Abstract
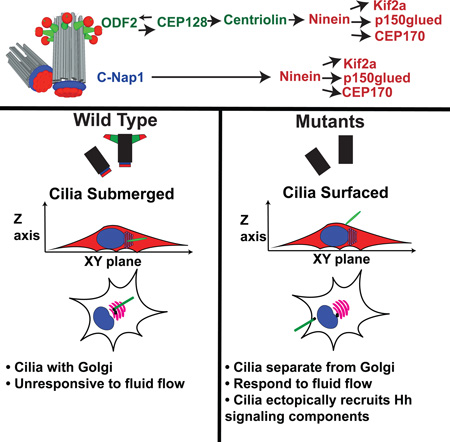
Vertebrate cells can initiate ciliogenesis from centrioles at the cell center, near the Golgi, forming primary cilia confined or submerged in a deep narrow pit created by membrane invagination. How or why cells maintain submerged cilia is unclear. Here, by characterizing centriole sub-distal appendages (sDAP) in cells exclusively growing submerged cilia, we found that a group of sDAP components localize to the centriole proximal end through the cohesion factor C-Nap1, and that sDAP functions redundantly with C-Nap1 for submerged cilia maintenance. Loss of sDAP and C-Nap1 has no effect on cilia assembly, but disrupts stable Golgi-cilia association, and allows normally submerged cilia to fully surface, losing the deep membrane invagination. Intriguingly, unlike submerged cilia (stationary), surfaced cilia actively respond to mechanical stimuli with motions, and can ectopically recruit hedgehog signaling components in the absence of agonist. We propose that spatial control of ciliogenesis uncouples or specifies sensory properties of cilia.
Graphical Abstract
Etoc blurb
Cilia often form submerged in a membrane pit. Mazo et al. show that centriole subdistal appendages (sDAP) function redundantly with C-Nap1 to maintain submerged cilia. Loss of both disrupts Golgi-cilia association, allowing submerged cilia to surface. Surfaced cilia respond to mechanical stimuli with motions and can ectopically recruit hedgehog-signaling components.
INTRODUCTION
Cilia are membrane-bound, hair-like structures projecting from the cell surface. At the cell surface, cilia can produce motility, or carry out sensory functions to detect stimuli that include light, and various chemical and mechanical signals (Goetz and Anderson, 2010). Cilia are nucleated from a microtubule-based structure known as the basal body or centriole, which anchors cilia to the plasma membrane. In vertebrates, centrioles also form the core of the centrosome or microtubule-organizing center (MTOC), while simultaneously nucleating ciliogenesis. The centrosome, i.e. the “central body”, is located near the cell center, often far away from the plasma membrane (Boveri, 1887; Burakov, 2003). As such, cilia formed from the centrally positioned centrosome are unusually situated: They are trapped or tightly confined in a deep narrow pit created by membrane invagination, presumably sensing the environment through the narrow opening at the end of the structure (Sorokin, 1962). We hereafter called these cilia “submerged cilia”. The literature has described the cavity or membrane curvature created by membrane invagination around the cilia base as the “ciliary pocket” (Benmerah, 2013). The pocket, however, is not a feature unique to submerged cilia, nor animal cells. In many cell types, a shallow ciliary pocket can be seen, morphologically resembling the flagellar pocket of ciliated protozoans such as Trypanosomes (Field and Carrington, 2009). Cilia or flagella with a shallow pocket, however, are nearly fully surfaced so are free to produce or sense motion, in contrast to submerged cilia. Thus, while both surfaced and submerged cilia can carry a ciliary pocket at their base, their maintenance or function may be fundamentally different. To avoid confusion, here we use the term “deep membrane invagination” or “deep ciliary pit” to specifically describe the pronounced structure in which submerged cilia are trapped in vertebrate cells.
Submerged cilia can be easily found in non-polarized stromal cells including fibroblasts and smooth muscle cells that carry centrally located centrosomes (Fisher and Steinberg, 1982; Rattner et al., 2010; Sorokin, 1962). Polarized epithelia, however, often grow surfaced cilia using centrosomes that are asymmetrically positioned near the apical cortex or cell surface (Sorokin, 1968). Interestingly, some fully polarized tissues such as retinal pigment epithelia form and maintain submerged cilia despite having apically located centrosomes (Allen, 1965; Fisher and Steinberg, 1982). Cultured cell lines that generally form submerged cilia can be coaxed into forming surfaced cilia under some conditions (Pitaval et al., 2010). This suggests that cells have a mechanism to regulate spatial configuration of their cilia. However, neither the purpose nor the mechanism for maintaining cilia in a submerged configuration is understood.
To facilitate the formation of submerged cilia, vertebrate centrioles may have acquired additional structural complexity. Prior to ciliogenesis, vertebrate centrioles are heavily decorated or modified with many accessory structures, including the distal and sub-distal appendages that project radially from the distal part of centrioles, and less distinct structures such as the pericentriolar material (PCM) or the centrosome cohesion linkers that attach to the proximal end of centrioles (Paintrand et al., 1992). In contrast, neither the appendage structures nor the cohesion linkers are seen in the centriole of some lower animals like Drosophila or C. elegans (Callaini et al., 1997; Gottardo et al., 2015; Hagan and Palazzo, 2006), where no submerged cilia have been detected. The distal appendages (DAP) have been reported to mediate the docking of centrioles with membrane vesicles, a step particularly important for ciliogenesis to occur at centrioles distant from the cell surface (Schmidt et al., 2012; Tanos et al., 2013). However, loss of DAP proteins abolishes all cilia assembly, surfaced or submerged, suggesting that DAP are broadly involved in centriole-to-membrane interaction during various modes of ciliogenesis (Graser et al., 2007; Schmidt et al., 2012; Tanos et al., 2013). Unlike DAP, a link of subdistal appendages (sDAP) to submerged cilia formation has not been explored. sDAP appear not essential for cilia assembly, but are required for proper alignment of basal bodies at the cell cortex in postmitotic, multiciliated epithelia (Kunimoto et al., 2012), which exclusively grow surfaced cilia. Proteins that localize to the sDAP have been reported to help maintain stable microtubule anchorage (Dammermann and Merdes, 2002; Delgehyr et al., 2005; Guarguaglini et al., 2005; Mogensen et al., 2000; Quintyne et al., 1999). How the sDAP may contribute to primary ciliogenesis has not been fully elucidated, and seems to vary among cell types (Hung et al., 2016; Ibi et al., 2011; Ishikawa et al., 2005; Kunimoto et al., 2012; Miyamoto et al., 2015; Soung et al., 2009; Soung et al., 2006). DAP and sDAP are distinct structures composed of different core components, including CEP83, CEP89, SCLT1, CEP164 and FBF1 for DAPs, and ODF2, CNTRL and NIN for sDAPs. DAP components are well preserved in all vertebrates and other deuterostomes, but are highly conserved only in some protostome lineages such as molluscs and annelids (Hodges et al., 2010; Tanos et al., 2013). A similar evolutionary profile is also seen for sDAP components CNTRL and NIN (Hodges et al., 2010), suggesting that DAP and sDAP may somehow be co-selected for specific functions. Perhaps one such function could be to maintain submerged cilia. To explore the idea, here we systematically characterized the assembly and function of sDAP in the retinal pigment epithelial cell line (RPE1), which, comparable to their in vivo counterpart (Allen, 1965; Fisher and Steinberg, 1982), nearly exclusively grow submerged cilia.
RESULTS
Identification of subdistal appendage components
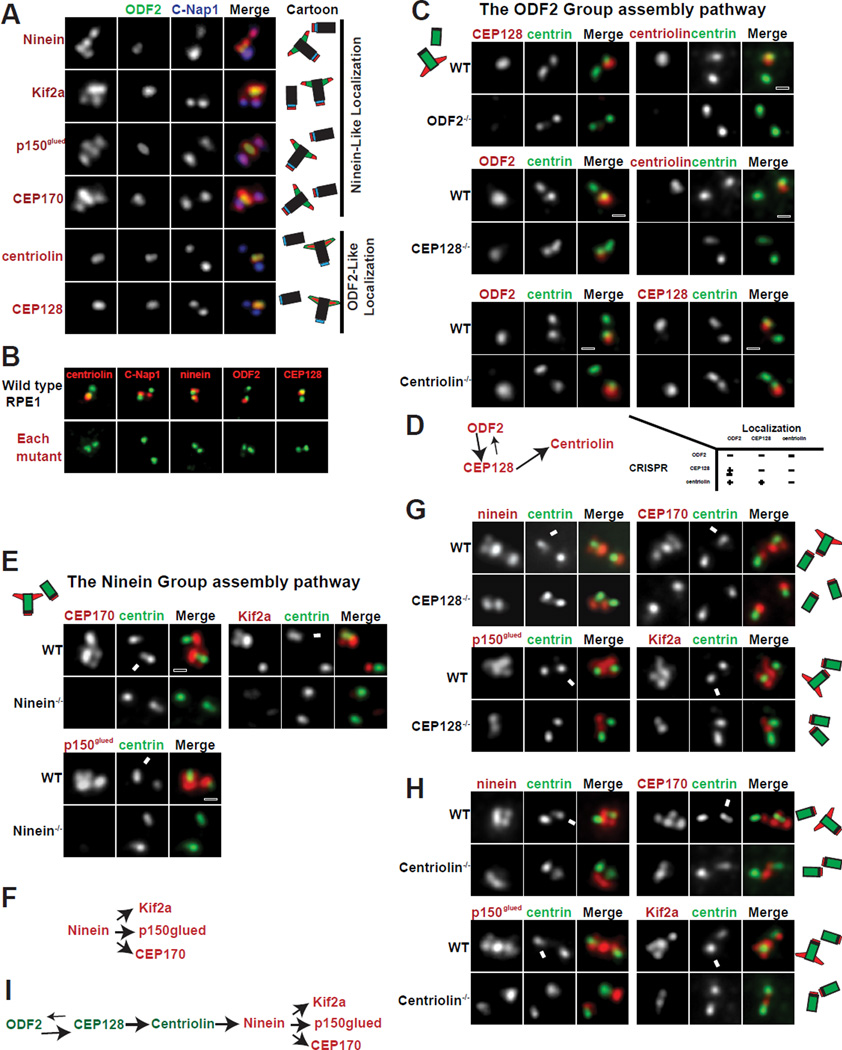
sDAP are structures located toward the distal end of the mature mother centriole. To systematically screen for components of sDAP, we focused on mother centriole specific proteins using our quantitative proteomic approach described previously (Tanos et al., 2013; Wang et al., 2013). We checked the subcellular localization of candidate proteins for ones with localization patterns similar to the known sDAP components: ODF2 or ninein (Figure 1A). Including the novel and those previously known (Guarguaglini et al., 2005; Hehnly et al., 2012; Ishikawa et al., 2005; Kodani et al., 2013; Ou et al., 2002), we identified seven proteins or protein complexes associated with sDAP, including ODF2, CEP128, centriolin (CNTRL), ninein (NIN), p150glued/dynactin complex, CEP170, and Kif2a. These components were further divided into two groups, based on their detailed localization patterns. ODF2 appears as a bar or focus near the distal end of the mature mother centriole where sDAP are located (Ishikawa et al., 2005). ODF2, CEP128 and centriolin exhibit a similar localization pattern, and were thus referred to as members of the ODF2 Group (Figure 1A). Ninein, however, localizes to sDAP of mother centrioles and to the proximal end of both mother and daughter centrioles. When the orientation of both centrioles is longitudinally parallel to the focal plane, ninein appears as a total of four foci, two for the sDAP and two for the centriole proximal ends (Ishikawa et al., 2005; Ou et al., 2002). We refer to proteins with a ninein-like localization as members of the Ninein Group. The Ninein Group includes ninein, CEP170, Kif2a and p150glued/dynactin complex (Figure 1A).
