Abstract
The human mutilating disease chromoblastomycosis is caused by melanized members of the order Chaetothyriales. To assess population diversity among 123 clinical strains of agents of the disease in Brazil we applied sequencing of the rDNA internal transcribed spacer region, and partial cell division cycle and β-tubulin genes. Strains studied were limited to three clusters divided over the single family Herpotrichiellaceae known to comprise agents of the disease. A Fonsecaea cluster contained the most important agents, among which F. pedrosoi was prevalent with 80% of the total set of strains, followed by 13% for F. monophora, 3% for F. nubica, and a single isolate of F. pugnacius. Additional agents, among which two novel species, were located among members of the genus Rhinocladiella and Cyphellophora, with frequencies of 3% and 1%, respectively.
Author Summary
Chromoblastomycosis, a skin disease found among rural populations in tropical and subtropical regions, is caused by melanized fungi related to the black yeasts. The present study evaluates the species distribution among 123 clinical strains from endemic areas in Brazil based on multilocus sequence data, and describes two new agents of the disease which proved to be affiliated to Rhinocladiella and Cyphellophora.
Introduction
Chromoblastomycosis is a chronic granulomatous infection of the skin caused by melanized fungi. It has a worldwide distribution mainly in tropical and subtropical climate zones, with a preference for humid climates with dense forestation, with Cladophialophora carrionii being the only species that is restricted to semi-arid areas with Cactaceae as main vegetation. Endemic areas are in Japan, Southeast Asia, Australia, Madagascar, as well as South and Central America [1–7]. In Brazil, the infection is observed in all states, with an estimated prevalence of 1/196 thousand inhabitants, but in some hyperendemic regions a considerably higher prevalence is noted [8]. Infection is assumed to occur through accidental inoculation of the fungus via contaminated plant debris, being favored by agricultural activities denoting an occupational nature of the disease. Chromoblastomycosis is one of the most frequent implantation mycoses found among rural populations [2, 8–11].
Clinically, the disease is characterized by pseudoepitheliomatous hyperplasia with epidermal microabscesses and dermal granulomata [12, 13]. The disease has a slow evolution, but finally may result in disfigurement of affected body sites [8]. The initial lesion is noted as a small pink papule at the site of inoculation which gradually enlarges. Development of superficial erythematous plaques with scaly or warty appearance probably takes several months or years. As a result of acanthosis these lesions may develop into large papillomatous and verrucous warts.
Species of the humid climate are particularly members of the genus Fonsecaea, with F. pedrosoi, F. monophora and F. nubica as prevalent agents [14–17]. Recently another species, F. pugnacius was described [18]. Rhinocladiella aquaspersa is an uncommon species of humid as well as of dry climates [19]. Other reported agents such as Phialophora verrucosa and Exophiala dermatitidis [17, 20] are extremely rare and mostly cause other types of infections. Fonsecaea pedrosoi is nearly exclusively isolated from chromoblastomycosis, while F. monophora repeatedly causes brain infection and F. pugnacius combines the two disorders by starting as chromoblastomycosis with cerebral dissemination in a single patient [18]. Species distinction of agents of the disease is clinically significant because of the differences in prognosis of the infection.
The present study evaluates the diversity of agents in endemic areas of Brazil based on multilocus sequence data, clinical aspects, direct mycological examination and culture. An enumeration of currently proven cases with molecular support in endemic areas in Brazil is provided.
Results
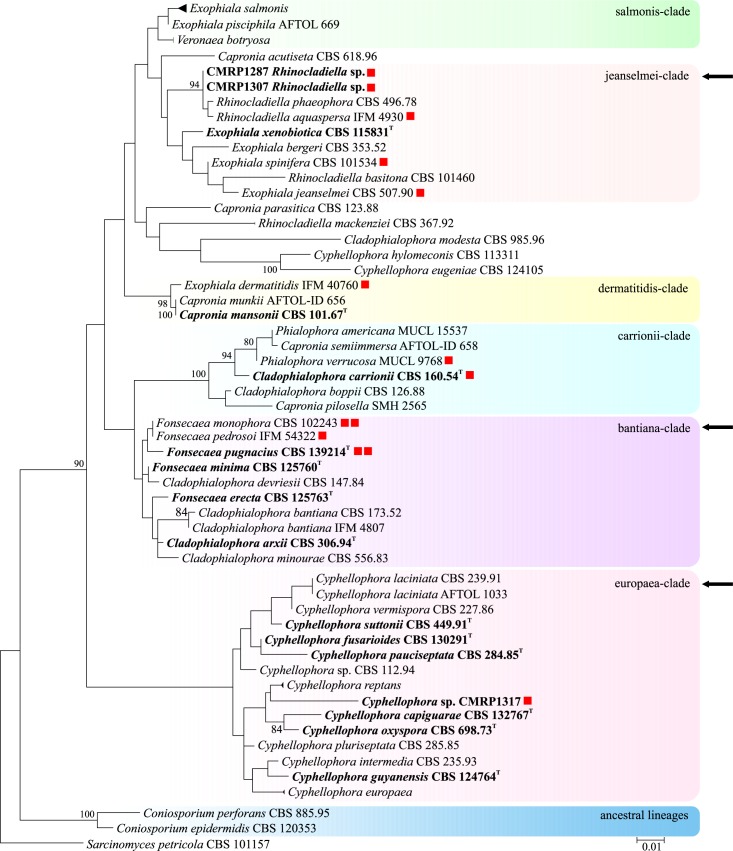
A set of 123 clinical strains from cases of chromoblastomycosis from different endemic areas in Brazil was analyzed. Judging from a reference set of partial LSU rDNA sequences of members of Chaetothyriales available at CBS, agents of chromoblastomycosis were polyphyletic within the order, being dispersed in three different clades: jeanselmei-, bantiana- and europaea-clades (Fig 1, arrows).
Fig 1. Phylogeny of a representative selection of species in Chaetothyriales, based on confidently aligned LSU sequences.
Constructed with Maximum likelihood implemented in MEGA 7. Bootstrap values > 80% from 100 resampled datasets are shown with branches. Coloured boxes represent species complexes taken from de Hoog et al. [21], Feng et al. [22], and Vicente et al. [23]. Clades with species causing chromoblastomycosis analysed in this study are indicated with arrows. Type strain in bold.
Multilocus sequence analyses using ITS, and partial BT2 and CDC42 genes were performed for identification and for elucidation of species identities (S1 Fig). The 123 clinical strains clustered in different clades within the Chaetothyriales, being centred around members of the genera Fonsecaea, Rhinocladiella, and Cyphellophora (Table 1). The Brazilian clinical isolates used in this study all were derived from humid climate zones; members of the Cladophialophora carrionii group were not encountered.
Table 1. Strains analysed.
