Abstract
The mixed lineage leukaemia (MLL) family of proteins (including MLL1–MLL4, SET1A and SET1B) specifically methylate histone 3 Lys4, and have pivotal roles in the transcriptional regulation of genes involved in haematopoiesis and development. The methyltransferase activity of MLL1, by itself severely compromised, is stimulated by the three conserved factors WDR5, RBBP5 and ASH2L, which are shared by all MLL family complexes. However, the molecular mechanism of how these factors regulate the activity of MLL proteins still remains poorly understood. Here we show that a minimized human RBBP5–ASH2L heterodimer is the structural unit that interacts with and activates all MLL family histone methyltransferases. Our structural, biochemical and computational analyses reveal a two-step activation mechanism of MLL family proteins. These findings provide unprecedented insights into the common theme and functional plasticity in complex assembly and activity regulation of MLL family methyltransferases, and also suggest a universal regulation mechanism for most histone methyltransferases.
Methylation of histone H3 Lys4 (H3K4), which is predominantly associated with actively transcribed genes1–3, is mainly mediated by MLL family histone lysine methyltransferases (HKMTs). Mammalian MLL family HKMTs contain six members (MLL1–MLL4, SET1A and SET1B)2–4, each of which has crucial yet non-redundant roles in cells4–6. MLL1 has been the most intensively studied because of its involvement by chromosomal translocations in a variety of acute lymphoid and myeloid leukaemias6,7. Recently, inactivating mutations in MLL3 (also known as KMT2C) and MLL4 (KMT2D) have been identified in several types of human tumours and in Kabuki syndrome8–12.
In contrast to most SET [SU(VAR)3–9, E(Z) and TRX]-domain-containing methyltransferases, MLL1 protein alone exhibits poor HKMT activity13,14. The crystal structure of the MLL1 SET domain (MLL1SET) reveals an open conformation that is not efficient for the methyl transfer from the cofactor S-adenosyl-L-methionine (AdoMet) to the target lysine15. The optimal HKMT activity of MLL1 requires additional factors, WDR5, RBBP5 and ASH2L, which are shared core components of all MLL complexes and also evolutionarily conserved from yeast to humans13,16. Depletion of any of these components results in the global loss of H3K4 methylation to varying degrees16–18. Despite the importance of WDR5, RBBP5 and ASH2L, it is still unclear how these factors stimulate the HKMT activity of MLL proteins. In this work, our biochemical and structural analyses reveal how RBBP5–ASH2L binds and activates MLL family methyltransferases through a conserved mechanism.
RBBP5–ASH2L binds and activates MLLs
We first examined the effects of individual components (WDR5, RBBP5 and ASH2L) and their combinations on the HKMT activities of MLL family methyltransferases. We selected the carboxy-terminal conserved regions of MLL proteins containing both the WIN (WDR5-interaction) motif and the SET domain in activity assays14 (Fig. 1a). For simplicity, hereafter we use ‘MLL’ to represent the MLL WIN-SET fragment, and ‘MLLSET’ to represent the MLL SET domain unless stated otherwise. Consistent with previous observations14,15,19, activity assays showed that the RBBP5–ASH2L heterodimer substantially upregulated the HKMT activity of MLL1, and this activity was further stimulated by the addition of WDR5 (Fig. 1b and Extended Data Fig. 1a–c). By contrast, MLL2–MLL4, SET1A and SET1B can be fully activated by just RBBP5–ASH2L, and WDR5 was dispensable for activity regulation (Fig. 1b and Extended Data Fig. 1a–c). The stimulatory effect of RBBP5–ASH2L on MLL HKMT activities indicated a possible direct interaction between RBBP5–ASH2L and MLL proteins20–22. Indeed, a glutathione S-transferase (GST) pull-down assay clearly showed that all MLL proteins directly interact with RBBP5–ASH2L (Fig. 1c and Extended Data Fig. 2a). Among them, MLL2–MLL4 could be efficiently pulled down by GST–ASH2L–RBBP5, whereas SET1A and SET1B maintained a medium level of interaction with RBBP5–ASH2L (Fig. 1c and Extended Data Fig. 2a). By contrast, MLL1 only exhibited a very weak interaction with RBBP5–ASH2L under low-salt buffer conditions (Fig. 1c and Extended Data Fig. 2a). Fluorescence polarization analysis also revealed that MLL proteins interact with RBBP5–ASH2L with very different binding affinities ranging from ~100 nM (for MLL3) to more than 100 μM (for MLL1) (Extended Data Fig. 2b). Formation of the RBBP5–ASH2L heterodimer is a prerequisite for MLL binding, as neither RBBP5 nor ASH2L alone can stably associate with MLL proteins (Extended Data Fig. 2c, d). Notably, MLL proteins can also stabilize the RBBP5–ASH2L interaction when high-salt buffer was used in the pull-down assay (Extended Data Fig. 2a), consistent with the observation that the RBBP5–ASH2L interaction is highly sensitive to ionic strength (Extended Data Fig. 2e).
Figure 1. RBBP5–ASH2L interacts and activates MLL proteins.
a, Domain organization of human ASH2L, RBBP5, WDR5 and MLL proteins. Only the C-terminal domain of MLL is shown. DBM, DPY30 binding motif; PHD-WH, plant homeodomain-winged helix. Shaded areas denote the interacting domains among these proteins. b, The normalized HKMT activities determined by a 3H-methyl-incorporation assay. Mean ± s.d. (n = 3) are shown. c, GST pull-down assay shows MLL proteins directly interact with the RBBP5–ASH2LSPRY heterodimer. d, The normalized HKMT assays revealed that an activation segment of RBBP5 (residues 330–344) is crucial for the stimulation of MLL3 activity. FL, full-length. Mean ± s.d. (n = 3) are shown.
Because MLL1 only maintained a weak direct interaction with RBBP5–ASH2L, we proposed that full activation of MLL1 by RBBP5–ASH2L requires the bridging molecule WDR5 that can interact with both MLL1 and RBBP5–ASH2L simultaneously. Consistent with this idea, the stimulatory effect of WDR5 on MLL1 HKMT activity is minimized when the protein concentration was increased in the assay (Extended Data Fig. 2f). Furthermore, the fusion of RBBP5 and MLL1 together achieved a robust HKMT activity that cannot be further stimulated by the addition of WDR5 (Extended Data Fig. 2g), suggesting that WDR5 per se is not directly involved in the MLL HKMT enzymatic reaction. Collectively, we conclude that RBBP5–ASH2L is the major functional unit that binds and activates MLL proteins. Conversely, WDR5 may have an indirect role in promoting HKMT activity by acting as a bridging molecule to facilitate the formation of MLL complexes under certain assay conditions, and this may explain the apparent discrepancy in reports about the role of WDR5 in the activity regulation of MLL complexes19–22.
Complex structure of MLL3–RBBP5–ASH2L
To determine the structural basis of how RBBP5–ASH2L activates MLL proteins, we first dissected the interactions among RBBP5, ASH2L and the SET domains of MLL proteins. Consistent with previous studies23,24, the ASH2L C-terminal SPRY (splA and ryanodine receptor) domain is sufficient to form a heterodimer with RBBP5 to stimulate the HKMT activity of MLL proteins (Fig. 1d, compare lanes 1 and 5). Three adjacent short motifs of RBBP5 were identified for the stimulation of MLL HKMT activity (residues 330–344, activation segment, AS), the interaction with ASH2L (residues 344–363, ASH2L-binding motif, ABM), and the association with WDR5 (residues 369–381, WDR5-binding motif, WBM)24,25 (Fig. 1a). A preformed RBBP5AS-ABM–ASH2LSPRY complex can stimulate MLL3SET HKMT activity to levels of ~70% of full-length RBBP5–ASH2L (Fig. 1d, compare lanes 2, 4 and 6), indicating that this minimized RBBP5AS-ABM–ASH2LSPRY heterodimer is essential for the stimulation of MLL3SET activity, and that other regions of RBBP5 might have a minor role in this process. We determined the crystal structure of this minimized ternary complex composed of MLL3SET, RBBP5AS-ABM and ASH2LSPRY (hereafter referred to as M3RA) in the presence of S-adenosyll-homocystein (AdoHcy) and a substrate peptide (H3 residues 1–9) (Fig. 2a, Extended Data Table 1 and Extended Data Fig. 3a). Notably, we crystallized the M3RA complex both with and without the H3 peptide in one asymmetric unit (Extended Data Fig. 3b).