Figure 1. The assembly hierarchy of subdistal appendage components.
(A) The localization patterns of sDAP-associated proteins. Localization of respective proteins (red) was shown in relation to centrosome markers for proximal ends (C-Nap1 in blue) and subdistal appendages (ODF2 in green). Results are summarized in diagram and shown in respective colors.
(B) CRISPR-mediated knockout cell lines for each sDAP component (red). Mutant RPE1 cell lines were stained with indicated antibodies (red). Proteins targeted by CRISPR are shown in red along with centrosome markers in green.
(C), Wild-type (WT) or clonal RPE1 cells lines knocked out (KO) of indicated proteins by CRISPR are stained with indicated antibodies. Loss of ODF2 disrupts localization of CEP128 and centriolin at centrioles. Loss of CEP128 disrupts localization of centriolin from centrioles and reduces ODF2 levels at centrioles. Loss of centriolin has no effect on the localization of CEP128 nor ODF2.
(D) A schematic of the assembly hierarchy of the ODF2 group members.
(E) Loss of ninein impairs the localization of CEP170, Kif2a and p150glued to the centrosome.
(F) A schematic of the assembly hierarchy of the Ninein Group.
(G and H) The centrioles of wild-type RPE1 cells have 4 foci positive for each of the Ninein Group members. In CEP128 depleted or centriolin-depleted cells, the foci at the sDAP disappear, leaving only two foci. Arrows indicate the mature mother centrosome. (I) A schematic of the assembly pathway at the sDAP including both ODF2 Group and Ninein Group members.
See also Figure S1
The assembly hierarchy of sDAP components
We next examined the assembly hierarchy of sDAP components. Experiments were done in clonal RPE1 cell lines permanently depleted of specific sDAP components by the CRISPR-Cas 9 method (Figure 1B; Table S1) (Mali et al., 2013). RNAi experiments were also performed in HeLa cells to confirm the results. We first checked the ODF2 Group members. ODF2 mutation impairs the recruitment of both CEP128 and centriolin to centrioles (Figure 1C; centriolin lost in 98%, n>100). CEP128 loss abolishes centriolin recruitment while having a partial effect on the level of ODF2 recruitment (ODF2 lost entirely from 47% while intensity diminished in rest, n>100). In contrast, centriolin loss does not affect the recruitment of ODF2 nor CEP128 (Figure 1C; normal intensity in 98% and 96%, n>100). Thus, ODF2 Group assembly follows a hierarchy in the order of ODF2, CEP128 and centriolin (Figure 1D), further confirming that CEP128 and centriolin are bonafide sDAP components. We next examined the Ninein Group members. Ninein loss prevents the proper recruitment of all other Ninein Group members including CEP170, Kif2a and p150glued (Figures 1E and F; lost in 94%, 91% and 98% respectively, diminished in rest, n>100). This suggests that ninein acts as a scaffold for the rest of the Ninein Group. Similar results were seen in HeLa cells using RNAi approaches (Figure S1).
Previous work has shown that ODF2 loss eliminates ninein from the subdistal appendages while having no effect on ninein localization at centriole proximal ends, leading to a reduction in the number of ninein foci associated with centrioles (Ishikawa et al., 2005; Kunimoto et al., 2012). We found that ODF2 depletion by RNAi in HeLa cells has precisely the same effect on CEP170, Kif2a and p150glued localization (Figure S1). We next determined whether deletion of the other ODF2 Group members by CRISPR could produce a similar effect on ninein localization. In WT RPE1 cells each member of the Ninein Group appears as 4 foci (Figures 1G and H), three associated with the mature mother (indicated by arrow) and one with the other centriole. In CEP128−/− or centriolin−/− cells, all Ninein Group members appear as two foci total, one at the end of each centriole (Figures 1G and H; >90%, n>100), indicating that while Ninein Group members required ODF2/CEP128/centriolin to localize to sDAP, their recruitment to centriole proximal ends is independent of the ODF2 Group. The same results were obtained using CEP128 siRNA in HeLa cells (Figure S1). Thus, for the sDAP, our results are consistent with an assembly hierarchy with ODF2 being the most upstream, followed by CEP128, centriolin, ninein and the rest of the Ninein Group members (Figure 1I).
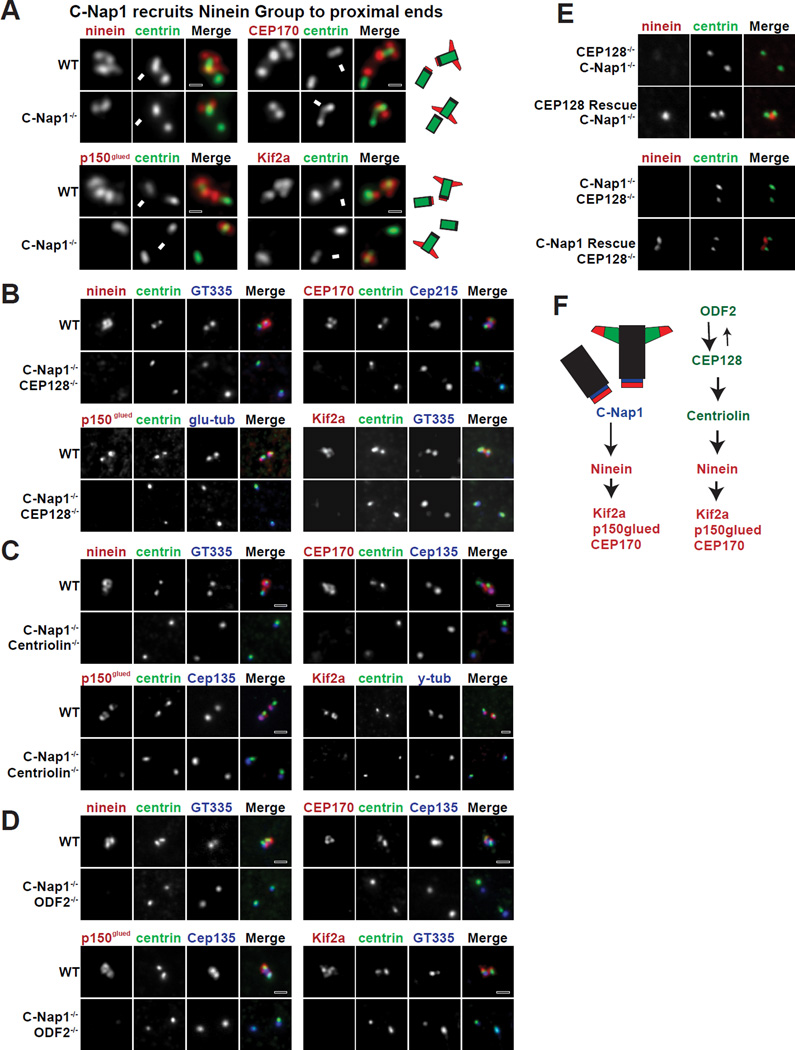
sDAP associated proteins are independently targeted to centriole proximal ends through C-Nap1
We next examined the mechanism responsible for recruitment of ninein to centriole proximal ends. We reasoned that a protein located exclusively at the proximal ends of centrioles should mediate ninein localization. Among the candidates, we found that C-Nap1, which localizes to proximal ends of both centrioles (Fry et al., 1998; Mayor et al., 2000), is essential for ninein recruitment. C-Nap1 depletion in CRISPR knockout cells resulted in displacement of all Ninein Group members from the proximal ends of centrioles without affecting their localization at sDAP (Figure 2A; >90%, n>100). C-Nap1 RNAi produced a similar effect in HeLa cells (Figure S1). To remove the Ninein Group from both ends of mother centrioles, we subsequently performed CRISPR targeting of each ODF2 Group protein in C-Nap1 knockout background. Loss of both C-Nap1 and any one ODF2 Group member resulted in displacement of the entire Ninein Group from both centrioles (Figure 2B–D). Note that ninein localization could be rescued (Figure 2F), confirming the specificity of our results. Taken together, our results suggest that sDAP-associated proteins, such as the Ninein Group and perhaps other molecules not yet identified, are independently targeted to both the distal and proximal ends of the mature mother centriole (Figure 2E). The underlying function, however, is unclear.
Figure 2. Removal of sDPA-associated components from both ends of centrioles.
(A) C-Nap1 is necessary to target each member of the Ninein Group to the proximal ends of centrioles. WT or C-Nap1 knockout (KO) cells were stained with indicated antibodies. Arrows indicate the mature mother centrosome.
(B, C and D) The localization of each Ninein Group protein in RPE1 cell lines depleted of indicated proteins by CRISPR is shown.
(E) Rescue of ninein localization. CEP128 was exogenously expressed in RPE1 cells that lacked both endogenous CEP128 and C-Nap1. Also shown included the restoration of C-Nap1 expression in CEP128 and C-Nap1 double KO cells by a second round of CRISPR-mediated gene editing (see Experimental Procedures).
(F) A full schematic of the assembly hierarchy of sDAP components.
