been applied within a clinical trial selecting patients with ERBB4-mutant melanoma for lapatinib treatment. With the increasing emergence of low-frequency oncogenes affected by heterogeneous activating mutations located in different exons and regions this method will provide a mean to translate the promise of recently obtained genetic knowledge into clinical genotype-directed targeted therapy trials.
Keywords: Metastatic melanoma, Genomic landscape, Somatic mutations, DNA isolation, Sanger sequencing, ERBB4 gene, Marker predictive for treatment, Mutation validation, Treatment target
1. Introduction
Melanoma has become a “poster-child” during recent years for genetic discoveries and how such improved understanding of the dysfunctional genetic makeup of a solid organ cancer can translate into novel treatments for patients afflicted by this disease. It has been recognized early on that cutaneous malignant melanoma harbors as an environmentally caused solid organ cancer, a unique and complex mutation profile [1, 2]. Ultraviolet irradiation causes C → T and with less frequency CC → TT transitions, which give rise to a specific “UV mutational signature” in these skin cancers [3, 4]. UV mutational gene aberrations are more prevalent in melanoma arising from chronically sun-exposed areas (e.g., trunk, scalp) versus, e.g., acral melanomas. This unique mutation pattern suggests a “dose–response” relation of UV irradiation-induced skin damage and melanoma formation; however, recent whole-genome sequencing (WGS) efforts of acral melanoma have now also identified mutation patterns consistent with UV-induced DNA damage in melanomas arising in non-sun-exposed locations [5, 6]. Consistent with a causal function of UV irradiation-induced genetic changes, nearly all large-scale sequencing efforts carried out to date have found that melanoma, despite a large variation between individual tumors, harbors the largest number of genetic mutations (20–30 mutations/MB) among any solid organ cancer [1, 4, 7]. Interestingly, lung cancer, another environmentally induced malignancy, ranks second [7].
Since many of the large number of genetic variants identified in melanoma are “bystander” mutations and not involved in melanogenesis, the initial evaluation of newly discovered genetic variants is usually subject to a strict algorithm to determine if a particular mutation is a “driver” or a “passenger” mutation (reviewed in Walia et al. [7]). While there is no final agreement on the best approach to determine “driver” mutations without functional testing, the following three features are most commonly applied as surrogates for formal in vitro and in vivo testing:
“Hotspot” or cluster formation: One of the strongest indicators for a genetic variant to be a “driver” mutation is its detection at the same nucleotide or amino acid location in a larger specimen cohort. Such nonrandom, recurrent selection strongly suggests a growth advantage. Prime examples in melanoma are BRAFV600E, NRASQ61L, or K642E and L576P c-KIT mutations.
The N:S ratio: This ratio calculates the ratio of non-synonymous (NS) mutations which cause amino acid substitutions to synonymous (S) changes which do not. If a gene or a gene locus is more frequently affected by NS mutations than by S mutations compared to the known background mutation frequency in a particular melanoma sample, it is more likely that these NS mutations are due to positive selection and represent “driver” mutations.
Biochemical impact of NS mutations: The functional impact of an NS mutation can be further validated by bioinformatic analysis of its impact on protein structure and function. For example, mutations affecting evolutionary conserved regions or functionally preserved domains like kinase or ligand-binding domains are more likely to be involved in cancer progression. The scale-invariant feature transform (SIFT) score is one of the most commonly used tools to rank the impact of amino acid changes on protein function.
Prior to the introduction of exomic and next-generation sequencing technologies, the genomic landscape of melanoma started to unfold with the extension of sequencing efforts from candidate genes onto whole candidate gene families [7]. The protein kinase family was the first gene family to be systematically interrogated using Sanger sequencing: since melanoma samples were underrepresented in the initial screening effort, a repeat genetic screen focusing on the protein tyrosine kinase family was carried out [1, 8]. The frequent involvement of activating mutations in tyrosine kinase genes in other cancers and the early success of targeting these mutated tyrosine kinase cancer genes with small-molecule inhibitors, like the L858R mutation-harboring non-small-cell lung cancer treated with erlotinib or gefitinib, provided a sound rationale for this effort [9, 10]. This sequencing effort yielded a number of new and therapeutically exploitable genes affected by oncogenic mutations, most notably the receptor tyrosine kinase ERBB4, or HER4, gene, a novel major oncogene in metastatic melanoma (Subheading 1.2). Other sequencing efforts directed against candidate gene families included screens in tyrosine phosphatases, matrix metalloproteinases, or the “A disintegrin and metalloproteinase” (ADAM) gene family [11–14]. While results of these sequencing efforts still await the identification of novel dysregulated targets and pathways which might become candidates for treatments, results of the recently completed sequencing of the coding regions of the G-protein-coupled receptor (GPCR) family have elucidated novel biological pathways involved in melanoma development and progression which might open new avenues for novel treatments in the future: using effective exon capture, the GPCRs GPR98 and GRM3 were frequently affected by somatic mutations in 27.5 and 16.3 % of cases in a cohort of 80 melanoma samples [15]. GRM3, which has a mutational hotspot at Glu870Lys found in four independent individuals, was shown in a number of in vitro and in vivo studies to function as an oncogenic driver mediating activation of the MAPK pathway [15]. The GRM3 receptor belongs to the metabotropic glutamate receptor family which is expressed in the brain and in gliomas [16]. Further findings from a recent exomic analysis performed by the same group showed that the N-methyl-d-aspartate (NMDA) receptor unit ε-1G GRIN2A, which