| Name | Strain number | Geograph/ Brazil | Chromoblastomycose lesions | ITS; BT2; CDC42; LSU |
| Fonsecaea monophora | CMRP1290 | MA, Northeast | Polymorphic lesions | KR732306; KR732312; KR732318 |
| Fonsecaea monophora | CMRP1329 | PR, North | Plaque | KX434631; KX583711 |
| Fonsecaea monophora | CMRP1330 | PR, North | Nodular | KX434632 |
| Fonsecaea monophora | CMRP1359 | AM, North | NI | KX434633 |
| Fonsecaea monophora | CMRP1360 | AM, North | NI | KX434634 |
| Fonsecaea monophora | CMRP1335 | MG, Southeast | NI | KX434639 |
| Fonsecaea monophora | CMRP1366 | PR, South | Polymorphic lesions | KX434635 |
| Fonsecaea monophora | CMRP1367 | RS, South | NI | KX434636 |
| Fonsecaea monophora | CMRP1368 | RS, South | NI | KX434637 |
| Fonsecaea monophora | CMRP1369 | RS, South | NI | KX434638 |
| Fonsecaea monophora | CMRP1371 | PR, South | NI | KX434640 |
| Fonsecaea monophora | CMRP1383 | PR, South | NI | KX434641 |
| Fonsecaea monophora | CBS 102242 | PR, South | Verrucous | EU938583; EU938549 |
| Fonsecaea monophora | CBS 102243 | PR, South | Scarring | EU938579; EU938542 |
| Fonsecaea monophora | CBS 102246 | PR, South | NI | AY366928; EU938543 |
| Fonsecaea monophora | CBS 102248 | PR, South | NI | AY366926; EU938550 |
| Fonsecaea nubica | IAL 4004 | BA, Northeast | Verrucous | KU892412 |
| Fonsecaea nubica | IAL 3994 | RO, North | Verrucous | KU881739 |
| Fonsecaea nubica | IAL 3992 | MG, Southeast | Nodular and tumoral | KU881735 |
| Fonsecaea nubica | IAL 3999 | SP, Southeast | Polymorphic lesions | KU8811742 |
| Fonsecaea pedrosoi | CMRP1342 | MA, Northeast | NI | KX434642 |
| Fonsecaea pedrosoi | CMRP1271 | MA, Northeast | Polymorphic lesions | KR732301; KR732307; KR732313 |
| Fonsecaea pedrosoi | CMRP1272 | MA, Northeast | Nodular and plaque | KR732302; KR732308; KR732314 |
| Fonsecaea pedrosoi | CMRP1273 | MA, Northeast | Plaque | KR732303; KR732309; KR732315 |
| Fonsecaea pedrosoi | CMRP1274 | MA, Northeast | Nodular and plaque | KR732304; KR732310; KR732316 |
| Fonsecaea pedrosoi | CMRP1275 | MA, Northeast | Plaque | KR732305; KR732311; KR732317 |
| Fonsecaea pedrosoi | CMRP1276 | MA, Northeast | Scarring, nodular and plaque | KX434643; -; KX583684 |
| Fonsecaea pedrosoi | CMRP1277 | MA, Northeast | Plaque | KX434644; KX583712; KX583685 |
| Fonsecaea pedrosoi | CMRP1278 | MA, Northeast | Scarring and plaque | KX434645; KX583713; KX583686 |
| Fonsecaea pedrosoi | CMRP1279 | MA, Northeast | Plaque | KX434646; -; KX583687 |
| Fonsecaea pedrosoi | CMRP1280 | MA, Northeast | Nodular and plaque | KX434647; KX583714; KX583688 |
| Fonsecaea pedrosoi | CMRP1281 | MA, Northeast | Nodular and plaque | KX434648 |
| Fonsecaea pedrosoi | CMRP1282 | MA, Northeast | Nodular and plaque | KX434649; -; KX583689 |
| Fonsecaea pedrosoi | CMRP1283 | MA, Northeast | Nodular and plaque | KX434650; -; KX583690 |
| Fonsecaea pedrosoi | CMRP1284 | MA, Northeast | Plaque | KX434651; KX583715; KX583691 |
| Fonsecaea pedrosoi | CMRP1285 | MA, Northeast | Plaque | KX434652; -; KX583692 |
| Fonsecaea pedrosoi | CMRP1286 | MA, Northeast | Scarring, nodular and plaque | KX434653; KX583716 |
| Fonsecaea pedrosoi | CMRP1288 | MA, Northeast | Scarring and plaque | KX434654; KX583717; KX583693 |
| Fonsecaea pedrosoi | CMRP1289 | MA, Northeast | Plaque | KX434655; -; KX583694 |
| Name | Strain number | Geograph/ Brazil | Chromoblastomycose lesions | ITS; BT2; CDC42; LSU |
| Fonsecaea pedrosoi | CMRP1291 | MA, Northeast | Plaque | KX434656; KX583718; KX583695 |
| Fonsecaea pedrosoi | CMRP1292 | MA, Northeast | Plaque | KX434657; KX583719; KX583696 |
| Fonsecaea pedrosoi | CMRP1293 | MA, Northeast | Plaque | KX434658; KX583720; KX583697 |
| Fonsecaea pedrosoi | CMRP1294 | MA, Northeast | Scarring and plaque | KX434659; KX583721; KX583698 |
| Fonsecaea pedrosoi | CMRP1295 | MA, Northeast | Scarring and plaque | KX434660; -; KX583699 |
| Fonsecaea pedrosoi | CMRP1296 | MA, Northeast | Scarring, nodular and plaque | KX434661; -; KX583700 |
| Fonsecaea pedrosoi | CMRP1298 | MA, Northeast | Plaque | KX434662; KX583722 |
| Fonsecaea pedrosoi | CMRP1299 | MA, Northeast | Plaque | KX434663; KX583723 |
| Fonsecaea pedrosoi | CMRP1300 | MA, Northeast | Scarring, nodular and plaque | KX434664 |
| Fonsecaea pedrosoi | CMRP1301 | MA, Northeast | Plaque | KX434665; KX583724; KX583701 |
| Fonsecaea pedrosoi | CMRP1302 | MA, Northeast | Scarring, nodular and plaque | KX434666; KX583725; KX583702 |
| Fonsecaea pedrosoi | CMRP1303 | MA, Northeast | Plaque | KX434667 |
| Fonsecaea pedrosoi | CMRP1304 | MA, Northeast | Polymorphic lesions | KX434668; KX583726; KX583703 |
| Fonsecaea pedrosoi | CMRP1305 | MA, Northeast | Nodular and plaque | KX434669; KX583727; KX583704 |
| Fonsecaea pedrosoi | CMRP1306 | MA, Northeast | Nodular, verrucous and plaque | KX434670; KX583728; KX583705 |
| Fonsecaea pedrosoi | CMRP1308 | MA, Northeast | Plaque | KX434671; KX583729; KX583706 |
| Fonsecaea pedrosoi | CMRP1309 | MA, Northeast | Scarring and plaque | KX434672; KX583730; KX583707 |
| Fonsecaea pedrosoi | CMRP1310 | MA, Northeast | Plaque | KX434673 |
| Fonsecaea pedrosoi | CMRP1311 | MA, Northeast | Plaque | KX434674; KX583731 |
| Fonsecaea pedrosoi | CMRP1313 | MA, Northeast | Plaque | KX434675; KX583732 |
| Fonsecaea pedrosoi | CMRP1314 | MA, Northeast | NI | KX434676; KX583733 |
| Fonsecaea pedrosoi | CMRP1315 | MA, Northeast | Nodular, tumoral and plaque | KX434677; KX583734 |
| Fonsecaea pedrosoi | CMRP1316 | MA, Northeast | Plaque | KX434678 |
| Fonsecaea pedrosoi | CMRP1318 | MA, Northeast | NI | KX434679 |
| Fonsecaea pedrosoi | IAL 3991 | BA, Northeast | Verrucous | KU881737 |
| Fonsecaea pedrosoi | IAL 3997 | BA, Northeast | Verrucous | KU881741 |
| Fonsecaea pedrosoi | IAL 3996 | BA, Northeast | Verrucous | KU881745 |
| Fonsecaea pedrosoi | CMRP1331 | PA, North | Nodular | KX434680 |
| Fonsecaea pedrosoi | CMRP1332 | PA, North | Nodular | KX434681; KX583735 |
| Fonsecaea pedrosoi | CMRP1333 | PA, North | Nodular | KX434682 |
| Fonsecaea pedrosoi | CMRP1334 | PA, North | Nodular | KX434683; KX583736 |
| Fonsecaea pedrosoi | CMRP1337 | PA, North | Nodular | KX434684 |