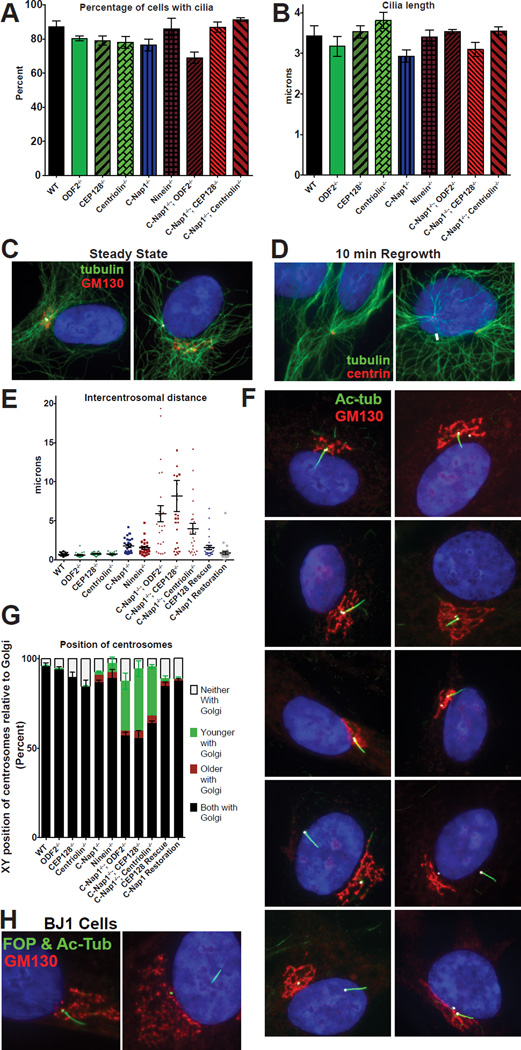
sDAP at mother centrioles are not essential for cilia assembly or microtubule aster formation
Previous work has suggested some involvements of sDAP in cilia assembly and cilia length control, although inconsistent results were also reported (Hehnly et al., 2013; Ishikawa et al., 2005; Kuhns et al., 2013; Kunimoto et al., 2012; Soung et al., 2009). We thus examined the functions of subdistal appendages and sDAP-associated proteins using our CRISPR knockout cell lines. Surprisingly, in mutant cells where the ODF2 Group, Ninein Group, or both groups are removed from mother centrioles, cilia grew efficiently and had a similar average length to controls (Figures 3A–B). These results indicate that unlike the DAP which is absolutely required for ciliogenesis, the presence of sDAP or sDAP-associated proteins at mother centrioles is not essential for cilia to assemble nor reach proper length. sDAP components have also been previously implicated in mediating microtubule anchorage/organization at the centrosome (Dammermann, 2002; Delgehyr, 2005; Guarguaglini et al., 2005; Mogensen et al., 2000; Quintyne et al., 1999). Therefore, we expected to see a dramatic defect in microtubule organization in our sDAP mutant cells. However, to our surprise, grossly normal looking microtubule asters were seen to associate with nearly all centrosomes in our mutant cells, both at steady state and after a microtubule regrowth procedure (Figures 3C–D). Thus, while we cannot rule out the presence of subtle defects, these results suggest that at least in RPE1 cells, sDAP do not seem to play the “predominant” role in the formation of microtubule asters at the centrosome.
Figure 3. C-Nap1 and sDAP depletion do not affect cilia formation or MTOC activity but disrupts centrosome cohesion and Golgi-cilia association.
(A) The percentage of cells with cilia for each genotype is depicted. RPE1 cells were serum starved for 48 hours prior to fixation and staining with anti-acetylated tubulin antibodies. Bars represent average of three experiments (n>100 cells each) and error bars represent the standard deviation of the three experiments.
(B) Cilia length for each genotype. Bars represent average cilia length between three experiments (n>50 cilia each) and error bars represent the standard deviation.
(C) Microtubule arrays in wild type and C-Nap1−/−; CEP128−/− double KO cells. White arrows point at the location of the centrosomes.
(D) After microtubule regrowth, microtubule arrays (green) are focused on the two centrosomes in both wild type and C-Nap1−/−; CEP128−/− double KO cells. White arrows point to the location of the centrosomes.
(E) The intercentrosomal distances for 25 cells of each genotype are shown. Error bars represent the standard error of the mean. RPE1 cells were serum starved and stained with anti gamma-tubulin antibodies to mark centrosomes for quantification.
(F) Cilia position in RPE1 cells of each genotype indicated is shown with acetylatedtubulin (green), GM130 (red), Cep135 (white) and DAPI (blue).
(G) Quantification of centrosome positions in the X-Y axis relative to the Golgi in each RPE1 lines. The relative position in the Z-axis is ignored. RPE1 cells were serum starved for 24 hours, and stained with markers against the centrosomes, the Golgi and distal appendage (as a marker of the older centrosome). >100 cells in each of three experiments were classified. Bars represent average of the three experiments and error bars represent the standard deviation of the three.
(H) Wild-type or mutant BJ1 cells knocked out of CEP128 and C-Nap1 by CRISPR were stained with indicated antibodies. Note that wild-type BJ1 cells proliferate poorly from a single cell, and thus could not survive the clonal selection process. C-Nap1−/−; CEP128−/− BJ1 cells were examined in the mixed population 6 days after CRISPR treatment. FOP (green) marks the centrosomes, acetylated tubulin (also green) marks the cilia, GM130 (red) marks the Golgi.
See also Figure S2.
sDAP and C-Nap1 work together to establish intact centrosome cohesion
In vertebrate cells, the two centrosomes during interphase are closely associated (about 1 µm apart) through centrosome cohesion, mediated in part by C-Nap1 and ninein (Graser et al., 2007; Mayor et al., 2000). Consistently, loss of C-Nap1 or ninein in our knockout cells caused weakened centrosome cohesion allowing centrosomes to separate mildly (~2 µm apart; Figure 3E). Centrosomes are described as “split” when the distance between them exceeds 2 microns (Mayor et al., 2000). The mild separation phenotype seen in C-Nap1 or ninein knockout cells suggests that other redundant mechanisms may exist to bring centrosomes together. Intriguingly, while loss of any member ODF2 Group alone had no effect on centrosome cohesion (Figure 3E), when both C-Nap1 and any member of the ODF2 Group were mutated, centrosome cohesion was completely disrupted, leading to extreme centrosome separations (Figure 3E). In some cases, centrosomes were over 10 microns apart in the two-dimensional XY-plane alone, placing them in opposite side of the cell or nucleus (Figures 3E–G). The extreme separation phenotype is specific, as we were able to rescue centrosome cohesion by exogenously expressing full length CEP128 in cells lacking both C-Nap1 and CEP128 (Figure 3E), or by restoring endogenous C-Nap1 expression through a second run of CRISPR gene editing that corrects the translational frameshift created by the 1st run (Figure 3E; see Experimental Procedures). These results show that sDAP and C-Nap1 function together to ensure tight centrosome cohesion.
C-Nap1 and sDAP are required for stable association of ciliated centrosomes with the Golgi
Vertebrate centrosomes are intimately associated with the Golgi apparatus (Kellogg et al., 1994), in turn bringing the two centrosomes in close vicinity. The purpose of this association is unknown but it might enhance the trafficking between cilia/centrosome and Golgi, facilitating cilia growth and maintenance. We reasoned that when two centrosomes are great distances apart, it would be difficult for an intact Golgi to associate with both centrosomes. We thus examined if centrosomes remain associated with the Golgi in our mutant cell lines. Centrosome-Golgi association remained normal when the ODF2 Group, C-Nap1 or ninein was lost individually (Figures 3F, G). In contrast, when C-Nap1 and ODF2 were both absent, one centrosome frequently remained with the otherwise intact Golgi while the other centrosome was completely separated (Figures 3F, G). More interestingly, in 90% of the cases where the Golgi is associated with only one centrosome, it was the older, ciliated centrosome that was distantly positioned away from the Golgi (Figures 3F, G), while the other (younger/nonciliated) centrosome remained associated the Golgi. The same pattern held true when CEP128 or centriolin was eliminated in the C-Nap1 knockout background (Figures 3F, G). Restoring either C-Nap1 or CEP128 expression in C-Nap1−/−; CEP128−/− double knockout cell lines completely rescued normal centrosome-Golgi association (Figures 3F, G). Depletion of the same proteins in non-transformed BJ1 cells using CRISPR resulted in a similar Golgi-cilia separation phenotype, indicating that the function is not cell line specific (Figure 3H). Thus, our results indicate that loss of both C-Nap1 and any member of the ODF2 Group specifically breaks the stable cilia-Golgi association. It is known that ciliary vesicles derived from the Golgi play a key role in ciliogenesis (Hehnly et al., 2012; Lu et al., 2015; Sorokin, 1962; Westlake et al., 2011), providing a potential reason why the Golgi is often closely associated with cilia. To our surprise, at least in RPE1 cells, the close cilia-Golgi association is neither essential for cilia formation nor for cilia length control.
Previous studies showed that expression of dominant-negative fragments of AKAP450, a protein localizing to both the Golgi and centrosome, impairs Golgi organization and centrosome-to-Golgi attachment (Hurtado et al., 2011). We found that AKAP450 localized normally to the Golgi apparatus in all of our mutant cells, regardless of whether or not the Golgi is stably associated with cilia (Figure S2A). Moreover, we found that while C-Nap1 was required for centrosomal localization of AKAP450, loss of C-Nap1 alone had no effect on Golgi-cilia association (Figure S2A and B; Figures 3F, G). Contrary to published work (Panic et al., 2015), Golgi size was no larger in C-Nap1 mutant cells than controls (Figure S2C). Thus, the Golgi-cilia detachment phenotype seen in our double mutant cells is not a result of non-specific disruption of the Golgi organization through AKAP450 or through C-Nap1.
