Abstract
Centrioles are critical for many cellular processes including cell division and cilia assembly. The number of centrioles within a cell is under strict control, and deregulation of centriole copy number is a hallmark of cancer. The molecular mechanisms that halt centriole amplification have not been fully elucidated. Here, we found that centrosomal protein of 76 kDa (Cep76), previously shown to restrain centriole amplification, interacts with cyclin-dependent kinase 2 (CDK2) and is a bona fide substrate of this kinase. Cep76 is preferentially phosphorylated by cyclin A/CDK2 at a single site S83, and this event is crucial to suppress centriole amplification in S phase. A novel Cep76 mutation S83C identified in a cancer patient fails to prevent centriole amplification. Mechanistically, Cep76 phosphorylation inhibits activation of polo-like kinase 1 (Plk1), thereby blocking premature centriole disengagement and subsequent amplification. Cep76 can also be acetylated, and enforced acetylation at K279 dampens the protein’s ability to inhibit amplification and precludes S83 phosphorylation. Acetylation of Cep76 normally occurs in G2 phase and correlates with loss of protein function. Our data suggest that temporal changes in posttranslational modifications of Cep76 during the cell cycle regulate its capacity to suppress centriole amplification, and its deregulation may contribute to malignancy.
Keywords: CDK2, cyclin A, Cep76, centriole amplification, phosphorylation, acetylation, cancer
Introduction
Centrosomes are well-known for their role in cell division (1). They are composed of a pair of centrioles surrounded by a pericentriolar matrix from which microtubules emanate and elongate. A single centrosome present in G1 phase of the cell cycle undergoes duplication in S phase. The duplicated centrosomes, once separated, establish the mitotic spindle at the onset of mitosis, ensuring faithful segregation of chromosomes into daughter cells. Centrosome duplication, like DNA replication, occurs once and only once per cell cycle and is under stringent control (2). Too few or too many centrosomes could jeopardize cell division, leading to cancer development (3). Indeed, centrosome amplification has been documented in many types of cancers (4–6) and is thought to be an important contributor to malignant transformation (5, 7–9) owing to its capacity to induce chromosome mis-segregation and cell invasion (10, 11). It is therefore of paramount importance to decipher the poorly understood mechanisms governing the fidelity of centrosome duplication.
Cyclin-dependent kinase 2 (CDK2) is a critical regulator of the cell cycle associated with cancer development. Although CDK2 is dispensable for viability in mice (12, 13) and its loss can be compensated by CDK1 (14), chemical genetics reveals a requirement of this kinase in cell cycle progression and cell proliferation (15). CDK2 also regulates centrosome duplication (16–19) by phosphorylating several key centrosomal substrates. Centrosome duplication begins when nucleophosmin/B23, a protein that associates with unduplicated centrosomes, dissociates from this organelle upon CDK2 phosphorylation (20). CDK2 phosphorylates Mps1 and CP110, two centrosomal proteins essential for duplication and amplification (21, 22). Furthermore, CDK2 cooperates with polo-like kinase 4 to regulate centrosome amplification, although the two proteins do not appear to interact (23, 24). Elegant studies using knockout mouse embryonic fibroblasts have demonstrated that whereas CDK2 is not required for normal centrosome duplication, it is needed for amplification (25, 26). Thus, current evidence favors the notion that CDK2 activity supports centrosome duplication/amplification. It is not entirely clear if CDK2 substrates always exert a positive effect on duplication/amplification, since a complete set of centrosomal CDK2 substrates have not been identified (27).
Centrosomal protein of 76 kDa (Cep76) was originally identified as a novel component of the centrosome using mass spectrometry-based proteomics (28). Previous studies indicate that this protein plays a pivotal role in restraining centriole amplification (29). A loss of Cep76 induces accumulation of supernumerary centrioles primarily during S phase (29). In addition, ectopic expression of Cep76 specifically suppresses centriole and centrosome amplification induced by hydroxyurea (HU), a drug that inhibits S phase progression and promotes multiple rounds of centriole duplication (29, 30). Interestingly, Cep76 protein levels peak in S phase (29), coinciding with elevated CDK2 activity and centriole duplication (31). The relationship between Cep76 and CDK2, if any, is unknown. Moreover, precisely how Cep76 suppresses centriole amplification at a mechanistic level has not been characterized.