belongs to the ionotropic glutamate receptor family, is affected in 25.2 % of cases by somatic mutations and functions as a tumor suppressor [17]. This independently discovered high mutation rate in both the GRM3 and GRIN2A receptors as well as previously identified mutated genes in melanoma which are involved in glutamate signalings strongly implies glutamate signaling as a novel mechanism in melanoma progression (reviewed in Prickett TD, Samuels Y. 2012) [16, 17]. Other genes involved in glutamate signal transduction and affected by somatic mutations include NRG-1-induced ERBB4 signaling, components of post-synaptic density complex (PSD complex) genes like PYK2, and PLCB4, a protein immediately downstream of NMDA receptors; or the ephrin receptors (in particular ephrin receptors B2 and B6) [18–20]. These genotyping results from multiple independent studies do open a plethora of potential novel therapeutic approaches which are not limited to direct downstream NDMA receptor signal transduction inhibition, like MEK inhibition, but for instance also include disruption or indirect manipulation of glutamate signaling using NMDA receptor agonists (glutamate analogs), inhibitors of metabotropic glutamate receptors like BAY 36-7620 or LY341495, or agents modulating the paracellular homeostasis of glutamate like riluzole [16, 21–23]. The recent exomic study also revealed a striking oncogenic hotspot in the TRAPP gene [17]. The Ser722Phe hotspot of the TRAPP gene, which regulates c-MYC- and E2F1-mediated transcription, was shown to function as a “gain-of-function” oncogene in melanoma. However, when extending this analysis onto additional specimens only 6 out of 167 (~4 %) melanomas harbored this mutation [17]. This increasing detection of low-frequency or “cancer-hill” genes, in recent candidate gene family and exomic sequencing efforts, is currently posing one of several unresolved problems with respect to clinical translation of these sequencing studies [24]. WGS on the other hand offers the unique advantage of integrating point mutations and genomic aberrations like amplifications, deletions, and chromosomal translocations into comprehensive maps of the entire tumor genome. The results of the first large-scale whole-genome analysis on 25 metastatic melanoma and matched normal specimens emphasize the comprehensiveness of the WGS approach: first, the study elegantly showed that the mutation rate in metastatic melanoma is more accurately determined as an average mutation rate of ~30 per MB with enrichment of C → T transitions consistent with UV-induced genetic changes and, secondly, identified several new genes affected by somatic mutations or chromosomal rearrangements [4]. One of the novel findings of the study is the discovery that different genetic mechanisms can lead to PREX2 dysregulation in melanoma. PREX2, a regulator of PTEN and the PI3K-Akt pathway, was found to harbor somatic mutations in 14 % of cases, and mutated variants of PREX2 functioned as tumor suppressors in an in vivo model of melanoma [4, 25]. Additionally, several rearrangements and chromosomal translocations at the PREX2 locus were identified suggesting a positive selection for PREX2 dysregulation on a genomic level, which, when combined with the findings of somatic PREX2 mutations, suggests that multiple different genetic causes of tumor-driving mechanism mediated by PREX2 exist in melanoma.
1.1. Mutations Within the Tyrosine Kinase Family in Melanoma
One of the early gene discoveries from candidate gene family sequencing with potential successful translation into the clinic stems from the genetic screen of the protein tyrosine kinase family in melanoma [8]. In the initial discovery set of 29 melanomas, 19 out of 86 tyrosine kinases were found to have mutations in the kinase domain-encoding exons [8]. Tyrosine kinase genes affected by kinase domain mutations in the initial screen were then comprehensively sequenced in an additional cohort of 79 specimens. The tyrosine kinase Erb receptor family member ERBB4, or HER4, was found to be the most frequently mutated gene affected by somatic mutations in 19 % of cases. Other frequently mutated genes included FLT3 in 10 %, PTK2B in 10 %, the ephrin receptor B2 and B6 genes in 9 %, and TIE2 in 8 %. The availability of additional independent mutation analyses obtained from additional specimens on different platforms now allows the buildup of detailed, in-depth mutation maps of melanoma oncogenes like recently performed for the BRAF, NRAS, and CDKN2A gene [26]. This allows both independent validation of initial sequencing results and, biologically more important, gives insight into positive selection of melanoma-driving genetic events when examining recurrence and location patterns or biophysical properties of the evolving somatic mutation pattern. The ERBB4 gene is a good example demonstrating the value of such comparative genetic screening. The mutational ERBB4 profile is quite different from other oncogenes including other ERBB receptor family members currently targeted in the clinic: to date, targeted therapy efforts focus predominantly on “hotspots” of oncogenes, e.g., vemurafenib or trametinib for BRAFV600 [27, 28], imatinib mesylate for c-KIT exon 11 and 13 mutations in melanoma [29], erlotinib or gefitinib for EGFR L858R mutations in lung cancer [10], or soon GDC-0941 for PI3KCA E545K, E542K (both exon 9), or H1047R (exon 20) mutations in breast or colon cancer [30, 31] to name a few. In the original genetic screen of melanoma samples, only the E452K mutation was found twice in the ERBB4 gene (“mini-hotspot”); all other identified mutations were spread across multiple different functional domains of the genes (see Fig. 1) raising initially doubt about both the validity and future clinical applicability of these mutations as genetic markers. However, when adding recent findings from the above mentioned WGS efforts as well as sequencing results obtained from patients with metastatic melanoma enrolled onto a clinical trial (NCI-11-C-0048, NCT01264081; A Phase II Study of Lapatinib for the Treatment of Stage IV Melanoma Harboring ERBB4 Mutations (Subheading 1.3)) new patterns of recurrence as well as clustering of several of the originally identified missense mutations were identified indicating positive selection during melanoma progression [4, 17, 32, 33].
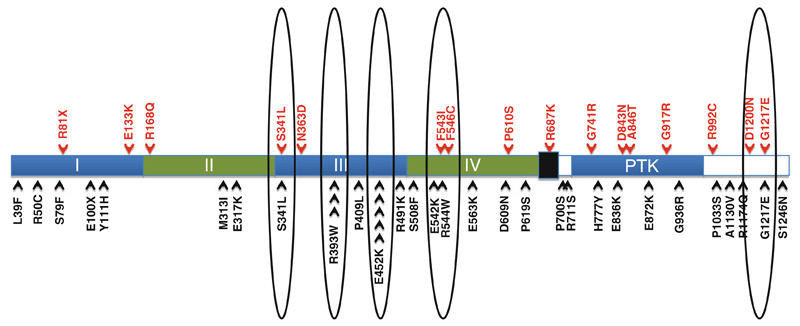
Fig. 1.