| Fonsecaea pedrosoi | CMRP1338 | PA, North | Nodular | KX434685; KX583737 |
| Fonsecaea pedrosoi | CMRP1339 | PA, North | Nodular | KX434686; KX583738 |
| Fonsecaea pedrosoi | CMRP1340 | PA, North | Nodular | KX434687; KX583739 |
| Fonsecaea pedrosoi | CMRP1341 | PA, North | Nodular | KX434688; KX583752 |
| Fonsecaea pedrosoi | CMRP1344 | PA, North | Nodular | KX434689; KX583740 |
| Fonsecaea pedrosoi | CMRP1345 | PA, North | Nodular | KX434690 |
| Fonsecaea pedrosoi | CMRP1346 | PA, North | Nodular | KX434691; KX583741 |
| Fonsecaea pedrosoi | CMRP1347 | PA, North | Nodular | KX434692 |
| Fonsecaea pedrosoi | CMRP1348 | PA, North | Nodular | KX434693; KX583742 |
| Fonsecaea pedrosoi | CMRP1349 | PA, North | Nodular | KX434694 |
| Fonsecaea pedrosoi | CMRP1350 | PA, North | Nodular | KX434695; KX583743 |
| Fonsecaea pedrosoi | CMRP1351 | PA, North | Plaque | KX434696; KX583744 |
| Fonsecaea pedrosoi | CMRP1352 | PA, North | Nodular | KX434697; KX583745 |
| Fonsecaea pedrosoi | CMRP1353 | PA, North | Nodular | KX434698; KX583746 |
| Fonsecaea pedrosoi | CMRP1354 | PA, North | Nodular | KX434699 |
| Fonsecaea pedrosoi | CMRP1355 | PA, North | Nodular | KX434700; KX583747 |
| Fonsecaea pedrosoi | CMRP1356 | PA, North | Nodular | KX434701 |
| Fonsecaea pedrosoi | CMRP1357 | PA, North | Nodular | KX434702; KX583748 |
| Name | Strain number | Geograph/ Brazil | Chromoblastomycose lesions | ITS; BT2; CDC42; LSU |
| Fonsecaea pedrosoi | CMRP1361 | PA, North | Nodular | KX434703 |
| Fonsecaea pedrosoi | IAL 3998 | RO, North | Nodular and tumoral | KU881736 |
| Fonsecaea pedrosoi | IAL 4001 | RO, North | Verrucous | KU881744 |
| Fonsecaea pedrosoi | IAL 4005 | RO, North | Polymorphic lesions | KU892413 |
| Fonsecaea pedrosoi | IAL 4007 | RO, North | Verrucous | KU892414 |
| Fonsecaea pedrosoi | CMRP1336 | MG, Southeast | NI | KX434709 |
| Fonsecaea pedrosoi | CMRP1362 | MS, Southeast | NI | KX434704 |
| Fonsecaea pedrosoi | CMRP1363 | MS, Southeast | NI | KX434705 |
| Fonsecaea pedrosoi | CMRP1364 | MS, Southeast | NI | KX434706 |
| Fonsecaea pedrosoi | CMRP1365 | MS, Southeast | NI | KX434707 |
| Fonsecaea pedrosoi | IAL 3993 | MG, Southeast | Nodular and tumoral | KU881738 |
| Fonsecaea pedrosoi | CMRP1372 | PR, South | Nodular and verrucous | KX434710 |
| Fonsecaea pedrosoi | CMRP1384 | PR, South | Nodular and verrucous | KX434717 |
| Fonsecaea pedrosoi | CMRP1373 | PR, South | Scarring | KX434711 |
| Fonsecaea pedrosoi | CMRP1374 | PR, South | Scarring and verrucous | KX434712 |
| Fonsecaea pedrosoi | CMRP1375 | PR, South | Nodular and verrucous | KX434713 |
| Fonsecaea pedrosoi | CMRP1376 | PR, South | Verrucous | KX434714 |
| Fonsecaea pedrosoi | CBS 102245 | PR, South | Scarring and verrucous | AY366918; EU938562 |
| Fonsecaea pedrosoi | CBS 102247 | PR, South | NI | AY366919; EU938566 |
| Fonsecaea pedrosoi | CMRP1377 | PR, South | Verrucous | KX434715 |
| Fonsecaea pedrosoi | CMRP1378 | PR, South | Plaque | KX434720 |
| Fonsecaea pedrosoi | CMRP1379 | PR, South | Polymorphic lesions | KX434721 |
| Fonsecaea pedrosoi | CMRP1380 | PR, South | Nodular and tumoral | KX434716 |
| Fonsecaea pedrosoi | CMRP1381 | PR, South | Nodular and tumoral | KX434718 |
| Fonsecaea pedrosoi | CMRP1382 | PR, South | NI | KX434719 |
| Fonsecaea pedrosoi | CMRP1370 | RS, South | NI | KX434708 |
| Fonsecaea pedrosoi | IAL 4000 | NI | Polymorphic lesions | KU881743 |
| Fonsecaea pedrosoi | IAL 4006 | NI | Verrucous | KU892409 |
| Fonsecaea pedrosoi | IAL 4002 | NI | Verrucous | KU892410 |
| Fonsecaea pugnacius | CBS 139214 | MA, Northeast | Plaque and disseminated to brain | KR706553; KR706547; KR706551 |
| Cyphellophora ludoviensis sp. nov. | CMRP1317 | MA, Northeast | Nodular and plaque | KX434722; KX583749; -; KX583708 |
| Rhinocladiella tropicalis sp. nov. | CMRP1287 | MA, Northeast | Polymorphic lesions | KX434723; KX583750; -; KX583709 |
| Rhinocladiella tropicalis | CMRP1307 | MA, Northeast | Plaque | KX434724; KX583751; -; KX583710 |
| Rhinocladiella tropicalis | IMT776 | SP, South | NI | KU854928 |
CMRP, Microbial Collections of Paraná Network- TAXon line; CBS, CBS-KNAW Fungal Biodiversity Centre, Utrecht, The Netherlands; IMT, Tropical Medicine Institute, SP, Brazil; IAL, Culture Collection of Adolfo Lutz Institute. BT2, partial beta-tubulin gene; ITS, internal transcribed spacer regions of the rDNA and intervening 5.8S nuclear ribosomal DNA (nrDNA); CDC42, partial cell division cycle gene. Brazil Sates: BA. Bahia; MA. Maranhão; MG. Minas Gerais; MS. Mato Grosso do Sul; PA. Para; RO. Rondônia; PR. Paraná; RS. Rio Grande do Sul; SP. São Paulo; NI. not informed, patients in treatment at SP.
Fonsecaea is nested in the ‘bantiana-clade’ (Table 1) and contains the prevalent agents of the disease. Of the studied isolates, 98 (80%) grouped as Fonsecaea pedrosoi, followed by 16 (13%) isolates of F. monophora, 4 (3%) isolates of F. nubica and one isolate of F. pugnacius. Isolates of the Rhinocladiella group and of phialophora-like species are considered as rare agents of chromoblastomycosis in the Americas. Rhinocladiella was nested in the ‘jeanselmei-clade’ and the phialophora-like agent clustered in the ‘europaea-clade’ (presently known as Cyphellophoraceae); two unnamed species were uncovered (Fig 1).
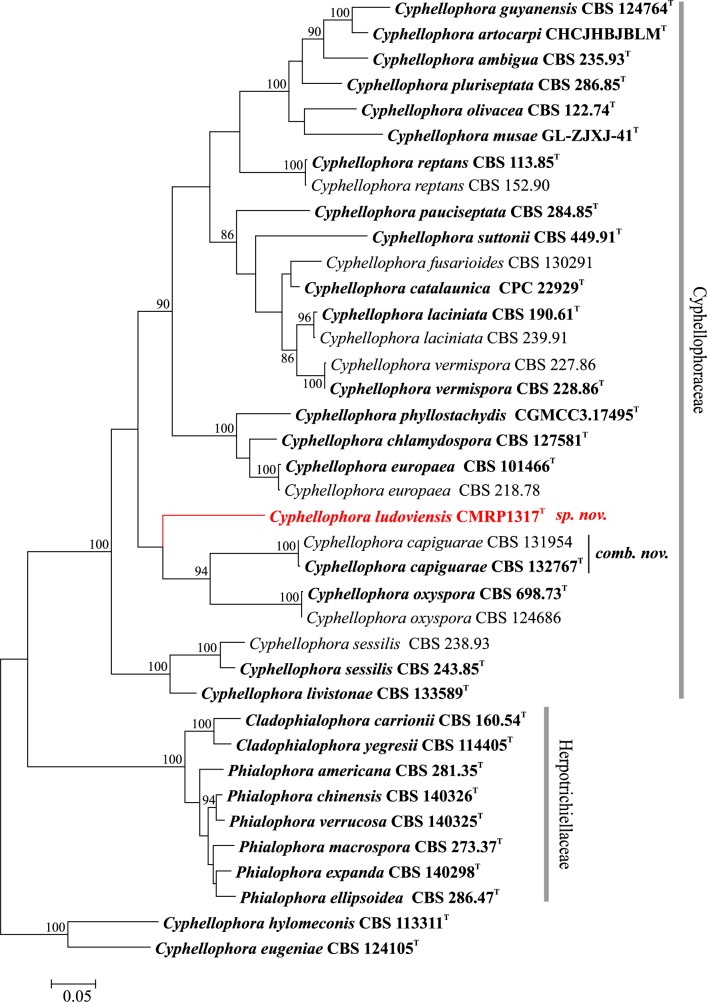
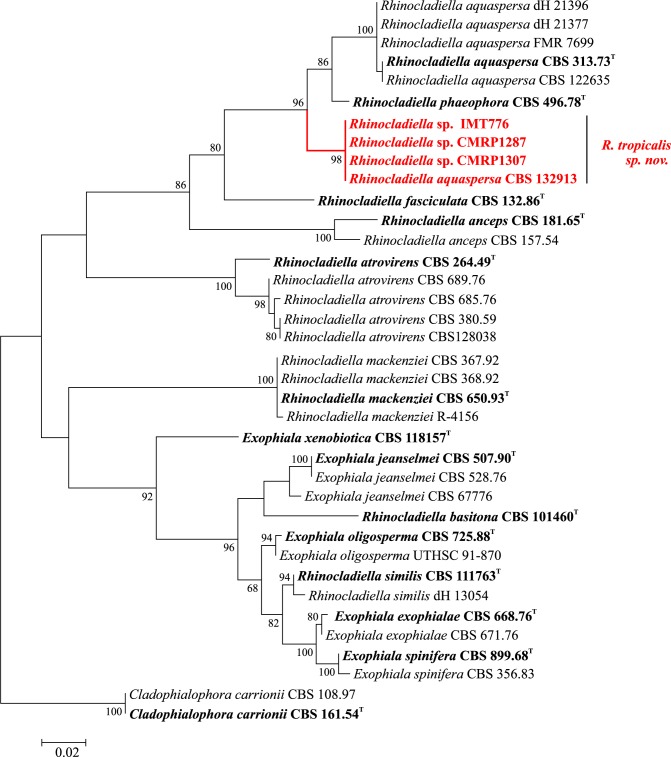
The analyzed ITS and BT2 regions and a tree resulting from combined loci revealed identical topologies in phylogenetic analyses. The unnamed strains CMRP1287, CMRP1307, IMT776 and CMRP1317 were found to be concordantly positioned in all trees and grouped with remaining members of Rhinocladiella and Cyphellophora causing chromoblastomycosis (Figs 2 and 3). Clinical strain CMRP1317 was located at a significant distance from all reference strains of Cyphellophora, i.e with combined ITS and BT2 data the distance was 23.4%. Strains CMRP1287, CMRP1307 and IMT776 were also located at significant distance from known species in Rhinocladiella (Table 2). Consequently the strains were judged to represent novel taxa in Cyphellophora and Rhinocladiella, respectively.