Figure 2. Crystal structure of the M3RA complex.
a, The crystal structure of MLL3SET–RBBP5AS-ABM–ASH2LSPRY in complex with cofactor product AdoHcy and the H3 peptide. b, MLL3SET shares the conserved features of SET-N, SET-I, SET-C and post-SET motifs. c, Comparison of the active centre of MLL3 and DIM-5 (PDB accession 1PEG) complex structures. d, The AdoHcy binding pocket in MLL3SET. e, The substrate H3 binding channel. Hydrogen bonds are indicated by dashed magenta lines; purple sphere denotes a water molecule.
In the M3RA complex, RBBP5AS-ABM adopts an extended conformation that consists sequentially of two β-strands (activation segment) and a rigid coil (ABM), which respectively mediate the interactions with MLL3SET and ASH2LSPRY (Fig. 2a). The overall fold of MLL3SET is similar to other SET-domain proteins, and shares the conserved features of N- and C-terminal regions (SET-N and SET-C), an insertion region (SET-I) and post-SET motifs15,26,27 (Fig. 2b). The active site residues of MLL3SET, the conformation of the target lysine and an invariant water molecule, are almost identical to those of the active site of DIM-5 (ref. 28), suggesting a catalytically active configuration of MLL3SET (Fig. 2c). The ‘U’-shaped cofactor product AdoHcy binds into a well-defined surface pocket on MLL3SET through an extensive network of highly conserved interactions as observed in other SET-domain structures (Fig. 2d and Extended Data Fig. 3c, d). The H3 substrate peptide sits in an opposite groove on the surface of MLL3SET, and an intricate network of hydrogen bonds stabilizes the binding (Fig. 2e and Extended Data Fig. 3e). The unique geometry of the H3-binding groove specifically recognizes Thr3H3 and Arg2H3, defining the substrate specificity of MLL3SET (Extended Data Fig. 3f, g). Since all the H3-peptide-binding residues in MLL3SET are highly conserved in other MLL proteins (Extended Data Fig. 4), we conclude that all MLL proteins achieve the substrate specificity towards H3K4 through the same recognition mechanism as observed in the M3RA complex.
Interfaces in the M3RA complex
The structure of the M3RA complex reveals extensive interactions among ASH2LSPRY, RBBP5AS-ABM and MLL3SET. ASH2LSPRY recognizes RBBP5ABM through extensive salt-bridge and hydrogen-bonding interactions; the C-terminal portion of RBBP5ABM adopts a coiled conformation sitting on two arginine residues (Arg343 and Arg367) at the centre of the basic pocket of ASH2LSPRY (Fig. 3a). Mutations of ASH2L Arg343 and its interacting residues in RBBP5ABM (Glu349 and Asp353) completely abrogated the RBBP5ABM–ASH2LSPRY interaction (Extended Data Fig. 5a) and impaired the HKMT activity of the MLL3 complex (Extended Data Fig. 5b). The primary feature of the RBBP5AS–MLL3SET interaction is the inter-molecular β-sheet interactions involving two strands of the L-shaped RBBP5AS paring with β4 and an induced strand β7 immediately before helix αC of MLL3SET (Fig. 3b). Mutations of residues on this L-shaped motif partially decreased the HKMT activity of the MLL3 complex (Extended Data Fig. 5c). In addition to these binary contacts, the side chain of the conserved Arg4806 of MLL3SET sticks outside towards an acidic pocket formed by both RBBP5ABM and ASH2LSPRY, forming five salt-bridge and hydrogen-bonding interactions with Glu347RBBP5, Tyr313ASH2L and Gln354ASH2L (Fig. 3c). This extensive electrostatic network functions as an anchor point to fix the relative position of MLL3SET to ASH2LSPRY, and is crucial for assembly of the MLL3–RBBP5–ASH2L complex.
Figure 3. Interfaces among MLL3SET, RBBP5AS-ABM and ASH2LSPRY.
a, Detailed view of the ASH2LSPRY–RBBP5ABM interface. b, The interface between MLL3SET and RBBP5AS. c, MLL3 Arg4806 forms an extensive salt-bridge and hydrogen-bonding network with ASH2L and RBBP5. d, Mutations of the conserved arginine in MLL proteins disrupt interactions with RBBP5–ASH2L, as shown by the GST pull-down assay in 300 mM NaCl buffer. e, Arginine mutations impair the HKMT activities of MLL family proteins. Activities of all complexes are normalized to the activity of wild-type MLL1–WDR5–RBBP5–ASH2L, and shown as mean ± s.d. (n = 3).
Because the RBBP5AS-ABM–ASH2LSPRY-interacting residues are highly conserved in MLL-family proteins (Extended Data Fig. 4), we proposed that all MLLSET domains including MLL1SET should interact with RBBP5–ASH2L through the same molecular surface as observed in the M3RA complex. In support of this idea, alanine substitutions of the conserved arginine residues in all MLL proteins, which do not affect the overall fold of MLL proteins (Extended Data Fig. 5d), abolished the interaction between MLLSET and RBBP5–ASH2L (Fig. 3d), and substantially decreased the HKMT activities of all MLL complexes (Fig. 3e). Accordingly, mutations of the arginine-interacting residues on RBBP5 and ASH2L (RBBP5 Glu347 and ASH2L Gln354) also weakened the association of MLLSET with RBBP5–ASH2L, and reduced the HKMT activities of MLL complexes (Extended Data Fig. 5e, f). Together, our data confirmed that the electrostatic network observed at the MLL3SET–RBBP5–ASH2L interface is essential for the interaction between RBBP5–ASH2L and all MLL proteins. Interestingly, an inactivating mutation of the same key arginine residue in MLL4 (Arg5432Trp) that was identified in patients with non-Hodgkin lymphoma9 also disrupted the interaction between MLL4 and RBBP5–ASH2L and abolished the HKMT activity (Fig. 3d, e), indicating that loss of a stable MLL4–RBBP5–ASH2L association leads to lymphomagenesis.
Difference between MLL1 and other MLL proteins
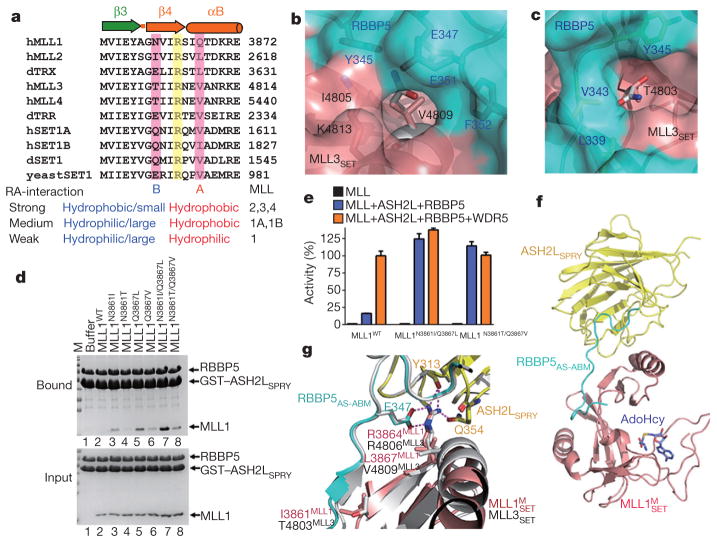
The structure of the M3RA complex revealed that subtle sequence differences in the RBBP5–ASH2L-binding region (residues 4804–4814 in MLL3) are probably responsible for the ability of RBBP5–ASH2L to distinguish MLL1 from other MLL proteins (Fig. 4a). Most notably, the side chain of Val4809 in the SET-I motif of MLL3SET fits snugly in a shallow pocket formed by both RBBP5 and MLL3SET (Fig. 4b), which can also accommodate the corresponding residues of MLL2, MLL4, SET1A and SET1B at the equivalent positions, but not for the bulky residue Gln3867 of MLL1 (Fig. 4a, b). In addition, the side-chain methyl group of MLL3SET Thr4803 is surrounded by three hydrophobic resides of RBBP5AS (Leu339, Val343 and Tyr345) (Fig. 4c). By contrast, a large hydrophilic residue Asn3861 at this position in MLL1 is incompatible with RBBP5 binding (Fig. 4c). Thus, we proposed that two residues (Asn3861 and Gln3867) in MLL1 weaken the otherwise stable interaction between RBBP5–ASH2L and MLL1. Indeed, both MLL1-to-MLL2 (Asn3861Ile/Gln3867Leu) and MLL1-to-MLL3 (Asn3861Thr/Gln3867Val) double mutants of MLL1 re-gained stable interactions with RBBP5–ASH2L (Fig. 4d), and WDR5 had no further stimulatory effect on the HKMT activities of these mutants (Fig. 4e). Therefore, mutations of these two residues restore the strong RBBP5–ASH2L binding ability of MLL1 and thus bypass the requirement of WDR5 as the bridging molecule for the optimal HKMT activity of the MLL1 complex. This idea is further supported by the crystal structure of the complex (hereafter referred to as M1MRA, in which ‘M’ denotes mutant) (Fig. 4f and Extended Data Table 2). The structure of M1MRA highly resembles that of M3RA, with an identical interface as the one between MLL3SET and RBBP5–ASH2L (Fig. 4g), strongly indicating that the RBBP5–ASH2L heterodimer interacts with and activates all MLL proteins through a conserved mechanism. Notably, the equivalent residues of MLL1 Asn3861 in SET1A (Gln1600) and SET1B (Gln1816) also have large hydrophilic side chains and therefore are not optimal for RBBP5–ASH2L binding (Fig. 4a). This is consistent with the medium levels of interaction of SET1A and SET1B with RBBP5–ASH2L observed in the pull-down and fluorescence polarization assays (Extended Data Fig. 2a, b).
