Abstract
Objective
The objective of this study was to observe the prognosis of pediatric patients with E2A-PBX1-positive acute lymphoblastic leukemia (ALL) from the treatment with the CCLG-ALL2008 protocol.
Design and methods
Three hundred and forty-nine Chinese pediatric patients with pre-B-cell ALL were enrolled in this study from December 2008 to September 2013. Of these, 20 patients with E2A-PBX1 expression and 223 without the gene expression were stratified into two cohorts. Clinical and biological characteristics and 5-year event-free survival (EFS), relapse-free survival (RFS), and overall survival (OS) were analyzed and compared between these two groups.
Results
The E2A-PBX1 fusion transcript was detected in 20 of 349 (5.7%) patients. Compared with the gene-negative subgroup, patients with E2A-PBX1 were younger in age but did not show significant differences in white blood cell (WBC) count or gender distribution at primary diagnosis. Moreover, there were more inferior karyotypes detected in the E2A-PBX1 subgroup (P=0.035). With the CCLG-ALL2008 treatment protocol, patients with E2A-PBX1 showed a favorable treatment response with lower minimal residual disease (MRD) levels (<10−4) at time point 1 (TP1, P=0.039) but no superior steroid response or histological remission. We also observed a promising survival outcome, with a 5-year EFS reaching 95.0%±4.9% versus 66.3%±3.9% in the gene-negative group (P=0.039). However, we did not find significant differences in RFS (P=0.061) and OS (P=0.113).
Conclusion
Our data provided clinical observation of Chinese pediatric patients. Patients with E2A-PBX1-positive ALL benefited well from the CCLG-ALL2008 protocol, a risk-based intensified treatment trial, with lower levels of MRD and longer RFS duration though they had no favorable characteristics at primary diagnosis.
Keywords: pediatric, acute lymphoblastic leukemia, E2A-PBX1 gene transcript, prognosis, CCLG-ALL2008 protocol
Introduction
Acute lymphoblastic leukemia (ALL) is one of the most common childhood malignancies, accounting for 75%–80% of cases of acute leukemia among this age group.1 Currently, the long-term survival of pediatric ALL is >80%, whereas it was 30% in the late 1960s.2 These impressive achievements were attributed to a comprehensive understanding of the biological characteristics and genetic diversity of ALL, affiliating the development of therapeutic strategies. ALL is strongly associated with acquired chromosomal abnormalities, resulting in the generation of fusion genes that have an important role in diagnosis and prognosis.3 In this regard, detection of a specific gene rearrangement allows the identification of prognostically relevant subgroups and helps to select an appropriate treatment to maintain a high remission rate.
There are four common gene rearrangements, TEL-AML1-t(12;21), E2A-PBX1-t(1;19), BCR-ABL1-t(9;22), and MLL-AF4 t(4;11), which had been widely studied in pediatric B-ALL. The (1;19)(q23;p13) translocation, leading to the production fusion transcript E2A/PBX1, is one of the most common translocations in pediatric B-ALL.4 The E2A-PBX1 chimeric transcription factor contains the N-terminal transactivation domain of E2A (TCF3) fused to the C-terminal DNA-binding homeodomain of PBX1 and was observed in 5%–7% of pediatric ALL cases.5 Patients with E2A-PBX1 gene expression usually had a more aggressive disease course and received an intensive chemotherapy.6 However, benefiting from the investigation of the pathological mechanism of E2A-PBX1-positive B-ALL and appropriate treatment till now, patients obtained an exciting survival outcome.7,8
In the Chinese Children’s Leukemia Group-acute lymphoblastic leukemia 2008 (CCLG-ALL2008) protocol, E2A-PBX1 is regarded as an intermediate risk (IR) index. In our retrospective study, we aimed to provide more information on the prevalence and prognostic significance of E2A-PBX1 in B-ALL.
