Abstract
The development of rapid, sensitive, and accurate mass spectrometric methods for measuring peptides, proteins, and even intact protein assemblies has made mass spectrometry (MS) an extraordinarily enabling tool for structural biology. Here, we provide a personal perspective of the increasingly useful role that mass spectrometric techniques are exerting during the elucidation of higher order protein structures. Areas covered in this brief perspective include MS as an enabling tool for the high resolution structural biologist, for compositional analysis of endogenous protein complexes, for stoichiometry determination, as well as for integrated approaches for the structural elucidation of protein complexes. We conclude with a vision for the future role of MS-based techniques in the development of a multi-scale molecular microscope.
Keywords: Mass spectrometry, Higher order structure, Structural biology, X-ray crystallography, Electrospray ionization, MALDI, Folding state in solution, Protein complex, Protein assembly, Protein composition, Stoichiometry, Native mass spectrometry, Endogenous protein complexes, Molecular microscope
Graphical Abstract

Introduction
Living cells are highly dynamic systems that utilize a large variety of proteins to organize their intricate architectures and to carry out their many functions. The spatial organization of these proteins and the structures that they form are hierarchical in nature [1] (Figure 1), beginning with the unique order of amino acids within each different protein (termed their primary structures). These primary arrangements of amino acids fold into forms that include alpha helices, beta sheets, and turns (their secondary structures), which in turn fold or are organized into highly specific three-dimensional shapes (their tertiary structures). Finally, these proteins can assemble with other proteins, DNA, RNA, and lipids to form higher order assemblies that we loosely term protein complexes. Such protein complexes can be stable or highly dynamic, and can form yet higher order assemblies, as in cellular machines such as the replisome responsible for DNA replication [5–7] or the nuclear pore complex responsible for controlling molecular traffic between the cytoplasm and the nucleus [8–10]. This hierarchy continues through still larger substructures, which incorporate other biomolecular components including lipids, to finally the entire cells themselves (Figure 1). Since cellular function is governed by the forms and interactions of its constituents, it is crucial to have available tools for characterizing the dynamic arrangements of proteins at each level within this hierarchy. Here we provide a personal perspective on the increasingly effective role of mass spectrometry (MS) as a tool for probing higher order protein structures. Along the way, we highlight key analytical challenges that still lie ahead of us.
Figure 1.
Hierarchy of protein structures within cells. Mass Spectrometry is playing an increasingly important role in revealing aspects of higher order protein structures as well as the architectures of protein assemblies at all relevant scales within cellular systems. For illustrative purposes, we show a yeast cell nucleus. Embedded in the nuclear envelope at its periphery are nuclear pore complexes (NPCs), the sole mediators of transport into and out of the nucleus. The yeast NPC is an assembly consisting of some 500 protein subunits that are formed through association of a host of protein complexes [2]. One of these, the seven-membered Nup84 protein complex, is shown as an example [3]. There are 16 copies of the Nup84 complex in the yeast NPC [4]
Ideally, biologists would like to investigate proteins and protein assemblies in their natural environment within living cells in full atomic detail. Since this goal remains largely beyond present analytical capabilities, it has been necessary to devise workarounds. Thus, structures are mostly determined using proteins that are recombinantly overexpressed and removed from their endogenous context before being subjected to high resolution analytical procedures such as X-ray crystallography and NMR spectroscopy [11]. Though clearly not ideal, these remarkable procedures have provided most of the information on protein structures currently available at atomic resolutions. Deriving such high resolution protein structures are formidable multidisciplinary undertakings, requiring: appropriate protein construct design and expression; successful folding and purification of the protein; crystallization trials and phase determination in the case of X-ray crystallography or careful tuning of solution conditions in the case of NMR; and finally accurate structure determination and interpretation. Modern MS methods can greatly facilitate these obligate tasks, especially since the advent of effective methods for ionizing proteins such as matrix-assisted laser desorption ionization (MALDI) and electrospray ionization (ESI) [12–14].
MS as an Enabling Tool for the High Resolution Structural Biologist
We and many others have shown that MS can be used to considerable advantage at virtually each of the steps described above that are needed to elucidate the high resolution structure of a protein [15–17]. We illustrate this extraordinary utility, using as an example the first X-ray structure determination of a transmembrane chloride ion transporter [18, 19] (Figure 2). One of the keys to successfully growing diffraction grade protein crystals is to initiate the crystal trials with homogenous starting material. In the early stages of this work, MS proved useful for detecting heterogeneity at the amino-terminal end of the protein resulting from an unanticipated second translation initiation site 26 residues downstream of the main initiator methionine. This heterogeneity was readily detected in the MALDI mass spectrum of the Salmonella ClC chloride transporter expressed in E. coli, and was eliminated by mutating methione-26 to a leucine residue. It is noteworthy that this microheterogeneity would not have been detected with standard analysis techniques, such as SDS-PAGE and size exclusion chromatography, which are commonly used by structural biologists, but lack the resolution of MS. Another widely used strategy to improve the chances of acquiring diffraction grade crystals is to design protein constructs that incorporate the more tightly folded domains while eliminating or reducing the size of other more flexible parts of the protein. Again, this proved to be the case for the ClC chloride transporter, which we determined could be truncated to homogeneity by appropriate digestion with Lys-C endoproteinase. MALDI MS analysis of the digestion products as a function of time determined optimal parameters for obtaining the most homogenous product, which ultimately yielded diffraction grade crystals. Because it is generally not possible to predict the detailed buffer, salt, detergent, and precipitant mixtures that yield high grade crystals, structural biologists standardly assay a wide array of conditions in experimental crystallization trials. Although the number of trials that are needed can sometimes be very large before conditions are found that promote pristine crystal growth, this number can be reduced by using MS to judiciously limit some of the parameters. For example, we tested the sensitivity to trypsin cleavage of the ClC chloride transporter in different detergents, reasoning that improved resistance to proteolysis might provide evidence for enhanced detergent stabilization. Indeed, we found that the transporter was more resistant to proteolysis in octylmaltoside (OM) than it was in decylmaltoside (DM), and ultimately crystals grown in OM yielded diffraction to 3.5 Å resolution [18]. Once crystals that diffract well are available, it is necessary to solve the phase problem [11]. This is generally done by heavy element incorporation—in the present example through incorporation of selenomethionine. To assess the completeness of this incorporation, we measured the mass difference between the methionine-containing protein and that in which selenomethionine had been incorporated, and were able to confirm that the selenomethionine incorporation was essentially complete. During the model building stage, we examined the results of partial proteolysis experiments on the E. coli channel in the light of a simple topology prediction, which indicated 12 putative transmembrane helical segments of approximately equal length [19]. It was immediately apparent from the observed positions of cleavage that this topology model was likely incorrect. Mapping of the cleavage sites onto the actual helical arrangement determined by X-ray diffraction showed that the actual topology was considerably more intricate [18], with the complex orientations of the polarized helices turning out to be a key contributor to the chloride transporter mechanism of action. From studies like these, we and others have observed that the rapid, informative feedback afforded by MS makes it an extraordinarily valuable tool for many high resolution structural biology studies.
Figure 2.
