Abstract
Mammalian evolution entailed multiple innovations in gene regulation, including the emergence of genomic imprinting, an epigenetic regulation leading to the preferential expression of a gene from its maternal or paternal allele. Genomic imprinting is highly prevalent in the brain, yet, until recently, its central roles in neural processes have not been fully appreciated. Here, we provide a comprehensive survey of adult and developmental brain functions influenced by imprinted genes, from neural development and wiring to synaptic function and plasticity, energy balance, social behaviors, emotions, and cognition. We further review the widespread identification of parental biases alongside monoallelic expression in brain tissues, discuss their potential roles in dosage regulation of key neural pathways, and suggest possible mechanisms underlying the dynamic regulation of imprinting in the brain. This review should help provide a better understanding of the significance of genomic imprinting in the normal and pathological brain of mammals including humans.
Keywords: genomic imprinting, parental bias, gene dosage, neural development, energy balance, behavior
INTRODUCTION
Genomic imprinting, a unique form of epigenetic regulation found in mammals and flowering plants, leads to the preferential expression of the maternal or the paternal allele of certain genes, with widespread implications for the development and function of the brain. The surprising concept that the two parentally inherited genomes of mammalian cells are functionally distinct originates from pronuclear transplantations in which embryos with uniparental (solely maternally or paternally derived) diploid genomes failed to develop normally (McGrath & Solter 1984, Surani et al. 1984), thus uncovering the requirement for distinct epigenetic imprints acquired in parental gametes. Following the discovery of the first imprinted genes (Barlow et al. 1991, Bartolomei et al. 1991, DeChiara et al. 1991), the search for the identity and functions of imprinted genes has thrived (Morison et al. 2005). Early studies implicated many imprinted genes with the control of developmental processes, with maternally and paternally imprinted genes often seen to promote and oppose growth, respectively, providing support for interesting evolutionary theories (see the sidebar, Evolutionary Theories of Genomic Imprinting).
EVOLUTIONARY THEORIES OF GENOMIC IMPRINTING.
Genomic imprinting waives the diploid advantage of buffering recessive mutations accumulating throughout the development of multicellular organisms (Otto & Gerstein 2008). Several theories have attempted to formalize the fitness advantages associated with the evolution of genomic imprinting in mammals and flowering plants. These have been thoroughly discussed elsewhere (Haig 2014, Patten et al. 2014, Spencer & Clark 2014), and we provide here a short summary of evolutionary ideas particularly relevant to genomic imprinting in the brain.
In mammals, mothers are the main nurturers of offspring, and progeny likely originates from multiple fathers owing to promiscuous mating systems. Based on these observations, the kinship theory proposes that a conflict between the parental genomes arises in offspring over the distribution of maternal resources (Wilkins & Haig 2003), such that maternally and paternally expressed genes tend to reduce and increase demand of maternal resources, respectively. Indeed, genes involved in acquiring resources (such as the Igf2 growth factor) are paternally expressed, whereas genes involved in conserving resources (such as the Igf2r growth repressor) are maternally expressed. The kinship theory also predicts that genes will be imprinted in organs that are engaged in social interactions, both during pregnancy (e.g., in the placenta) and postnatally (e.g., in the brain). However, the biological functions of many imprinted genes in the developing and adult brain are difficult to explain based solely on these predictions.
In a different theory, Keverne (2013) addressed more specifically the role of genomic imprinting in the brain, suggesting that this phenomenon helps coordinate the functions of the placenta, maternal hypothalamus, and developing fetal hypothalamus to ensure optimal embryonic and postnatal nurturing. This idea is based primarily on the observations that deletion of the paternally expressed Peg3 leads to impaired hypothalamic function in pups (e.g., poor suckling ability) and poor maternal care in rearing mothers (e.g., impaired milk letdown) (Curley et al. 2004). Furthermore, programs of gene expression appear to be synchronized in the placenta and developing hypothalamus at E12.5, which is controlled partially by the transcription factor PEG3 (Broad & Keverne 2011).
Wolf & Hager (2006) introduced a model suggesting that genomic imprinting evolved to optimize mother:offspring coadaptation. Matching (or mismatching) the offspring’s expressed allele with its mother’s allele in pleiotropic loci affecting both maternal and offspring traits benefits their interaction. This view is supported by maternal Grb10 expression, which in pups reduces nutrient demand and in mammary glands of lactating mothers enhances milk supply, thereby achieving an optimal effect on pup growth (Cowley et al. 2014). This theory, however, rationalizes the function of a limited set of imprinted genes, some of which are also explained by the kinship theory (Úbeda & Gardner 2015).
Additional hypotheses about the evolution of imprinting have been suggested (Patten et al. 2014, Spencer & Clark 2014). However, their explanatory power of the actual function of imprinted genes is often vague and fails to interpret key aspects of imprinting, such as parent of origin–specific expression, uniqueness to mammals, and postnatal and brain-specific functions.
The relevance of genomic imprinting to the brain emerged when two disorders with significant developmental and neurological deficits, Prader–Willi syndrome (PWS) and Angelman syndrome (AS), were linked to paternally and maternally transmitted mutations, respectively, on human 15q11–13 (Nicholls et al. 1989). This notion was reinforced by the discovery that chimeric embryos containing gynogenetic (two maternal genomes) cells develop abnormally large brains in contrast to chimeric embryos containing androgenetic (two paternal genomes) cells, which develop abnormally small brains (Keverne et al. 1996), pointing to an essential role for parent-of-origin information in brain development. Moreover, gynogenetic cells were observed exclusively in the cortex, striatum, and hippocampus, whereas androgenetic cells occupied hypothalamic regions, indicating distinct roles for maternally and paternally inherited information in various brain regions. The central role of genomic imprinting in brain function has been supported further by the extensive neural and behavioral phenotypes of mutants of imprinted genes and by the widespread expression of imprinted genes throughout the brain (Gregg et al. 2010, Wilkinson et al. 2007). Thus, the brain has emerged as a main target of genomic imprinting, generating great interest on how this epigenetic regulation provides stable transcriptional control of neural development and behavior.
We summarize here the vast literature on the role of genomic imprinting in the developing and mature brain, where imprinted genes often serve as control hubs in regulatory networks of brain functions. Although many of these findings came from studies in the mouse, strong correlates exist in humans as inferred from disorders associated with abnormal imprinting (see the sidebar, Human Brain Disorders Associated with Imprinting Dysregulation). Traits encoded by imprinted genes follow non-Mendelian inheritance, such that heterozygous mutants are often indistinguishable from controls if mutations are carried by the silenced parental allele but are similar to homozygotes when mutations are carried by the expressed parental allele. In addition, imprinted genes are commonly dosage sensitive, such that phenotypes are observed following alteration of the expressed allele as well as in loss of imprinting seen in derepression of the normally silenced allele.
HUMAN BRAIN DISORDERS ASSOCIATED WITH IMPRINTING DYSREGULATION.
In humans, dysregulation of imprinted gene expression resulting from null mutations, deletions, duplications, uniparental disomies, or alterations in epigenetic marks often affects brain function and behavior. Other imprinting dysregulations leading to disorders without strong neurological or behavioral components are not detailed here, even though some of the involved genes are expressed in the brain (Eggermann et al. 2015).
The 15q11–13 interval includes several paternally expressed noncoding RNAs and protein coding genes, as well as UBE3A, which exhibits brain-specific maternally biased expression (Angulo et al. 2015). Prader-Willi syndrome (PWS) results from loss of the 15q11–13 paternally expressed genes, which most frequently includes the SNORD116 small nucleolar RNAs (snoRNAs) cluster that is hence regarded as the critical contributor to the PWS phenotype (Sahoo et al. 2008). PWS runs a biphasic clinical course, starting with reduced fetal activity followed by postnatal poor muscle tone and diminished swallowing and suckling. By age two, these features are replaced with hyperphagia, food foraging behavior, and lack of satiety leading to obesity. Intellectual disability and behavioral patterns such as temper tantrums, stubbornness, skin picking, and obsessive compulsions are later observed. PWS also features sleeping abnormalities, hypogonadism due to impaired puberty, and short stature due to hypothalamic growth hormone deficiency. Angelman syndrome (AS), which results from null mutations or loss of expression of UBE3A, has a strikingly different clinical manifestation, characterized by an abnormally happy demeanor with inappropriate laughter and excitability, severe motor and intellectual disability, autism spectrum disorder–like speech impairment, epilepsy, sleep disorders, and seizures (Bird 2014).
The GNAS locus produces several isoforms with different parent-of-origin expression biases in which null mutations lead to pseudohypoparathyroidism type 1a (PHP1a) when maternally transmitted and pseudopseudohypoparathyroidism (PHPP) when paternally transmitted (Turan & Bastepe 2013). PHP1a features resistance to various hormones, skeletal abnormalities, obesity, and cognitive impairments (Mouallem et al. 2008), likely due to the tissue specificity of maternal GNAS isoform expression. PHPP is characterized mainly by neonatal metabolic and growth defects.
The 7q21–31 interval harbors several imprinted genes and is linked with autism susceptibility (Schanen 2006). Paternally transmitted mutations in the sarcoglycan-ε (SGCE) gene cause myoclonus-dystonia syndrome (Zimprich et al. 2001), characterized by early adolescence onset of myoclonic jerks (Asmus et al. 2002) and psychiatric symptoms such as obsessive-compulsive disorder, panic attacks, anxiety, depression, agoraphobia, and alcohol dependence (Peall et al. 2013).
The 8q24 interval includes the maternally expressed potassium channel KCNK9, which is expressed primarily in the brain and for which maternally transmitted null mutations lead to Birk-Barel syndrome, characterized by moderate to severe intellectual disability, hyperactivity, low muscle tone and feeding difficulties in infancy, and unique facial dysmorphism (Barel et al. 2008). 8q24 also includes TRAPPC9 for which recessive mutations cause intellectual disability (Mochida et al. 2009). Trappc9 isoform–specific imprinting has been demonstrated in mouse (Gregg et al. 2010), and it was recently reported to be imprinted in humans (Babak et al. 2015).
Genomic imprinting has classically been defined as the full silencing of one of the two parental alleles (Bartolomei & Ferguson-Smith 2011). We discuss new evidence from mice and humans showing that genomic imprinting encompasses a wide range of parental expression biases with important functional significance, supporting a conceptual expansion of genomic imprinting to the parent-of-origin regulation of gene dosage.
IMPRINTED GENES AND EPIGENETIC CONTROL OF NEURAL DEVELOPMENT
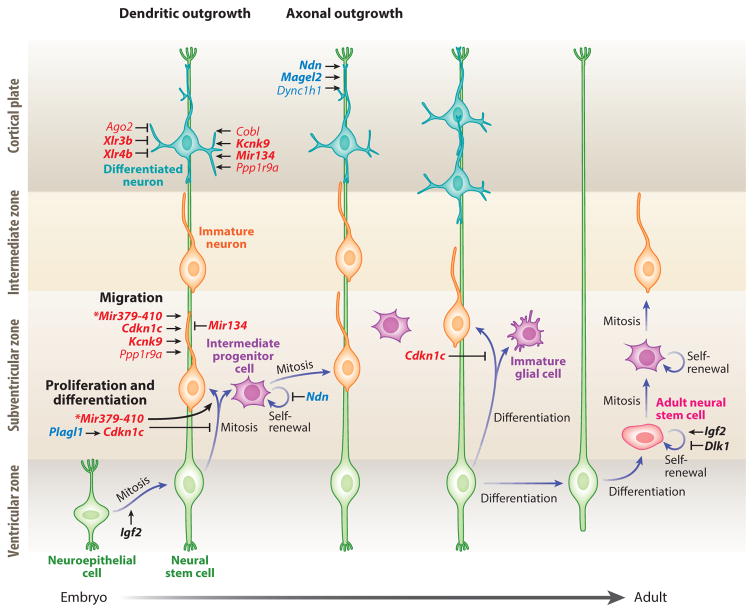
Studies of mouse chimeras containing either gynogenetic or androgenetic cells provided initial evidence for the differential contributions of the maternal and paternal genomes in the developing brain (Keverne et al. 1996). Since then, extensive evidence has implicated genomic imprinting in key neurodevelopmental processes including neural progenitor expansion, migration and differentiation, and cell polarization. We describe here how imprinted genes regulate these essential neurodevelopmental processes (Figure 1).
Figure 1.
Roles of imprinted genes during cortical neurogenesis. Genes preferentially expressed from the maternal and paternal allele appear in red and blue, respectively. In black are imprinted genes that are biallelically expressed in this context. Strongly biased and monoallelically expressed imprinted genes are in bold. Lines with arrowheads indicate enhancement, and lines with notched ends indicate reduction of the biological function. The asterisks by Mir379–410 indicate that the corresponding biological functions are regulated by three miRNAs of this cluster: Mir369-3p, Mir496, and Mir543.
Neurogenesis
Neural stem cells (NSCs) of the ventricular zone proliferate asymmetrically to produce differentiated neurons or glia or to generate intermediate progenitor cells (IPCs), which further divide symmetrically to produce either IPCs or neurons (Kriegstein & Alvarez-Buylla 2009). NSC fate decisions are highly regulated by imprinted genes through intricate control pathways. Cortical NSCs are maintained through N-cadherin-mediated adhesion. N-cadherin expression is repressed by the maternally expressed miRNAs—miR369-3p, miR496, and miR543 of the MiR379–410 cluster—thereby promoting the shift from NSC proliferation to cell differentiation and migration (Rago et al. 2014). NSCs strongly express PLAGL1 (Valente et al. 2005), a paternally expressed zinc finger protein, which induces expression of the maternally expressed cyclin-dependent kinase inhibitor Cdkn1c. In turn, Cdkn1c promotes NSC cell cycle arrest and subsequent differentiation by inhibiting cyclin-dependent kinases (Cdks) (Schmidt-Edelkraut et al. 2014); later, Cdkn1c promotes a shift from proneural to proglial NSC differentiation ( Joseph et al. 2009). The self-renewal of neuroepithelial progenitors and NSCs is promoted by Igf2, which is biallelically expressed in the choroid plexus and vascular compartments (yet preferentially maternally expressed in other regions of the brain and paternally expressed outside the brain), and is secreted into the cerebrospinal fluid (DeChiara et al. 1991, Lehtinen et al. 2011). Finally, the proliferative capacity of IPCs is repressed by the paternally expressed NDN, which inhibits Cdk1 expression (Minamide et al. 2014). In the adult brain, NSCs persist in unique niches of the subventricular zone (SVZ) and the subgranular zone (SGZ) of the dentate gyrus. In the SVZ, a soluble DLK1 isoform is secreted by niche astrocytes and signals through a membrane-tethered alternative isoform of DLK1 expressed by NSCs to maintain NSC self-renewal (Ferrón et al. 2012). Interestingly, this paracrine mode of DLK1 signaling requires derepression of its maternal allele to achieve biallelic expression in both astrocytes and NSCs. Similarly, derepression of the Igf2 maternal allele to achieve biallelic expression in the SVZ is also required for NSC self-renewal postnatally, suggesting a remarkable mechanism of transcriptional dosage control in adult neurogenesis through imprinting regulation (Ferrón et al. 2015). In the SGZ, the maternally expressed CDKN1C facilitates the maintenance of NSC quiescence (Furutachi et al. 2013), and in contrast to the SVZ, IGF2 is paternally expressed and operates in an autocrine manner to regulate NSC survival (Bracko et al. 2012, Ferrón et al. 2015).
Neuronal differentiation requires the induction of multiple transcription factors to activate neuron-specific transcriptional programs. The roles of imprinted genes during this process are widespread. In the developing cerebellum, transcription of the paternally expressed Plagl1 is restricted to the ventricular zone and external granule layer of specific lobules, where it promotes differentiation of GABAergic interneurons and Golgi cells (Chung et al. 2011). Paternally expressed DIO3 protects the developing cerebellum from premature stimulation by thyroid hormone, which controls granule-cell formation in the external germinal layer, their migration to the internal layer, cerebellar foliation, and dendritic arborization of Purkinje cells (Peeters et al. 2013). Accordingly, Dio3 deletions show accelerated external layer disappearance, premature expanded molecular layer, and locomotor defects.
The midbrain dopaminergic (mdDA) system plays a critical role in the control and modulation of emotional, motivational, and cognitive behaviors as well as voluntary movements. The development of this system is targeted particularly by imprinted genes. The transcription factors LMX1A and NURR1 are essential for early and terminal differentiation of mdDA neurons, respectively (Hoekstra et al. 2013, Jacobs et al. 2009), by inducing transcription of the paternally expressed Grb10 and of Dlk1, and of the maternally expressed Cdkn1c. Interestingly, enhanced maternal care during early life increases Cdkn1c expression in the maturing ventral tegmental area accompanied by enhanced survival of mdDA neurons (Peña et al. 2014). Survival of mdDA neurons also requires expression of the paternally biased Bcl-x, which protects from developmental and Parkinson’s disease–related apoptosis (Savitt et al. 2005). Finally, loss of the maternally expressed UBE3A in AS patients results in fewer substantia nigra mdDA neurons and Parkinson’s-like motor impairments (Mulherkar & Jana 2010) but increased nucleus accumbens (NAc) dopamine transmission (Riday et al. 2012).
Neuronal Migration
In the developing brain, neuronal migration to appropriate sites is essential for the establishment of proper identity and functional connectivity. As discussed below, it is mediated by cellular processes that are influenced highly by genomic imprinting. Actin polymerization, which is critical for cell motility within the cortical plate, is promoted by CDKN1C and the maternally biased PPP1R9A (Causeret et al. 2007, Tury et al. 2012). Additionally, silencing of DCX by the maternally expressed miR134 inhibits cortical neuron migration (Gaughwin et al. 2011). Tangential migration of differentiating GABAergic interneurons from the basal forebrain to the cortex requires enhanced transcriptional activity of the DLX2-NDN complex (Kuwajima et al. 2006). Accordingly, the cortical GABAergic system is impaired in Ndn paternal deletions (Kuwajima et al. 2010). Migration of gonadotropin-releasing hormone (GnRH) neurons from the olfactory placode to the medial preoptic area (mPOA) also depends on DLX proteins and is reduced in Ndn paternal deletions (Miller et al. 2009). The majority of PWS patients lack NDN and exhibit hypogonadism and impaired puberty (Cassidy et al. 2012), a phenotype consistent with defects in GnRH neurons. Neuronal activity is also critical for neuronal migration. In particular, the maternally expressed KCNK9 controls resting potentials and neuronal excitability, and maternally transmitted mutations that cause Birk-Barel syndrome in humans (see the sidebar, Human Brain Disorders Associated with Imprinting Dysregulation) impair neuronal migration and delay dendritic maturation (Bando et al. 2014).