Design and methods
Patients and treatment protocol
During December 2008 to September 2013, 349 pediatric patients aged 3 months to 16 years (median, 4 years) were diagnosed as pre-B-cell ALL according to morphology, immunophenotype, cytogenetics, and molecular biology criteria and received treatment with the protocol of CCLG-ALL08 in our single institution. Before October 2010, patients were risk grouped and treated based on only traditional prognostic factors, but after that, minimal residual disease (MRD) was incorporated for risk stratification and treatment modulation. Follow-up observations extended through November 2014. Details of the stratification and treatment regimen of the protocol are outlined in the literature9 (Tables 1 and 2). The CCLG-ALL2008 protocol was approved by the Children’s Hospital of Soochow University Institutional Ethics Committee. The written informed consents were signed by the parents or guardians of each patient.
Table 1.
Different risk stratification criteria
| Risk degree | Risk stratification
|
||
|---|---|---|---|
| Traditional risk | MRD-based risk | MRD-combined risk | |
| SR | (All the factors) None of the HR factor; B-precursor ALL; age ranged 1–10 years; WBC counts <50×109/L; the absolute value of spinal fluid blasts <5×106/L; good BM histological remission (M1 or M2) on day 15; histological CR (normal BM cellularity with <5% undifferentiated cells) achieved on day 33 of induction remission phase. |
MRD <10−4 at TP1 | Traditional SR plus MRD <10−4 at TP1 |
| IR | None of the HR factor; BM showed M3 after SR induction or M1/M2 after IR induction on day 15 (At least one factor) Age ≥10 years; age <1 year without MLL rearrangements; WBC counts ≥50×109/L; the absolute value of spinal fluid blasts >5×106/L; T-ALL; E2A-PBX1 fusion gene or t(1;19) chromosomal translocation. |
MRD <10−2 at TP1 | Traditional IR plus MRD <10−2 at TP1 |
| HR | (At least one factor) MLL gene rearrangement (t[4;11] chromosomal translocation and/or MLL-AF4 gene fusion or other MLL rearrangement); BCR/ABL fusion gene or t(9;22) chromosomal translocation; early resistance to the 1-week steroid induction (the absolute value of peripheral blasts >1,000/µL); poor BM blast clearance (M3) after IR induction on day 15; no histological CR (normal BM cellularity with >5% undifferentiated cells) achieved on day 33 of induction remission phase. |
MRD ≥10−2 at TP1 or MRD ≥10−3 at TP2 | Traditional HR or MRD ≥10−2 at TP1 or MRD ≥10−3 at TP2 |
Notes: TP1, at the end of induction around day 33; TP2, before consolidation around week 12; M1, BM cellularity with <5% undifferentiated cells; M2, BM cellularity with ≥5% but <25% undifferentiated cells; M3, BM cellularity with ≥25% undifferentiated cells.
Abbreviations: CR, complete response; MRD, minimal residual disease; SR, standard risk; HR, high risk; ALL, acute lymphoblastic leukemia; WBC, white blood cell; BM, bone marrow; TP1, time point 1; IR, intermediate risk; TP2, time point 2.
Table 2.
CCLG-2008 treatment protocol
| CCLG-2008 | Prednisoneinduction | Remission induction | Early intensification | Consolidation | Delay intensification I | Maintenance I | Delay intensification II | Maintenance II |
|---|---|---|---|---|---|---|---|---|
| Standard risk | P (1 week) | VDLD (DNR ×2) | CAM | HD-MTX (2 g/m2) ×4 | VDLD + CAM | – | – | 6-MP + MTX/VD + IT |
| Intermediate risk | P (1 week) | VDLD (DNR ×4) | CAM ×2 | HD-MTX (5 g/m2) ×4 | VDLD + CAM | 6-MP + MTX | VDLD + CAM | 6-MP + MTX/VD + TIT |
| High risk | P (1 week) | VDLD (DNR ×4) | CAM ×2 | I′II′III′ ×2 | VDLD + CAM | – | – | 6-MP + MTX/CA/VD + IT |
Notes: I′, BFM high-risk block-1′; II′, BFM high-risk block-2′; III′, BFM high-risk block-3′.
Abbreviations: CCLG, Chinese Childhood Leukemia Group; VDLD, vincristine + daunorubicin + l-asparaginase + dexamethasone; CAM, cyclophosphamide + cytarabine + 6-mercaptopurine; HD-MTX, high-dose methotrexate; 6-MP, 6-mercaptopurine; VD, vincristine + dexamethasone; IT, intrathecal injection with dexamethasone and methotrexate; CA, cyclophosphamide + cytarabine; TIT, intrathecal injection with dexamethasone, methotrexate, and cytarabine; BFM, Berlin-Frankfurt-Münster; DNR, daunorubicin.
