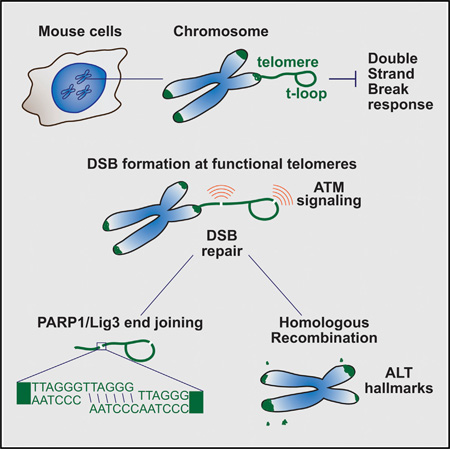
SUMMARY
Shelterin protects chromosome ends from the DNA damage response. Although the mechanism of telomere protection has been studied extensively, the fate of double-strand breaks (DSBs) inside telomeres is not known. Here, we report that telomere-internal FokI-induced DSBs activate ATM kinase-dependent signaling in S-phase but are well tolerated and repaired efficiently. Homologous recombination contributes to repair, leading to increased telomere length heterogeneity typical of the alternative lengthening of telomeres (ALT) pathway. Furthermore, cells accumulate extra chromosomal telomeric signals (ECTS), a second hallmark of ALT. Telomere-internal DSBs are also repaired by a PARP1- and Ligase3-dependent reaction, suggesting alternative non-homologous end- joining (alt-NHEJ), which relies on microhomology at DSBs. However, as resected telomere-internal DSBs have perfect homology, their PARP1/Lig3-dependent end-joining may be more akin to single strand break repair. We conclude that shelterin does not repress ATM kinase signaling or DSB repair at telomere-internal sites, thereby allowing DNA repair to maintain telomere integrity.
Graphical Abstract
INTRODUCTION
Telomeres protect chromosome ends from the DNA damage response (DDR). Mammalian telomeres comprise several kilobases of TTAGGG repeats, bound by the six-subunit shelterin complex (Palm and de Lange, 2008). Shelterin represses a multitude of DNA damage response pathways, and distinct shelterin subunits are dedicated to different signaling and repair pathways. For instance, when telomeres are deprived of TRF2, a double-stranded telomeric DNA binding protein in shelterin, ATM-dependent signaling is activated and telomeres fuse due to Lig4-dependent non-homologous end-joining (NHEJ) (van Steensel et al., 1998; Karlseder et al., 1999; Smogorzewska et al., 2002; Celli and de Lange, 2005; Denchi and de Lange, 2007). In contrast, when one of the single-stranded DNA binding proteins in shelterin, POT1a, is removed, the ATR signaling pathway is activated at telomeres, but ATM signaling remains repressed. Repression of homologous recombination between telomeres, giving rise to telomere sister chromatid exchanges (T-SCEs), requires both Rap1 and the presence of either of the two POT1 proteins at telomeres. A third type of DSB repair, the PARP1- and Lig3-dependent alternative lengthening of telomeres (alt)-NHEJ pathway, is repressed in a partially redundant fashion by TRF2 and another shelterin protein. Both homologous recombination (HR) and alt-NHEJ are only prominent when shelterin is compromised in Ku70/80-deficient cells, consistent with the ability of the Ku70/80 heterodimer to compete with these repair pathways at genome-wide DSBs.
Despite this wealth of information on how shelterin represses the DDR at chromosome ends, it is unclear to what extent the presence of shelterin on the telomeric DNA affects the repair of DNA damage occurring within telomeres. Were shelterin to repress HR, alt-NHEJ, and c-NHEJ at telomere-internal positions, the telomeric DNA would be more vulnerable to DNA damaging agents than other parts of the genome. Indeed, it was inferred that DNA damage in telomeric DNA persists longer than at other sites (Fumagalli et al., 2012), and diminished repair was reported for DSBs in subtelomeric regions (Miller et al., 2011). The only setting in which the repair of telomere internal DSBs has been monitored directly involves studies in cells that use the ALT pathway, which bear telomeres with altered function (Cho et al., 2014).
In order to gain insight into the response to damage within the telomeric repeat array, we introduced DSBs with the FokI endonuclease fused to the shelterin protein TRF1 (Cho et al., 2014; Tang et al., 2013). The telomere-internal DSBs activated the ATM kinase-dependent signaling pathway and were repaired through a PARP1- and Lig3-dependent joining reaction as well as by HR. These data indicate that the ability of shelterin to repress ATM signaling, alt-NHEJ, and HR at chromosome ends is not apparent at telomere-internal DSBs.
RESULTS
Activation of the DDR Response at Telomeric DSBs
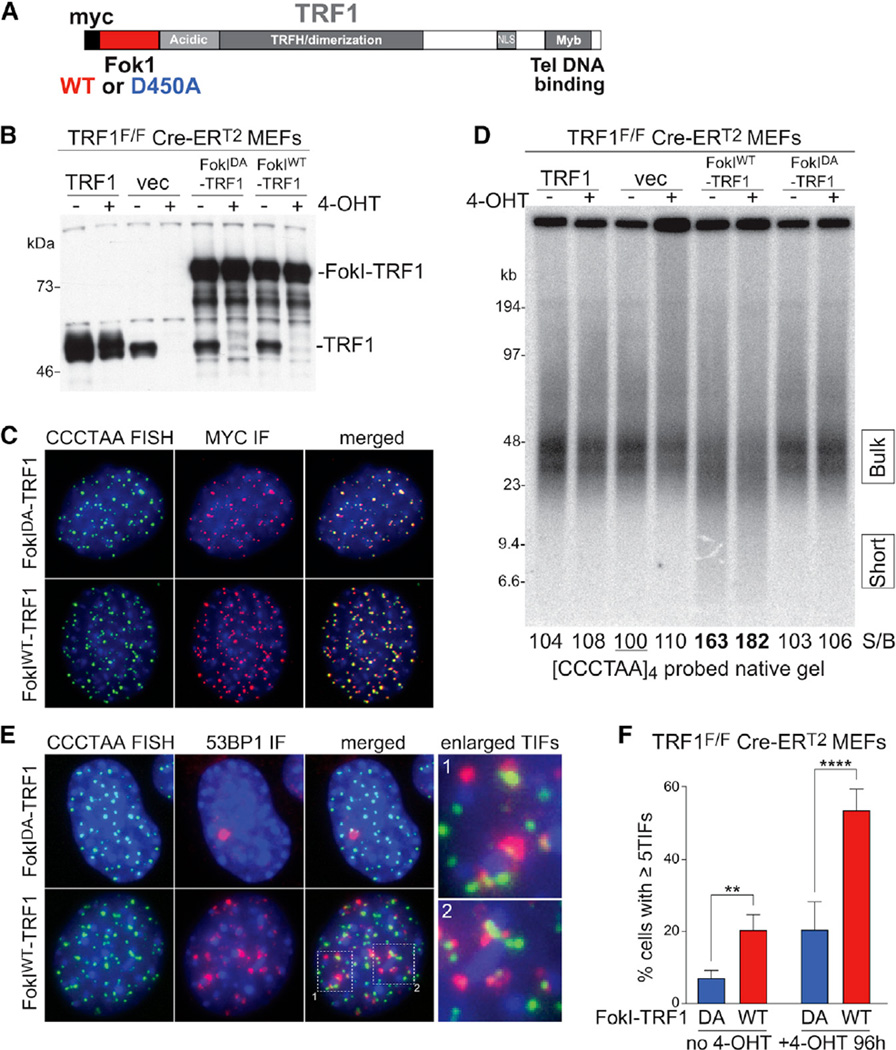
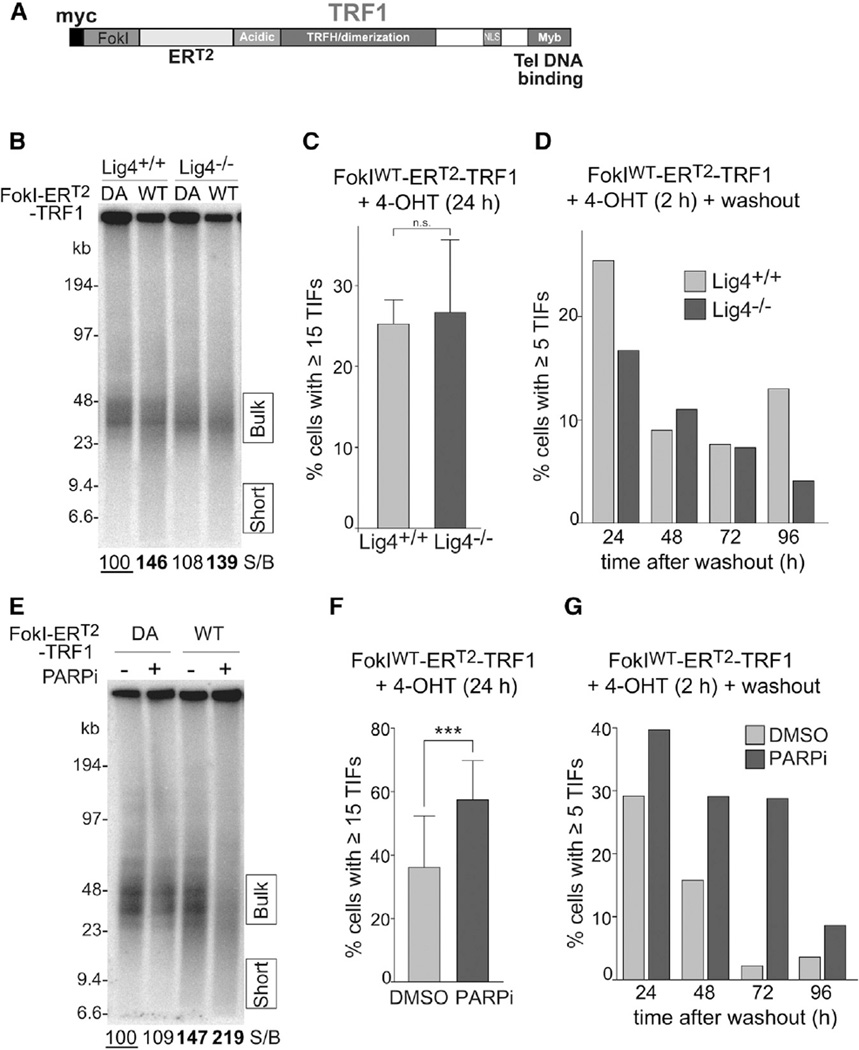
To generate DSBs within the telomeric repeat array at mouse chromosome ends, we fused the nuclease domain of FokI to the N terminus of the shelterin component TRF1 (FokIWT-TRF1) (Figure 1A), analogous to a strategy previously used to cut human telomeres (Tang et al., 2013; Cho et al., 2014). FokIWT-TRF1 or a nuclease-dead control allele (FokIDA-TRF1) was introduced by retroviral infection in SV40LT-immortalized TRF1F/F Rs-Cre-ERT2 mouse embryonic fibroblast (MEFs), where deletion of the endogenous TRF1 can be induced with 4-OHTamoxifen (4-OHT) (Figure 1B). The fusion proteins were overexpressed ~4- to 5-fold compared to endogenous TRF1 (Figures 1B, S1A, and S1B) and localized to telomeres (Figure 1C). The FokIDA-TRF1 version could fully complement the telomere replication defects associated with the loss of TRF1 (Sfeir et al., 2009; Martínez et al., 2009; Zimmermann et al., 2014) (Figures S1D–S1F). PFGE analysis of telomeric restriction fragments showed that FokIWT-TRF1-expressing cells contained shorter telomeric fragments, indicative of DSB formation in the telomeres, whereas the profile of the nuclease-dead control, FokIDA-TRF1, was indistinguishable from that of cells infected with the empty vector (Figures 1D and S1G).
