Abstract
Zika virus (ZIKV) is a mosquito-borne virus (arbovirus) in the family Flaviviridae, and the symptoms caused by ZIKV infection in humans include rash, fever, arthralgia, myalgia, asthenia and conjunctivitis. Codon usage bias analysis can reveal much about the molecular evolution and host adaption of ZIKV. To gain insight into the evolutionary characteristics of ZIKV, we performed a comprehensive analysis on the codon usage pattern in 46 ZIKV strains by calculating the effective number of codons (ENc), codon adaptation index (CAI), relative synonymous codon usage (RSCU), and other indicators. The results indicate that the codon usage bias of ZIKV is relatively low. Several lines of evidence support the hypothesis that translational selection plays a role in shaping the codon usage pattern of ZIKV. The results from a correspondence analysis (CA) indicate that other factors, such as base composition, aromaticity, and hydrophobicity may also be involved in shaping the codon usage pattern of ZIKV. Additionally, the results from a comparative analysis of RSCU between ZIKV and its hosts suggest that ZIKV tends to evolve codon usage patterns that are comparable to those of its hosts. Moreover, selection pressure from Homo sapiens on the ZIKV RSCU patterns was found to be dominant compared with that from Aedes aegypti and Aedes albopictus. Taken together, both natural translational selection and mutation pressure are important for shaping the codon usage pattern of ZIKV. Our findings contribute to understanding the evolution of ZIKV and its adaption to its hosts.
Introduction
Zika virus (ZIKV) is classified as a mosquito-borne arbovirus of the family Flaviviridae, genus Flavivirus [1]. This virus was first isolated from a blood sample of a Rhesus monkey in Uganda in 1947 and, before its outbreak in Oceania in 2007, it was confined to Africa and Southeast Asia [1]. Since then, ZIKV has been circulating in the Americas, and in May 2015, the first case of ZIKV originating from the Americas was reported in Brazil. Thus far, ZIKV has expanded from South America to more than 28 countries and has aroused the attention of the World Health Organization (WHO) as well as that of many governments [2, 3]. Clinical presentation of ZIKV fever is non-specific; the most common symptoms are rash, fever, arthralgia, myalgia, asthenia, and conjunctivitis. ZIKV is thought to be transmitted to humans mainly by Aedes aegypti and Aedes albopictus. The genome of ZIKV is a 10794-bp linear single strand of RNA that contains a large open-reading frame (ORF) encoding a polyprotein, which can be spliced into capsid protein (C), pro-envelope protein (prM), envelop protein (E), and seven nonstructural (NS) proteins [4]. The ZIKV genome has been detected in the blood, saliva, urine, amniotic liquid, and tissue samples [5–7]. Although ZIKV infection is often asymptomatic, symptomatic infections have also been described, and these patients usually report mild symptoms [2]. Importantly, ZIKV infection in pregnant women may lead to the fetal malformation. Specifically, ZIKV has been linked with the occurrence of microcephaly in the babies [8]. Currently, there is no effective medicine or vaccine against this virus.
All amino acids, except methionine (Met) and tryptophane (Trp), are coded by more than one synonymous codon. The alternative synonymous codons do not occur equally; they instead follow a special codon usage pattern, a phenomenon termed codon usage bias [9]. Several factors are known to contribute to codon usage bias, such as mutational bias, translational and transcriptional selection, protein structure, tRNA abundance, RNA stability, GC content, gene expression level, and gene length [10–12]. The codon usage bias is regarded as a consequence of the balance between the mutation and translational selection [13]. Analysis of codon usage bias can provide useful insights into the molecular evolution of species and their genes.
The complete genome sequencing of ZIKV has been completed [14, 15]. However, the extensive studies on the codon usage bias of ZIKV are rare. Here, we analyzed the codon usage bias of the ZIKV polyprotein-coding region and explored factors that might be related to this codon usage bias. A comprehensive analysis of ZIKV codon usage bias will be important for understanding its molecular evolution, and will provide clues for its prevention and treatment.
Materials and methods
Ethics Statement
This article does not contain any studies with human participants or that were performed on animals.
Sequence data
The 46 available nucleotide sequences of ZIKV polyprotein-coding regions (Table 1) were downloaded from the NCBI GenBank database (http://www.ncbi.nlm.nih.gov/). Detailed information about these ZIKV strains is listed in Table 1.
Table 1. The accession number, origin and isolation year, and indices of polyprotein-coding region of each ZIKV isolate.
| Accession | GC3sc | GCd | Mononucleotide frequency [%] | GC12k | GC3l | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No | number | Species | CAIa | ENcb | [%] | [%] | U3se | C3sf | A3sg | G3sh | Gravyi | Aromoj | [%] | [%] |
| 1 | LC002520 | Uganda(1947) | 0.737 | 52.94 | 50.80 | 50.70 | 24.97 | 31.64 | 34.21 | 31.23 | -0.140 | 0.080 | 49.07 | 53.94 |
| 2 | KF383116 | Senegal(1968) | 0.739 | 52.58 | 51.20 | 51.00 | 24.78 | 31.71 | 33.85 | 31.73 | -0.142 | 0.079 | 49.22 | 54.38 |
| 3 | EU545988 | Micronesia(1968) | 0.735 | 53.64 | 51.80 | 51.00 | 25.28 | 31.93 | 32.65 | 32.28 | -0.148 | 0.081 | 49.11 | 54.88 |
| 4 | KJ776791 | Polynesia(2013) | 0.735 | 53.36 | 52.00 | 51.10 | 24.84 | 32.19 | 32.85 | 32.27 | -0.147 | 0.081 | 49.11 | 55.08 |
| 5 | KU312312 | Suriname(2015) | 0.735 | 53.40 | 52.00 | 51.10 | 24.79 | 32.24 | 32.86 | 32.24 | -0.147 | 0.081 | 49.15 | 55.08 |
| 6 | KU321639 | Brazil(2015) | 0.736 | 53.28 | 52.00 | 51.10 | 24.85 | 32.24 | 32.82 | 32.23 | -0.148 | 0.081 | 49.11 | 55.11 |
| 7 | KF383119 | Senegal(2001) | 0.738 | 52.92 | 50.80 | 50.90 | 25.00 | 31.70 | 34.07 | 31.19 | -0.139 | 0.079 | 49.24 | 54.03 |
| 8 | KF383118 | Senegal(2001) | 0.741 | 52.67 | 50.80 | 50.80 | 24.99 | 31.52 | 34.12 | 31.32 | -0.151 | 0.078 | 49.14 | 54.03 |
| 9 | KF383115 | CAF m(1968) | 0.738 | 52.56 | 50.00 | 50.50 | 25.68 | 30.97 | 34.42 | 31.03 | -0.140 | 0.080 | 48.98 | 53.13 |
| 10 | KF268949 | CAF(1980) | 0.737 | 52.69 | 49.60 | 50.40 | 25.74 | 31.09 | 34.90 | 30.34 | -0.142 | 0.080 | 49.08 | 52.87 |
| 11 | KF268948 | CAF(1979) | 0.739 | 52.64 | 49.90 | 50.40 | 25.67 | 31.24 | 34.59 | 30.57 | -0.141 | 0.080 | 49.07 | 53.17 |
| 12 | KF268950 | CAF(1976) | 0.739 | 52.67 | 49.90 | 50.40 | 25.67 | 31.24 | 34.61 | 30.55 | -0.140 | 0.080 | 49.07 | 53.14 |
| 13 | KF383117 | Senegal(1997) | 0.735 | 52.77 | 50.30 | 50.60 | 25.66 | 30.72 | 34.03 | 31.68 | -0.141 | 0.080 | 49.04 | 53.42 |
| 14 | KU509998 | Haiti(2014) | 0.736 | 53.30 | 52.00 | 51.10 | 24.85 | 32.24 | 32.82 | 32.22 | -0.148 | 0.081 | 49.12 | 55.11 |
| 15 | KU501217 | Guatemala(2015) | 0.735 | 53.15 | 51.90 | 51.10 | 24.82 | 32.23 | 33.07 | 32.03 | -0.144 | 0.081 | 49.11 | 54.91 |