Fig 2. Multilocus tree of Cyphellophora based on ITS and partial BT2 sequences.
Constructed with maximum likelihood implemented in MEGA 7. Bootstrap values of >80% from 100 resampled data sets are shown with branches. Cladophialophora yegresii and C. carrionii comprised the outgroup. Novel species causing chromoblastomycosis are indicated with red branches. Type strain in bold.
Fig 3. Multilocus tree of Rhinocladiella based on ITS and partial BT2 sequences.
Constructed with maximum likelihood implemented in MEGA 7. Bootstrap values of >80% from 100 resampled data sets are shown with branches. Cladophialophora yegresii and C. carrionii comprised the outgroup. Novel species causing chromoblastomycosis are indicated with red branches. Type strain in bold.
Table 2. Estimates of Evolutionary Divergence of Rhinocladiella tropicalis.
Distance R. aquaspersa to R. phaeophora ITS (4.5) and BT2 (5.6); Lengths of alignments of ITS: 548 bp and BT2: 349 bp. Analyses were conducted using the Kimura 2-parameter model. All ambiguous positions were removed for each sequence pair. Evolutionary analyses were conducted in MEGA 7.
| Locus | R. aquaspersa | R. phaeophora | R. fasciculata | R. atrovirens | R. mackenzie | R. anceps | R. basitona | R. similis |
|---|---|---|---|---|---|---|---|---|
| ITS | 5.1 | 5.4 | 11.4 | 18.6 | 19.7 | 20.4 | 21.7 | 22.0 |
| BT2 | 9.7 | 10.7 | ---- | ---- | ---- | ---- | 34.8 | 36.5 |
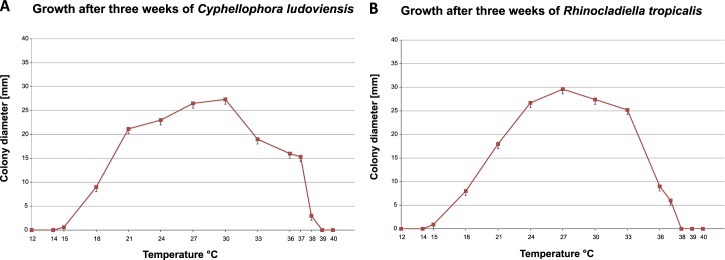
We report the clinical cases caused by the novel Cyphellophora and Rhinocladiella species, which are named below as C. ludoviensis and R. tropicalis, respectively. The species showed optimal development at 30°C and 27°C, respectively, with a wide growth range between 18°C and 37°C and with residual growth at 15°C and 38°C and maximum growth temperature at 37°C; no growth was observed at 40°C (Fig 4).
Fig 4. Cardinal temperatures of strains described.
(A) Cyphellophora ludoviensis with optimal growth temperature at 30°C and maximum at 37°C. (B) Rhinocladiella tropicalis with optimal development at 27°C and maximum at 37°C.
Cyphellophora ludoviensis C.M.P.S. Azevedo, R.R. Gomes, V.A. Vicente & de Hoog, sp. nov. ‒ MycoBank MB 817305 (Figs 2 and 5).
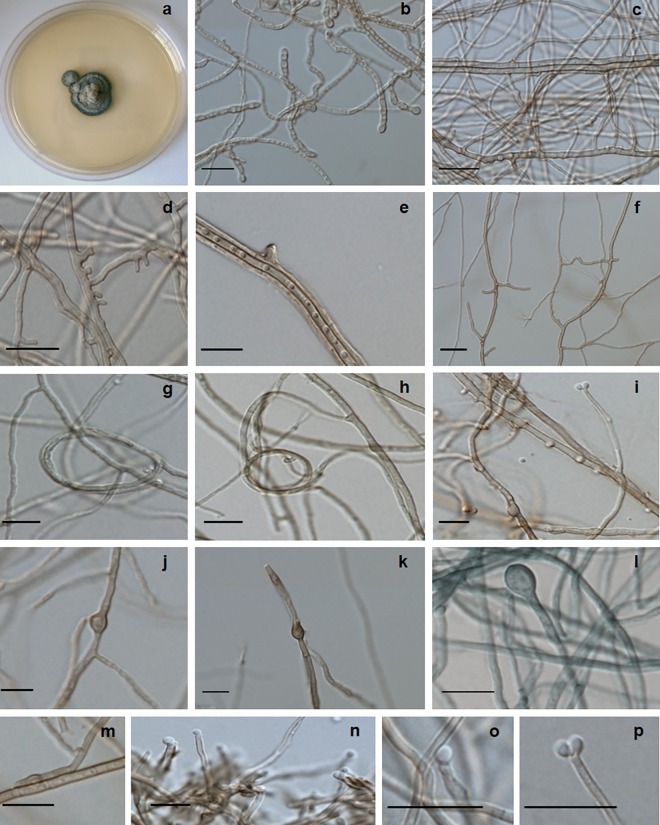
Fig 5. Cyphellophora ludoviensis microscopic morphology.
(A) colonies on SGA; (B-E) hyphae with chlamydospores and lateral extensions; (F) anastomosis; (G-H) spirally twisted hyphae; (I) poorly differentiated phialide producing conidia; (J-P) chlamydospores and conidia. Scale bars 10 μm.
Etymology: named after the city where the case was first diagnosed, San Luis in Maranhão State, Brazil.
Holotype: Maranhão State, Brazil, from skin lesion of human patient, dried holotype UPCB 85592 at Department of Botany Herbarium at Federal University of Paraná (UPCB); type strain CMRP1317 = LMICRO356 = CBM47. Additional information listed in Table 1.
Description of CMRP51317 after two weeks incubation on MEA at 28°C: Colonies moderately expanding, greyish olive to olivaceus black, with olivaceus black reverse. Hyphae pale olivaceous brown, 1.2–6 μm wide, septate every 9–25 μm, occasionally bearing scarce phialides and conidia. Phialides poorly differentiated, producing sub-spherical, hyaline conidia, 1.8−2.5 μm. Creeping hyphae producing lateral outgrowths which become septate, pale brown conidiophores 1.5−2.0 μm wide and with frequent anastomoses; chlamydospores developing intercalarily or terminally on hyphae. Chlamydospores ovoidal, brown, 4.5−6.5 × 4.5 μm, with irregularly thickened walls. Thickened terminal hyphae and spirally twisted hyphae frequently present. Teleomorph unknown. Cardinal temperatures: minimum 18°C, optimum 30°C, maximum 37°C, with residual growth at 15°C and 38°C.