Figure 1. Telomere-Internal DSB Formation and DDR Signaling.
(A) Schematic of the FokI-TRF1 fusion proteins.
(B) Western blot analysis of FokI-TRF1 expression. The indicated constructs were introduced by retroviral infection in SV40-LT immortalized TRF1F/F Rs-Cre-ERT2 MEFs. After selection, 4-OHT was added to induce Cre-mediated deletion of the endogenous TRF1 gene. Samples were taken 96 hr after 4-OHT.
(C) Immunofluorescence (IF)-FISH analysis of cells as in (B). The FokI-TRF1 alleles were detected with MYC IF and telomeres with FISH using a [TTAGGG]3 probe.
(D) PFGE analysis of mouse telomeric DNA. Genomic DNA from cells as in (B) was digested with MboI, telomeric fragments were separated in PFGE, and hybridized under native conditions with a 32P-[CCCTAA]4 probe that recognizes the 3′ overhang. The numbers at the bottom indicate the ratio between short versus bulk telomere signal, quantified as the amount of signal in the areas indicated by the boxes to the right of the gel. The values are reported relative to the vector samples not treated with 4-OHT, which was set at 100.
(E) IF-FISH analysis on cells as in (B). The 53BP1 was detected by IF and telomeres by FISH with a [TTAGGG]3 probe.
(F) Quantification of TIF-positive cells as in (B). The bars represent means with SDs from three independent experiments (~100 cells each) for no 4-OHT samples and six independent experiments (~100 cells each) for +4-OHT samples. p value from two-tailed paired t test.
FokIWT-TRF1 expression resulted in telomere dysfunction induced foci (TIFs; Takai et al., 2003), detected based on the co-localization of the DDR marker 53BP1 with fluorescence in situ hybridization (FISH)-labeled telomeres (Figures 1E and 1F). The TIF data are consistent with previous observations showing γH2AX and 53BP1 at FokI-cut human telomeres (Tang et al., 2013; Cho et al., 2014). TIFs were also induced in the presence of the endogenous TRF1 (Figure 1F), although the lower level of the TIF response suggested that the endogenous TRF1 competes with the FokIWT-TRF1 fusion protein. However, most telomeres do not contain 53BP1 foci, suggesting that FokI-induced DSBs are not present in all telomeres. This finding is consistent with the persistence of full-length telomeric restriction fragments after FokIWT-TRF1 expression (Figures 1D and S1G).
Induction of DSBs within the mouse telomeres did not lead to an obvious growth defect. When monitored over 20 days, the culture expressing FokIWT-TRF1 proliferated at the same rate as cells expressing the nuclease-dead control (Figure S1C).
Telomeric DSBs Activate the DDR in S-phase
Unexpectedly, EdU labeling showed that around 80%–90% of the TIF-positive cells were in S-phase (Figure S1H). However, FokI-TRF1 is present at telomeres in >70% of the cells, indicating no preferential accumulation in S-phase. Attempts to induce DSBs at telomeres in non-cycling cells were thwarted by the strongly reduced expression of the fusion protein in G0 (Figure S1I). One explanation for the S-phase specific occurrence of TIFs is the possibility that FokI-induced breaks are held together by shelterin positioned on both sides of the DSB, tethering the ends and denying access to DDR factors. If replication fork progression disrupts shelterin binding, the ends would become exposed and TIFs would preferentially occur in S-phase.
DSBs at the Telomeric DNA Activate ATM Kinase Signaling
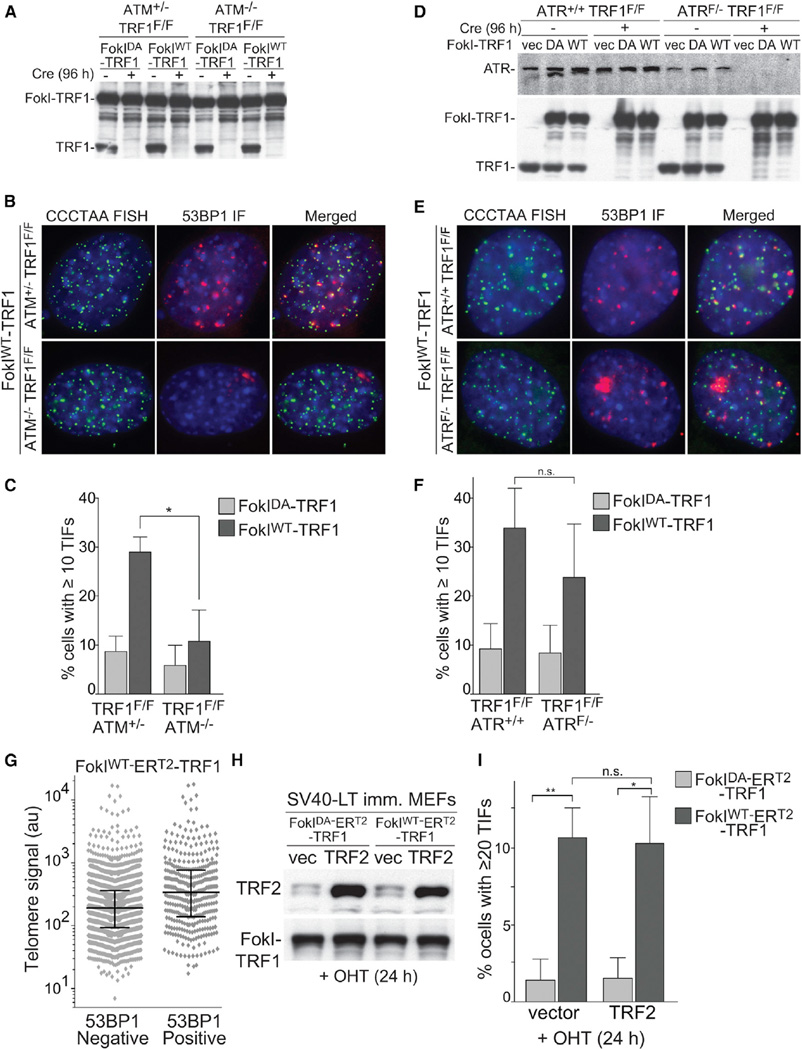
As 53BP1 foci could be due to activation of either ATM or ATR kinase signaling, we analyzed the effect of the absence of ATM or ATR on the FokIWT-TRF1-induced TIFs (Figure 2). The accumulation of shorter telomeric fragments was not affected by deficiency in either of the DDR kinases (Figures S2A and S2B), indicating similar DSB induction. However, the TIF response was strongly diminished in the absence of ATM (Figures 2A–2C), pointing to an ATM-dependent DDR in this context. EdU labeling showed that the effect of ATM deficiency was not due to a change in the S-phase index (Figures S3A and S3B). The ATM-dependence of the TIF response was also confirmed in the presence of the endogenous TRF1 as well as with an ATM inhibitor (Figures S3C and S3D). In contrast to ATM deficiency, absence of ATR appeared to have a modest effect on the appearance of TIFs, although the difference was not statistically significant (Figures 2D–2F).