MS helped facilitate several of the obligatory steps that were required during the X-ray structure determination of the ClC chloride transporter [18, 19]. See text for details
MS as a Tool for the Compositional Analysis of Endogenous Protein Complexes
Although solving the structures of recombinant and heterologously expressed proteins is enormously informative, this only provides a narrow snapshot of the hugely complex and dynamic living interactome; hence, we still need to gain as much information as possible about the structure and behavior of proteins in concert with their normal partners and in the context of their native cellular environment. As discussed above, cellular systems are rich assemblies of dynamic, heterogeneous macromolecular complexes. Given this extraordinary complexity, it remains challenging to elucidate their composition and stoichiometry. Here again, modern MS methods are playing an increasingly important role. In particular, high sensitivity qualitative MS approaches are proving especially enabling for identifying and defining the substituents of protein complexes [4, 20–22], while quantitative MS-based approaches are becoming increasingly powerful for determining subunit stoichiometry [23–30]. Indeed, bottom-up MS-based protein identification [31–34] has become a method of choice in the initial discovery mode to determine the composition of an assembly. As an illustrative example, we used this approach in our initial determination of the components that make up the yeast nuclear pore complex (NPC) (Figure 1), a large (~50 MDa) protein-aceous machine embedded in the nuclear envelope [8–10]. The NPC acts as a selective gate for molecular traffic between the cytoplasm and the nuclear interior, but also organizes many genetic processes in the nucleus [35]. At the time that we initiated these studies (mid 1990s), many of the components of the NPC (so-called nucleoporins) were already known, but the full complement had not been elucidated. Thus, the first problem was to define what should be considered to be a bona fide nucleoporin. This was not a trivial exercise because a vast number of macromolecules interact either directly or indirectly with the NPC, including the transport factors the help ferry the myriad proteins, ribosomal subunits, messenger RNA, and other cargo between the nucleus and cytoplasm as well as, for example, interactions with proximal chromosomal components. The problems that we encountered in this study remain classic challenges in most proteomic studies. They include (1) differentiating specific from nonspecific interactions, (2) maintaining the stability of the assembly during biochemical isolation procedures in the face of dissociation and exchange, and (3) differentiating proximal relevant interactions from more distal indirect interactions. Figure 3 shows our separation of the isolated NPC into its component proteins together with their identification by MS whereby we identified 174 different proteins [4]. The major challenge then, as it remains today, was to determine which of these proteins prove to be actual components of the NPC. We used several approaches to address this problem. First, we reasoned that because nucleoporins were present in modest amount (~2000 copies/cell) and because most of the nucleoporins were found exclusively in the NPC, abundant proteins with known function (e.g., ribosomal proteins) could be excluded. Candidates other than these were then genomically tagged, and each of the resulting strains assayed to determine if the tag localized as punctate patterns at the nuclear periphery. In this way, we were able to pare down the 174 co-isolating proteins to 30 putative nucleoporins [4]. Since this analysis was published in 2000, we are now able to assess whether the strategy was accurate and complete. Over the intervening years, all of the putative nucleoporins have been repeatedly verified [10], showing that the accuracy of this type of analysis can be high. It is worth pointing out that at the time that we carried out this analysis, our mass spectrometer had a much lower dynamic range than is available today from the best new instruments. Thus, we were able to detect only the most abundant proteins. Fortunately our NPC preparation was relatively pure (estimated at ~70%) and the components approximately stoichiometric, so that the substituents of interest should have been readily detected. If, however, a preparation is less pure, analysis with the current generation of high dynamic range mass spectrometers will lead to the detection of literally thousands of proteins. So, especially in cases where the bona fide substituents are substoichiometric, it becomes increasingly difficult to discern which of these proteins actually belong to the assembly of interest.
Figure 3.
Identification of proteins in the highly enriched nuclear pore complex (NPC) fraction [4]. Proteins in the highly enriched NPC fraction were separated by hydroxyapatite HPLC and SDS-PAGE. The number above each lane indicates the corresponding fraction number. Proteins were visualized with Coomassie blue; the approximate molecular mass of proteins as estimated from standards shown on the left side. Bands analyzed by MALDI-TOF mass spectrometry are indicated by the adjacent numbers. The proteins identified in each band are shown in the top panel. Proteins known to directly associate with the NPC are colored blue, whereas proteins believed not to be NPC-associated are colored red. On the top left are listed proteins not found in this separation but identified by MS/MS of reversed phase HPLC-separated peptides (RP/HPLC) or peptide microsequencing (PROT SEQ). Figure adapted from [4] with permission
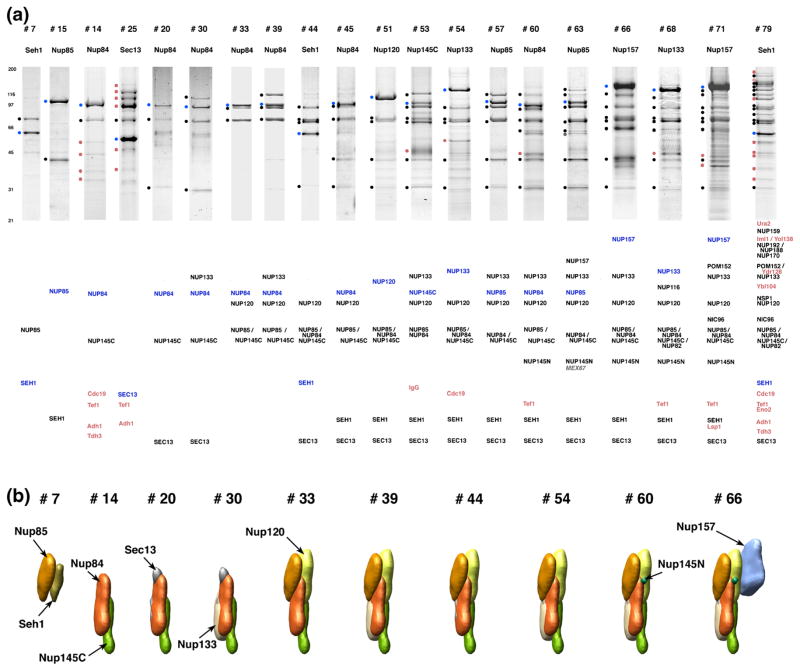
There is a second approach for determining the composition of protein assemblies and learning about their architectures. This involves the use of affinity capture to isolate complexes from an assembly [20, 21, 36–40]. Bottom-up MS readout of these isolated sub-complexes enables identification of the protein constituents as well as information about their proximity to one another. Figure 4 provides an example of this strategy, where we have genomically tagged each of the seven members of a subcomplex within the NPC (termed the Nup84 complex shown in Figure 1) and affinity captured components of the subcomplex at different levels of stringency [41]. The resulting data can be used to provide restraints on an architectural model of the subcomplex. Extending this strategy to include similar data on all the components of the NPC allowed us to obtain the first molecular model of this 50 MDa machine, comprised of some 500 protein subunits [2] (see Figure 1). Again, differentiating specific from nonspecific associations can be highly problematic. Ideally, one would use extraction and affinity capture buffers that co-isolate just the relevant proteins without contaminants. In practice, choosing such optimal buffer systems represents a problem not dissimilar to choosing conditions for crystalizing proteins—a job that we now believe is better left to screening strategies [22]. However, even with the best buffer systems, contaminants will inevitably be present at some level. There are several possible ways of discerning such contaminants from bona fide components of the assembly of interest: first, analyses of large collections of datasets have revealed so-called “CRAPomes,” typically composed of high abundance or sticky proteins that are repeatedly observed in many unrelated experiments [42]. With care, these “CRAPomes” can be exploited to eliminate likely contaminants. Variants of this include computational tools using MS spectral counting to derive the probability of a co-isolating protein being a true interactor [43–45]. Second, if the affinity isolation is relatively clean and the assembly under investigation approximately stoichiometric, minor components can be rationally eliminated, such as when they arise from known highly abundant or sticky proteins [42]. Third, various more specific quantitative approaches can be devised for differentiating specific from nonspecific interactors, as for example in our own development of I-DIRT [46] as well as other useful strategies [47]. In the I-DIRT approach, one strain of the organism of interest is grown in heavy isotope-containing medium, while a second is grown in light medium. The only difference between the two strains is that the one grown in the heavy medium contains an epitope-tagged version of the target protein to be fished out together with its partners in the complex. The cells are mixed and affinity isolation performed on the resulting lysate. Proteins that co-isolate with the heavy epitope-tagged protein and do not exchange during isolation will contain exclusively heavy isotopes—thus unambiguously identifying these interactors as being specific. Contaminants (as well as rapidly exchanging proteins) will contain a 50:50 mix of heavy and light isotopes.