Axonal and Dendritic Outgrowth
High motility of the axonal growth cone allows navigation in extracellular environments and identification of synaptic targets. PPP1R9A-mediated actin polymerization regulates growth-cone motility and directionality (Nakanishi et al. 1997). Furthermore, centrosome-based cytoskeletal rearrangements that promote axonal outgrowth are regulated by FEZ1 and BBS4, the proteasomal degradation of which is prevented by the paternally expressed NDN and MAGEL2 (Lee et al. 2005). In addition, both axonal extension and growth-cone motility rely on active transport across microtubules, mediated by motor proteins including the paternally biased dynein subunit DYNC1H1 (Ori-McKenney & Vallee 2011). Growing axons require signaling cues for proper guidance. In sympathetic neurons, axonal pathfinding and tissue innervation is regulated by the maternally biased endothelin-3, which is secreted by the carotid vasculature (Makita et al. 2008).
The signals received by a neuron are determined by the growth of its dendrites, as well as by the density, shape, and synaptic targets of its dendritic spines. Imprinted genes both promote and oppose these processes. Downregulation of the maternally biased Ago2 in the absence of the transcription cofactor NCOA3 results in abnormally high dendritogenesis (Störchel et al. 2015). miR134 regulates the abundance of the pumilio2 RNA-binding protein, and its over- or underexpression reduces dendritogenesis (Fiore et al. 2009). Dendrite arborization is promoted by both maternally biased PPP1R9A and COBL through their regulation of actin filaments (Ahuja et al. 2007, Zito et al. 2004). Spine growth is negatively regulated by the X-linked chromatin remodeling maternally expressed genes Xlr3b and Xlr4b, which are repressed by the CUX1 and CUX2 transcription factors (Cubelos et al. 2010). Finally, alternative splicing and allele-specific expression of protocadherins provide single-neuron diversity that is essential for dendritic self-avoidance and neuronal wiring (Chen & Maniatis 2013). Interestingly, three beta-protocadherins, Pcdhβ20, Pcdhβ12, and Pcdhβ10, were discovered to be imprinted (the former paternally and the latter two maternally biased) in the developing cerebellum (Perez et al. 2015).
Apoptosis
In the developing nervous system, apoptosis serves to control the overall neuronal pool and matching of pre- and postsynaptic neurons. Internal and external stimuli regulate apoptosis by shifting the balance between proapoptotic and antiapoptotic proteins. In a proapoptotic state, mitochondrial-membrane permeability is increased, and the release of cytochrome c leads to caspase activation and subsequent cell death. Generally, paternally expressed genes are mostly antiapoptotic, whereas maternally expressed genes are more pleiotropic (Perez et al. 2015). The paternally biased Bcl-xL is a major antiapoptotic Bcl-2 family member in the brain, as its deletion results in massive death of postmitotic immature neurons (Motoyama et al. 1995). Remarkably, deletion of the paternal but not maternal Bcl-x allele reduces brain size significantly, suggesting increased apoptosis (Perez et al. 2015). Absence of paternally expressed Peg3 results in increased P53-dependent apoptosis in forebrain, striatal, amygdalar, and hypothalamic regions (Broad et al. 2009). Interestingly, these abnormalities vary between males and females, suggesting that Peg3 function regulates the establishment of sexual dimorphisms. Absence of Peg3 or Magel2 also reduces hypothalamic oxytocin neurons (Li et al. 1999, Schaller et al. 2010). NDN prevents apoptosis in cerebellar granule cells by blocking E2F transactivation of proapoptotic targets (Kurita et al. 2006). During ischemia and hypoxia, however, PEG3 and BCL-XL induce apoptosis (Ofengeim et al. 2012, Yamaguchi et al. 2002), whereas NDN prevents apoptosis (Hasegawa & Yoshikawa 2008). On the maternal side, granule cell death, which contributes to numerically matching with their postsynaptic Purkinje cell targets, is mediated by KCNK9-dependent apoptosis (Patel & Lazdunski 2004). The maternally expressed MEG3 may also play a proapoptotic role in the brain based on its pituitary tumor suppression activity, transcriptional upregulation of p53, and downregulation of P53-degrading genes (Zhou et al. 2012). Brain-specific deletions of Cdkn1c show massive P53-dependent apoptosis in the developing brain, thought to result from hyperactivation of E2F1 (Matsumoto et al. 2011), whereas in nonbrain tissues it promotes apoptosis (Yan et al. 1997). UBE3A may also play an antiapoptotic role by ubiquitinating P53 (Mishra & Jana 2008), as its maternal deletions reduce brain weight ( Jiang et al. 1998).
FUNCTIONS OF IMPRINTED GENES IN THE POSTNATAL BRAIN
Phenotypic studies of mouse mutants and human disorders have uncovered the vast significance of genomic imprinting in the postnatal brain, encompassing all essential adult neural functions from key elements of synaptic transmission and plasticity (Figure 2) to the control of energy balance and metabolism, as well as emotional, social, and cognitive behaviors (Figure 3).
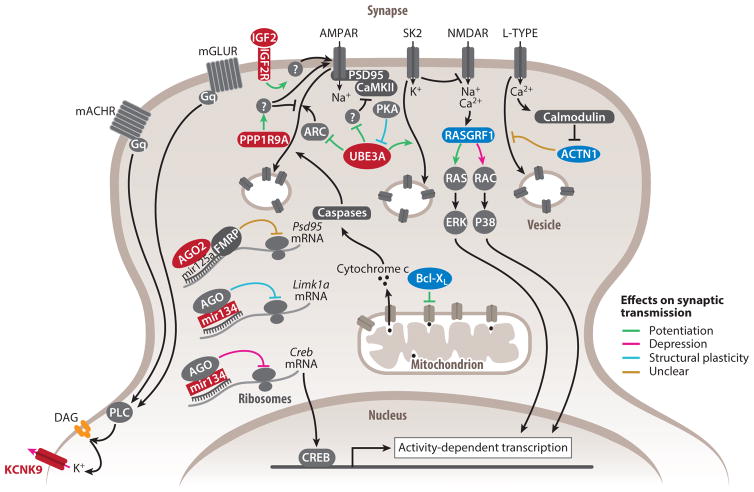
Figure 2.
Functions of imprinted genes in synaptic transmission and plasticity. Products of genes preferentially expressed from the maternal and paternal allele appear in red and blue, respectively. Products of biallelically expressed genes appear in dark gray. Products of strongly biased and monoallelically expressed imprinted genes are in bold. Lines with arrowheads represent stimulatory molecular interactions, whereas lines with notched ends represent inhibitory molecular interactions. The overall role of imprinted genes in synaptic plasticity is shown in green to indicate synaptic potentiation or activation, in pink to indicate synaptic depression or inhibition, in blue to indicate the induction of structural changes, and in brown when the contribution to synaptic transmission or plasticity remains unclear. White-filled circles with beige borders represent vesicles containing membrane receptors or channels.
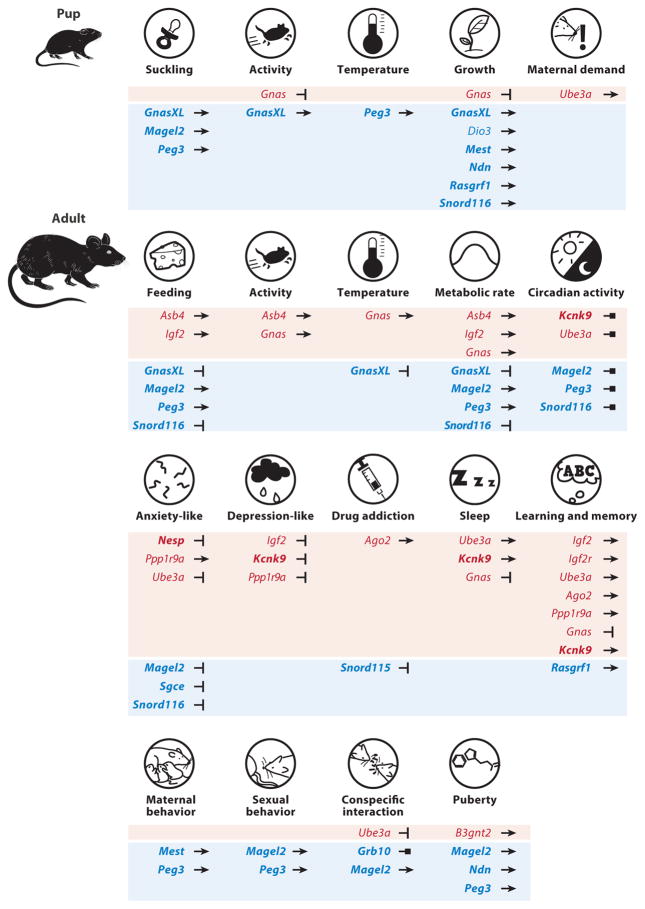
Figure 3.
Mouse phenotypes influenced by imprinted genes. Genes associated with specific metabolic, social, emotional, and cognitive phenotypes are listed. Genes preferentially expressed from the maternal and paternal allele appear in red and blue, respectively. Strongly biased and monoallelically expressed imprinted genes are in bold. Lines with arrowheads and notched ends indicate stimulation and inhibition, respectively, of the given phenotype, whereas lines with square ends indicate functions that cannot be defined as enhancement or reduction.
Synaptic Transmission
Synaptic transmission underlies the propagation of information throughout the brain and directs proper wiring of neuronal circuits. Multiple imprinted genes participate in baseline transmission and activity dependent modifications of neuronal excitability.
Action potentials
Voltage-insensitive K+ leak channels set resting potentials and thus influence the likelihood of action potentials and neuronal firing patterns. The maternally expressed KCNK9 potassium channel hyperpolarizes neurons according to extracellular pH, bivalent cation concentration, and phospholipase C signaling (Bista et al. 2015). Interestingly, KCNK9 deletion reduces the membrane resting potential and changes firing patterns from burst to tonic firing (Brickley et al. 2007, Musset et al. 2006). Initiation of action potentials occurs in the axon initial segment where densely clustered voltage-gated Na+ channels power depolarization. In Ube3a maternal deletions, hippocampal action potentials exhibit increased amplitudes associated with increased α1-NaKA channel levels (Kaphzan et al. 2011). Crossing Ube3a maternal deletions with α1-NaKA null heterozygotes restores normal action potentials and synaptic plasticity (Kaphzan et al. 2013).
Presynaptic regulation
The number of available presynaptic vesicles determines in part the strength of postsynaptic stimulation. Ca2+ import induces BCL-XL translocation into clathrin-coated vesicles, enhancing formation and replenishment of the vesicle pool (Li et al. 2013). Importantly, the endocytic rate is increased and decreased by BCL-XL overexpression and knockdown, respectively. Ube3a maternal deletions exhibit reduced capacity to replenish the presynaptic vesicle pool in L4 inhibitory interneurons after repeated stimulation, resulting in weaker inhibitory postsynaptic currents (Wallace et al. 2012). Presynaptic deficits are also observed when Ube3a is overexpressed (Smith et al. 2011).
Postsynaptic regulation
Mechanisms of excitatory synaptic plasticity such as long-term potentiation (LTP) and long-term depression (LTD) are dependent on N-methyl-D-aspartate (NMDA) and α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) glutamate receptors (NMDARs and AMPARs, respectively), which initiate signaling cascades of immediate as well as long-term adaptations. Most imprinted genes known to regulate synaptic plasticity are maternally expressed and for the most part promote synaptic potentiation (Figure 2). Several imprinted genes mediate immediate synaptic responses. UBE3A promotes LTP by preventing inhibition of NMDARs (Sun et al. 2015) and of the Ca2+-regulated kinase CaMKII (Weeber et al. 2003) and also by preventing activity-regulated cytoskeletal-associated protein mediated internalization of membrane AMPARs (Greer et al. 2010). Furthermore, PPP1R9A prevents inhibitory dephosphorylation of the AMPAR GLUR1 subunit (Wu et al. 2008). In the postsynapse, Ca2+ is the main second messenger regulating both LTP and LTD. The paternally biased ACTN1 binds to L-type Ca2+ channels to prevent their endocytic internalization. This interaction is relieved by activity-dependent Ca2+ influx and thus represents a negative-feedback response to Ca2+ levels (Hall et al. 2013). Recently, an essential and sufficient role of apoptotic genes in LTD induction was identified ( Jiao & Li 2011, Z. Li et al. 2010), such that proapoptotic genes induce modest levels of active caspases in the postsynapse, which in turn induce AMPAR internalization. BCL-XL inhibits caspases activation and thus AMPAR internalization and LTD.
Local protein synthesis (LPS) operates at intermediate timescales in synaptic plasticity and provides neurons with effective dosage control of proteins, such as channels, receptors, and scaffolds, in spatially and temporally restricted domains. The maternally biased AGO2 regulates mRNA-miRNA interactions that prevent LPS in an activity-dependent manner. For instance, prior to membrane depolarization, LPS of the scaffold-encoding Psd95 is prevented by interactions between AGO2, miR125a, and FMRP (Muddashetty et al. 2011). Additionally, AGO2 is part of dendrite-specific, P-body-like structures potentially involved in mRNA transport toward active synapses (Cougot et al. 2008). Recently, profiles of ribosome-linked dendritic RNAs have suggested activity-dependent translation of >1,000 genes, including several imprinted genes such as Ago2 and Ube3a and paternally expressed Bcl-x and Rasgrf1 (Ainsley et al. 2014). Additionally, pharmacological inhibition of NMDAR leads to LPS of the paternally expressed Nnat (Oyang et al. 2011).
Long-lasting synaptic potentiation relies on activity-induced transcription by cAMP response element-binding protein (CREB) and other transcription factors. Paternally expressed Rasgrf1 encodes a Ca2+-dependent GTP exchange factor that activates small G proteins RAS and RAC, which in turn activate transcription through MAP kinases (Ye & Carew 2010). RASGRF1 induces RAS-dependent LTP and RAC-dependent LTD in the amygdala and hippocampus, respectively (Brambilla et al. 1997, Li et al. 2006). Overexpression of the maternally expressed MiR134 prevents CREB synthesis, thereby reducing hippocampal LTP (Gao et al. 2010). Lasting potentiation often induces modifications in dendritic spine size and number (Bailey et al. 2015). Loss of MiR134, which normally silences the spine growth–inducing kinase LIMK1A, results in spine overgrowth (Schratt et al. 2006). Spine density is reduced in Ube3a maternal deletions (Dindot et al. 2008) but augmented when UBE3A ligase activity is increased (Yi et al. 2015).
Homeostatic plasticity
Intrinsic mechanisms to maintain excitatory-inhibitory balance within a circuit require widespread adaptations of the underlying synapses. Increasing the ratio of excitation over inhibition in cultured neurons induces synaptic changes in protein levels and structural plasticity, which are impaired in the absence of either the maternally expressed MiR134 or MiR485 (Cohen et al. 2011, Fiore et al. 2014).
Critical periods
Critical periods are temporal windows of increased plasticity when sensory experiences shape cortical circuits. Monocular depravation during the critical period of the visual cortex fails to induce ocular dominance plasticity in Ube3a maternal deletions, indicating a role in experience-dependent plasticity (Sato & Stryker 2010, Yashiro et al. 2009).
Learning and Memory
The role of imprinted genes in synaptic function and plasticity suggests a more general role in learning and memory. Manipulations of multiple imprinted genes lead to deficits in hippocampus-based spatial and contextual memories. For instance, Ube3a maternal deletions perform poorly in paradigms of contextual memory ( Jiang et al. 1998, Sun et al. 2015), but they can be rescued by constitutively dephosphorylated CaMKII (van Woerden et al. 2007), α1-NaKA reductions (Kaphzan et al. 2013), and SK2 channel inhibition (Sun et al. 2015). Deficiencies in contextual memory are also observed when Ago2, Ppp1r9a, or Rasgrf1 are absent (Batassa et al. 2010, d’Isa et al. 2011, Giese et al. 2001, Wu et al. 2008).
After a memory is acquired, it remains labile for several hours during its consolidation. In the rat hippocampus, IGF2 is increased hours after training, which is necessary for memory consolidation (Chen et al. 2011). Moreover, administration of IGF2 after training enhances memory consolidation. Memory retrieval within a few days of learning returns it to a labile state, allowing its modification (reconsolidation), which is also enhanced by IGF2 administration. Mechanisms by which IGF2 affects memory require protein synthesis and are accompanied by increments in synaptic AMPARs, suggesting a synaptic effect. IGF2 action is dependent on IGF2R, which, in the brain, can signal through the protein kinase C pathway (Hawkes et al. 2006). This is in remarkable contrast to the IGF2R-mediated lysosomal degradation of IGF2, observed in other tissues where Igf2 and Igf2r are exclusively expressed from the paternal and maternal alleles, respectively. In the brain, however, Igf2 is maternally biased, whereas Igf2r fluctuates between maternally biased and biallelic expression (Perez et al. 2015, Yamasaki et al. 2005). Accordingly, upregulation of IGF2 during the process of memory reconsolidation is contributed mainly by the maternal allele (Ye et al. 2015). Finally, enhanced contextual memory is also observed when Ndn is paternally deleted (Muscatelli et al. 2000).
Sleep
Sleep serves multiple homeostatic functions including enhancement of memory consolidation. Ube3a maternal deletions show reduced sleep induction after extended wakefulness and overall reductions in rapid eye movement (REM) sleep (Colas et al. 2005), recapitulating AS hallmarks (Bird 2014). Fragmented sleep is also observed in Kcnk9 deletions, accompanied by reduced REM and increased non-REM sleep (Gotter et al. 2011, Pang et al. 2009). In contrast, overexpression of maternally biased Gnas decreases REM and increases non-REM sleep (Lassi et al. 2012). Consistent with the role of sleep in memory, both Kcnk9 deletion and Gnas overexpression are accompanied with impaired contextual memory (Lassi et al. 2012, Linden et al. 2007).
Working memory
Working memory represents an interface between incoming and previously stored information, which mediates activity and behavior. This cortex-regulated process is impaired in Kcnk9 deletions specifically during phases of general inactivity (Gotter et al. 2011, Linden et al. 2007). Strikingly, theta II oscillations, characteristic of immobile animals processing sensory stimuli relevant to motor responses, are completely absent in Kcnk9 mutants (Pang et al. 2009). The neuronal mechanisms underlying this effect are unknown.
Neural Regulation of Energy Balance
Genomic imprinting plays a key role in the regulation of growth and energy balance during embryonic and placental development, as well as in nonneuronal tissues (Charalambous et al. 2007, Tunster et al. 2013). Ultimately, the brain controls energy balance and metabolism of the entire organism, and this homeostatic function is performed primarily by the hypothalamus, which senses internal energy states and orchestrates visceral responses. Here we describe key roles played by imprinted genes in this process in neonates and adults.
Energy balance in early life
The brain regulates nutrient acquisition by neonates through milk suckling, a behavior targeted by multiple paternally expressed genes. For instance, mice carrying paternal deletions of Magel2 fail to suckle and thus die, a phenotype associated with reduced oxytocin levels that can be rescued by a single oxytocin injection (Schaller et al. 2010). Paternal Peg3 deletion also reduces suckling, resulting in perinatal death (Curley et al. 2004). The developing brain controls neonatal metabolism and growth through thyroid-hormone regulation. DIO3, which inactivates the thyroid hormone T3, is paternally biased in a brain-region specific manner (Martinez et al. 2014). Interestingly, hypothalamic paternal biases of DIO3 decrease with age (Martinez et al. 2014), correlated with its reduced activity (Hernandez et al. 2006). Pups carrying Dio3 deletions exhibit growth retardation, associated with abnormally fluctuating T3 levels due to hypothalamic, pituitary, and thyroid impairments (Hernandez et al. 2007). Additionally, the paternally expressed NDN participates in hypothalamic negative feedback responses to increased T3 (Hasegawa et al. 2012).