Figure 2. Telomere-Internal DSBs Induce ATM Kinase Signaling.
(A) Western blot analysis of FokI-TRF1 fusion proteins in the indicated cell lines. After retroviral infection and selection, cells were infected with Cre to induce deletion of the endogenous TRF1 and samples were analyzed 96 hr after Cre.
(B) IF-FISH analysis to detect TIFs in cells treated as in (A). The 53BP1 was detected by IF and telomeres by PNA-FISH with a [TTAGGG]3 probe.
(C) Quantification of TIF-positive cells from three independent experiments as in (A). The bars represent means with SDs (~100 cells per experiment). p value from two-tailed paired t test.
(D) Western blot analysis of FokI-TRF1 and ATR, analogous to (A) except that Cre treatment induced deletion of both ATR (top) and TRF1 (bottom).
(E) IF-FISH analysis to detect TIFs in cells shown in (D). The 53BP1 was detected by IF and telomeres by PNA-FISH with a [TTAGGG]3 probe.
(F) Quantification of TIF-positive cells from three independent experiments as shown in (D). The bars represent means with SDs (~100 cells per sample). p value from two-tailed paired t test.
(G) Telomere signal intensity distributions. FokI-ERT2-TRF1 was introduced into SV40-LT immortalized MEFs by retroviral infection. After selection, 4-OHT was added to induce FokI-ERT2-TRF1 and samples were analyzed after 24 hr. The 53BP1 was detected by IF and telomeres by FISH with a [TTAGGG]3 probe. Telomeric signal intensities were determined in FIJI/ImageJ. The background was subtracted using a rolling ball algorithm with r = 10 pixel. A circular selection was created for each telomeric signal and the integrated density was measured. To correct for size differences in the selected telomeres, a background value was obtained by measuring the integrated density in 4–5 telomere-free areas, multiplied for the area of each selection, and subtracted to the corresponding integrated density value. There were 1,986 telomeres without 53BP1 signals and 360 telomeres with a 53BP1 signal that were counted in 20 cells with ≥5 TIFs each. The corresponding values are reported as a scatter dot plot with bars representing median with interquartile range.
(H) Western blot for TRF2 overexpression. The TRF2 (or empty vector) and FokI-ERT2-TRF1 retroviruses were introduced into SV40-LT immortalized MEFs. After selection for the second retrovirus, 4-OHT was added to induce FokI-ERT2-TRF1 and samples were analyzed after 24 hr.
(I) Quantification of TIF-positive cells from the experiment described in (H). The bars represent mean with SD from three experimental replicates. p values from two-tailed unpaired t test.
We considered the possibility that the TIF response only occurred at those telomeres rendered critically short by the nuclease. To address this possibility, we compared fluorescence signal intensities of telomeres that co-localized with 53BP1 versus those that did not contain 53BP1. The 53BP1-positive telomeres had a median TTAGGG FISH signal ~70% greater than 53BP1-negative telomeres (Figure 2G), arguing against the possibility that ATM signaling is only activated at telomeres rendered critically short by the FokI-induced breaks. The greater signal intensity of the 53BP1-positive telomeres is consistent with the TIF response primarily occurring at telomeres that are in the process of being replicated.
Since TRF2 is required to repress ATM signaling at telomeres, we considered the possibility that TIFs are due to limiting levels of TRF2 and/or its displacement from the TTAGGG repeats. However, the TIF response was not diminished when TRF2 was grossly overexpressed (Figures 2H and 2I). Therefore, the results indicate that the presence of TRF2 on the telomeric DNA is not sufficient to block the activation of ATM signaling at a telomere-internal DSB.
Telomere-Internal DSBs Are Processed by HR
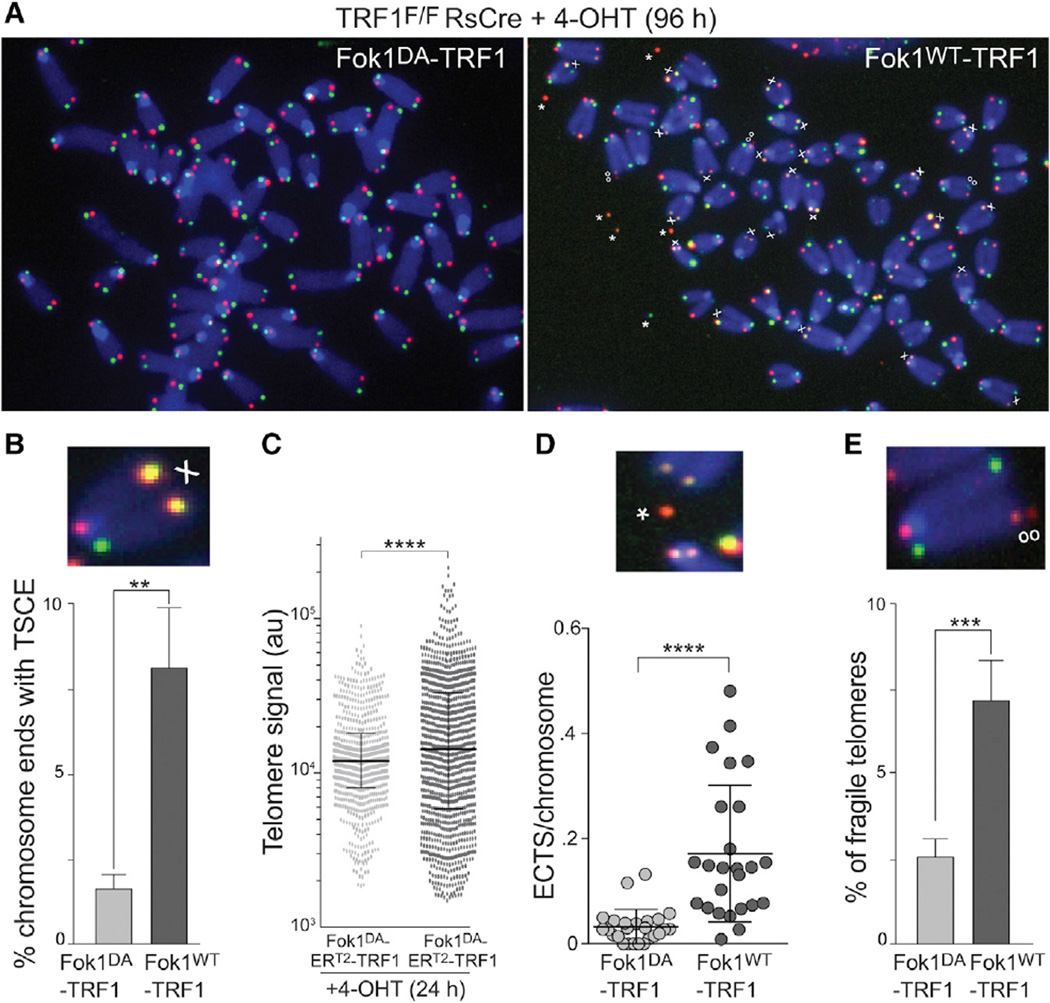
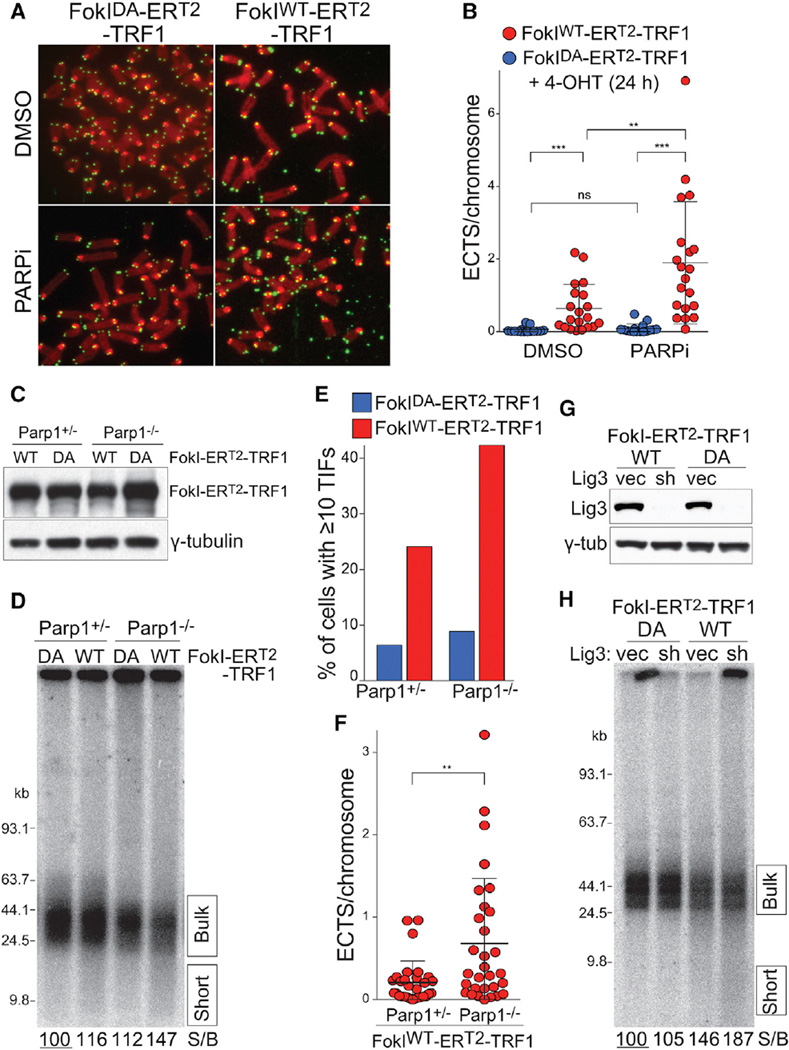
HR at telomeres leads to exchanges between T-SCEs that can be visualized by chromosome orientation (CO)-FISH (Bailey et al., 1996). HR-induced T-SCEs are prominent at telomeres upon deletion of either POT1a/b or Rap1 in a Ku-deficient setting. However, FokIWT-TRF1 expression alone induced T-SCEs in Ku70/80-proficient cells (Figures 3A and 3B). It is likely that preferential detection of the FokI-induced DSBs in S-phase cells contributes to this prevalence of HR. Consistent with frequent HR-mediated repair, the cells expressing FokIWT-TRF1 showed a considerable increase in the heterogeneity of telomeric signals (Figures 3A and 3C). Interestingly, DSB induction was also associated with the accumulation of extra chromosomal telomeric signals (ECTS) in metaphase spreads (Figures 3A and 3D). Since T-SCEs, telomere length heterogeneity, and ECTS are hallmarks of ALT cells, the results indicate that formation of DSBs in the telomeric DNA is sufficient to unleash recombination and ALT-like phenotypes in otherwise wild-type MEFs.
Figure 3. Telomere-Internal DSBs Are Processed by HR.
(A) CO-FISH on metaphase spreads with and without FokI induced DSBs. The experimental procedure is as in Figure 1B. The leading strand telomeres were labeled by PNA-FISH with a Cy3-[TTAGGG]3 probe (red) and lagging strand telomeres with a FITC-[CCCTAA]3 probe (green). The examples from FokIWT-TRF1 or FokIDA-TRF1 expressing cells are shown. X, T-SCEs; oo, fragile telomeres; *, ECTS.