Figure 4.
Protein interactions of the Nup84 complex [41]. (a) Sample of affinity purifications containing Nup84 complex proteins. Affinity-purified protein A (PrA)-tagged proteins and interacting proteins were resolved by SDS-PAGE and visualized with Coomassie blue. The name of the PrA-tagged protein is indicated above each lane. Molecular mass standards (kDa) are indicated to the left of the panel. The bands marked by filled circles at the left of the gel lanes were identified by MS. The identity of the co-purifying proteins is indicated in order below each lane; PrA-tagged proteins are indicated in blue, co-purifying nucleoporins in black, NPC-associated proteins in grey, and other proteins (including contaminants) in red. Each individual gel image was differentially scaled along its length so that its molecular mass standards aligned to a single reference set of molecular mass standards, and contrast-adjusted to improve visibility. (b) The mutual arrangement of the Nup84-complex-associated proteins as visualized by their calculated localization volumes. Figure adapted from [41] with permission
MS for Stoichiometry Determination
To elucidate the structure of any endogenous assembly, it is necessary to know the stoichiometry of its subunits. Although a variety of approaches have been developed to address this analytical task, they can be difficult to implement and sometimes do not provide sufficient accuracy to be definitive [48–53]. The high inherent precision and sensitivity of MS make it an attractive alternative for extracting accurate stoichiometry measurements. Two types of MS approaches for obtaining stoichiometry have been used. The first approach entails isolation of the assembly, enzymatic digestion of its substituent proteins into peptides, followed by measurement of the relative amounts of selected peptides from each of the proteins, sometimes achieving higher accuracies by calibration with the addition of heavy isotopically labeled internal standards [24, 25, 26, 54–60]. A second method for stoichiometry determination arose through the development of MS techniques for measuring the masses of intact multi-subunit protein assemblies [61–77]. This class of methods is a major focus of the present issue of JASMS, being particularly attractive because it can directly examine the composition, stoichiometry, and even connectivity of protein complexes in their near-native states. Figure 5 provides some landmarks in the development of “native mass spectrometry.”
Figure 5.
Selection of landmarks in the development of “native MS.” (a) Mass spectra of cytochrome c electrosprayed from variously acidified aqueous solutions. The observed charge distributions reflect the folding state of cytochrome c in the ESI solution [78]; (b) (top panel) early observation of noncovalent ternary complex between the dimeric enzyme HIV-1 protease and a substrate-based inhibitor obtained under native spray conditions [79]; (bottom panel) only the protease monomer was observed when a high amount of collisional energy was injected into the system; (c) m/z spectrum of a mixture of four compounds obtained with a collisional damping interface for an orthogonal injection ESI time-of-flight mass spectrometer [80], showing that it was possible to measure masses ranging from 1000 to 1,000,000 Da in a single mass spectrum; (d) m/z spectrum of the intact rotary ATPase from T. thermophiles, a membrane-embedded molecular machine with mass 659,202 Da [81]. Peaks are assigned to the intact ATPase complex (stars) as well as to losses of the indicated subcomplex and subunits; (e) (left panel) ESI mass spectrum of ~18 MDa Prohead-1 particles from bacteriophage HK97 capsids. A partially resolved series of charge states is observed, allowing the accurate mass calculation indicated [82]; (right panel) charge detection MS, which simultaneously measures the charge and the m/z of individual ions, of bacteriophage P22 procapsid distributions [83]. Shown is the charge versus mass scatter plot for individual P22 procapsid ions centered at mass 23.60 MDa as well as a lower mass cluster of ions centered at 19.84 MDa, attributed to empty capsids. Figures adapted from the indicated references with permission
One of the early indicators that native MS stoichiometry determination might prove possible had its origins in our observation that the MS charge distributions of proteins produced by ESI reflect (to some extent) the folding state of the proteins in the electrosprayed solution [78] (Figure 5a). Not long thereafter, it was observed that stable cofactors could survive dissociation from proteins during the transition from the solution to the gas phase in the ESI process [84–86]. Remarkably, it was found that even noncovalent protein complexes could maintain their interactions into the gas phase when produced by ESI from nondenaturing solutions [79] (Figure 5b). Because non-denatured proteins and their complexes were considerably less charged than denatured proteins (Figure 5a), one limiting factor during this early period was the low m/z range of the quadrupole mass analyzers then in use (typically 2000–3000 m/z units). This problem was initially overcome with time-of-flight mass spectrometry having specially designed orthogonal injection interfaces that incorporated collisional cooling [80] (Figure 5c). In the intervening years, an impressive array of noncovalent assemblies have been determined by native MS [81–83, 87] (Figure 5d and e), with accumulating evidence that the molecular masses of the measured assemblies can be used to accurately deduce their stoichiometry [61–77]. One of the obvious advantages of this approach is its simplicity and directness—here a relatively precise mass determination, taken together with the identities and masses of the substituents, can often be enough to unambiguously assign the stoichiometry of a protein complex. Another significant advantage is its ability to deal with mixtures of complexes. This is key, for example, when using affinity capture techniques because the isolated material is never guaranteed to be a single pristine complex but could, instead, be a composite of complexes centered about the targeted protein. The high sensitivity of native MS even allows for analysis of endogenous protein assemblies isolated from their native cellular context, eliminating the need for the time-consuming expression of the constituents in heterologous systems and for ensuring native reconstitution. It also allows one to examine native heterogeneity that can be important for normal cellular function [88]. Unfortunately, to date the much needed application of native MS to the analysis of endogenous protein complexes has been limited. The main limitations have been the difficulty in capturing sufficiently pristine cellular protein complexes and preparing them at sufficiently high concentrations in ESI-compatible solutions to obtain useful MS spectra. In one attempt to address this need, we have presented a robust workflow that couples rapid and efficient affinity isolation of endogenous protein complexes with a sensitive native MS readout [89] (Figure 6).
Figure 6.