Evolvement of clustering and the development of recurrence patterns (“mini-hotspot” mutations; circles) of somatic mutations affecting the ERBB4 gene in melanoma upon combining sequencing results from candidate gene family sequencing and exomic and whole-genome sequencing studies (previously identified mutations listed as black arrows on bottom, mutations identified as part of the NIH, Clinical Genomic Core profiling (clinical trial samples), listed in red on top (unpublished))
Thus, as the majority of cancer genes currently discovered have a dispersed mutation profile without a clear recurrence pattern a robust algorithm applicable to clinical specimens will be necessary in order for these findings to be used for targeted therapy approaches in the future. In this respect, the presented ERBB4 mutation testing method which was designed to capture all “non-hotspot” mutations in all 28 exons of the ERBB4 gene should be applicable to a wider number of newly discovered oncogenes affected by heterogeneous mutations dispersed across multiple regions.
1.2. Somatic Mutations in the ERBB4 Gene
The distribution pattern of mutations affecting the ERBB4 gene is unique and substantially different from the mutation profile of other members of the Erb receptor family, e.g., EGFR or HER2. While activating somatic mutations of the EGFR, and to a lesser degree the HER2 receptor, predominantly affect the kinase domain of the gene, the majority of ERBB4 mutations in melanoma, or in lung cancer, are located in exons encoding the extracellular domains of the receptor [34, 35]. This mutation profile appeared initially more consistent with that of a tumor-suppressor gene than an oncogene. However, thorough functional assessment of eight independent ERBB4 mutants showed that all examined ERBB4 gene mutations drive melanoma biology and function as a “gain-of-function” oncogene [8]. Studies on ERBB4 mutants included kinase assays showing an increase in basal ERBB4 autophosphorylation, increase in cell transformation ability, and increase in foci formation. Most importantly, loss of ERBB4 signaling led to selective cell death in cells harboring mutated, but not wild-type, ERBB4 receptors indicating true gain of function and “oncogene” addiction to mutated ERBB4 signaling in these melanomas. To explain the fact that all examined ERBB4 mutations, which are dispersed across various regions of the gene, cause the same phenotype of “oncogene addiction” to mutant ERBB4 signaling, a closer look at the structure–function relationship between location of these mutations and the functional impact of the induced amino acid changes onto the involved receptor domains was taken. The described S341L and R393W mutations of the ERBB4 receptor, for example, involve structurally highly conserved motifs of the receptor L2 domain [36, 37]. These amino acid alterations are likely to either directly alter the kinetics of ligand binding or expose differently charged residues due to a different arrangement of the helices or β sheets which indirectly enhance the affinity of the ligand to the receptor possibly leading to receptor activation and increased signal transduction. Another mechanism of how the identified ERBB4 mutations might cause receptor activation might be the release of the intramolecular “tether” of the inactive receptor leading to exposure of the dimerization domain and activation of ERB signaling [35]. The dimerization domain is facing in the non-active state of the receptor facing towards domain III/IV of the receptor. Ligand binding triggers a major rearrangement of all extracellular domains involving a 270° outward twist of the dimerization region of receptor domain II [35]. Multiple residues have been identified to mediate this tether: for instance, Arg405 in EGFR forms a salt bond with Glu293, and it is conceivable that R393W or the previously identified P409L mutation, which involves functionally homologous regions of the ERBB4 molecule involved in tether regulation, might alter the equilibrium towards a more open and consecutively active form of the receptor [35, 37].
While to date for many of the confirmed activating mutations like the C-terminal G1217E or the domain III E452K “mini-hotspots” no structural correlate has been identified, these mutations have potential clinical value (Subheading 1.3) and should not be disregarded. Considering the heterogeneity of both the actual genetic variants as well as their response to small-molecule inhibition genetic testing must employ a technique which captures all mutations in these oncogenes. As many other genes mediating tumor progression harbor a similar mutation profile like the ERBB4 gene, the presented sequencing strategy should be applicable to the rapidly enlarging pool of novel genes involved in melanoma progression.
1.3. Mutations in the ERBB4 Gene Predict Sensitivity to Treatment with the Lapatinib in Melanoma
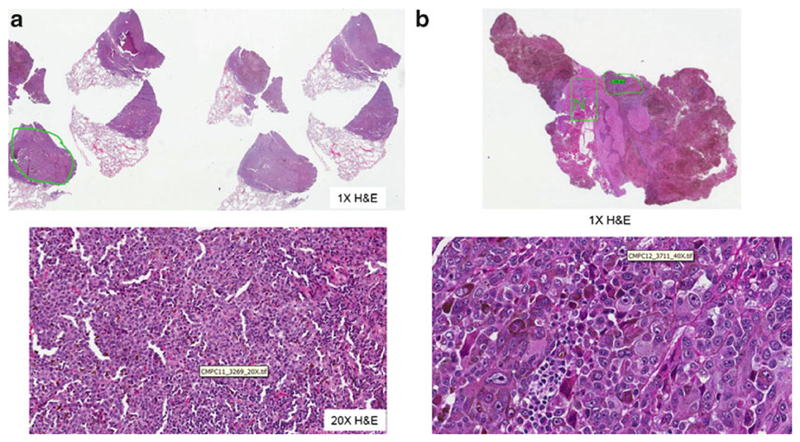