Results
Cep76 preferentially interacts with cyclin A/CDK2
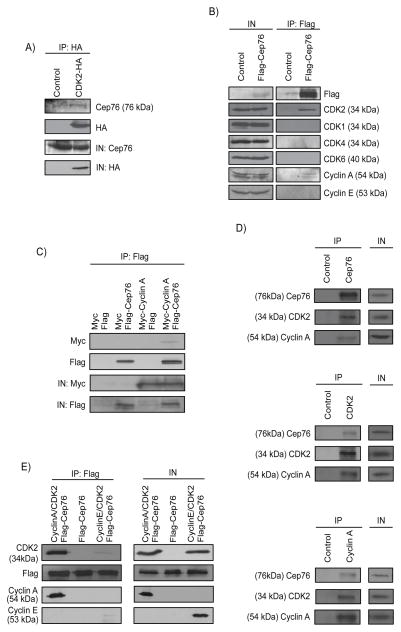
In an effort to identify potential upstream and downstream regulators of Cep76, we performed a proteomic screen for components of Cep76-containing complexes. Two centrosomal proteins, CP110 and Cep97, previously shown to interact with Cep76 were found (29), confirming the success of the screen. In addition, we identified CDK2, an S/G2 phase CDK (32, 33), as a novel Cep76-interacting partner from two independent experiments. To determine whether Cep76 and CDK2 interact in cells, we transfected a plasmid expressing HA-CDK2 in HEK293 cells, performed immunoprecipitations with anti-HA antibody, and showed that both HA-CDK2 and endogenous Cep76 were co-precipitated (Figure 1A). When Flag-Cep76 was expressed, endogenous CDK2 was likewise detected in Flag-Cep76 immunoprecipitates (Figure 1B). Flag-Cep76 specifically associated with CDK2 but not with other CDKs, including CDK1, CDK4 and CDK6, involved in cell cycle progression (Figure 1B). CDK2 is known to pair with cyclin E in late G1 and cyclin A in S/G2 phase (32), and we found that Flag-Cep76 interacts with cyclin A only (Figure 1B). The interaction between Cep76 and cyclin A was confirmed in other cell lines including U2OS and Saos-2 (Figure 1C and data not shown). Endogenous Cep76, cyclin A and CDK2 also interacted with each other (Figure 1D). Furthermore, in vitro binding experiments using purified proteins revealed that while Cep76 associates weakly with cyclin E/CDK2, it binds strongly to cyclin A/CDK2 (Figure 1E), in close agreement with our immunoprecipitation results (Figure 1B). Taken together, these findings suggest that Cep76 preferentially interacts with cyclin A/CDK2.
Figure 1.
Cep76 interacts with cyclin A/CDK2. (A) HA (control) or CDK2-HA was expressed in HEK293 cells and immunoprecipitated from lysate with an anti-HA antibody. The resulting immunoprecipitates were Western blotted with anti-HA or anti-Cep76 antibodies. IN, input. (B) Flag (control) or Flag-Cep76 were expressed in HEK293 cells and immunoprecipitated with anti-Flag beads. Immunoprecipitates were Western blotted with the indicated antibodies. IN, input. (C) Flag or Flag-Cep76 and myc or myc-cyclin A were co-expressed in U2OS cells and Flag proteins were immunoprecipitated. Flag-Cep76 and myc-cyclin A were detected after western blotting the resulting immunoprecipitates. IN, input. (D) Western blotting of endogenous Cep76, CDK2 and Cyclin A after immunoprecipitation of U2OS cell extracts with anti-Flag (control), anti-Cep76, anti-CDK2 or anti-Cyclin A antibodies. IN, input. (E) Purified Flag-tagged Cep76 protein was mixed with purified cyclin A/CDK2 (left lane), cyclin E/CDK2 (right lane) or an irrelevant protein (middle lane). Proteins were immunoprecipitated with anti-Flag beads, followed by Western blotting of Flag, CDK2, cyclin A and cyclin E. IN, input.