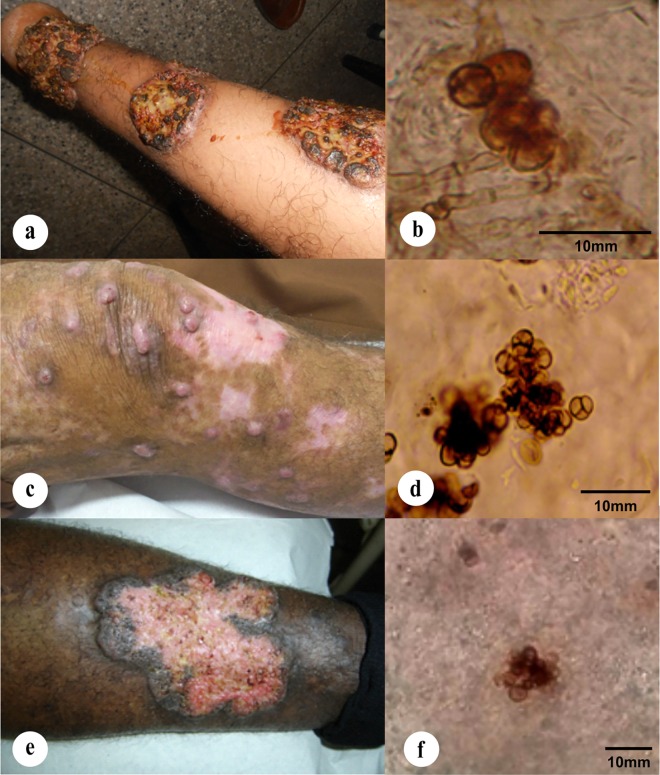
Case report: Patient, a 57-year-old Caucasian male from Icatu, Brazil, was diagnosed in 2008 with lower limb injuries with nodular appearance and plaque. Moderate disease progressed with 3 years of evolution; muriform cells were observed in tissue (Fig 6A and 6B). Patient was treated orally with itraconazole (200 mg/day), leading to improvement of the lesions during treatment within three months. Patient abandoned treatment after 12 months and returned in 2011 with recurred lesions spreading throughout the lower limb, following a lymphatic path, with secondary infections, presenting verrucous type injuries, ulceration and crusting at the surface. The fungus was re-isolated from the recurred lesions. The patient was treated using itraconazole (200 mg/day) combined with cryosurgery.
Fig 6. Clinical case pictures.
(A, B) Nodular and verrucous lesions and muriform cells from skin tissue biopsy of arm lesion caused by Cyphellophora ludoviensis (CMRP1317); (C-E) polymorphic infiltrative plaque lesions caused by different strains of Rhinocladiella tropicalis affecting the legs, (C) with nodular and cicatricial and (E) verrucous lesions; (D-F) muriform cells from skin tissue biopsy of lesions caused by R. tropicalis strains CMRP1287 and CMRP1307, respectively.
Notes: The strain clustered with members of Chaetothyriales variously classified in Cyphellophora or Phialophora. Several of these have been reported from mild infections of human skin but without presence of muriform cells. Cyphellophora ludoviensis is the only species in the ‘europaea-clade’ (Cyphellophoraceae) where muriform cells were observed in the lesions, and is distant from remaining species causing this disease, as well as from the generic type of Phialophora, P. verrucosa.
Rhinocladiella tropicalis C.M.P.S. Azevedo, R.R. Gomes, V.A. Vicente & de Hoog, sp. nov. ‒ MycoBank MB 817306 (Figs 3 and 7).
Fig 7. Rhinocladiella tropicalis, microscopic morphology.
(A) Colonies on SGA; (B, C) twisted hyphae and conidia; (D-G) conidiophores with conidia produced in sympodial order; (H-O) conidial apparatus with conidia. Scale bars 10 μm.
Etymology: Named after its occurrence in tropical South America.
Holotype: Maranhão State, Brazil, from skin lesion of human patient, dried holotype UPCB 85593 at Department of Botany Herbarium at Federal University of Paraná (UPCB); type strain CMRP1287 = LMICRO326 = CBM17. Additional material examined listed in Table 1.
Description of CMRP1287 after two weeks incubation on MEA at 28°C: Colonies growing moderate rapidly, velvety, elevated, olivaceus black with dark reverse. Mycelium partly immersed but mainly aerial, composed of branched hyphae which are pale brown, occasionally reddish brown, 1.5–2.5 μm wide, regularly septate every 7–18 μm, with integrated conidiogenous cells which are somewhat differentiated from vegetative hyphae. Conidiophores erect, straight, thick-walled, brown to dark brown, up to 19 μm high, bearing small, pigmented denticles on which conidia are produced in sympodial order. Conidiogenous cells terminal or lateral, often becoming intercalary, cylindrical in the apical part with numerous flat scars. Conidia one-celled, hyaline to pale brown, ellipsoidal to clavate, 2.5−5.0 × 1.5−2.5 μm; scars slightly prominent, approx. 0.4 μm diam, pigmented. Conidia usually 1-septate, 5−8 × 1.5−3.0 μm. Budding cells developing from single conidia may be present. Teleomorph unknown. Cardinal temperatures: minimum 18°C, optimum 27°C, maximum 37°C, with residual growth at 15°C.
Case reports: Patient infected by R. tropicalis CMRP1287 was a 65-year-old caucasian male from Pinheiro, Maranhão, Brazil, diagnosed in 2004 with lower limb injuries showing polymorphic lesions with plaques and nodular and cicatrized lesions. Muriform cells were observed in tissue (Fig 6C and 6D). The infection evolved during a 6-year period. Treatment was started with itraconazole (200 mg/day), irregularly until 2012; subsequently regular treatment was installed but with low response. From then onwards additional cryosurgery with liquid nitrogen was applied once a week. After 4 years of therapy still some lesions with a fibrotic aspect and with few murifom cells were observed. Patient infected by R. tropicalis CMRP1307 strain was a 78-year-old afrodescendant male, from Icatu, Maranhão, Brazil, diagnosed in 2002 with lower limb injuries showing polymorphic lesions with plaques and infiltration and muriform cells in tissue (Fig 6E and 6F). The diseased showed moderate development over a 3-year period of evolution. Patient was treated with oral itraconazole (200 mg/day), initiated in 2010. Some treatment interruptions occurred because of the distance to the patient’s living place and the low response to itraconazole, with worsening of the symptoms.
Both patients are still under treatment in 2016. The patient infected by R. tropicalis CMRP1287 is currently treated using itraconazole (200 mg/day) combined with cryosurgery and the patient infected by R. tropicalis CMRP1307 uses the antifungals itraconazole (400 mg/day) and terbinafin (250 mg twice daily) in combination.
During our studies we noticed strain IMT766, isolated in 1970 from a case of chromoblastomycosis and deposited in the Institute of Tropical Medicine, São Paulo, Brazil, and supplementary material from a patient infected by strain CBS 132913, a 63-year-old male construction worker from Venezuela with an asymptomatic and localized skin lesion of the hand with a scaly, crusted, dull-red appearance, friable with hemorrhagic dots [19].
The response to treatment has been evaluated by clinical, mycological and histopathological criteria. A complete clinical response is being accompanied assuming for cure criteria after two years follow up without recurrence, with complete healing of the lesions and disappearance of itching and local pain. The patients must also be monitored by three to four consecutive biopsies to access the mycological and histopathological criteria of cure. A mycological response will be achieved after no observation of fungal elements upon direct examination and failure to isolate the causal agent from tissue fragments.
Notes: With LSU rDNA the novel species clusters close to Rhinocladiella aquaspersa and R. phaeophora in the ‘jeanselmei clade’ in Herpotrichiellaceae (Fig 1). Members of this cluster are morphologically outstanding by stiff, erect conidiophores packed with sympodial, non-catenate conidia. With ITS (Fig 3), distances of 5.1% were noted between R. tropicalis and R. aquaspersa, and 5.4% with R. phaeophora (Table 2), underlining that a complex of sibling species is concerned.
Discussion
In this study, we describe two novel species of black fungi causing chromoblastomycosis infections. Among a set of 123 strains from cases of this disease, representatives of three genera were recognized, i.e. Fonsecaea, Rhinocladiella, and Cyphellophora. No member of Cladophialophora was encountered, which is explained by climatic conditions, as C. carrionii is prevalent in arid environmental conditions. In Brazil C. carrionii is rarely reported [24, 25]. The prevalent species in humid tropical climates of South America remains Fonsecaea pedrosoi, followed by F. monophora [2], while the latter species is predominant in southern China [26]. All four species of genus Fonsecaea related to chromoblastomycosis were found in the Brazilian endemic area. Judging from literature data, F. pedrosoi and F. nubica seem to be pathogens that are strictly associated with chromoblastomycosis, while F. monophora and F. pugnacius show some degree of neurotropism eventually leading to dissemination to the brain and other organs [14, 27].
The genus Cyphellophora is characterized by phialides producing sickle-shaped, septate conidia [28]. The group clusters with some phialidic species with small, one-celled conidia. Although the generic type species of Phialophora, P. verrucosa clusters in the ‘carrionii-clade’ distant from Cyphellophora, Feng et al. [22] classified them in Phialophora on morphological criteria. Later Réblová et al. [29] took phylogeny as the leading principle and reclassified all species of the ‘europaea-clade’ in Cyphellophora, the only genus of the newly established family Cyphellophoraceae. Since the taxonomy of Chaetothyriales is in a flux with numerous species to be added, we judge establishment of categories above the species level premature. Currently, phylogenetic data do not match with any other criterion established thus far in Chaetothyriales [21], and therefore phylogenetic genera and families in this order inevitably will remain counterintuitive.