The clinical importance of the described sequencing protocol for ERBB4 mutations stems from the exquisite sensitivity of melanomas harboring ERBB4 mutations to the dual-tyrosine kinase inhibitor lapatinib. Melanomas harboring ERBB4 mutations have 10–250-fold lower inhibitory 50 concentrations to lapatinib, a dual-EGFR/HER2 small-molecule inhibitor which also has activity against ERBB4, than melanomas with wild-type ERBB4 receptors. While the exact mechanism of lapatinib-mediated mutated ERBB4 signal inhibition remains to be elucidated, treatment of ERBB4 mutant melanoma leads to decreased ERBB4 receptor autophosphorylation, decreased Akt signaling, and induction of apoptosis. Melanomas harboring mutant ERBB4 showed a wide range of sensitivity to lapatinib suggesting possibly several different mechanisms to drug inhibition, for example inhibition of HER2 mutant ERBB4 heterodimer formation. Several key observations with respect to results of preclinical lapatinib testing in ERBB4 mutant cell lines emphasize the need to employ a comprehensive sequencing strategy: first, all identified ERBB4 mutations which mediated addiction to ERBB4 signaling and functioned as a gain-of-function oncogene also showed sensitivity to lapatinib confirming that the somatic mutation status of the ERBB4 gene is a suitable biomarker for selecting patients for lapatinib. Therefore, based on currently available data, no ERBB4 mutation should be missed in order to erroneously exclude patients from possible tyrosine kinase inhibitor treatment. Secondly, inhibition of cell growth in ERBB4 mutant melanomas was achieved at clinically achievable concentrations of lapatinib with the exception of melanomas harboring two or more synchronous mutations (e.g., melanoma 7T or 71T in the original report) [8]. While melanomas with multiple ERBB4 mutations still show sensitivity to lapatinib treatment, lapatinib is less active in these cases and patients with two or more mutations in their ERBB4 gene were excluded from the above phase II clinical trial. Overall, the heterogeneity of ERBB4 mutations, which do not form classical “hotspot” mutations, as well as a select sensitivity profile to lapatinib demand a sequencing method which reliably captures somatic mutations in any of the 28 exons of the ERBB4 gene and has a turnaround time which allows its use in patients frequently presenting with dire needs for additional treatment options. Subheading 3 describes a protocol which is optimized to perform such whole candidate gene sequencing. As for other clinical molecular pathology testing described in this series, a detailed histopathological review is required to confirm diagnosis of metastatic melanoma and actual harvesting of melanoma-harboring tissue. Figure 2 shows examples of histopathological reviews of metastatic lesions from the lung and brain prior to “macrodissection” and isolation of genomic DNA.
Fig. 2.
Histopathological review and “macrodissection” of metastatic melanoma and matched normal tissue from surgically resected (a) lung metastasis and (b) brain metastasis
2. Materials
2.1. Equipment
ABI 3730 DNA Sequencer (Applied Biosystems).
Eppendorf Microcentrifuge or similar.
Eppendorf Thermomixer (24 × 2.0 mL).
Nanodrop Spectrophotometer ND-1000 to quantify and assess the quality of the DNAs.
VWR High-Speed Microplate Shaker.
DNA Engine Tetrad Thermal Cycler.
SORVALL Legend RT Plus Centrifuge.
2.2. Processing of Formalin-Fixed Paraffin-Embedded Tissue Blocks
Disposable biopsy punch with plunger (2 mm) (Miltex).
Ethyl alcohol (200 proof).
Micro tubes with screw cap (1.5 mL).
Sterile disposable safety scalpel.
Xylene substitute for deparafinizing formalin-fixed paraffin-embedded (FFPE) samples.
Tris–EDTA (TE) buffer (10 mM Tris-Cl, pH 7.5, 1 mM EDTA).
2.3. Genomic DNA Isolation
Proteinase K (600 AU/mL).
RNase A (7,000 U/mL).
QIAquick PCR Purification Kit contains QIAquick spin columns, Collection tubes, and PE Wash Buffer (Qiagen).
DNeasy Blood and Tissue Kit contains ATL tissue Lysis Buffer and AE Elution Buffer (Qiagen).
PM: Binding Buffer (Qiagen).
2.4. Array Comparative Genome Hybridization
Genomic DNA high-throughput ULS labeling kit (Agilent Technologies).
Oligo array comparative genome hybridization (aCGH)/ChiP-on-Chip Hybridization Kit (Agilent Technologies).
SurePrint G3 Hmn CGH 4x180K Oligo Microarray kit (Agilent Technologies).
2.5. Polymerase Chain Reaction (PCR Amplification) and PCR Cleanup
Titanium® Taq DNA Polymerase (Clontech).
MgCl2 solution (25 mM).
Nuclease-free water.
Exo-Sap It for cleanup of PCR products (USB/Affymetrix).
MicroAmp DNA/RNA/RNAse free adhesive film (Applied Biosystem).
2 % Polyacrylamide gel.
2.6. Sanger Sequencing
Ultrapure deionized H2O.
BigDye® Terminator v3.1 Cycle Sequencing Kit for PCR products including SAM solution (Applied Biosystems).
BigDye® Terminator v1.1, v3.1 5× Sequencing Buffer (Applied Biosystems).
Universal M13-F forward tgtaaaacgacggccagt and reverse M13-R caggaaacagctatgacc primers for DNA sequencing (IDT).
BigDye XTerminatorPurification Kit for DNA sequencing (Applied Biosystems).
3. Methods
3.1. Tissue Harvest of Metastatic Melanoma Deposits and Matched Normal
Manufacture Hematoxylin and Eosin (H&E)-stained slides from FFPE tissue blocks of metastatic melanoma lesions harvested surgically from any site.
Confirm diagnosis of melanoma, and select areas harboring tumor and normal surrounding tissue (see Fig. 2).
Using the marked H&E slide as a “template,” microdissect corresponding areas of the FFPE block using a biopsy punch or scalpel.
Transfer paraffin cores to 1.5 mL micro tubes with screw caps and deparaffinize using xylene substitute. Add 1 mL of xylene substitute to each tube that contains the dissected specimen and incubate at 65 °C for 15 min.
Centrifuge tubes at maximum speed in an Eppendorf microcentrifuge for 1 min, and carefully discard the xylene substitute.
Repeat processes once for a total of two xylene substitute incubation steps. After removing most xylene substitute from each tube, wash residual xylene substitute away with two ethanol washes.
Add 1 mL of 100 % ethanol to each tube at room temperature, mix well, and centrifuge at maximum speed before discarding ethanol.