Cep76 is phosphorylated by cyclin A/CDK2 at a single site S83
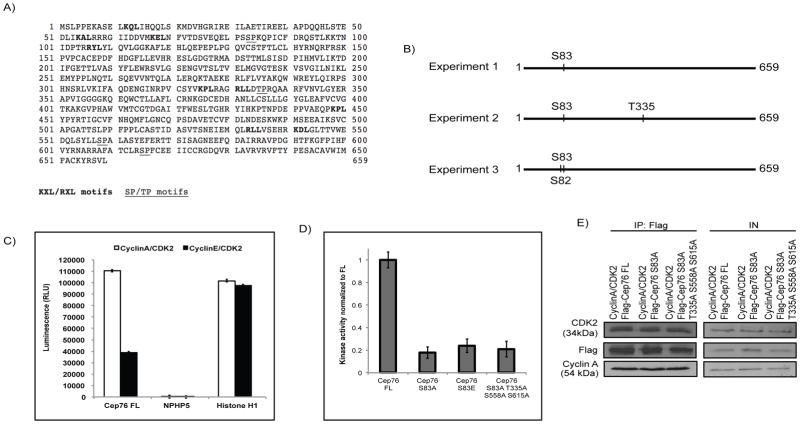
Cyclin/CDKs are known to phosphorylate substrates possessing the consensus sequence (S/T)-P (34–36). Sequence analysis of Cep76 revealed the presence of four putative consensus phosphorylation sites (Figure 2A). To determine whether Cep76 is a cyclin/CDK substrate, we first immunoprecipitated Flag-Cep76 from HEK293 cells, excised the band corresponding to the recombinant protein from a SDS-PAGE gel, and analyzed phosphorylation by mass spectrometry. In three independent experiments, we reproducibly detected phosphorylation of one single amino acid, serine 83 (S83) (Figure 2B), which overlaps with one of the four predicted CDK consensus sites of Cep76 (Figure 2A). Phosphorylation on S83 can be attenuated by the addition of a CDK inhibitor roscovitine (Figure S1), indicating that Cep76 is phosphorylated by CDK in vivo. Next, we performed in vitro kinase assays using purified cyclin/CDK and Cep76 and asked if the former directly phosphorylates the latter. In accordance with earlier results that Cep76 strongly interacts with cyclin A/CDK2, this protein is robustly phosphorylated by cyclin A/CDK2 (Figure 2C). Cyclin E/CDK2, on the other hand, exhibited negligible kinase activity on Cep76 (Figure 2C). In comparison, a known cyclin/CDK substrate histone H1 and a non-substrate NPHP5 are good and poor substrate for both kinases, respectively (Figure 2C). Moreover, we generated two Cep76 phosphorylation site mutants, one in which S83 was mutated to alanine (S83A) and the other in which all four putative CDK consensus sites were mutated to alanine (S83AT3335AS558AS615A; quadruple mutant), and tested their ability to undergo cyclin A/CDK2-dependent phosphorylation. We found that phosphorylation of S83A was drastically reduced compared to wild type protein, and the extent of diminution was comparable to the quadruple mutant (Figure 2D). The failure of cyclin A/CDK2 to phosphorylate mutant proteins is not a consequence of impaired binding, since wild type, S83A and the quadruple mutant bound equally well to the kinase (Figure 2E). S83, therefore, appears to be the only CDK consensus site that becomes phosphorylated by cyclin A/CDK2.
Figure 2.
Cyclin A/CDK2 phosphorylates Cep76 at S83. (A) Amino acid sequence of human Cep76. Putative cyclin-binding motifs (KXL/RXL) and CDK2 phosphorylation sites (SP/TP) are bold and underlined, respectively. Numbers denote amino acid positions. (B) Detection of phosphorylated residues by mass spectrometry. (C) In vitro kinase assays using purified Cep76 (Cep76 FL), NPHP5 or Histone H1 and purified cyclin A/CDK2 or cyclin E/CDK2. RLU, relative light units. Error bars represent standard errors. (D) In vitro kinase assays using Cep76 wild type (Cep76 FL), mutant S83A, S83E or S83AT335AS558AS615A protein and cyclin A/CDK2. (E) Purified Flag-tagged Cep76 protein (Cep76 FL) or mutant protein (Cep76 S83A or Cep76 S83AT335AS558AS615A) was mixed with purified cyclin A/CDK2. Proteins were immunoprecipitated with anti-Flag beads, followed by Western blotting of Flag, CDK2 and cyclin A. IN, input.