Synchronization of growth by the neonatal brain relies on hypothalamic secretion of growth hormone (GH)-releasing hormone (GHRH) and somatostatin, which stimulate and inhibit GH and thus IGF1 secretion, respectively. Expression of Rasgrf1 is detected mainly in the brain and changes from exclusively paternal before weaning to preferentially paternal afterward (Drake et al. 2009). Mice with Rasgrf1 paternal deletions are normal at birth but growth retarded by weaning (Itier et al. 1998), and loss of Rasgrf1 imprinting increases growth and weight after birth (Drake et al. 2009). Importantly, levels of both GH and IGF1 decrease or increase when Rasgrf1 is missing or overexpressed, respectively. Because Rasgrf1 is expressed in the hypothalamus but not in the pituitary, these effects are likely due to hypothalamic impairments. Indeed, hypothalamic expression of GHRH and somatostatin is increased with Rasgrf1 loss of imprinting (biallelic expression). Similarly, pups lacking the paternal Snord116 exhibit drastically reduced IGF1 levels and are smaller with reduced organ sizes.
Thermoregulation is another key aspect of energy expenditure essential to early-life survival. Pups perform nonshivering thermogenesis through sympathetic control of energy breakdown in brown adipose tissue. Pups with paternal deletions of Peg3 have a lower temperature that decreases further after maternal separation (Curley et al. 2004), whereas Dlk1 and Dio3 overexpression results in abnormal thermoregulation, failure to adapt to postweaning life, and eventually death (Charalambous et al. 2012). Although expression of these genes is enriched in the hypothalamus (Meister et al. 2013), a brain origin of this phenotype has not been demonstrated.
The Gnas locus is a complex transcriptional unit responsible for remarkably distinct defects in neonate physiology according to the affected parental allele. Gnas encodes the ubiquitously expressed G-protein α-subunit Gsα, which is maternally biased in a tissue-specific manner, as well as GnasXL, which is translated into a distinct G-protein α-subunit called XLαs and exhibits a brain region–specific paternal expression pattern (Krechowec et al. 2012, Peters & Williamson 2007). Maternal deletions affecting Gsα result in pup overgrowth, hyperactivity, and early death (Yu et al. 1998). In contrast, paternal deletions affecting GnasXL result in impaired suckling, growth retardation, inactivity, and early death (Plagge et al. 2004). Remarkably, overexpression of Gnas recapitulates some phenotypes observed in the absence of GnasXL, whereas overexpression of GnasXL recapitulates some phenotypes observed in the absence of Gnas (Ball et al. 2013). However, matched dosages of Gnas and GnasXL, even if both are overexpressed, results in normal development indicating that balanced expression of both proteins is essential. Interestingly, mutations specific to GnasXL’s first exon, which encodes ALEX, alter suckling, suggesting a role of ALEX in this process (Eaton et al. 2012, Kelly et al. 2009). In humans, children with maternally transmitted GNAS mutations are obese, whereas paternally transmitted GNAS mutations exhibit perinatal growth retardation, severe feeding difficulties, and reduced adiposity (Bastepe 2012).
Circuits regulating energy balance are still developing around weaning, and imprinted genes of the PWS cluster are thought to play a role in the transition to adult metabolic programs. A hallmark of PWS, associated primarily with paternal deletions of the SNORD116 locus, is the early transition from weak and hypophagic neonates to hyperphagic and obese infants (Sahoo et al. 2008). Similarly, newborn mice carrying Snord116 paternal deletions fail to gain weight and are substantially leaner than wild types by weaning (Ding et al. 2008). As adults, these mice develop hyperphagia but, contrary to humans, their increased metabolism maintains a leaner phenotype. In mouse neonates carrying mutations equivalent to human PWS, pro-opiomelanocortin (Pomc) mRNA levels are elevated tenfold compared to wild types (Bittel et al. 2007). As discussed below, the effects of multiple imprinted genes converge in the function of adult POMC-expressing neurons.
Energy balance in the adult brain
Regulation of adult weight by imprinted genes generally involves maternally expressed genes contributing to reduced weight as a function of metabolic rate and food intake, whereas paternally expressed genes contribute to increased weight. Paradoxically, imprinted genes that decrease metabolism also decrease food intake, but those that increase metabolism also increase food intake. The actions of multiple imprinted genes in adult metabolism appear to converge into the regulation of POMC-expressing neurons of the arcuate nucleus. POMC neurons are stimulated by leptin to induce satiety and energy expenditure through regulation of melanocortin 4 receptor (MC4R)-expressing neurons in the paraventricular nucleus (PVN) (Gao & Horvath 2007). In POMC neurons, expression of maternally biased Asb4 decreases during fasting, whereas localized leptin administration increases Asb4 expression (J.Y. Li et al. 2005, 2010). Conditional overexpression of Asb4 in these neurons increases ambulatory activity and energy expenditure as well as food intake, resulting in leaner animals resistant to high-fat diet–induced obesity. These animals also exhibit increased levels of Pomc mRNA (J.Y. Li et al. 2010). Together, these results indicate that ASB4 action on POMC neurons accelerates metabolic rate. By contrast, paternal deletions of Magel2 exhibit reduced food intake and increased weight, indicative of hypometabolism (Bischof et al. 2007). Magel2 knockout mice display elevated leptin and develop leptin resistance, which is associated with fewer adult POMC neurons (Mercer et al. 2013). Moreover, leptin stimulation of remaining POMC neurons is absent, whereas normal inhibition by leptin is observed in AGRP neurons (Pravdivyi et al. 2015). Similarly, Peg3 paternal deletions exhibit reduced food intake but slower metabolism and increased adiposity (Curley et al. 2005) in addition to elevated leptin and leptin resistance.
MC4R neurons in the PVN are essential for the neuroendocrine control of visceral responses, including the metabolic regulation by sympathetic nervous system (SNS) activity (Gao & Horvath 2007). As in neonates, the opposed effects of the maternally and paternally expressed Gnas and GnasXL, respectively, are maintained in adulthood with far-reaching metabolic consequences. Maternal deletions of Gnas slow metabolism and increase weight (Chen et al. 2005), whereas its overexpression reduces weight and increases ambulatory activity (Xie et al. 2008). Circulating levels of norepinephrine are positively correlated with Gnas expression, suggesting alterations in SNS activity. Accordingly, brain-specific Gnas deletions exhibit increased weight, hypometabolism, and impaired thermoregulation due to lagging SNS activity (Chen et al. 2009). Furthermore, PVN-specific deletions recapitulate some but not all the effects observed in whole-brain deletions, indicating that Gnas imprinting regulates metabolism across multiple areas (Chen et al. 2012). Consistently, adult obesity is common in human maternally transmitted GNAS mutations (Turan & Bastepe 2013). Similarly, paternal deletions of GnasXL result in adult hyperphagia, hypermetabolism, and increased body temperature (Plagge et al. 2004, Xie et al. 2006), a phenotype caused by increased SNS activity (Nunn et al. 2013). Remarkably, the increased metabolic rate observed after Gnas overexpression is rescued by GnasXL complementary overexpression (Ball et al. 2013). Together, these results indicate a delicate balance between Gnas and GnasXL activities in central metabolic control.
Finally, a homozygous deletion of a brain-specific enhancer reduces Igf2 expression drastically in the brain and results in increased weight and decreased food intake, indicating reduced metabolic rate ( Jones et al. 2001). Thus, contrary to its growth-promoting role during gestation, Igf2, which switches from paternally expressed in most tissues to maternally biased in the adult brain (Gregg et al. 2010, Perez et al. 2015), appears to decrease adipose storage.
Circadian rhythms
Animals synchronize their general activity and behaviors to the circadian light/dark cycle. In mammals, the suprachiasmatic nucleus (SCN) is the master pacemaker that coordinates circadian oscillations in the activity of the brain and peripheral tissues. Rhythmicity of individual SCN neurons is established by a delayed negative feedback loop in the activity of the transcription factors CLOCK and BMAL1, which orchestrates periodic changes in gene transcription, among which are the imprinted paternally expressed Magel2 and Peg3 and the maternally expressed Blcap and Calcr (Panda et al. 2002). Magel2 expression is restricted to arginine-vasopressin positive cells of the SCN and is highest during the day yet absent at night. When placed in constant darkness, mice with Magel2 paternal deletions exhibit abnormal general activity amplitudes across the 24-h period (Kozlov et al. 2007). Abnormal periodicity and accelerated adjustment to light/dark changes was documented in Ube3a maternal deletions and is associated with lack of UBE3A-mediated degradation of BMAL1 (Shi et al. 2015). Deletions of Peg3 or Kcnk9 cause increased nocturnal activity (Curley et al. 2005, Linden et al. 2007).
Although synchronized by the SCN, functionally significant circadian oscillations in transcription are autonomous properties of individual cells across the body (Mohawk et al. 2012). These cell-autonomous oscillations alter the expression of genes to enable different tissues to coordinate their unique functions according to the circadian cycle. In neurons, splicing of a long noncoding RNA (lncRNA) at the paternally expressed Snord116 locus produces intronic 116 small nucleolar RNAs (snoRNAs) and the spliced 116 host gene (116HG). 116HG RNA accumulates in its transcription site, creating a cloud-like structure that condenses and expands during the light and dark phase, respectively (Powell et al. 2013). Deletion of the Snord116 locus causes phase-specific differential expression of thousands of genes, including core components of the cell autonomous clock such as cryptochrome and period genes. Interestingly, Snord116 deletions have increased fat metabolism during the light period, but it is currently unclear whether the 116 snoRNAs or 116HG is responsible for this phenotype. Cloud-like structures are also observed for maternally expressed host genes of snoRNA in the Dlk1-Dio3 cluster (Vitali et al. 2010), where expression of multiple transcripts undergoes circadian regulation (Labialle et al. 2008).
Imprinted Genes in the Social Brain
Increments in social interactions were likely a key driving force in the evolution of the size and processing capacity of mammalian brains (Dunbar & Shultz 2007). By virtue of their parent-of-origin regulation, parents influence their progeny through imprinted genes in parallel to the social relation that is parenting. Thus, genomic imprinting may have either contributed to, or been shaped by, the evolution of social behaviors.
Mother-pup interactions
Parenting in mammals is essential for infant survival. Mutations affecting the paternally expressed Mest and Peg3 result in a striking lack of maternal care (Curley et al. 2004, Lefebvre et al. 1998, Li et al. 1999), which supports the notion that genomic imprinting is associated with the evolution of social behaviors. In turn, pup behavior influences maternal responses strongly. Maternally inherited deletions encompassing Ube3a and the neighboring Atp10a and Gabrb3 increase ultrasonic vocalizations (USVs), specifically in the presence of maternal but not stranger’s bedding ( Jiang et al. 2010). This behavior has been interpreted as increased demand for maternal attention, which may be analogous to the happy disposition of AS patients (Isles 2011).
Reproductive behavior and puberty
The control of reproductive behavior and its development is an important target of genomic imprinting. Inbreeding is believed to be selected against to avoid harmful costs of homozygosity. Male and female hybrids of inbred as well as wild-derived mice avoid urine of their maternal strains and prefer to mate with their paternal strains (Isles et al. 2002, Montero et al. 2013), indicating a parental effect of unknown origin. Indeed, mate recognition is under the control of several imprinted genes. Males with Peg3 paternal deletions have impaired odor preferences for estrous females (Swaney et al. 2008), reduced sexual behavior, and hypothalamic responses to female cues (Swaney et al. 2007). Similarly, males with Magel2 paternal deletions show impaired preference for female odors (Mercer & Wevrick 2009).
Several aspects of sexual development are influenced strongly by imprinted genes. In humans, a genome-wide association study found significant parent-of-origin effects on late-onset menarche in polymorphic sites near the paternally expressed NDN, MAGEL2, and MKRN3 (supporting abnormal PWS puberty and hypogonadism) and within the maternally expressed KCNK9 and paternally expressed DLK1 (Perry et al. 2014). Male hypogonadism and reduced testosterone levels are observed in human paternally transmitted MAGEL2 null mutations and in mouse paternal Magel2 deletions, respectively (Schaaf et al. 2013). In female mice, Magel2 deletions also delay sexual maturation (Mercer & Wevrick 2009). Interestingly, Peg3 deletions in female mice also delay sexual maturation (Curley et al. 2004). Both MKRN3 null mutations (Abreu et al. 2013) and chromosome 14 maternal uniparental disomies exhibiting abnormal expression of DLK1, among other genes, cause human precocious puberty (Kotzot 2004). MKRN3 likely represses the onset of puberty, as its mouse hypothalamic expression declines at this time point (Abreu et al. 2013). In contrast, Dlk1 mouse hypothalamic expression rises at this time point, similar to the major activator of GnRH secretion: Kisspeptin (Villanueva et al. 2012). Finally, GnRH neuron migration to the mPOA is facilitated both by NDN (Miller et al. 2009) and by glycoconjugates synthesized by the maternally biased B3gnt2-encoded enzyme (Bless et al. 2006).
Social interactions and autism
Imprinted genes are involved in the regulation of social behavior between adult conspecifics. Mice with Grb10 paternal deletions are less likely to back down to opponents in tube tests and to remove the facial hair of their cage mates, which has been interpreted as increased social dominance (Curley 2011, Garfield et al. 2011).
Impaired reciprocal social interactions and other autistic traits are typically observed in mutations affecting the PWS and AS 15q11–13 interval. For instance, compulsive skin scraping is a common repetitive behavior in PWS individuals (Dykens et al. 2011) and is recapitulated in mice lacking Ndn (Muscatelli et al. 2000). Similarly, truncating mutations of human MAGEL2 result in autistic features (Schaaf et al. 2013), and male mice carrying paternal deletions of Magel2 show reduced social interactions and novel object exploration (Meziane et al. 2015). These mice exhibit multiple defects including reduced oxytocin and oxytocin receptor binding sites in the lateral septum. Remarkably, injecting oxytocin during the first postnatal week or 24 h before testing restores normal social behavior.
In humans, both paternal and maternal duplications or triplications of 15q11–13 result in neurodevelopmental deficits and, frequently, autism (Hogart et al. 2010). In mice, paternal duplications of the homologous region result in reduced social interactions, failure to adjust to novel conditions in multiple memory tests, and abnormal pup USVs (Nakatani et al. 2009). In addition, triplications of Ube3a normal expression in mice causes autism-like phenotypes, including reduced social recognition and interactions and abnormal USVs during social interactions, whereas doubling Ube3a’s dosage impairs these behaviors mildly (Smith et al. 2011). This is consistent with more pronounced autistic features observed in humans with 15q11–13 maternal triplications than duplications, suggesting that the brain is highly sensitive to UBE3A dosage (Hogart et al. 2010, Noor et al. 2015). Recently, a mutation enhancing UBE3A ubiquitin ligase activity has been implicated in autism (Yi et al. 2015). Because UBE3A self-ubiquitinates, its protein levels were actually reduced in individuals and mice carrying this mutation, suggesting that besides dosage, spatiotemporal control of UBE3A activity is critical. In support of potentially different etiologies between Ube3a overexpression and ligase hyperactivity, mice with Ube3a triplication had impaired pre-and postsynaptic transmission but normal spine density (Smith et al. 2011), whereas the missense mutation substantially increased spine density (Yi et al. 2015).
Emotional and Cognitive Behaviors
A growing body of evidence implicates imprinted genes in the regulation of complex brain functions, including emotions and cognition, relevant to human neuropsychiatric disorders. Studies in rodents may recapitulate specific neurobiological and behavioral components of these complex disorders (Donaldson & Hen 2015).
Anxiety and depression
In humans, anxiety disorders are highly comorbid with major depression, and the two are thought to affect common neural substrates (Ressler & Mayberg 2007). Increased hypothalamic-pituitary-adrenal axis activity under chronic stress diminishes glucocorticoid receptor (GR) action in the hippocampus and is associated with anxiety and depression (Arnett et al. 2015). UBE3A interacts with GR to induce its transcriptional activity while also inducing GR proteasomal degradation (Godavarthi et al. 2012). Importantly, Ube3a maternal deletions exhibit downregulation of GR and GR-targeted genes in the hippocampus, which is associated with increased corticosterone and anxiety-like behavior. Neuronal circuits of monoamine neurotransmitters are associated strongly with human anxiety and depression. Mutations in the human paternally expressed sarcoglycan-ε (SGCE) cause myoclonus dystonia (see the sidebar, Human Brain Disorders Associated with Imprinting Dysregulation) and are accompanied by high incidence of anxiety (Peall et al. 2013). Both motor deficits and anxiety- and depression-like behaviors are observed in mice with paternal Sgce deletions (Yokoi et al. 2006). Importantly, dopamine levels are increased in these animals, likely due to reductions in the dopamine autoreceptor D2R (Zhang et al. 2012). Paternal duplication of the PWS genes increases depression- and anxiety-like behaviors, which is associated with reductions in serotonin levels (Nakatani et al. 2009, Tamada et al. 2010). Maternally expressed KCNK9 regulates resting potentials of serotonergic neurons (Washburn et al. 2002) and some of their downstream neurons responsible for the mammalian startle reflex (Weber et al. 2008). Importantly, Kcnk9 deletions also show decreased depression-like behavior (Gotter et al. 2011).
Adult hippocampal neurogenesis is reduced by chronic stress, impaired in rodent models of depression, and increased by antidepressant treatment (Miller & Hen 2015). IGF2, which promotes adult neurogenesis (Bracko et al. 2012, Ferrón et al. 2015), is downregulated in animals exhibiting depression-like behavior after chronic stress (Luo et al. 2014). Remarkably, overexpression of Igf2 in the dentate gyrus reverses these behaviors, but whether this effect is dependent on adult neurogenesis remains to be tested.
Addiction
Drug addiction involves neuronal plasticity in the mesolimbic reward pathway, which is influenced by several imprinted genes. The paternally expressed SNORD115 is thought to prevent editing of the serotonin receptor Htr2c mRNA (Kishore & Stamm 2006). Repeated cocaine administration in mice reduces Snord115 expression in the NAc, whereas NAc overexpression of either Snord115 or Htr2c attenuates cocaine addiction significantly (Chen et al. 2014). Conditional knockouts of the maternally biased Ago2 in dopamine receptor D2-expressing neurons fail to develop addiction in a cocaine self-administration paradigm (Schaefer et al. 2010). The knockout additionally revealed downregulation of many miRNAs, the majority of which are maternally expressed. Administration of cocaine leads to upregulation of the maternally expressed miR369, which targets key addiction-implicated genes, including FosB.
