Abstract
Recently, accumulating evidence has suggested that long noncoding RNAs (lncRNAs) play crucial roles in carcinogenesis and cancer progression. Hyaluronan-mediated motility receptor (HMMR) is an essential cancer-related gene in basal-like breast cancer (BLBC). In our study, HMMR antisense RNA 1 (HMMR-AS1) was analyzed in BLBC patients through polymerase chain reaction analysis. Here, we found that the expression of HMMR was positively correlated with HMMR-AS1 (RP11-80G.1). When HMMR-AS1 (RP11-80G.1) was knocked down, the expression of HMMR markedly reduced. Furthermore, in MDA-MB-231 and MDA-MB-468 breast cancer cells, the proliferation and migration abilities were remarkably suppressed via knocking down HMMR-AS1 (RP11-80G.1) in vitro. The results showed that lncRNA HMMR-AS1 (RP11-80G.1) influenced the progression of BLBCs through regulating HMMR, suggesting that HMMR-AS1 (RP11-80G.1) could be regarded as a novel biomarker and therapeutic target in the treatment of BLBCs in future.
Keywords: HMMR antisense RNA 1, long noncoding RNA, hyaluronan mediated motility receptor, HMMR, basal-like breast cancer cells
Introduction
Basal-like breast cancers (BLBCs) could express the gene signatures of basal cells in the mammary glands, which belong to the aggressive malignancies. Among various human breast cancers, the basal-like subtype has been identified as a distinct entity along with poor prognosis. Moreover, the present targeted systemic therapy for BLBCs still remains ineffective due to the underexpression of estrogen receptor (ER), progesterone receptor (PR), and HER2.1–3 Hence, further evidence associated with the molecular basis of this disease is urgently required to improve the diagnosis and therapy of BLBCs.
In recent years, emerging evidence has showed that long noncoding RNAs (lncRNAs) play a pivotal role in the development of various cancers.4–6 As promising treatment targets, it is very necessary to further investigate the functions of lncRNAs in BLBCs.
Commonly, lncRNAs are regarded as important regulators associated with various molecular modulation mechanisms.7–9 A previous study revealed that many lncRNAs were functionally linked with their adjacent messenger RNAs (mRNAs) as “lncRNA–mRNA pairs” in the regulatory network.10
In our previous study, by analyzing the lncRNA microarray data (Affymetrix, Santa Clara, CA, USA) in human tumor tissues, we have primarily characterized a number of cancer-related lncRNAs (data not published). Among these diversely expressed lncRNAs, we noticed an lncRNA, HMMR antisense RNA 1 (HMMR-AS1; RP11-80G.1), which is located at 5p34 and is the antisense transcript of hyaluronan-mediated motility receptor (HMMR). In addition, transcript factors chip-seq data from Encyclopedia of DNA Elements (ENCODE) showed that many cancer-related transcript factors interact with the HMMR-AS1 locus. However, the function of HMMR-AS1 (RP11-80G.1) is still unelucidated. A survey within the GENCODE project indicated that the expression of about 3% of lncRNAs shows high positive correlation with their neighboring mRNA.11
HMMR-AS1 is transcribed on the strand opposite to the adjacent HMMR gene which codes for receptor for hyaluronan-mediated motility (RHAMM), a mitotic spindle-binding protein. On the cell surface, RHAMM interacts with the hyaluronan receptor CD44 to enhance signaling over the extracellular receptor kinase (ERK1/ERK2) pathway.12
The existing evidence showed that the expression of HMMR was found to be potentially connected with the progression of cancer.13 Notably, it has been reported that BRCA1 and HMMR act upon each other to regulate epithelial apicobasal polarization. Moreover, common genetic variation in HMMR may be correlated with the risk of breast cancer.14,15 Therefore, it is tempting to speculate that HMMR-AS1 may have a connection with BLBCs. As we know, HMMR is now confirmed as a crucial biomarker of BLBCs. However, the function of HMMR-AS1 in BLBCs remains largely unknown.
In the present study, we mainly explored the functions of HMMR-AS1 in MDA-MB-231 and MDA-MB-468 human BLBC cells in vitro.