Cytogenetic abnormalities and fusion transcript analysis
Conventional cytogenetic analysis was performed at the time of initial diagnosis. Chromosomes were R-banded on bone marrow (BM) cells from direct and/or 24-hour unstimulated cultures.10 Cytogenetic analysis was considered successful if a clonal chromosomal abnormality was detected or at least 20 metaphases were analyzed. Total RNA was extracted using Trizol (Thermo Fisher Scientific, Waltham, MA, USA), according to the manufacturer’s instructions. A multiplex reverse transcription polymerase chain reaction system was adapted with modifications. The system was able to detect simultaneously the following fusion transcripts of ALL: BCR-ABL1, E2A-PBX1, TEL-AML1, SIL-TAL1, TLS-ERG, HOX11, and MLL rearrangements.
MRD analysis
The MRD analysis was performed using flow Cytometry, and multiple markers were used to identify leukemia-specific immunophenotypes. Detection of a leukemia cell was feasible among at least 10,000 normal cells (10−4). The MRD level was analyzed at two check points, which were time point 1 (TP1; at the end of induction around day 33 after beginning chemotherapy) and time point 2 (TP2; before consolidation around week 12 after beginning chemotherapy).
Criteria for assessment of response to treatment and risk stratification
In the CCLG-ALL08 protocol, the clinical presentation (age, white blood cell [WBC] count, ALL type, and gene aberrations), early treatment response (prednisone response and histological remission status of BM), and MRD measurement were used for risk classification (Table 1). In this study, we separated these factors for conventional risk stratification, MRD-based risk stratification, and MRD-combined risk stratification. The detailed information is listed in Table 1.
Relapse was defined as the reappearance of leukemic cells in BM (>25% blasts). Central nervous system relapse was defined as more than five blasts in the cerebrospinal fluid. Testicular relapse was diagnosed clinically and confirmed with ultrasonography.
Statistical analysis
October 31, 2014, was chosen as the reference date for the end of data collection for statistical analysis purposes. Comparisons of pretreatment characteristics and response to treatment between E2A-PBX1-positive group and gene-negative group were evaluated by chi-square test. Relapse-free survival (RFS) was defined from the time of diagnosis to the date of disease relapse; event-free survival (EFS) was defined from the date of diagnosis to the date of relapse, death, or treatment failure, whichever came first, or the last contact with patients in continuous hematological complete response; overall survival (OS) was referred to the date of diagnosis till the date of death or last follow-up. RFS, EFS, and OS were estimated using the Kaplan–Meier procedure. Survival comparisons between these two groups were performed with the log-rank test. All tests were two sided and considered statistically significant at P<0.05. SPSS 16.0 software (Statistical Product and Service Solutions Inc., Chicago, IL, USA) was used for all statistical analyses.
Results
Clinical features
According to gene expressions detected in the total 349 patients, E2A-PBX1 expression was found in 20 cases, accounting for 5.7%. Two hundred and twenty-three cases were without gene expression. In the E2A-PBX1-positive group, the mean age was 45 months (range 21–89 months) and the ratio of boys versus girls was 1.2:1. In the gene-negative group, the mean age was 62 months (range 5–199 months) and the gender ratio was 1.5:1. It seemed that patients with E2A-PBX1 expression were younger than gene-negative ones, but significant differences were not found whether in age (P=0.074) or in gender (P=0.675). We also compared the WBC counts at initial diagnosis between these two cohorts, but there was no significant difference (P=0.280). The risk distributions of most of the karyotypes in each of the groups were favorable or intermediate. In our single retrospective study, more intermediate karyotypes were found in the E2A-PBX1-positive group (P=0.021). Details of the clinical characteristics comparison are listed in Table 3.
Table 3.