In addition to CDK consensus sites, a number of cyclin/CDK substrates possess KXL/RXL motifs known to facilitate interactions with cyclins, especially cyclin A, allowing for efficient substrate-kinase interaction and substrate phosphorylation (37–39). Cep76 contains nine putative KXL/RXL motifs (Figures 2A and S2A) and exhibits preferential binding to cyclin A (Figures 1B and 1E); thus, we sought to examine the potential role of these motifs in mediating interaction with cyclin A. We mutated each of the nine KXL/RXL motifs individually by replacing arginine and leucine with alanine (AXA mutants, Figure S2A), and found that these mutants are still able to interact with cyclin A (Figure S2B). A limited set of double and triple AXA mutants were tested and they could also bind cyclin A (data not shown). Furthermore, AXA mutant proteins appeared to be fully functional, since they localized to the centrosome and were able to suppress centriole amplification (Figures S2C and S3; see below for functional assays). From these results, we conclude that other determinants apart from the KXL/RXL motifs are likely responsible for the interaction between Cep76 and cyclin A.
Phosphorylation of Cep76 suppresses centriole amplification in S phase
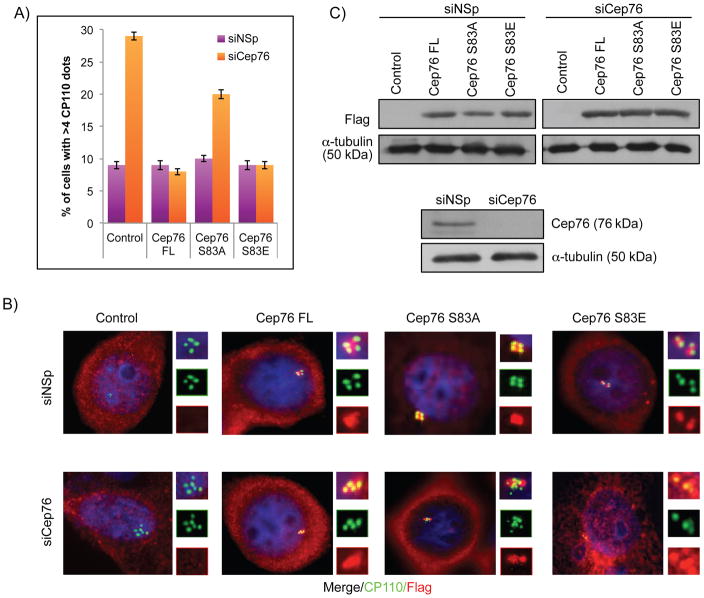
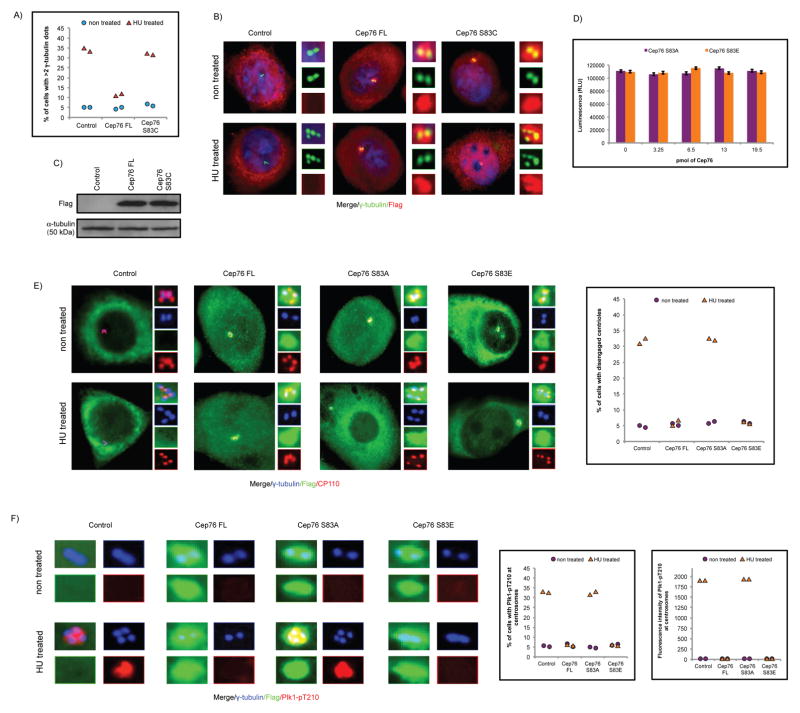
To assess the functional significance of Cep76 phosphorylation, we conducted rescue experiments in which we introduced recombinant wild type Cep76 or the S83A mutant in U2OS cells depleted of endogenous Cep76, using a siRNA oligo that targets the 3′UTR of Cep76 mRNA. A phosphomimetic mutant S83E was also tested in the same assay. A loss of Cep76 specifically leads to the production of supernumerary centrioles in S phase (29), and we indeed confirmed an earlier report that Cep76 depletion led to a 3–4 fold increase in the number of cells with >4 centriolar CP110 dots or >2 dots of C-Nap1 which stains the proximal ends of mother and daughter centrioles (Figures 3A–B and data not shown). Remarkably, this phenotype was rescued by wild type Cep76 and S83E, but not S83A mutant expression (Figures 3A–B, S4A and data not shown), despite comparable levels of protein expression (Figure 3C). Furthermore, both S83A and S83E mutants were properly localized to the centrosome (Figure 3B). These data suggest that the phosphomimetic mutant is functional and that phosphorylation at S83 is critical for Cep76 to suppress centriole amplification.