Materials and methods
Patient samples
We obtained the specimens from 52 patients diagnosed with breast cancers and 52 normal samples at the Hubei Cancer Hospital (Wuhan, People’s Republic of China) between 2007 and 2013. Clinical classification was performed by immunehistochemical studies for ER, PR, HER2, cytokeratin 5/6 and epidermal growth factor receptor.16,17 The 52 tumor samples were derived from each group of the molecular subtypes based on their immunohistochemical surrogate: 13 luminal A-like (ER+/PR+/HER2−), seven luminal B-like (ER+/PR+/HER2+), six HER2-enriched (ER−/PR−/HER2+), and 26 basal-like (ER−/PR−/HER2+/CK5/6+ or EGFR+). The utilization of material for this study was approved by the Ethical Committee of Hubei Cancer Hospital. Written informed consent was obtained from the patients or their families.
Cell lines and cell culture
MDA-MB-231 and MDA-MB-468 human BLBC cell lines were collected from the American Type Culture Collection (Manassas, VA, USA). MDA-MB-231 cell lines were cultured in Dulbecco’s modified Eagle’s medium (Hyclone, Logan, UT, USA), which contained 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Waltham, MA), and MDA-MB-468 cell lines were incubated in Roswell Park Memorial Institute-1640 medium (Hyclone) added with 10% FBS at 37°C with 5% CO2.
siRNA transfection
The sequences of one negative control small interfering RNA (NC siRNA) and two HMMR-AS1 (RP11-80G.1) siRNA were purchased from GenePharma (Shanghai, People’s Republic of China). Total sequences are shown in Table 1. siRNA transfection was carried out with X-tremeGENE transfection reagent (Roche, Mannheim, Germany) following the manufacturer’s instructions.
Table 1.
Primers for real-time polymerase chain reaction analysis
| Gene name | Forward | Reverse |
|---|---|---|
| β-Actin | 5′-CCACTGGCATCGTGATGGA-3′ | 5′-CGCTCGGTGAGGATCTTCAT-3′ |
| HMMR | 5′-ATGATGGCTAAGCAAGAAGGC-3′ | 5′-TTTCCCTTGAGACTCTTCGAGA-3′ |
| HMMR-AS1 | 5′-CACAACTCCTGGTTCCTG-3′ | 5′-GGATGGGTATTGGTCTGT-3′ |
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
RNA was isolated from breast cancer tumor tissues, adjacent normal tissues, and breast cancer cells using TRIzol Total RNA reagent (Thermo Fisher Scientific). All these primers were derived from Sheng Gong (Shanghai, People’s Republic of China). All the sequences are shown in Table 2. RT-qPCR was conducted using SYBR PrimeScript RT-PCR kit (Takara, Kusatsu, Japan). Expression levels of lncRNA and mRNA were analyzed using comparative Ct method.
Table 2.
Sequences for small interfering RNA analysis
| Gene name | Sense (5′–3′) | Antisense (5′–3′) |
|---|---|---|
| HMMR-si1 | 5′-CAAGAAUCAUGGAAGUAAACAUCTG-3′ | 5′-CAGAUGUUUACUUCCAUGAUUCUUGAC-3′ |
| HMMR-AS1-si2 | 5′-GUAGAAGCCACUAGAACUAUCUGTA-3′ | 5′-UACAGAUAGUUCUAGUGGCUUCUACUU-3′ |
Cell proliferation assays
Cell proliferation was carried out using a CellTiter 96® (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt; MTS) assay kit (Promega Corporation, Fitchburg, WI, USA) according to the manufacturer’s instructions. Each group of MDA-MB-231 and MDA-MB-468 cells was incubated in 96-well plates. Each culture medium (100 µL) was supplemented with MTS reagent (20 µL). Each plate was cultured for 2 h at 37°C in a humidified, 5% CO2 atmosphere. All experiments were performed three times.
Matrigel invasion assays
Cell invasion assay was conducted using modified Boyden Chambers (BD Biosciences, San Jose, CA, USA). The culturing medium supplemented with 10% FBS was added in the lower chamber. Migrated cells were stained and counted through the filter, and the average migration rate was examined. All experiments were performed three times.
Statistical analysis
Student’s t-test was used to analyze the group differences. Also, Pearson’s correlation was estimated to analyze the interaction between gene expressions. Further, statistical analyses were conducted using SPSS version 18.0 (SPSS Inc., Chicago, IL, USA). For all statistical analyses, P<0.05 was considered statistically significant.