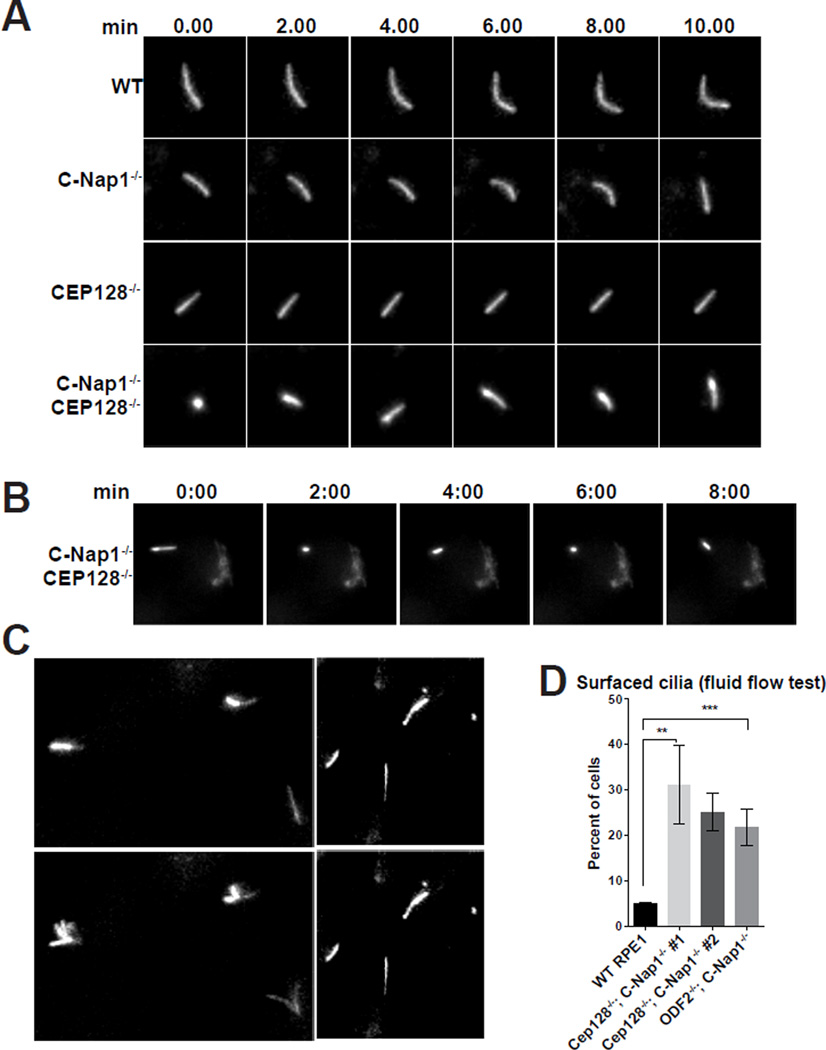
C-Nap1−/−; CEP128−/− double mutant cells grow surfaced cilia capable of responding to fluid flow with motion
Cilia form in normal frequency and length in C-Nap1−/−; CEP128−/− double mutant cells, despite their stable association with the Golgi being disrupted. To examine if these cilia behave differently in ways that cannot be easily detected in fixed cells, live cell imaging was performed. Wild-type or mutant cells were induced to express Arl13b-GFP to mark the cilia, and filmed by time lapse microscopy for hours. Cilia in wild-type, C-Nap1−/− knockout, or CEP128−/− knockout cells behaved similarly, exhibiting a confined, leisurely motion, rarely changing direction and remained parallel to the XY plane (Figure 4A; Movie S1), consistent with them being submerged. On the contrary, cilia in C-Nap1−/−; CEP128−/− double mutant cells had frequent episodes of erratic motion in which the cilia changed direction randomly, often pointing along the Z-axis (Figure 4A; Movie S2), a pattern nearly unseen in wild-type RPE1 cells. Moreover, live-cell imaging also confirmed that the wide-range motion of cilia occurred while cilia were completely detached from the Golgi (Figure 4B; Movie S3). These observations suggest that cilia formed in double mutant cells are not trapped in a deep membrane invagination. To further test the idea, we examined the bending of the cilium in response to fluid flow, a physical property specific to surfaced cilia. As expected, cilia in wild-type RPE1 cells were stationary, showing no bending when fluid was pumped over them, consistent with being submerged (Figures 4C–D; Movie S4 and S5). In contrast, at any given time, about 25–32% of cilia in C-Nap1−/−; CEP128−/− cells and 23% in C-Nap1−/−; ODF2−/− cells flutter violently in response to fluid flow (Figures 4C–D; Movie S5), strongly suggesting that these cilia project from the apical cell surface. Note that similar behaviors were seen in independently derived clones of cells (Figure 4D).
Figure 4. Cilia mobility and position are altered upon depletion of sDAP and C-Nap1.
(A) Still images extracted from long time-lapse movies of indicated RPE1 cell lines expressing Arl13b-GFP. Row labels indicate genotypes of the imaged cells. Column labels indicate the time intervals (in minutes). Each row represents time lapse. The movie for WT or C-Nap1−/−; CEP128−/− mutant cells was provided (see Movie S1 and S2).
(B) Still frames from long time-lapse movies of C-Nap1−/−; CEP128−/− double knockout cell lines expressing both Arl13b-GFP and GalT-GFP, a Golgi marker (see Movie S3). Note that cilia showed wide-range motion while detach from the Golgi.
(C) Fluid was pumped over the cultured cells during the time lapse imaging of Arl13-GFP expressing RPE1 cells. Frames from a short time-lapse movie (10 seconds) are shown. Genotypes of the imaged cells are indicated. Images taken before and during fluid flow are shown. Note that cilia fluttered upon flow activation (see Movie S4 and S5).
(D) The percentage of cilia that respond to fluid flow in WT or indicated mutant cell lines. The average of 3–4 independent experiments is shown. Two independently derived clones of C-Nap1−/−; CEP128−/− mutant cells (#1 & #2) were analyzed. 100–150 cells were scored to determine percentage in each of 3–4 separate experiments. Error bars represent the standard deviations. Significance determined by unpaired two-tailed t-test with Welch’s correction (p<0.01** p<0.001***).
C-Nap1−/−; CEP128−/− double mutant cells grow cilia not trapped in a deep membrane invagination
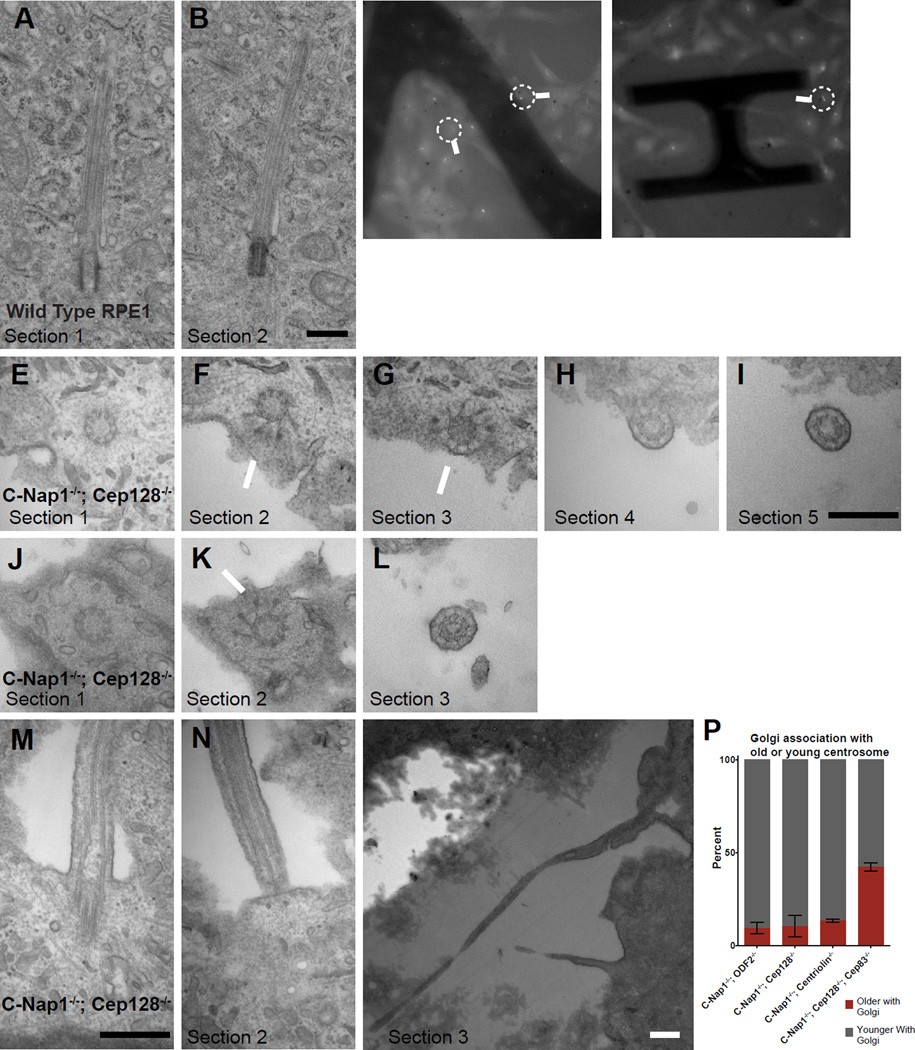
To further characterize the spatial configuration of cilia in detail, serial sectioning transmission electron microscopy (TEM) was performed. TEM analyses confirmed that wild-type RPE1 cilia are submerged in a deep membrane invagination (or pit) surrounded by cytoplasm (100%, n=16) (Figures 5A–B). In C-Nap1−/−; CEP128−/− double mutant cells, however, some cilia respond to flow and others do not. We thus used correlated light and electron microscopy (LM/EM) to explore why these cilia behave differently (Figures 5C–D; Movie S6 and S7). LM/EM analyses showed that flow-sensitive cilia indeed projected from the apical cell surface (n=5/5), lacking an intact ciliary pit, with the centriole docked to the plasma membrane via distal appendages (Figures 5E–I and 5J–L) and the axoneme growing from the edge of the cell into the environment (Figures 5E–I and 5J–L). Surprisingly, in addition to flow-sensitive cilia, we found that cilia that are non-responsive to flow also lacked intact ciliary pits (n=2/2) (Figures 5M–O); these cilia, instead of protruding from the apical surface, were found to grow out of the basal surface, where they were trapped in a cavity devoid of cytoplasm underneath the cell (Figures 5M–O), consistent with their static behavior in response to fluid flow. Moreover, the physical anchoring of the ciliated centrosome to the plasma membrane at the cell surface can perhaps explain why only the non-ciliated (or daughter) centrosome can “freely” stay at the cell center with the Golgi (Figure 3F, G). Indeed, consistent with this prediction, when ciliogenesis (membrane docking) was disrupted by abolishing the DAP component CEP83 in C-Nap1−/−; CEP128−/−; CEP83−/− triple knockout cells, we found that the biased association between the Golgi and daughter centrosomes disappeared (Figure 5P). Together, these results indicate that factors from both distal and proximal ends of centrioles are required to maintain a deep membrane invagination for proper spatial configuration of cilia.
Figure 5. Loss of the deep ciliary pit in CEP128−/−; C-Nap1−/− double knockout cells.
(A and B) Two serial TEM sections of a wild type RPE1 cell carrying a submerged cilium buried in a deep membrane invagination. Scale bar represents 500nm.
(C and D) Still light microscopy images of C-Nap1−/−; CEP128−/− double knockout cells carrying either flow-sensitive or insensitive cilia, extracted from time-lapse movies (see Movie S6 and S7). Arrows mark the cells that were processed for TEM and shown in panels below as indicated.
(E, F, G and I) LM/EM analyses of a flow responsive cilium from C-Nap1−/−; CEP128−/− double mutant cells. A series of successive EM sections revealed that the cilium was at the apical surface. Section 1 contained the centriole. Section 2 and 3 showed visible distal appendages (arrows) with tip of centriole clearly on edge of the cell while the immediate next two sections showed the ciliary axoneme clearly outside of the cell. Scale bar represents 500nm.