Figure 4. Difference between MLL1 and other MLL proteins.
a, Sequence alignment of the RBBP5–ASH2L binding fragments from MLL family proteins. Two key residues that explain the RBBP5–ASH2L binding affinity difference between MLL1 and other MLL proteins are indicated by A and B sites. d, Drosophila; h, human. RA denotes RBBP5–ASH2L. b, The MLL3–RBBP5 interface around MLL3 Val4809. MLL1 Gln3867 (grey) cannot fit into this pocket. c, The MLL3–RBBP5 interface around MLL3 Thr4803, which is not compatible with MLL1 Asn3861 (grey). d, GST pull-down assay for the interactions of RBBP5–ASH2L with MLL1SET and its mutants. e, The normalized HKMT activities of MLL1WT and MLL1N3861I/Q3867L in the presence of full-length RBBP5–ASH2L and WDR5–RBBP5–ASH2L. Mean ± s.d. (n = 3) are shown. f, The overall structure of the M1MRA complex. g, Superposition of the structures of M1MRA and M3RA shows conserved interfaces between MLLSET and RBBP5–ASH2L.
Activation mechanism of MLL complexes
Next we asked why MLL proteins by themselves are catalytically inactive, and how RBBP5–ASH2L stimulates their HKMT activities. One prevailing model suggests that the SET domain of MLL adopts an open conformation, and the interaction with regulatory factors induces MLL SET domain into a closed conformation15. To test this model, we crystallized apo MLL3SET and determined its structure in complex with AdoHcy (Extended Data Fig. 6a). Surprisingly, the apo structure of MLL3SET was almost indistinguishable from the active conformation of MLL3SET in the M3RA complex (Fig. 5a). In addition, we also determined the crystal structure of , the SET-I motif of which exhibits an even more closed conformation than that in the M1MRA complex (Fig. 5b and Extended Data Fig. 6b). Nevertheless, our data clearly showed that both MLL3 and MLL1M by themselves are catalytically inactive (Fig. 4e and Extended Data Fig. 1). This apparent discrepancy between the low enzymatic activity and the closed conformation of or MLL3SET led us to propose that, in the absence of RBBP5–ASH2L, MLLSET might be highly dynamic, and the configuration of MLL SET-I motif captured in the crystal structure is a snapshot of a spectrum of conformations of a mobile motif. In support of this idea, normal mode analysis revealed a highly dynamic motion of the SET-I motif in apo and MLL3SET, which is substantially suppressed upon the association with RBBP5–ASH2L (Supplementary Videos 1–5). To test this model experimentally, we use 19F-NMR (fluorine-19 nuclear magnetic resonance) to probe the structural dynamics of MLL3SET in solution. The 19F-NMR spectrum of Phe4827, a key residue at the substrate-binding site in the SET-I motif, displayed two peaks at different chemical shifts, defining at least two different conformations or states with dynamic exchanges (Fig. 5c). With titration of RBBP5–ASH2L, the 19F-NMR spectrum showed prominent changes with conformational equilibrium towards a single active state, indicating that RBBP5–ASH2L reduced the flexibility of SET-I to lock it in an active state (Fig. 5c). By contrast, Tyr4762 that is located in the SET-N motif exhibited no peak shift upon the addition of RBBP5–ASH2L (Fig. 5c).
Figure 5. Activation mechanism of MLL proteins by RBBP5–ASH2L.
a, Structural comparison of the apo MLL3SET and MLL3SET in the M3RA complex. The structures are superimposed according to AdoHcy. b, Structural comparison of the apo and in the M1MRA complex. c, One-dimensional 19F-NMR measurements of MLL3SET with substitution of F4827tfmF (top) or Y4762tfmF (bottom) in the absence or presence of RBBP5–ASH2L. The locations of these two residues on MLLSET are shown. tfmF, l-4-trifluoromethylphenylalanine. d, Root mean square fluctuation (RMSF) of the SET domains in apo MLL3SET (black line) and in the M3RA complex (red line). e, RBBP5 Phe336 together with MLL3 Arg4845, Tyr4846 and Tyr4825 maintain a configuration that favours cofactor binding. f, The most highly correlated residues (correlation coefficients greater than 0.55) of SET-I in molecular dynamics simulation are indicated by red lines. g, Structural superimposition of the M3RA and M3RA–H3 complexes by the SET-I motifs highlights the local rearrangement of loop LB5. h, A working model for the activation of MLL family methyltransferases.
To provide further insight into this dynamic process, we carried out molecular dynamics simulation to investigate how RBBP5–ASH2L affects the structures of MLL3SET and . Results showed that RBBP5–ASH2L reduces the root mean square fluctuation of helix αB and strand β7 of MLL3SET substantially (Fig. 5d). This coincides with our observation that the most variable element in apo MLLSET is the αB helix, illustrated by the superimposition of four apo MLLSET structures (Extended Data Fig. 6c). Furthermore, a flexible loop in apo MLL3SET (L6C) is induced to form strand β7 by pairing with strand β1 of RBBP5AS (Fig. 5e). Other than the structural variation of individual residues, molecular dynamics simulation also clearly showed that the cross-correlation within the SET-I motif was greatly enhanced upon RBBP5–ASH2L association (Fig. 5f and Extended Data Fig. 6d–f). The reduced flexibility of the SET-I motif may help cofactor binding and substrate recognition. Indeed, isothermal titration calorimetry (ITC) analysis showed that the binding affinities of cofactor to MLL3SET and are markedly increased in the presence of RBBP5–ASH2L (Extended Data Fig. 7a, b). Furthermore, the association with RBBP5–ASH2L also facilitates MLL3 binding with the H3 substrate peptide (Extended Data Fig. 7c). Notably, Phe336 at the beginning of β1 in RBBP5AS stacks together with the side chains of Arg4845/Arg3903, Tyr4846/Phe3904 and Tyr4825/Tyr3883, and the latter makes a direct hydrogen-bonding interaction with AdoHcy (Fig. 5e). Molecular dynamics simulation revealed an obvious stabilizing effect of RBBP5–ASH2L on this network of interactions, which could explain the enhanced cofactor-binding ability for the M3RA complex (Extended Data Fig. 7d). Quantum mechanics/molecular mechanics (QM/MM) investigations further indicated that the presence of RBBP5–ASH2L facilitated the methyl transfer process from the cofactor AdoMet to the target lysine by lowering the energy barrier (Extended Data Fig. 7e). Taken together, we conclude that the RBBP5–ASH2L-induced conformational constraints on the SET-I motif help to stabilize MLLSET in a conformation competent for cofactor binding and substrate recognition.