Cognition and associated disorders
Executive functions rely on the neuronal underpinnings of attention, which are frequently impaired in neuropsychiatric disorders. PWS patients manifest deficits in attentional switching, working memory, and organization and planning skills ( Jauregi et al. 2007, Woodcock et al. 2009). Furthermore, patients with PWS specifically due to maternal uniparental disomy, who lose paternal expression of PWS genes and have doubled Ube3a expression, develop psychosis and schizophrenia (Boer et al. 2002). A mouse model of the maternal uniparental disomy shows reduced performance in attentional tasks and reduced prepulse inhibition, endophenotypes commonly observed in schizophrenics (Relkovic et al. 2010). Finally, a mouse model of DiGeorge syndrome (DS), a disorder with increased risk for schizophrenia, shows reduced adult hippocampal neurogenesis accompanied by lower Igf2 expression (Ouchi et al. 2013). Strikingly, IGF2 injections in the dentate gyrus restore neurogenesis and behavioral defects of DS mice, supporting IGF2 as a promising treatment for multiple neuropsychiatric disorders.
FROM MONOALLELIC EXPRESSION TO PARENTAL BIAS
Initial studies of genomic imprinting identified this phenomenon as the silencing of one of the parental alleles and the monoallelic expression of the other. However, imprinted genes such as Ube3a, Phlda2, and Gnas were shown to display parental allelic bias rather than strict monoallelic expression (Albrecht et al. 1997, Qian et al. 1997, Yu et al. 1998). Furthermore, the number of reported imprinted genes displaying parental biases has increased dramatically (Figure 4a), and specific genes display monoallelic or parentally biased expression, depending on the tissue or developmental stage (Martinez et al. 2014). Thus, genomic imprinting clearly manifests itself as a continuum from monoallelic to weakly biased parental expression, and we review here the functional and mechanistic implications of these findings.
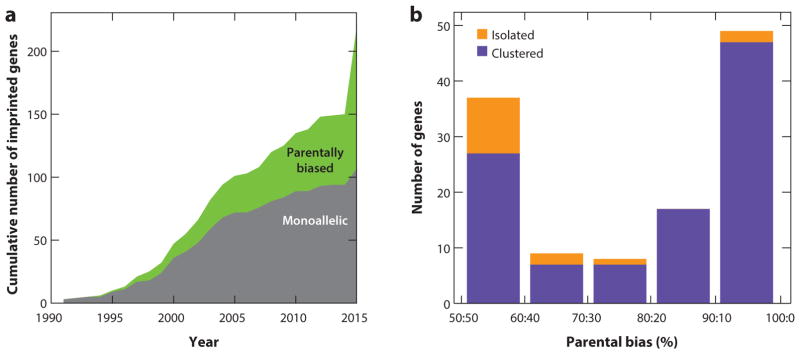
Figure 4.
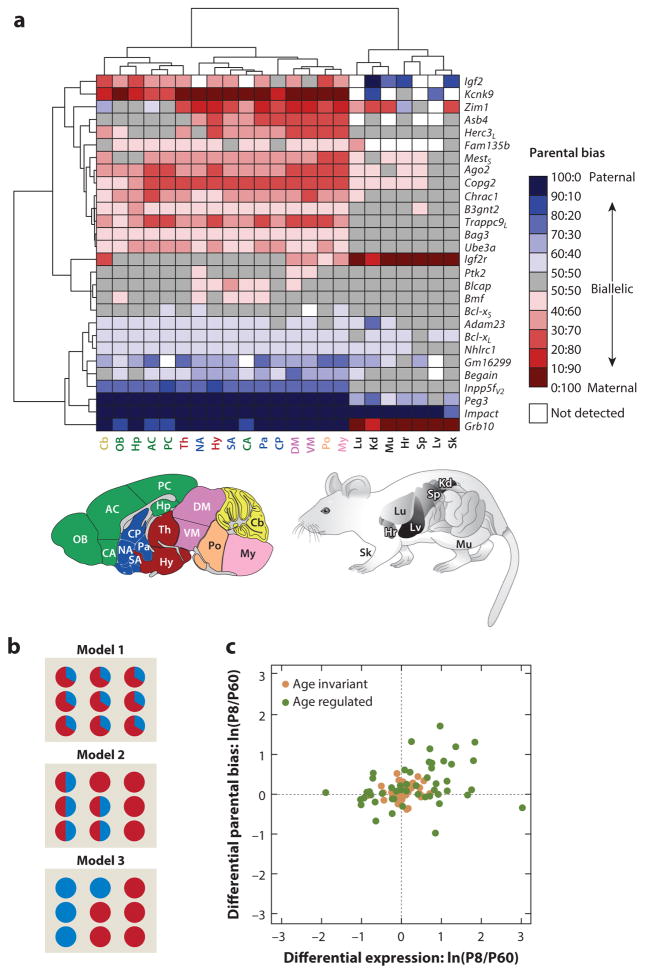
Monoallelic and parentally biased genes in mouse. (a) Cumulative increase in the number of imprinted genes reported in the literature. (b) The distribution of parental allelic expression ratios of clustered and isolated imprinted genes in the developing and adult cerebellum. Figure adapted from Perez et al. (2015).
Studies Across Organs and Organisms
Species exhibiting genomic imprinting show evidence of both parentally biased and monoallelically expressed genes and many parentally biased genes are shared across species. For instance, Dio3 and Igf2 are parentally biased in mouse and rat brains (Gregg et al. 2010, Martinez et al. 2014, Sittig & Redei 2014, Ye et al. 2015), IGF2R is parentally biased in humans and macaques (Babak et al. 2015, Cheong et al. 2015), and GNAS and BLCAP are parentally biased in human and mouse (Babak et al. 2015). Genes may also exhibit various degrees of parental biases in different species. For instance, L3mbtl exhibits a paternal bias in the mouse hypothalamus (Bonthuis et al. 2015), whereas its human ortholog is paternally expressed in most tissues (Babak et al. 2015). Finally, in flowering plants, the only other organisms outside mammals exhibiting parent-of-origin imprinting, most imprinted genes display parental biases rather than monoallelic expression (Gehring 2013). Because flowering plants and mammals are thought to share similar imprinting epigenetic machinery, a continuum from parental bias to monoallelic expression may reflect general properties of this epigenetic regulation and indicate selective advantages or constraints of partial, rather than complete, silencing of certain genes.
Highly sensitive expression measurements, such as RNA sequencing (RNA-seq), applied over many biological samples, analyzed with robust statistical methods, and independently validated, have led to accurate detection of a large spectrum of parental allelic biases (Bonthuis et al. 2015, Crowley et al. 2015, Perez et al. 2015) (Figure 4a). In whole brain, specific adult and developing brain regions, and muscle (Bonthuis et al. 2015, Crowley et al. 2015, Perez et al. 2015), the distribution of parental allelic expression ratios displays a clear bimodal shape, with one mode showing strong bias to monoallelic expression, whereas the other mode exhibits a weak bias (Figure 4b). This pattern was observed neither in the liver, which expresses very few imprinted genes (Bonthuis et al. 2015), nor in the placenta, where the latter is likely due to contamination from the maternal decidua (Finn et al. 2014, Okae et al. 2012).
Recent studies show that parental biases, in contrast to monoallelic expression, tend to be regulated dynamically according to tissue and developmental stage, particularly in the brain (Figure 5a) (Perez et al. 2015). The majority of biased genes either shift between parentally biased and biallelic expression in different tissues, brain regions, or developmental stages, or they display changes in the strength of parental bias in various tissues or brain regions. For some genes, the preferentially expressed parental allele even switches according to tissue. Fluctuations in parental biases can be shared among genes both within and among different clusters, suggesting coregulation of parental biases. These widespread tissue-specific and developmental changes in parental biases highlight the existence of intricate programs of imprinting regulation in the brain and raise the following important questions: (a) Are they a property of individual cells, or do they result from mixes of cells exhibiting monoallelic and biallelic expression? (b) Are their underlying epigenetic mechanisms common with those of monoallelic expression? And ultimately, (c) Is parental bias functionally relevant to the fitness of the organism, as is assumed for monoallelic expression?
Figure 5.
Regulation of parentally biased imprinted genes in mouse. (a) Spatial regulation of imprinting represented by a hierarchically clustered heat map of the deviation from biallelic gene expression. Panel adapted from Perez et al. (2015). L and S subscripts in the gene names indicate long and short isoforms, respectively, and V2 indicates variant 2 splice form. Acronyms of brain regions (color coded according to their broad developmental relatedness): AC, anterior cortex; CA, cortical amygdala; Cb, cerebellum; CP, caudate putamen; DM, dorsal midbrain; Hp, hippocampus; Hy, hypothalamus; My, medulla; NA, nucleus accumbens; OB, olfactory bulb; Pa, pallidum; PC, posterior cortex; Po, pons; SA, striatum-like amygdala; Th, thalamus; VM, ventral midbrain. Acronyms of nonbrain tissues: Hr, heart; Kd, kidney; Lu, lung; Lv, liver; Mu, muscle; Sk, skin; Sp, spleen. (b) Models providing the single cell–level bases for the parental biases observed in tissue homogenates. Each circle represents a single cell within the homogenized tissue. Red represents maternal expression, and blue represents paternal expression. In model 1, parental biases are identically present in each individual cell. In model 2, parental biases result from a mixture of cells exhibiting monoallelic expression with cells exhibiting biallelic expression. In model 3, parental biases result from a mixture of cells that monoallelically express either the maternal or paternal allele. (c) Age fold change {developing (P8) and adult (P60) cerebella [ln(P8/P60)]} in the magnitude of parental biases versus their age fold change in overall expression level. Panel adapted from Perez et al. (2015). Most genes exhibiting significant age-dependent changes in parental biases also show significant changes in overall expression, with the largest effects for both categories observed during development (Pearson correlation coefficient = 0.33; P = 4 × 10−4). Age-regulated imprinted genes are represented by green circles (those located near the origin show weak yet statistically significant age effects), and age-invariant imprinted genes are represented by brown circles.
Cellular Basis of Parental Biases
Although researchers have investigated and validated parental biases thoroughly in dissected tissues, their manifestation at the cellular level is still largely unknown (Gregg 2014). One model postulates that parental biases are present in each individual cell (Figure 5b), such that a chromatin state is able to partially repress expression. This model is consistent with the phenotype of heterozygous mutants for the maternally biased Gnas, which, in maternal versus paternal deletion, shows reduced but still detectable expression in cells of renal proximal tubules (Yu et al. 1998) and PVN (Chen et al. 2009). Similarly, a paternal Commd1 duplication due to chromosome 11 uniparental disomy exhibits fainter, yet similarly patterned, expression as wild types, suggesting its preferential-maternal rather than exclusive-maternal expression in all cells (Wang et al. 2004).
In a second model, parental biases result from coexisting cells exhibiting either monoallelic or biallelic expression (Figure 5b). Maternal transmission of a Ube3a-Yfp reporter shows strong labeling in both glia and neurons, whereas paternal transmission shows strong glial labeling yet faint neuronal signaling (Dindot et al. 2008), suggesting biallelic and nearly exclusive maternal expression in glia and neurons, respectively. Consistently, Ube3a is maternally expressed in neuronal cultures but biallelic in glial cultures (Yamasaki et al. 2003). Similarly, Snx14 appears biallelicaly expressed in cortex homogenates but is paternally expressed in sorted neurons (Huang et al. 2014).
A third model postulates that parental biases result from a mixture of cells with monoallelic expression, with the parental identity of the expressed allele different from cell to cell (Figure 5b). The parental identity of the expressed allele may be cell-type-specific, such as with Grb10, which, through alternative promoter usage, is paternally expressed in neurons but maternally expressed in glia (Yamasaki-Ishizaki et al. 2007). In another scenario, a cell selects only one allele for expression but with a probability that is higher for one parent of origin than the other (a biased coin flip). Parental biases of the beta-protocadherin genes in the brain support this model. Similar to other clustered protocadherins, the promoter of one beta-protocadherin is believed to be selected at each allele by an enhancer, resulting in monoallelic expression of 2 of the 22 Pcdhβs (one from each allele) in most neurons (Hirano et al. 2012). In homogenized cerebella, Pcdhβ12 and Pcdhβ10 show maternal biases, whereas Pcdhβ20 is paternally biased (Perez et al. 2015), suggesting that the probability of promoter selection, with respect to each, is higher for the preferentially expressed allele.
High-resolution in situ hybridization (ISH) studies appear well suited to interrogate parental biases at the single-cell level. Attempts in culture so far have highlighted significant variation in the allele-specific expression among single cells, making the results difficult to interpret (Herzing et al. 2002, Jouvenot et al. 1999). In mouse brain sections, Bonthuis et al. (2015) reported monoallelic ISH nascent mRNA signals in a large fraction of cells for several parentally biased genes. However, because parental identity of the alleles was unavailable, monoallelic signals due to transcriptional bursts unsynchronized between the alleles cannot be ruled out. Thus, the nature of imprinting in single cells remains largely unresolved and will benefit from future progress in quantitative allele-specific expression analyses at the single-cell level.
Functional Significance of Parental Biases
Perhaps the most important question raised by the existence of widespread parental biases is that of their functional relevance to the organism. Genetic analyses of behavioral and neurological defects highlight that normal brain development and function require a fine regulation of gene dosages, such that even small deviations may disturb the balance of the biological pathways they control, leading to striking neural dysfunctions. This has been observed for nonimprinted genes such as MECP2 in Rett syndrome and MECP2 duplication disorder (Chao & Zoghbi 2012), as well as variation in posttranscriptional regulation of MECP2 levels (Gennarino et al. 2015). In fragile X intellectual disability (FXS), FMRP mutations lead to increased metabotropic glutamate receptor (mGLUR) signaling, and reductions in mGluR dosage in an FXS mouse model was shown to rescue cognitive phenotypes (Dölen et al. 2007). In an intervention that may be analogous to dosage regulation, chronic treatment of FXS mice with an mGLUR inhibitor also rescued the cognitive defects (Michalon et al. 2012).
Similarly, are changes in the parental biases of certain imprinted genes regulating the overall dosage of a gene? Remarkably, developmental variations in the parental biases of many imprinted genes in the mouse cerebellum were shown to be associated with overall expression changes (Perez et al. 2015) (Figure 5c). For most genes, both the parental bias and overall expression are higher during development, and a downregulation of the preferentially expressed allele is observed from development to adulthood, suggesting a tight control of imprinted gene dosages during critical developmental points, similar to what is seen with Mecp2 and mGluRs. These transcriptional changes may occur within individual cells or may reflect changes in cell types populating the cerebellum at different developmental stages.
Pathways regulating apoptosis and monoamine neurotransmission appear preferentially enriched among parentally biased and monoallelically imprinted genes (Figure 6). Recent genetic analyses of these pathways suggest that even moderate parental biases exert key roles in assuring normal brain development and function.
Figure 6.
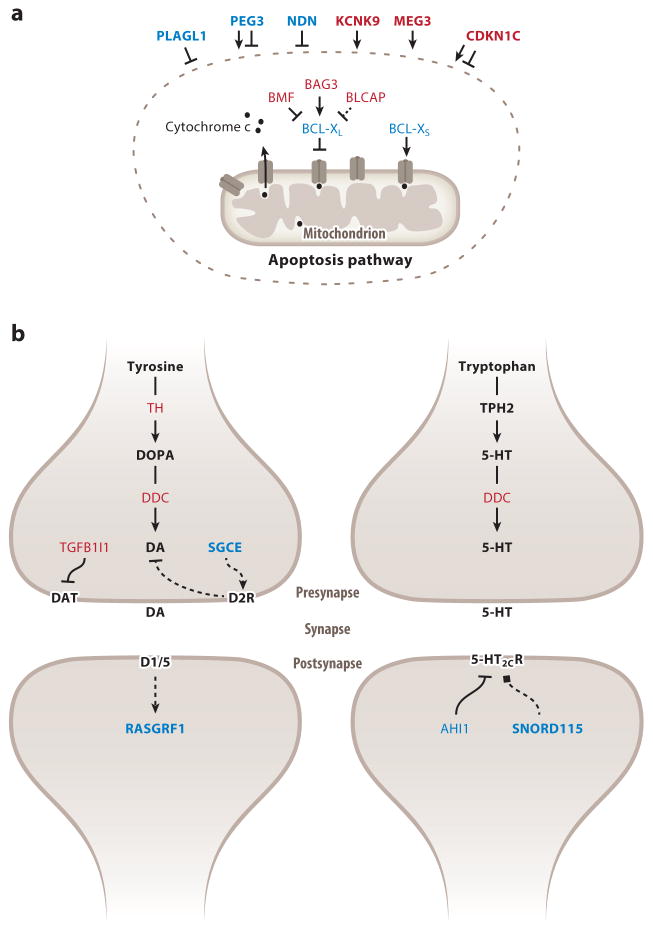
Parentally biased and strongly imprinted genes are enriched in two biological pathways in the brain. (a) Regulation of apoptosis. Within the circle are products of imprinted genes that interact directly with the apoptosis machinery, and outside the circle are products of imprinted genes that influence pro- or antiapoptosis. L and S subscripts in the gene names indicate long and short isoforms, respectively. (b) Brain monoaminergic pathway. Genes preferentially or exclusively expressed from the maternal and paternal allele appear in red and blue, respectively. Products of strongly biased or monoallelically expressed genes are in bold. Lines with arrowheads and lines with notched ends indicate stimulation and inhibition, respectively, of the given phenotype, whereas lines with square ends indicate functions that cannot be defined as enhancement or reduction. Full lines indicate direct interactions, whereas dashed lines indicate indirect effect. Abbreviations: 5-HT, serotonin; 5-HT2CR, 5-HT2C receptor; D1/5, dopamine receptor 1 or 5; D2R, dopamine receptor 2; DA, dopamine; DAT, dopamine transporter; DDC, DOPA decarboxylase; TH, tyrosine hydroxylase.
The paternally biased Bcl-xL produces an essential antiapoptotic signal during brain development (Motoyama et al. 1995). Paternal but not maternal deletion of Bcl-x was shown to reduce brain size and, in the cortex, led to specific loss of excitatory neurons, thus demonstrating the functional significance of this gene’s parental biases during brain development (Perez et al. 2015). The antiapoptotic Bag3 shows stronger maternal biases in brain regions where Bcl-xL shows weaker paternal biases and vice versa, suggesting that parental biases of these two genes may influence apoptosis in distinct brain regions. The proapoptotic Bmf and Blcap also exhibit spatially regulated maternal biases in the brain (Perez et al. 2015). These observations support the notion that the life-death balance of brain cells is regulated partly by parental biases in pro- and antiapoptotic imprinted genes (Figure 6a).
In the brain monoaminergic system (Figure 6b), maternally biased genes generally increase monoamine levels, whereas paternally biased genes may selectively reduce their postsynaptic effects (Bonthuis et al. 2015). Most notably, tyrosine hydroxylase (TH), which limits the rate of dopamine and norepinephrine synthesis, shows a weak maternal bias in monoaminergic nuclei. Maternal deletion of Th was shown to exhibit lower anxiety- and anhedonia-like behavior than paternal deletions, thus supporting the functional significance of this gene’s maternal bias (Bonthuis et al. 2015). Interestingly, DOPA decarboxylase, an enzyme necessary for the synthesis of all monoamines, is maternally biased in monoaminergic nuclei. Tgfb1i1, which encodes a dopamine transporter inhibitor (Carneiro et al. 2002).is also maternally biased in all monoaminergic nuclei (Bonthuis et al. 2015). Finally, Ahi1, whose encoded protein increases feeding by downregulating 5-HT2CR in the arcuate nucleus (ARN) (Wang et al. 2012), shows paternal biases in the ARN and monoaminergic nuclei. Considering the extensive effects of brain monoamines, these results suggest widespread influences of parental biases on brain function.