Cyphellophora ludoviensis is not effective to produce conidia on common mycological media; it was placed in the genus based on DNA sequence analyses. Members of the genus Cyphellophora colonize different habitats including, in addition to humans, plant debris, ant nests and abiotic substrates [22]. Our species is genetically close to C. capiguarae, described as living in association with ants (Atta capiguara), and to C. oxyspora from decaying leaves [30, 31]. Other members of Cyphellophora originate from mild cutaneous infections in humans, mostly from skin and nails [22]. Cyphellophora europaea in particular has been encountered globally as an agent of mild skin disease and onychomycosis [31] and was noted co-occurring with dermatophytes, mainly Trichophyton rubrum affecting the skin of diabetic patients [32]. Environmental strains of this species were found in indoor wet cells, such as bathrooms and washing machines [33, 34]. Cyphellophora ludoviensis is the first species in the Cyphellophoraceae that had muriform cells in tissue and showed acanthosis rather than necrosis; on the basis of these features the infection was classified as chromoblastomycosis.
The new species Rhinocladiella tropicalis is a cryptic species close to R. phaeophora and R. aquaspersa. Rhinocladiella phaeophora was known from a single strain recovered from maize field soil in Colombia [35]. It has recently been reported from a case of human chromoblastomycosis, but the sequence of this strain was not available for comparison [36]. This report nevertheless suggests that all members of the cluster consistently are able to cause chromoblastomycosis when inoculated into human skin. Rhinocladiella aquaspersa is a classical agent of chromoblastomycosis, nearly all cases having been reported from the American continent [19], but the majority of historical clinical cases have not been verified by sequence data. Strain CBS 132913 was originally reported as R. aquaspersa but was found to be 100% identical with R. tropicalis. The three species are phenotypically and clinically similar, but their sequence diversity interferes with molecular recognition of R. aquaspersa as a single species. According to Chen et al. [37] the term ‘species complex’ could be applied used to indicate closely related species do not seem to differ in clinically relevant parameters. The present rhinocladiella-like lineages have sufficient molecular distance to be recognized as species, but additional studies of antifungal susceptibility, clinical course, virulence and physiology are needed to verify significance of distinction of the three agents as individual species in hospital routine.
Chromoblastomycosis is clinically highly variable, with six different clinical types [8, 38]. Despite the polymorphic nature of lesions, common factor at the patient side is the absence of necrosis and often even hyper-growth of dermal tissues, which distinguishes the disease from its clinical counterpart, phaeohyphomycosis. At the fungal side, the invasive form is the muriform cell, which is likely to be the cause of the growth-promoting dermal response. As such the disease is polyphyletic within the Chaetothyriales, but has not or extremely rarely been observed outside this order. In both cases caused by these two new species here described it was encountered polymorphic lesions frequently related to clinical cases of chromoblastomycosis [8].
In the State of Maranhão, Brazil, five chaetothyrialean agents of chromoblastomycosis are endemic. A potential source of infection has been suggested [39] to be the harvest of babassu coconuts from a wild palm tree (Orbignya phalerata). A large part of the local rural population is involved in the collection of nuts to extract babassu oil, an important component for local and international beauty product manufacturers. Members of Chaetothyriales are indeed enriched on babassu shell fragments, which are considered a risk factor for developing chromoblastomycosis after trauma sustained at work [39, 40]. A direct link between shells and agents of the disease has however not unambiguously been established. In other Brazilian regions the number of new cases of chromoblastomycosis is decreasing [8, 41, 42]. This is thought to reflect changes in agricultural practices, the extensive use of agricultural antifungals, especially azole derivatives, and the progressive mechanization of plantations resulting in a reduction of the risk of occupational exposure [8, 43, 44].
Our results showed that C. ludoviensis and R. tropicalis had their optimal development at 30 and 27°C, respectively, the maximum growth temperature of all strains analyzed being at 37°C. The chronic nature of the infection corresponds with borderline survival of the fungus in tissue. Epidermal temperatures are usual below 37°C, allowing infection by fungi that barely support this temperature. This agrees with the clinical observation that members of the Cyphellophoraceae (‘europaea-clade’), which have their maximum growth at 36°C, cause only mild, superficial infections, having a very low degree of invasive ability and virulence [22].
According to the World Health Organization (WHO) [45] the Neglected Tropical Diseases include a series of endemic diseases that prevail in tropical or subtropical areas worldwide. The prevalence of causative microbes is linked to poverty and disadvantage. However, fungal diseases were still not included in this list, except mycetoma, another implantation mycosis [46]. Chromoblastomycosis has been reported in the literature as an overlooked disease [11]. Its global burden is comparable to or greater than that of mycetoma. Considering to its global distribution, its impact on the impoverished, and its refractoriness, it also should be considered a true neglected disease as defined by WHO.
Materials and methods
Strains studied
Strains analyzed comprised 123 clinical isolates (Table 1) from different cases and several endemic areas in Brazil, with viable cultures for molecular epidemiology studies and with detailed registration data deposited in medical file systems of the institutions involved in this study. All the clinical strains were deposited at Microbial Collections of Paraná Network- TAXon line at Federal University of Paraná, the register numbers, others nomenclature references and additional information were informed in the Table 1. The Holotype number was provided from Department of Botany Herbarium at Federal University of Paraná (UPCB), TAXon line collections network (http://taxonline.nerdweb.com.br/), register number at http://www.splink.org.br/.
This work was approved by the Research Ethics Committee-CEP-HUUFMA (University Hospital of the Federal University of Maranhão), according to Brazilian Resolution -Approval number: 1.276.342. All samples were anonymized. The set was supplemented with reference strains from the Centraalbureau voor Schimmelcultures (CBS/KNAW) Fungal Biodiversity Centre, Utrecht, Netherlands and the strain previously described as R. aquaspersa CBS 132913 was included in the phylogenetic analysis. Stock cultures were maintained in slants of 2% malt extract agar (MEA) and oatmeal agar (OA) at 24°C. For morphological studies, MEA and potato dextrose agar (PDA) slide cultures were prepared and mounted in aniline blue.
Physiology
Cardinal growth temperatures were determined on 2% MEA. Plates were incubated in the dark for 3 weeks at a 3–36°C temperature range with intervals of 3°C; growth was also recorded at 14, 37, 38, 39 and 40°C. Growth rates per species were obtained by calculation of the average growth of all isolates proven to belong to that species, including the respective standard deviations. Results were plotted with temperature (°C) versus colony diameter (mm) as parameters. Optimum range (= average ± standard deviation) and maximum growth temperatures were determined using the type strains of each species with three replicates averages of three measurements were calculated. Different temperatures were tested, at 28°C was observed the largest number of structures to Chyphellophora and it was used to describe the species morphology.
DNA extraction and amplification
DNA extraction and quality tests were performed using glass beads (Sigma G9143) according to protocols described previously [23] and when it was required, the purification of DNA was undertaken using the UltraClean™ Microbial DNA Kit (MO Bio, Carlsbad, CA, USA) according to manufacturer’s instructions. Colonies were cultivated on Sabouraud’s glucose agar (SGA).
The partial large subunit of the nuclear ribosomal RNA gene (LSU) was amplified using primers NL1 and LR5 [22] for phylogenetic assessment. Three gene regions were chosen for species delimitation: rDNA Internal Transcribed Spacer (ITS), and the partial genes cell division cycle gene (CDC42) and β-tubulin (BT2). ITS amplicons were generated with primers V9G and LS266 [47, 48] and were sequenced with primers ITS1 and ITS4. CDC42 amplification and sequencing was generated with cdc42w and cdc42f [49] and BT2 amplification and sequencing was generated with Bt-2a and T2 [50]. PCR was performed in a 12.5 μL volume of a reaction mixture containing 1× PCR buffer, 2.0 mM MgCl2, 25 μM dNTPs, 0.5 μM of each forward and reverse primers, 1 U of BioTaq DNA polymerase, and 10 ng of genomic DNA.
Amplification was performed in an ABI PRISM 2720 (Applied Biosystems, Foster City, USA) thermocycler as follows: 95°C for 4 min, followed by 35 cycles consisting of 95°C for 45s, 52°C for 30s and 72°C for 2 min, and a delay at 72°C for 7 min. Annealing temperatures were changed to 52°C, 55°C and 58°C for ITS, CDC42 and BT2 respectively. Amplicons were cleaned with Exonuclease I and Shrimp Alkaline Phosphatase (SAP) according to manufacturer’s instructions. Amplicons were sequenced with a BigDye Terminator Cycle Sequencing Kit v. 3.1 (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions, reactions were purified with Sephadex G-50 fine (GE Healthcare Bio-Sciences, Uppsala, Sweden) and sequences were analysed on an ABI Prism 3700 DNA Sequencer (Perkin-Elmer, Norwalk, Foster City, CA, USA).