Comparison of clinical characteristics and treatment response between E2A-PBX1 subgroup and gene-negative subgroup
| Characteristics | E2A-PBX1 | Gene-negative | P-value |
|---|---|---|---|
| Age (years) | 0.142 | ||
| <10 | 20 | 197 | |
| >10 | 0 | 26 | |
| Gender | 0.657 | ||
| Boys | 11 | 134 | |
| Girls | 9 | 89 | |
| WBC (×109/L) | 0.280 | ||
| <50 | 16 | 179 | |
| 50–100 | 1 | 29 | |
| >100 | 3 | 15 | |
| Karyotype | 0.035 | ||
| Favorable | 2 | 71 | |
| Intermediate | 16 | 113 | |
| Adverse | 0 | 2 | |
| Steroid response | 0.443 | ||
| Sensitive | 17 | 201 | |
| Resistant | 3 | 22 | |
| BM remission on day 15 | 0.148 | ||
| M1 | 15 | 118 | |
| M2 | 3 | 60 | |
| M3 | 2 | 45 | |
| BM remission on day 33 | 0.606 | ||
| CR | 20 | 211 | |
| NCR | 0 | 12 | |
| MRD level at TP1 | 0.039 | ||
| <10−4 | 13 | 97 | |
| >10−4 | 5 | 111 | |
| MRD level at TP1 | 0.081 | ||
| <10−3 | 17 | 157 | |
| >10−3 | 1 | 51 | |
| MRD level at TP2 | 0.304 | ||
| <10−4 | 12 | 106 | |
| >10−4 | 6 | 90 |
Abbreviations: WBC, white blood cell; BM, bone marrow; MRD, minimal residual disease; TP1, time point 1; TP2, time point 2; CR, complete response; NCR, not complete response.
Treatment response
Early sensitivity to the 1-week steroid induction was defined as the absolute value of peripheral blasts ≤1,000/µL. According to our statistical analysis, the early treatment response to steroid showed no difference between these two groups (P=0.443). Moreover, histological responses in BM also showed no differences on day 15 (P=0.148) and day 33 (P=0.606) during remission induction. Furthermore, we evaluated the MRD, a more sensitive index, to find any underlying difference between the E2A-PBX1-positive group and the gene-negative group. In total, 208 patients and 196 patients in the gene-negative group had MRD records at TP1 and TP2 respectively, and in the E2A-PBX1 group, 18 cases had MRD records. We categorized the MRD into different range levels. Patients with E2A-PBX1 expression received a favorable response with lower MRD level (<10−4) at TP1 (P=0.037). Though no significant difference was found with MRD division level of 10−3, more cases need to be enrolled for observation and analysis. At TP2, six of the 18 patients in the E2A-PBX1 group had an MRD level <10−3 but >10−4, and all the remaining patients had <10−4. However, no significant difference existed when compared with the gene-negative group (P=0.304). The treatment responses between these two groups are shown in Table 3.
Survival outcome
Figures 1 and 2 show the influence of gene aberrations to survival outcomes of EFS and OS in B-ALL patients. The survival curves showed favorable outcomes in the E2A-PBX1-positive group. The 18 patients with MRD record in the E2A-PBX1-positive cohort were grouped to IR (15 cases) and high risk (HR; three cases) according to conventional risk stratification. However, according to MRD-based risk stratification, they were either regrouped to MRD-SR (13 cases) or MRD-IR (five cases). In both the risk stratifications, the E2A-PBX1-positive group had significant differences compared with the gene-negative group (P<0.001 and P=0.035, Table 4). The 5-year EFS, RFS, and OS of the E2A-PBX1-positive group compared with the gene-negative group were 95.0%±4.9% versus 66.3%±3.9% (P=0.039, Figure 3), 95.0%±4.9% versus 68.9%±3.9% (P=0.061, Figure 4), and 95.0%±4.9% versus 73.6%±3.9% (P=0.113, Figure 5), respectively. Although the P-value is >0.05 for RFS and OS, the trends are significant.
Figure 1.