Structural comparison of the M3RA complex structures with and without the H3 peptide revealed a role of the substrate peptide in further stabilizing the active conformation of MLL3SET, which has been observed in other SET-domain methyltranferases28. After H3 binding, a marked local structural rearrangement occurs to loop LB5 between helix αB and strand β5 in the SET-I motif, leading to the completion of a narrow hydrophobic channel that orients the H3 Lys4 side chain for catalysis (Fig. 5g). Remarkably, the side chain of MLL3 Val4824 shifts ~4.1 Å and rotates ~50° relative to its position in the H3-peptide-free structure, enclosing the target lysine access channel (Extended Data Fig. 7f). Collectively, our studies suggest a novel two-step mechanism for MLLSET activation: interaction with the RBBP5–ASH2L heterodimer reduces the inherent flexibility of MLLSET and favours formation of a catalytically competent conformation; and then H3 substrate binding induces a local conformational change in the SET-I motif of MLLSET to achieve the fully active configuration that facilitates the methyl transfer process (Fig. 5h).
Implications for other methyltransferases
Structural comparison of the M1MRA and M3RA complexes with a large group of SET domain proteins reveals a striking similarity with other intrinsic active methyltransferases. In all SUV39- and SET2-family proteins, a short fragment amino-terminal to the pre-SET region (referred to as the activation segment) interacts with the SET-I motif in the same manner as RBBP5AS binding to MLL3SET (Extended Data Fig. 8a, b). Deletion of this activation segment from DIM-5 did not affect the overall fold of the protein but completely abrogated the HKMT activity of DIM-5, underscoring the importance of this segment in DIM-5 activity regulation (Extended Data Fig. 8c, d). Such an activation segment is also found in the EZH2 complex structure29, further supporting a conserved activation mechanism for a subset of SET-domain-containing methyltransferases.
In summary, the present structural, biochemical and computational analyses provide new insights into the assembly and regulation mechanism of MLL family complexes. Our results suggest that a minimized RBBP5–ASH2L heterodimer is the structural unit to interact with and activate all MLL family histone methyltransferases. By contrast, WDR5 is not directly involved in the enzymatic stimulation of MLL complexes. WDR5 may serve as a recruitment module or crosstalk mediator to regulate H3K4 methylation in vivo30–34.
METHODS
No statistical methods were used to predetermine sample size.
Protein expression and purification
The SET domains of MLL family proteins (with or without the WIN motif), RBBP5, ASH2L, WDR5 and their truncations or mutants were purified as described before19. Escherichia coli Rosetta cells bearing expression plasmids were induced for 16 h with 0.1 mM IPTG at 18 °C, and the cells were collected by centrifugation. For MLL expression, 10 μM ZnSO4 was included in the media. The cell pellets were resuspended in lysis buffer (50 mM Tris-HCl, pH 8.0, 400 mM NaCl, 10% glycerol, 2 mM 2-mercaptoethanol, and home-made protease inhibitor cocktail). The cells were broken by sonication and cleared by ultracentrifugation at 100,000g for 30 min. The proteins were purified using Ni-NTA agarose beads (Qiagen) for His-tagged proteins or Glutathione Sepharose 4B beads (GE) for GST-tagged proteins, followed by enzyme digestion to remove the tags and gel-filtration chromatography. MLLSET, ASH2LSPRY, WDR5 and RBBP5 fragments were separated on Hiload Superdex 75, while full-length proteins of ASH2L and RBBP5 were separated on Hiload Superdex 200. The buffer for gel-filtration chromatography contains 25 mM Tris-HCl, pH 8.0, 150 mM NaCl except for MLLSET (which is in buffer 50 mM Tris-HCl, 300 mM NaCl and 10% glycerol, pH 8.0). The purified proteins were concentrated to 10–20 mg ml−1 and store at −80 °C. RBBP5 peptides were separated on Hiload Superdex 75 after tag digestion in buffer (100 mM NH4HCO3) and the peptide-containing fractions were lyophilized. The MLLSET–RBBP5AS-ABM–ASH2LSPRY complex was obtained by step-wise gel-filtration chromatography; binary complex of RBBP5AS-ABM–ASH2LSPRY was first purified, and then mixed with or MLL3SET, followed by separation on Hiload Superdex 75. Mutations were introduced by PCR-based site-directed mutagenesis, and mutated proteins were purified using the same protocol as described above.
Crystallization, data collection and structural determination
For structural studies, more than 50 different combinations of numerous RBBP5 fragments, ASH2LSPRY constructs, and SET domains from different MLL proteins were tested for crystallization. MLL3SET was crystallized in 100 mM Tris-HCl, pH 8.5, 3 M NaCl at 4 °C in the presence of 1 mM AdoHcy. Zinc single-wavelength anomalous dispersion (SAD) and native data sets of MLL3SET were collected at SSRF (Shanghai Synchrotron Radiation Facility in China) beamline BL17U at wavelengths of 1.2818 Å and 0.9793 Å, respectively. Data were indexed, integrated, and scaled using program HKL2000 (ref. 35). Crystals belong to space group P4132 and contain one MLL3SET per asymmetric unit. Zinc SAD phase determination, density modification and automatic model building were carried out using SHARP36. The initial model was further refined using the native data set diffracted at 2.8 Å. After several rounds of refinement in PHENIX package37 with manual rebuilding in COOT38, the final model has good stereochemistry with an R value of 18.0% and an Rfree of 22.9%.
The MLL3SET–ASH2LSPRY–RBBP5AS-ABM complex was crystallized in 100 mM sodium cacodylate, pH 6.5, 10% PEG-3350, 0.1 M MgCl2 at 4 °C in the presence of 1 mM AdoHcy. The co-crystal with H3 peptide (ARTKQTARK) was obtained by soaking crystals in reservoir solution with 1 mM H3 peptide for 2 h before collection. A data set of 2.4 Å resolution was collected at Advanced Photon Source beamline 21ID-D at wavelength of 1.1272 Å. The crystal belongs to space group P21212 with cell dimension a = 80.342 Å, b = 236.076 Å, c = 44.416 Å. The complex structure was solved by molecular replacement using PHASER39 with ASH2LSPRY structure (PDB accession 3TOJ) and the MLL3 structure SET-N, SET-I, and SET-C motifs as search models. There are two MLL3SET–ASH2LSPRY–RBBP5AS-ABM complexes in one asymmetrical unit, and we can only observed H3 peptide in the density map of one complex. The model was further refined using PHENIX with manually rebuilding in COOT.
crystals were grown by sitting drop vapour diffusion method at 4 °C in a solution containing 35% (v/v) tacsimate, pH 7.0, in the presence of 2 mM AdoHcy, and the crystals were cryo-protected in the same reservoir solution supplemented with 20% glycerol. Data sets were screened and collected at SSRF BL18U and BL19U. The structures were solved by molecular replacement (starting model PDB accession 2W5Y). The complex was crystallized at 200 mM NaCl, 20% PEG3350 in the presence of 2 mM AdoHcy. A data set of 1.9 Å resolution was collected at SSRF BL17U at wavelength of 0.9792 Å. The structures were solved by molecular replacement and further refined with PHENIX. All structure figures were generated using PyMOL (The PyMOL Molecular Graphics System, version 1.4.1 Schrödinger, LLC.).
Histone methyltransferase assay
In vitro methyltransferase assays were performed using an H3 peptide as the substrate. Two assay systems were used. The first one is the 3H-methyl-incorporation assay that measured the incorporation of 3H from [3H]AdoMet (S-adenosyl-L-[methyl-3H]-methionine) into the H3 peptide (9 mer: ARTKQTARK). Reactions were carried out at 22 °C for 1 h in the buffer containing 20 mM HEPES, pH 7.8, 5% glycerol, 5 mM dithiothreitol (DTT), 0.5 mM EDTA, 1 μCi [3H]AdoMet as previously described19. Unmodified H3 K4 peptides (0.25 mM) and 1 μM of WDR5, RBBP5, ASH2L and MLL proteins were used, except for SET1A (5 μM). For all activity assays, full-length WDR5, RBBP5 and ASH2L were used unless stated otherwise. MLL constructs containing both the WIN motif and SET domain were used. Each assay was performed in triplicate, and the mean ± s.d. was reported. The second assay system is to monitor the methylation kinetics of the H3 peptide substrate using MALDI–TOF (matrix-assisted laser desorption ionization–time-of-flight) mass spectrometry.