Results
HMMR-AS1 expression was upregulated in BLBC specimens
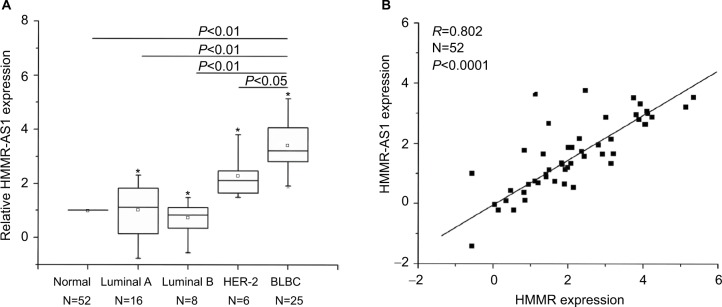
The results suggested that the expression of HMMR-AS1 in BLBC tissue specimens was higher than that in non-tumorous tissue specimens. The HMMR-AS1 expression was markedly higher in BLBCs in contrast to those in non-BLBC subtypes (Figure 1A, P<0.01 or P<0.05). Besides, the expression levels of HMMR-AS1 and HMMR in the tumor tissue samples were positively correlated (Figure 1B, R=0.802, P<0.0001).
Figure 1.
Expression levels of HMMR-AS1 (RP11-80G.1) long noncoding RNA were analyzed by RT-qPCR in 52 breast cancer tissue samples. (A) Expression levels of HMMR in breast cancer tissues were found by RT-qPCR to be significantly lower than those in adjacent normal tissues. (B) The expression levels of HMMR were significantly correlated with those of HMMR-AS1 (RP11-80G.1) lncRNA (R=0.802, P<0.0001). *Maximum value; □ mean value.
Abbreviations: HMMR-AS1, HMMR antisense RNA 1; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; HMMR, hyaluronan-mediated motility receptor; lncRNA, noncoding RNA; BLBC, basal-like breast cancer.
The expression of HMMR was decreased after treatment with HMMR-AS1 siRNA in BLBC cell lines
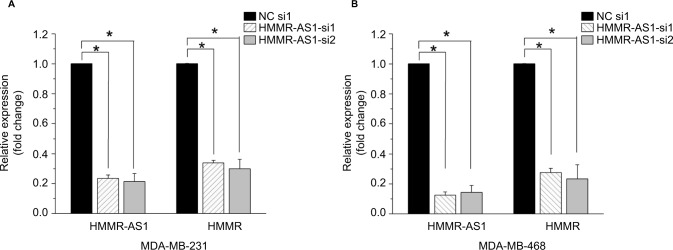
To explore the association between the expression of HMMR-AS1 and HMMR, RNA interference analysis was conducted in MDA-MB-231 and MDA-MB-468 cells. The expression levels of HMMR and lncRNA HMMR-AS1 were analyzed using RT-qPCR. The data suggested that expression of both HMMR-AS1 and HMMR was significantly downregulated in HMMR-AS1 siRNA group compared with NC siRNA group (Figure 2, P<0.05).
Figure 2.
The expression levels of HMMR mRNA and HMMR-AS1 long noncoding RNA in (A) MDA-MB-231 and (B) MDA-MB-468 cells after siRNA transfection. The results showed that the expression levels of HMMR-AS1 and HMMR were significantly reduced in HMMR-AS1 siRNA groups. *P<0.05 compared with the siRNA control group.
Abbreviations: HMMR, hyaluronan-mediated motility receptor; HMMR-AS1, HMMR antisense RNA1; siRNA, small interfering RNA; NC, negative control; mRNA, messenger RNA.
Knockdown of HMMR-AS1 in cells impaired the proliferation ability of MDA-MB-231 and MDA-MB-468 cells
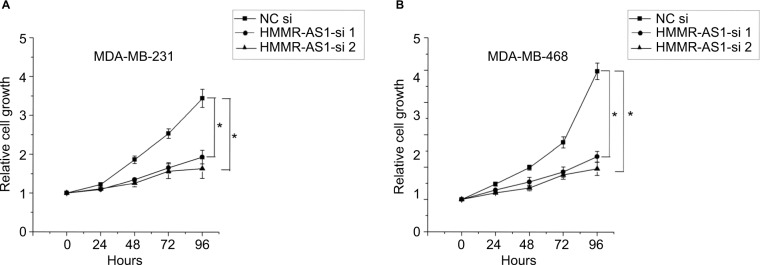
HMMR-AS1 in the MDA-MB-231 and MDA-MB-468 BLBC cell lines was knocked down to further investigate the regulatory mechanism of HMMR-AS1, and then MTS assay was conducted to explore the cell proliferation ability. Consequently, we found that the proliferation ability of cells in HMMR-AS1 siRNA groups was lower than that in NC siRNA group (Figure 3, P<0.05).