(J, K and L) A series of EM sections of another C-Nap1−/−; CEP128−/− double mutant cilium that had been shown to respond to fluid flow under time-lapse microscopy. Sections 1 and 2 showed the distal end of the centriole while section 3 showed an axoneme outside the apical cell surface. Scale bars represent 500nm.
(M and O) Serial EM sections of a C-Nap1−/−; CEP128−/− double mutant cilium that could not respond to flow. Section 1 and 2 showed that the deep ciliary pit is disrupted, while sections 3 at a lower magnification showed that the rest of the cilium was trapped below the cell, outside of the basal cell surface. All scale bars are 500nm.
(P) The biased separation of the older/ciliated centrosome from the Golgi depends on CEP83. Quantification shows the percentage of the indicated mutant cell lines in which the older or younger centrosome associates with the Golgi. Numbers were collected from cells in which the two centrosomes are distantly separated and only one is associated with the Golgi. Older centrosomes in C-Nap1−/−; CEP128−/−; CEP83−/− triple KO cells were identified by ODF2 staining. Error bars represent standard deviations. Data of C-Nap1−/−; ODF2−/−, C-Nap1−/−; CEP128−/− and C-Nap1−/−; CNTRL−/− double KO cells are extracted from Figure 3F.
Loss of deep ciliary pit allows multiple intact cilia to form individually in the same cell
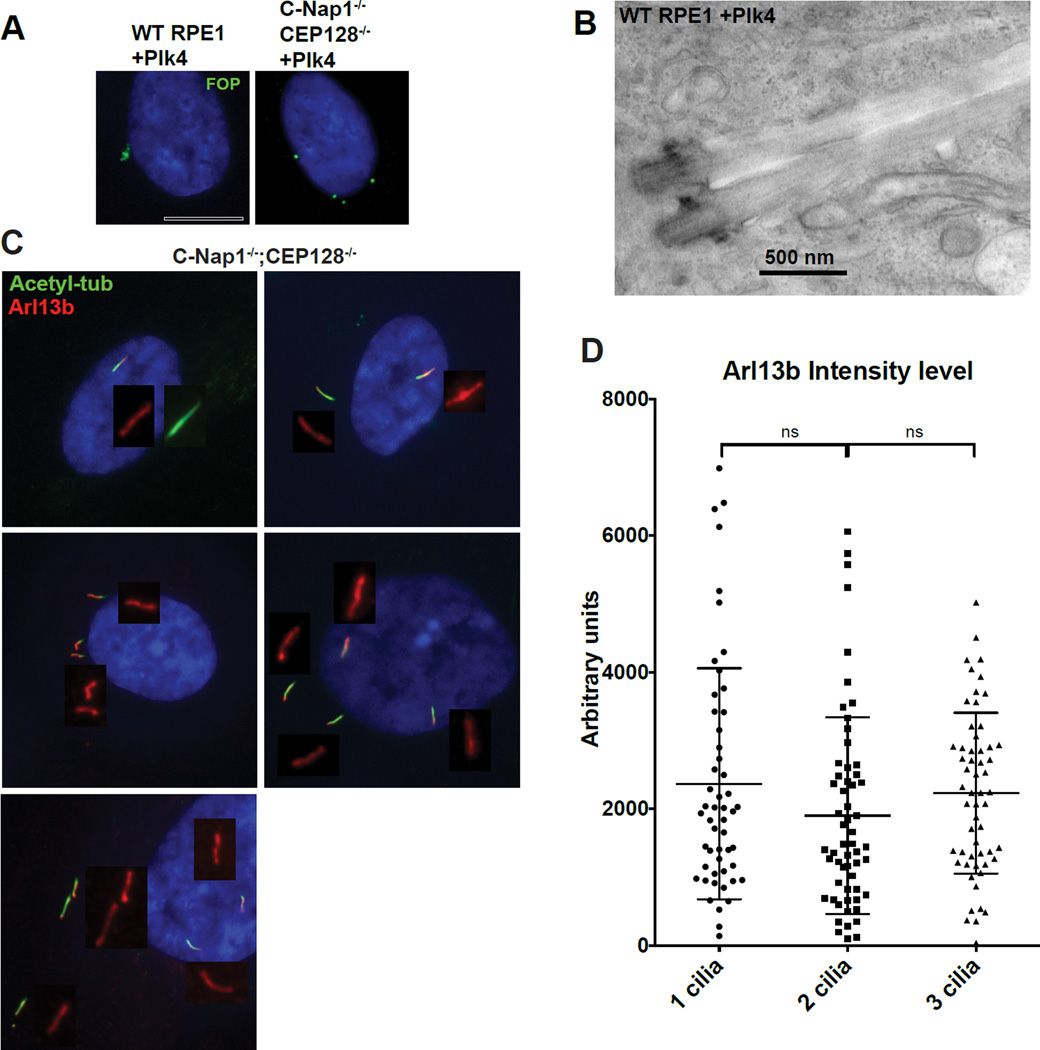
Over-duplication of centrosomes in RPE1 cells can lead to productions of multiple cilia that are clustered in the same membrane invagination (Mahjoub and Stearns, 2012). These clustered cilia are diluted of membrane-bound components and therefore functionally compromised as compared to a solitary cilia that is not in a cluster (Mahjoub and Stearns, 2012) (Figure S3). This reveals that the deep ciliary pit, while important in some cell types, can potentially cause serious problems for multi-ciliated cells (Mahjoub and Stearns, 2012). To explore if the loss of the ciliary pit can liberate cilia, allowing multiple cilia to form with undiluted membrane components, we induced centrosome over-duplication in C-Nap1−/−; CEP128−/− double mutant cells (which lack a proper ciliary pit) (Figures 6A and B). Intriguingly, while overexpression of PLK4 in wild-type RPE-1 cells drove the formation of clustered centrosomes and cilia (Figure 6A left; Figure 6B), in C-Nap1−/−; CEP128−/− cells, multiple individual centrosomes or cilia at distinct locations were formed and maintained (Figure 6A right and Figure 6C). The distance between these cilia indicates that they are not physically linked through specific structures. Furthermore, cilia growing in separate locations in the same cell had full levels of Arl13b (Figures 6C–D), similar to that seen in the single primary cilium. Together, our observations suggest that forming a deep membrane invagination is not essential for ciliary trafficking of membrane components (such as Arl13b), neither for cilia assembly nor for length control. Instead, we propose that C-Nap1 and sDAP function in specific cell types to confine primary cilia in deep pits. Confinement of multiple cilia in a shared, deep cavity may negatively impact the function of multi-ciliated cells or of cells that carry more than one non-motile cilia such as olfactory neurons.
Figure 6. Loss of the deep ciliary pit allows multiple intact cilia to form with undiluted membrane composition in one cell.
(A) Indicated RPE1 cell lines were infected with lentiviral vectors carrying tet-inducible PLK4 expression construct and induced to overexpress PLK4. Doxycycline was applied to cells for 2 days before examination. FOP (green) marks centrosomes. Wild type RPE1 cells have centrosomes cluster in a small area. In contrast, centrosomes are well separated in the CEP128−/−; C-Nap1−/− double KO cells.
(B) EM images of two clustered cilia that grew from two centrosomes in the same ciliary pit. PLK4 was expressed in wild type RPE1 cells to induce formation of extra centrosomes before serum starvation and preparation for TEM.
(C) Multiple cilia in CEP128−/−; C-Nap1−/− double knockout cells were distantly separated with undiluted Arl13b. Multiple cilia formation in the same cell was induced by overexpression of the PLK4 kinase, followed by 48 hours of serum starvation. Cells were stained with indicated antibodies.
(D) Quantification of Arl13b intensity. Mean value of Arl13b intensity in regions surrounding cilia was taken. Plot displays Arl13b intensity per cilia in bi-ciliated or triciliated cells. Error bars represent standard deviations.
(See also Figure S3)
Abnormally positioned cilia at the apical cell surface results in ectopic accumulation of Hedgehog signaling components
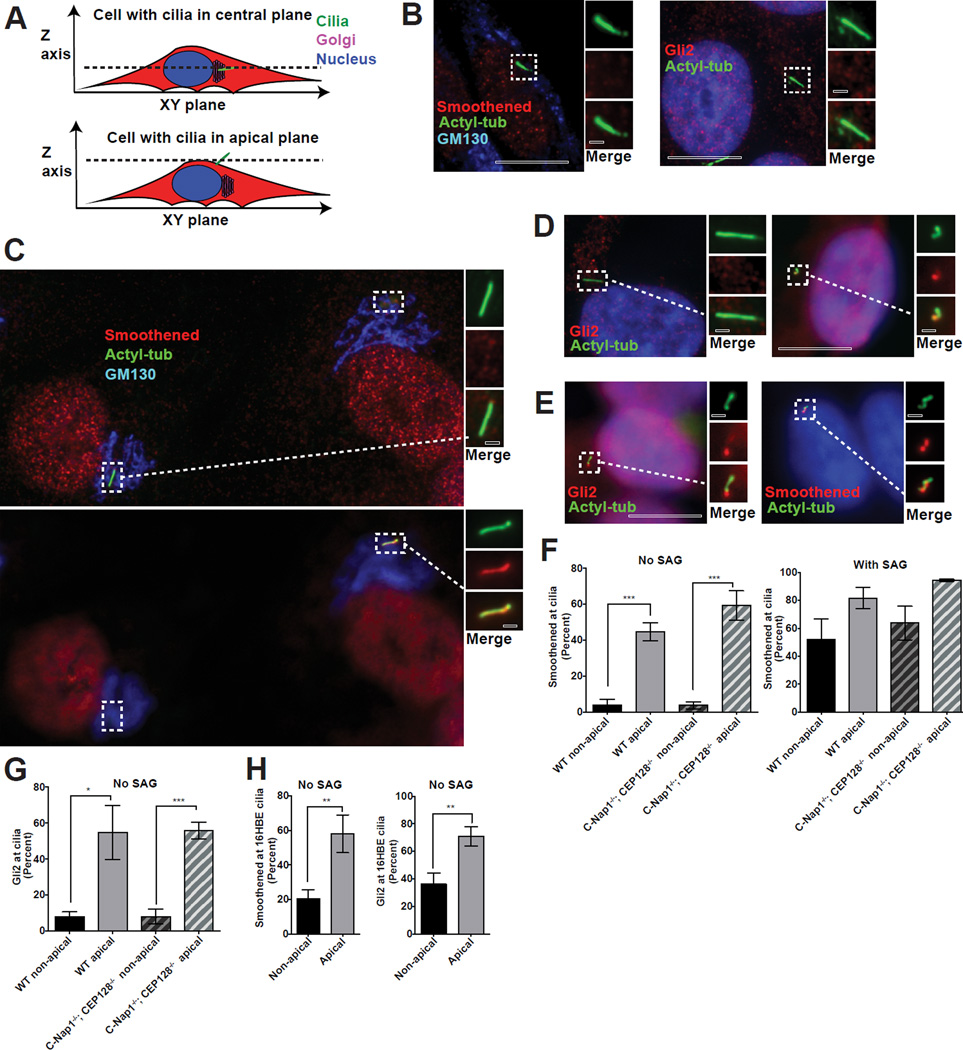
We next examined if cilia may alter their biochemical properties upon changes in their spatial configuration. Submerged cilia are found in the same focal plane as the Golgi and nucleus while surfaced cilia are often at the top of cells (Figure 7A), making it possible to distinguish likely surfaced from submerged cilia. Using this method, we found that submerged cilia in both wild-type and C-Nap1−/−; CEP128−/− mutant RPE1 cells, were nearly all devoid of Hedgehog signaling components Smoothened and Gli2 (Figures 7B, 7C and 7F), unless treated with the Smoothened agonist (SAG) (Figure 7F). In striking contrast, apically surfaced cilia in C-Nap1−/−; CEP128−/− mutant cells, as revealed by their location at a focal plane above the nucleus and Golgi (Figures 7C–D), frequently accumulated Smoothened and Gli2 even in the absence of SAG (Figures 7C, D, and F). Moreover, in about 1–2% of wild-type RPE1 cells in which cilia were apically surfaced (Figure 7E), as revealed by their apical location above the Golgi and nucleus, similar accumulations of Smoothened or Gli2 were observed (Figures 7E–G). To further confirm the link between apical cilia and Smoothened accumulation, we examined 16HBE cells, a human bronchial epithelial cell line. Prior to forming a fully polarized monolayer, 16HBE cells frequently grew submerged cilia that were mostly devoid of Smoothened or Gli2 (Figure 7H). In contrast, after polarization, 16HBE cells were found to form surfaced cilia that accumulated high levels of Smoothened or Gli2 in the absence of SAG (Figure 7H).