| 16 | KU365777 | Brazil(2015) | 0.735 | 53.27 | 51.90 | 51.10 | 24.83 | 32.22 | 32.95 | 32.14 | -0.147 | 0.081 | 49.14 | 54.99 |
| 17 | KU501216 | Guatemala(2015) | 0.735 | 53.18 | 51.80 | 51.00 | 24.85 | 32.20 | 33.07 | 32.03 | -0.144 | 0.081 | 49.11 | 54.88 |
| 18 | KU501215 | PRn (2015) | 0.735 | 53.25 | 52.10 | 51.10 | 24.65 | 32.40 | 32.95 | 32.12 | -0.147 | 0.081 | 49.12 | 55.14 |
| 19 | KU365780 | Brazil(2015) | 0.735 | 53.27 | 51.90 | 51.10 | 24.87 | 32.18 | 32.95 | 32.14 | -0.147 | 0.081 | 49.14 | 54.96 |
| 20 | KU365779 | Brazil(2015) | 0.735 | 53.29 | 51.90 | 51.10 | 24.87 | 32.18 | 32.95 | 32.14 | -0.147 | 0.081 | 49.12 | 54.96 |
| 21 | KU365778 | Brazil(2015) | 0.735 | 53.34 | 51.90 | 51.10 | 24.83 | 32.22 | 33.03 | 32.05 | -0.147 | 0.081 | 49.14 | 54.94 |
| 22 | KU647676 | Martinique(2015) | 0.735 | 53.21 | 51.90 | 51.10 | 24.82 | 32.24 | 33.07 | 32.00 | -0.146 | 0.081 | 49.12 | 54.94 |
| 23 | KU866423 | China(2016) | 0.735 | 53.32 | 52.00 | 51.10 | 24.70 | 32.27 | 32.96 | 32.24 | -0.147 | 0.081 | 49.14 | 55.11 |
| 24 | KU870645 | USA(2016) | 0.734 | 53.29 | 51.90 | 51.10 | 24.89 | 32.12 | 32.96 | 32.16 | -0.142 | 0.080 | 49.14 | 54.91 |
| 25 | KU926310 | Brazil(2016) | 0.735 | 53.28 | 52.00 | 51.10 | 24.72 | 32.32 | 32.98 | 32.12 | -0.147 | 0.081 | 49.12 | 55.05 |
| 26 | KU926309 | Brazil(2016) | 0.736 | 53.22 | 52.10 | 51.20 | 24.67 | 32.31 | 32.86 | 32.30 | -0.146 | 0.081 | 49.12 | 55.17 |
| 27 | KU922960 | Mexico(2016) | 0.736 | 53.21 | 51.90 | 51.10 | 24.83 | 32.23 | 33.05 | 32.03 | -0.147 | 0.081 | 49.10 | 54.94 |
| 28 | KU820898 | China(2016) | 0.736 | 53.27 | 51.90 | 51.10 | 24.73 | 32.27 | 33.09 | 32.04 | -0.146 | 0.081 | 49.14 | 54.96 |
| 29 | KU853013 | Italy(2016) | 0.736 | 53.23 | 52.00 | 51.10 | 24.72 | 32.33 | 33.04 | 32.01 | -0.146 | 0.081 | 49.17 | 54.99 |
| 30 | KU853012 | Italy(2016) | 0.736 | 53.27 | 52.00 | 51.10 | 24.74 | 32.35 | 32.99 | 32.04 | -0.147 | 0.081 | 49.15 | 55.02 |
| 31 | KU955591 | Senegal(1984) | 0.740 | 52.67 | 51.20 | 51.00 | 24.91 | 31.58 | 33.73 | 31.91 | -0.143 | 0.080 | 49.21 | 54.41 |
| 32 | KU955592 | Senegal(1984) | 0.740 | 52.67 | 51.20 | 50.90 | 24.94 | 31.54 | 33.69 | 31.96 | -0.143 | 0.080 | 49.19 | 54.41 |
| 33 | KU955593 | Cambodia(2010) | 0.735 | 53.53 | 51.60 | 51.00 | 25.40 | 31.72 | 32.74 | 32.31 | -0.148 | 0.081 | 49.17 | 54.73 |
| 34 | KU955594 | Uganda(1947) | 0.738 | 52.89 | 50.90 | 50.80 | 24.79 | 31.80 | 34.20 | 31.25 | -0.139 | 0.080 | 49.09 | 54.09 |
| 35 | KU955595 | Senegal(1984) | 0.740 | 52.68 | 51.30 | 51.00 | 24.87 | 31.61 | 33.69 | 31.96 | -0.143 | 0.079 | 49.19 | 54.47 |
| 36 | KX056898 | China(2016) | 0.735 | 53.27 | 51.70 | 51.10 | 24.94 | 32.06 | 33.08 | 32.05 | -0.147 | 0.081 | 49.17 | 54.82 |
| 37 | KU681082 | Philippines(2012) | 0.735 | 53.55 | 51.50 | 51.00 | 25.29 | 31.69 | 33.04 | 32.07 | -0.149 | 0.081 | 49.14 | 54.59 |
| 38 | KU681081 | Thailand(2014) | 0.735 | 53.56 | 51.90 | 51.10 | 24.77 | 32.34 | 33.00 | 31.96 | -0.146 | 0.081 | 49.17 | 55.02 |
| 39 | KU761564 | China(2016) | 0.736 | 53.25 | 51.90 | 51.10 | 24.76 | 32.26 | 33.03 | 32.09 | -0.146 | 0.081 | 49.14 | 54.99 |
| 40 | KU729218 | Brazil(2015) | 0.735 | 53.33 | 52.00 | 51.10 | 24.93 | 32.09 | 32.81 | 32.31 | -0.148 | 0.081 | 49.14 | 54.99 |
| 41 | KU744693 | China(2016) | 0.737 | 53.29 | 52.10 | 51.20 | 24.77 | 32.43 | 32.75 | 32.07 | -0.147 | 0.081 | 49.24 | 55.20 |
| 42 | KU729217 | Brazil(2015) | 0.736 | 53.21 | 52.00 | 51.10 | 24.71 | 32.31 | 32.99 | 32.16 | -0.146 | 0.081 | 49.15 | 55.02 |
| 43 | KU720415 | Uganda(1947) | 0.738 | 52.91 | 50.90 | 50.70 | 24.86 | 31.75 | 34.21 | 31.23 | -0.140 | 0.080 | 49.07 | 54.03 |
| 44 | KU497555 | Brazil(2015) | 0.736 | 53.26 | 52.00 | 51.10 | 24.68 | 32.32 | 33.04 | 32.11 | -0.145 | 0.081 | 49.11 | 55.05 |
| 45 | KU707826 | Brazil(2015) | 0.735 | 53.29 | 51.90 | 51.10 | 24.87 | 32.18 | 32.95 | 32.14 | -0.147 | 0.081 | 49.12 | 54.96 |
| 46 | KU527068 | Brazil(2015) | 0.735 | 53.38 | 52.00 | 51.10 | 24.75 | 32.28 | 32.94 | 32.18 | -0.144 | 0.081 | 49.13 | 55.05 |
a CAI represents codon adaptation index.
b ENc represents the effective number of codons.
c GC3s represents the frequency of the nucleotides G+C at the third positions of synonymous codons.
d GC represents the G+C content.
e U3s represents the frequency of the nucleotide U at the third positions of codons.
f C3s represent the frequency of the nucleotide C at the third positions of codons.
g A3s represents the frequency of the nucleotide A at the third positions of codons.
h G3s represents the frequency of the nucleotide G at the third positions of codons.
i Gravy represents the hydrophobicity of protein.
j Aromo represents the aromaticity of protein.
k GC12 represents the G+C content at the first and second positions of codons.
l GC3 represents the G+C content at the third positions of codons.
m represents Central African Republic.
n represents Puerto Rico.
Phylogenetic analysis
A phylogenetic tree was drawn based on the ZIKV polyprotein-coding region using the neighbor-joining (NJ) method with a bootstrap value of 1000 replicates on MEGA6 software [16].
Analysis of codon usage in the ZIKV polyprotein-coding region
We calculated several indicators to analyze the codon usage of the ZIKV polyprotein-coding region. The codon adaptation index (CAI) [17] was calculated by EMBOSS CAI program using human data set as a reference [18]. The relative synonymous codon usage (RSCU) [19], effective number of codons (ENc) [20], GC content, and GC3s content, hydrophobicity (GRAVY), and aromaticity (AROMO) were calculated using the CodonW 1.4.2 program (http://codonw.sourceforge.net/).
The value of relative abundance of dinucleotides (RAD) in the polyprotein-coding regions of ZIKV was calculated by computing the relevant odds ratio described by Chris Burge [21, 22]. The odds ratio , where f x denotes the frequency of the nucleotide X, f y denotes the frequency of the nucleotide Y, and f xy the frequency of the dinucleotide XY. As a criterion, If ρxy >1.23 or <0.78, the XY dinucleotide is considered to be over-represented or under-represented compared with a random association of mononucleotides [22].
Comparison between the codon usage pattern in ZIKV and those in its hosts
RSCU
RSCU was employed to investigate the overall synonymous codon usage bias among the genes, and this value was defined as the ratio of the observed codon usage to the expected value [23]. Codons with a RSCU value of >1.6 were regarded as over-represented, while codons with a RSCU value of <0.6 were regarded as under-represented. Codons used at an average level (no bias) have the RSCU values of 1 [24]. In our comparison of the ZIKV codon usage pattern with those of its hosts, if the RSCU value for the polyprotein-coding region of ZIKV and that of the same codon for the host were both <0.6, >1.6, or between 0.6 and 1.6, their codon usage patterns were judged to be similar [25]. The codon usage data of ZIKV’s hosts, including human (Homo sapiens) and mosquitoes (A. aegypti and A. albopictus) was retrieved from the codon usage database (http://www.kazusa.or.jp/codon).