(B) Quantification of chromosome ends with T-SCEs detected by CO-FISH. The bars represent mean with SD from five independent experiments, analogous to the one described in Figure 1B, except that the endogenous TRF1 was not deleted. p value from two-tailed paired t test.
(C) Telomere length heterogeneity measured using quantitative FISH analysis on metaphase spreads. FokI-ERT2-TRF1 constructs were introduced into SV40-LT immortalized MEFs by retroviral infection and selection. The samples were analyzed 24 hr after 4-OHT. The telomere signal intensity was measured as in Figure 2D. The ten metaphases, ~800 telomeres, were analyzed per sample, and the values are reported as a scatter dot plot with bars representing median with interquartile range. p value from F-test of variance.
(D) Quantification of ECTS from the experiment described in (A). There were 25 metaphases per sample that were analyzed. p value from two-tailed unpaired t test.
(E) Quantification of fragile telomeres. The fragile telomeres were scored in four independent experiments as described in (C). The bars represent means with SDs. p value from two-tailed paired t test.
Metaphase spreads from cells expressing FokIWT-TRF1 also showed a significant increase in telomere fragility (Figures 3A and 3E). The cause of this fragility is not clear. It is unlikely to be due to displacement of the endogenous TRF1 since the FokIDA-TRF1 fusion fully represses the fragile telomere phenotype associated with TRF1 loss, indicating that the fusion proteins are competent in terms of this TRF1 function. One possibility is that the fragility observed in metaphase spreads reflects DNA condensation problems of those telomeres engaged in post-replicative HR repair of the FokI-induced DSBs.
Telomere-Internal DSBs Are Not Processed by c-NHEJ
We examined the effect of ablating c-NHEJ on the generation of smaller telomeric DNA fragments and TIFs. These experiments employed a conditional FokI-ERT2-TRF1 version that includes a 4-OHT controlled estrogen receptor domain (Figure 4A). Induction of FokIWT-ERT2-TRF1 in wild-type and c-NHEJ-deficient Lig4−/− MEFs revealed no difference in the accumulation of shorter telomeric fragments detected by PFGE analysis and did not reveal an altered TIF response (Figures 4B and 4C). Transient (2 hr) induction of FokIWT-ERT2-TRF1 followed by removal of 4-OHT showed that in c-NHEJ proficient cells, the TIFs persisted for ~24 hr and then returned to preinduction levels over 48 hr (Figure S4A). This dissipation rate is roughly consistent with the half-life TRF1 (Sfeir et al., 2009). Importantly, Lig4−/− MEFs showed the same rate of dissipation of the FokI-induced TIFs as Lig4-proficient cells (Figure 4D). In a control experiment, the Lig4−/− MEFs showed the expected delay in the clearance of genome-wide DDR foci induced by IR (Figure S4B). These results were not affected by differences in the levels of FokIWT-ERT2-TRF1 expression or changes in the S-phase index (Figures S4C–S4E). Therefore, in this setting, the Lig4-dependent c-NHEJ pathway does not contribute to the repair of DSBs within the telomeric repeats.
Figure 4. Telomere-Internal DSBs Are Repaired by alt-NHEJ.
(A) Schematic of the conditional FokI-ERT2-TRF1 fusion proteins.
(B) PFGE analysis of telomeric DNA as in Figure 1D. The FokI-ERT2-TRF1 constructs were introduced into SV40-LT-immortalized TRF2F/+ Lig4+/+ and TRF2F/FLig4−/− MEFs by retroviral infection. After selection, 4-OHT was added to induce FokI-ERT2-TRF1 and samples were analyzed after 24 hr. The values below the lanes are reported relative to the Lig4+/+ cells with FokIDA-ERT2-TRF1, which was set to 100.
(C) Quantification of TIF-positive cells from three independent experiments, analogous to the one described in (A). The 53BP1-telomere co-localization counting was automated with a Fiji/ImageJ macro described in the Experimental Procedures. The bars represent means with SDs from three independent experiments (~100 cells each).
(D) Time course of TIF clearance after transient FokIWT-ERT2-TRF1 induction. The cells, infected with FokIWT-ERT2-TRF1, were treated with 4-OHT for 2 hr, washed with PBS, and fresh media without 4-OHT was added. The samples were collected at the indicated time points and TIFs were quantified using IF-FISH for 53BP1 and telomeres.
(E) PFGE analysis of telomeric DNA. The FokI-ERT2-TRF1 constructs were introduced into SV40-LT-immortalized MEFs by retroviral infection and 4-OHT was added together with 2 µM Olaparib. The samples were collected 18 hr after 4-OHT and analyzed by PFGE as described in (A).
(F) Quantification of TIF-positive cells from five independent experiments (~100 cells each) as described in (D). The bars represent means with SDs. p value from two-tailed paired t test.
(G) Time course of TIF clearance after transient FokIWT-ERT2-TRF1 induction. The SV40-LT-immortalized MEFs expressing FokIWT-ERT2-TRF1 were treated with 4-OHT and 2 µM Olaparib for 2 hr followed by washout as in (C) with or without 2 µM Olaparib in the media. The samples were collected at the indicated time points, and TIFs were quantified using IF-FISH for 53BP1 and telomeres.