Mass spectrometry analysis of the methylation process
Methylation reactions were carried out in 20 mM HEPES, pH 7.8, 10 mM NaCl, 5 mM DTT, 250 μM AdoMet, 10 μM histone peptide (ARTKQTARKS) and 1 μM MLL complexes at 22 °C. The reaction was quenched at different time points by addition of trifluoroacetate (TFA) to 0.5%. Reaction mixture was diluted in 10 mg ml−1 CHCA (α-cyano-4-hydroxycinnamic acid) matrix and was spotted onto sample plate and air-dried. The molecular mass was measure by MALDI–TOF (AB SCIEX TOF/TOF 5800) operated in reflectron mode. Final spectra were the average of 200 shots per position at 200 different positions chosen randomly on each spot. To estimate the pseudo-first-order rate constants, we fit the decrease in the relative intensity of the unmodified peptide over time using a model for a single irreversible reaction [Lys4]t = [Lys4]0e−kt, in which [Lys4]0 is the initial concentration of the unmodified peptide, [Lys4]t represents the concentrations of the unmodified peptide at time t and k is the pseudo-first-rate constant.
GST pull-down assays
GST-fusion proteins and interacting partners were incubated with glutathione Sepharose 4B beads for 2 h at 4 °C in 100 μl buffer (50 mM Tris-HCl, 300 mM NaCl and 2 mM DTT, pH 8.0). After extensive wash with the same buffer, the bound proteins were eluted in elution buffer (50 mM Tris-HCl, pH 8.0, 300 mM NaCl and 15 mM reduced glutathione). The input samples and eluted samples were visualized on 12% SDS–PAGE by Coomassie blue staining. Initially, different pull-down buffers were tested and it was found that the interaction between ASH2L and RBBP5 could be disrupted by high ionic strength used in the pull-down assay, whereas the formation of the MLL–RBBP5–ASH2L trimeric complex is relatively insensitive to salt concentration. Thus, in most pull-down assays, buffer with 300 mM NaCl was chosen to assure undisrupted RBBP5–ASH2L interaction and also keep protein stable through pull-down experiments unless stated otherwise.
Isothermal titration calorimetry
The equilibrium dissociation constants of cofactor binding to MLLSET or MLLSET–RBBP5AS-ABM–ASH2LSPRY were determined by an ITC200 calorimeter (GE healthcare). The binding of proteins (20–200 μM) and cofactor AdoMet (0.5–2mM) were measure in the 25 mM Tris-HCl, pH 8.0, 300 mM NaCl at 20 °C. ITC data were analysed and fit using Origin 7 (OriginLab) using one-site model. Owing to instability of apo MLL protein during ITC experiments, curve fitting errors for apo MLL titration are relatively large, so the binding parameters of apo MLL proteins are rough estimations.
Fluorescence polarization assay
Different MLL proteins were diluted in 20 mM HEPES, pH 7.8, 150 mM NaCl, 10% glycerol, 0.5 mg ml−1 BSA to a serial of concentrations from 25 nM to 50 μM. The FAM-labelled RBBP5 peptide (residues 330–363) was mixed with ASH2LSPRY and used at a final concentration of 100 nM. The final volume was brought up to 100 μl with dilution buffer (20 mM HEPES, pH 7.8, 150 mM NaCl, 10% glycerol and 0.5 mg ml−1 BSA) and incubated in dark for 30 min. The fluorescence polarization values were measured using Synergy Neo Multi-Mode Reader (Bio-Tek) at 27 °C. Excitation wavelength was 485 nm and emission was detected at 528 nm. Fluorescence was quantitated with GEN 5 software and date was analysed with Prism 6. For MLL1, SET1A and SET1B, the binding is not saturated even at the highest protein concentration, so the calculated Kd should be an estimated lower limit of Kd value.
19F-NMR spectra measurements
Expression of 19F-labelled proteins was achieved by an established protocol by incorporation of non-natural amino acid tfmF (l-4-trifluoromethylphenylalanine) into specific sites using genetic code TAG40. The 19F-labelled MLL3SET proteins were purified using the same protocol as for wild-type MLL3SET protein. The 19F-NMR spectra were obtained on a Bruker DMX Avance-500 MHz spectrometer equipped with a 5 mM PABBO room temperature probe. The spectra of 0.3 mM MLL3 F4827tfmF or 0.35 mM MLL3SET Y4762tfmF with or without RBBP5–ASH2L were collected at 293 K. The observation channel was tuned to 19F (470.54 MHz), with 512 free induction decay accumulations in every 3-s recycling delay. Each one-dimensional 19F-spectrum was acquired with a standard pulse program with a 90° pulse width of 16.75 μs and power at 35.9 W. 19F-chemical shifts were referenced to an external standard TFA. The free induction decay accumulations, which consisted of 20,480 complex points, was linear predicted to 40,560 points, backward linear predicated three points, and apodized with 20 Hz Lorentzian filter. All spectral processing was performed with Topspin 3.2 software.
Normal mode analysis
Normal modes were calculated using the NOMAD-Ref method41. For all MLL3SET, , M3RA and M1MRA structures, default parameters in the method were used, including the analysis of ‘all atoms’, ‘default distance weight parameters for elastic constant’ of 5.0 Å, ‘ENM cutoff values’ of 1 Å, ‘average RMSD in output trajectories’ of 1.0 Å and ‘output’ of the lowest 16 modes. The first six trivial normal modes are discarded because they represent only translation and rotation. The motion patterns under certain mode and angle monitoring were achieved by using PyMOL.
Molecular dynamics simulation
To delineate how RBBP5–ASH2L modulates the dynamic behaviour of MLLSET domain, we performed molecular dynamic simulations (100 ns) of MLL3SET and MLL1SET in the presence or absence of RBBP5–ASH2L, respectively. The complex structure of MLLSET with RBBP5–ASH2L were centred into a 115 × 115 × 115 Å3 cubic box and dissolved with TIP3P waters. 0.1 M NaCl ions were used to neutralize the net charge of the system. While for the systems of MLLSET alone, we just removed the RBBP5–ASH2L from the complex to make sure the identical conformations of MLLSET before performing molecular dynamics simulations. The same procedures were used in setting up the MLLSET domains without RBBP5–ASH2L except for a smaller cubic box (83 × 83 × 83 Å3). All molecular dynamics simulations were performed using Gromacs 5.0.4 with Charmm36 force field. After the energy minimization of the whole system using the steepest descent algorithm, we first gradually heated the system to 300 K under NVT condition. Then we equilibrated the solvent and ions around the protein using NPT ensemble. In the equilibrations, the backbone of the protein was constrained with a harmonic potential of 1,000 kJ mol−1. The leap-frog integrator was used with an integration time-step of 2 fs. The Berendsen barostat was used to control the pressure at 1 bar with a coupling constant of 2 ps and the modified Berendsen (V-rescale) thermostat was employed to control the temperature of the systems at 300 K with a time constant of 0.1 ps. The Particle Mesh Ewald method was used to compute the electrostatic interactions with a real-space cut-off distance of 1 nm. The same cutoff value was chosen for treating the van der Waals interactions. After a 5 ns equilibration, we conducted the production molecular dynamics by changing the pressure and the thermostat coupling to Parrinello–Rahman and Nose–Hoover with coupling constants of 5 ps and 1 ps, respectively.
The dynamical network analysis of MLL3SET and were performed using networkSetup in VMD. Cα atoms of MLLSET were defined as the node domains and the dynamical contact was drawn if two nodes were within a cutoff distance of 4.5 Å for at least 75% of the molecular dynamics trajectory. The cross correlation data were also calculated to weight edges in the dynamical network. The edge distances dij, which define the probability of information transfer across a given edge: dij = −log(|Cij|), were derived from pairwise correlations (Cij) using program Carma.