D(A,B)
To determine the influence of the overall codon usage of hosts on that of ZIKV, the similarity index D(A,B) [26] was calculated as follows:
where R(A,B) is termed as a cosine value of an included angle between A and B spatial vectors and represents the extent of similarity between ZIKV and hosts in the aspect of overall codon usage pattern. ai is defined as the RSCU value for a specific codon among 59 synonymous codons in ZIKV polyprotein-coding region. bi is defined as the RSCU value for the same codon of ZIKV’s hosts. D(A,B) represents the potential effect of the overall codon usage of the hosts on that of ZIKV, and its value varies from 0 to 1 [26]. The higher D(A,B) means the stronger influence of environment related synonymous codon usage patterns of hosts on that of ZIKV.
tRNA adaptation index
tRNA adaptation index (tAI) is used to estimate tRNA usage for the coding sequences of a species [27]. It represents the levels of co-adaption between a special codon and a corresponding tRNA pool and has greater correlations with protein abundance compared with other indicators [28]. The tAI value of ZIKV polyprotein-coding region based on the tRNA copy number of H. sapiens was calculated by Visual Gene Developer [29].
Effect of mutation pressure and translational selection on the codon usage bias
ENc-GC3s plot
A ENc-GC3s plot was used to investigate the influence of the GC3s content on codon usage [20]. The expected ENc values for each GC3s were calculated using the following formula:
where s represents the GC3s value.
Parity rule 2 (PR2) plot
A Parity rule 2 (PR2) plot was used to assess the influence of mutation pressure and translational selection on the codon usage of genes [30]. This plot is shown by the value of AU-bias [A3/(A3+U3)] as the ordinate and GC-bias [G3/(G3+C3)] as the abscissa at the third codon position of the four-codon amino acids. The center of the plot, where both coordinates are 0.5, is the position where A = U and G = C (PR2), with no bias between influence of mutation and translational selection rates.
Neutrality plot (GC12 Vs GC3)
Analysis of the correlation between the GC contents at the first and second codon positions (GC12) and that at the third codon position (GC3) is useful to examine the effect of mutation pressure and translational selection on the base composition [31]. Therefore, GC12 and GC3 were calculated by using the EMBOSS CUSP program [32] and then subjected to correlation analysis.
Correspondence analysis (CA)
Correspondence analysis (CA) is a useful multivariate statistical method for studying the internal relationship between variables and samples [33]. The mathematics procedure of CA transforms the RSCU values into a series of dimensional factors, and the results can be used to analyze the major trend in codon usage patterns among different samples. Each gene is represented with 59 dimensional variables, and each dimension matches the RSCU value of one codon, with the exclusions of AUG, UGG and stop codons. The CA was performed using the CodonW 1.4.2 program. The first two axes of CA (Axis 1 and Axis 2) were subjected to a correlation analysis.
Statistical analysis
The statistical analyses were performed using SPSS 13.0 software package (SPSS Inc., Chicago, USA). Correlation analyses were carried out using Spearman’s rank correlation test. The P-values less than 0.05 were considered statistically significant.
Results and Discussion
Phylogenetic analysis of ZIKV based on polyprotein-coding region
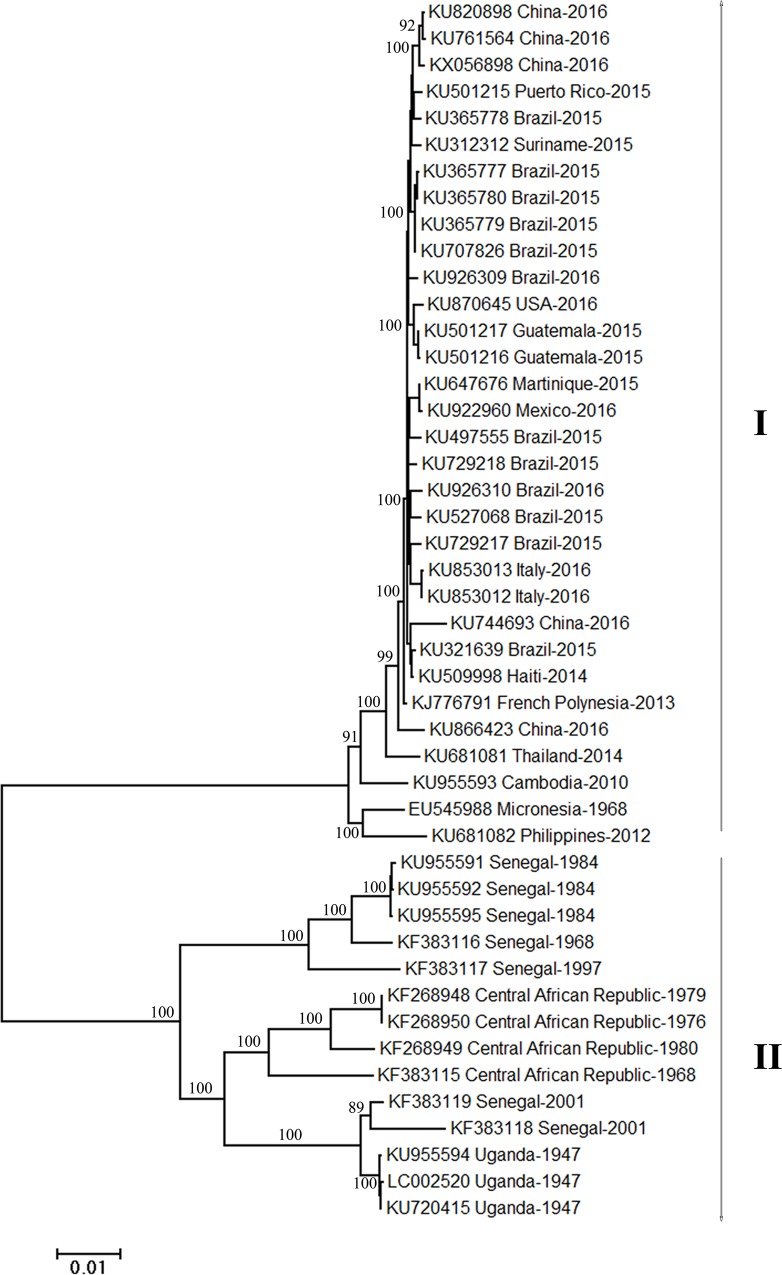
To determine the phylogenetic relationship of different ZIKV strains, a phylogenetic tree was drawn (Fig 1). The results show that 46 strains of ZIKV can be divided into two genera (I, II) and the strains isolated from the same geographic regions cluster together (Fig 1). It can be seen that the members isolated from Africa, including Senegal, Central African Republic and Uganda, firstly cluster together and form a separate branch, and subsequently cluster with the members isolated from other countries all over the world (Fig 1).
Fig 1. Phylogenetic tree based on the polyprotein-coding regions of 46 ZIKV strains.
The tree was generated by the neighbor-joining (NJ) method using MEGA6 software. The bootstraps values were calculated with 1000 replicates.
Nucleotide composition analysis
The GC3s content is a useful indicator of the extent of the base composition bias, representing the frequency of the nucleotides G+C at the synonymous third codon position, excluding Met, Trp, and the termination codons. The mean value of the GC contents in the 46 tested strains of ZIKV is 50.98% (50.40–51.20%; SD, 0.216), while the average value of their GC3s contents is 51.53 (49.60–52.10%; SD, 0.685) (Table 1).
An analysis of nucleotide composition at the third position of synonymous codons (G3s, A3s, U3s, C3s) indicates that the mean values of C3s (31.97%) and A3s (33.32%) are higher compared with those of G3s (31.87%) and U3s (24.95%) in ZIKV polyprotein-coding region (Table 1). Moreover, it was found that the G and A nucleotides are abundant with mean values of 29.16% and 27.50%, respectively, while the average values of U and C nucleotide were 21.52% and 21.82%, respectively (data not shown in Tables). The G and A contents are significantly higher compared with U and C contents (Student’s t test, p<0.01). These results highlight that there is a GA-rich composition in ZIKV polyprotein-coding region.
The synonymous codon usage characteristics of the ZIKV polyprotein-coding region
ENc was used to quantify the codon usage bias of each gene [20]. ENc values can range from 20 to 61, and lower values of ENc represent higher levels of codon usage bias. To measure whether or not ZIKV strains show similar codon usage biases, the ENc values of 46 different strains were calculated. The ENc values of the ZIKV polyprotein-coding regions vary from 52.13 to 55.00, with a mean value of 53.32 and a standard deviation (SD) of 0.81, showing that the codon usage bias of ZIKV is low (Table 1).
The CAI value is a universal measure of the synonymous codon usage of genes in different organisms and can be used to analyze the adaption of a species to its hosts [17]. CAI values can range from 0 to 1 and higher CAI values signify higher levels of codon usage bias. We found that, in relation to human, the CAI values of ZIKV polyprotein-coding regions range from 0.734 to 0.741, with an average value of 0.740 and a SD of 0.002 (Table 1).
This study on 46 ZIKV strains revealed that the codon usage bias in the polyprotein-coding region of ZIKV is low as the mean ENc value of ZIKV polyprotein-coding regions is 53.32 (>40). This result is analogous to those of previous studies, which found that some RNA viruses, such as hepatitis A virus, bovine viral diarrhea virus, SARS-coronavirus, Newcastle disease virus, Marburg virus, and swine fever virus, also show a weak codon usage bias [22, 34–38]. A possible explanation for this is that the low codon usage bias may be beneficial for the efficient transcription and translation of virus genes in host cells [22]. In addition, ZIKV shows the high CAI value (0.740) for H. sapiens, suggesting that natural selection from H. sapiens can affect the codon usage of ZIKV and the evolution of codon usage in ZIKV has made it to utilize the translation resource of H. sapiens more efficiently. This is similar to Marburg virus, which also has a higher CAI value for H. sapiens but shows low codon usage bias [37].