Telomere-Internal DSBs Are Repaired by a PARP1/Lig3-Dependent Pathway
The alt-NHEJ pathway relies on the poly (ADP-ribose) polymerase 1 PARP1 and microhomology to repair DSBs using either Ligase 1 or 3 (reviewed in Frit et al., 2014). After 5′ end resection, a DSB in the telomeric DNA is expected to have perfect homology of the two 3′ protrusions (the G- and C-telomeric repeat strands), providing an excellent substrate for alt-NHEJ. In the presence of the PARP1 inhibitor Olaparib, FokIWT-ERT2-TRF1 expression resulted in a substantially greater accumulation of shortened telomeric DNA fragments (Figure 4E) and there was a significant increase in the TIF response (Figures 4F and S4F). Upon removal of 4-OHT, the TIF response persisted much longer, corroborating the contribution of alt-NHEJ to the repair of the FokI-induced DSBs (Figure 4G). The effect of PARP inhibition was also apparent in metaphase spreads, which showed an increase in ECTS (Figures 5A and 5B). The dependence of the DSB repair on PARP1 was verified using Parp1−/− MEFs, which showed an increase in shortened telomeric fragments and a greater TIF response upon induction of FokIWT-ERT2-TRF1 (Figures 5C–5F). The effect of PARP1 deficiency is not as striking as the effect of Olaparib. This finding is consistent with the view that absence of PARP1 is less of an impediment to repair than the inability of PARP1 to dislodge from a DNA end due to inhibition of its activity (Helleday, 2011; Murai et al., 2014). Consistent with the involvement of PARP1 in the repair of the DSBs, short hairpin (sh)RNA-mediated knockdown of DNA Ligase3 showed an increase in shorter telomeric DNA fragments upon induction of the DSBs (Figures 5G and 5H). The effect of Lig3 knockdown on repair was modest compared to that of PARP1 inhibition, likely reflecting the redundant roles of Lig3 and Lig1 in alt-NHEJ (Paul et al., 2013; Masani et al., 2016). Taken together, these results indicate that a pathway with similarities to alternative NHEJ is a major mechanism by which S-phase DSBs in the telomeric repeat array are repaired.
Figure 5. PARP1 and Ligase3 Contribute to the Repair of Telomere-Internal DSBs.
(A) ECTS accumulation after PARP inhibition. The FokI-ERT2-TRF1 constructs were introduced into SV40LT-immortalized MEFs and 4-OHT and Olaparib (2 µM) were added for 24 hr. The representative metaphase spreads are shown. Telomere-FISH is in green, and DAPI is false-colored in red.
(B) Quantification of ECTS detected as in (A). There were 20–25 metaphases per sample that were analyzed. p value from two-tailed unpaired t test.
(C) Western blot analysis FokI-ERT2-TRF1 expression in PARP1+/− and PARP1−/− MEFs. After the selection of retrovirally infected MEFs, 4-OHT was added and the samples were analyzed after 24 hr.
(D) PFGE analysis of telomeric DNA from the experiment described in (C)
(E) Quantification of TIF-positive cells from the experiment described in (C).
(F) Quantification of extra chromosomal telomeric signals from the experiment described in (C). There were 20–25 metaphases per each sample that were analyzed. p value from two-tailed unpaired t test.
(G) Western blot to detect Lig3 knockdown with shRNA. The FokI-ERT2-TRF1 constructs were introduced into SV40-LT-immortalized MEFs. After selection, the cells were infected three times at 12 hr intervals with the Lig3 shRNA lentivirus and selected for the shRNA construct. The samples were analyzed 24 hr after 4-OHT induction.
(H) PFGE analysis of telomeric DNA from the experiment described in (G).
DISCUSSION
Telomeric DSBs Induce Homologous Recombination and an ALT-like Phenotype
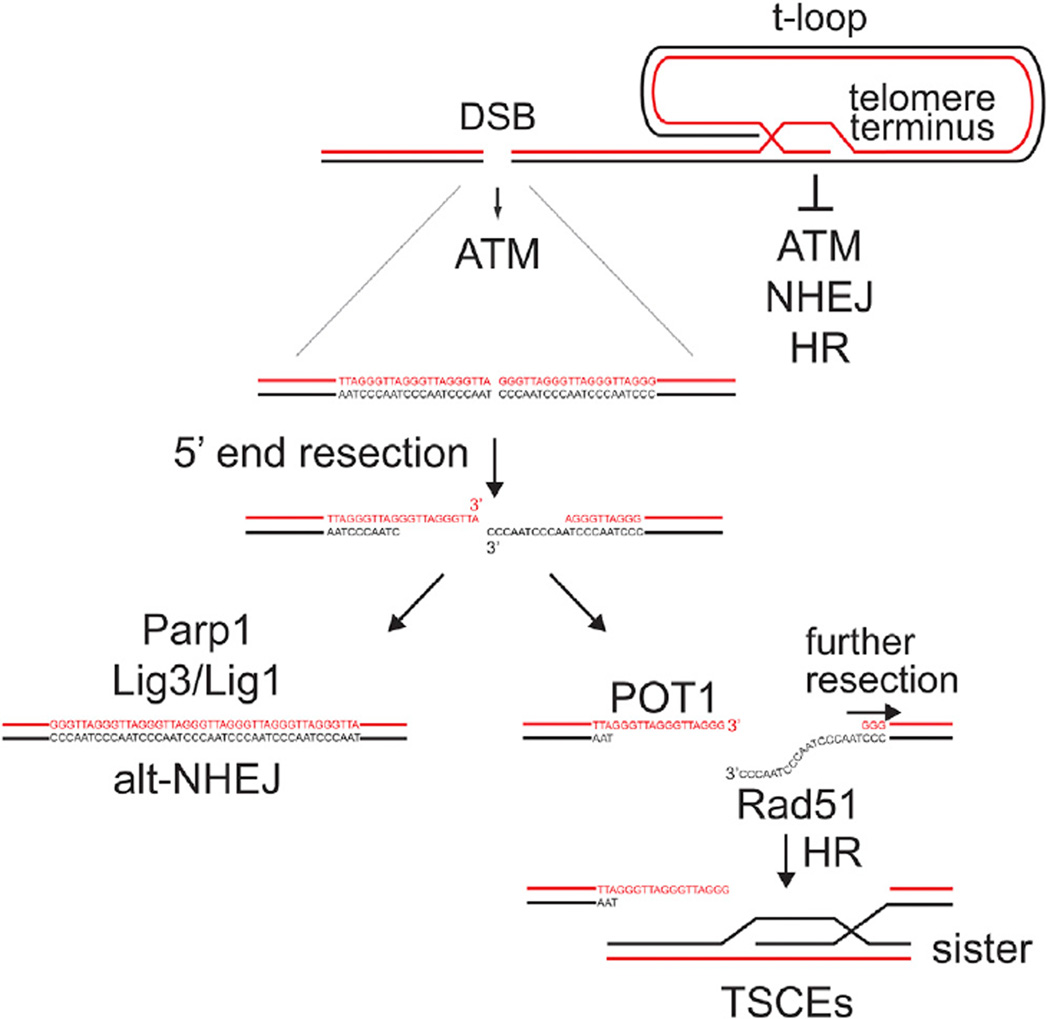
The DSBs within the telomeric repeats induce telomere recombination in the presence of a functional shelterin complex. These data suggest that recombination between sister telomeres is not repressed throughout the telomeric DNA. At functional telomeres, repression of recombination requires both Rap1 and one of the two POT1 proteins (POT1a or POT1b), which bind to the ss TTAGGG overhang (Palm et al., 2009; Sfeir et al., 2010). In the setting of the telomere-internal DSBs, one end could be protected by POT1a or POT1b since resection would expose their bindings sites in the G-rich 3′ overhang (Lei et al., 2004; Loayza et al., 2004). However, the other end of the break will carry the C-rich telomeric repeats after resection. Since CCCTAA repeats do not bind POT1 (Baumann and Cech, 2001), they could become a substrate for Rad51 and invade the sister telomere, leading to T-SCEs (Figure 6).
Figure 6. DDR Pathways at Natural Chromosome Ends versus Telomere-Internal DSB.
Schematic representation of the DDR pathways responding to DSBs inside the telomeric repeats and possible outcomes of HR and alt-NHEJ activities at these breaks. The same pathways are repressed at the natural ends of chromosomes. The telomere-internal FokI-induced DSB activate ATM kinase signaling and processed by 5′ end resection, resulting in one end that has a G-strand overhang and thus resembles the natural telomere terminus. This end can load POT1a/b and would therefore be protected from further resection. The other end has the C-rich telomeric strand exposed and hence is not protected from further resection. Annealing of the two ends will lead to alt-NHEJ. Because of lack of POT1 binding, the C-strand overhang becomes a substrate for Rad51-mediated strand-invasion into the sister telomere, initiating HR.