Native mass spectrometry for determining stoichiometry and subunit connectivity of endogenous protein complexes [89]. (a) Cryomilling, affinity isolation, protease elution, and subsequent native MS analysis of the endogenous Nup84 complex from budding yeast. (b) SDS-PAGE separation and Coomassie staining to assess the post-elution sample handling steps (elution was achieved by cleavage with the HRV 3C protease, later removed by filtration; subsequent buffer exchange was performed with 500 mM ammonium acetate, 0.01% Tween-20). Also shown is the native MS spectrum of the Nup84 complex with (c) low in-source activation and (d) high in-source activation. The structural model for the Nup84 holocomplex, based on integrative structural studies (see text) is also shown. Figure adapted from [89] with permission
MS for Structural Elucidation of Protein Complexes
As structural biologists turn their focus on increasingly larger and more complex macromolecular assemblies, it has been necessary to develop hybrid approaches that computationally integrate data generated by a large variety of experimental techniques [2, 41, 90–97]. These include X-ray crystallography, NMR spectroscopy, homology modeling, electron microscopy, co-affinity isolation, as well as any other biochemical methods able to generate spatial restraints that may be useful for modeling these systems. Ideally, such integrative modeling approaches should objectively generate an ensemble of solutions that sample the space of all possible models, and these models should optimally satisfy the available experimental data. Thus, for example, we used a variety of proteomic data, including the determination of the spatial proximities of all the different subunits in the NPC (as determined by affinity capture experiments with MS readout of protein identities), to determine the molecular architecture of the yeast NPC [2, 41] (Figure 1). This architecture, albeit obtained at relatively low resolution, revealed new symmetries and a degree of modularity not previously appreciated, providing clues to the functionality and evolutionary origins of the NPC.
We and others have also been developing means to generate higher resolution integrative models for systems where limited conventional high resolution data are available. A key tool that is enabling us to accomplish this goal is chemical crosslinking with MS readout [17, 88, 98–113]. It provides data on subnanometer scale proximities within and between substituents of protein complexes—data that can be readily translated into spatial restraints for integrative modeling [90–92, 94–96]. In the first example shown, we used this approach to model the structure of a seven member subcomplex from the NPC [3]. The affinity capture data shown in Figure 4 originally provided us with a very low resolution model for this complex (precision ~6 nm) [2, 41]. Integrative modeling of this complex, incorporating additional higher resolution restraints including those generated from crosslinking experiments (Figure 7a), yielded the subnanometer resolution structure shown in Figure 7b [3]. By modeling the effect of adding increasing numbers of crosslinks, we deduce that 4–5 crosslinks are sufficient to optimally define the docking interface of two interacting proteins, at least in cases where we can use the simplifying assumption that they interact as non-deformable rigid bodies [3] (Figure 7c).
Figure 7.
Integrated modeling approaches that incorporate MS for the structural elucidation of protein complexes. (a) Chemical crosslinking with MS readout (CX-MS) maps of the Nup84 complex by disuccinimidyl suberate (DSS) crosslinker (top) and 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC) crosslinker (bottom). (b) The Nup84 complex molecular architecture revealed by CX-MS integrative pipeline. (c) Correlation between the number of crosslinks and the accuracy of dimer models as compared to a part of the structure for which a crystallographic dimer structure is available (see text). Accuracy of dimer models as a function of the number and type of crosslinks. Each symbol displays the first and third quartile (lower and upper side of the boxes), the median (red line), as well as minimum and maximum (lower and upper limit of the dashed whiskers, respectively) of the Cα dRMSD with respect to the crystallographic structure for the 100 best-scoring models. (d) (left) Subunit proximities within S. cerevisiae replisome determined by CX-MS. (d) (right) Architecture of the eukaryotic replisome with the proposed DNA path indicated. Red and black lines illustrate possible leading- and lagging-strand DNA. The blue arrow indicates the direction of replisome movement on DNA. The diagram indicates a long path of leading-strand DNA through the entire Mcm ring and then bending back up to Pol ε, requiring about 40 nucleotides of ssDNA. Leading ssDNA is illustrated as going completely through the Mcm2–7 complex and then bending up through the second ‘accessory’ channel of CMG, but this path is speculative. Other DNA paths are possible. Figure adapted from [3, 7] with permission
Even in cases where we do not use the full integrative modeling strategy outlined above, MS crosslinking data can provide extraordinarily useful architectural information on specific features of protein assemblies. This proved to be the case for the architectural elucidation of a eukaryotic replisome, the protein machine responsible for unwinding the DNA helix and then creating duplicate helices for cell division [7]. A helicase enzyme within the replisome separates the double-stranded DNA into two single strands, which are each replicated by a different DNA polymerase. In contrast to the long held belief that the two DNA polymerases trail behind the helicase as it unzips the strands, our new pictures of the replisome obtained by single-particle EM appeared to show that one polymerase sits above the helicase (Figure 7d, right). We used crosslinking with MS readout to identify that the top polymerase is Pol-ε (Figure 7d, left). These new approaches are therefore providing an exciting new window into the high resolution architectures and functions of macromolecular assemblies.
Future Directions
This brief Perspective provides illustrative examples of the broad utility of MS as a tool for defining aspects of higher order protein structures. Indeed, we feel that in a sense, MS is contributing to the development of a “multiscale molecular microscope.” While still largely out of reach, this microscope would ultimately be comprised of suites of tools that allow the full hierarchy of endogenous protein assemblies to be defined in both space and time at all meaningful scales and resolutions (Figure 8).
Figure 8.
Vision for a multiscale molecular microscope to define cellular protein assemblies in space and time
Acknowledgments
B.T.C. gratefully acknowledges continuous ongoing support by the NIH (through grants PHS RR00862, GM103314, and GM109824) and The Rockefeller University. In addition, B.T.C. express his enormous gratitude to all members who have worked in his laboratory during the past 36 years and who have contributed so significantly to the work summarized in this personal perspective.