Figure 3.
Ectopic expression of Cep76 or a phosphomimetic mutant rescues centriole amplification induced by loss of endogenous Cep76. (A) U2OS cells transfected with control siRNA (siNSp) or siRNA targeting Cep76 3′UTR (siCep76) and plasmid expressing an irrelevant Flag-tagged protein (control), Flag-Cep76 wild type (Cep76 FL) or mutant (Cep76 S83A or Cep76 S83E). The percentages of transfected cells with >4 centrioles were determined by using CP110 as a marker. At least 75 transfected cells were scored per condition, and average of three independent experiments is shown. Error bars represent standard errors. (B) Cells were stained with DAPI (blue) and antibodies against Flag (red) and CP110 (green). (C) (Bottom) Western blotting of Cep76 in U2OS cells treated with control siRNA (siNSp) or siRNA targeting Cep76 3′UTR and (top) of Flag in U2OS cells transfected with the indicated siRNA oligos and plasmids expressing the indicated Flag-tagged proteins. α-tubulin was used as a loading control.
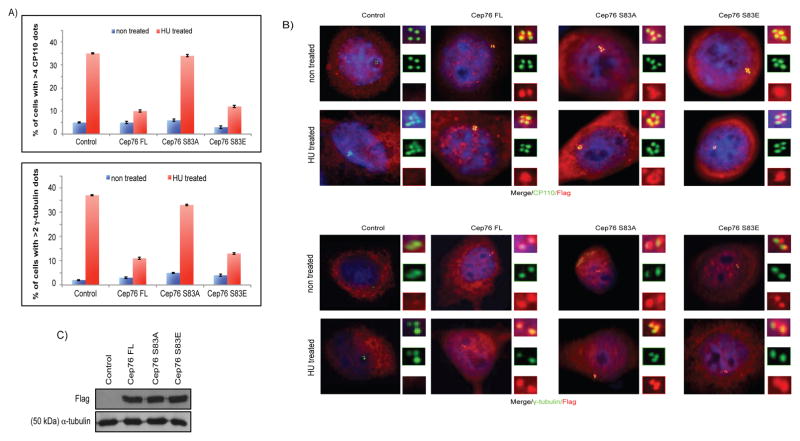
In addition, we tested the ability of wild type Cep76, S83A and S83E mutants to suppress HU-induced centriole amplification. The percentages of HU-treated control cells possessing >4 CP110 (centriolar) dots and >2 γ-tubulin (centrosomal) dots were ~30–35% and ~35–40%, respectively. These percentages dropped to ~5–15% and ~10–15% in HU-treated cells expressing wild type Cep76 or the S83E mutant (Figures 4A–B and S4B), indicating that either protein inhibits amplification. In contrast, expression of the S83A mutant in HU-arrested cells did not suppress amplification, since the percentages of cells with >4 CP110 dots and >2 γ-tubulin dots (~34% and 33%, respectively) were similar to control (Figures 4A–B). Finally, we verified that wild type Cep76, S83A and S83E proteins were expressed at similar levels (Figure 4C). Our rescue experiments and HU overduplication assay altogether revealed that phosphorylation of Cep76 at S83 prevents centriole amplification in S phase.