Figure 3.
Effects of HMMR-AS1 knockdown on the proliferation of MDA-MB-231 and MDA-MB-468 cells. HMMR-AS1 siRNAs evidently inhibited the growth of (A) MDA-MB-231 and (B) MDA-MA-468 compared with NC siRNA in an MTS assay (*P<0.05).
Abbreviations: HMMR-AS1, HMMR antisense RNA1; siRNA, small interfering RNA; NC, negative control; MTS, 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium.
Knockdown of HMMR-AS1 in cells weakened the invasion ability of MDA-MB-231 and MDA-MB-468 cells
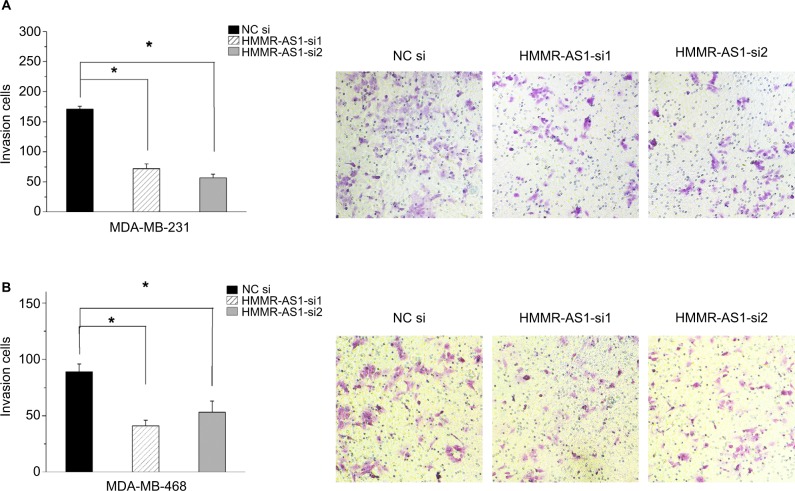
Matrigel invasion assay was carried out to explore the cell invasion ability. The data suggested that the invasion ability of the MDA-MB-231 and MDA-MB-468 cells was distinctly suppressed by HMMR-AS1 siRNA (Figure 4, P<0.05).
Figure 4.
Effects of HMMR-AS1 knockdown on the migration ability of MDA-MB-231 and MDA-MB-468 cells. The capacity of cell invasion in the (A) MDA-MB-231 and (B) MDA-MB-468 cells was apparently impaired by HMMR-AS1 siRNA. *P<0.05 compared with that of the NC siRNA control.
Abbreviations: HMMR-AS1, HMMR antisense RNA1; siRNA, small interfering RNA; NC, negative control.
Expression of HMMR-AS1 correlated with that of other BLBC-related genes
Furthermore, we investigated the correlation between the expression of HMMR-AS1 and that of BRCA1, BRCA2, and PTEN. The data showed that HMMR-AS1 expression was positively associated with two tumor promoter genes, BRCA1 (R=0.57, P<0.05) and BRCA2 (R=0.62, P<0.05), whereas negatively correlated with the tumor suppressor gene PTEN (R=−0.57, P<0.05) (Table 3).
Table 3.
Correlations of HMMR-AS1 with other cancer-related genes
| Gene name | Correlation coefficients with HMMR-AS1 |
|---|---|
| BRCA1 | R = 0.57, P<0.05 |
| BRCA2 | R = 0.62, P<0.05 |
| PTEN | R = 0.62, P<0.05 |
Abbreviation: HMMR-AS1, hyaluronan-mediated motility receptor antisense RNA 1.
Discussion
High-throughput screening advances in assay technologies allow the biological classification of breast cancers into subtypes with significant gene expression profiles. BLBCs are considered as one of the most aggressive subtypes, with basal cells containing a distinct gene expression pattern.