References
- 1.Kuriyan J, Konforti B, Wemmer D. Garland Science. The Molecules of Life: Physical and Chemical Principles. 1. Taylor and Francis Group; New York; London: 2013. [Google Scholar]
- 2.Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, Sali A, Rout MP. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- 3.Shi Y, Fernandez-Martinez J, Tjioe E, Pellarin R, Kim SJ, Williams R, Schneidman-Duhovny D, Sali A, Rout MP, Chait BT. Structural characterization by crosslinking reveals the detailed architecture of a coatomer-related heptameric module from the nuclear pore complex. Mol Cell Proteomics. 2014;13:2927–2943. doi: 10.1074/mcp.M114.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao YM, Chait BT. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson A, O’Donnell M. Cellular DNA replicases: components and dynamics at the replication fork. Annu Rev Biochem. 2005;74:283–315. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- 6.Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 7.Sun J, Shi Y, Georgescu RE, Yuan Z, Chait BT, Li H, O’Donnell ME. The architecture of a eukaryotic replisome. Nat Struct Mol Biol. 2015;22:976–982. doi: 10.1038/nsmb.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandez-Martinez J, Rout MP. A jumbo problem: mapping the structure and functions of the nuclear pore complex. Curr Opin Cell Biol. 2012;24:92–99. doi: 10.1016/j.ceb.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hurt E, Beck M. Towards understanding nuclear pore complex architecture and dynamics in the age of integrative structural analysis. Curr Opin Cell Biol. 2015;34:31–38. doi: 10.1016/j.ceb.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 10.Wente SR, Rout MP. The nuclear pore complex and nuclear transport. Cold Spring Harb Perspect Biol. 2010;2:73–91. doi: 10.1101/cshperspect.a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brändén C-I, Tooze J. Introduction tp protein structure. 2. Garland Publications; New York: 1999. [Google Scholar]
- 12.Karas M, Hillenkamp F. Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal Chem. 1988;60:2299–2301. doi: 10.1021/ac00171a028. [DOI] [PubMed] [Google Scholar]
- 13.Hillenkamp F, Karas M, Beavis RC, Chait BT. Matrix assisted laser desorption ionization mass spectrometry of biopolymers. Anal Chem. 1991;63:A1193–A1202. doi: 10.1021/ac00024a002. [DOI] [PubMed] [Google Scholar]
- 14.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 15.Chait BT. Mass-spectrometry—a useful tool for the protein X-ray crystallographer and NMR spectroscopist. Structure. 1994;2:465–467. doi: 10.1016/s0969-2126(00)00047-2. [DOI] [PubMed] [Google Scholar]
- 16.Cadene M, Chait BT. A robust, detergent-friendly method for mass spectrometric analysis of integral membrane proteins. Anal Chem. 2000;72:5655–5658. doi: 10.1021/ac000811l. [DOI] [PubMed] [Google Scholar]
- 17.Cohen SL, Chait BT. Mass spectrometry as a tool for protein crystallography. Annu Rev Biophys Biomol Struct. 2001;30:67–85. doi: 10.1146/annurev.biophys.30.1.67. [DOI] [PubMed] [Google Scholar]
- 18.Dutzler R, Campbell EB, Cadene M, Chait BT, MacKinnon R. X-ray structure of a CIC chloride channel at 3.0 angstrom reveals the molecular basis of anion selectivity. Nature. 2002;415:287–294. doi: 10.1038/415287a. [DOI] [PubMed] [Google Scholar]
- 19.Gabant G, Cadene M. Mass spectrometry of full-length integral membrane proteins to define functionally relevant structural features. Methods. 2008;46:54–61. doi: 10.1016/j.ymeth.2008.10.021. [DOI] [PubMed] [Google Scholar]
- 20.Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dümpelfeld B, Edelmann A, Heurtier MA, Hoffman V, Hoefert C, Klein K, Hudak M, Michon AM, Schelder M, Schirle M, Remor M, Rudi T, Hooper S, Bauer A, Bouwmeester T, Casari G, Drewes G, Neubauer G, Rick JM, Kuster B, Bork P, Russell RB, Superti-Furga G. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 21.Krogan NJ, Cagney G, Yu HY, Zhong GQ, Guo XH, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, Punna T, Peregrín-Alvarez JM, Shales M, Zhang X, Davey M, Robinson MD, Paccanaro A, Bray JE, Sheung A, Beattie B, Richards DP, Canadien V, Lalev A, Mena F, Wong P, Starostine A, Canete MM, Vlasblom J, Wu S, Orsi C, Collins SR, Chandran S, Haw R, Rilstone JJ, Gandi K, Thompson NJ, Musso G, St Onge P, Ghanny S, Lam MH, Butland G, Altaf-Ul AM, Kanaya S, Shilatifard A, O’Shea E, Weissman JS, Ingles CJ, Hughes TR, Parkinson J, Gerstein M, Wodak SJ, Emili A, Greenblatt JF. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 22.Hakhverdyan Z, Domanski M, Hough LE, Oroskar AA, Oroskar AR, Keegan S, Dilworth DJ, Molloy KR, Sherman V, Aitchison JD, Fenyö D, Chait BT, Jensen TH, Rout MP, LaCava J. Rapid, optimized interactomic screening. Nat Methods. 2015;12:553–560. doi: 10.1038/nmeth.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ong SE, Mann M. Mass spectrometry-based proteomics turns quantitative. Nat Chem Biol. 2005;1:252–262. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- 24.Beynon RJ, Doherty MK, Pratt JM, Gaskell SJ. Multiplexed absolute quantification in proteomics using artificial QCAT proteins of concatenated signature peptides. Nat Methods. 2005;2:587–589. doi: 10.1038/nmeth774. [DOI] [PubMed] [Google Scholar]
- 25.Ding C, Li Y, Kim BJ, Malovannaya A, Jung SY, Wang Y, Qin J. Quantitative analysis of cohesin complex stoichiometry and SMC3 modification-dependent protein interactions. J Proteome Res. 2011;10:3652–3659. doi: 10.1021/pr2002758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holzmann J, Pichler P, Madalinski M, Kurzbauer R, Mechtler K. Stoichiometry determination of the MP1-p14 complex using a novel and cost-efficient method to produce an equimolar mixture of standard peptides. Anal Chem. 2009;81:10254–10261. doi: 10.1021/ac902286m. [DOI] [PubMed] [Google Scholar]
- 27.Smits AH, Jansen PW, Poser I, Hyman AA, Vermeulen M. Stoichiometry of chromatin-associated protein complexes revealed by label-free quantitative mass spectrometry-based proteomics. Nucleic Acids Res. 2013;41:e28. doi: 10.1093/nar/gks941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Politis A, Stengel F, Hall Z, Hernandez H, Leitner A, Walzthoeni T, Robinson CV, Aebersold R. A mass spectrometry-based hybrid method for structural modeling of protein complexes. Nat Methods. 2014;11:403–406. doi: 10.1038/nmeth.2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wohlgemuth I, Lenz C, Urlaub H. Studying macromolecular complex stoichiometries by peptide-based mass spectrometry. Proteomics. 2015;15:862–879. doi: 10.1002/pmic.201400466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X, Liang Y, Liu L, Shi J, Zhu H-J. Targeted absolute quantitative proteomics with SILAC internal standards and unlabeled full-length protein calibrators (TAQSI) Rapid Commun Mass Spectrom. 2016;30:5–561. 53. doi: 10.1002/rcm.7482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 32.Bogdanov B, Smith RD. Proteomics by FTICR mass spectrometry: top down and bottom up. Mass Spectrom Rev. 2005;24:168–200. doi: 10.1002/mas.20015. [DOI] [PubMed] [Google Scholar]