Figure 7. Apically surfaced cilia can ectopically recruit Smoothened and Gli2 in the absence of agonist.
(A) Cartoon depicting submerged or apically surfaced cilia in fixed RPE1 cells guided by the relative position between the cilia, Golgi, and nucleus. Only the cilia positioned at an apical focal plane not overlapping with the Golgi or nucleus were considered “apically surfaced”. Cilia at the same focal plane with the Golgi and nucleus were considered submerged. Wild-type RPE1 cilia are mostly submerged, with only 1–2% surfaced apically. In contrast, apical cilia can be easily found in CEP128−/−; C-Nap1−/− double knockout cells.
(B) Wild-type RPE1 cilia are devoid of Smoothened (left) and Gli2 (right) in the absence of SAG.
(C) Two CEP128−/−; C-Nap1−/− mutant cells in the same field, one carrying apically surfaced cilia (right) and the other submerged cilia (left), were stained with Smoothened, cilia, and Golgi antibodies as indicated. At the central focal plane (top panel), submerged cilia, the Golgi and nucleus were in focus. At the apical focal plane, only surfaced cilia was in focus (bottom panel, right).
(D) CEP128−/−; C-Nap1−/− mutant cells carrying either submerged (left) or apically surfaced (right) cilia were stained with Gli2 antibodies.
(E) Rare wild-type RPE1 cells carrying apically surfaced cilia were stained with Smoothened or Gli2 antibodies as indicated.
(F) Quantification of the ciliary accumulation of Smoothened for each indicated genotype and cilia position in the presence or absence of SAG. Mean and standard deviation are depicted as bars and error bars respectively. 30–60 cilia were scored for each of the three repeats, except for the rare surfaced cilia in wild type cells, which were 10 cilia per repeat. Significance determined by unpaired two-tailed t-test with Welch’s correction (p<0.001 corresponds to ***).
(G) Quantification of the ciliary accumulation of Gli2 for each indicated genotype and cilia position in the absence of SAG. Mean and standard deviation are depicted. 30–60 cilia were scored for each of the three repeats, except for the rare surfaced cilia in wild type cells, which were 10 cilia per repeat. Significance determined by unpaired two-tailed t-test with Welch’s correction (p<0.05 for *) (p<0.001 for ***).
(H) Plot depicting the percentage of 16HBE cells with Smoothened or Gli2 accumulation in cilia for each cilia position. 20–40 cilia were scored for each of three repeats. Mean percentage of the 3 experiments is shown. Error bars correspond to standard deviation. Significance determined by unpaired two-tailed t-test with Welch’s correction (p<0.01**). See also Figure S4.
The accumulations of Smoothened and Gli2 in surfaced cilia were also seen in C-Nap1−/− ; ODF2−/− cells as well as in BJ5, a fibroblast cell line in which about 10% of cilia are apically surfaced (Figure S4A–B). Importantly, the Smoothened enrichment at surfaced cilia is seen in Triton-extracted, methanol fixed cells, suggesting that the pattern is unlikely due to a difference in antigen accessibility (Figure S4D). To further rule out this possibility, we expressed Smoothened-GFP in C-Nap1−/−; CEP128−/− mutant cells. Under these conditions, overexpressed Smoothened-GFP was seen in most cilia, but more enriched in surfaced cilia (Figure S4C), consistent with our observations with the endogenous Smoothened. Thus, the biochemical properties of primary cilia can be altered through spatial control of ciliogenesis, a type of regulation previously not recognized. Together, our studies identified redundant functions of sDAP and the proximal end factor C-Nap1 in centrosome cohesion, cilia-to-Golgi association, and ciliary pit maintenance, critical for spatial control of ciliogenesis.
DISCUSSION
In this study, we have identified molecules required in human cells to maintain primary cilia in submerged configurations, a process involving redundant functions of accessory structures located at both the distal and proximal ends of centrioles. Removal of these structures from centrioles has no effect on cilia assembly per se, but produces multiple defects in cilia/centrosome position. Their removal allows normally submerged cilia to fully surface, losing the deep ciliary pit, and leads to wide separation of the Golgi from ciliated centrosomes. Importantly, surfaced cilia respond to fluid flow with motions, a process important for mechanosensation, and gain abilities to recruit signaling molecules that are normally not seen in submerged cilia, revealing a potential link between the sensation functionality and spatial configuration of cilia. Moreover, our studies support the published model that the deep ciliary pit can trap a group of cilia together (Mahjoub and Stearns, 2012), and argue that abolishing the clustering of cilia within a shared pit could be an important process to ensure proper function of cells that have multiple surfaced cilia, such as multi-ciliated epithelia or olfactory neurons. Further research would be required to determine the mechanical mechanisms by which sDAP maintain submerged cilia, checking particularly whether it involves the activity of sDAP in interacting with cytoplasmic microtubules (Delgehyr et al., 2005; Quintyne et al., 1999).
The cilium is historically described either as a cellular antenna that receives extracellular signals or as a hair-like structure that produces and senses motions. Yet vertebrate cilia are often maintained in a submerged configuration (Fisher and Steinberg, 1982; Sorokin, 1968), where they certainly cannot generate nor detect motions, and perhaps might even be shielded from properly receiving chemical signals, raising many interesting ideas and questions for future studies. For example, is it possible that placing the cilium in a submerged configuration is simply a more efficient way to temporarily or periodically silence the cilia function (a substitute to the costly cilia disassembly and reassembly processes)? Alternatively, cilia may be buried to shut off only certain signaling activities (e.g. mechanosensation) and thereby enhance or specify others. Another interesting idea is that while diffusible ligands can reach submerged cilia, but when they do, the ciliary pit may serve as a trap to retain these stimuli that otherwise quickly diffuse away, allowing “effective signal transduction” to occur selectively in some cells. These ideas or possibilities are beyond the scope of the current study. A vertebrate model allowing manipulation of the spatial configuration of ciliogenesis is certainly required to address some of these questions.
EXPERIMENTAL PROCEDURES
Cloning and Plasmids
Full-length HA tagged CEP128, Smoothened-GFP (from Addgene: 25395 (Taipale et al., 2000)), Arl13b-GFP, GalT-GFP (A Golgi marker from Addgene:11929 (Cole et al., 1996)) and Arl13b-mCherry were cloned into pLVX-Tight-Puro vector (Clontech). For constant Arl13b and GalT expression, CMV promoter was cloned into pLVX vector to replace the tetracycline-inducible promoter in Arl13b-GFP, GalT-GFP and Arl13b-mCherry constructs. Both tetracycline-inducible and non-inducible Arl13b constructs were used in this work.
CRISPR
CRISPR/Cas9 mediated gene targeting was used to inactivate C-Nap1 and various sDAP components as described previously (Izquierdo et al., 2014). The targeting sequences of gRNAs used for CRISPR are as follows: C-Nap1 gRNA4 (5’-GATACTACAGACCCAGCTCCAGG-3’), ODF2 gRNA1 (5’-GAGGGAACAGCACTGCAAAGAGG-3’), ODF2 gRNA2 (5’-GAGTGTCCGGGTGAAAACCAAGG-3’), Ninein gRNA0 (5’-GCTCAGCCCAAATATGTTAGAGG-3’), CEP128 gRNA2 (5’-GCTGCCAGATCAACGCACAGGG-3’), CEP128 gRNA4 (5’-GAGTCAGCTCTGAGATCTGAAGG-3’), CEP128 gRNA5 (5’-GCAGCTGAACTTCAGCGCAATGG-3’), Centriolin gRNA6 (5’-AGTGGGTTGCAAGAATACCTGG-3’), Centriolin gRNA9 (5’-GTGCCTATGAAGCTGAGCTAGAGG-3’), Cep83 gRNA1 (5’-GGTGGAGACAGTGGATTGACAGG-3’), Cep83 gRNA2 (5’-GATATTAACTCCACAAAAATTGG-3’) Frame shift mutations in each target gene were confirmed by sequence analyses of clonal cell lines (see Table S1). To achieve complete protein depletion, multiple gRNAs targeting different exons were used for CEP128 and centriolin as shown above. For rescuing C-Nap1 expression, a second lesion near the site of the 1st lesion was introduced by CRISPR to put the sequence back into frame, with the targeting sequence (5’-GAGCCTCCTGGAATCCCAGTGG-3’).
Cell culture
hTERT-RPE1 cells were cultured in DME/F-12 10% FBS, 1% penicillin/streptomycin. hTERT-BJ1 cells were cultured in DMEM medium supplemented with 199 media (1:4 ratio) 10% FBS, 1% penicillin/streptomycin. 16HBE cells were cultured in MEM supplemented with Glutamax (Gibco), 10% FBS, 1% penicillin/streptomycin. All other cells were cultured in DMEM medium, 10% FBS, 1% penicillin/streptomycin. Stable expression of various gene constructs was achieved with the Lentiviral pLVX-tight-Puro vector.