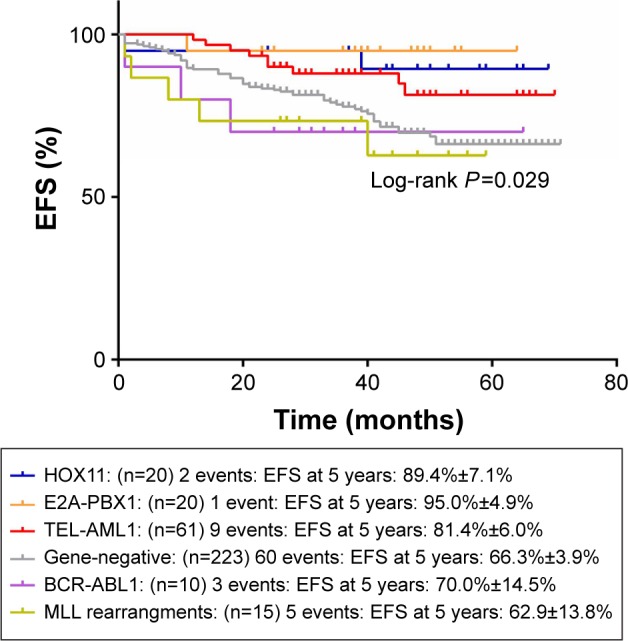
The influence of gene aberrations to EFS in B-ALL patients.
Abbreviation: EFS, event-free survival.
Figure 2.
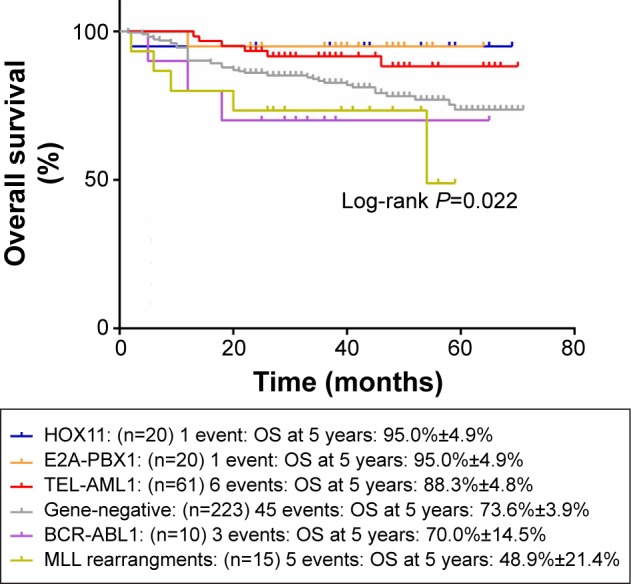
The influence of gene aberrations to OS in B-ALL patients.
Abbreviation: OS, overall survival.
Table 4.
The risk distributions of gene-negative group and E2A-PBX1-positive group according to either traditional risk stratification or MRD-based risk stratification
| Risk degree | E2A-PBX1 | Gene-negative | P-value |
|---|---|---|---|
| Traditional risk stratification | <0.001 | ||
| SR | 0 | 115 | |
| IR | 15 | 51 | |
| HR | 3 | 32 | |
| MRD-based risk stratification | 0.035 | ||
| SR | 13 | 86 | |
| IR | 5 | 77 | |
| HR | 0 | 35 | |
Abbreviations: SR, standard risk; IR, intermediate risk; HR, high risk; MRD, minimal residual disease.
Figure 3.
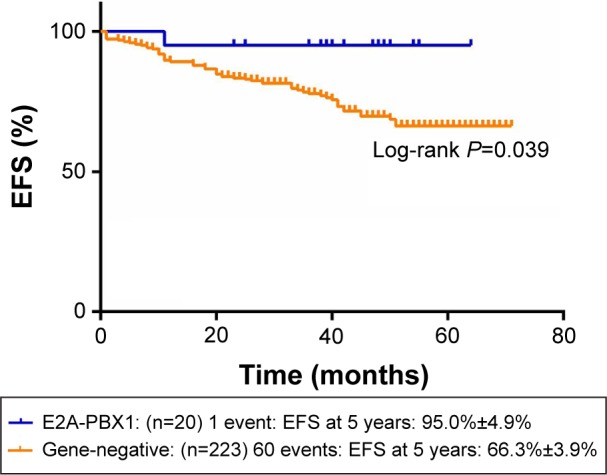
The 5-year EFS of gene-negative group and E2A-PBX1-positive group.