Recently, numerous studies have proved that the lncRNAs play a pivotal role in some malignant tumors.4–6 In various breast cancers, it has been reported that several lncRNAs could serve as novel biomarkers and therapeutic targets, such as HOTAIR, BC200, and CCAT2.18–20 However, the expression and functional roles of tumor-associated lncRNAs in BLBCs are still not clear.
In our study, a novel lncRNA HMMR-AS1 has been found. It may be a tumor promoter gene in BLBCs. Through RT-qPCR analysis, we found that the expression level of lncRNA HMMR-AS1 was significantly higher in BLBCs than in other non-BLBC subtypes. lncRNA HMMR-AS1 was overexpressed in BLBCs compared with negative control group, suggesting that lncRNA HMMR-AS1 may serve as a novel biomarker in BLBCs.
Furthermore, our study demonstrated that knockdown of HMMR-AS1 impaired the proliferation ability of MDA-MB-231 and MDA-MB-468 cells. The result showed that HMMR-AS1 may be a diagnostic marker in BLBCs and serve as an essential regulator working on the breast cancer cell, like the lncRNA HOTAIR.18
Some lncRNAs associated with cancers could participate in the regulating activities through various mechanisms. These lncRNAs may exert their functional roles through regulating their neighboring protein-coding genes, which is similar to lncRNA PVT1 and protein-coding gene c-MYC.21,22 lncRNA HMMR-AS1 is a neighboring gene of the HMMR. These lncRNAs and their neighboring protein-coding genes form “lncRNA–mRNA gene pairs”, which interplay with each other in the process of tumors.23–25 It was reported that the protein-coding gene HMMR could interplay with the critical cancer driver genes BRCA1/2 in BLBC15 and the function of HMMR is involved in the progression of breast cancer, colorectal cancer, and glioma.26–28 Thus, it is tempting to speculate that lncRNA HMMR-AS1 and HMMR may be correlated with each other to effect the progression of BLBCs. Through the RT-qPCR data, we found that the expression of lncRNA HMMR-AS1 and HMMR was positively correlated. Using RNA interference analysis, we found that the lncRNA HMMR-AS1 knockdown evidently suppressed the expression of HMMR. Meanwhile, both cell growth rate and migration abilities were correspondingly suppressed. However, the concrete mechanism behind the contribution of lncRNA HMMR-AS1 and HMMR to the progression of BLBC still needs to be further investigated. Intriguingly, our study also indicated that the expression of HMMR-AS1 was also positively linked with oncogenes BRCA1 and BRCA2, and negatively linked with the tumor suppressor gene PTEN.
Conclusion
This study may be the first to identify the expression and functional role of lncRNA HMMR-AS1 and its correlation with the adjacent mRNA HMMR in BLBCs. The results suggested that HMMR-AS1 may be regarded as a potential biomarker and therapeutic target in the future.
To exactly clarify the modulating method between HMMR-AS1 and HMMR, it is necessary to further explore additional pathways which may also play a part in HMMR-AS1, HMMR as well as cancer-related genes in BLBC (such as BRCA1/2 and PTEN).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Kreike B, van Kouwenhove M, Horlings H, et al. Gene expression profiling and histopathological characterization of triple-negative/basal-like breast carcinomas. Breast Cancer Res. 2007;9(5):R65. doi: 10.1186/bcr1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rakha EA, Reis-Filho JS, Ellis IO. Basal-like breast cancer: a critical review. J Clin Oncol. 2008;26(15):2568–2581. doi: 10.1200/JCO.2007.13.1748. [DOI] [PubMed] [Google Scholar]
- 3.Carey LA, Perou CM, Livasy CA, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295(21):2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 4.Huarte M, Rinn JL. Large non-coding RNAs: missing links in cancer? Hum Mol Genet. 2010;19(R2):R152–R161. doi: 10.1093/hmg/ddq353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31(43):4577–4587. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsai MC, Spitale RC, Chang HY. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res. 2011;71(1):3–7. doi: 10.1158/0008-5472.CAN-10-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okazaki Y, Furuno M, Kasukawa T, et al. FANTOM Consortium. RIKEN Genome Exploration Research Group Phase I & II Team Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420(6915):563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 8.Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perez DS, Hoage TR, Pritchett JR, et al. Long, abundantly expressed non-coding transcripts are altered in cancer. Hum Mol Genet. 2008;17(5):642–655. doi: 10.1093/hmg/ddm336. [DOI] [PubMed] [Google Scholar]