In 10%–15% of cancers, the HR-based ALT pathway ensures telomere maintenance in the absence of telomerase (Henson and Reddel, 2010). ALT is thought to require de-regulation of recombination at telomeres, but the molecular basis is poorly understood (Henson et al., 2002; Draskovic and Londono Vallejo, 2013). Induction of DSBs at telomeres has been shown to stimulate interchromosomal recombination and telomere clustering in ALT cells, but not in telomerase-positive cells (Cho et al., 2014). Here, we show that induction of telomeric DSBs in telomerase-positive cells is sufficient to induce telomere recombination, telomere length heterogeneity, and extra chromosomal telomeric DNA, all hallmarks of ALT. These data suggest that telomeric damage may be a driver of ALT as suggested previously (Doksani and de Lange, 2014). While identification of a single pathway to ALT has proven difficult, it is possible that there are many different pathways that can drive ALT via a common mechanism: induction of telomere damage. Indeed, ALT telomeres have frequent nicks and gaps (Nabetani and Ishikawa, 2009) and conditions that may induce telomeric damage have been associated with the onset of ALT (O’Sullivan et al., 2014; Hu et al., 2012). Based on this consideration, it is possible that accumulation of telomeric damage promotes telomere maintenance in the absence of telomerase reactivation in human cancers.
Control of Non-homologous End-Joining Pathways at Telomeres
We did not detect a contribution of the Lig4-dependent c-NHEJ pathway in the repair of telomeric DSBs. Since these DSBs will transiently generate telomere ends that are not engaged in t-loops, this might suggest the existence of a TRF2-dependent, but t-loop-independent, mechanism of repression of c-NHEJ at telomeres (Ribes-Zamora et al., 2013; Fumagalli et al., 2012; Bae and Baumann, 2007; Cesare et al., 2009; Benarroch-Popivker et al., 2016). However, considering that FokI-induced DSBs are preferentially expressed S-phase cells, this result could also be attributed to the prevalence of 5′ end resection that promotes alt-NHEJ and HR over c-NHEJ. Future experiments aimed to address the mechanisms that control c-NHEJ at telomeres will require a system that allows induction of telomeric DSBs specifically in G1.
We found a substantial contribution of the PARP1-dependent alt-NHEJ pathway in the repair of telomeric DSBs. This result distinguishes telomeres from the rest of the genome, where the alt-NHEJ repair pathway becomes relevant only in c-NHEJ-defective backgrounds (Iliakis et al., 2015; Chiruvella et al., 2013). This prevalence of the alt-NHEJ pathway cannot be attributed to the 4-nt 5′ overhangs generated by the FokI nuclease because repair of DSBs with 5′ overhangs has shown a similar dependency on c-NHEJ as do DSB with 3′ overhangs (Budman and Chu, 2005; Liang et al., 2016). Moreover, at least part of the FokI-induced breaks must be resected to generate 3′ overhangs that initiate the observed HR. Given that telomeres are made of short tandem repeats of a 6 nt motif, every end-joining reaction of minimally resected telomeric DSBs will be guided by perfect homology of the broken ends (which technically challenges the definition of non-homologous end-joining inside the telomeric repeats). In the presence of telomeric DSBs, the microhomology-directed repair would impose a head to tail orientation in the end-joining of broken telomeric fragments that favors correct repair over generation of the head to head telomere-telomere fusions, potentially resulting from c-NHEJ.
Relevance to Telomere Protection by TRF2
TRF2 has been reported to have a number of features that could account for its role in end protection. TRF2 is required for formation/maintenance of t-loops, which have been proposed to sequester the chromosome end via strand invasion of the 3′ telomeric overhang into the duplex telomeric repeats, thus making the telomere terminus inaccessible to the initiating factors of ATM signaling and classical c-NHEJ, MRN, and Ku70/80, respectively (Griffith et al., 1999; Doksani et al., 2013). An alternative explanation for the repression of ATM signaling was based on the finding that TRF2 interacts with ATM and can inhibit ATM activation by IR-induced DSBs when it is overexpressed (Karlseder et al., 2004). In addition, the iDDR domain of TRF2 (when placed in TRF1) has been shown to block the propagation of ATM signaling by inhibiting the RNF168 ubiquitination cascade (Okamoto et al., 2013). Finally, it has been proposed that TRF2 and other shelterin proteins enforce compaction of the telomeric chromatin as a mechanism to exclude DNA damage response factors from entering the telomeric domain (Bandaria et al., 2016).
These models for telomere protection by TRF2 fall into two broad categories: one that depends on the formation of t-loop structures, where DDR inhibition by TRF2 only occurs at the physiological ends of chromosomes; and a second category of models in which TRF2 directly inhibits the DDR response throughout the telomeric domain. While it is possible that TRF2 has multiple mechanisms for end protection, the lack of inhibition of ATM signaling at a telomere-internal DSB argues against models in which TRF2 directly inhibits ATM throughout the telomeric chromatin.
EXPERIMENTAL PROCEDURES
Sequences and Cloning
The FokI nuclease domain was amplified from a TALEN plasmid (pTAL015349) with the following oligos: BglII-FokIND-Fw: GGCCAGATCTCAGCTGGTGAAGTCCGAGCTG and BamHI-FokIND-Rev: GGCCGGATCCGTCGGCCGCGAAGTTGATCTC. The PCR product was digested with BamHI/BglII and inserted into the BamHI-linearized pLPC-NMYC-mTRF1 (Addgene #64162). The FokID450A-TRF1 nuclease-dead control was produced by site-directed mutagenesis using the oligos: FokID450A-Fw: CAGGAAGCCCGCCGGCGC CATCTAC and FokID450A-Rev: GAGCCGCCCAGGTGCTTGCCCCTGT. The ERT2 domain was amplified from pCAG-ERT2-Cre-ERT2 (Addgene #13777), with: BamHI-ERT2-Fw: GGCCGGATCCGCTGGAGACATGAGAGCTGCC and BamHI-ERT2-Rev: GGCCGGATCCGTCGACAGCTGTGGCAGGGAA. The PCR product was digested with BamHI and inserted into the BamHI-linearized FokIWT-TRF1 or FokID450A-TRF1 plasmids.
Cell Culture, Viral Infections, and Inhibitors
TRF1F/F, TRF1F/F ATRF/−, TRF1F/F ATM+/−, and TRF1F/F ATM−/− MEFs were described previously (Sfeir et al., 2009). TRF2F/+Lig4−/− and TRF2F/+Lig4+/+ were obtained by standard mouse crosses. Primary MEFs isolated from embryonic day (E) 12.5 or E13.5 embryos were cultured in DMEM (Cellgro) with 0.1 mM β-mercaptoethanol (Sigma-Aldrich), 1 mM sodium pyruvate (Sigma-Aldrich), 100 U/mL penicillin (Gibco), 0.1 µg/mL streptomycin (Gibco), 0.2 mM L-glutamine (Gibco), 0.1 mM non-essential amino acids (Gibco), and 15% fetal bovine serum (Gibco). Genotyping was carried out by Transnetyx. MEFs were immortalized at passage two using infection with pBabe-SV40LT (a gift from Greg Hannon) and maintained in the same media without β-mercaptoethanol and sodium pyruvate, as described (Celli et al., 2006). Cre recombinase was introduced by two retroviral infections with Hit & Run Cre in pMMP at 12 hr intervals (Celli et al., 2006; Sfeir and de Lange, 2012). For the Cre-ERT2 system, Cre was induced with 0.5 µM 4-OHT (Sigma-Aldrich) for 6 hr; cells were washed with PBS twice, and the media were exchanged to fresh media without 4-OHT. Time point t = 0 was set at 12 hr after the first infection or at the time of the addition of media without 4-OHT. The FokI-TRF1 or FokI-ERT2-TRF1 constructs in pLPC were introduced by two retroviral infections at 12 hr intervals and selected in 2–3 µg/mL puromycin for 2–3 days. FokI-ERT2-TRF1 construct was induced with 0.5 µM 4-OHT at 24 hr before harvest. The TRF2 construct in pWZL-Hygro was introduced before FokI-ERT2-TRF1 by two retroviral infections at 12 hr intervals and selected in 90 µg/mL Hygromycin for 4–5 days. The Lig3 shRNA (CCAGACTTCAAACGTCTCAAA) in pLKO.1-Hygro was introduced after FokI-ERT2-TRF1 by three lentiviral infections at 12 hr intervals following selection in 90 µg/mL Hygromycin for 4–5 days. There were 5 µM of the ATM inhibitor KU55933 that was added 48 hr before harvest and replaced with fresh inhibitor after 24 hr. The PARP inhibitor Olaparib, 2 µM, was added 16–24 hr before harvest. DMSO was used as the negative control. EdU-labeling and detection was performed according to manufacturer’s instructions (Click-iT, Invitrogen).
Western Blot Analysis
Immunoblotting was performed as described previously (Celli et al., 2006). The following primary antibodies were used: TRF1 (1449), TRF2 (1255), MYC (9E10, Cell Signaling), ATR (N-19, Santa Cruz), LIG3 (611876; BD Transduction), and γ-tubulin (GTU-88, Sigma-Aldrich). The chemiluminescent signals were detected using enhanced chemiluminescence western blotting detection reagents (GE Healthcare) and BioMax MR film or XAR film (Kodak) according to the manufacturer’s protocol.