Phylogenetic analysis
Consensus sequences of the ITS region, BT2, CDC42, and the LSU were visually inspected using MEGA v.7 software [51]. The alignment of obtained sequences was performed using the online MAFFT interface [52]. The genes ITS, CDC42 and BT2 were first analyzed separately and for analysis of multilocus (S1 Fig). We did the LSU analyses to assess the phylogenetic position of the species analyzed in this study. The phylogenetic analyses of the small subunit (SSU) and LSU groups previously recognized in the Herpotrichiellaceae by de Hoog et al. [21], Feng et al. [22] and Vicente et al. [23] were taken as a basis for clade delimitation. Trees were constructed with 100 bootstrap replicates using the Maximum Likelihood Implemented in Mega v. 7 software [51], with the best evolutionary model to this dataset. Conflicts were estimated using the partition homogeneity test available in PAUP* v. 4.Ob10 [53]. To elucidated and explore a more detailed the clustering unnamed species, their sequences were compared to those deposited at GenBank and the CBS-KNAW sequence data sets.
Supporting Information
Constructed with maximum likelihood implemented in MEGA 7.0. Bootstrap values of <80% from 1,000 resampled data sets are shown with branches. Cladophialophora yegresii (CBS 114406, CBS 114407 and CBS 114405) and C. carrionii (CBS 161.54, CBS 406.96, CBS 165.54 and CBS 108.97) comprised the outgroup. Fonsecaea species causing chromoblastomycosis are indicated in red. Type strain in bold.
(EPS)
Acknowledgments
We thank the Bert Gerrits van den Ende from the Centraalbureau voor Schimmelcultures KNAW Fungal Biodiversity Centre, Utrecht, The Netherlands for technical assistance.
Data Availability
All relevant data are within the paper and its Supporting Information files. Nomenclatural novelties and descriptions were deposited in MycoBank (www.MycoBank.org), accession number: MB 817305 and MB 817306. Sequence data are available in GenBank (http://www.ncbi.nlm.nih.gov/genbank/), accession numbers in Table 1.
Funding Statement
This work was supported by Brazilian government by financial support (Special Visiting Researcher Project; grant number 059/2012PVE-CAPES) from the Brazilian Federal Agency for Support and Evaluation of Graduate: Education Coordination for the Improvement of Higher Education Personnel—CAPES (www.capes.gov.br) with fellowship for RRG, GH and GSdH. The authors VAV and CGS were supported by fellowship from National Counsel of Technological and Scientific Development (http://cnpq.br/), Brasilia, Brazil. The author DPA received fellowship from Fundação Amazônia de Amparo a Estudos e Pesquisas- FAPESPA (http://www.fapespa.pa.gov.br) and CGS received financial support from CAPES /PROAMAZONIA grant number 3288/2013. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Deng S, Tsui CKM, Gerrits van den Ende AHG, Yang L, Najafzadeh M J, Badali H, et al. (2015) Global spread of human chromoblastomycosis is driven by recombinant Cladophialophora carrionii and predominantly clonal Fonsecaea species. PLoS Negl Trop Dis 10: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Queiroz-Telles F, Santos DW (2013) Challenges in the therapy of chromoblastomycosis. Mycopathologia. 175: 477–488. 10.1007/s11046-013-9648-x [DOI] [PubMed] [Google Scholar]
- 3.Silva JP, de Souza W, Rozental S (1998) Chromoblastomycosis: a retrospective study of 325 cases on Amazonic Region (Brazil). Mycopathologia 143:171–175. [DOI] [PubMed] [Google Scholar]
- 4.Bayles MA (1986) Chromomycosis In Hay RJ (ed.), Baillie`re’s Clinical Tropical Medicine and Comunicable Diseases. Tropical Fungal Infections. London: WB Saunders, 1986: 45–70. [Google Scholar]
- 5.Esterre P, Pecarrère JL, Raharisolo C, Huerre M (1999) Squamous cell carcinoma arising from chromomycosis. Report of two cases. Ann Pathol 19:516–520. [PubMed] [Google Scholar]
- 6.Bonifaz A, Carrasco-Gerard E, Saul A (2001) Chromoblastomycosis: clinical and mycologic experience of 51 cases. Mycoses 44:1–7. [DOI] [PubMed] [Google Scholar]
- 7.Attapattu MC (1997). Chromoblastomycosis a clinical and mycological study of 71 cases from Sri Lanka. Mycopathologia 137:145–151. [DOI] [PubMed] [Google Scholar]
- 8.Queiroz-Telles F, Santos DW, Salgado C, Vicente VA, Bonifaz AB, Roilides E, et al. (2016). Submitted at Clin Microbiol Rev. control number CMR00032-16.
- 9.Martínez LR, Tovar MLJ (2007) Chromoblastomycosis. Clin Dermatol 25:188–194. 10.1016/j.clindermatol.2006.05.007 [DOI] [PubMed] [Google Scholar]
- 10.Torres-Guerrero E, Isa-Isa R, Isa M, Arenas R (2012) Chromoblastomycosis. Clin Dermatol 30:403–408. 10.1016/j.clindermatol.2011.09.011 [DOI] [PubMed] [Google Scholar]
- 11.Queiroz-Telles F (2015) Chromoblastomycosis: a neglected global disease. Rev Inst Med Trop Sao Paulo. 57: 46–50. 10.1590/S0036-46652015000700009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elder DE, Elenitsas R, Johnson BL Jr, Murphy GF, Xu X (2009) In: Lever's histopathology of the skin 10th ed. Philadelphia: Lippincott-Raven; 2009. pp. 607–608. [Google Scholar]
- 13.Salgado CG, Silva MB, Yamano SSP, Salgado UI, Diniz JAP, Silva JP (2009) Cutaneous localized annular chromoblastomycosis. Journal of Cutaneous Pathology 36:257–261. 10.1111/j.1600-0560.2008.01025.x [DOI] [PubMed] [Google Scholar]
- 14.Najafzadeh MJ, Gueidan C, Badali H, Gerrits van den Ende AHG, Xi L, de Hoog GS (2009) Genetic diversity and species delimitation in the opportunistic genus Fonsecaea. Med Mycol 47:17–25. 10.1080/13693780802527178 [DOI] [PubMed] [Google Scholar]
- 15.Najafzadeh MJ, Sun J, Vicente VA, Xi L, Gerrits van den Ende AHG, de Hoog GS (2010) Fonsecaea nubica sp. nov., a new agent of human chromoblastomycosis revealed using molecular data. Med Mycol 48:800–806. 10.3109/13693780903503081 [DOI] [PubMed] [Google Scholar]
- 16.de Hoog GS, Zeng JS, Harrak MJ, Sutton DA (2006) Exophiala xenobiotica sp. nov. an opportunistic black yeast inhabiting environments rich in hydrocarbons. Antonie van Leeuwenhoek 90:257–268. 10.1007/s10482-006-9080-z [DOI] [PubMed] [Google Scholar]
- 17.Badali H, Bonifaz A, Barron-Tapia T, Vázquez-González D, Estrada-Aguilar L, Oliveira NM, Sobral Filho JF, et al. (2010) Rhinocladiella aquaspersa, proven agent of verrucous skin infection and a novel type of chromoblastomycosis. Med Mycol 48:696–703. 10.3109/13693780903471073 [DOI] [PubMed] [Google Scholar]
- 18.Azevedo CMPS Marques SG, Santos DWCL Silva RR, Silva NF Santos DA, et al. (2015) Squamous cell carcinoma derived from chronic chromoblastomycosis in Brazil. Clin Infect Dis 60: 1500–1504. 10.1093/cid/civ104 [DOI] [PubMed] [Google Scholar]
- 19.González GM, Rojas OC, González JG, Kang Y, de Hoog GS (2013) Chromoblastomycosis caused by Rhinocladiella aquaspersa. Med Mycol Case Rep 2:148–151. 10.1016/j.mmcr.2013.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hofmann H, Choi SM, Wilsmann-Theis D, Horré R, Bieber T, Hoog GS (2005) Phialophora verrucosa causing invasive chromoblastomycosis and sinusitis in a child from northern Africa. Mycoses 48: 456–461. 10.1111/j.1439-0507.2005.01150.x [DOI] [PubMed] [Google Scholar]
- 21.de Hoog GS, Vicente VA, Najafzadeh MJ, Harrak MJ, Seyedmousavi S (2011) Waterborne Exophiala species causing disease in coldblooded animals. Persoonia 27:46–72. 10.3767/003158511X614258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng P, Lu Q, Najafzadeh MJ, Gerrits van den Ende AHG, Sun J, Li R, et al. (2012) Cyphellophora and its relatives in Phialophora: biodiversity and possible role in human infection. Fungal Divers 65:17–45. [Google Scholar]