Antibodies
Mouse monoclonal antibodies used in this study are anti-centrin2 (clone 20H5; 04-1624, Millipore), anti-centrin3 (clone 3E6; H00001070-M01; dilution 1:200), anti-FOP (clone 2B1, H00011116-M01; dilution 1:1000) (Abnova), anti-Cep170 (72-413-1; dilution 1:200; Invitrogen Antibodies), anti-ODF2 (1a1, H00004957-M01; dilution 1:200; Novus Biologicals), anti-GM130 (610822; dilution 1:2000), anti-AKAP450 (611518; dilution 1:500), anti-p150glued (610473; dilution 1:1000)(BD Transduction Laboratories), anti-Gli2 (C-10, sc-271786; dilution 1:200), anti-gamma tubulin (TU-30, sc-51715; 1:500 dilution), anti-Centriolin (C9, sc-365521; dilution 1:200)(Santa Cruz Biotechnology), anti-acetylated alpha tubulin (clone 6-11B-1, T7451; dilution 1:1000), anti-alpha tubulin (clone DM1A, T9026; dilution 1:2,000) (Sigma-Aldrich). Rabbit polyclonal antibodies used in this work include anti-ODF2 (HPA001874; dilution 1:500), anti-CEP128 (HPA001116; dilution 1:500) (Sigma-Aldrich/Atlas), anti-Ninein (A301-504; dilution 1:3000; Bethyl Labs), anti-FBF1 (11521-1-AP; dilution 1:500; Proteintech Group), anti-Kif2a (ab37005; dilution 1:3000), anti-CEP135 (ab75005; dilution 1:1000) and anti-Smoothened (ab38686; dilution 1:200) (Abcam). A rabbit polyclonal antibody against the human C-Nap1 was produced as previously described (Tsou and Stearns, 2006). Anti-Arl13b rabbit polyclonal antibody was a gift from the Kathryn Anderson lab as was the Smoothened Antibody used in S4. A rat monoclonal anti-tubulin (clone Yl1/2, MCA77G; dilution 1:500; AbD Serotec) was also used in this work. Secondary antibodies Alexa-Fluor 405, 488, 594, 680 were from Molecular Probes.
Immunofluorescence
Cells were washed with phosphate-buffered saline (PBS) or extracted in PTEM (20 mM PIPES pH 6.8, 0.2% Triton X-100, 10 mM EGTA, 1 mM MgCl2) for 2 minutes, before fixation with methanol at −20°C or with 4% paraform aldyhyde for 10 minutes. Fixed cells were blocked in 3% BSA 0.1% TritonX100 in PBS before incubation with antibodies. DNA was visualized using 4’,6-diamidino-2-phenylindole (DAPI). An upright microscope (Axio imager; Carl Zeiss) with a 100× objective (NA 1.4) and a camera (ORCA ER; HamamatsuPhotonics) was used to collect still images. For p150glued or Kif2a staining at centrosomes, microtubules were depolymerized by cold treatment for 20–30 minutes to remove microtubule associated staining (centrosomal Kif2a and p150glued were not affected by the cold treatment). For microtubule regrowth assay, the same cold treatment described above was followed by a 10-minute incubation at 37C to allow microtubule aster formation, before fixation and examination. To induce cilia growth, RPE1 cells were switched to media without FBS. SAG treatments were done for 20 hours with 200nM SAG. ImageJ and Adobe Illustrator software were used to prepare images for figures. Graphs, plots and statistical analysis were done in Prism software. P-values and significance were determined by unpaired t-test with Welch’s correction.
Arl13b level and Smoothened-GFP quantification
Cells were induced to express modest amounts of Smoothened-GFP under tetracycline-inducible promoter. Regions of interest were drawn around the cilia in ImageJ using the wand tool and measured.
To produce the Arl13b ratios used in Figure S3, the following formulas were applied Arl13b Level = ((‘Mean Intensity’−‘Background Level Mean’) * ‘Area’) / (‘Number Cilia in Roi’ * ’Major Axis Length’)
Ratio= ‘Average Arl13b Level in cells containing 2 cilia’ / ‘Average Arl13b Level in cells containing 1 cilia’
Time-lapse microscopy and generation of fluid flow
For live cell imaging, cells expressing fluorescent protein-labeled Arl13b or Golgi marker (GalT-GFP) were grown in glass bottom plates. A Zeiss Axiovert microscope equipped with 40× objective, a motorized temperature-controlled stage, an environmental chamber and a CO2 enrichment system was used for time-lapse microscopy. Images in time-lapse movies were acquired and processed by an electron-multiplying charge-coupled device (EMCCD) camera (from Hamamatsu Photonics) and axiovision software (Zeiss). To generate fluid flow, holes were melted into the lid of glass bottom plate using a soldering iron. Silicone tubing (1/16”, Fisher Scientific) was filled with media, put though the openings, submerged in the media, and positioned near the field of view of the objective. The other end of the silicone tubing was attached to a 6V peristaltic pump with a maximum flow rate of ~40ml/min.
Correlated light and Electron Microscopy
Aclar sheets with Carbon patterns were glued onto glass bottom plates (Cellvis). Cells expressing tet-inducible GFP-Arl13b were split onto the aclar sheets and grown for 4 days. Cells were then serum starved and treated with 1ug/ml doxycycline for two more days. Using the same microscope and flow apparatus as described above, we imaged cilia movements on the patterned aclar sheets at 20×. We then fixed rapidly by adding 8% Paraformaldehyde, 4% Glutaraldehyde, 4mM CaCl2, 40mM Sodium Cacodylate PH7.4 buffer to an equal volume of media. We incubated for 45 minutes. We then changed to 4% Paraformaldehyde, 2% Glutaraldehyde, 2mM CaCl2 in 0.1M Sodium Cacodylate buffer with 0.2% tanic acid for a 2h treatment. Plate was left in 4% Paraformaldehyde, 2% Glutaraldehyde, 2mM CaCl2 in 0.1M Sodium Cacodylate buffer at 4C for storage. Cells were post-fixed in 1% reduced OsO4 with 1.1% potassium ferrocynide in 0.1M sodium cacodylate buffer for 60 min on ice, stained with 1% uranyl acetate for 30min, dehydrated in a graded series of ethanol, infiltrated with Eponate12 resin (Electron Microscopy Sciences) and then embedded in the resin. Based on live cell imaging, serial sections (70-nm thickness) in regions containing cells of interest were cut on a microtome (Ultracut E; Leica). Samples were examined on JOEL 100CX transmission electron microscope (TEM) with the digital imaging system (XR41-C, Advantage Microscopy Technology Corp, Denver, MA) at 80kV or 100kV in the electron microscopy resource center in The Rockefeller University.
Supplementary Material
Highlights.
Depletion of both sDAP and C-Nap1 produces several synthetic phenotypes in cilia
Such mutants lose cilia-Golgi association and often have cilia on the cell surface.
Surfaced cilia are able to ectopically recruit hedgehog-signaling components
Multiple unclustered cilia of complete composition can form in these mutants
Acknowledgments
We thank Dr. Kathryn Anderson for the gift of an anti-Arl13b antibody aliquot and a Smoothened Antibody Aliquot. We thank Dr. Michael Overholtzer for the gift of an anti-GM130 antibody aliquot. We also thank Dr. Hui-Ju Yang for preliminary work that helped lead to this project. This work was supported by the National Institute of Health grant GM088253 and American Cancer Society grant RSG-14-153-01-CCG to MFBT. MFBT was also supported by Geoffrey Beene Cancer Research Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
N. Soplop, and K. Uryu performed correlated Light and Elecron Microscopy shown in Figures 5. W-J Wang provided some EM images for Figure 5A, 5B & 6B and made Arl13b-GFP construct. G. Mazo and M-F B. Tsou planned and conducted all other works, and wrote the manuscript.