Figure 4.
Ectopic expression of Cep76 or a phosphomimetic mutant suppresses HU-induced centriole amplification. U2OS cells were transiently transfected with plasmid expressing an irrelevant Flag-tagged protein (control), Flag-Cep76 wild type (Cep76 FL) or mutant (Cep76 S83A or Cep76 S83E) and either left untreated or treated with HU. (A) The percentages of transfected cells with >4 CP110 or >2 γ-tubulin dots were determined. At least 75 transfected cells were scored per condition, and average of three independent experiments is shown. Error bars represent standard errors. (B) Cells were stained with DAPI (blue) and antibodies against Flag (red) and CP110 or γ-tubulin (green). (C) Western blotting of Flag in U2OS cells expressing the indicated Flag-tagged proteins. α-tubulin was used as a loading control.
Significance of Cep76 phosphorylation in human cancer
Since Cep76 is critical for protection against centriole amplification, a hallmark of cancer cells, a loss of protein function may cause a predisposition to cancer. A database search for somatic mutations in human cancer uncovered the Cep76 S83C mutation, identified in one of 307 cases of cervical squamous cell carcinoma and endocervical adenocarcinoma. It is currently unknown whether the mutation is present in one or both gene copies. The S83C mutation destroys a CDK2 phosphorylation site, and accordingly, we predicted that it would cripple protein function. Indeed, mutant S83C protein was unable to suppress centriole amplification induced by HU (Figures 5A–C) or depletion of Cep76 (Figures S5A–B).
Figure 5.
Characterization of a cancer-associated Cep76 S83C mutation and the mechanism that suppresses centriole amplification. (A) U2OS cells were transfected with plasmid expressing an irrelevant Flag-tagged protein (control), Flag-Cep76 wild type (Cep76 FL) or disease mutant (Cep76 S83C) and either left untreated or treated with HU. The percentages of transfected cells with >2 γ-tubulin dots were determined. (B) Cells were stained with DAPI (blue) and antibodies against Flag (red) and γ-tubulin (green). (C) Western blotting of Flag in U2OS cells expressing the indicated Flag-tagged proteins. α-tubulin was used as a loading control. (D) In vitro kinase assay with purified histone H1 as a substrate and cyclin A/CDK2 as a kinase. Increasing amounts of purified Cep76 were added to reactions. RLU, relative light units. Error bars represent standard errors. (E, F) U2OS cells were transfected with plasmid expressing an irrelevant Flag-tagged protein (control), Flag-Cep76 wild type (Cep76 FL) or mutant (Cep76 S83A or Cep76 S83E) and either left untreated or treated with HU. (E) (Left) Cells were stained with antibodies against Flag (green), CP110 (red) and γ-tubulin (blue). (Right) The percentages of transfected cells with disengaged centrioles were determined. (F) (Left) Cells were stained with antibodies against Flag (green), Plk1-pT210 (red) and γ-tubulin (blue). (Right) The percentages of transfected cells with Plk1-pT210 at centrosomes and the fluorescence intensities of centrosomal Plk1-pT210 were measured. In (A, E and F), at least 75 transfected cells were scored per condition, and two independent experiments were conducted.
Mechanism underlying suppression of centriole amplification by Cep76
We sought to address the mechanism by which Cep76 prevents centriole amplification. Because CDK2 promotes centriole amplification (16–19) and certain CDK2 substrates are known to regulate the kinase (40–42), we first asked whether the activity of cyclin A/CDK2 is inhibited by Cep76. Cyclin A/CDK2 activity was monitored using histone H1 as a substrate in the presence of the S83A or S83E mutant, neither of which is phosphorylated by the kinase (Figure 2D). Excessive amounts of S83A or S83E did not substantially modulate cyclin A/CDK2 kinase activity towards histone H1 (Figure 5D), suggesting that Cep76 is not an inhibitor of cyclin A/CDK2. Next, we examined if Cep76 regulates targeting of the kinase to the centrosome and hence, its ability to phosphorylate centrosomal substrates. Although CDK2 and cyclin A were both reported to be associated with centrosomes (43–46), depletion of Cep76 slightly influenced the centrosomal amount of cyclin A and CDK2 (Figures S6A–B). Thus, Cep76 has minimal effect on the abundance and activity of cyclin A/CDK2 at the centrosome.