Repeat ethanol wash step once with 50 % ethanol (50 % 1× TE). After discarding most of the 50 % ethanol, leave tubes open at room temperature for 10 min to let residual ethanol evaporate.
3.2. Genomic DNA Extraction
Lyze the deparaffinized tissues in 100 μL of ATL lysis buffer and 20 μL of proteinase K at 65 °C overnight (18–24 h) in an Eppendorf thermo mixer.
Add an additional 20 μL of proteinase K on the next day, and continue the lysis at 65 °C overnight (additional 18–24 h) in the Eppendorf thermo mixer.
Inspect the specimens to ensure complete lysis.
If undigested tissues are observed, add an additional 20 μL of proteinase K and incubate at 65 °C for an additional 2–4 h or until the tissues are completely digested.
Remove the tubes from the thermo mixer, add 2 μL of RNAse A to the lysates, and incubate at room temperature for 5 min.
Add buffer PM, three times the volume, to the lysates and pipette mix.
Apply the mixture to the QIAquick columns and centrifuge at 9,300 × g in an Eppendorf microcentrifuge for 1 min.
Reapply the flow-through to the columns, repeat step 7 to increase DNA yield, and then discard the flow-through.
Add 720 μL of PE wash buffer to the columns, centrifuge at 16,100 × g for 1 min to purify DNA that are selectively bound to the Qiaquick silica-based membrane, and then discard flow-through.
Add 720 μL of 80 % ethanol, centrifuge at 16,100 × g for 1 min for an additional purification step, and then discard flow-through.
Transfer the columns to new collection tubes and centrifuge at 16,100 × g for 5 min.
Transfer the columns to pre-labeled 1.5 mL Eppendorf tubes and allow to air-dry with open caps for 5 min at room temperature to rid all residual ethanol.
Apply 70 μL 10 % buffer AE to the columns, wait for 2 min, and then centrifuge at 16,100 × g for 1 min to elute the bound DNA.
Reapply the eluted DNA to the columns, wait for 2 min, and then centrifuge at 16,100×g for 1 min to increase DNA elution.
Measure the absorbance of the eluted DNAs at 230, 260, and 280 nm using NanoDrop to quantify and assess the quality of the DNAs.
(Optional) To determine the tumor fraction, label 500 μg of sample and reference genomic DNA using Agilent’s Genomic DNA High-Throughput ULS Labeling Kit and hybridize to Agilent’s SurePrint G3 Hmn CGH 4x180K Microarrays following the manufacturer’s instructions. Analyze the results with software Nexus 6. Detection of gross chromosomal aberrations is indicative of satisfactory tumor fraction for the acceptance of a negative (wild type) ERBB4 mutation result.
3.3. PCR Amplification of 28 Exons of ERBB4 Gene
A total of 31 pairs of primers were custom designed to cover all 28 exons of the ERBB4 gene, including their splice junctions, using Primer3, a primer design software publically available online (http://frodo.wi.mit.edu/) (Table 1). The primers all contained tagged M13 forward or reverse sequences for downstream Sanger sequencing reaction (Subheading 2.5). An additional primer pair to amplify exon 15 of the BRAF gene was also included in the assay as an internal control.
Table 1. PCR primers used for ERBB4 sequencing.
| Gene | Exon | Primer name | aForward primer | Primer name | aReverse primer |
|---|---|---|---|---|---|
| ERBB4 | 1 | ERBB4-X1b-F | GGGGATATGCCATTTGGAC | ERBB4-X1b-R | CGGAGTGCCAGAAGGAAC |
| ERBB4 | 2.1 | ERBB4-X2.1c-F | TAAAGGTTATGTAATTTCACGTGTATC | ERBB4-X2.1c-R | GCTGGTTATCTCCAGGTTGC |
| ERBB4 | 2.2 | ERBB4-X2.2c-F | AACAGCAGTACCGAGCCTTG | ERBB4-X2.2c-R | GCCACTGTCCATTCACAAAG |
| ERBB4 | 3 | ERBB4-X3b-F | GGCAACTGTTTGTGTCTTTTCA | ERBB4-X3b-R | AAGCATATTTGCCATTTTGGA |
| ERBB4 | 4 | ERBB4-X4b-F | TTCATCAACAAGCAGTTTGACA | ERBB4-X4b-R | TCGCCACATAGGGTAGAACA |
| ERBB4 | 5 | ERBB4-X5-F | AAATCCTCATAAAGGAGCAGGAG | ERBB4-X5-R | CCAAAGCAAATCAACCACAAG |
| ERBB4 | 6 | ERBB4-X6-F | TGAATTGAGTCAAAGACAGGGTG | ERBB4-X6-R | GGAATGACTTTGAGGAGGGC |
| ERBB4 | 7 | ERBB4-X7b-F | TGAAAGTAATATTTGCTGTGTTGC | ERBB4-X7b-R | TTCTTTTGATTTCAAATAATGACCT |
| ERBB4 | 8 | ERBB4-X8b-F | TGTTTTGAGCTTGTTTGCTGA | ERBB4-X8b-R | AAACCTTGTTATATAGGCCCAGTTC |
| ERBB4 | 9 | ERBB4-X9b-F | TTGGCCAAAAATAAGTTTCTCAA | ERBB4-X9b-R | CACTTTGTAAAATAACTTGCACAAAAA |