- 33.Han X, Aslanian A, Yates JR., III Mass spectrometry for proteomics. Curr Opin Chem Biol. 2008;12:483–490. doi: 10.1016/j.cbpa.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cox J, Mann M. Quantitative, High-Resolution Proteomics for Data-Driven Systems Biology. In: Kornberg RD, Raetz CRH, Rothman JE, Thorner JW, editors. Annual Review of Biochemistry. Vol. 80. Palo Alto, CA: Annual Reviews; pp. 273–299. [DOI] [PubMed] [Google Scholar]
- 35.Ibarra A, Hetzer MW. Nuclear pore proteins and the control of genome functions. Genes Dev. 2015;29:337–349. doi: 10.1101/gad.256495.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cristea IM, Williams R, Chait BT, Rout MP. Fluorescent proteins as proteomic probes. Mol Cell Proteomics. 2005;4:1933–1941. doi: 10.1074/mcp.M500227-MCP200. [DOI] [PubMed] [Google Scholar]
- 37.Selbach M, Mann M. Protein interaction screening by quantitative im-munoprecipitation combined with knockdown (QUICK) Nat Methods. 2006;3:981–983. doi: 10.1038/nmeth972. [DOI] [PubMed] [Google Scholar]
- 38.Gingras AC, Gstaiger M, Raught B, Aebersold R. Analysis of protein complexes using mass spectrometry. Nat Rev Mol Cell Biol. 2007;8:645–654. doi: 10.1038/nrm2208. [DOI] [PubMed] [Google Scholar]
- 39.Dunham WH, Mullin M, Gingras AC. Affinity-purification coupled to mass spectrometry: basic principles and strategies. Proteomics. 2012;12:1576–1590. doi: 10.1002/pmic.201100523. [DOI] [PubMed] [Google Scholar]
- 40.LaCava J, Molloy KR, Taylor MS, Domanski M, Chait BT, Rout MP. Affinity proteomics to study endogenous protein complexes: pointers, pitfalls, preferences, and perspectives. Biotechniques. 2015;58:103–119. doi: 10.2144/000114262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, Rout MP, Sali A. Determining the architectures of macromolecular assemblies. Nature. 2007;450:683–694. doi: 10.1038/nature06404. [DOI] [PubMed] [Google Scholar]
- 42.Mellacheruvu D, Wright Z, Couzens AL, Lambert JP, St-Denis NA, Li T, Miteva YV, Hauri S, Sardiu ME, Low TY, Halim VA, Bagshaw RD, Hubner NC, Al-Hakim A, Bouchard A, Faubert D, Fermin D, Dunham WH, Goudreault M, Lin ZY, Badillo BG, Pawson T, Durocher D, Coulombe B, Aebersold R, Superti-Furga G, Colinge J, Heck AJ, Choi H, Gstaiger M, Mohammed S, Cristea IM, Bennett KL, Washburn MP, Raught B, Ewing RM, Gingras AC, Nesvizhskii AI. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat Methods. 2013;10:730–736. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sardiu ME, Cai Y, Jin J, Swanson SK, Conaway RC, Conaway JW, Florens L, Washburn MP. Probabilistic assembly of human protein interaction networks from label-free quantitative proteomics. Proc Natl Acad Sci U S A. 2008;105:1454–1459. doi: 10.1073/pnas.0706983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skarra DV, Goudreault M, Choi H, Mullin M, Nesvizhskii AI, Gingras AC, Honkanen RE. Label-free quantitative proteomics and SAINT analysis enable interactome mapping for the human Ser/Thr protein phosphatase 5. Proteomics. 2011;11:1508–1516. doi: 10.1002/pmic.201000770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nesvizhskii AI. Computational and informatics strategies for identification of specific protein interaction partners in affinity purification mass spectrometry experiments. Proteomics. 2012;12:1639–1655. doi: 10.1002/pmic.201100537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tackett AJ, DeGrasse JA, Sekedat MD, Oeffinger M, Rout MP, Chait BT. I-DIRT, a general method for distinguishing between specific and nonspecific protein interactions. J Proteome Res. 2005;4:1752–1756. doi: 10.1021/pr050225e. [DOI] [PubMed] [Google Scholar]
- 47.Hubner NC, Bird AW, Cox J, Splettstoesser B, Bandilla P, Poser I, et al. Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J Cell Biol. 2010;189:739–754. doi: 10.1083/jcb.200911091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hendil KB, Welinder KG, Pedersen D, Uerkvitz W, Kristensen P. Subunit stoichiometry of human proteosomes. Enzyme Protein. 1993;47:232–240. doi: 10.1159/000468682. [DOI] [PubMed] [Google Scholar]
- 49.Mullins RD, Stafford WF, Pollard TD. Structure, subunit topology, and actin-binding activity of the Arp2/3 complex from Acanthamoeba. J Cell Biol. 1997;136:331–343. doi: 10.1083/jcb.136.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Farrar SJ, Whiting PJ, Bonnert TP, McKernan RM. Stoichiometry of a ligand-gated ion channel determined by fluorescence energy transfer. J Biol Chem. 1999;274:10100–10104. doi: 10.1074/jbc.274.15.10100. [DOI] [PubMed] [Google Scholar]
- 51.Leake MC, Chandler JH, Wadhams GH, Bai F, Berry RM, Armitage JP. Stoichiometry and turnover in single, functioning membrane protein complexes. Nature. 2006;443:355–358. doi: 10.1038/nature05135. [DOI] [PubMed] [Google Scholar]
- 52.Ji W, Xu P, Li Z, Lu J, Liu L, Zhan Y, Chen Y, Hille B, Xu T, Chen L. Functional stoichiometry of the unitary calcium release activated calcium channel. Proc Natl Acad Sci U S A. 2008;105:13668–13673. doi: 10.1073/pnas.0806499105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Slotboom DJ, Duurkens RH, Olieman K, Erkens GB. Static light scattering to characterize membrane proteins in detergent solution. Methods. 2008;46:73–82. doi: 10.1016/j.ymeth.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 54.Kito K, Ota K, Fujita T, Ito T. A synthetic protein approach toward accurate mass spectrometric quantification of component stoichiometry of multiprotein complexes. J Proteome Res. 2007;6:792–800. doi: 10.1021/pr060447s. [DOI] [PubMed] [Google Scholar]
- 55.Nanavati D, Gucek M, Milne JLS, Subramaniam S, Markey SP. Stoichiometry and absolute quantification of proteins with mass spectrometry using fluorescent and isotope-labeled concatenated peptide standards. Mol Cell Proteomics. 2008;7:442–447. doi: 10.1074/mcp.M700345-MCP200. [DOI] [PubMed] [Google Scholar]
- 56.Schmidt C, Lenz C, Grote M, Luehrmann R, Urlaub H. Determination of protein stoichiometry within protein complexes using absolute quantification and multiple reaction monitoring. Anal Chem. 2010;82:2784–2796. doi: 10.1021/ac902710k. [DOI] [PubMed] [Google Scholar]
- 57.Cheung CSF, Anderson KW, Wang M, Turko IV. Natural flanking sequences for peptides included in a quantification concatamer internal standard. Anal Chem. 2015;87:1097–1102. doi: 10.1021/ac503697j. [DOI] [PubMed] [Google Scholar]
- 58.Warnken U, Schleich K, Schnolzer M, Lavrik I. Quantification of high-molecular weight protein platforms by AQUA mass spectrometry as exemplified for the CD95 death-inducing signaling complex (DISC) Cells. 2013;2:476–495. doi: 10.3390/cells2030476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ori A, Andres-Pons A, Beck M. The Use of Targeted Proteomics to Determine the Stoichiometry of Large Macromolecular Assemblies. Methods in Cell Biology. 2014;122:117–146. doi: 10.1016/B978-0-12-417160-2.00006-0. [DOI] [PubMed] [Google Scholar]