Relationships between the codon usage pattern of ZIKV and that of its hosts
To investigate the synonymous codon usage pattern, the RSCU values of 59 codons (excluding Met, Trp, and the termination codons) in ZIKV polyprotein-coding regions were calculated. Among 18 preferable codons, 13 have an end base of A or C, while only five have an end base of U or G; therefore, the codons with end bases of A and C are prone to be preferentially utilized in the ZIKV genome (Table 2).
Table 2. The relative synonymous codon usage frequency (RSCU) of ZIKV and its natural hosts.
| AAa | Codon | RSCUb | |||
|---|---|---|---|---|---|
| ZIKV | Homo sapiens | Aedes aegypti | Aedes albopictus | ||
| Phe | UUU | 1.008 | 0.87 | 0.56 | 0.48 |
| UUC | 0.992 | 1.13 | 1.44 | 1.52 | |
| Leu | UUA | 0.330 | 0.39 | 0.35 | 0.23 |
| UUG | 1.323 | 0.73 | 1.34 | 1.11 | |
| CUU | 0.801 | 0.73 | 0.67 | 0.49 | |
| CUC | 1.000 | 1.21 | 0.81 | 0.87 | |
| CUA | 0.659 | 0.40 | 0.54 | 0.57 | |
| CUG | 1.892 | 2.53 | 2.28 | 2.73 | |
| Ile | AUU | 0.864 | 1.03 | 1.00 | 0.74 |
| AUC | 1.167 | 1.52 | 1.59 | 1.86 | |
| AUA | 0.967 | 0.44 | 0.40 | 0.40 | |
| Val | GUU | 0.843 | 0.69 | 1.05 | 0.88 |
| GUC | 1.099 | 1.00 | 1.09 | 1.30 | |
| GUA | 0.387 | 0.42 | 0.60 | 0.51 | |
| GUG | 1.668 | 1.90 | 1.26 | 1.31 | |
| Ser | UCU | 0.854 | 1.11 | 0.67 | 0.54 |
| UCC | 0.985 | 1.39 | 1.20 | 1.40 | |
| UCA | 1.515 | 0.84 | 0.68 | 0.48 | |
| UCG | 0.420 | 0.33 | 1.41 | 1.70 | |
| AGU | 0.997 | 0.84 | 0.93 | 0.79 | |
| AGC | 1.228 | 1.50 | 1.11 | 1.08 | |
| Pro | CCU | 0.671 | 1.12 | 0.67 | 0.35 |
| CCC | 1.129 | 1.35 | 0.83 | 1.13 | |
| CCA | 1.809 | 1.07 | 1.20 | 1.07 | |
| CCG | 0.391 | 0.46 | 1.30 | 1.44 | |
| Thr | ACU | 0.980 | 0.94 | 0.80 | 0.64 |
| ACC | 1.133 | 1.52 | 1.48 | 1.79 | |
| ACA | 1.478 | 1.07 | 0.70 | 0.58 | |
| ACG | 0.409 | 0.46 | 1.01 | 0.99 | |
| Ala | GCU | 1.132 | 1.09 | 1.09 | 0.99 |
| GCC | 1.290 | 1.64 | 1.48 | 1.81 | |
| GCA | 1.125 | 0.85 | 0.75 | 0.59 | |
| GCG | 0.453 | 0.42 | 0.69 | 0.62 | |
| Tyr | UAU | 0.761 | 0.84 | 0.64 | 0.55 |
| UAC | 1.239 | 1.16 | 1.36 | 1.45 | |
| His | CAU | 0.863 | 0.81 | 0.84 | 0.75 |
| CAC | 1.137 | 1.19 | 1.16 | 1.25 | |
| Gln | CAA | 1.112 | 0.51 | 0.81 | 0.60 |
| CAG | 0.888 | 1.49 | 1.19 | 1.40 | |
| Asn | AAU | 0.692 | 0.89 | 0.79 | 0.64 |
| AAC | 1.308 | 1.11 | 1.21 | 1.36 | |
| Lys | AAA | 0.854 | 0.82 | 0.79 | 0.58 |
| AAG | 1.146 | 1.18 | 1.21 | 1.42 | |
| Asp | GAU | 0.903 | 0.89 | 1.12 | 0.96 |
| GAC | 1.097 | 1.11 | 0.88 | 1.04 | |
| Glu | GAA | 0.930 | 0.81 | 1.15 | 1.11 |
| GAG | 1.070 | 1.19 | 0.85 | 0.89 | |
| Cys | UGU | 0.941 | 0.86 | 0.83 | 0.69 |
| UGC | 1.060 | 1.14 | 1.17 | 1.31 | |
| Arg | CGU | 0.450 | 0.51 | 1.36 | 1.5 |
| CGC | 0.600 | 1.20 | 1.25 | 1.32 | |
| CGA | 0.253 | 0.63 | 1.17 | 0.97 | |
| CGG | 0.559 | 1.20 | 1.05 | 1.22 | |
| AGA | 2.443 | 1.20 | 0.64 | 0.58 | |
| AGG | 1.692 | 1.26 | 0.53 | 0.41 | |
| Gly | GGU | 0.529 | 0.64 | 1.10 | 1.24 |
| GGC | 0.672 | 1.40 | 1.04 | 1.07 | |
| GGA | 1.793 | 0.98 | 1.49 | 1.21 | |
| GGG | 1.006 | 0.98 | 0.37 | 0.47 | |
a AA represents amino acid.
b The “RSCU” value represents the pattern of relative synonymous codon usage.
To determine if the codon usage pattern of ZIKV is influenced by that of its hosts, the codon usage pattern of ZIKV was compared with the codon usage patterns of its natural hosts, including H. sapiens, A. aegypti, and A. albopictus. We found that 47 of 59 synonymous codons between ZIKV and H. sapiens are equivalently selected while 40 or 30 of 59 synonymous codons between ZIKV and A. aegypti or A. albopictus, respectively, are similarly selected (Table 2). In general, the similarity in the degree of codon usage between ZIKV and H. sapiens is higher than that between ZIKV and A. aegypti or A. albopictus. Specifically, CUG for leucine (Leu), AGC for serine (Ser), GCC for alanine (Ala), UAC for tryptophan (Tyr), CAC for histidine (His), AAC for asparagine (Asn), AAG for lysine (Lys), and UGC for cysteine (Cys) have high similarity between ZIKV and its natural hosts. Additionally, the RSCU values of several codons showed a strong discrepancy between ZIKV and its hosts, such as CUA for Leu, AUA for isoleucine (Ile), CCA for proline (Pro), and CGA/CGG/AGA for arginine (Arg).
These results suggest that the selection pressure from the hosts may influence the codon usage pattern of ZIKV, which may assist it in adapting to the cellular environment of the hosts and allow it to replicate efficiently in the hosts [24, 31]. Interestingly, the role of the translational selection from H. sapiens in shaping the codon usage pattern of ZIKV is different from that of its insect hosts (A. aegypti and A. albopictus). Compared with the codon usage pattern of A. aegypti or A. albopictus, the codon usage pattern of ZIKV is more similar to that of H. sapiens. This discrepancy of similarity in the degree of codon usage between ZIKV and its hosts may be caused by the various defense mechanisms from different hosts against ZIKV infection. Indeed, a recent study indicated that skin immune cells, including fibroblasts, epidermal keratinocytes, and immature dendritic cells, are highly permissive to ZIKV infection and replication, which can lead to the activation of an antiviral innate immune response [39]. Another study found that although A. aegypti and A. albopictus are susceptible to ZIKV infection, they are both low-competent vectors for ZIKV [40]. It is presumed that the evolution of the flavivirus genome sequence involved in anti-host countermeasures may be faster than that of other flavivirus sequence [41]. This may be one reason why the codon usage pattern of ZIKV tends to show more similarities to that of H. sapiens.
Assessing effects of the overall codon usage of hosts on that of ZIKV
To determine how the overall codon usage of ZIKV’s hosts has contributed to virus codon usage bias, the similarity index analysis was carried out. The results indicated that all of the average values of D (A,B) among three hosts are slightly low, suggesting that ZIKV has adapted to self-replicate efficiently with strong independence of overall codon usage of its hosts during the long-term evolution. In particular, the average value of D (A,B) in A. albopictus (0.0696±0.0017) or A. aegypti (0.0528±0.0012) is higher compared with that in H. sapiens (0.0307±0.0015). This phenomenon also can be seen in the Marburg virus, in which Rousettus aegyptiacus exerts a more dominant effect on forming virus codon usage compared with that of H. sapiens [37].
Relationship between dinucleotide biases and codon usage in ZIKV
Previous studies found that dinucleotide compositional constraints of genomes can affect the codon usage bias [33]. Therefore, we determined the relative abundance of 16 dinucleotides in ZIKV polyprotein-coding regions. The results show that the occurrences of dinucleotides in ZIKV are not randomly distributed and no dinucleotide is present at the expected frequency (Table 3). Specially, the dinucleotides UG and CA are over-represented (ρxy > 1.23) while UA and CG are markedly under-represented (ρxy < 0.78). These data is consistent with previous study, which suggested that the dinucleotides UA and CG are under-represented in many sequence sets [21]. Moreover, the analysis of RSCU values of the eight codons containing CG (UCG, CCG, ACG, GCG, CGU, CGC, CGA, and CGC) suggests that these codons are not preferentially used. Meanwhile, in case of UA containing codons, most of codons are not preferentially selected, except for UAC. Taken together, the composition of dinucleotides plays a role in the synonymous codon usage pattern of ZIKV.