The Regulation of Parentally Biased Expression
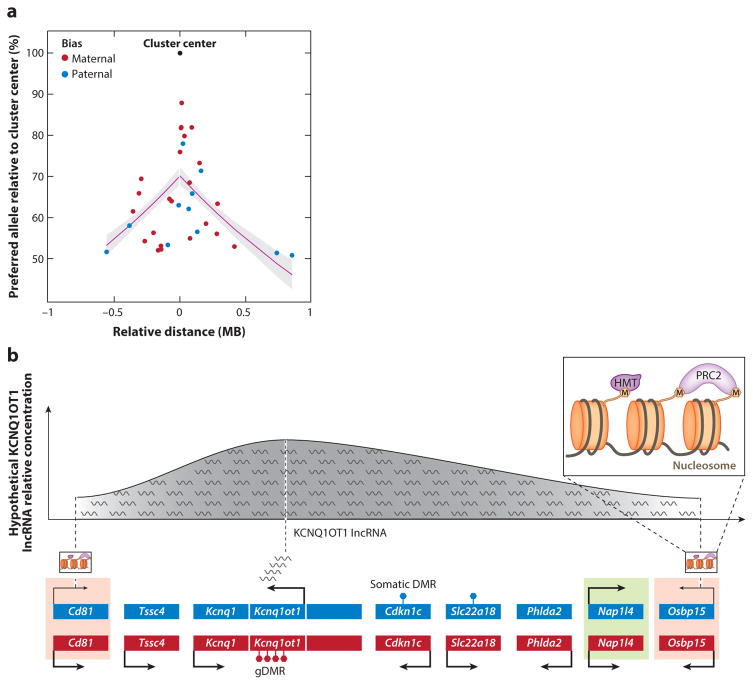
Differential DNA methylation of regulatory regions in one parental gamete but not the other instructs the chromatin machinery to silence alleles in cis, leading to parent-of-origin expression in imprinted genes (Bartolomei & Ferguson-Smith 2011). Most known monoallelically expressed genes are clustered around at least one germ-line differentially methylated region (gDMR), whereas approximately 70% of parentally biased genes localize to the periphery of these clusters. The remaining 30% of parentally biased genes are isolated, among which four genes appear near a gDMR (Perez et al. 2015). The location of parentally biased genes at the periphery of imprinted clusters suggests that they may experience cis acting signals differently than monoallelic genes. Consistently, and although the three-dimensional effects in the nucleus are not accounted for, the strength of parental bias decays as a function of the genomic distance from the gDMR (Figure 7a) (Perez et al. 2015). Moreover, the synteny of weakly biased genes in imprinted clusters is strongly conserved in mammals, suggesting selective constraints on this genomic organization.
Figure 7.
Mechanistic models of parentally biased expression. (a) The decay of parental bias of imprinted genes as a function of their relative linear genomic distance from an imprinted cluster center (which is defined as the location of the monoallelically expressed imprinted genes in the cluster) in the cerebellum. The regression line is shown in magenta, and the gray shading indicates corresponding standard errors (Perez et al. 2015). (b) Known mechanisms of genomic imprinting in the mouse Kcnq1 imprinted cluster and their potential extrapolation to mechanisms of parental biases. The Kcnq1 intergenic, maternally methylated gDMR induces paternal-specific expression of Kcnq1ot1, an lncRNA that orchestrates imprinting of multiple genes at both ends of this cluster (Mancini-Dinardo et al. 2006, Terranova et al. 2008). In the placenta, genes closer to the gDMR, such as Cdkn1c, are monoallelically expressed in a DNA methylation–dependent manner (indicated by filled hexagons on top of gene bodies), whereas genes at the cluster periphery, such as Cd81 and Osbpl5, exhibit DNA methylation–independent maternal biases (red shading) (Lewis et al. 2004, Umlauf et al. 2004). Another peripheral gene, Nap1l4, is biallelically expressed ( green shading). Imprinting in these maternally biased genes is dependent on interactions between their respective promoters and Kcnq1ot1 transcripts, which recruit HMTs and PRC2, forming a contracted nuclear domain (Pandey et al. 2008, Terranova et al. 2008). Importantly, KCNQ1OT1 lncRNA does not interact with the Nap1l4 promoter. We suggest a potential model explaining parentally biased expression in this cluster (depicted on top) where the effective concentration or activity of the KCNQ1OT1 lncRNA across the length of the cluster may exhibit a normal distribution centered in its transcriptional site that decreases gradually toward the ends. In this model, the differences in KCNQ1OT1 lncRNA concentration or activity may explain its different actions in central and peripheral genes. Abbreviations: gDMR, germ-line differentially methylated region; HMT, histone methyltransferase; lncRNA, long noncoding RNA; PRC2, polycomb repressive complex 2.
Mechanistically, the function of the maternal gDMR within the Kcnq1 cluster suggests a model underlying the spread of imprinting regulation initiated at gDMRs and the generation of parental biases (Figure 7b). The gDMR of this cluster restricts expression of Kcnq1ot1, an lncRNA, to the paternal allele. KCNQ1OT1 spreads peripherally and recruits repressive chromatin machinery, including polycomb repressive complexes (PRCs), to promoters of nearby genes (Mancini-Dinardo et al. 2006, Terranova et al. 2008). The diffusion rate and/or half-life of the KCNQ1OT1 lncRNA (Figure 7b), the silencing capacity of the recruited repressive chromatin, and the resulting three-dimensional position in the nucleus may thus affect the degree of repression. Mechanisms reminiscent of position effect variegation, which in flies consists of the stochastic spread of heterochromatin to adjacent genes (Elgin & Reuter 2013), may represent alternative models.
However, the fact that most parentally biased isolated genes are not associated with gDMRs, suggests mechanisms independent of DNA-methylation. PRC2 represents an attractive candidate because its genomic position can be conserved after cell division (Steffen & Ringrose 2014). Indeed, PRC2-associated histone modifications are present in Smfbt2 and Gab1 maternal alleles, two isolated paternally expressed genes in the placenta which lack DMRs (Okae et al. 2012, Wang et al. 2011).
Finally, genome-wide methylation profiles of mouse and human cortical neurons have revealed that non-CG DNA cytosines accumulate methylation during postnatal development (He & Ecker 2015, Lister et al. 2013). It was additionally found that intragenic and near–transcription start site non-CG as well as CG methylation levels are higher in cells in which the corresponding genes are not expressed (Mo et al. 2015). This pattern is paralleled by accumulation of hydroxymethyl CGs (hmCs), the oxidized form of methylated CGs catalyzed by TET hydroxylase proteins, thought to represent an intermediate between methylated and nonmethylated cytosines (He & Ecker 2015, Lister et al. 2013). Although Xie et al. (2012) observed higher levels of non-CG methylation in the silenced allele at a few imprinted loci in the mouse brain, the relationships between this type of methylation as well as that of hmC with imprinting and parental bias are not yet known, and they represent an interesting avenue for future research.
CONCLUDING REMARKS
Although early studies of genomic imprinting highlighted its roles during embryonic and placental growth, its pleiotropic influences on brain development and function have emerged more recently. These include positive and negative controls by both maternally and paternally preferentially expressed genes of the generation, wiring, and survival of cells in the developing and adult brain. Consistent with the kinship theory of genomic imprinting (see the sidebar, Evolutionary Theories of Genomic Imprinting), paternally expressed genes promote suckling and growth in newborns, and to a lesser extent, maternally expressed genes reduce growth and increase metabolic rate in both newborns and adults. These results are also consistent with the roles of imprinted genes in embryonic development (Tunster et al. 2013). Paternally expressed genes also appear to contribute to the regulation of puberty as well as to reproductive and maternal behaviors. Interestingly, maternally expressed genes appear to be the main contributors of positive and negative regulation of emotional and cognitive behaviors.
The importance of imprinted genes in brain function is evidenced by the devastating neurological and behavioral conditions resulting from mutations in these loci in humans and mice. Strikingly, recent advances suggest that imprinting disorders may be amenable for therapeutic intervention and that their pathological manifestations could be, at least partially, rescued by reactivating the usually unaffected repressed allele. Indeed, reactivation in a mouse model of AS of the repressed Ube3a paternal allele rescued cognitive features, even in adults (Huang et al. 2012, Meng et al. 2014). However, the rescue of other AS phenotypes may require embryonic intervention (Silva-Santos et al. 2015).
An emerging theme is that many imprinted genes exhibit various degrees of parental expression biases. Many of these biases were evidenced recently thanks to the increased resolution afforded by RNA-seq, suggesting that additional imprinted genes may still be discovered, particularly within tissues of high cellular and functional diversity such as the brain. Consequently, single cell–resolution studies will be necessary for a better mechanistic and biological understanding of parental biases in the brain and other complex tissues. Finally, researchers have proposed that imprinted genes form a regulatory network influencing common biological functions (Al Adhami et al. 2015, Varrault et al. 2006), possibly impinging on nonimprinted genes (Mott et al. 2014). Such a network may be impaired by dosage alterations in specific genes and compensated by titration in other genes, therefore representing a unique opportunity to study brain function at a polygenic level.
Acknowledgments
We thank David Haig for critically reading the manuscript. We also thank Harvard Molecular and Cellular Biology Graphics team for their invaluable assistance in producing the figures. C.D. is an investigator of the Howard Hughes Medical Institute, and some of the work described in this review has been supported by the National Institute of Mental Health’s Silvio O. Conte Center Grant (P50MH094271) for Basic and Translational Research.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- Abreu AP, Dauber A, Macedo DB, Noel SD, Brito VN, et al. Central precocious puberty caused by mutations in the imprinted gene MKRN3. N Engl J Med. 2013;368(26):2467–75. doi: 10.1056/NEJMoa1302160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahuja R, Pinyol R, Reichenbach N, Custer L, Klingensmith J, et al. Cordon-bleu is an actin nucleation factor and controls neuronal morphology. Cell. 2007;131(2):337–50. doi: 10.1016/j.cell.2007.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainsley JA, Drane L, Jacobs J, Kittelberger KA, Reijmers LG. Functionally diverse dendritic mRNAs rapidly associate with ribosomes following a novel experience. Nat Commun. 2014;5:4510. doi: 10.1038/ncomms5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Adhami H, Evano B, Le Digarcher A, Gueydan C, Dubois E, et al. A systems-level approach to parental genomic imprinting: The imprinted gene network includes extracellular matrix genes and regulates cell cycle exit and differentiation. Genome Res. 2015;25:353–67. doi: 10.1101/gr.175919.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht U, Sutcliffe JS, Cattanach BM, Beechey CV, Armstrong D, et al. Imprinted expression of the murine Angelman syndrome gene, Ube3a, in hippocampal and Purkinje neurons. Nat Genet. 1997;17(1):75–78. doi: 10.1038/ng0997-75. [DOI] [PubMed] [Google Scholar]
- Angulo MA, Butler MG, Cataletto ME. Prader-Willi syndrome: a review of clinical, genetic, and endocrine findings. J Endocrinol Investig. 2015;38(12):1249–63. doi: 10.1007/s40618-015-0312-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnett MG, Muglia LM, Laryea G, Muglia LJ. Genetic approaches to hypothalamic-pituitary-adrenal axis regulation. Neuropsychopharmacology. 2015 doi: 10.1038/npp.2015.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asmus F, Zimprich A, Tezenas Du Montcel S, Kabus C, Deuschl G, et al. Myoclonus-dystonia syndrome: ε-sarcoglycan mutations and phenotype. Ann Neurol. 2002;52(4):489–92. doi: 10.1002/ana.10325. [DOI] [PubMed] [Google Scholar]
- Babak T, Deveale B, Tsang EK, Zhou Y, Li X, et al. Genetic conflict reflected in tissue-specific maps of genomic imprinting in human and mouse. Nat Genet. 2015;47:544–49. doi: 10.1038/ng.3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey CH, Kandel ER, Harris KM. Structural components of synaptic plasticity and memory consolidation. Cold Spring Harb Perspect Biol. 2015;7(7):a021758. doi: 10.1101/cshperspect.a021758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball ST, Kelly ML, Robson JE, Turner MD, Harrison J, et al. Gene dosage effects at the imprinted Gnas cluster. PLOS ONE. 2013;8(6):e65639. doi: 10.1371/journal.pone.0065639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bando Y, Hirano T, Tagawa Y. Dysfunction of KCNK potassium channels impairs neuronal migration in the developing mouse cerebral cortex. Cereb Cortex. 2014;24(4):1017–29. doi: 10.1093/cercor/bhs387. [DOI] [PubMed] [Google Scholar]
- Barel O, Shalev SA, Ofir R, Cohen A, Zlotogora J, et al. Maternally inherited Birk Barel mental retardation dysmorphism syndrome caused by a mutation in the genomically imprinted potassium channel KCNK9. Am J Hum Genet. 2008;83(2):193–99. doi: 10.1016/j.ajhg.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow DP, Stöger R, Herrmann BG, Saito K, Schweifer N. The mouse insulin-like growth factor type-2 receptor is imprinted and closely linked to the Tme locus. Nature. 1991;349(6304):84–87. doi: 10.1038/349084a0. [DOI] [PubMed] [Google Scholar]
- Bartolomei MS, Ferguson-Smith AC. Mammalian genomic imprinting. Cold Spring Harb Perspect Biol. 2011;3(7):a002592. doi: 10.1101/cshperspect.a002592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomei MS, Zemel S, Tilghman SM. Parental imprinting of the mouse H19 gene. Nature. 1991;351(6322):153–55. doi: 10.1038/351153a0. [DOI] [PubMed] [Google Scholar]
- Bastepe M. Relative functions of Gαs and its extra-large variant XLαs in the endocrine system. Horm Metab Res. 2012;44(10):732–40. doi: 10.1055/s-0032-1316331. [DOI] [PubMed] [Google Scholar]
- Batassa EM, Costanzi M, Saraulli D, Scardigli R, Barbato C, et al. RISC activity in hippocampus is essential for contextual memory. Neurosci Lett. 2010;471(3):185–88. doi: 10.1016/j.neulet.2010.01.038. [DOI] [PubMed] [Google Scholar]
- Bird LM. Angelman syndrome: review of clinical and molecular aspects. Appl Clin Genet. 2014;7:93–104. doi: 10.2147/TACG.S57386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof JM, Stewart CL, Wevrick R. Inactivation of the mouse Magel2 gene results in growth abnormalities similar to Prader-Willi syndrome. Hum Mol Genet. 2007;16(22):2713–19. doi: 10.1093/hmg/ddm225. [DOI] [PubMed] [Google Scholar]
- Bista P, Cerina M, Ehling P, Leist M, Pape H-C, et al. The role of two-pore-domain background K+ (K2P) channels in the thalamus. Pflügers Arch Eur J Physiol. 2015;467(5):895–905. doi: 10.1007/s00424-014-1632-x. [DOI] [PubMed] [Google Scholar]
- Bittel DC, Kibiryeva N, McNulty SG, Driscoll DJ, Butler MG, White RA. Whole genome microarray analysis of gene expression in an imprinting center deletion mouse model of Prader-Willi syndrome. Am J Med Genet. 2007;143A(5):422–29. doi: 10.1002/ajmg.a.31504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bless E, Raitcheva D, Henion TR, Tobet S, Schwarting GA. Lactosamine modulates the rate of migration of GnRH neurons during mouse development. Eur J Neurosci. 2006;24(3):654–60. doi: 10.1111/j.1460-9568.2006.04955.x. [DOI] [PubMed] [Google Scholar]
- Boer H, Holland A, Whittington J, Butler J, Webb T, Clarke D. Psychotic illness in people with Prader Willi syndrome due to chromosome 15 maternal uniparental disomy. Lancet. 2002;359(9301):135–36. doi: 10.1016/S0140-6736(02)07340-3. [DOI] [PubMed] [Google Scholar]
- Bonthuis PJ, Huang W-C, Hörndli CNS, Ferris E, Cheng T, Gregg C. Noncanonical genomic imprinting effects in offspring. Cell Rep. 2015;12(6):979–91. doi: 10.1016/j.celrep.2015.07.017. [DOI] [PubMed] [Google Scholar]
- Bracko O, Singer T, Aigner S, Knobloch M, Winner B, et al. Gene expression profiling of neural stem cells and their neuronal progeny reveals IGF2 as a regulator of adult hippocampal neurogenesis. J Neurosci. 2012;32(10):3376–87. doi: 10.1523/JNEUROSCI.4248-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambilla R, Gnesutta N, Minichiello L, White G, Roylance AJ, et al. A role for the Ras signalling pathway in synaptic transmission and long-term memory. Nature. 1997;390(6657):281–86. doi: 10.1038/36849. [DOI] [PubMed] [Google Scholar]
- Brickley SG, Aller MI, Sandu C, Veale EL, Alder FG, et al. TASK-3 two-pore domain potassium channels enable sustained high-frequency firing in cerebellar granule neurons. J Neurosci. 2007;27(35):9329–40. doi: 10.1523/JNEUROSCI.1427-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broad KD, Curley JP, Keverne EB. Increased apoptosis during neonatal brain development underlies the adult behavioral deficits seen in mice lacking a functional paternally expressed gene 3 (Peg3) Dev Neurobiol. 2009;69(5):314–25. doi: 10.1002/dneu.20702. [DOI] [PubMed] [Google Scholar]
- Broad KD, Keverne EB. Placental protection of the fetal brain during short-term food deprivation. PNAS. 2011;108(37):15237–41. doi: 10.1073/pnas.1106022108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro AM, Ingram SL, Beaulieu J-M, Sweeney A, Amara SG, et al. The multiple LIM domain-containing adaptor protein Hic-5 synaptically colocalizes and interacts with the dopamine transporter. J Neurosci. 2002;22(16):7045–54. doi: 10.1523/JNEUROSCI.22-16-07045.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy SB, Schwartz S, Miller JL, Driscoll DJ. Prader-Willi syndrome. Genet Med. 2012;14(1):10–26. doi: 10.1038/gim.0b013e31822bead0. [DOI] [PubMed] [Google Scholar]
- Causeret F, Jacobs T, Terao M, Heath O, Hoshino M, Nikolic M. Neurabin-I is phosphorylated by Cdk5: implications for neuronal morphogenesis and cortical migration. Mol Biol Cell. 2007;18(11):4327–42. doi: 10.1091/mbc.E07-04-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao H-T, Zoghbi HY. MeCP2: Only 100% will do. Nat Neurosci. 2012;15(2):176–77. doi: 10.1038/nn.3027. [DOI] [PubMed] [Google Scholar]
- Charalambous M, da Rocha ST, Ferguson-Smith AC. Genomic imprinting, growth control and the allocation of nutritional resources: consequences for postnatal life. Curr Opin Endocrinol Diabetes Obes. 2007;14(1):3–12. doi: 10.1097/MED.0b013e328013daa2. [DOI] [PubMed] [Google Scholar]