Centriole disengagement is a prerequisite for a new round of duplication (47) and reduplication induced by HU (48). We therefore asked if Cep76 suppresses amplification by restraining centriole disengagement. Centriole disengagement was determined by examining the spacing of CP110/γ-tubulin dots and the ratio of CP110 to γ-tubulin dots. When the two centrioles are engaged, two closely spaced CP110 dots are associated with a single γ-tubulin dot and a 2:1 ratio of CP110:γ-tubulin is expected. Centriole disengagement causes centrioles to move further apart, and in this scenario, each CP110 dot is associated with a single γ-tubulin dot and a 1:1 ratio of CP110:γ-tubulin is expected. We noticed a 2:1 ratio, but rarely a 1:1 ratio, of CP110:γ-tubulin dots in HU-treated cells expressing wild type Cep76 or S83E (Figure 5E). In striking contrast, a 1:1 ratio of CP110:γ-tubulin dots was frequently observed in a subpopulation of HU-treated control cells or cells expressing S83A which have not undergone amplification (Figure 5E). In addition, we found that activated polo-like kinase 1 (Plk1) or Plk1-pT210, reportedly associated with disengaged centrioles in HU-treated control cells (48), is present at centrioles in HU-treated cells expressing S83A only (Figure 5F). From these results, we conclude that Cep76 inhibits centriole disengagement and consequently amplification by blocking Plk1 activation at the centrosome.
To further interrogate the relationship between Cep76 and Plk1, we found that centriole amplification due to loss of Cep76 can be prevented by the addition of a Plk1 inhibitor BI 2536 (Figure S7A). Moreover, while centriole amplification is suppressed by enforced expression of Cep76 in HU-treated cells, this phenotype can be overridden by co-expression of constitutively active Plk1 (Plk1-T210D) (Figure S7B). Taken together, these data suggest that Cep76 inhibits Plk1 at the centrosome.
Acetylation of Cep76 at K279 cannot suppress centriole amplification
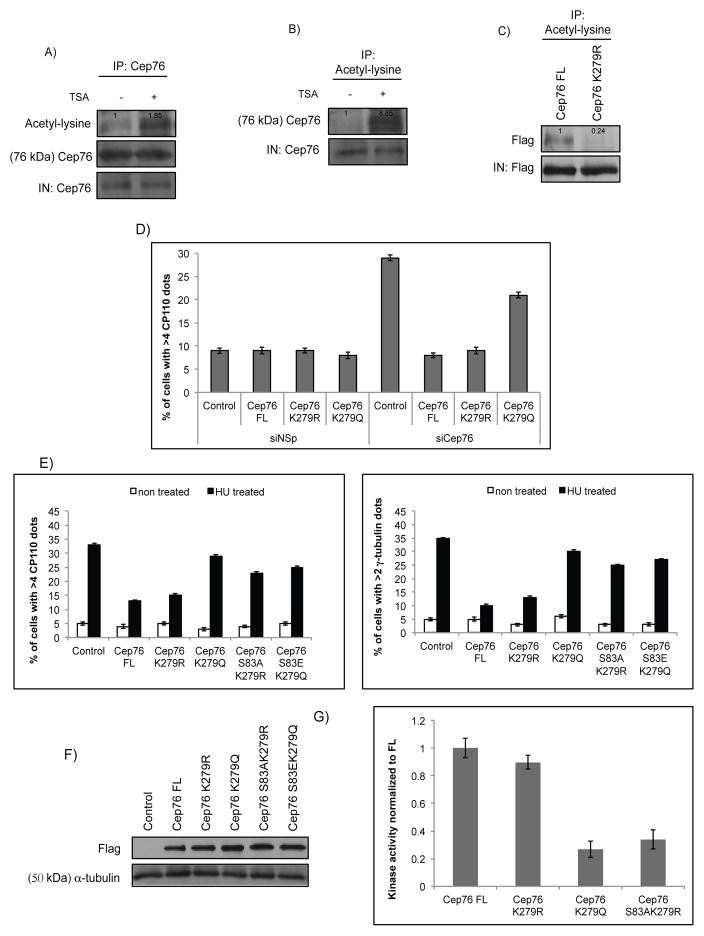
So far, we have demonstrated that phosphorylation at S83 is crucial for Cep76 function in S phase. Because protein function is often controlled by different types of posttranslational modification (PTM) (49), we first examined if Cep76 could be regulated by other modifications. A recent proteomic study identified lysine 279 (K279) as an acetylation site on Cep76 (50), although its biological relevance is unknown. To determine if Cep76 is acetylated, we immunoprecipitated Cep76 and showed that the endogenous protein is highly acetylated when cells were treated with a histone deacetylase inhibitor trichostatin A (TSA) (Figure 6A). Likewise, reciprocal immunoprecipitations with an anti-acetyl-lysine antibody revealed a marked increase in endogenous Cep76 acetylation upon TSA treatment (Figure 6B). Acetylation of recombinant Flag-Cep76 could also be detected (Figures S8A–B). We subsequently confirmed that acetylation takes place at K279, since an acetylation site mutant in which K279 was mutated to an arginine (K279R) exhibited considerably less acetylation compared to wild type protein (Figure 6C). Next, we asked if acetylation impacts Cep76 function. For this purpose, the ability of the acetylmimetic mutant K279Q, along with K279R, to suppress centriole amplification was investigated. We found that centriole overduplication induced by depletion of Cep76 or prolonged HU treatment was suppressed by K279R but not K279Q expression (Figures 6D–E). Both mutants localize to the centrosome and showed similar levels of protein expression compared to wild type protein (Figure 6F and data not shown). Thus, Cep76 can be acetylated at K279, and acetylation likely dampens the protein’s capacity to suppress centriole amplification.