Abbreviation: EFS, event-free survival.
Figure 4.
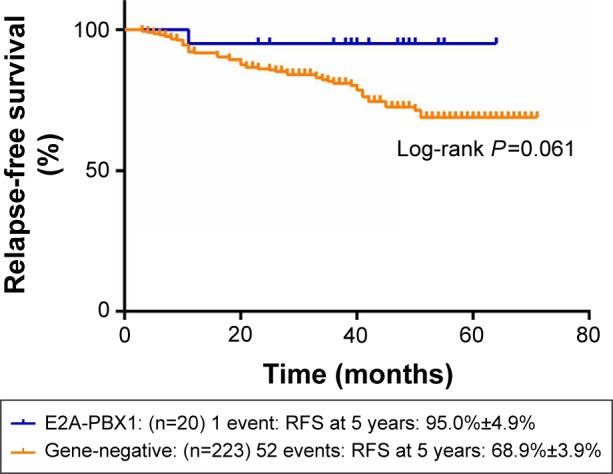
The 5-year RFS of gene-negative group and E2A-PBX1-positive group.
Abbreviation: RFS, relapse-free survival.
Figure 5.
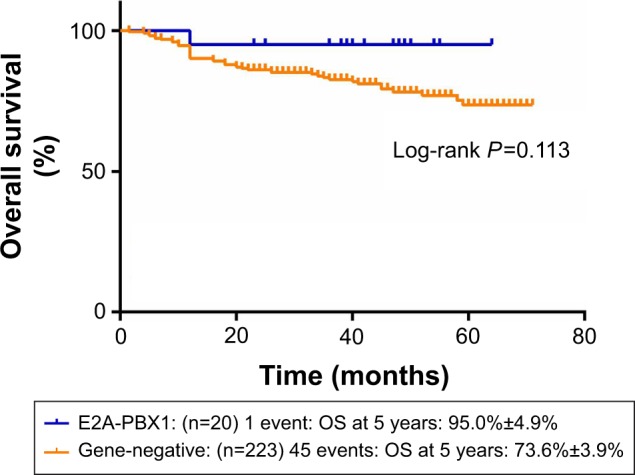
The 5-year OS of gene-negative group and E2A-PBX1-positive group.
Abbreviation: OS, overall survival.
Discussion
In childhood ALL, translocations are detected in approximately one-half of all patients, and the presence of a specific translocation has been evident that plays a central role in the process of malignant transformation.11 The t(1;19)(q23;p13) is one of the most common recurring translocations in childhood ALL. It was first described by Carroll et al12 in 1983 as a nonrandom translocation associated with pre- B-cell ALL. Subsequently, Mellentin et al13 demonstrated that the E2A gene is consistently located at the breakpoint of the t(1;19). The t(1;19) leads to juxtaposition of the E2A gene from chromosome 19 with PBXl, a novel homeobox gene on chromosome 1.14 Hunger et al15 then further demonstrated the consistent fusion of E2A- and PBXl-coding sequences resulting from t(1;19) and suggested that site-specific fusion of E2A and PBXl is an important pathogenic event in t(1;19) ALL.
Since E2A-PBX1 was demonstrated as a key transcript fusion in pediatric B-ALL, reports focusing on this type of ALL had provided people with important insights. Studies of the Pediatric Oncology Group (POG) showed that children with pre-B-cell ALL had a significantly poorer response to treatment than other children and that the subgroup with the t(1;19) was responsible for the adverse prognosis.16 In a 4-year consecutive study, the E2A-PBX1 fusion transcript was detected in 5.8% of the total 261 patients diagnosed as pre-B-cell ALL in Mexican population. Unexpectedly, patients of this subgroup showed a hematologic remission duration similar to the known HR molecular subgroups such as BCR-ABL or MLL-AF9, in addition, a more aggressive disease at diagnosis such as higher WBC counts and older age.17 Given to the adverse effect of E2A-PBX1, this subgroup of patients usually received a more intensive chemotherapy or hematopoietic stem cell transplantation. In an Australian cohort, the implementation of early-dose-intensified remission induction to the ALL-Berlin-Frankfurt-Münster (BFM) trial was highly effective in children with E2A-PBX1, with the 5-year EFS reaching 90%±5%.7 Felice et al8 also reported a BFM-based protocol, with which the probability of event-free survival (pEFS) of patients with t(1;19)/E2A-PBX1 was 85% and significantly superior to that of patients without t(1;19)/E2A-PBX1 (P <0.0001).