Reference
- Allen RA. Isolated cilia in inner retinal neurons and in retinal pigment epithelium. Journal of Ultrastructure Research. 1965;12:730–747. doi: 10.1016/s0022-5320(65)80058-2. [DOI] [PubMed] [Google Scholar]
- Benmerah A. The ciliary pocket. Current Opinion in Cell Biology. 2013;25:78–84. doi: 10.1016/j.ceb.2012.10.011. [DOI] [PubMed] [Google Scholar]
- Boveri T. Über den Antheil des Spermatozoon an der Theilung des Eies. Sitzungsber. d. Ges. f. Morph, u. Phys. München, BD. 1887:3. [Google Scholar]
- Burakov A. Centrosome positioning in interphase cells. The Journal of Cell Biology. 2003;162:963–969. doi: 10.1083/jcb.200305082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaini G, Whitfield WGF, Riparbelli MG. Centriole and Centrosome Dynamics during the Embryonic Cell Cycles That Follow the Formation of the Cellular Blastoderm in Drosophila. Experimental Cell Research. 1997;234:183–190. doi: 10.1006/excr.1997.3618. [DOI] [PubMed] [Google Scholar]
- Cole NB, Smith CL, Sciaky N, Terasaki M, Edidin M, Lippincott-Schwartz J. Diffusional mobility of Golgi proteins in membranes of living cells. Science (New York, NY) 1996;273:797–801. doi: 10.1126/science.273.5276.797. [DOI] [PubMed] [Google Scholar]
- Dammermann A. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. The Journal of Cell Biology. 2002;159:255–266. doi: 10.1083/jcb.200204023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammermann A, Merdes A. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. The Journal of Cell Biology. 2002;159:255–266. doi: 10.1083/jcb.200204023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgehyr N. Microtubule nucleation and anchoring at the centrosome are independent processes linked by ninein function. Journal of Cell Science. 2005;118:1565–1575. doi: 10.1242/jcs.02302. [DOI] [PubMed] [Google Scholar]
- Delgehyr N, Sillibourne J, Bornens M. Microtubule nucleation and anchoring at the centrosome are independent processes linked by ninein function. Journal of Cell Science. 2005;118:1565–1575. doi: 10.1242/jcs.02302. [DOI] [PubMed] [Google Scholar]
- Field MC, Carrington M. The trypanosome flagellar pocket. Nat Rev Micro. 2009;7:775–786. doi: 10.1038/nrmicro2221. [DOI] [PubMed] [Google Scholar]
- Fisher SK, Steinberg RH. Origin and organization of pigment epithelial apical projections to cones in cat retina. The Journal of Comparative Neurology. 1982;206:131–145. doi: 10.1002/cne.902060204. [DOI] [PubMed] [Google Scholar]
- Fry AM, Mayor T, Meraldi P, Stierhof YD, Tanaka K, Nigg EA. C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase Nek2. The Journal of Cell Biology. 1998;141:1563–1574. doi: 10.1083/jcb.141.7.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nature reviews Genetics. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottardo M, Callaini G, Riparbelli MG. The Drosophila centriole – conversion of doublets into triplets within the stem cell niche. Journal of Cell Science. 2015;128:2437–2442. doi: 10.1242/jcs.172627. [DOI] [PubMed] [Google Scholar]
- Graser S, Stierhof Y-D, Lavoie SB, Gassner OS, Lamla S, Le Clech M, Nigg EA. Cep164, a novel centriole appendage protein required for primary cilium formation. The Journal of Cell Biology. 2007;179:321–330. doi: 10.1083/jcb.200707181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarguaglini G, Duncan PI, Stierhof YD, Holmström T, Duensing S, Nigg EA. The forkhead-associated domain protein Cep170 interacts with Polo-like kinase 1 and serves as a marker for mature centrioles. Molecular Biology of the Cell. 2005;16:1095–1107. doi: 10.1091/mbc.E04-10-0939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan IM, Palazzo RE. Warming up at the poles. Workshop on Centrosomes and Spindle Pole Bodies. 2006;7:364–371. doi: 10.1038/sj.embor.7400660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehnly H, Chen C-T, Powers Christine M, Liu H-L, Doxsey S. The Centrosome Regulates the Rab11- Dependent Recycling Endosome Pathway at Appendages of the Mother Centriole. Current Biology. 2012;22:1944–1950. doi: 10.1016/j.cub.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehnly H, Hung H-F, Doxsey S. One among many: ODF2 isoform 9, a.k.a. Cenexin-1, is required for ciliogenesis. Cell cycle (Georgetown, Tex) 2013;12:1021-1021. doi: 10.4161/cc.24330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges ME, Scheumann N, Wickstead B, Langdale JA, Gull K. Reconstructing the evolutionary history of the centriole from protein components. Journal of Cell Science. 2010;123:1407–1413. doi: 10.1242/jcs.064873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung H-F, Hehnly H, Doxsey S. The Mother Centriole Appendage Protein Cenexin Modulates Lumen Formation through Spindle Orientation. Current Biology. 2016;26:793–801. doi: 10.1016/j.cub.2016.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado L, Caballero C, Gavilan MP, Cardenas J, Bornens M, Rios RM. Disconnecting the Golgi ribbon from the centrosome prevents directional cell migration and ciliogenesis. The Journal of Cell Biology. 2011;193:917–933. doi: 10.1083/jcb.201011014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibi M, Zou P, Inoko A, Shiromizu T, Matsuyama M, Hayashi Y, Enomoto M, Mori D, Hirotsune S, Kiyono T, et al. Trichoplein controls microtubule anchoring at the centrosome by binding to Odf2 and ninein. Journal of Cell Science. 2011;124:857–864. doi: 10.1242/jcs.075705. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Kubo A, Tsukita S, Tsukita S. Odf2-deficient mother centrioles lack distal/subdistal appendages and the ability to generate primary cilia. Nat Cell Biol. 2005;7:517–524. doi: 10.1038/ncb1251. [DOI] [PubMed] [Google Scholar]
- Izquierdo D, Wang W-J, Uryu K, Tsou M-Fu B. Stabilization of Cartwheel-less Centrioles for Duplication Requires CEP295-Mediated Centriole-to-Centrosome Conversion. Cell Reports. 2014;8:957–965. doi: 10.1016/j.celrep.2014.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg DR, Moritz M, Alberts BM. The centrosome and cellular organization. Annual Review of Biochemistry. 1994;63:639–674. doi: 10.1146/annurev.bi.63.070194.003231. [DOI] [PubMed] [Google Scholar]
- Kodani A, Sirerol-Piquer MSe, Seol A, Garcia-Verdugo JM, Reiter JF. Kif3a interacts with Dynactin subunit p150Glued to organize centriole subdistal appendages. The EMBO Journal. 2013;32:597–607. doi: 10.1038/emboj.2013.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhns S, Schmidt KN, Reymann J, Gilbert DF, Neuner A, Hub B, Carvalho R, Wiedemann P, Zentgraf H, Erfle H, et al. The microtubule affinity regulating kinase MARK4 promotes axoneme extension during early ciliogenesis. The Journal of Cell Biology. 2013;200:505–522. doi: 10.1083/jcb.201206013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunimoto K, Yamazaki Y, Nishida T, Shinohara K, Ishikawa H, Hasegawa T, Okanoue T, Hamada H, Noda T, Tamura A, et al. Coordinated Ciliary Beating Requires Odf2-Mediated Polarization of Basal Bodies via Basal Feet. Cell. 2012;148:189–200. doi: 10.1016/j.cell.2011.10.052. [DOI] [PubMed] [Google Scholar]
- Lu Q, Insinna C, Ott C, Stauffer J, Pintado PA, Rahajeng J, Baxa U, Walia V, Cuenca A, Hwang Y-S, et al. Early steps in primary cilium assembly require EHD1/EHD3-dependent ciliary vesicle formation. Nat Cell Biol. 2015;17:228–240. doi: 10.1038/ncb3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahjoub Moe R, Stearns T. Supernumerary Centrosomes Nucleate Extra Cilia and Compromise Primary Cilium Signaling. Current Biology. 2012;22:1628–1634. doi: 10.1016/j.cub.2012.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-Guided Human Genome Engineering via Cas9. Science (New York, NY) 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor T, Stierhof YD, Tanaka K, Fry AM, Nigg EA. The centrosomal protein C-Nap1 is required for cell cycle-regulated centrosome cohesion. The Journal of Cell Biology. 2000;151:837–846. doi: 10.1083/jcb.151.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto T, Hosoba K, Ochiai H, Royba E, Izumi H, Sakuma T, Yamamoto T, Dynlacht Brian D, Matsuura S. The Microtubule-Depolymerizing Activity of a Mitotic Kinesin Protein KIF2A Drives Primary Cilia Disassembly Coupled with Cell Proliferation. Cell Reports. 2015;10:664–673. doi: 10.1016/j.celrep.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen MM, Malik A, Piel M, Bouckson-Castaing V, Bornens M. Microtubule minus-end anchorage at centrosomal and non-centrosomal sites: the role of ninein. 2000:1–11. doi: 10.1242/jcs.113.17.3013. [DOI] [PubMed] [Google Scholar]
- Ou YY, Mack GJ, Zhang M, Rattner JB. CEP110 and ninein are located in a specific domain of the centrosome associated with centrosome maturation. Journal of Cell Science. 2002;115:1825–1835. doi: 10.1242/jcs.115.9.1825. [DOI] [PubMed] [Google Scholar]
- Paintrand M, Moudjou M, Delacroix H, Bornens M. Centrosome organization and centriole architecture: Their sensitivity to divalent cations. Journal of Structural Biology. 1992;108:107–128. doi: 10.1016/1047-8477(92)90011-x. [DOI] [PubMed] [Google Scholar]
- Panic M, Hata S, Neuner A, Schiebel E. The Centrosomal Linker and Microtubules Provide Dual Levels of Spatial Coordination of Centrosomes. PLoS genetics. 2015;11:e1005243. doi: 10.1371/journal.pgen.1005243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitaval A, Tseng Q, Bornens M, Thery M. Cell shape and contractility regulate ciliogenesis in cell cycle-arrested cells. The Journal of Cell Biology. 2010;191:303–312. doi: 10.1083/jcb.201004003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintyne NJ, Gill SR, Eckley DM, Crego CL, Compton DA, Schroer TA. Dynactin is required for microtubule anchoring at centrosomes. The Journal of Cell Biology. 1999;147:321–334. doi: 10.1083/jcb.147.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattner JB, Sciore P, Ou Y, van der Hoorn FA, Lo IK. Primary cilia in fibroblast-like type B synoviocytes lie within a cilium pit: a site of endocytosis. Histology and histopathology. 2010;25:865–875. doi: 10.14670/HH-25.865. [DOI] [PubMed] [Google Scholar]
- Schmidt KN, Kuhns S, Neuner A, Hub B, Zentgraf H, Pereira G. Cep164 mediates vesicular docking to the mother centriole during early steps of ciliogenesis. The Journal of Cell Biology. 2012;199:1083–1101. doi: 10.1083/jcb.201202126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin S. Centrioles and the formation of rudimentary cilia by fibroblasts and smooth muscle cells. The Journal of Cell Biology. 1962;15:363–377. doi: 10.1083/jcb.15.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin SP. Reconstructions of centriole formation and ciliogenesis in mammalian lungs. Journal of Cell Science. 1968;3:207–230. doi: 10.1242/jcs.3.2.207. [DOI] [PubMed] [Google Scholar]
- Soung N-K, Park J-E, Yu L-R, Lee KH, Lee J-M, Bang JK, Veenstra TD, Rhee K, Lee KS. Plk1-Dependent and -Independent Roles of an ODF2 Splice Variant, hCenexin1, at the Centrosome of Somatic Cells. Developmental Cell. 2009;16:539–550. doi: 10.1016/j.devcel.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soung NK, Kang YH, Kim K, Kamijo K, Yoon H, Seong YS, Kuo YL, Miki T, Kim SR, Kuriyama R, et al. Requirement of hCenexin for Proper Mitotic Functions of Polo-Like Kinase 1 at the Centrosomes. Molecular and Cellular Biology. 2006;26:8316–8335. doi: 10.1128/MCB.00671-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale J, Chen JK, Cooper MK, Wang B, Mann RK, Milenkovic L, Scott MP, Beachy PA. Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature. 2000;406:1005–1009. doi: 10.1038/35023008. [DOI] [PubMed] [Google Scholar]
- Tanos BE, Yang HJ, Soni R, Wang W-J, Macaluso FP, Asara JM, Tsou MFB. Centriole distal appendages promote membrane docking, leading to cilia initiation. Genes & Development. 2013;27:163–168. doi: 10.1101/gad.207043.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou MF, Stearns T. Mechanism limiting centrosome duplication to once per cell cycle. Nature. 2006;442:947–951. doi: 10.1038/nature04985. [DOI] [PubMed] [Google Scholar]
- Wang W-J, Tay HG, Soni R, Perumal GS, Goll MG, Macaluso FP, Asara JM, Amack JD, Tsou M-FB. CEP162 is an axoneme-recognition protein promoting ciliary transition zone assembly at the cilia base. Nature. 2013;15:591–601. doi: 10.1038/ncb2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westlake CJ, Baye LM, Nachury MV, Wright KJ, Ervin KE, Phu L, Chalouni C, Beck JS, Kirkpatrick DS, Slusarski DC, et al. Primary cilia membrane assembly is initiated by Rab11 and transport protein particle II (TRAPPII) complex-dependent trafficking of Rabin8 to the centrosome. Proceedings of the National Academy of Sciences. 2011;108:2759–2764. doi: 10.1073/pnas.1018823108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.