To investigate how RBBP5–ASH2L affects methyl transfer process from the cofactor AdoMet to the target lysine of the H3 substrate, we performed QM/MM simulations to calculate the potentials of mean force for the methyl transfer reaction along the reaction coordinate (RC) of r(CM − Sδ) − r(CM − Nη1). Initial structures of the QM/MM simulations were derived from the snapshots of molecular dynamics trajectories in the presence of AdoMet and H3 substrates, simultaneously. Then each structure was solvated in an equilibrated 25 Å spherical water box represented by the TIP3P water model. The water box was centred at the centre of mass of the target lysine residue accepting the methyl group. In total, 20 atoms were selected as the QM zone, including the sulfur atom and the to-be-transferred methyl group on the peptide as well as the lysine residue. The simulation was performed in NVT ensemble at 300 K. The hybrid QM/MM method was used in the simulation. QM interactions are calculated using semi-empirical AM1 method and three GHO atoms (C4′ and CB, which connect the sulfur atom to the other two methyl group, and CD of the lysine residue) were selected as the boundary between QM and mM regions. The solvent boundary potential was treated by the generalized solvent boundary potential method and all atoms out of the water box were fixed. The umbrella sampling method was used to model the reaction process, with the reaction coordinate set as the difference between the sulfur atom on the peptide and the nitrogen atom on the QM lysine. The whole reaction process was distributed into 46 windows and the corresponding reaction coordinate ranged from −1.5 to 2.0 Å with an interval of 0.1 Å. Systems were restrained to each window with a force constant of 500 kcal mol−1 Å−2.
Extended Data
Extended Data Figure 1. Methyltransferase activity of MLL1–MLL4, SET1A and SET1B with the different combinations of WDR5, RBBP5 and ASH2L.
a, HKMT activities determined by the 3H-methylin-corporation assay. MLL constructs were chosen to contain both the WIN motif and the SET domain. Full-length WDR5, RBBP5 and ASH2L were used. The HKMT activities are normalized to the activity of the MLL–WDR5–RBBP5–ASH2L complexes setting at 100%. Mean ± s.d. (n = 3) are shown. b, Representative MALDI–TOF spectra at different time points for MLL complexes and apo MLL proteins clearly revealed that MLL complexes have much higher HKMT activities than apo MLL proteins. The peaks for unmodified (un) and mono-, di- and tri-methylated products are labelled. The minor peaks are sodium adducts of major peaks (+22 Da). Asterisks denote the adduct of un-peaks; filled circles denote the adduct of mono-peaks; and filled squares denote the adduct of di-peaks. c, Comparison of the overall rates of the methylation reactions catalysed by different MLL proteins in the presence of WDR5–ASH2L–RBBP5 or ASH2L–RBBP5. The overall rates were derived by fitting the decrease in the relative intensity of the unmodified H3 peptide peaks in MALDI–TOF mass spectra using one-phase exponential decay model [Lys4]t = [Lys4]0e−kt.
Extended Data Figure 2. Interactions between MLL proteins and RBBP5–ASH2L.
a, GST pull-down assays showed direct interactions between MLL proteins and RBBP5–ASH2L. ASH2L C-terminal SPRY domain has been previously shown to interact with RBBP5. GST-fused ASH2LSPRY was incubated with full-length RBBP5 and different MLLSET proteins in the GST pull-down assay. Bound proteins were eluted and separated by SDS–PAGE. Three different salt concentration buffers were tested. b, Fluorescence polarization assay shows that MLL proteins can interact with RBBP5AS-ABM–ASH2LSPRY with different affinities. For MLL1, SET1A and SET1B, lower limits of the Kd values are reported because saturation of the binding could not be achieved in fluorescence polarization assays. c, GST–RBBP5 alone cannot pull down MLL proteins in the buffer with 300 mM NaCl. d, GST–ASH2LSPRY alone cannot pull down MLL proteins in the buffer with 300 mM NaCl. e, The RBBP5–ASH2L interaction is highly dependent on the salt concentration used in the assay. ITC measurements were carried out using ASH2LSPRY and RBBP5AS-ABM under buffer conditions with different salt concentrations. f, The requirement of WDR5 in methyltransferase activity of the MLL1 complex is sensitive to protein concentration. MLL1 (5 μM) could be markedly stimulated by equal amounts of ASH2L–RBBP5, and WDR5 had a minor stimulation effect. g, HKMT activities of RBBP5–MLL1 fusion proteins in the presence of ASH2L or ASH2L and WDR5. Full-length RBBP5 was fused to MLL1 (residues 3754–3969) with a GGSGGS linker. The addition of ASH2L substantially stimulated the HKMT activity of the RBBP5–MLL1 fusion protein, while further addition of WDR5 only had a marginal effect.
Extended Data Figure 3. The overall structure of the MLL3SET–RBBP5AS-ABM–ASH2LSPRY–H3 complex.
a, The overall structure of the MLL3SET–RBBP5AS-ABM–ASH2LSPRY–H3 complex in cartoon diagram. ASH2L is in yellow-orange, RBBP5 in cyan, MLL3SET in salmon, the H3 peptide in yellow, and cofactor product (AdoHcy) in blue. The electron density (2Fo − Fc) map, contoured at 1σ, is shown for the RBBP5 fragment, the H3 peptide and AdoHcy. b, The electron density (2Fo − Fc) map, contoured at 1σ, is shown around the substrate-binding channel. There are two complexes in one asymmetric unit. One complex has clear electron density for H3 residues 2–7 (left), while the other exhibits no extra density in the substrate channel (right). c, Cofactor interaction network. Residues important for the AdoHcy–MLL3SET interaction are shown in stick models. Hydrogen bonds are indicated by dashed magenta lines. d, The space-filling model of MLL3SET shows that AdoHcy and H3 bind to the opposite surfaces on MLL3SET. The distance between the sulfur atom and ε-amine of Lys4 is shown. e, The binding interface between MLL3SET and H3. f, MLL3SET is in surface representation and coloured according to its electrostatic potential. Thr3 of H3 sits snugly on a shallow hydrophobic depression, which cannot accommodate residues with a large side chain. Arg2 is involved in electrostatic interactions with MLL3SET. g, Sequence alignment of histone methylation sites. Residues are numbered relative to the target lysine. Because only the Lys4 site of H3 contains a large basic residue and a small residue occupying the −2 and −1 positions respectively, Arg(−2) and Thr(−1) define the substrate specificity of MLL complexes.
Extended Data Figure 4. Sequence alignment of MLL homologues from human, Drosophila and Saccharomyces cerevisiae.
The WDR5-interacting motif (WIN) and SET domain are aligned. Secondary structure assignments based on the MLL3 structure are shown as cylinders (α-helices) and arrows (β-strands) above the sequences. The WIN motif is coloured in blue, SET-N in green, SET-I in orange, SET-C in purple and post-SET in magenta. Conserved residues important for RBBP5–ASH2L interactions are highlighted in magenta. Four Zn-binding cysteine residues are highlighted in pale yellow. Residues important for cofactor binding are in brown; residues important for substrate H3 binding and maintenance of the active centre are in grey. Two glycine residues, which serve as the hinge for SET-I motif rotation, are indicated by blue dots. The residues with the corresponding MLL4 mutations found in Kabuki syndrome and non-Hodgkin lymphoma are indicated by stars.
Extended Data Figure 5. The ternary interaction interface among MLL, RBBP5 and ASH2L.
a, Mutations of RBBP5 and ASH2L disrupted the interaction between ASH2LSPRY and RBBP5. Left, GST–RBBP5330–381 was used to pull down ASH2LSPRY and its mutants. Right, GST–ASH2LSPRY was used to pull down full-length RBBP5 and its mutants. Several control mutations (such as ASH2L(Q354A) and RBBP5(E347A)), which are not on the RBBP5–ASH2L interface, did not affect the interaction between ASH2L and RBBP5. b, ASH2L and RBBP5 mutants that disrupted the RBBP5–ASH2L interaction decreased the HKMT activities of the MLL3 complex. The activities of the mutant proteins are normalized to the wild-type MLL3–RBBP5–ASH2L complex. Mean ± s.d. (n = 3) are shown. c, Mutations of RBBP5AS residues decreased the HKMT activity of the MLL3 complex. d, Representative gel-filtration profiles for MLL and MLL mutant proteins indicate MLL mutant proteins have a similar fold to wild-type protein. e, GST–MLL3SET was used to pull down full-length RBBP5, ASH2LSPRY and their mutants. Mutations of RBBP5 Glu347 and ASH2L Gln354 in the ternary interface impaired the interaction with MLL3SET. Mutations of RBBP5AS residues (Phe336Ala, Glu338Ala/Leu339Ala) also decreased the interaction with MLL3SET to different degrees. f, RBBP5(Glu347Ala) and ASH2L(Gln354Ala) compromised the HKMT activities of all MLL complexes, indicating that RBBP5–ASH2L regulates MLL family proteins through the same mechanism. Activities of mutant complexes are normalized to the activity of wild-type MLLSET–RBBP5–ASH2L, setting at 100%. Mean ± s.d. (n = 3) are shown.