- Charalambous M, Ferrón SR, da Rocha ST, Murray AJ, Rowland T, et al. Imprinted gene dosage is critical for the transition to independent life. Cell Metab. 2012;15(2):209–21. doi: 10.1016/j.cmet.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen DY, Stern SA, Garcia-Osta A, Saunier-Rebori B, Pollonini G, et al. A critical role for IGF-II in memory consolidation and enhancement. Nature. 2011;469(7331):491–97. doi: 10.1038/nature09667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Qiang H, Fan K, Wang S, Zheng Z. The snoRNA MBII-52 regulates cocaine-induced conditioned place preference and locomotion in mice. PLOS ONE. 2014;9(6):e99986. doi: 10.1371/journal.pone.0099986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Berger A, Kablan A, Zhang J, Gavrilova O, Weinstein LS. Gsαdeficiency in the paraventricular nucleus of the hypothalamus partially contributes to obesity associated with Gsαmutations. Endocrinology. 2012;153(9):4256–65. doi: 10.1210/en.2012-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Gavrilova O, Liu J, Xie T, Deng C, et al. Alternative Gnas gene products have opposite effects on glucose and lipid metabolism. PNAS. 2005;102(20):7386–91. doi: 10.1073/pnas.0408268102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Wang J, Dickerson KE, Kelleher J, Xie T, et al. Central nervous system imprinting of the G protein Gsα and its role in metabolic regulation. Cell Metab. 2009;9(6):548–55. doi: 10.1016/j.cmet.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WV, Maniatis T. Clustered protocadherins. Development. 2013;140(16):3297–302. doi: 10.1242/dev.090621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong CY, Chng K, Ng S, Chew SB, Chan L, Ferguson-Smith AC. Germline and somatic imprinting in the nonhuman primate highlights species differences in oocyte methylation. Genome Res. 2015;25:611–23. doi: 10.1101/gr.183301.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S-H, Marzban H, Aldinger K, Dixit R, Millen K, et al. Zac1 plays a key role in the development of specific neuronal subsets in the mouse cerebellum. Neural Dev. 2011;6(1):25. doi: 10.1186/1749-8104-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JE, Lee PR, Chen S, Li W, Fields RD. MicroRNA regulation of homeostatic synaptic plasticity. PNAS. 2011;108(28):11650–55. doi: 10.1073/pnas.1017576108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colas D, Wagstaff J, Fort P, Salvert D, Sarda N. Sleep disturbances in Ube3a maternal-deficient mice modeling Angelman syndrome. Neurobiol Dis. 2005;20(2):471–78. doi: 10.1016/j.nbd.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Cougot N, Bhattacharyya SN, Tapia-Arancibia L, Bordonné R, Filipowicz W, et al. Dendrites of mammalian neurons contain specialized P-body-like structures that respond to neuronal activation. J Neurosci. 2008;28(51):13793–804. doi: 10.1523/JNEUROSCI.4155-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley M, Garfield AS, Madon-Simon M. Developmental programming mediated by complementary roles of imprinted Grb10 in mother and pup. PLOS Biol. 2014;12(2):e1001799. doi: 10.1371/journal.pbio.1001799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley JJ, Zhabotynsky V, Sun W, Huang S, Pakatci IK, et al. Analyses of allele-specific gene expression in highly divergent mouse crosses identifies pervasive allelic imbalance. Nat Genet. 2015;47:353–60. doi: 10.1038/ng.3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubelos B, Sebastián-Serrano A, Beccari L, Calcagnotto ME, Cisneros E, et al. Cux1 and Cux2 regulate dendritic branching, spine morphology, and synapses of the upper layer neurons of the cortex. Neuron. 2010;66(4):523–35. doi: 10.1016/j.neuron.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curley JP. Is there a genomically imprinted social brain? BioEssays. 2011;33(9):662–68. doi: 10.1002/bies.201100060. [DOI] [PubMed] [Google Scholar]
- Curley JP, Barton S, Surani A, Keverne EB. Coadaptation in mother and infant regulated by a paternally expressed imprinted gene. Proc R Soc B. 2004;271(1545):1303–9. doi: 10.1098/rspb.2004.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curley JP, Pinnock SB, Dickson SL, Thresher R, Miyoshi N, et al. Increased body fat in mice with a targeted mutation of the paternally expressed imprinted gene Peg3. FASEB J. 2005;19(10):1302–4. doi: 10.1096/fj.04-3216fje. [DOI] [PubMed] [Google Scholar]
- DeChiara TM, Robertson EJ, Efstratiadis A. Parental imprinting of the mouse insulin-like growth factor II gene. Cell. 1991;64(4):849–59. doi: 10.1016/0092-8674(91)90513-x. [DOI] [PubMed] [Google Scholar]
- Dindot SV, Antalffy BA, Bhattacharjee MB, Beaudet AL. The Angelman syndrome ubiquitin ligase localizes to the synapse and nucleus, and maternal deficiency results in abnormal dendritic spine morphology. Hum Mol Genet. 2008;17(1):111–18. doi: 10.1093/hmg/ddm288. [DOI] [PubMed] [Google Scholar]
- Ding F, Li HH, Zhang S, Solomon NM, Camper SA, et al. SnoRNA Snord116 (Pwcr1/MBII-85) deletion causes growth deficiency and hyperphagia in mice. PLOS ONE. 2008;3(3):e1709. doi: 10.1371/journal.pone.0001709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Isa R, Clapcote SJ, Voikar V, Wolfer DP, Giese P, et al. Mice lacking Ras-GRF1 show contextual fear conditioning but not spatial memory impairments: convergent evidence from two independently generated mouse mutant lines. Front Behav Neurosci. 2011;5:00078. doi: 10.3389/fnbeh.2011.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dölen G, Osterweil E, Rao BSS, Smith GB, Auerbach BD, et al. Correction of fragile X syndrome in mice. Neuron. 2007;56(6):955–62. doi: 10.1016/j.neuron.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson ZR, Hen R. From psychiatric disorders to animal models: a bidirectional and dimensional approach. Biol Psychiatry. 2015;77(1):15–21. doi: 10.1016/j.biopsych.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake NM, Park YJ, Shirali AS, Cleland TA, Soloway PD. Imprint switch mutations at Rasgrf1 support conflict hypothesis of imprinting and define a growth control mechanism upstream of IGF1. Mamm Genome. 2009;20(9–10):654–63. doi: 10.1007/s00335-009-9192-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbar RIM, Shultz S. Evolution in the social brain. Science. 2007;317(5843):1344–47. doi: 10.1126/science.1145463. [DOI] [PubMed] [Google Scholar]
- Dykens EM, Lee E, Roof E. Prader-Willi syndrome and autism spectrum disorders: an evolving story. J Neurodev Disord. 2011;3(3):225–37. doi: 10.1007/s11689-011-9092-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton SA, Williamson CM, Ball ST, Beechey CV, Moir L, et al. New mutations at the imprinted Gnas cluster show gene dosage effects of Gsα in postnatal growth and implicate XLαs in bone and fat metabolism but not in suckling. Mol Cell Biol. 2012;32(5):1017–29. doi: 10.1128/MCB.06174-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggermann T, de Nanclares GP, Maher ER, Temple IK, Tümer Z, et al. Imprinting disorders: a group of congenital disorders with overlapping patterns of molecular changes affecting imprinted loci. Clin Epigenet. 2015;7(1):123. doi: 10.1186/s13148-015-0143-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin SCR, Reuter G. Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. Cold Spring Harb Perspect Biol. 2013;5(8):a017780. doi: 10.1101/cshperspect.a017780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrón SR, Charalambous M, Radford E, Mcewen K, Wildner H, et al. Postnatal loss of Dlk1 imprinting in stem cells and niche astrocytes regulates neurogenesis. Nature. 2012;475(7356):381–85. doi: 10.1038/nature10229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrón SR, Radford EJ, Domingo-Muelas A, Kleine I, Ramme A, et al. Differential genomic imprinting regulates paracrine and autocrine roles of IGF2 in mouse adult neurogenesis. Nat Commun. 2015;6:8265. doi: 10.1038/ncomms9265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn EH, Smith CL, Rodriguez J, Sidow A, Baker JC. Maternal bias and escape from X chromosome imprinting in the midgestation mouse placenta. Dev Biol. 2014;390(1):80–92. doi: 10.1016/j.ydbio.2014.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore R, Khudayberdiev S, Christensen M, Siegel G, Flavell SW, et al. Mef2-mediated transcription of the miR379–410 cluster regulates activity-dependent dendritogenesis by fine-tuning Pumilio2 protein levels. EMBO J. 2009;28(6):697–710. doi: 10.1038/emboj.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore R, Rajman M, Schwale C, Bicker S, Antoniou A, et al. MiR-134-dependent regulation of Pumilio-2 is necessary for homeostatic synaptic depression. EMBO J. 2014;33(19):2231–46. doi: 10.15252/embj.201487921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutachi S, Matsumoto A, Nakayama KI, Gotoh Y. p57 controls adult neural stem cell quiescence and modulates the pace of lifelong neurogenesis. EMBO J. 2013;32(7):970–81. doi: 10.1038/emboj.2013.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Wang W-Y, Mao Y-W, Gräff J, Guan J-S, et al. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. 2010;466(7310):1105–9. doi: 10.1038/nature09271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q, Horvath TL. Neurobiology of feeding and energy expenditure. Annu Rev Neurosci. 2007;30(1):367–98. doi: 10.1146/annurev.neuro.30.051606.094324. [DOI] [PubMed] [Google Scholar]
- Garfield AS, Cowley M, Smith FM, Moorwood K, Stewart-Cox JE, et al. Distinct physiological and behavioural functions for parental alleles of imprinted Grb10. Nature. 2011;469(7331):534–38. doi: 10.1038/nature09651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaughwin P, Ciesla M, Yang H, Lim B, Brundin P. Stage-specific modulation of cortical neuronal development by Mmu-miR-134. Cereb Cortex. 2011;21(8):1857–69. doi: 10.1093/cercor/bhq262. [DOI] [PubMed] [Google Scholar]
- Gehring M. Genomic imprinting: insights from plants. Annu Rev Genet. 2013;47(1):187–208. doi: 10.1146/annurev-genet-110711-155527. [DOI] [PubMed] [Google Scholar]
- Gennarino VA, Alcott CE, Chen C-A, Chaudhury A, Gillentine MA, et al. NUDT21-spanning CNVs lead to neuropsychiatric disease and altered MeCP2 abundance via alternative polyadenylation. eLife. 2015;4:e10782. doi: 10.7554/eLife.10782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giese KP, Friedman E, Telliez JB, Fedorov NB, Wines M, et al. Hippocampus-dependent learning and memory is impaired in mice lacking the Ras-guanine-nucleotide releasing factor 1 (Ras-GRF1) Neuropharmacology. 2001;41(6):791–800. doi: 10.1016/s0028-3908(01)00096-x. [DOI] [PubMed] [Google Scholar]
- Godavarthi SK, Dey P, Maheshwari M, Jana NR. Defective glucocorticoid hormone receptor signaling leads to increased stress and anxiety in a mouse model of Angelman syndrome. Hum Mol Genet. 2012;21(8):1824–34. doi: 10.1093/hmg/ddr614. [DOI] [PubMed] [Google Scholar]
- Gotter AL, Santarelli VP, Doran SM, Tannenbaum PL, Kraus RL, et al. TASK-3 as a potential antide-pressant target. Brain Res. 2011;1416:69–79. doi: 10.1016/j.brainres.2011.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer PL, Hanayama R, Bloodgood BL, Mardinly AR, Lipton DM, et al. The Angelman syndrome protein Ube3A regulates synapse development by ubiquitinating arc. Cell. 2010;140(5):704–16. doi: 10.1016/j.cell.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg C. Known unknowns for allele-specific expression and genomic imprinting effects. F1000Prime Rep. 2014;6:75. doi: 10.12703/P6-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg C, Zhang J, Weissbourd B, Luo S, Schroth GP, et al. High-resolution analysis of parent-of-origin allelic expression in the mouse brain. Science. 2010;329(5992):643–48. doi: 10.1126/science.1190830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haig D. Coadaptation and conflict, misconception and muddle, in the evolution of genomic imprinting. Heredity. 2014;113(2):96–103. doi: 10.1038/hdy.2013.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall DD, Dai S, Tseng P-Y, Malik Z, Nguyen M, et al. Competition between α-actinin and Ca2-calmodulin controls surface retention of the L-type Ca2+ channel Ca(V)1.2. Neuron. 2013;78(3):483–97. doi: 10.1016/j.neuron.2013.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa K, Kawahara T, Fujiwara K, Shimpuku M, Sasaki T, et al. Necdin controls Foxo1 acetylation in hypothalamic arcuate neurons to modulate the thyroid axis. J Neurosci. 2012;32(16):5562–72. doi: 10.1523/JNEUROSCI.0142-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa K, Yoshikawa K. Necdin regulates p53 acetylation via Sirtuin1 to modulate DNA damage response in cortical neurons. J Neurosci. 2008;28(35):8772–84. doi: 10.1523/JNEUROSCI.3052-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkes C, Jhamandas JH, Harris KH, Fu W, MacDonald RG, Kar S. Single transmembrane domain insulin-like growth factor-II/mannose-6-phosphate receptor regulates central cholinergic function by activating a G-protein-sensitive, protein kinase C-dependent pathway. J Neurosci. 2006;26(2):585–96. doi: 10.1523/JNEUROSCI.2730-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Ecker JR. Non-CG methylation in the human genome. Annu Rev Genom Hum Genet. 2015;16(1):15–77. doi: 10.1146/annurev-genom-090413-025437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez A, Martinez ME, Fiering S, Galton VA, St Germain D. Type 3 deiodinase is critical for the maturation and function of the thyroid axis. J Clin Investig. 2006;116(2):476–84. doi: 10.1172/JCI26240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez A, Martinez ME, Liao X-H, Van Sande J, Refetoff S, et al. Type 3 deiodinase deficiency results in functional abnormalities at multiple levels of the thyroid axis. Endocrinology. 2007;148(12):5680–87. doi: 10.1210/en.2007-0652. [DOI] [PubMed] [Google Scholar]
- Herzing LBK, Cook EH, Ledbetter DH. Allele-specific expression analysis by RNA-FISH demonstrates preferential maternal expression of UBE3A and imprint maintenance within 15q11–q13 duplications. Hum Mol Genet. 2002;11(15):1707–18. doi: 10.1093/hmg/11.15.1707. [DOI] [PubMed] [Google Scholar]
- Hirano K, Kaneko R, Izawa T. Single-neuron diversity generated by Protocadherin-β cluster in mouse central and peripheral nervous systems. Front Mol Neurosci. 2012;5:90. doi: 10.3389/fnmol.2012.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra EJ, von Oerthel L, van der Linden AJA, Schellevis RD, Scheppink G, et al. Lmx1a is an activator of Rgs4 and Grb10 and is responsible for the correct specification of rostral and medial mdDA neurons. Eur J Neurosci. 2013;37(1):23–32. doi: 10.1111/ejn.12022. [DOI] [PubMed] [Google Scholar]
- Hogart A, Wu D, LaSalle JM, Schanen NC. The comorbidity of autism with the genomic disorders of chromosome 15q11.2–q13. Neurobiol Dis. 2010;38(2):181–91. doi: 10.1016/j.nbd.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H-S, Allen JA, Mabb AM, King IF, Miriyala J, et al. Topoisomerase inhibitors unsilence the dormant allele of Ube3a in neurons. Nature. 2012;481(7380):185–89. doi: 10.1038/nature10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H-S, Yoon B-J, Brooks S, Bakal R, Berrios J, et al. Snx14 regulates neuronal excitability, promotes synaptic transmission, and is imprinted in the brain of mice. PLOS ONE. 2014;9(5):e98383. doi: 10.1371/journal.pone.0098383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isles AR. Genomic imprinting; the cost of mother’s care. BioEssays. 2011;33(12):924–26. doi: 10.1002/bies.201100115. [DOI] [PubMed] [Google Scholar]
- Isles AR, Baum MJ, Ma D, Szeto A, Keverne EB, Allen ND. A possible role for imprinted genes in inbreeding avoidance and dispersal from the natal area in mice. Proc R Soc B. 2002;269(1492):665–70. doi: 10.1098/rspb.2001.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itier JM, Tremp GL, Léonard JF, Multon MC, Ret G, et al. Imprinted gene in postnatal growth role. Nature. 1998;393(6681):125–26. doi: 10.1038/30120. [DOI] [PubMed] [Google Scholar]
- Jacobs FMJ, van der Linden AJA, Wang Y, von Oerthel L, Sul HS, et al. Identification of Dlk1, Ptpru and Klhl1 as novel Nurr1 target genes in meso-diencephalic dopamine neurons. Development. 2009;136(14):2363–73. doi: 10.1242/dev.037556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauregi J, Arias C, Vegas O, Alén F, Martinez S, et al. A neuropsychological assessment of frontal cognitive functions in Prader-Willi syndrome. J Intellect Disabil Res. 2007;51(Pt. 5):350–65. doi: 10.1111/j.1365-2788.2006.00883.x. [DOI] [PubMed] [Google Scholar]
- Jiang Y-H, Armstrong D, Albrecht U, Atkins CM. Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron. 1998;21(4):799–811. doi: 10.1016/s0896-6273(00)80596-6. [DOI] [PubMed] [Google Scholar]
- Jiang Y-H, Pan Y, Zhu L, Landa L, Yoo J, et al. Altered ultrasonic vocalization and impaired learning and memory in Angelman syndrome mouse model with a large maternal deletion from Ube3a to Gabrb3. PLOS ONE. 2010;5(8):e12278. doi: 10.1371/journal.pone.0012278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao S, Li Z. Nonapoptotic function of BAD and BAX in long-term depression of synaptic transmission. Neuron. 2011;70(4):758–72. doi: 10.1016/j.neuron.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones BK, Levorse J, Tilghman SM. Deletion of a nuclease-sensitive region between the Igf2 and H19 genes leads to Igf2 misregulation and increased adiposity. Hum Mol Genet. 2001;10(8):807–14. doi: 10.1093/hmg/10.8.807. [DOI] [PubMed] [Google Scholar]
- Joseph B, Andersson ER, Vlachos P, Södersten E, Liu L, et al. p57Kip2 is a repressor of Mash1 activity and neuronal differentiation in neural stem cells. Cell Death Differ. 2009;16(9):1256–65. doi: 10.1038/cdd.2009.72. [DOI] [PubMed] [Google Scholar]
- Jouvenot Y, Poirier F, Jami J, Paldi A. Biallelic transcription of Igf2 and H19 in individual cells suggests a post-transcriptional contribution to genomic imprinting. Curr Biol. 1999;9(20):1199–202. doi: 10.1016/S0960-9822(00)80026-3. [DOI] [PubMed] [Google Scholar]