Table 3. Relative abundance of the 16 dinucleotides in polyprotein-coding region of 46 ZIKV strains.
| Dinucleotides | Range | Mean + SD |
|---|---|---|
| AA | 0.944–1.004 | 0.987 ± 0.020 |
| AU | 0.969–1.000 | 0.981 ± 0.010 |
| AC | 0.904–0.984 | 0.967 ± 0.012 |
| AG | 1.032–1.077 | 1.049 ± 0.013 |
| UA | 0.514–0.553 | 0.535 ± 0.009 |
| UU | 0.965–1.064 | 0.999 ± 0.016 |
| UC | 0.985–1.036 | 1.015 ± 0.012 |
| UG | 1.414–1.492 | 1.434 ± 0.023 |
| CA | 1.286–1.357 | 1.308 ± 0.019 |
| CU | 1.213–1.277 | 1.256 ± 0.014 |
| CC | 1.099–1.148 | 1.112 ± 0.010 |
| CG | 0.373–0.454 | 0.431 ± 0.025 |
| GA | 1.049–1.159 | 1.123 ± 0.018 |
| GU | 0.818–0.857 | 0.831 ± 0.008 |
| GC | 0.990–0.992 | 0.931 ± 0.013 |
| GG | 0.445–1.079 | 1.046 ± 0.091 |
The relative abundance of dinucleotides has been shown to influence the codon usage in some RNA viruses [42]. In our study, we found the relative low abundances of CpG and UpA in ZIKV, which may be beneficial for the virus to escape the host anti-viral immune response and complete virus transcription reaction efficiently [43]. The unmethylated CpG can be recognized by the host innate immune system as a pathogen signature, and activates various immune response pathways [42, 44]. UpA deficiency was proposed to avail virus by reducing the risk of nonsense mutations, minimizing the improper transcription and decreasing the opportunities of cleavage by RNase L [45].
Correspondence analysis and correlation analysis: compositional properties of the ZIKV polyprotein-coding region
The A, U, C, G, and GC contents were compared with the A3s, U3s, C3s, G3s, and GC3s contents, respectively (Table 4). The results show that correlations in nucleotide compositions are complicated. Specifically, both the G and GC contents have a significant negative correlation with the content of A3s or U3s, as well as a significant positive correlation with the content of C3s, G3s, or GC3s. The A content has a significant negative correlation with the content of C3s, G3s or GC3s and a significant positive correlation with the content of A3s or U3s. The U content has a significant negative correlation with the content of C3s or GC3s as well as a significant positive correlation with A3s or U3s content, except for the insignificant correlation between U and G3s contents. The C content has a significant negative correlation with A3s, U3s, or C3s content as well as a significant positive correlation with G3s or GC3s content. These data shows that the nucleotide compositional constraint may also affect the codon usage of ZIKV.
Table 4. The correlation analysis between the A, U, C, G contents and A3s, U3s, C3s, G3s contents in all selected ZIKV strainsa.
| A3s | U3s | C3s | G3s | GC3s | |
|---|---|---|---|---|---|
| A | 0.856** | 0.560** | -0.777** | -0.793** | -0.847** |
| U | 0.300* | 0.705** | -0.593** | -0.258 | -0.579** |
| C | -0.653** | -0.764** | -0.920** | 0.596** | 0.882** |
| G | -0.475** | -0.376** | 0.343* | 0.418** | 0.459** |
| GC | -0.738** | -0.725** | 0.856** | 0.704** | 0.911** |
a Value in this table is the r value of correlation analysis.
** represents P-value < 0.01.
* represents 0.01 < P-value < 0.05.
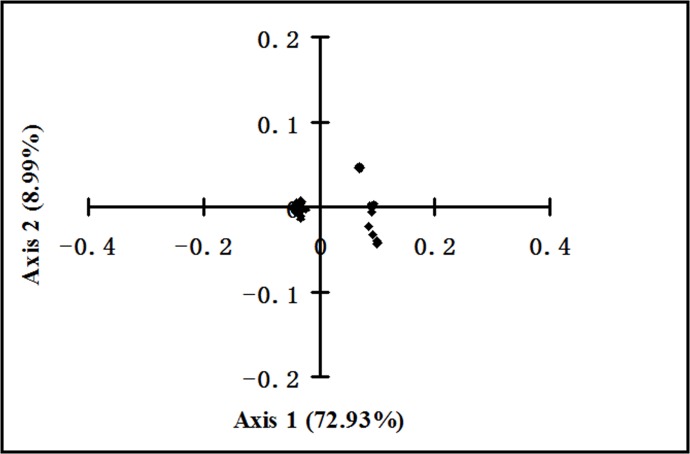
A correspondence analysis was performed to determine the main trends in the codon usage variation and the distribution of each gene along the continuous axes. The positions of each polyprotein-coding region defined by the first axis (Axis 1) and second axis (Axis 2) are shown in Fig 2. The first axis accounts for 72.93% of the total variation, and the second, third and fourth axes account for 8.99%, 6.33%, and 3.08%, respectively, of the total variation in synonymous codon usage. A correlation analysis also showed that, except G content, the Axis1 is positively correlated with the contents of A, U, A3s, U3s, whereas it is negatively correlated with the GC3s, GC, ENc, C, C3s and G3s (Table 5). Meanwhile, Axis2 is only negatively correlated with the G content (Table 5). Overall, these results suggest that mutation pressure from the base composition plays a role in constructing the codon usage pattern of ZIKV.
Fig 2. A plot of values of the first axis (Axis 1) and the second axis (Axis 2) of each polyprotein-coding region of ZIKV in correspondence analysis.
The first axis accounts for 72.93% of total variation, and the second axis accounts for 8.99% of total variation.
Table 5. Correlation analysis between the first two axes and different nucleotide-relevant indices.
| GC3s | GC | ENc | A | U | C | G | A3s | U3s | C3s | G3s | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Axis1 | |||||||||||
| r | -0.693** | -0.716** | -0.589** | 0.748** | 0.339* | -0.691** | -0.106 | 0.642** | 0.444** | -0.624** | -0.661** |
| P | 0.000 | 0.000 | 0.000 | 0.000 | 0.021 | 0.000 | 0.483 | 0.000 | 0.002 | 0.000 | 0.000 |
| Axis2 | |||||||||||
| r | -0.072 | -0.027 | -0.105 | 0.123 | -0.043 | -0.053 | -0.447** | 0.145 | 0.017 | 0.047 | -0.197 |
| P | 0.633 | 0.859 | 0.487 | 0.417 | 0.777 | 0.726 | 0.002 | 0.337 | 0.913 | 0.756 | 0.191 |
** represents P-value < 0.01.
* represents 0.01 < P-value < 0.05.
The effect of translational selection on the codon usage of ZIKV
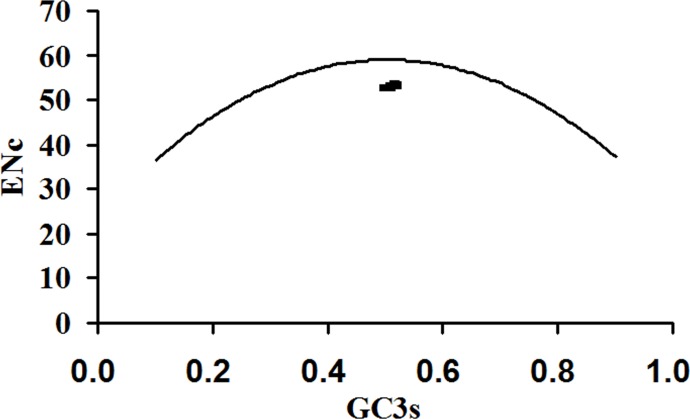
A plot of the ENc values against the GC3s values was constructed to check the heterogeneity of codon usage [20]. If a gene is subject to the GC compositional constraints, it will lie on or near the theoretical fitting curve that represents random codon usage. In contrast, if a gene is subject to translational selection, it will lie considerably below the expected curve [46]. Here, the ENc value of each polyprotein- coding region of ZIKV was plotted against the corresponding GC3s content (Fig 3). The resulting points lie considerably below the solid curve, implying that, in addition to mutation pressure, other factors, such as translational selection, also influence the codon usage pattern of ZIKV. This result is generally similar to the related plot in previous study [34].
Fig 3. Graph showing the relationship between GC3s and effective number of codon (ENc) for the polyprotein-coding regions of ZIKV.
The solid curve shows the expected ENc value if the codon usage is only determined by the variation in the GC3s.