Immunofluorescence-FISH
Cells grown on coverslips were fixed for 10 min in 4% formaldehyde at room temperature and permeabilized for 5 min in 0.5% Triton X-100 buffer. Coverslips were incubated in blocking buffer (1 mg/mL BSA and 2% goat serum, in PBS) for 30 min, followed by incubation with primary antibodies for 2 hr, overnight. The antibodies used were 53BP1 (ab175933, Abcam), MYC (9E10, Cell Signaling), and γH2AX (JBW301, Millipore). Coverslips were then washed for three times in PBS-T and incubated for 30 min with secondary antibodies raised against mouse or rabbit and labeled with Alexa 488 or Alexa 555 (Molecular Probes; 1:1,000). Coverslips were washed with PBS-T, fixed for 10 min in 4% formaldehyde at room temperature, dehydrated in 70%, 90%, and 100% ethanol for 5 min each, and allowed to air dry. A Cy3-OO-[TTAGGG]3 labeled PNA probe (PNA Bio) was added in a buffer containing 70% formamide, 1 mg/mL blocking reagent (Roche), and 10 mM Tris-HCl pH 7.2, and the coverslips were denatured on a heat block (5 min at 80°C) and incubated for 4 hr in the dark. The coverslips were washed twice with 70% formamide, 10 mM Tris-HCl pH 7.2 for 15 min each, and three times in PBS-T for 5 min each. The DNA was counterstained by including DAPI in the second PBS-T wash. Coverslips were dehydrated in 70%, 90%, and 100% ethanol for 5 min each and allowed to air dry prior to mounting in ProLong Gold antifade (Sigma). Digital images were captured on a Zeiss Axioplan II microscope with a Hamamatsu C4742-95 camera using Volocity software.
Automated Foci and Co-localization Analysis
Co-localization of foci was quantified by the automated foci and co-localization analysis macro generated by Leonid Timashev (Rockefeller University) in FIJI running ImageJ 1.49v. The script removes the background using a median filter with a radius of 10 pixels and identifies foci using user-entered contrasting values and a mean local grayscale distribution thresholding algorithm with a constant value that is adjusted by the user. The same constant value in the thresholding algorithm was applied to all the samples within a set of experiment. After foci are identified in all channels of interest, they are overlaid and overlap is counted based on a minimum and maximum number of pixels (default 2–1,000). Next, DAPI stained nuclei are identified using the same method described above and watershed to split touching nuclei. The number of foci and co-localizations meeting the criteria are counted and overlaid onto nuclear DAPI staining. Only the foci or co-localizations within the nucleus are scored.
CO-FISH
Cells were labeled with BrdU:BrdC (3:1, final concentration: 10 µM) for 14–16 hr prior to harvesting and 0.2 µg/mL colcemid during the last hr. The cells were harvested by trypsinization, resuspended in 0.075 M KCl at 37°C for 30 min, and fixed overnight in methanol/acetic acid (3:1) at 4°C. The cells were dropped onto glass slides and the slides were air dried overnight. Slides were rehydrated with PBS for 15 min, treated with 0.5 mg/mL RNase A (in PBS, DNase free) for 10 min at 37°C, incubated with 0.5 mg/mL Hoechst 33258 (Sigma) in 2XSSC for 15 min at room temperature (rt), and exposed to 365-nm UV light (Stratalinker 1800 UV irradiator) for 30 min. The slides were then digested with 800 U Exonuclease III (Promega) at 37°C for 30 min, washed with PBS, and dehydrated through an ethanol series of 70%, 95%, and 100%. After air drying, slides were hybridized with a Cy3-OO-[TTAGGG]3 PNA probe in hybridization solution (70% formamide, 1 mg/mL blocking reagent [Roche], and 10 mM TrisHCl pH 7.2) for 2 hr at rt, washed for a few s with 70% formamide/10 mM Tris-HCl pH 7.2, and incubated with a FITC-OO-(CCCTAA)3 PNA probe in hybridization solution for 2 hr. Slides were washed, mounted, and imaged as described for FISH.
PFGE Analysis of Telomere Restriction Fragments
At the indicated time points, 1 × 106 cells were harvested by trypsinization, suspended in PBS, mixed with 2% agarose (1:1 ratio), and casted in a plug mold. Plugs were digested overnight in proteinase K digestion buffer (10 mM Tris-HCl pH 8.0, 250 mM EDTA, 0.2% sodium deoxycholate, and 1% sodium lauryl sarcosine) at 55°C. After extensive washes with Tris EDTA (TE), plugs were incubated with 60 U MboI overnight at 37°C. Digested DNA was resolved on a 1% agarose/0.5XTBE gel using a CHEF-DRII PFGE apparatus (Bio-Rad) for 24 hr. The gels were then dried at room temperature and hybridized overnight at 50°C with γ-32P-ATP end-labeled (AACCCT)4 probe in Church mix (0.5 M sodium phosphate, pH 7.2, 1 mM EDTA, 0.7% SDS, and 0.1% BSA). The gel was washed at 50°C three times in 4XSSC (30 min each), once in 4XSSC/0.1% SDS (30 min), and exposed to a PhosphoImager screen. After capturing the single-stranded telomere signal, the DNA was denatured in situ with 0.5 M NaOH/1.5 M NaCl for 30 min, neutralized with two 30 min washes in 0.5 M Tris-HCl pH 7.5/3 M NaCl, prehybridized in Church mix for 30 min at 55°C, and hybridized overnight with the same probe at 55°C. The next day, the denatured gel was washed as described above and exposed to capture the total telomere signal. Signals were quantified using ImageJ.
Statistical Analysis
Quantification of TIF-positive cells, T-SCEs, and fragile telomeres was based on at least three independent experiments where ~100 cells or ~25 metaphase spreads were analyzed for each condition. p value was calculated from two-tailed paired t test. Telomere length heterogeneity was measured from ten metaphases, ~800 telomeres per condition, and the p value was calculated from the F-test of variance.
Supplementary Material
Highlights.
Telomere-internal DSBs activate the ATM kinase-dependent signaling
Telomere-internal DSBs are repaired by HR and PARP1-dependent end-joining
Induction of DSBs at functional telomeres unleashes ALT-like phenotypes
Shelterin does not repress the DNA damage response at telomere-internal DSBs
In Brief.
Shelterin represses the DNA double-strand break (DSB) response at telomere ends. Doksani and de Lange introduce DSBs inside functional telomeres and show that they activate ATM kinase-dependent signaling and are repaired by homologous recombination (HR) and PARP1/Lig3-dependent end-joining.
Acknowledgments
We are grateful to Leonid Timashev for generating the automated foci and colocalization analysis macro. We thank Isabelle Schmutz for generating the PARP1−/− MEFs and Devon White for the invaluable help with generating MEFs. Tatsuya Kibe is thanked for performing a western blot analysis for this paper. Members of the T.d.L. lab are thanked for their technical assistance, insightful discussions, and comments. For part of this work, Y.D. was an Ellison Medical Foundation/AFAR Fellow of the Life Sciences Research Foundation. This work was supported by a grant from the NIH to T.d.L. (5R01AG016642).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes four figures and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2016.10.008.
AUTHOR CONTRIBUTIONS
Y.D. and T.d.L. designed the experiments, Y.D. performed the experiments, and Y.D. and T.d.L. wrote the paper.