Extended Data Figure 6. Activation mechanism of MLL proteins.
a, The structure of MLL3SET is shown in cartoon diagram. The electron density (2Fo − Fc) maps (contoured at 1σ) of AdoHcy are shown. b, The structure of is shown in cartoon diagram. The electron density (2Fo − Fc) maps (contoured at 1σ) of AdoHcy are shown. c, Structural comparison of MLL1SET (PDB 2W5Y), , MLL3SET and MLL4SET (PDB 4Z4P) suggests that the SET-I motif is intrinsically flexible, and can be captured in different configurations by crystallization. There are two highly conserved glycine residues serving as hinge points that connect the SET-I motif to the rest of MLLSET. The rotation of helix αB in the SET-I motif refers to an axis defined by the two hinge points of SET-I as indicated. d, Dynamic cross-correlation matrix for motions of all Cα atoms in apo MLL3SET and MLL3SET in the M3RA complex over the course of the simulation. The right panel shows enlarged cross-correlation maps of the SET-I motif. e, Dynamic cross-correlation matrix for motions of all Cα atoms in apo and in the M1MRA complex over the course of the simulation. The right panel shows enlarged cross-correlation maps of the SET-I motif. f, The most highly correlated residues of the SET-I motif by molecular dynamics simulation are indicated by red lines. Left panel is for apo and right panel is for in the MLL1MRA complex. Red lines are connected Cα atoms for pairs of residues with calculated correlation coefficients greater than 0.55.
Extended Data Figure 7. Association of RBBP5–ASH2L increases the binding affinities of MLL to cofactor and substrate peptide.
a, ITC measurement of interactions of AdoMet with MLL3SET alone (blue) and the M3RA complex (red). The insets show the ITC titration data. b, Equilibrium dissociation constants between cofactor and MLL proteins obtained from ITC measurements. c, Fluorescence polarization assay shows that RBBP5–ASH2L increases the binding affinity between MLL3 and the H3 peptide substrate. d, Molecular dynamics simulation show dynamics of the cofactor binding pocket. Top, the distance between AdoHcy and Tyr4825; bottom, the distance between Arg4845 and Tyr4825. These distances are almost fixed in the M3RA complex, while the distances in apo MLL3 have large variations, explaining why the MLL3 complex has a higher binding affinity to cofactor than apo MLL3 does. e, The potentials of mean force for the methyl transfer reaction along the reaction coordinate range from −1.5 to 2.0 Å with an interval of 0.1 Å. It clearly shows that the MLLSET–RBBP5–ASH2L complex is more energetic favourable for the methyl transfer reaction than MLLSET alone. f, The space-filling surface model shows that the Ly4H3 binding channel exhibits open and closed conformations in the M3RA and M3RA–H3 structures.
Extended Data Figure 8. A conserved activation mechanism for SET-domain-containing HKMTs.
a, Structural comparison of MLL3SET in the M3RA–H3–AdoHcy complex, and the SET domains of CLR4 (PDB 1MVH), DIM-5 (PDB 1PEG), EZH2 (PDB 5CH1), ASH1 (PDB 3OPE) and NSD1 (PDB 3OOI). Histone H3 peptide and AdoHcy in the CLR4 structure were modelled based on the M3RA–H3–AdoHcy complex structure. RBBP5AS and the corresponding activation segments in these proteins are almost identical in overall conformation (coloured in cyan). The recently reported EZH2 complex structure also revealed such an activation segment. Most notably, an aromatic residue (shown as stick model), equivalent to Phe336 in RBBP5, stacks with another two aromatic residues to form an aromatic cage to sandwich a conserved arginine. Another conserved hydrophobic residue (shown as stick model) is also important for the stable association between the activation segment and the SET-I motif. b, Sequence alignment of the activation segments of RBBP5 and several representative HKMTs, including members from the SUV39 and SET2 families. c, Gel-filtration profiles and SDS–PAGE for DIM-5 and DIM-5ΔAS show that activation segment does not affect protein folding. DIM-5ΔAS denotes DIM-5 (residues 51–319) that does not contain the activation segment. d, HKMT activities of DIM-5 and its mutants. Activities of mutant proteins are normalized to the activity of the wild-type protein setting at 100%. Mean ± s.d. (n = 3) are shown.
Extended Data Table 1.
Data collection and refinement statistics for MLL3SET and the MLL3SET–RBBP5AS-ABM–ASH2LSPRY complex
| MLL3SET Native† | MLL3SET Peak (Zn-SAD)† | MLL3SET-RBBP5AS-ABM-ASH2LSPRY† | |
|---|---|---|---|
| Data collection | |||
| Space group | P4132 | P4132 | P21212 |
| Cell dimensions | |||
| a, b, c (Å) | 129.056 | 129.323 | 80.342,236.076,44.416 |
| α, β, γ (°) | 90 | 90 | 90, 90, 90 |
| Wavelength (Å) | 0.9793 | 1.2818 | 1.1272 |
| Resolution (Å) | 50-2.8 | 50-3.4 | 100-2.4 |
| Rmerge | 0.135(0.530)* | 0.204(0.563) | 0.110 (0.654) |
| l/σl | 31.6(7.0) | 31.0(12.3) | 32.6 (5.1) |
| Completeness (%) | 99.9(100) | 99.9(100) | 99.9(100) |
| Redundancy | 10.1(10.6) | 13.3(13.9) | 7.1 (7.3) |
| Refinement | |||
| Resolution (Å) | 38.9-2.8 | 44.4-2.4 | |
| No. reflections | 9521 | 33458 | |
| Rwork/Rfree (%) | 18.0/22.9 | 18.0/22.8 | |
| No. atoms | |||
| Protein | 1198 | 5570 | |
| Ligand | 27 | 54 | |
| Water | 52 | 220 | |
| B-factors (Å2) | |||
| Protein | 36.22 | 46.65 | |
| Ligand | 33.13 | 52.53 | |
| Water | 32.30 | 41.28 | |
| R.m.s deviations | |||
| Bond lengths (Å) | 0.011 | 0.003 | |
| Bond angles (°) | 1.039 | 0.654 | |
Values in parentheses are for the highest-resolution shell.
The data are collected from one crystal.
Extended Data Table 2.
Data collection and refinement statistics for and complex
|
|
|
|||
|---|---|---|---|---|
| Data collection | ||||
| Space group | P3221 | C2 | ||
| Cell dimensions | ||||
| a, b, c (Å) | 54.547,54.574,104.656 | 74.966,44,410, 117.792 | ||
| α, β, γ (°) | 90,90,122 | 90,106.157,90 | ||
| Wavelength (Å) | 0.9785 | 0.9792 | ||
| Resolution (Å) | 50-1.8 | 50-1.9 | ||
| Rmerge | 0.090(0.421)* | 0.158 (0.548) | ||
| l/σl | 36.5(4.1) | 11.3(2.4) | ||
| Completeness (%) | 100(100) | 99.7(99.8) | ||
| Redundancy | 9.6(9.9) | 3.6 (3.3) | ||
| Refinement | ||||
| Resolution (Å) | 28.1-1.8 | 37.5-1.9 | ||
| No. reflections | 17284 | 29678 | ||
| Rwork/Rfree (%) | 20.2/23.6 | 16.6/21.3 | ||
| No. atoms | ||||
| Protein | 1156 | 2804 | ||
| Ligand | 27 | 27 | ||
| Water | 142 | 361 | ||
| B-factors (Å2) | ||||
| Protein | 45.34 | 20.31 | ||
| Ligand | 33.19 | 27.14 | ||
| Water | 45.77 | 29.99 | ||
| R.m.s deviations | ||||
| Bond lengths (Å) | 0.011 | 0.008 | ||
| Bond angles (°) | 1.105 | 1.044 | ||
The data are collected from one crystal.
Values in parentheses are for highest-resolution shell.