| ERBB4 | 10 | ERBB4-X10b-F | AAATTTGGCTACATCTCTTCTTGA | ERBB4-X10b-R | AAATTATATTGTTCATAGCGCAACA |
| ERBB4 | 11 | ERBB4-X11-F | CCTTTCTCACTTCCCAACTTTC | ERBB4-X11-R | TACCTCACACCATCATCGGAG |
| ERBB4 | 12.1 | ERBB4-X12.1c-F | TTCATGGTTCCGTTTTCTCA | ERBB4-X12.1c-R | AACACAGGTTGCTGTTGTCAG |
| ERBB4 | 12.2 | ERBB4-X12.2c-F | AATCAGCGCAGGAAACATCT | ERBB4-X12.2c-R | GGTCCAAAGAAGAATGGGAAA |
| ERBB4 | 13 | ERBB4-X13b-F | TCCCCTTGATTTTGGTGTTT | ERBB4-X13b-R | ATGAGGTGAAGGCAACCCTA |
| ERBB4 | 14 | ERBB4-X14-F | TGATGCTCCTGGCACATAGAG | ERBB4-X14-R | CCCATGGCATCCTGTAAGTAG |
| ERBB4 | 15 | ERBB4-X15c-F | GGAAATGACACCACTTTTCTTTTT | ERBB4-X15c-R | CCTTGGCCAGCAAGAATG |
| ERBB4 | 16 | ERBB4-X16c-F | GGATGGAGAATGATGTTTTCTG | ERBB4-X16c-R | TGAGCGACAAAATGGAAACA |
| ERBB4 | 17 | ERBB4-X17c-F | TTTTTCTCATTCTGGTTGTCAG | ERBB4-X17c-R | TCCCCAAACACATGAAGAGG |
| ERBB4 | 18 | ERBB4-X18b-F | TTCTTCTTTCCGCTTTGCAG | ERBB4-X18b-R | TCCATTGGCTATTATTTTCTAAACA |
| ERBB4 | 19 | ERBB4-X19b-F | TGTAACAGGTGCTAAATAAACATTTG | ERBB4-X19b-R | TGATTGCCTGGGTGTCTGTA |
| ERBB4 | 20 | ERBB4-X20b-F | TTGAGTTGAAATCATGGTATTGC | ERBB4-X20b-R | TTCCATAGAAATTGACAGGCACT |
| ERBB4 | 21 | ERBB4-X21b-F | GGGAAAACCTGGGCATTAAC | ERBB4-X21b-R | TCAAGCAAGATTGCTCTCAAAA |
| ERBB4 | 22 | ERBB4-X22c-F | CCAGCCCAAAGACTCACATT | ERBB4-X22c-R | CAAGCTTTAATTCGCAAAGAAGA |
| ERBB4 | 23 | ERBB4-X23b-F | TTGGTGTTTGGATTGACCTG | ERBB4-X23b-R | TGATGGTGATAACATTATTTTGCAG |
| ERBB4 | 24 | ERBB4-X24-F | GAGTCGTTTCTTTCACTAGCTTGC | ERBB4-X24-R | TGTTTGTGGTCCTTTCCACAG |
| ERBB4 | 25 | ERBB4-X25c-F | TGTGTCTGATGGGCAATCTT | ERBB4-X25c-R | TTATTTTGAAATGTTAGTGCTTATGAA |
| ERBB4 | 26 | ERBB4-X26b-F | CCATCATTCCATTTTTCTTTCC | ERBB4-X26b-R | AAGCAAAGACCGAAAATCCT |
| ERBB4 | 27 | ERBB4-X27b-F | ACAACGCCTTCTCTCCACAT | ERBB4-X27b-R | AATGGCGATCGTTTCTGAAT |
| ERBB4 | 28.1 | ERBB4-X28.1c-F | TTTTCCAGAAACTAGAGGTTAGCTG | ERBB4-X28.1c-R | CAGGGTTGTCAAACGCTTTC |
| ERBB4 | 28.2 | ERBB4-X28.2c-F | TGAAGAACAACATACTGTCAATGC | ERBB4-X28.2c-R | CAGGTGTGTCTCTCCACCTAAA |
| BRAF | 15 | BRAF-X15-F | TCATCCTAACACATTTCAAGCC | BRAF-X15-R | TTTGTGAATACTGGGAACTATGAAA |
Note: Every PCR primer contains a 5′ M13 forward (tgtaaaacgacggccagt) or reverse (caggaaacagctatgacc) tag
Adjust sample DNAs and a positive control DNA control containing at least one previously identified variant to a DNA concentration of 10 ng/μL, and prepare a minimum total volume of 65 μL for each sample.
Aliquot 2 μL of normalized DNA into 32 wells (four columns) in a 96-well reaction plate for each sample and the positive control.
Aliquot 2 μL of PCR-grade water into 32 wells for the notemplate control.
Aliquot 2 μL of 32 (2 μM) individual forward and reverse primer mixes into the separate wells for each sample and control.
Prepare reagent master mix containing the components listed in the table below, excluding DNA and primers, enough for all PCR reactions.
- Aliquot 16 μL the reagent mix into each individual well.
Reagent/material Volume (μL) Final concentration 10× Titanium Taq buffer 2 1× 50× dNTP mix (10 mM) 0.4 0.2 mM 50× Taq polymerase 0.4 1× MgCl2 (25 mM) 0.08 3.6 mMa Genomic DNA (10 ng/μL) 2 1 ng/μL Primer mix (2 μM) 2 0.2 μM PCR-grade water 13.9 – Total 20 aNote: In addition to MgCl2 in 10× buffer for final concentration of 3.6 mM Seal the PCR plate with a PCR-compatible DNA/RNA/RNase-free adhesive film.
Lightly vortex the PCR plate for 5 s.
Centrifuge the 96-well reaction plates for 30 s at maximum speed of 600 × g.
- Run the PCR reactions on a PTC-225 DNA Engine Tetrad Thermal Cycler with the following cycling conditions:
Step Temperature Time and cycles 1 95 °C 3 min 2 95 °C 30 s 3 61 °C 30 s 4 – Repeat steps 2–3 for a total of 5 cycles 5 95 °C 30 s 6 68 °C 1 min 7 – Repeat steps 5–6 for a total of 34 cycles 8 72 °C 5 min 9 4 °C Hold Upon completion of the PCR reaction, run 5 μL of each PCR amplification products on a 2 % gel electrophoresis.
Inspect gel image for the presence of single bands (expected size of 250–500 bp) indicative of successful PCR reactions (see Note 1).
3.4. PCR Product Cleanup
Add 6 μL of ExoSAP-IT into each remaining 15 μL of PCR products and pipette mix.
Seal the 96-well reaction plate and centrifuge for 30 s at maximum speed of 600 × g.