- Kaphzan H, Buffington SA, Jung JI, Rasband MN, Klann E. Alterations in intrinsic membrane properties and the axon initial segment in a mouse model of Angelman syndrome. J Neurosci. 2011;31(48):17637–48. doi: 10.1523/JNEUROSCI.4162-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaphzan H, Buffington SA, Ramaraj AB, Lingrel JB, Rasband MN, et al. Genetic reduction of the α1 subunit of Na/K-ATPase corrects multiple hippocampal phenotypes in Angelman syndrome. Cell Rep. 2013;4(3):405–12. doi: 10.1016/j.celrep.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly ML, Moir L, Jones L, Whitehill E, Anstee QM, et al. A missense mutation in the non-neural G-protein α-subunit isoforms modulates susceptibility to obesity. Int J Obes Relat Metab Disord. 2009;33(5):507–18. doi: 10.1038/ijo.2009.30. [DOI] [PubMed] [Google Scholar]
- Keverne EB. Importance of the matriline for genomic imprinting, brain development and behaviour. Philos Trans R Soc B. 2013;368(1609):20110327. doi: 10.1098/rstb.2011.0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keverne EB, Fundele R, Narasimha M, Barton SC, Surani MA. Genomic imprinting and the differential roles of parental genomes in brain development. Brain Res Dev Brain Res. 1996;92(1):91–100. doi: 10.1016/0165-3806(95)00209-x. [DOI] [PubMed] [Google Scholar]
- Kishore S, Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. 2006;311(5758):230–32. doi: 10.1126/science.1118265. [DOI] [PubMed] [Google Scholar]
- Kotzot D. Maternal uniparental disomy 14 dissection of the phenotype with respect to rare autosomal recessively inherited traits, trisomy mosaicism, and genomic imprinting. Ann Génét. 2004;47(3):251–60. doi: 10.1016/j.anngen.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Kozlov SV, Bogenpohl JW, Howell MP, Wevrick R, Panda S, et al. The imprinted gene Magel2 regulates normal circadian output. Nat Genet. 2007;39(10):1266–72. doi: 10.1038/ng2114. [DOI] [PubMed] [Google Scholar]
- Krechowec SO, Burton KL, Newlaczyl AU, Nunn N, Vlatković N, Plagge A. Postnatal changes in the expression pattern of the imprinted signalling protein XLαs underlie the changing phenotype of deficient mice. PLOS ONE. 2012;7(1):e29753. doi: 10.1371/journal.pone.0029753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein A, Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci. 2009;32(1):149–84. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurita M, Kuwajima T, Nishimura I, Yoshikawa K. Necdin downregulates CDC2 expression to attenuate neuronal apoptosis. J Neurosci. 2006;26(46):12003–13. doi: 10.1523/JNEUROSCI.3002-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwajima T, Hasegawa K, Yoshikawa K. Necdin promotes tangential migration of neocortical interneurons from basal forebrain. J Neurosci. 2010;30(10):3709–14. doi: 10.1523/JNEUROSCI.5797-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwajima T, Nishimura I, Yoshikawa K. Necdin promotes GABAergic neuron differentiation in cooperation with Dlx homeodomain proteins. J Neurosci. 2006;26(20):5383–92. doi: 10.1523/JNEUROSCI.1262-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labialle S, Yang L, Ruan X, Villemain A, Schmidt JV, et al. Coordinated diurnal regulation of genes from the Dlk1–Dio3 imprinted domain: implications for regulation of clusters of non-paralogous genes. Hum Mol Genet. 2008;17(1):15–26. doi: 10.1093/hmg/ddm281. [DOI] [PubMed] [Google Scholar]
- Lassi G, Ball ST, Maggi S, Colonna G, Nieus T, et al. Loss of Gnas imprinting differentially affects REM/NREM sleep and cognition in mice. PLOS Genet. 2012;8(5):e1002706. doi: 10.1371/journal.pgen.1002706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Walker CL, Karten B, Kuny SL, Tennese AA, et al. Essential role for the Prader-Willi syndrome protein necdin in axonal outgrowth. Hum Mol Genet. 2005;14(5):627–37. doi: 10.1093/hmg/ddi059. [DOI] [PubMed] [Google Scholar]
- Lefebvre L, Viville S, Barton SC, Ishino F, Keverne EB, Surani MA. Abnormal maternal behaviour and growth retardation associated with loss of the imprinted gene Mest. Nat Genet. 1998;20(2):163–69. doi: 10.1038/2464. [DOI] [PubMed] [Google Scholar]
- Lehtinen MK, Zappaterra MW, Chen X, Yang YJ, Hill AD, et al. The cerebrospinal fluid provides a proliferative niche for neural progenitor cells. Neuron. 2011;69(5):893–905. doi: 10.1016/j.neuron.2011.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis A, Mitsuya K, Umlauf D, Smith P, Dean W, et al. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat Genet. 2004;36(12):1291–95. doi: 10.1038/ng1468. [DOI] [PubMed] [Google Scholar]
- Li H, Alavian KN, Lazrove E, Mehta N, Jones A, et al. A Bcl-xL–Drp1 complex regulates synaptic vesicle membrane dynamics during endocytosis. Nature. 2013;15(7):773–85. doi: 10.1038/ncb2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JY, Chai B-X, Zhang W, Wang H, Mulholland MW. Expression of ankyrin repeat and suppressor of cytokine signaling box protein 4 (Asb-4) in proopiomelanocortin neurons of the arcuate nucleus of mice produces a hyperphagic, lean phenotype. Endocrinology. 2010;151(1):134–42. doi: 10.1210/en.2009-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JY, Kuick R, Thompson RC, Misek DE, Lai YM, et al. Arcuate nucleus transcriptome profiling identifies ankyrin repeat and suppressor of cytokine signalling box-containing protein 4 as a gene regulated by fasting in central nervous system feeding circuits. J Neuroendocrinol. 2005;17(6):394–404. doi: 10.1111/j.1365-2826.2005.01317.x. [DOI] [PubMed] [Google Scholar]
- Li LL, Keverne EB, Aparicio SA, Ishino F, Barton SC, Surani MA. Regulation of maternal behavior and offspring growth by paternally expressed Peg3. Science. 1999;284(5412):330–33. doi: 10.1126/science.284.5412.330. [DOI] [PubMed] [Google Scholar]
- Li S, Tian X, Hartley DM, Feig LA. Distinct roles for Ras-guanine nucleotide-releasing factor 1 (Ras-GRF1) and Ras-GRF2 in the induction of long-term potentiation and long-term depression. J Neurosci. 2006;26(6):1721–29. doi: 10.1523/JNEUROSCI.3990-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Jo J, Jia J-M, Lo S-C, Whitcomb DJ, et al. Caspase-3 activation via mitochondria is required for long-term depression and AMPA receptor internalization. Cell. 2010;141(5):859–71. doi: 10.1016/j.cell.2010.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden AM, Sandu C, Aller MI, Vekovischeva OY, Rosenberg PH, et al. TASK-3 knockout mice exhibit exaggerated nocturnal activity, impairments in cognitive functions, and reduced sensitivity to inhalation anesthetics. J Pharmacol Exp Ther. 2007;323(3):924–34. doi: 10.1124/jpet.107.129544. [DOI] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y-W, Xu Y, Cao W-Y, Zhong X-L, Duan J, et al. Insulin-like growth factor 2 mitigates depressive behavior in a rat model of chronic stress. Neuropharmacology. 2014;89C:318–24. doi: 10.1016/j.neuropharm.2014.10.011. [DOI] [PubMed] [Google Scholar]
- Makita T, Sucov HM, Gariepy CE, Yanagisawa M, Ginty DD. Endothelins are vascular-derived axonal guidance cues for developing sympathetic neurons. Nature. 2008;452(7188):759–63. doi: 10.1038/nature06859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini-Dinardo D, Steele SJS, Levorse JM, Ingram RS, Tilghman SM. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev. 2006;20(10):1268–82. doi: 10.1101/gad.1416906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez ME, Charalambous M, Saferali A, Fiering S, Naumova AK, et al. Genomic imprinting variations in the mouse type 3 deiodinase gene between tissues and brain regions. Mol Endocrinol. 2014;28(11):1875–86. doi: 10.1210/me.2014-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto A, Susaki E, Onoyama I, Nakayama K, Hoshino M, Nakayama KI. Deregulation of the p57-E2F1-p53 axis results in nonobstructive hydrocephalus and cerebellar malformation in mice. Mol Cell Biol. 2011;31(20):4176–92. doi: 10.1128/MCB.05370-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath J, Solter D. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell. 1984;37(1):179–83. doi: 10.1016/0092-8674(84)90313-1. [DOI] [PubMed] [Google Scholar]
- Meister B, Perez-Manso M, Daraio T. Delta-like 1 homologue is a hypothalamus-enriched protein that is present in orexin-containing neurones of the lateral hypothalamic area. J Neuroendocrinol. 2013;25(7):617–25. doi: 10.1111/jne.12029. [DOI] [PubMed] [Google Scholar]
- Meng L, Ward AJ, Chun S, Bennett CF, Beaudet AL, Rigo F. Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature. 2014 doi: 10.1038/nature13975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer RE, Michaelson SD, Chee MJS, Atallah TA, Wevrick R, Colmers WF. Magel2 is required for leptin-mediated depolarization of POMC neurons in the hypothalamic arcuate nucleus in mice. PLOS Genet. 2013;9(1):e1003207. doi: 10.1371/journal.pgen.1003207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer RE, Wevrick R. Loss of magel2, a candidate gene for features of Prader-Willi syndrome, impairs reproductive function in mice. PLOS ONE. 2009;4(1):e4291. doi: 10.1371/journal.pone.0004291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meziane H, Schaller F, Bauer S, Villard C, Matarazzo V, et al. An early postnatal oxytocin treatment prevents social and learning deficits in adult mice deficient for Magel2, a gene involved in Prader-Willi syndrome and autism. Biol Psychiatry. 2015;78(2):85–94. doi: 10.1016/j.biopsych.2014.11.010. [DOI] [PubMed] [Google Scholar]
- Michalon A, Sidorov M, Ballard TM, Ozmen L, Spooren W, et al. Chronic pharmacological mGlu5 inhibition corrects fragile X in adult mice. Neuron. 2012;74(1):49–56. doi: 10.1016/j.neuron.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BR, Hen R. The current state of the neurogenic theory of depression and anxiety. Curr Opin Neurobiol. 2015;30:51–58. doi: 10.1016/j.conb.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller NLG, Wevrick R, Mellon PL. Necdin, a Prader-Willi syndrome candidate gene, regulates gonadotropin-releasing hormone neurons during development. Hum Mol Genet. 2009;18(2):248–60. doi: 10.1093/hmg/ddn344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamide R, Fujiwara K, Hasegawa K, Yoshikawa K. Antagonistic interplay between necdin and Bmi1 controls proliferation of neural precursor cells in the embryonic mouse neocortex. PLOS ONE. 2014;9(1):e84460. doi: 10.1371/journal.pone.0084460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra A, Jana NR. Regulation of turnover of tumor suppressor p53 and cell growth by E6-AP, a ubiquitin protein ligase mutated in Angelman mental retardation syndrome. Cell Mol Life Sci. 2008;65(4):656–66. doi: 10.1007/s00018-007-7476-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo A, Mukamel EA, Davis FP, Luo C, Henry GL, et al. Epigenomic signatures of neuronal diversity in the mammalian brain. Neuron. 2015;86(6):1369–84. doi: 10.1016/j.neuron.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida GH, Mahajnah M, Hill AD, Basel-Vanagaite L, Gleason D, et al. A truncating mutation of TRAPPC9 is associated with autosomal-recessive intellectual disability and postnatal microcephaly. Am J Hum Genet. 2009;85(6):897–902. doi: 10.1016/j.ajhg.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. Annu Rev Neurosci. 2012;35(1):445–462. doi: 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero I, Teschke M, Tautz D. Paternal imprinting of mating preferences between natural populations of house mice (Mus musculus domesticus) Mol Ecol. 2013;22(9):2549–62. doi: 10.1111/mec.12271. [DOI] [PubMed] [Google Scholar]
- Morison IM, Ramsay JP, Spencer HG. A census of mammalian imprinting. Trends Genet. 2005;21(8):457–65. doi: 10.1016/j.tig.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Motoyama N, Wang F, Roth KA, Sawa H, Nakayama K, et al. Massive cell death of immature hematopoietic cells and neurons in Bcl-x-deficient mice. Science. 1995;267(5203):1506–10. doi: 10.1126/science.7878471. [DOI] [PubMed] [Google Scholar]
- Mott R, Yuan W, Kaisaki P, Gan X, Cleak J, et al. The architecture of parent-of-origin effects in mice. Cell. 2014;156(1–2):332–42. doi: 10.1016/j.cell.2013.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouallem M, Shaharabany M, Weintrob N, Shalitin S, Nagelberg N, et al. Cognitive impairment is prevalent in pseudohypoparathyroidism type Ia, but not in pseudopseudohypoparathyroidism: possible cerebral imprinting of Gsα. Clin Endocrinol. 2008;68(2):233–39. doi: 10.1111/j.1365-2265.2007.03025.x. [DOI] [PubMed] [Google Scholar]
- Muddashetty RS, Nalavadi VC, Gross C, Yao X, Xing L, et al. Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Mol Cell. 2011;42(5):673–88. doi: 10.1016/j.molcel.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulherkar SA, Jana NR. Loss of dopaminergic neurons and resulting behavioural deficits in mouse model of Angelman syndrome. Neurobiol Dis. 2010;40(3):586–92. doi: 10.1016/j.nbd.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Muscatelli F, Abrous DN, Massacrier A, Boccaccio I, Le Moal M, et al. Disruption of the mouse Necdin gene results in hypothalamic and behavioral alterations reminiscent of the human Prader-Willi syndrome. Hum Mol Genet. 2000;9(20):3101–10. doi: 10.1093/hmg/9.20.3101. [DOI] [PubMed] [Google Scholar]
- Musset B, Meuth SG, Liu GX, Derst C, Wegner S, et al. Effects of divalent cations and spermine on the K+ channel TASK-3 and on the outward current in thalamic neurons. J Physiol. 2006;572(Pt. 3):639–57. doi: 10.1113/jphysiol.2006.106898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi H, Obaishi H, Satoh A, Wada M, Mandai K, et al. Neurabin: a novel neural tissue–specific actin filament–binding protein involved in neurite formation. J Cell Biol. 1997;139(4):951–61. doi: 10.1083/jcb.139.4.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani J, Tamada K, Hatanaka F, Ise S, Ohta H, et al. Abnormal behavior in a chromosome-engineered mouse model for human 15q11–13 duplication seen in autism. Cell. 2009;137(7):1235–46. doi: 10.1016/j.cell.2009.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls RD, Knoll JH, Butler MG, Karam S, Lalande M. Genetic imprinting suggested by maternal heterodisomy in nondeletion Prader-Willi syndrome. Nature. 1989;342(6247):281–85. doi: 10.1038/342281a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor A, Dupuis L, Mittal K, Lionel AC, Marshall CR, et al. 15q11.2 duplication encompassing only the UBE3A gene is associated with developmental delay and neuropsychiatric phenotypes. Hum Mutat. 2015;36(7):689–93. doi: 10.1002/humu.22800. [DOI] [PubMed] [Google Scholar]
- Nunn N, Feetham CH, Martin J, Barrett-Jolley R, Plagge A. Elevated blood pressure, heart rate and body temperature in mice lacking the XLαs protein of the Gnas locus is due to increased sympathetic tone. Exp Physiol. 2013;98(10):1432–45. doi: 10.1113/expphysiol.2013.073064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofengeim D, Chen Y-B, Miyawaki T, Li H, Sacchetti S, et al. N-terminally cleaved Bcl-xL mediates ischemia-induced neuronal death. Nat Neurosci. 2012;15(4):574–80. doi: 10.1038/nn.3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okae H, Hiura H, Nishida Y, Funayama R, Tanaka S, et al. Re-investigation and RNA sequencing-based identification of genes with placenta-specific imprinted expression. Hum Mol Genet. 2012;21(3):548–58. doi: 10.1093/hmg/ddr488. [DOI] [PubMed] [Google Scholar]
- Ori-McKenney KM, Vallee RB. Neuronal migration defects in the Loa dynein mutant mouse. Neural Dev. 2011;6(1):26. doi: 10.1186/1749-8104-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto SP, Gerstein AC. The evolution of haploidy and diploidy. Curr Biol. 2008;18(24):R1121–24. doi: 10.1016/j.cub.2008.09.039. [DOI] [PubMed] [Google Scholar]
- Ouchi Y, Banno Y, Shimizu Y, Ando S, Hasegawa H, et al. Reduced adult hippocampal neurogenesis and working memory deficits in the Dgcr8-deficient mouse model of 22q11.2 deletion-associated schizophrenia can be rescued by IGF2. J Neurosci. 2013;33(22):9408–19. doi: 10.1523/JNEUROSCI.2700-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyang EL, Davidson BC, Lee W, Poon MM. Functional characterization of the dendritically localized mRNA neuronatin in hippocampal neurons. PLOS ONE. 2011;6(9):e24879. doi: 10.1371/journal.pone.0024879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109(3):307–20. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32(2):232–46. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- Pang DSJ, Robledo CJ, Carr DR, Gent TC, Vyssotski AL, et al. An unexpected role for TASK-3 potassium channels in network oscillations with implications for sleep mechanisms and anesthetic action. PNAS. 2009;106(41):17546–51. doi: 10.1073/pnas.0907228106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AJ, Lazdunski M. The 2P-domain K+ channels: role in apoptosis and tumorigenesis. Pflügers Arch Eur J Physiol. 2004;448(3):261–73. doi: 10.1007/s00424-004-1255-8. [DOI] [PubMed] [Google Scholar]
- Patten MM, Ross L, Curley JP, Queller DC, Bonduriansky R, Wolf JB. The evolution of genomic imprinting: theories, predictions and empirical tests. Heredity. 2014;113(2):119–28. doi: 10.1038/hdy.2014.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peall KJ, Smith DJ, Kurian MA, Wardle M, Waite AJ, et al. SGCE mutations cause psychiatric disorders: clinical and genetic characterization. Brain. 2013;136(Pt. 1):294–303. doi: 10.1093/brain/aws308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters RP, Hernandez A, Ng L, Ma M, Sharlin DS, et al. Cerebellar abnormalities in mice lacking type 3 deiodinase and partial reversal of phenotype by deletion of thyroid hormone receptor α1. Endocrinology. 2013;154(1):550–61. doi: 10.1210/en.2012-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña CJ, Neugut YD, Calarco CA, Champagne FA. Effects of maternal care on the development of mid-brain dopamine pathways and reward-directed behavior in female offspring. Eur J Neurosci. 2014;39(6):946–56. doi: 10.1111/ejn.12479. [DOI] [PubMed] [Google Scholar]