REFERENCES
- Bae NS, Baumann P. A RAP1/TRF2 complex inhibits nonhomologous end-joining at human telomeric DNA ends. Mol. Cell. 2007;26:323–334. doi: 10.1016/j.molcel.2007.03.023. [DOI] [PubMed] [Google Scholar]
- Bailey SM, Goodwin EH, Meyne J, Cornforth MN. CO-FISH reveals inversions associated with isochromosome formation. Mutagenesis. 1996;11:139–144. doi: 10.1093/mutage/11.2.139. [DOI] [PubMed] [Google Scholar]
- Bandaria JN, Qin P, Berk V, Chu S, Yildiz A. Shelterin protects chromosome ends by compacting telomeric chromatin. Cell. 2016;164:735–746. doi: 10.1016/j.cell.2016.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P, Cech TR. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001;292:1171–1175. doi: 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
- Benarroch-Popivker D, Pisano S, Mendez-Bermudez A, Lototska L, Kaur P, Bauwens S, Djerbi N, Latrick CM, Fraisier V, Pei B, et al. TRF2-mediated control of telomere DNA topology as a mechanism for chromosome-end protection. Mol. Cell. 2016;61:274–286. doi: 10.1016/j.molcel.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budman J, Chu G. Processing of DNA for nonhomologous end-joining by cell-free extract. EMBO J. 2005;24:849–860. doi: 10.1038/sj.emboj.7600563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli GB, de Lange T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat. Cell Biol. 2005;7:712–718. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- Celli GB, Denchi EL, de Lange T. Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat. Cell Biol. 2006;8:885–890. doi: 10.1038/ncb1444. [DOI] [PubMed] [Google Scholar]
- Cesare AJ, Kaul Z, Cohen SB, Napier CE, Pickett HA, Neumann AA, Reddel RR. Spontaneous occurrence of telomeric DNA damage response in the absence of chromosome fusions. Nat. Struct. Mol. Biol. 2009;16:1244–1251. doi: 10.1038/nsmb.1725. [DOI] [PubMed] [Google Scholar]
- Chiruvella KK, Liang Z, Wilson TE. Repair of double-strand breaks by end joining. Cold Spring Harb. Perspect. Biol. 2013;5:a012757. doi: 10.1101/cshperspect.a012757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho NW, Dilley RL, Lampson MA, Greenberg RA. Interchromosomal homology searches drive directional ALT telomere movement and synapsis. Cell. 2014;159:108–121. doi: 10.1016/j.cell.2014.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- Doksani Y, de Lange T. The role of double-strand break repair pathways at functional and dysfunctional telomeres. Cold Spring Harb. Perspect. Biol. 2014;6:a016576. doi: 10.1101/cshperspect.a016576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doksani Y, Wu JY, de Lange T, Zhuang X. Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation. Cell. 2013;155:345–356. doi: 10.1016/j.cell.2013.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draskovic I, Londono Vallejo A. Telomere recombination and alternative telomere lengthening mechanisms. Front. Biosci. (Landmark Ed.) 2013;18:1–20. doi: 10.2741/4084. [DOI] [PubMed] [Google Scholar]
- Frit P, Barboule N, Yuan Y, Gomez D, Calsou P. Alternative end-joining pathway(s): bricolage at DNA breaks. DNA Repair (Amst.) 2014;17:81–97. doi: 10.1016/j.dnarep.2014.02.007. [DOI] [PubMed] [Google Scholar]
- Fumagalli M, Rossiello F, Clerici M, Barozzi S, Cittaro D, Kaplunov JM, Bucci G, Dobreva M, Matti V, Beausejour CM, et al. Telomeric DNA damage is irreparable and causes persistent DNA-damage-response activation. Nat. Cell Biol. 2012;14:355–365. doi: 10.1038/ncb2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- Helleday T. The underlying mechanism for the PARP and BRCA synthetic lethality: clearing up the misunderstandings. Mol. Oncol. 2011;5:387–393. doi: 10.1016/j.molonc.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson JD, Reddel RR. Assaying and investigating alternative lengthening of telomeres activity in human cells and cancers. FEBS Lett. 2010;584:3800–3811. doi: 10.1016/j.febslet.2010.06.009. [DOI] [PubMed] [Google Scholar]
- Henson JD, Neumann AA, Yeager TR, Reddel RR. Alternative lengthening of telomeres in mammalian cells. Oncogene. 2002;21:598–610. doi: 10.1038/sj.onc.1205058. [DOI] [PubMed] [Google Scholar]
- Hu J, Hwang SS, Liesa M, Gan B, Sahin E, Jaskelioff M, Ding Z, Ying H, Boutin AT, Zhang H, et al. Antitelomerase therapy provokes ALT and mitochondrial adaptive mechanisms in cancer. Cell. 2012;148:651–663. doi: 10.1016/j.cell.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliakis G, Murmann T, Soni A. Alternative end-joining repair pathways are the ultimate backup for abrogated classical non-homologous end-joining and homologous recombination repair: Implications for the formation of chromosome translocations. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2015;793:166–175. doi: 10.1016/j.mrgentox.2015.07.001. [DOI] [PubMed] [Google Scholar]
- Karlseder J, Broccoli D, Dai Y, Hardy S, de Lange T. p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science. 1999;283:1321–1325. doi: 10.1126/science.283.5406.1321. [DOI] [PubMed] [Google Scholar]
- Karlseder J, Hoke K, Mirzoeva OK, Bakkenist C, Kastan MB, Petrini JH, de Lange T. The telomeric protein TRF2 binds the ATM kinase and can inhibit the ATM-dependent DNA damage response. PLoS Biol. 2004;2:E240. doi: 10.1371/journal.pbio.0020240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, Podell ER, Cech TR. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat. Struct. Mol. Biol. 2004;11:1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- Liang Z, Sunder S, Nallasivam S, Wilson TE. Overhang polarity of chromosomal double-strand breaks impacts kinetics and fidelity of yeast non-homologous end joining. Nucleic Acids Res. 2016;44:2769–2781. doi: 10.1093/nar/gkw013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loayza D, Parsons H, Donigian J, Hoke K, de Lange T. DNA binding features of human POT1: a nonamer 5′-TAGGGTTAG-3′ minimal binding site, sequence specificity, and internal binding to multimeric sites. J. Biol. Chem. 2004;279:13241–13248. doi: 10.1074/jbc.M312309200. [DOI] [PubMed] [Google Scholar]
- Martínez P, Thanasoula M, Muñoz P, Liao C, Tejera A, McNees C, Flores JM, Fernández-Capetillo O, Tarsounas M, Blasco MA. Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice. Genes Dev. 2009;23:2060–2075. doi: 10.1101/gad.543509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masani S, Han L, Meek K, Yu K. Redundant function of DNA ligase 1 and 3 in alternative end-joining during immunoglobulin class switch recombination. Proc. Natl. Acad. Sci. USA. 2016;113:1261–1266. doi: 10.1073/pnas.1521630113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D, Reynolds GE, Mejia R, Stark JM, Murnane JP. Subtelomeric regions in mammalian cells are deficient in DNA double-strand break repair. DNA Repair (Amst.) 2011;10:536–544. doi: 10.1016/j.dnarep.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai J, Zhang Y, Morris J, Ji J, Takeda S, Doroshow JH, Pommier Y. Rationale for poly(ADP-ribose) polymerase (PARP) inhibitors in combination therapy with camptothecins or temozolomide based on PARP trapping versus catalytic inhibition. J. Pharmacol. Exp. Ther. 2014;349:408–416. doi: 10.1124/jpet.113.210146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabetani A, Ishikawa F. Unusual telomeric DNAs in human telomerase-negative immortalized cells. Mol. Cell. Biol. 2009;29:703–713. doi: 10.1128/MCB.00603-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan RJ, Arnoult N, Lackner DH, Oganesian L, Haggblom C, Corpet A, Almouzni G, Karlseder J. Rapid induction of alternative lengthening of telomeres by depletion of the histone chaperone ASF1. Nat. Struct. Mol. Biol. 2014;21:167–174. doi: 10.1038/nsmb.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Bartocci C, Ouzounov I, Diedrich JK, Yates JR, 3rd, Denchi EL. A two-step mechanism for TRF2-mediated chromosome-end protection. Nature. 2013;494:502–505. doi: 10.1038/nature11873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- Palm W, Hockemeyer D, Kibe T, de Lange T. Functional dissection of human and mouse POT1 proteins. Mol. Cell. Biol. 2009;29:471–482. doi: 10.1128/MCB.01352-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul K, Wang M, Mladenov E, Bencsik-Theilen A, Bednar T, Wu W, Arakawa H, Iliakis G. DNA ligases I and III cooperate in alternative non-homologous end-joining in vertebrates. PLoS ONE. 2013;8:e59505. doi: 10.1371/journal.pone.0059505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribes-Zamora A, Indiviglio SM, Mihalek I, Williams CL, Bertuch AA. TRF2 interaction with Ku heterotetramerization interface gives insight into c-NHEJ prevention at human telomeres. Cell Rep. 2013;5:194–206. doi: 10.1016/j.celrep.2013.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, de Lange T. Removal of shelterin reveals the telomere end-protection problem. Science. 2012;336:593–597. doi: 10.1126/science.1218498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, Kabir S, van Overbeek M, Celli GB, de Lange T. Loss of Rap1 induces telomere recombination in the absence of NHEJ or a DNA damage signal. Science. 2010;327:1657–1661. doi: 10.1126/science.1185100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, Karlseder J, Holtgreve-Grez H, Jauch A, de Lange T. DNA ligase IV-dependent NHEJ of deprotected mammalian telomeres in G1 and G2. Curr. Biol. 2002;12:1635–1644. doi: 10.1016/s0960-9822(02)01179-x. [DOI] [PubMed] [Google Scholar]
- Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr. Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- Tang J, Cho NW, Cui G, Manion EM, Shanbhag NM, Botuyan MV, Mer G, Greenberg RA. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat. Struct. Mol. Biol. 2013;20:317–325. doi: 10.1038/nsmb.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- Zimmermann M, Kibe T, Kabir S, de Lange T. TRF1 negotiates TTAGGG repeat-associated replication problems by recruiting the BLM helicase and the TPP1/POT1 repressor of ATR signaling. Genes Dev. 2014;28:2477–2491. doi: 10.1101/gad.251611.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.