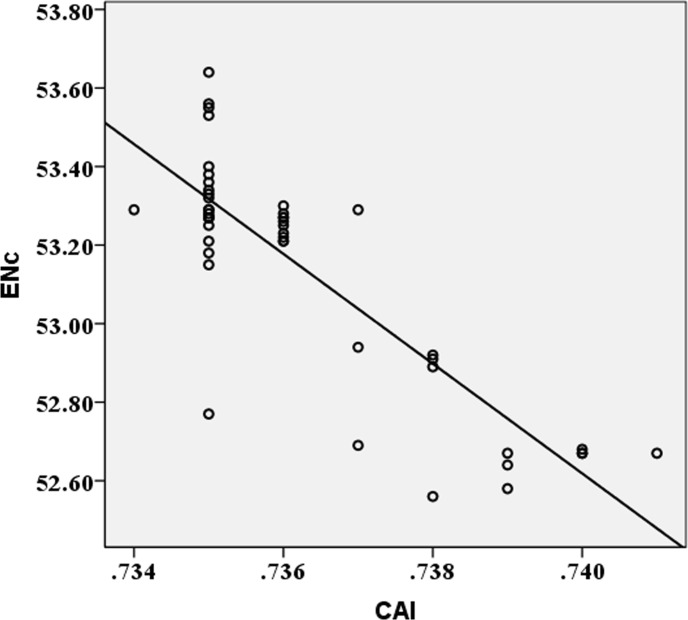
The base composition and codon usage bias of the ORFs of a species with an A/U-rich genome may be different from those species with G/C-rich genomes. Previous studies have employed a correlation between CAI values and ENc values to demonstrate the effect of mutation and translational selection on the codon usage bias [37, 47]. If the correlation (r) between the two indices approaches –1, this suggests that the translational selection is preferred over mutation. Otherwise, if the r value approaches 0 (no correlation), mutation may be more influential than translational selection. Our results showed that the CAI value of ZIKV is significantly positively correlated with the ENc value (r = -0.749, P<0.01) (Fig 4). This result reflects the influence of both translational selection and mutation pressure on the codon usage pattern of ZIKV.
Fig 4. Correlation between codon adaptation index (CAI) and effective number of codons (ENc).
The line represents the correlation curve produced by correlation analysis.
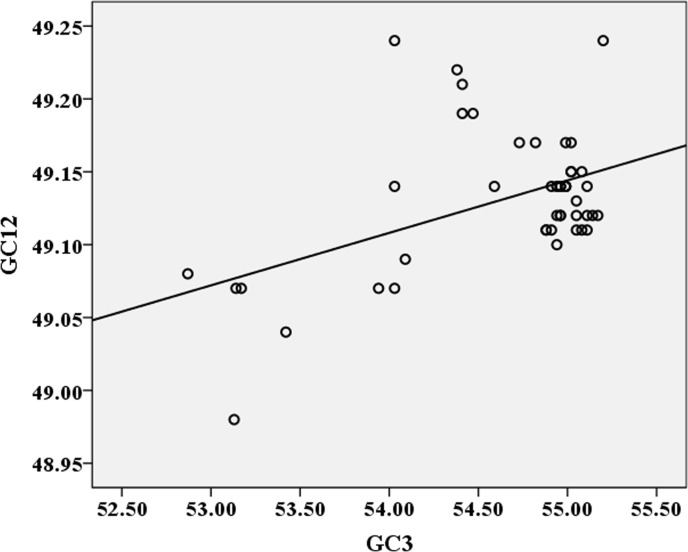
A significant correlation between the GC12 and GC3 values is regarded to indicate that the mutation pressure dominates over the translational selection pressure in shaping the codon usage bias [22, 46]. If the correlation between GC12 and GC3 is significant, the mutation pressure is regarded as the main force forming the codon usage bias. To further determine the role of mutation pressure and translational selection in shaping the codon usage bias of ZIKV, a correlation analysis was performed to analyze the relationship between GC12 and GC3. There was no significant correlation observed between them (r = 0.25, P>0.05), suggesting that both translational selection and the mutation pressure are involved in shaping the codon usage pattern of ZIKV (Fig 5).
Fig 5. Correlation between the GC content at the first and second codon positions (GC12) and that at the third codon position (GC3).
The line represents the correlation curve produced by correlation analysis.
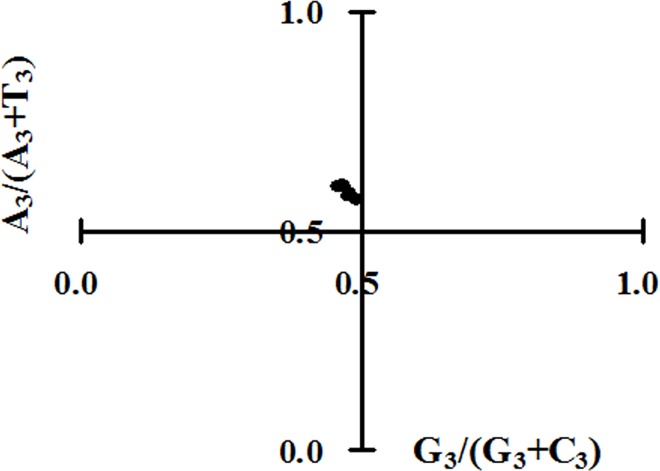
To determine whether the biased codon selection are restricted in highly biased coding sequences, the relationship between pyrimidines (C and U) and purines (A and G) contents in four-fold degenerate codon families (alanine, arginine, glycine, leucine, proline, serine, threonine and valine) are analyzed by PR2 bias plot. It can be seen that A and C are more frequently used than U and G in ZIKV in four-fold degenerate codon families (Fig 6). This result shows that the codon usage pattern of ZIKV is shaped by mutation pressure and other factors including translational selection.
Fig 6. Parity rule 2 (PR2) plot [A3/(A3+U3) against G3/(G3+C3)].
PR2 bias plot was calculated for each polyprotein-coding region of ZIKV.
To confirm whether translation selection from the hosts plays a role in shaping the codon usage pattern of ZIKV, the tAI values were calculated based on the tRNA copy numbers of H. sapiens. The results indicated that the tAI values of 46 ZIKV strains range from 0.329 to 0.347, with an average value of 0.344 and a SD of 0.004. Moreover, the positive correlation between tAI and CAI values (r = 0.457, P<0.01) in ZIKV highlights the importance of translational selection in the formation of synonymous codon usage pattern.
Compared with translational selection, mutation bias seems to have a stronger effect on the codon usage bias of some viruses [48, 49]. However, for ZIKV, the translational selection pressure also takes part in shaping the codon usage bias. Our result is consistent with a previous study that showed that recent Asian lineage spread is linked to the codon usage adaptation of the NS1 protein to human housekeeping genes [50]. During the preparation of this manuscript, two papers were published employing some ZIKV strains to analyze the codon usage [51, 52]. They concluded that mutation pressure is an important determinant of the codon usage bias of ZIKV mainly based on the result of a GC3s-ENc analysis [51]. The reasons that they do not mention the role of translational selection in the codon usage of ZIKV may be due to the lack of application of other codon usage analysis methods in their studies.
Effect of other factors on codon usage
GRAVY and AROMO may also be related to the codon usage pattern of viruses [53]. Our correlation analysis indicated that AROMO is positively correlated with GC3s, GC, and ENc, but it is negatively correlated with Axis 1. GRAVY showed a significant positive correlation with Axis 1, but it showed a significant negative correlation with GC, GC3s, and ENc, respectively (Table 6). Both GRAVY and AROMO do not show any correlation with Axis 2. These results indicated that the aromaticity and degree of protein hydrophobicity are linked to the codon usage variation in ZIKV, emphasizing the importance of natural translational selection on forming the codon usage pattern [34].
Table 6. Correlation analysis among AROMO, GRAVY, the first two axes, GC3s, ENc and GC in the polyprotein-coding region of ZIKV isolates.
| Axis1 | Axis2 | GC3s | GC | ENc | |
|---|---|---|---|---|---|
| AROMO | |||||
| r | -0.768** | -0.038 | 0.773** | 0.787** | 0.747** |
| P | 0.000 | 0.803 | 0.000 | 0.000 | 0.000 |
| GRAVY | |||||
| r | 0.528** | 0.101 | -0.550** | -0.515** | -0.669** |
| P | 0.000 | 0.506 | 0.000 | 0.000 | 0.000 |
** represents P-value < 0.01.
The involvement of aromaticity and hydrophobicity in the construction of codon usage bias has been revealed in some RNA viruses, such as bovine viral diarrhea virus, classical swine fever virus, and duck hepatitis A virus [34, 35, 54]. This study found that Axis 1 has a significant role in shaping the ZIKV codon usage pattern and is significantly correlated with aromaticity and hydrophobicity indices, implying that the aromaticity and hydrophobicity of proteins are related to the codon usage pattern of ZIKV. Aromaticity and hydrophobicity are known to play a role in peptide self-assembly and protein aggregation rates [55, 56]. A recent study showed that the structure of ZIKV particles is thermally stable, and this feature may help the virus to survive in the harsh conditions of semen, saliva, and urine [57].