- 23.Vicente VA, Najafzadeh MJ, Sun J, Gomes RR, Robl D, Marques SG, et al. (2013) Environmental siblings of black agents of human chromoblastomycosis. Fungal Divers 62:1–17. [Google Scholar]
- 24.Mouchalouat MF, Galhardo MCG, Fialho PCM, Coelho JMCO, Zancopé-Oliveira RM, Valle ACF (2008) Cladophialophora carrionii: a rare agent of chromoblastomycosis in Rio de Janeiro State, Brazil. Rev Inst Med trop S. Paulo 50:351–353. [DOI] [PubMed] [Google Scholar]
- 25.Bona E, Canton LM, Fuentefria AM (2010) Chromoblastomycosis in Santa Catarina state, Brazil. Rev Cubana de Med Trop 62:254–256. http://hdl.handle.net/10183/63175. [PubMed] [Google Scholar]
- 26.Xi L, Sun J, Lu C, Liu H, Xie Z, Fukushima K, et al. (2009) Molecular diversity of Fonsecaea (Chaetothyriales) causing chromoblastomycosis in southern China. Med Mycol 47:27–33. 10.1080/13693780802468209 [DOI] [PubMed] [Google Scholar]
- 27.Azevedo CMPS Gomes RR, Vicente VA Santos DWCL, Marques SG Nascimento MMF, et al. (2015) Fonsecaea pugnacius, a Novel Agent of Disseminated Chromoblastomycosis. Journal of Clinical Microbiology 53:2674–2685. 10.1128/JCM.00637-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Hoog GS, Guarro J, Gené J, Figueras MJ (2000) Atlas of clinical fungi, 2nd ed. Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands: and Universitat Rovira i Virgili, Reus, Spain. 1126 p. [Google Scholar]
- 29.Réblová M, Untereiner WA, Réblová K (2013) Novel Evolutionary Lineages Revealed in the Chaetothyriales (Fungi) Based on Multigene Phylogenetic Analyses and Comparison of ITS Secondary Structure. Plos One. 8:1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Attili-Angelis D, Duarte APM, Pagnocca FC, Nagamoto NS, Vries M, et al. (2014) Novel Phialophora species from leaf-cutting ants (tribe Attini). Fungal Divers 65: 65–75. [Google Scholar]
- 31.Gao L, Ma Y, Zhao W, Wei Z, Gleason ML, Chen H, et al. (2015) Three New Species of Cyphellophora (Chaetothyriales) Associated with Sooty Blotch and Flyspeck. PLoS ONE 10(9): e0136857 10.1371/journal.pone.0136857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eckhard M, Lengler A, Liersch J, Bretzel RG, Mayser P (2007) Fungal foot infections in patients with diabetes mellitus-Results of two independent investigations. Mycoses. 50S14–19. [DOI] [PubMed] [Google Scholar]
- 33.Lian X, de Hoog GS (2010) Indoor wet cells harbour melanized agents of cutaneous infection. Med Mycol 48:622–628. 10.3109/13693780903405774 [DOI] [PubMed] [Google Scholar]
- 34.Hamada N, Abe N (2010) Comparison of fungi found in bathrooms and sinks. Biocontrol Sci. 15:51–56. [DOI] [PubMed] [Google Scholar]
- 35.Veerkamp J, Gams W (1983) Los hongos de Colombia—VIII: Some new species of soil fungi from Colombia. Caldasia. 13:709717. [Google Scholar]
- 36.Kampirapap K, Reangchainam S, Ornpaew P, Tresukosol P (2015) Chromoblastomycosis masquerading as dermatophytosis, with the description of a new opportunistic species. Southeast Asian J Trop Med Public Health 46:105–109. [PubMed] [Google Scholar]
- 37.Chen M, Zeng J, De Hoog GS, Stielow B, Gerrits Van Den Ende AH, Liao W, Lackner M (2015) The 'species complex' issue in clinically relevant fungi: A case study in Scedosporium apiospermum. Fungal Biol 120(2):137–46. 10.1016/j.funbio.2015.09.003 [DOI] [PubMed] [Google Scholar]
- 38.Badali H, Gueidan C, Najafzadeh MJ, Bonifaz A, Gerrits van den Ende AHG, de Hoog GS (2008) Biodiversity of the genus Cladophialophora. Stud Mycol 61:175–191. 10.3114/sim.2008.61.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marques SG, Silva SMP, Saldanha PC, Rezende PC, Vicente MA, Queiroz-Telles F, et al. (2006) Isolation of Fonsecaea pedrosoi from the shell of Babassu coconut (Orbignya phalerata Martius) in the Amazon Region of Maranhao, Brazil. Jpn J Med Mycol 47: 305–311. [DOI] [PubMed] [Google Scholar]
- 40.Vicente VA, Attili-Angelis D, Pie MR, Queiroz-Telles F, Cruz LM, Najafzadeh MJ, et al. (2008) Environmental isolation of black yeast-like fungi involved in human infection. Stud Mycol 61:137–144. 10.3114/sim.2008.61.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Londero AT, Ramos CD (1976) Chromomycosis: a clinical and mycologic study of thirty-five cases observed in the hinterland of Rio Grande do Sul, Brazil. Am J Trop Med Hyg 25:132–135. [DOI] [PubMed] [Google Scholar]
- 42.Minotto R, Bernardi CD, Mallmann LF, Edelweiss MI, Scroferneker ML (2001) Chromoblastomycosis: a review of 100 cases in the state of Rio Grande do Sul, Brazil. J Am Acad Dermatol. 44:585–492. 10.1067/mjd.2001.112220 [DOI] [PubMed] [Google Scholar]
- 43.Ono MA, Itano EN, Mizuno LT, Mizuno EH, Camargo ZP (2002) Inhibition of Paracoccidioides brasiliensis by pesticides: is this a partial explanation for the difficulty in isolating this fungus from the soil? Med Mycol 40:493–499. [DOI] [PubMed] [Google Scholar]
- 44.Queiroz-Telles F (2008) Influence of alternating coffee and sugar cane agriculture in the incidence of paracoccidioidomycosis in Brazil. Biomedica 28: (Suppl 1):129. [Google Scholar]
- 45.World Health Organization (2010) Working to overcome the global impact of neglected tropical diseases First report on neglected tropical diseases. Geneva, Switzerland: World Heath Organization; Available from: http://apps.who.int/iris/bitstream/10665/44440/1/9789241564090_eng.pdf [Google Scholar]
- 46.van de Sande WWJ, Maghoub ES, Fahal AH, Goodfellow M, Welsh O, et al. (2014) The mycetoma knowledge gap: identification of research priorities. PLoS Negl Trop Dis 8: e2667 10.1371/journal.pntd.0002667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Hoog GS, Gerrits van den Ende AHG (1998) Molecular diagnostics of clinical strains of filamentous basidiomycetes. Mycoses 41:183–189. [DOI] [PubMed] [Google Scholar]
- 48.Masclaux F, Guého E, de Hoog GS, Christen R (1995) Phylogenetic relationships of human-pathogenic Cladosporium (Xylohypha) species inferred from partial LS rRNA sequences. J Med Vet Mycol 33:327–338. [DOI] [PubMed] [Google Scholar]
- 49.Sun J, Najafzadeh MJ, Gerrits van den Ende AH, Vicente VA, Feng P, Xi L, et al. (2012) Molecular characterization of pathogenic members of the genus Fonsecaea using multilocus analysis. Plos One 7:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Glass N, Donaldson G (1995) Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous Ascomycetes. Appl Environ Microbiol 61:1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 22; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Katoh K, Toh H (2008) Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform 9:286–298. 10.1093/bib/bbn013 [DOI] [PubMed] [Google Scholar]
- 53.Swofford D (2003) PAUP*. Phylogenetic Analysis using parsimony. Version 4 Sinauer Associates, Sunderland, Massachusetts: http://paup.csit.fsu.edu/about.html. Acessed 20 April 2016. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Constructed with maximum likelihood implemented in MEGA 7.0. Bootstrap values of <80% from 1,000 resampled data sets are shown with branches. Cladophialophora yegresii (CBS 114406, CBS 114407 and CBS 114405) and C. carrionii (CBS 161.54, CBS 406.96, CBS 165.54 and CBS 108.97) comprised the outgroup. Fonsecaea species causing chromoblastomycosis are indicated in red. Type strain in bold.
(EPS)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. Nomenclatural novelties and descriptions were deposited in MycoBank (www.MycoBank.org), accession number: MB 817305 and MB 817306. Sequence data are available in GenBank (http://www.ncbi.nlm.nih.gov/genbank/), accession numbers in Table 1.