- 60.von Appen A, Kosinski J, Sparks L, Ori A, DiGuilio AL, Vollmer B, Mackmull MT, Banterle N, Parca L, Kastritis P, Buczak K, Mosalaganti S, Hagen W, Andres-Pons A, Lemke EA, Bork P, Antonin W, Glavy JS, Bui KH, Beck M. In situ structural analysis of the human nuclear pore complex. Nature. 2015;526:140–143. doi: 10.1038/nature15381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Loo JA. Studying noncovalent protein complexes by electrospray ionization mass spectrometry. Mass Spectrom Rev. 1997;16:1–23. doi: 10.1002/(SICI)1098-2787(1997)16:1<1::AID-MAS1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 62.Veenstra TD. Electrospray ionization mass spectrometry in the study of biomolecular non-covalent interactions. Biophys Chem. 1999;79:63–79. doi: 10.1016/s0301-4622(99)00037-x. [DOI] [PubMed] [Google Scholar]
- 63.Krutchinsky AN, Ayed A, Donald LJ, Ens W, Duckworth HW, Standing KG. Studies of noncovalent complexes in an electrospray ionization/time-of-flight mass spectrometer. Methods Mol Biol (Clifton, NJ) 2000;146:239–249. doi: 10.1385/1-59259-045-4:239. [DOI] [PubMed] [Google Scholar]
- 64.Loo JA, Berhane B, Kaddis CS, Wooding KM, Xie YM, Kaufman SL, Chernushevich IV. Electrospray ionization mass spectrometry and ion mobility analysis of the 20S proteasome complex. J Am Soc Mass Spectrom. 2005;16:998–1008. doi: 10.1016/j.jasms.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 65.Hernandez H, Dziembowski A, Taverner T, Seraphin B, Robinson CV. Subunit architecture of multimeric complexes isolated directly from cells. EMBO Rep. 2006;7:605–610. doi: 10.1038/sj.embor.7400702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hernandez H, Robinson CV. Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nat Protoc. 2007;2:715–726. doi: 10.1038/nprot.2007.73. [DOI] [PubMed] [Google Scholar]
- 67.Vaughan CK, Gohlke U, Sobott F, Good VM, Ali MMU, Prodromou C, Robinson CV, Saibil HR, Pearl LH. Structure of an Hsp90-Cdc37-Cdk4 complex. Mol Cell. 2006;23:697–707. doi: 10.1016/j.molcel.2006.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Heck AJR. Native mass spectrometry: a bridge between interactomics and structural biology. Nat Methods. 2008;5:927–933. doi: 10.1038/nmeth.1265. [DOI] [PubMed] [Google Scholar]
- 69.Kitagawa N, Mazon H, Heck AJR, Wilkens S. Stoichiometry of the peripheral stalk subunits E and G of yeast V-1-ATPase determined by mass spectrometry. J Biol Chem. 2008;283:3329–3337. doi: 10.1074/jbc.M707924200. [DOI] [PubMed] [Google Scholar]
- 70.Taverner T, Hernandez H, Sharon M, Ruotolo BT, Matak-Vinkovic D, Devos D, Russell RB, Robinson CV. Subunit architecture of intact protein complexes from mass spectrometry and homology modeling. Acc Chem Res. 2008;41:617–627. doi: 10.1021/ar700218q. [DOI] [PubMed] [Google Scholar]
- 71.Barrera NP, Isaacson SC, Zhou M, Bavro VN, Welch A, Schaedler TA, Seeger MA, Miguel RN, Korkhov VM, van Veen HW, Venter H, Walmsley AR, Tate CG, Robinson CV. Mass spectrometry of membrane transporters reveals subunit stoichiometry and interactions. Nat Methods. 2009;6:585–587. doi: 10.1038/nmeth.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Morton VL, Stockley PG, Stonehouse NJ, Ashcroft AE. Insights into virus capsid assembly from noncovalent mass spectrometry. Mass Spectrom Rev. 2008;27:575–595. doi: 10.1002/mas.20176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Laganowsky A, Reading E, Hopper JTS, Robinson CV. Mass spectrometry of intact membrane protein complexes. Nat Protoc. 2013;8:639–651. doi: 10.1038/nprot.2013.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Barrera NP, Robinson CV. Advances in the Mass Spectrometry of Membrane Proteins: From Individual Proteins to Intact Complexes. In: Kornberg RD, Raetz CRH, Rothman JE, Thorner JW, editors. Annual Review of Biochemistry. Vol. 80. Palo Alto, CA: Annual Reviews; pp. 247–271. [DOI] [PubMed] [Google Scholar]
- 75.Schreiber A, Stengel F, Zhang Z, Enchev RI, Kong EH, Morris EP, Robinson CV, da Fonseca PC, Barford D. Structural basis for the subunit assembly of the anaphase-promoting complex. Nature. 2011;470:227–232. doi: 10.1038/nature09756. [DOI] [PubMed] [Google Scholar]
- 76.Hilton GR, Benesch JLP. Two decades of studying noncovalent bio-molecular assemblies by means of electrospray ionization mass spectrometry. J Royal Soc Interface. 2012;9:801–816. doi: 10.1098/rsif.2011.0823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rose RJ, Damoc E, Denisov E, Makarov A, Heck AJR. High-sensitivity Orbitrap mass analysis of intact macromolecular assemblies. Nat Methods. 2012;9:1084–1086. doi: 10.1038/nmeth.2208. [DOI] [PubMed] [Google Scholar]
- 78.Chowdhury SK, Katta V, Chait BT. Probing conformational changes in proteins by mass spectrometry. J Am Chem Soc. 1990;112:9012–9013. [Google Scholar]
- 79.Baca M, Kent SBH. Direct observation of a ternary complex between the dimeric enzyme HIV-1 protease and a substrate-based inhibitor. J Am Chem Soc. 1992;114:3992–3993. [Google Scholar]
- 80.Krutchinsky AN, Chernushevich IV, Spicer VL, Ens W, Standing KG. Collisional damping interface for an electrospray ionization time-of-flight mass spectrometer. J Am Soc Mass Spectrom; Data first presented at the 43rd ASMS Conference on Mass Spectrometry and Allied Topics; 1995.1998. pp. 569–579. [Google Scholar]
- 81.Zhou M, Morgner N, Barrera NP, Politis A, Isaacson SC, Matak-Vinkovic D, Zhou M, Morgner N, Barrera NP. Mass spectrometry of intact V-type ATPases reveals bound lipids and the effects of nucleotide binding. Science. 2011;334:380–385. doi: 10.1126/science.1210148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Snijder J, Rose RJ, Veesler D, Johnson JE, Heck AJR. Studying 18 MDa virus assemblies with native mass spectrometry. Angew Chem Int Ed. 2013;52:4020–4023. doi: 10.1002/anie.201210197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Keifer DZ, Pierson EE, Hogan JA, Bedwell GJ, Prevelige PE, Jarrold MF. Charge detection mass spectrometry of bacteriophage P22 procapsid distributions above 20 MDa. Rapid Commun Mass Spectrom. 2014;28:483–488. doi: 10.1002/rcm.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Katta V, Chait BT. Observation of the heme globin complex in native myoglobin by electrospray-ionization mass-spectrometry. J Am Chem Soc. 1991;113:8534–8535. [Google Scholar]
- 85.Ganem B, Li YT, Henion JD. Observation of noncovalent enzyme substrate and enzyme product complexes by ion-spray mass spectrometry. J Am Chem Soc. 1991;113:7818–7819. [Google Scholar]
- 86.Ganem B, Li YT, Henion JD. Detection of noncovalent receptor ligand complexes by mass spectrometry. J Am Chem Soc. 1991;113:6294–6296. [Google Scholar]
- 87.Snijder J, Heck AJR. Analytical Approaches for Size and Mass Analysis of Large Protein Assemblies. In: Cooks RG, Pemberton JE, editors. Annual Review Analytical Chemistry. Vol. 7. Palo Alto: 2014. pp. 43–64. [DOI] [PubMed] [Google Scholar]