In the retrospective study from our single institution, we enrolled 349 patients diagnosed as pre-B-cell ALL; of these, 20 (5.7%) cases showed E2A-PBX1-positive expression. This was consistent with most studies. Patients in the E2A-PBX1 subgroup seemed younger than those in the gene-negative subgroup. However, clinical characteristics had no significant differences at initial diagnosis, with karyotype for exception. The gene-negative subgroup had more favorable karyotypes than the E2A-PBX1 subgroup. Both of these two groups received the CCLG-ALL2008 treatment protocol, which was implemented in our country since 2008. There are four points for treatment response assessment. Though no differences were observed whether on steroid response or on histological remission, patients in the E2A-PBX1 subgroup showed a lower level of MRD on day 33. The MRD is regarded as a more precise index for disease surveillance. An MRD <10−4 usually predicts a better prognosis, with less risk to relapse and longer survival duration. Our result was in contrast to that of Gao et al,18 who reported that in patients positive for MRD, the expression level of E2A-PBX1 was high at primary diagnosis. In another CCLG-ALL2008 treatment trial-based study, Mei et al19 reported that the EFS of E2A-PBX1 subgroup was 71.5%. Surprisingly, in accordance with MRD, patients with E2A-PBX1 expression in our cohort showed a 5-year EFS of 95.0%±4.9%, superior to those without gene expression.
Our result provides more clinical information about the subgroup of E2A-PBX1 ALL from Asian population. E2A-PBX1 fusion transcript is more common to be detected in younger pediatric population and usually accompanied by inferior karyotypes. However, in our single retrospective study, we proved that patients with E2A-PBX1 were well benefited from the CCLG-ALL2008 protocol, a risk-based intensified treatment trial.
Acknowledgments
This work was supported by the grants from the Natural Science Foundation of China (nos 81370627 and 81170513), Jiangsu Province key point project (no BL2013014), and a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions, and Suzhou Clinical Key Project (LCZX201507 and SZZX201504).
Footnotes
Author contributions
All authors contributed toward data analysis, drafting and revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Asselin BL, Gaynon P, Whitlock JA. Recent advances in acute lymphoblastic leukemia in children and adolescents: an expert panel discussion. Curr Opin Oncol. 2013;25(suppl 3):S1–S13. doi: 10.1097/CCO.0000000000000017. [DOI] [PubMed] [Google Scholar]
- 2.Bartram CR, Schrauder A, Kohler R, Schrappe M. Acute lymphoblastic leukemia in children: treatment planning via minimal residual disease assessment. Dtsch Arztebl Int. 2012;109(40):652–658. doi: 10.3238/arztebl.2012.0652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harrison CJ. Cytogenetics of paediatric and adolescent acute lymphoblastic leukaemia. Br J Haematol. 2009;144(2):147–156. doi: 10.1111/j.1365-2141.2008.07417.x. [DOI] [PubMed] [Google Scholar]
- 4.Piccaluga PP, Malagola M, Rondoni M, et al. Poor outcome of adult acute lymphoblastic leukemia patients carrying the (1;19)(q23;p13) translocation. Leuk Lymphoma. 2006;47(3):469–472. doi: 10.1080/10428190500331261. [DOI] [PubMed] [Google Scholar]