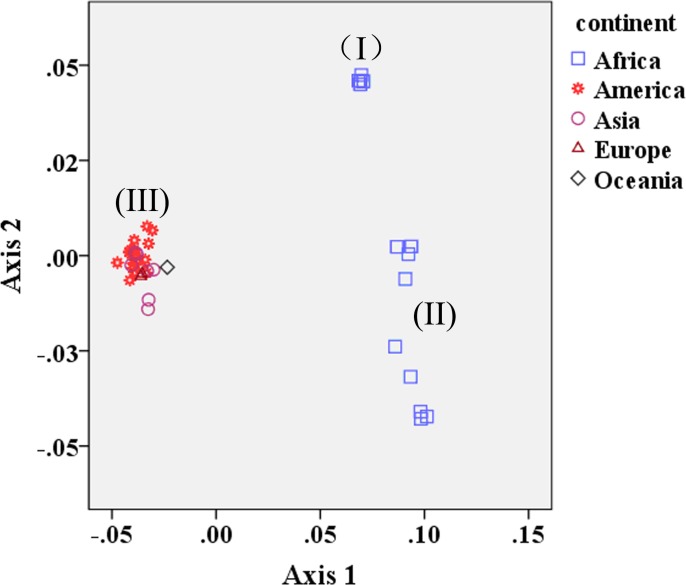
It has been reported that there is a significant correlation between the phylogroups of isolates and their geographic regions, and an obvious pattern of geographic clustering has been observed in ZIKV isolates [14]. To determine if geographic factors influence the evolution of ZIKV, a plot of Axis 1 and Axis 2 was drawn according to the geographic distribution of the tested ZIKV strains. The resulting coordinate spots are separated into three groups, classified as group I, II, and III (Fig 7). Some strains isolated in Uganda clustered together with the strains isolated from Senegal, and were classified as group I. Additionally, some strains isolated from Central African Republic also clustered together with the strains isolated from Senegal, and these were classified as group II. Most of the strains isolated, regardless of their isolation countries, tended to cluster together and were classified as group III. The codon usage pattern reflects the close relationship of ZIKV strains in different geographic regions.
Fig 7. A plot of the values of the first axis (Axis 1) and the second axis (Axis 2) of ZIKV strains isolated from different continents in correspondence analysis.
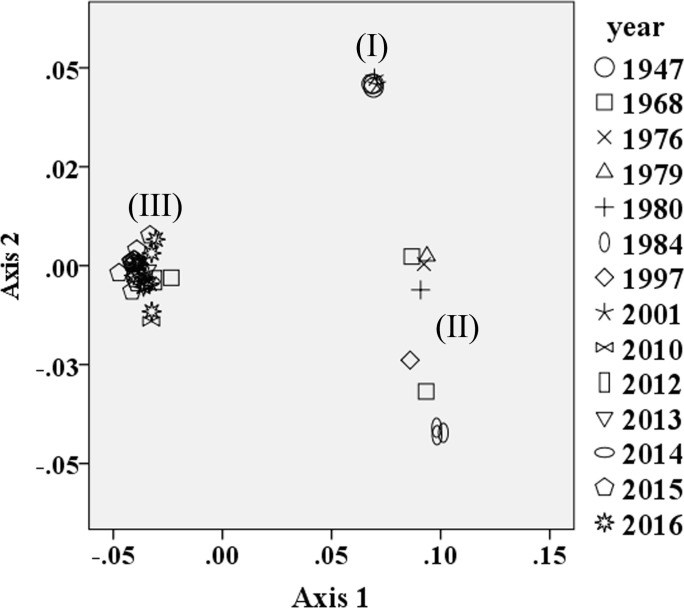
To investigate if the ZIKV codon usage pattern displays changes over time, a plot of Axis 1 and Axis 2 was drawn according to the outbreak time of the ZIKV strains. The 46 ZIKV isolates were divided into three groups, classified as group I, II and III (Fig 8). Most of the strains isolated from 2010 to 2016 tended to cluster together in group III. The strains isolated from 1968 to 1997 clustered together in group II, while the strains isolated in 1947 and 2001 clustered in group I. Interestingly, the strains isolated in 1968 exist in both group II and group III. These results indicated that ZIKV strains isolated in different time intervals show genetic variation in their codon usage patterns.
Fig 8. A plot of the values of the first axis (Axis 1) and the second axis (Axis 2) of ZIKV strains isolated in different years in correspondence analysis.
Previous studies showed that the Dengue virus strains occurring in the same continental region are more closely related to one another, forming a cluster when plotted by their codon usage biases, indicating that the viruses from a geographical group can show similar codon usage biases [58]. Andrew et al found that the geographic origin of the strains responsible for the ZIKV epidemics that occurred on Yap island in 2007 and in Cambodia in 2010 most likely originated in Southeast Asia [14]. In this study, we further found that most of the American ZIKV strains isolated in recent years cluster with some Asian, Europe and Oceania strains, supporting the idea that a close evolutionary relationship exists among Asian, Europe, Oceania and American strains.
Conclusions
Our findings reveal that the codon usage bias of ZIKV is weak and that, in addition to mutation pressure, translational selection also influences the codon usage bias. Other factors, such as base composition, aromaticity, and hydrophobicity, also have an effect on the codon usage pattern. Importantly, there are similarities between the codon usage patterns of ZIKV and its natural hosts. This study not only provides an understanding about the variation in ZIKV codon usage patterns, but it also contributes to understanding the factors that drive ZIKV evolution.
Acknowledgments
This work was supported by grants from the Plant Foundation for Young Scientists of Henan University (CX0000A40557), and the National Natural Science Foundation of China (31501701). We thank Yihui Yuan (Wuhan Institute of Virology, Chinese Academy of Sciences) for helpful discussions and editing of this manuscript.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by grants from the Plant Foundation for Young Scientists of Henan University (CX0000A40557) and the National Natural Science Foundation of China (31501701). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Didier M. Zika Virus Transmission from French Polynesia to Brazil. Emerg Infect Dis. 2015; 21(10): 1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Samarasekera U, Triunfol M. Concern over Zika virus grips the world. Lancet. 2016; 387(10018): 521–524. 10.1016/S0140-6736(16)00257-9 [DOI] [PubMed] [Google Scholar]
- 3.Bogoch II, Brady OJ, Kraemer MU, German M, Creatore MI, Kulkarni MA, et al. Anticipating the international spread of Zika virus from Brazil. Lancet. 2016; 387(10016): 335–336. 10.1016/S0140-6736(16)00080-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuno G, Chang GJJ. Full-length sequencing and genomic characterization of Bagaza, Kedougou, and Zika viruses. Arch Virol. 2007; 152(4): 687–696. 10.1007/s00705-006-0903-z [DOI] [PubMed] [Google Scholar]
- 5.Musso D, Roche C, Nhan TX, Robin E, Teissier A, Cao-Lormeau VM. Detection of Zika virus in saliva. J Clin Virol. 2015; 68: 53–55. 10.1016/j.jcv.2015.04.021 [DOI] [PubMed] [Google Scholar]
- 6.Gourinat AC, O'Connor O, Calvez E, Goarant C, Dupont-Rouzeyrol M. Detection of Zika virus in urine. Emerg Infect Dis. 2015; 21(1): 84–86. 10.3201/eid2101.140894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Triunfol M. A new mosquito-borne threat to pregnant women in Brazil. Lancet Infect Dis. 2015; 16(2): 156–157. 10.1016/S1473-3099(15)00548-4 [DOI] [PubMed] [Google Scholar]
- 8.Rubin EJ, Greene MF, Baden LR. Zika Virus and Microcephaly. N Engl J Med. 2016; 374(10): 984–985. 10.1056/NEJMe1601862 [DOI] [PubMed] [Google Scholar]
- 9.Babbitt GA, Alawad MA, Schulze KV, Hudson AO. Synonymous codon bias and functional constraint on GC3-related DNA backbone dynamics in the prokaryotic nucleoid. Nucleic Acids Res. 2014; 42(17): 10915–10926. 10.1093/nar/gku811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ermolaeva MD. Synonymous codon usage in bacteria. Curre Issues Mol Bio. 2001; 3(4): 91–97. [PubMed] [Google Scholar]
- 11.Wan XF, Xu D, Kleinhofs A, Zhou J. Quantitative relationship between synonymous codon usage bias and GC composition across unicellular genomes. Bmc Evol Biol. 2004; 4(3): 19–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plotkin JB, Dushoff J, Desai MM, Fraser HB. Codon Usage and Selection on Proteins. J Mol Evol. 2006; 63(5): 635–653. 10.1007/s00239-005-0233-x [DOI] [PubMed] [Google Scholar]
- 13.Bulmer M. The Selection-Mutation-Drift Theory of Synonymous Codon Usage. Genetics. 1991; 129(3): 897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haddow AD, Schuh AJ, Yasuda CY, Kasper MR, Heang V, Huy R, et al. Genetic Characterization of Zika Virus Strains: Geographic Expansion of the Asian Lineage. Plos Neglect Trop. 2012; 6(2): e1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuno G, Chang GJJ. Full-length sequencing and genomic characterization of Bagaza, Kedougou, and Zika viruses. Arch Virol. 2007; 152(4): 687–696. 10.1007/s00705-006-0903-z [DOI] [PubMed] [Google Scholar]
- 16.Lewis PO, Kumar S, Tamura K, Nei M. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013; 30(12): 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carbone A, Zinovyev AF. Codon adaptation index as a measure of dominating codon bias. Bioinformatics. 2003; 19(16): 2005–2015. [DOI] [PubMed] [Google Scholar]
- 18.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000; 16(6): 276–277. [DOI] [PubMed] [Google Scholar]
- 19.Sharp PM, Matassi G. Codon usage and genome evolution. Curr Opin Genet Dev. 1994; 4(6): 851–860. [DOI] [PubMed] [Google Scholar]
- 20.Wright F. The effective number of codon used in a gene. Gene. 1990; 87(1): 23–27. [DOI] [PubMed] [Google Scholar]