- Perez JD, Rubinstein ND, Fernandez DE, Santoro SW, Needleman LA, et al. Quantitative and functional interrogation of parent-of-origin allelic expression biases in the brain. eLife. 2015;4:e07860. doi: 10.7554/eLife.07860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry JRB, Day F, Elks CE, Sulem P, Thompson DJ, et al. Parent-of-origin-specific allelic associations among 106 genomic loci for age at menarche. Nature. 2014;514(7520):92–97. doi: 10.1038/nature13545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J, Williamson CM. Control of imprinting at the Gnas cluster. Epigenetics. 2007;2(4):207–13. doi: 10.4161/epi.2.4.5380. [DOI] [PubMed] [Google Scholar]
- Plagge A, Gordon E, Dean W, Boiani R, Cinti S, et al. The imprinted signaling protein XLαs is required for postnatal adaptation to feeding. Nat Genet. 2004;36(8):818–26. doi: 10.1038/ng1397. [DOI] [PubMed] [Google Scholar]
- Powell WT, Coulson RL, Crary FK, Wong SS, Ach RA, et al. A Prader–Willi locus lncRNA cloud modulates diurnal genes and energy expenditure. Hum Mol Genet. 2013;22(21):4318–28. doi: 10.1093/hmg/ddt281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pravdivyi I, Ballanyi K, Colmers WF, Wevrick R. Progressive postnatal decline in leptin sensitivity of arcuate hypothalamic neurons in the Magel2-null mouse model of Prader–Willi syndrome. Hum Mol Genet. 2015;24(15):4276–83. doi: 10.1093/hmg/ddv159. [DOI] [PubMed] [Google Scholar]
- Qian N, Frank D, O’Keefe D, Dao D, Zhao L, et al. The IPL gene on chromosome 11p15.5 is imprinted in humans and mice and is similar to TDAG51, implicated in Fas expression and apoptosis. Hum Mol Genet. 1997;6(12):2021–29. doi: 10.1093/hmg/6.12.2021. [DOI] [PubMed] [Google Scholar]
- Rago L, Beattie R, Taylor V, Winter J. miR379–410 cluster miRNAs regulate neurogenesis and neuronal migration by fine-tuning N-cadherin. EMBO J. 2014;33(8):906–20. doi: 10.1002/embj.201386591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relkovic D, Doe CM, Humby T, Johnstone KA, Resnick JL, et al. Behavioural and cognitive abnormalities in an imprinting centre deletion mouse model for Prader-Willi syndrome. Eur J Neurosci. 2010;31(1):156–64. doi: 10.1111/j.1460-9568.2009.07048.x. [DOI] [PubMed] [Google Scholar]
- Ressler KJ, Mayberg HS. Targeting abnormal neural circuits in mood and anxiety disorders: from the laboratory to the clinic. Nat Neurosci. 2007;10(9):1116–24. doi: 10.1038/nn1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riday TT, Dankoski EC, Krouse MC, Fish EW, Walsh PL, et al. Pathway-specific dopaminergic deficits in a mouse model of Angelman syndrome. J Clin Investig. 2012;122(12):4544–54. doi: 10.1172/JCI61888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo T, Del Gaudio D, German JR, Shinawi M, Peters SU, et al. Prader-Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nat Genet. 2008;40(6):719–21. doi: 10.1038/ng.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Stryker MP. Genomic imprinting of experience-dependent cortical plasticity by the ubiquitin ligase gene Ube3a. PNAS. 2010;107(12):5611–16. doi: 10.1073/pnas.1001281107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitt JM, Jang SS, Mu W, Dawson VL, Dawson TM. Bcl-x is required for proper development of the mouse substantia nigra. J Neurosci. 2005;25(29):6721–28. doi: 10.1523/JNEUROSCI.0760-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf CP, Gonzalez-Garay ML, Xia F, Potocki L, Gripp KW, et al. Truncating mutations of MAGEL2 cause Prader-Willi phenotypes and autism. Nat Genet. 2013;45(11):1405–8. doi: 10.1038/ng.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A, Im HI, Venø MT, Fowler CD, Min A, et al. Argonaute 2 in dopamine 2 receptor–expressing neurons regulates cocaine addiction. J Exp Med. 2010;207(9):1843–51. doi: 10.1084/jem.20100451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller F, Watrin F, Sturny R, Massacrier A, Szepetowski P, Muscatelli F. A single postnatal injection of oxytocin rescues the lethal feeding behaviour in mouse newborns deficient for the imprinted Magel2 gene. Hum Mol Genet. 2010;19(24):4895–4905. doi: 10.1093/hmg/ddq424. [DOI] [PubMed] [Google Scholar]
- Schanen NC. Epigenetics of autism spectrum disorders. Hum Mol Genet. 2006;15(Suppl 2):R138–50. doi: 10.1093/hmg/ddl213. [DOI] [PubMed] [Google Scholar]
- Schmidt-Edelkraut U, Daniel G, Hoffmann A, Spengler D. Zac1 regulates cell cycle arrest in neuronal progenitors via Tcf4. Mol Cell Biol. 2014;34(6):1020–30. doi: 10.1128/MCB.01195-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, et al. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439(7074):283–89. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Shi S, Bichell TJ, Ihrie RA, Johnson CH. Ube3a imprinting impairs circadian robustness in Angelman syndrome models. Curr Biol. 2015;25(5):537–45. doi: 10.1016/j.cub.2014.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Santos S, van Woerden GM, Bruinsma CF, Mientjes E, Jolfaei MA, et al. Ube3a reinstatement identifies distinct developmental windows in a murine Angelman syndrome model. J Clin Investig. 2015;125(5):2069–76. doi: 10.1172/JCI80554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittig LJ, Redei EE. Fine-tuning notes in the behavioral symphony: parent-of-origin allelic gene expression in the brain. Epigenetics Cancer Part A. 2014;86:93–106. doi: 10.1016/B978-0-12-800222-3.00005-X. [DOI] [PubMed] [Google Scholar]
- Smith SEP, Zhou Y-D, Zhang G, Jin Z, Stoppel DC, Anderson MP. Increased gene dosage of Ube3a results in autism traits and decreased glutamate synaptic transmission in mice. Sci Transl Med. 2011;3(103):103ra97. doi: 10.1126/scitranslmed.3002627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer HG, Clark AG. Non-conflict theories for the evolution of genomic imprinting. Heredity. 2014;113(2):112–18. doi: 10.1038/hdy.2013.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen PA, Ringrose L. What are memories made of? How Polycomb and Trithorax proteins mediate epigenetic memory. Nat Rev Mol Cell Biol. 2014;15(5):340–56. doi: 10.1038/nrm3789. [DOI] [PubMed] [Google Scholar]
- Störchel PH, Thümmler J, Siegel G, Aksoy-Aksel A, Zampa F, et al. A large-scale functional screen identifies Nova1 and Ncoa3 as regulators of neuronal miRNA function. EMBO J. 2015;34:2237–54. doi: 10.15252/embj.201490643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Zhu G, Liu Y, Standley S, Ji A, et al. UBE3A regulates synaptic plasticity and learning and memory by controlling SK2 channel endocytosis. Cell Rep. 2015;12(3):449–61. doi: 10.1016/j.celrep.2015.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surani M, Barton SC, Norris ML. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature. 1984;308(5959):548–50. doi: 10.1038/308548a0. [DOI] [PubMed] [Google Scholar]
- Swaney WT, Curley JP, Champagne FA, Keverne EB. Genomic imprinting mediates sexual experience-dependent olfactory learning in male mice. PNAS. 2007;104(14):6084–89. doi: 10.1073/pnas.0609471104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaney WT, Curley JP, Champagne FA, Keverne EB. The paternally expressed gene Peg3 regulates sexual experience-dependent preferences for estrous odors. Behav Neurosci. 2008;122(5):963–73. doi: 10.1037/a0012706. [DOI] [PubMed] [Google Scholar]
- Tamada K, Tomonaga S, Hatanaka F, Nakai N, Takao K, et al. Decreased exploratory activity in a mouse model of 15q duplication syndrome: implications for disturbance of serotonin signaling. PLOS ONE. 2010;5(12):e15126. doi: 10.1371/journal.pone.0015126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terranova R, Yokobayashi S, Stadler MB, Otte AP, van Lohuizen M, et al. Polycomb group proteins Ezh2 and Rnf2 direct genomic contraction and imprinted repression in early mouse embryos. Dev Cell. 2008;15(5):668–79. doi: 10.1016/j.devcel.2008.08.015. [DOI] [PubMed] [Google Scholar]
- Tunster SJ, Jensen AB, John RM. Imprinted genes in mouse placental development and the regulation of fetal energy stores. Reproduction. 2013;145(5):R117–37. doi: 10.1530/REP-12-0511. [DOI] [PubMed] [Google Scholar]
- Turan S, Bastepe M. The GNAS complex locus and human diseases associated with loss-of-function mutations or epimutations within this imprinted gene. Horm Res Paediatr. 2013;80(4):229–41. doi: 10.1159/000355384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tury A, Mairet-Coello G, DiCicco-Bloom E. The multiple roles of the cyclin-dependent kinase inhibitory protein p57KIP2 in cerebral cortical neurogenesis. Dev Neurobiol. 2012;72(6):821–42. doi: 10.1002/dneu.20999. [DOI] [PubMed] [Google Scholar]
- Úbeda F, Gardner A. Mother and offspring in conflict: Why not? PLOS Biol. 2015;13(3):e1002084. doi: 10.1371/journal.pbio.1002084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umlauf D, Goto Y, Cao R, Cerqueira F, Wagschal A, et al. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat Genet. 2004;36(12):1296–1300. doi: 10.1038/ng1467. [DOI] [PubMed] [Google Scholar]
- Valente T, Junyent F, Auladell C. Zac1 is expressed in progenitor/stem cells of the neuroectoderm and mesoderm during embryogenesis: differential phenotype of the Zac1-expressing cells during development. Dev Dyn. 2005;233(2):667–79. doi: 10.1002/dvdy.20373. [DOI] [PubMed] [Google Scholar]
- van Woerden GM, Harris KD, Hojjati MR, Gustin RM, Qiu S, et al. Rescue of neurological deficits in a mouse model for Angelman syndrome by reduction of αCaMKII inhibitory phosphorylation. Nat Neurosci. 2007;10(3):280–82. doi: 10.1038/nn1845. [DOI] [PubMed] [Google Scholar]
- Varrault A, Gueydan C, Delalbre A, Bellmann A, Houssami S, et al. Zac1 regulates an imprinted gene network critically involved in the control of embryonic growth. Dev Cell. 2006;11(5):711–22. doi: 10.1016/j.devcel.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Villanueva C, Jacquier S, de Roux N. DLK1 is a somato-dendritic protein expressed in hypothalamic arginine-vasopressin and oxytocin neurons. PLOS ONE. 2012;7(4):e36134. doi: 10.1371/journal.pone.0036134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitali P, Royo H, Marty V, Bortolin-Cavaillé M-L, Cavaillé J. Long nuclear-retained non-coding RNAs and allele-specific higher-order chromatin organization at imprinted snoRNA gene arrays. J Cell Sci. 2010;123(1):70–83. doi: 10.1242/jcs.054957. [DOI] [PubMed] [Google Scholar]
- Wallace ML, Burette AC, Weinberg RJ, Philpot BD. Maternal loss of Ube3a produces an excitatory/inhibitory imbalance through neuron type-specific synaptic defects. Neuron. 2012;74(5):793–800. doi: 10.1016/j.neuron.2012.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Huang Z, Huang L, Niu S, Rao X, et al. Hypothalamic Ahi1 mediates feeding behavior through interaction with 5-HT2C receptor. J Biol Chem. 2012;287(3):2237–46. doi: 10.1074/jbc.M111.277871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Chow J, Hong J, Smith A, Moreno C, et al. Recent acquisition of imprinting at the rodent Sfmbt2 locus correlates with insertion of a large block of miRNAs. BMC Genom. 2011;12(1):204. doi: 10.1186/1471-2164-12-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Joh K, Masuko S, Yatsuki H, Soejima H, et al. The mouse Murr1 gene is imprinted in the adult brain, presumably due to transcriptional interference by the antisense-oriented U2af1-rs1 gene. Mol Cell Biol. 2004;24(1):270–79. doi: 10.1128/MCB.24.1.270-279.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn CP, Sirois JE, Talley EM, Guyenet PG, Bayliss DA. Serotonergic raphe neurons express TASK channel transcripts and a TASK-like pH- and halothane-sensitive K+conductance. J Neurosci. 2002;22(4):1256–65. doi: 10.1523/JNEUROSCI.22-04-01256.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Schmitt A, Wischmeyer E, Döring F. Excitability of pontine startle processing neurones is regulated by the two-pore-domain K+ channel TASK-3 coupled to 5-HT2C receptors. Eur J Neurosci. 2008;28(5):931–40. doi: 10.1111/j.1460-9568.2008.06400.x. [DOI] [PubMed] [Google Scholar]
- Weeber EJ, Jiang Y-H, Elgersma Y, Varga AW, Carrasquillo Y, et al. Derangements of hippocampal calcium/calmodulin-dependent protein kinase II in a mouse model for Angelman mental retardation syndrome. J Neurosci. 2003;23(7):2634–44. doi: 10.1523/JNEUROSCI.23-07-02634.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins JF, Haig D. What good is genomic imprinting: the function of parent-specific gene expression. Nat Rev Genet. 2003;4(5):359–68. doi: 10.1038/nrg1062. [DOI] [PubMed] [Google Scholar]
- Wilkinson LS, Davies W, Isles AR. Genomic imprinting effects on brain development and function. Nat Rev Neurosci. 2007;8(11):832–43. doi: 10.1038/nrn2235. [DOI] [PubMed] [Google Scholar]
- Wolf JB, Hager R. A maternal-offspring coadaptation theory for the evolution of genomic imprinting. PLOS Biol. 2006;4:2238–43. doi: 10.1371/journal.pbio.0040380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock KA, Oliver C, Humphreys GW. Task-switching deficits and repetitive behaviour in genetic neurodevelopmental disorders: data from children with Prader-Willi syndrome chromosome 15 q11–q13 deletion and boys with Fragile X syndrome. Cogn Neuropsychol. 2009;26(2):172–94. doi: 10.1080/02643290802685921. [DOI] [PubMed] [Google Scholar]
- Wu L-J, Ren M, Wang H, Kim SS, Cao X, Zhuo M. Neurabin contributes to hippocampal long-term potentiation and contextual fear memory. PLOS ONE. 2008;3(1):e1407. doi: 10.1371/journal.pone.0001407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T, Chen M, Gavrilova O, Lai EW, Liu J, Weinstein LS. Severe obesity and insulin resistance due to deletion of the maternal Gsαallele is reversed by paternal deletion of the Gsαimprint control region. Endocrinology. 2008;149(5):2443–50. doi: 10.1210/en.2007-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T, Plagge A, Gavrilova O, Pack S, Jou W, et al. The alternative stimulatory G protein α-subunit XLαs is a critical regulator of energy and glucose metabolism and sympathetic nerve activity in adult mice. J Biol Chem. 2006;281(28):18989–99. doi: 10.1074/jbc.M511752200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Barr C, Kim A, Yue F, Lee A. Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell. 2012;148(4):816–31. doi: 10.1016/j.cell.2011.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi A, Taniguchi M, Hori O, Ogawa S, Tojo N, et al. Peg3/Pw1 is involved in p53-mediated cell death pathway in brain ischemia/hypoxia. J Biol Chem. 2002;277(1):623–29. doi: 10.1074/jbc.M107435200. [DOI] [PubMed] [Google Scholar]
- Yamasaki K, Joh K, Ohta T, Masuzaki H, Ishimaru T, et al. Neurons but not glial cells show reciprocal imprinting of sense and antisense transcripts of Ube3a. Hum Mol Genet. 2003;12(8):837–47. doi: 10.1093/hmg/ddg106. [DOI] [PubMed] [Google Scholar]
- Yamasaki Y, Kayashima T, Soejima H, Kinoshita A, Yoshiura K-I, et al. Neuron-specific relaxation of Igf2r imprinting is associated with neuron-specific histone modifications and lack of its antisense transcript. Air Hum Mol Genet. 2005;14(17):2511–20. doi: 10.1093/hmg/ddi255. [DOI] [PubMed] [Google Scholar]
- Yamasaki-Ishizaki Y, Kayashima T, Mapendano CK, Soejima H, Ohta T, et al. Role of DNA methylation and histone H3 lysine 27 methylation in tissue-specific imprinting of mouse Grb10. Mol Cell Biol. 2007;27(2):732–42. doi: 10.1128/MCB.01329-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Frisén J, Lee MH, Massagué J, Barbacid M. Ablation of the CDK inhibitor p57Kip2 results in increased apoptosis and delayed differentiation during mouse development. Genes Dev. 1997;11(8):973–83. doi: 10.1101/gad.11.8.973. [DOI] [PubMed] [Google Scholar]
- Yashiro K, Riday TT, Condon KH, Roberts AC, Bernardo DR, et al. Ube3a is required for experience-dependent maturation of the neocortex. Nat Neurosci. 2009;12(6):777–83. doi: 10.1038/nn.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Carew TJ. Small G protein signaling in neuronal plasticity and memory formation: the specific role of Ras family proteins. Neuron. 2010;68(3):340–61. doi: 10.1016/j.neuron.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Kohtz A, Pollonini G, Riccio A, Alberini CM. Insulin like growth factor 2 expression in the rat brain both in basal condition and following learning predominantly derives from the maternal allele. PLOS ONE. 2015;10(10):e0141078. doi: 10.1371/journal.pone.0141078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi JJ, Berrios J, Newbern JM, Snider WD, Philpot BD, et al. An autism-linked mutation disables phosphorylation control of UBE3A. Cell. 2015;162(4):795–807. doi: 10.1016/j.cell.2015.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoi F, Dang MT, Li J, Li Y. Myoclonus, motor deficits, alterations in emotional responses and monoamine metabolism in ε-sarcoglycan deficient mice. J Biochem. 2006;140(1):141–46. doi: 10.1093/jb/mvj138. [DOI] [PubMed] [Google Scholar]
- Yu S, Yu D, Lee E, Eckhaus M, Lee R, et al. Variable and tissue-specific hormone resistance in heterotrimeric Gs protein α-subunit (Gsα) knockout mice is due to tissue-specific imprinting of the Gsα gene. PNAS. 1998;95(15):8715–20. doi: 10.1073/pnas.95.15.8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Yokoi F, Parsons DS, Standaert DG, Li Y. Alteration of striatal dopaminergic neurotrans-mission in a mouse model of DYT11 myoclonus-dystonia. PLOS ONE. 2012;7(3):e33669. doi: 10.1371/journal.pone.0033669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhang X, Klibanski A. MEG3 noncoding RNA: a tumor suppressor. J Mol Endocrinol. 2012;48(3):R45–53. doi: 10.1530/JME-12-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimprich A, Grabowski M, Asmus F, Naumann M, Berg D, et al. Mutations in the gene encoding ε-sarcoglycan cause myoclonus–dystonia syndrome. Nat Genet. 2001;29(1):66–69. doi: 10.1038/ng709. [DOI] [PubMed] [Google Scholar]
- Zito K, Knott G, Shepherd GMG, Shenolikar S, Svoboda K. Induction of spine growth and synapse formation by regulation of the spine actin cytoskeleton. Neuron. 2004;44(2):321–34. doi: 10.1016/j.neuron.2004.09.022. [DOI] [PubMed] [Google Scholar]