Incubate at 37 °C for 30 min to enzymatically degrade residual primers and dephosphorylate access dNTPs from the PCR step, and then incubate at 85 °C for 15 min to deactivate the enzymes.
3.5. Sequencing Reaction
Make 1:50 dilutions of the cleaned-up PCR products by adding 3 μL of the cleaned-up PCR products to 147 μL of 10 % buffer AE (see Note 2).
Aliquot 2 µL each of the diluted PCR products to two separate 96-well reaction plates, for both the forward and reverse sequencing reactions.
- Each Sanger sequencing reaction would contain the following components:
Reagent/material Volume (μL) Final concentration BigDye 3.1 (2.5×) 1 0.25× BigDye buffer (5×) 1.5 0.75× M13 F or R (10 μM) 0.32 320 nM Cleaned-up PCR (1:50) 2 1:250 PCR-grade water 5.18 – Total 10 - Run the sequencing reaction on the PTC-200 Peltier Thermal cycler with the following cycling conditions:
Step Temperature Time and cycles 1 96 °C 1 min 2 96 °C 10 s 3 50 °C 30 s 4 60 °C 4 min 5 – Repeat steps 2–4 for a total of 25 cycles 6 4 °C Hold
3.6. Sanger Sequencing Reaction Purification
Purify the sequencing reaction products using the BigDye XTerminator Purification Kit by adding 45 μL of SAM Solution and 10 μL of BigDye XTerminator Solution to each well.
Seal the reaction plate, put it on a microplate shaker, and vortex for 30 min at 600 × g.
Centrifuge the plates at 1,000 × g for 2 min.
3.7. Capillary Electrophoresis
Load the centrifuged plates containing purified Sanger sequencing products on the Applied Biosystems 3730 DNA Analyzer for capillary electrophoresis following standard instrument protocol.
Wait for approximately 3 h for individual runs to complete.
3.8. Sequence Analysis
Move .abl sequence files that were generated into a designated folder.
Import the sequence files into the Variant Reporter software for subsequent sequence analysis.
Import ERBB4 reference sequence NM005235.2 and BRAF reference sequence NG_007873.2. 3. Use the default sequence quality and mixed base threshold for the automatic alignment and variant calling steps (see Note 3).
Click the Analyze button in the Variant Reporter software for alignment to reference and variant calling (see Note 4).
Manually inspect each aligned sequence trace and variant that was called.
Validate each variant by an additional validation assay starting from the PCR amplification step (see Note 5).
4. Notes
The agarose gel electrophoresis served as a quality control (QC) step for the PCR reaction. For any given PCR reaction, when a single visible band of expected size was absent or when multiple bands or smears appeared in the gel image, the individual PCR reaction would be repeated. When similar problems occur to all PCR products of a particular sample, the DNA would be re-extracted.
For the sequencing reactions, background noise within the sequence traces that reduce confidence in variant calling will result in repeat of the sequencing reaction. Unless the PCR failed QC, the original PCR products were used in the repeat sequencing reactions at three different dilutions: 1:25, 1:50, and 1:100.
To avoid false-negative results due to insufficient tumor DNA fraction, a positive finding at the BRAFV600 hotspot, which should be found in ~50 % of melanoma cases, in addition to the evidence of positive copy number changes in the tumor genome which were almost always seen in melanoma cases, was used to confirm true negatives.
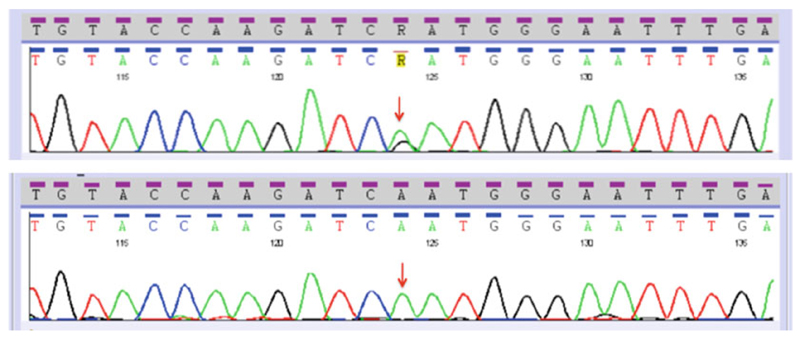
To determine if the identified variants were previously found in other cancers or if they were polymorphic genetic variations in the general population, they were searched against both the Catalogue of Somatic Mutations in Cancer (COSMIC) and the Single Nucleotide Polymorphism (dbSNP) databases. In addition, to determine whether they were somatic or germline variants, the assays starting from the PCR step were repeated using matched normal DNA samples for the particular ERBB4 exon harboring the identified variant (see Fig. 3).
To validate positive results when a variant is identified, an additional assay to confirm the result was run starting from the PCR amplification step. The positive result would be confirmed when the same variant was observed the second time.
Fig. 3.
Sanger sequencing traces for ERBB4 exon 9. Top panel shows the c.1185A>G p.N363D variant, and lower panel shows the matched normal tissue from the same patient. Arrow indicates the location of missense mutation
Acknowledgement
Yardena Samuels is supported by the ERC (StG-335377).