- 10.Han L, Zhang K, Shi Z, et al. LncRNA profile of glioblastoma reveals the potential role of lncRNAs in contributing to glioblastoma pathogenesis. Int J Oncol. 2012;40(6):2004–2012. doi: 10.3892/ijo.2012.1413. [DOI] [PubMed] [Google Scholar]
- 11.Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hatano H, Shigeishi H, Kudo Y, et al. Overexpression of receptor for hyaluronan-mediated motility (RHAMM) in MC3T3-E1 cells induces proliferation and differentiation through phosphorylation of ERK1/2. J Bone Miner Metab. 2012;30(3):293–303. doi: 10.1007/s00774-011-0318-0. [DOI] [PubMed] [Google Scholar]
- 13.Misra S, Hascall VC, Markwald RR, Ghatak S. Interactions between hyaluronan and its receptors (CD44, RHAMM) regulate the activities of inflammation and cancer. Front Immunol. 2015;6:201. doi: 10.3389/fimmu.2015.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maxwell CA, Benítez J, Gómez-Baldó L, et al. Interplay between BRCA1 and RHAMM regulates epithelial apicobasal polarization and may influence risk of breast cancer. PLoS Biol. 2011;9(11):e1001199. doi: 10.1371/journal.pbio.1001199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blanco I, Kuchenbaecker K, Cuadras D, et al. Assessing associations between the AURKA-HMMR-TPX2-TUBG1 functional module and breast cancer risk in BRCA1/2 mutation carriers. PLoS One. 2015;10(4):e0120020. doi: 10.1371/journal.pone.0120020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Preker P, Nielsen J, Kammler S, et al. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322(5909):1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- 17.Jacquier A. The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nat Rev Genet. 2009;10(12):833–844. doi: 10.1038/nrg2683. [DOI] [PubMed] [Google Scholar]
- 18.Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iacoangeli A, Lin Y, Morley EJ, et al. BC200 RNA in invasive and preinvasive breast cancer. Carcinogenesis. 2004;25(11):2125–2133. doi: 10.1093/carcin/bgh228. [DOI] [PubMed] [Google Scholar]
- 20.Redis RS, Sieuwerts AM, Look MP, et al. CCAT2, a novel long non-coding RNA in breast cancer: expression study and clinical correlations. Oncotarget. 2013;4(10):1748–1762. doi: 10.18632/oncotarget.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shtivelman E, Bishop JM. Effects of translocations on transcription from PVT. Mol Cell Biol. 1990;10(4):1835–1839. doi: 10.1128/mcb.10.4.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guan Y, Kuo WL, Stilwell JL, et al. Amplification of PVT1 contributes to the pathophysiology of ovarian and breast cancer. Clin Cancer Res. 2007;13(19):5745–5755. doi: 10.1158/1078-0432.CCR-06-2882. [DOI] [PubMed] [Google Scholar]
- 23.Wahlestedt C. Natural antisense and noncoding RNA transcripts as potential drug targets. Drug Discov Today. 2006;11:503–508. doi: 10.1016/j.drudis.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 24.Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol. 2009;1:637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yelin R, Dahary D, Sorek R, et al. Widespread occurrence of antisense transcription in the human genome. Nat Biotechnol. 2003;21:379–386. doi: 10.1038/nbt808. [DOI] [PubMed] [Google Scholar]
- 26.Akent’eva NP, Shushanov SS, Kotel’nikov AI. Effects of RHAMM/HMMR-selective peptides on survival of breast cancer cells. Bull Exp Biol Med. 2015;159(5):658–661. doi: 10.1007/s10517-015-3041-3. [DOI] [PubMed] [Google Scholar]
- 27.Koelzer VH, Huber B, Mele V, et al. Expression of the hyaluronan-mediated motility receptor RHAMM in tumor budding cells identifies aggressive colorectal cancers. Hum Pathol. 2015;46(11):1573–1581. doi: 10.1016/j.humpath.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 28.Tilghman J, Wu H, Sang Y, et al. HMMR maintains the stemness and tumorigenicity of glioblastoma stem-like cells. Cancer Res. 2014;74(11):3168–3179. doi: 10.1158/0008-5472.CAN-13-2103. [DOI] [PMC free article] [PubMed] [Google Scholar]