Acknowledgments
We thank staffs of beamlines BL18U, BL19U1 and 17U at the National Center for Protein Sciences Shanghai and Shanghai Synchrotron Radiation Facility for their assistance in data collection. We are grateful to protein expression, protein purification and mass spectrometry facilities at the National Center for Protein Sciences Shanghai for their instrument support and technical assistance. This work was supported by grants from the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB08010201 to M.L. and Y.C., XDB08030302 to C.T.), the Ministry of Science and Technology of China (2013CB910402 to M.L., 2013CB910401 to Y.C., 2012AA01A305 and 2012CB721002 to G.L., 2011CB910400 to C.T.), the National Science and Technology Major Project ‘Key New Drug Creation and Manufacturing Program’ of China (2014ZX09507002-005 to M.L.), the National Natural Science Foundation of China (31330040 to M.L., 31470737 to Y.C., and 91430110 to G.L.), the Basic Research Project of Shanghai Science and Technology Commission (14JC1407200 to Y.C.), the National Institutes of Health (R01 GM082856 to Y.D.), and Fundamental Research for the Central University (WK2340000064 to C.T.). Y.C. is a recipient of the Thousand Young Talents Program of the Chinese government.
Footnotes
Online Content Methods, along with any additional Extended Data display items and Source Data, are available in the online version of the paper; references unique to these sections appear only in the online paper.
Supplementary Information is available in the online version of the paper.
Author Contributions M.L. and Y.C. conceived and supervised the project. M.L. and Y.D. initiated the project. Y.L., J.H., C.H. and Y.C. purified the proteins, performed crystallization and determined the crystal structure. Y.L., J.H., F.C., C.H., J.W., Y.W. and Y.C. performed the biochemical assays. Z.L., P.S. and C.T. performed 19F-NMR experiments. S.L. and J.Z. performed normal mode analysis. Y.Z., L.C. and G.L. performed molecular dynamics and QM/MM simulation. D.L. and Y.D. contributed to manuscript preparation. G.L., Y.C. and M.L. analysed the data and wrote the manuscript.
The atomic coordinates have been deposited in the Protein Data Bank (PDB) under the following accessions: 5F59 (MLL3SET), 5F6K (MLL3SET–RBBP5AS-ABM–ASH2LSPRY), 5F5E ( ) and 5F6L ( ).
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of the paper.
References
- 1.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 2.Shilatifard A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr Opin Cell Biol. 2008;20:341–348. doi: 10.1016/j.ceb.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 4.Ansari KI, Mandal SS. Mixed lineage leukemia: roles in gene expression, hormone signaling and mRNA processing. FEBS J. 2010;277:1790–1804. doi: 10.1111/j.1742-4658.2010.07606.x. [DOI] [PubMed] [Google Scholar]
- 5.Wang P, et al. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol Cell Biol. 2009;29:6074–6085. doi: 10.1128/MCB.00924-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nature Rev Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 7.Ansari KI, Mishra BP, Mandal SS. MLL histone methylases in gene expression, hormone signaling and cell cycle. Front Biosci. 2009;14:3483–3495. doi: 10.2741/3466. [DOI] [PubMed] [Google Scholar]
- 8.Parsons DW, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331:435–439. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morin RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, et al. A mutation screen in patients with Kabuki syndrome. Hum Genet. 2011;130:715–724. doi: 10.1007/s00439-011-1004-y. [DOI] [PubMed] [Google Scholar]
- 11.Pleasance ED, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dalgliesh GL, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dou Y, et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nature Struct Mol Biol. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 14.Patel A, Dharmarajan V, Vought VE, Cosgrove MS. On the mechanism of multiple lysine methylation by the human mixed lineage leukemia protein-1 (MLL1) core complex. J Biol Chem. 2009;284:24242–24256. doi: 10.1074/jbc.M109.014498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Southall SM, Wong PS, Odho Z, Roe SM, Wilson JR. Structural basis for the requirement of additional factors for MLL1 SET domain activity and recognition of epigenetic marks. Mol Cell. 2009;33:181–191. doi: 10.1016/j.molcel.2008.12.029. [DOI] [PubMed] [Google Scholar]
- 16.Steward MM, et al. Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nature Struct Mol Biol. 2006;13:852–854. doi: 10.1038/nsmb1131. [DOI] [PubMed] [Google Scholar]
- 17.Dou Y, Hess JL. Mechanisms of transcriptional regulation by MLL and its disruption in acute leukemia. Int J Hematol. 2008;87:10–18. doi: 10.1007/s12185-007-0009-8. [DOI] [PubMed] [Google Scholar]
- 18.Wysocka J, et al. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 19.Cao F, et al. An Ash2L/RbBP5 heterodimer stimulates the MLL1 methyltransferase activity through coordinated substrate interactions with the MLL1 SET domain. PLoS ONE. 2010;5:e14102. doi: 10.1371/journal.pone.0014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shinsky SA, Monteith KE, Viggiano S, Cosgrove MS. Biochemical reconstitution and phylogenetic comparison of human SET1 family core complexes involved in histone methylation. J Biol Chem. 2015;290:6361–6375. doi: 10.1074/jbc.M114.627646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shinsky SA, Cosgrove MS. Unique role of the WD-40 repeat protein 5 (WDR5) subunit within the mixed lineage leukemia 3 (MLL3) histone methyltransferase complex. J Biol Chem. 2015;290:25819–25833. doi: 10.1074/jbc.M115.684142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang P, Lee H, Brunzelle JS, Couture JF. The plasticity of WDR5 peptide-binding cleft enables the binding of the SET1 family of histone methyltransferases. Nucleic Acids Res. 2012;40:4237–4246. doi: 10.1093/nar/gkr1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang P, et al. A phosphorylation switch on RBBP5 regulates histone H3 Lys4 methylation. Genes Dev. 2015;29:123–128. doi: 10.1101/gad.254870.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Y, Cao F, Wan B, Dou Y, Lei M. Structure of the SPRY domain of human Ash2L and its interactions with RbBP5 and DPY30. Cell Res. 2012;22:598–602. doi: 10.1038/cr.2012.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Odho Z, Southall SM, Wilson JR. Characterization of a novel WDR5-binding site that recruits RbBP5 through a conserved motif to enhance methylation of histone H3 lysine 4 by mixed lineage leukemia protein-1. J Biol Chem. 2010;285:32967–32976. doi: 10.1074/jbc.M110.159921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, et al. evolving catalytic properties of the MLL family set domain. Structure. 2015;23:1921–1933. doi: 10.1016/j.str.2015.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng X, Collins RE, Zhang X. Structural and sequence motifs of protein (histone) methylation enzymes. Annu Rev Biophys Biomol Struct. 2005;34:267–294. doi: 10.1146/annurev.biophys.34.040204.144452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X, et al. Structural basis for the product specificity of histone lysine methyltransferases. Mol Cell. 2003;12:177–185. doi: 10.1016/s1097-2765(03)00224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sirinupong N, Brunzelle J, Doko E, Yang Z. Structural insights into the autoinhibition and posttranslational activation of histone methyltransferase SmyD3. J Mol Biol. 2011;406:149–159. doi: 10.1016/j.jmb.2010.12.014. [DOI] [PubMed] [Google Scholar]
- 30.Wang KC, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ang YS, et al. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell. 2011;145:183–197. doi: 10.1016/j.cell.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gan Q, et al. WD repeat-containing protein 5, a ubiquitously expressed histone methyltransferase adaptor protein, regulates smooth muscle cell-selective gene activation through interaction with pituitary homeobox 2. J Biol Chem. 2011;286:21853–21864. doi: 10.1074/jbc.M111.233098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dou Y, et al. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 34.Thompson BA, Tremblay V, Lin G, Bochar DA. CHD8 is an ATP-dependent chromatin remodeling factor that regulates β-catenin target genes. Mol Cell Biol. 2008;28:3894–3904. doi: 10.1128/MCB.00322-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 36.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 37.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 38.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 39.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr D. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 40.Shi P, et al. Site-specific 19F NMR chemical shift and side chain relaxation analysis of a membrane protein labeled with an unnatural amino acid. Protein Sci. 2011;20:224–228. doi: 10.1002/pro.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lindahl E, Azuara C, Koehl P, Delarue M. NOMAD-Ref: visualization, deformation and refinement of macromolecular structures based on all-atom normal mode analysis. Nucleic Acids Res. 2006;34:W52–W56. doi: 10.1093/nar/gkl082. [DOI] [PMC free article] [PubMed] [Google Scholar]