- 88.Shi Y, Pellarin R, Fridy PC, Fernandez-Martinez J, Thompson MK, Li Y, Wang QJ, Sali A, Rout MP, Chait BT. A strategy for dissecting the architectures of native macromolecular assemblies. Nat Methods. 2015;12:1135–1138. doi: 10.1038/nmeth.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Olinares PD, Dunn AD, Padovan JC, Fernandez-Martinez J, Rout MP, Chait BT. A robust workflow for native mass spectrometric analysis of affinity-isolated endogenous protein assemblies. Anal Chem. 2016;88:2799–2807. doi: 10.1021/acs.analchem.5b04477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Robinson CV, Sali A, Baumeister W. The molecular sociology of the cell. Nature. 2007;450:973–982. doi: 10.1038/nature06523. [DOI] [PubMed] [Google Scholar]
- 91.Lasker K, Forster F, Bohn S, Walzthoeni T, Villa E, Unverdorben P, Beck F, Aebersold R, Sali A, Baumeister W. Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc Natl Acad Sci U S A. 2012;109:1380–1387. doi: 10.1073/pnas.1120559109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lasker K, Phillips JL, Russel D, Velazquez-Muriel J, Schneidman-Duhovny D, Tjioe E, Webb B, Schlessinger A, Sali A. Integrative structure modeling of macromolecular assemblies from proteomics data. Mol Cell Proteomics. 2010;9:1689–1702. doi: 10.1074/mcp.R110.000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fernandez-Martinez J, Phillips J, Sekedat MD, Diaz-Avalos R, Velazquez-Muriel J, Franke JD, Williams R, Stokes DL, Chait BT, Sali A, Rout MP. Structure-function mapping of a heptameric module in the nuclear pore complex. J Cell Biol. 2012;196:419–434. doi: 10.1083/jcb.201109008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Russel D, Lasker K, Webb B, Velazquez-Muriel J, Tjioe E, Schneidman-Duhovny D, Peterson B, Sali A. Putting the pieces together: integrative modeling platform software for structure determination of macromolecular assemblies. Plos Biol. 2012;10:e1001244. doi: 10.1371/journal.pbio.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ward AB, Sali A, Wilson IA. Integrative structural biology. Science. 2013;339:913–915. doi: 10.1126/science.1228565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yang Z, Lasker K, Schneidman-Duhovny D, Webb B, Huang CC, Pettersen EF, Goddard TD, Meng EC, Sali A, Ferrin TE. UCSF Chimera, MODELLER, and IMP: an integrated modeling system. J Struct Biol. 2012;179:269–278. doi: 10.1016/j.jsb.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Algret R, Fernandez-Martinez J, Shi Y, Kim SJ, Pellarin R, Cimermancic P, Cochet E, Sali A, Chait BT, Rout MP, Dokudovskaya S. Molecular architecture and function of the SEA complex, a modulator of the TORC1 pathway. Mol Cell Proteomics. 2014;13:2855–2870. doi: 10.1074/mcp.M114.039388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Young MM, Tang N, Hempel JC, Oshiro CM, Taylor EW, Kuntz ID, Gibson BW, Dollinger G. High throughput protein fold identification by using experimental constraints derived from intramolecular crosslinks and mass spectrometry. Proc Natl Acad Sci U S A. 2000;97:5802–5806. doi: 10.1073/pnas.090099097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Trester-Zedlitz M, Kamada K, Burley SK, Fenyo D, Chait BT, Muir TW. A modular crosslinking approach for exploring protein interactions. J Am Chem Soc. 2003;125:2416–2425. doi: 10.1021/ja026917a. [DOI] [PubMed] [Google Scholar]
- 100.Dihazi GH, Sinz A. Mapping low-resolution three-dimensional protein structures using chemical crosslinking and Fourier transform ion-cyclotron resonance mass spectrometry. Rapid Commun Mass Spectrom. 2003;17:2005–2014. doi: 10.1002/rcm.1144. [DOI] [PubMed] [Google Scholar]
- 101.Sinz A. Chemical crosslinking and mass spectrometry to map three-dimensional protein structures and protein-protein interactions. Mass Spectrom Rev. 2006;25:663–682. doi: 10.1002/mas.20082. [DOI] [PubMed] [Google Scholar]
- 102.Rinner O, Seebacher J, Walzthoeni T, Mueller LN, Beck M, Schmidt A, Mueller M, Aebersold R. Identification of crosslinked peptides from large sequence databases. Nat Methods. 2008;5:315–318. doi: 10.1038/nmeth.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chen ZA, Jawhari A, Fischer L, Buchen C, Tahir S, Kamenski T, Rasmussen M, Lariviere L, Bukowski-Wills JC, Nilges M, Cramer P, Rappsilber J. Architecture of the RNA polymerase II-TFIIF complex revealed by crosslinking and mass spectrometry. EMBO J. 2010;29:717–726. doi: 10.1038/emboj.2009.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Herzog F, Kahraman A, Boehringer D, Mak R, Bracher A, Walzthoeni T, Leitner A, Beck M, Hartl FU, Ban N, Malmström L, Aebersold R. Structural probing of a protein phosphatase 2a network by chemical crosslinking and mass spectrometry. Science. 2012;337:1348–1352. doi: 10.1126/science.1221483. [DOI] [PubMed] [Google Scholar]
- 105.Leitner A, Walzthoeni T, Kahraman A, Herzog F, Rinner O, Beck M, Aebersold R. Probing native protein structures by chemical crosslinking, mass spectrometry, and bioinformatics. Mol Cell Proteomics. 2010;9:1634–1649. doi: 10.1074/mcp.R000001-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Clifford-Nunn B, Showalter HD, Andrews PC. Quaternary diamines as mass spectrometry cleavable crosslinkers for protein interactions. J Am Soc Mass Spectrom. 2012;23:201–212. doi: 10.1007/s13361-011-0288-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yang B, Wu YJ, Zhu M, Fan SB, Lin J, Zhang K, Li S, Chi H, Li YX, Chen HF, Luo SK, Ding YH, Wang LH, Hao Z, Xiu LY, Chen S, Ye K, He SM, Dong MQ. Identification of crosslinked peptides from complex samples. Nat Methods. 2012;9:904–906. doi: 10.1038/nmeth.2099. [DOI] [PubMed] [Google Scholar]
- 108.Trnka MJ, Baker PR, Robinson PJJ, Burlingame AL, Chalkley RJ. Matching crosslinked peptide spectra: only as good as the worse identification. Mol Cell Proteomics. 2014;13:420–434. doi: 10.1074/mcp.M113.034009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kao AH, Chiu CL, Vellucci D, Yang YY, Patel VR, Guan SH, Randall A, Baldi P, Rychnovsky SD, Huang L. Development of a novel crosslinking strategy for fast and accurate identification of crosslinked peptides of protein complexes. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Weisbrod CR, Chavez JD, Eng JK, Yang L, Zheng CX, Bruce JE. In vivo protein interaction network identified with a novel real-time crosslinked peptide identification strategy. J Proteome Res. 2013;12:1569–1579. doi: 10.1021/pr3011638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Leitner A, Joachimiak LA, Unverdorben P, Walzthoeni T, Frydman J, Forster F, Aebersold R. Chemical crosslinking/mass spectrometry targeting acidic residues in proteins and protein complexes. Proc Natl Acad Sci U S A. 2014;111:9455–9460. doi: 10.1073/pnas.1320298111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tan D, Li Q, Zhang MJ, Liu C, Ma C, Zhang P, Ding YH, Fan SB, Tao L, Yang B, Li X, Ma S, Liu J, Feng B, Liu X, Wang HW, He SM, Gao N, Ye K, Dong MQ, Lei X. Trifunctional cross-linker for mapping protein-protein interaction networks and comparing protein conformational states. eLife. 2016;5:e12509. doi: 10.7554/eLife.12509. [DOI] [PMC free article] [PubMed] [Google Scholar]