References
- 1.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446(7132):153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pleasance ED, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463(7278):191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drobetsky EA, Grosovsky AJ, Glickman BW. The specificity of UV-induced mutations at an endogenous locus in mammalian cells. Proc Natl Acad Sci U S A. 1987;84(24):9103–9107. doi: 10.1073/pnas.84.24.9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger MF, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485(7399):502–506. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curtin JA, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353(20):2135–2147. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 6.Turajlic S, et al. Whole genome sequencing of matched primary and metastatic acral melanomas. Genome Res. 2012;22(2):196–207. doi: 10.1101/gr.125591.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walia V, et al. Delving into somatic variation in sporadic melanoma. Pigment Cell Melanoma Res. 2012;25(2):155–170. doi: 10.1111/j.1755-148X.2012.00976.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prickett TD, et al. Analysis of the tyrosine kinome in melanoma reveals recurrent mutations in ERBB4. Nat Genet. 2009;41(10):1127–1132. doi: 10.1038/ng.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mok TS, et al. Gefitinib or carboplatinpaclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361(10):947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 10.Rosell R, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009;361(10):958–967. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- 11.Solomon DA, et al. Mutational inactivation of PTPRD in glioblastoma multiforme and malignant melanoma. Cancer Res. 2008;68(24):10300–10306. doi: 10.1158/0008-5472.CAN-08-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palavalli LH, et al. Analysis of the matrix metalloproteinase family reveals that MMP8 is often mutated in melanoma. Nat Genet. 2009;41(5):518–520. doi: 10.1038/ng.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei X, et al. Mutational and functional analysis reveals ADAMTS18 metalloproteinase as a novel driver in melanoma. Mol Cancer Res. 2010;8(11):1513–1525. doi: 10.1158/1541-7786.MCR-10-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei X, et al. Analysis of the disintegrin-metalloproteinases family reveals ADAM29 and ADAM7 are often mutated in melanoma. Hum Mutat. 2011;32(6):E2148–E2175. doi: 10.1002/humu.21477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prickett TD, et al. Exon capture analysis of G protein-coupled receptors identifies activating mutations in GRM3 in melanoma. Nat Genet. 2011;43(11):1119–1126. doi: 10.1038/ng.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prickett TD, Samuels Y. Molecular pathways: dysregulated glutamatergic signaling pathways in cancer. Clin Cancer Res. 2012;18(16):4240–4246. doi: 10.1158/1078-0432.CCR-11-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei X, et al. Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat Genet. 2011;43(5):442–446. doi: 10.1038/ng.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hahn CG, et al. Altered neuregulin 1-erbB4 signaling contributes to NMDA receptor hypofunction in schizophrenia. Nat Med. 2006;12(7):824–828. doi: 10.1038/nm1418. [DOI] [PubMed] [Google Scholar]
- 19.Garcia RA, Vasudevan K, Buonanno A. The neuregulin receptor ErbB-4 interacts with PDZ-containing proteins at neuronal synapses. Proc Natl Acad Sci U S A. 2000;97(7):3596–3601. doi: 10.1073/pnas.070042497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delint-Ramirez I, et al. In vivo composition of NMDA receptor signaling complexes differs between membrane subdomains and is modulated by PSD-95 and PSD-93. J Neurosci. 2010;30(24):8162–8170. doi: 10.1523/JNEUROSCI.1792-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D’Onofrio M, et al. Pharmacological blockade of mGlu2/3 metabotropic glutamate receptors reduces cell proliferation in cultured human glioma cells. J Neurochem. 2003;84(6):1288–1295. doi: 10.1046/j.1471-4159.2003.01633.x. [DOI] [PubMed] [Google Scholar]
- 22.Namkoong J, et al. Metabotropic glutamate receptor 1 and glutamate signaling in human melanoma. Cancer Res. 2007;67(5):2298–2305. doi: 10.1158/0008-5472.CAN-06-3665. [DOI] [PubMed] [Google Scholar]
- 23.Yip D, et al. A phase 0 trial of riluzole in patients with resectable stage III and IV melanoma. Clin Cancer Res. 2009;15(11):3896–3902. doi: 10.1158/1078-0432.CCR-08-3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318(5853):1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 25.Fine B, et al. Activation of the PI3K pathway in cancer through inhibition of PTEN by exchange factor P-REX2a. Science. 2009;325(5945):1261–1265. doi: 10.1126/science.1173569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colombino M, et al. BRAF/NRAS mutation frequencies among primary tumors and metastases in patients with melanoma. J Clin Oncol. 2012;30(20):2522–2529. doi: 10.1200/JCO.2011.41.2452. [DOI] [PubMed] [Google Scholar]
- 27.Chapman PB, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364(26):2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flaherty KT, et al. Improved survival with MEK inhibition in BRAF-mutated melanoma. N Engl J Med. 2012;367(2):107–114. doi: 10.1056/NEJMoa1203421. [DOI] [PubMed] [Google Scholar]
- 29.Guo J, et al. Phase II, open-label, single-arm trial of imatinib mesylate in patients with metastatic melanoma harboring c-Kit mutation or amplification. J Clin Oncol. 2011;29(21):2904–2909. doi: 10.1200/JCO.2010.33.9275. [DOI] [PubMed] [Google Scholar]
- 30.Samuels Y, Waldman T. Oncogenic mutations of PIK3CA in human cancers. Curr Top Microbiol Immunol. 2010;347:21–41. doi: 10.1007/82_2010_68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turke AB, Engelman JA. PIKing the right patient. Clin Cancer Res. 2010;16(14):3523–3525. doi: 10.1158/1078-0432.CCR-10-1201. [DOI] [PubMed] [Google Scholar]
- 32.Dutton-Regester K, et al. A high-throughput panel for identifying clinically relevant mutation profiles in melanoma. Mol Cancer Ther. 2012;11(4):888–897. doi: 10.1158/1535-7163.MCT-11-0676. [DOI] [PubMed] [Google Scholar]
- 33.Wagle N, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol. 2011;29(22):3085–3096. doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Molina-Vila MA, et al. Screening for EGFR mutations in lung cancer. Discov Med. 2009;8(43):181–184. [PubMed] [Google Scholar]
- 35.Rudloff U, Samuels Y. A growing family: adding mutated Erbb4 as a novel cancer target. Cell Cycle. 2010;9(8):1487–1503. doi: 10.4161/cc.9.8.11239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garrett TP, et al. Crystal structure of a truncated epidermal growth factor receptor extracellular domain bound to transforming growth factor alpha. Cell. 2002;110(6):763–773. doi: 10.1016/s0092-8674(02)00940-6. [DOI] [PubMed] [Google Scholar]
- 37.Ogiso H, et al. Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains. Cell. 2002;110(6):775–787. doi: 10.1016/s0092-8674(02)00963-7. [DOI] [PubMed] [Google Scholar]