- 5.Diakos C, Xiao Y, Zheng S, Kager L, Dworzak M, Wiemels JL. Direct and indirect targets of the E2A-PBX1 leukemia-specific fusion protein. PLoS One. 2014;9(2):e87602. doi: 10.1371/journal.pone.0087602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vey N, Thomas X, Picard C, et al. Allogeneic stem cell transplantation improves the outcome of adults with t(1;19)/E2A-PBX1 and t(4;11)/MLL-AF4 positive B-cell acute lymphoblastic leukemia: results of the prospective multicenter LALA-94 study. Leukemia. 2006;20(12):2155–2161. doi: 10.1038/sj.leu.2404420. [DOI] [PubMed] [Google Scholar]
- 7.Kager L, Lion T, Attarbaschi A, et al. Incidence and outcome of TCF3-PBX1-positive acute lymphoblastic leukemia in Austrian children. Haematologica. 2007;92(11):1561–1564. doi: 10.3324/haematol.11239. [DOI] [PubMed] [Google Scholar]
- 8.Felice MS, Gallego MS, Alonso CN, et al. Prognostic impact of t(1;19)/TCF3-PBX1 in childhood acute lymphoblastic leukemia in the context of Berlin-Frankfurt-Munster-based protocols. Leuk Lymphoma. 2011;52(7):1215–1221. doi: 10.3109/10428194.2011.565436. [DOI] [PubMed] [Google Scholar]
- 9.Gao C, Liu SG, Zhang RD, et al. NOTCH1 mutations are associated with favourable long-term prognosis in paediatric T-cell acute lymphoblastic leukaemia: a retrospective study of patients treated on BCH-2003 and CCLG-2008 protocol in China. Br J Haematol. 2014;166(2):221–228. doi: 10.1111/bjh.12866. [DOI] [PubMed] [Google Scholar]
- 10.Howlader N, Noone AM, Krapcho M, et al. SEER Cancer Statistics Review, 1975–2010. Bethesda, MD: National Cancer Institute; 2013. [Google Scholar]
- 11.Williams DL, Harber J, Murphy SB, et al. Chromosomal translocations play a unique role in influencing prognosis in childhood acute lymphoblastic leukemia. Blood. 1986;68(1):205–212. [PubMed] [Google Scholar]
- 12.Carroll AM, Crist WM, Parmley RT, Roper MA, Finley WH. Pre-B acute lymphocytic leukemia and chromosome translocation 1;19. Am J Hum Genet. 1983;35:60A. [Google Scholar]
- 13.Mellentin JD, Murre C, Donlon TA, et al. The gene for enhancer binding proteins E12/E47 lies at the t(1;19) breakpoint in acute leukemias. Science. 1989;246(4928):379–382. doi: 10.1126/science.2799390. [DOI] [PubMed] [Google Scholar]
- 14.Kamps MP, Murre C, Sun XH, Baltimore D. A new homeobox gene contributes the DNA binding domain of the t(1;19) translocation protein in pre-B ALL. Cell. 1990;60(4):547–555. doi: 10.1016/0092-8674(90)90658-2. [DOI] [PubMed] [Google Scholar]
- 15.Hunger SP, Galili N, Carroll AJ, Crist WM, Link MP, Cleary ML. The t(1;19)(q23;p13) results in consistent fusion of E2A and PBX1 coding sequences in acute lymphoblastic leukemias. Blood. 1991;77(4):687–693. [PubMed] [Google Scholar]
- 16.Crist WM, Carroll AJ, Shuster JJ, et al. Poor prognosis of children with pre-B acute lymphoblastic leukemia is associated with the t(1;19) (q23;p13): a Pediatric Oncology Group study. Blood. 1990;76(1):117–122. [PubMed] [Google Scholar]
- 17.Martinez-Mancilla M, Rodriguez-Aguirre I, Tejocote-Romero I, Medina-Sanson A, Ocadiz-Delgado R, Gariglio P. Clinical relevance of the fusion transcripts distribution pattern in Mexican children with acute lymphoblastic leukemia. J Pediatr Hematol Oncol. 2013;35(3):170–173. doi: 10.1097/MPH.0b013e318286d241. [DOI] [PubMed] [Google Scholar]
- 18.Gao C, Li ZG, Zhao W, Wu MY. Correlation of E2a-pbx1 expression level with clinical characteristics and early response to treatment in children with acute lymphoblastic leukemia. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2008;16(3):569–573. Chinese. [PubMed] [Google Scholar]
- 19.Mei YY, Gao C, Cui L, et al. Evaluation of the efficacy of two successive protocols on pediatric acute lymphoblastic leukemia with E2A-PBX1 fusion gene. Zhonghua Er Ke Za Zhi. 2013;51(6):467–471. Chinese. [PubMed] [Google Scholar]