- 21.Burge C, Campbell AM, Karlin S. Over- and under-representation of short oligonucleotides in DNA sequences. Proc Natl Acad Sci U S A. 1992; 89(4): 1358–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang M, Liu YS, Zhou JH, Chen HT, Ma LN, Ding YZ, et al. Analysis of codon usage in Newcastle disease virus. Virus Genes. 2011; 42(2): 245–253. 10.1007/s11262-011-0574-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharp PM, Li WH. Codon usage in regulatory genes in Escherichia coli does not reflect selection for 'rare' codons. Nucleic Acids Res. 1986; 14(19): 7737–7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wong EH, Smith DK, Rabadan R, Peiris M, Poon LL. Codon usage bias and the evolution of influenza A viruses. Codon Usage Biases of Influenza Virus. Bmc Evol Biol. 2010; 10(1): 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma YP, Liu ZX, Hao L, Ma JY, Liang ZL, Li YG, et al. Analysing codon usage bias of cyprinid herpesvirus 3 and adaptation of this virus to the hosts. J Fish Dis. 2015; 38(7): 665–673. 10.1111/jfd.12316 [DOI] [PubMed] [Google Scholar]
- 26.Zhou JH, Zhang J, Sun DJ, Ma Q, Chen HT, Ma LN, et al. The distribution of synonymous codon choice in the translation initiation region of dengue virus. PLoS One. 2013; 8(10): 1175–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dos RM, Wernisch L, Savva R. Unexpected correlations between gene expression and codon usage bias from microarray data for the whole Escherichia coli K-12 genome. Nucleic Acids Res. 2003; 31(23): 6976–6985. 10.1093/nar/gkg897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma YP, Ke H, Liang ZL, Liu ZX, Hao L, Ma JY, et al. Multiple Evolutionary Selections Involved in Synonymous Codon Usages in the Streptococcus agalactiae Genome. Int J Mol Sci. 2016; 17(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jung SK, Mcdonald K. Visual gene developer: a fully programmable bioinformatics software for synthetic gene optimization. Bmc Bioinformatics. 2011; 12(1): 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sueoka N. Translation-coupled violation of Parity Rule 2 in human genes is not the cause of heterogeneity of the DNA G+C content of third codon position. Gene. 1999; 238(1): 53–58. [DOI] [PubMed] [Google Scholar]
- 31.Ma YP, Zhou ZW, Liu ZX, Hao L, Ma JY, Feng GQ, et al. Codon usage bias of the phosphoprotein gene of spring viraemia of carp virus and high codon adaptation to the host. Arch Virol. 2014; 159(7): 1841–1847. 10.1007/s00705-014-2000-z [DOI] [PubMed] [Google Scholar]
- 32.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends in Genetics Tig. 2000; 16(6): 276–277. [DOI] [PubMed] [Google Scholar]
- 33.Liu XS, Zhang YG, Fang YZ, Wang YL. Patterns and influencing factor of synonymous codon usage in porcine circovirus. Virol J. 2012; 9(6): 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Y, Chen YF. Analysis of synonymous codon usage patterns in duck hepatitis A virus: a comparison on the roles of mutual pressure and natural selection. Virusdisease. 2014; 25(3): 285–293. 10.1007/s13337-014-0191-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang M, Zhang J, Zhou JH, Chen HT, Ma LN, Ding YZ, et al. Analysis of codon usage in bovine viral diarrhea virus. Arch Virol. 2011; 156(1): 153–160. 10.1007/s00705-010-0848-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gu W, Zhou T, Ma J, Sun X, Lu Z. Analysis of synonymous codon usage in SARS Coronavirus and other viruses in the Nidovirales. Virus Res. 2004; 101(2): 155–161. 10.1016/j.virusres.2004.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nasrullah I, Butt AM, Tahir S, Idrees M, Tong Y. Genomic analysis of codon usage shows influence of mutation pressure, natural selection, and host features on Marburg virus evolution. Bmc Evol Biol. 2015; 15(1): 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeng Z, Liu Z, Wang W, Tang D, Liang H, Liu Z. Establishment and application of a multiplex PCR for rapid and simultaneous detection of six viruses in swine. J Virol Methods. 2014; 208: 102–106. 10.1016/j.jviromet.2014.08.001 [DOI] [PubMed] [Google Scholar]
- 39.Hamel R, Dejarnac O, Wichit S, Ekchariyawat P, Neyret A, Luplertlop N, et al. Biology of Zika Virus Infection in Human Skin Cells. J Virol. 2015; 89(17): 8880–8896. 10.1128/JVI.00354-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chouin-Carneiro T, Vega-Rua A, Vazeille M, Yebakima A, Girod R, Goindin D, et al. Differential Susceptibilities of Aedes aegypti and Aedes albopictus from the Americas to Zika Virus. Plos Neglect Trop. 2016; 10(3): e0004543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weaver SC, Costa F, Garcia-Blanco MA, Ko AI, Ribeiro GS, Saade G, et al. Zika virus: History, emergence, biology, and prospects for control. Antiviral Res. 2016; 130: 69–80. 10.1016/j.antiviral.2016.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Greenbaum BD, Levine AJ, Bhanot G, Rabadan R. Patterns of Evolution and Host Gene Mimicry in Influenza and Other RNA Viruses. Plos Pathog. 2008; 4(6):: e1000079 10.1371/journal.ppat.1000079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar N, Bera BC, Greenbaum BD, Bhatia S, Sood R, Selvaraj P, et al. Revelation of Influencing Factors in Overall Codon Usage Bias of Equine Influenza Viruses. PLoS One. 2016; 11(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng X, Virk N, Chen W, Ji S, Sun Y, Wu X. CpG usage in RNA viruses: data and hypotheses. PLoS One. 2013; 8(9): e74109 10.1371/journal.pone.0074109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Washenberger CL, Han J-Q, Kechris KJ, Jha BK, Silverman RH, Barton DJ. Hepatitis C virus RNA: Dinucleotide frequencies and cleavage by RNase L. Virus Res. 2007; 130(1–2): 85–95. 10.1016/j.virusres.2007.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jenkins GM, Holmes EC. The extent of codon usage bias in human RNA viruses and its evolutionary origin. Virus Res. 2003; 92(1): 1–7. [DOI] [PubMed] [Google Scholar]
- 47.Vicario S, Moriyama EN, Powell JR. Codon usage in twelve species of Drosophila. Bmc Evol Biol. 2006; 7(1627): 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prasert A. Composition bias and genome polarity of RNA viruses. Virus Res. 2005; 109(1): 33–37. 10.1016/j.virusres.2004.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shackelton LA, Parrish CR, Holmes EC. Evolutionary basis of codon usage and nucleotide composition bias in vertebrate DNA viruses. J Mol Evol. 2006; 62(5): 551–563. 10.1007/s00239-005-0221-1 [DOI] [PubMed] [Google Scholar]
- 50.Freire CCdM, Iamarino A, Neto DFdL, Sall AA, Zanotto PMdA. Spread of the pandemic Zika virus lineage is associated with NS1 codon usage adaptation in humans. Preprint Available: bioRxiv. 2015.
- 51.Hemert FV, Berkhout B. Nucleotide composition of the Zika virus RNA genome and its codon usage. Virol J. 2015; 13(1): 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cristina J, Fajardo A, Soñora M, Moratorio G, Musto H. A detailed comparative analysis of codon usage bias in Zika virus. Virus Res. 2016; 223: 147–152. 10.1016/j.virusres.2016.06.022 [DOI] [PubMed] [Google Scholar]
- 53.Das S, Paul S, Dutta C. Synonymous codon usage in adenoviruses: Influence of mutation, selection and protein hydropathy. Virus Res. 2006; 117(2): 227–236. 10.1016/j.virusres.2005.10.007 [DOI] [PubMed] [Google Scholar]
- 54.Tao P, Dai L, Luo M, Tang F, Tien P, Pan Z. Analysis of synonymous codon usage in classical swine fever virus. Virus Genes. 2009; 38(1): 104–112. 10.1007/s11262-008-0296-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Doran TM. Role of Hydrophobicity, Aromaticity, and Turn Nucleation in Peptide Self-Assembly. Dissertations & Theses—Gradworks, University of Rochester. 2011.
- 56.Tartaglia GG, Cavalli A, Pellarin R, Caflisch A. The role of aromaticity, exposed surface, and dipole moment in determining protein aggregation rates. Protein Sci. 2004; 13(7): 1939–1941. 10.1110/ps.04663504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kostyuchenko VA, Lim EXY, Zhang S, Fibriansah G, Ng T-S, Ooi JSG, et al. Structure of the thermally stable Zika virus. Nature. 2016; 533: 425–428. 10.1038/nature17994 [DOI] [PubMed] [Google Scholar]
- 58.Lararamírez EE, Salazar MI, Lópezlópez MJ, Salasbenito JS, Sánchezvarela A, Guo X. Large-scale genomic analysis of codon usage in dengue virus and evaluation of its phylogenetic dependence. Biomed Res Int. 2014; 2014(851425): 66–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.