Figure 6.
Enforced acetylation of Cep76 at K279 abrogates the protein’s ability to suppress centriole amplification. (A) U2OS cells were either left untreated or treated with TSA. Endogenous Cep76 was immunoprecipitated with an anti-Cep76 antibody, and the resulting immunoprecipitates were Western blotted with antibodies against acetyl-lysine and Cep76. IN, input. (B) Same as (A), except that endogenous proteins were immunoprecipitated with an anti-acetyl-lysine antibody. The resulting immunoprecipitates were Western blotted with an anti-Cep76 antibody. IN, input. Densitometric analyses were performed with ImageJ. (C) Flag-Cep76 wild type (Cep76 FL) or mutant (Cep76 K279R) protein expressed in U2OS cells were immunoprecipitated with an anti-acetyl-lysine antibody. The resulting immunoprecipitates were Western blotted with an anti-Flag antibody. IN, input. (D) U2OS cells transfected with control siRNA (siNSp) or siRNA targeting Cep76 3′UTR (siCep76) and plasmid expressing an irrelevant Flag-tagged protein (control), Flag-Cep76 wild type (Cep76 FL) or mutant (Cep76 K279R or Cep76 K279Q). The percentages of transfected cells with >4 CP110 dots were determined. (E) U2OS cells were transfected with plasmid expressing an irrelevant Flag-tagged protein (control), Flag-Cep76 wild type (Cep76 FL) or mutant (Cep76 K279R, K279Q, S83AK279R or S83EK279Q) and either left untreated or treated with HU. The percentages of transfected cells with (left) >4 CP110 or (right) >2 γ-tubulin dots were determined. (F) Western blotting of Flag in U2OS cells expressing the indicated Flag-tagged proteins. α-tubulin was used as a loading control. (G) In vitro kinase assays using purified Cep76 wide type (Cep76 FL), mutant K279R, K279Q or S83AK279R protein and cyclin A/CDK2. In (D and E), at least 75 transfected cells were scored per condition, and average of three independent experiments is shown. Error bars represent standard errors.
To gain molecular insight into why the K279R but not the K279Q mutant is proficient in suppressing centriole amplification and to explore the possibility of PTM crosstalk, we determined whether these mutant proteins could be phosphorylated by cyclin A/CDK2 in vitro. In sharp contrast to K279Q, the K279R mutant was robustly phosphorylated, reaching a level comparable to wild type protein (Figure 6G). Importantly, phosphorylation of K279R appears to occur on S83, since the S83AK279R double mutant exhibited markedly reduced phosphorylation (Figure 6G). The functionality of the K279R mutant is likewise dependent on S83, since the S83AK279R mutant could no longer suppress HU-induced amplification (Figures 6E–F). Remarkably, we found that the S83EK279Q double mutant, similar to the K279Q single mutant, was also unable to suppress centriole amplification induced by HU (Figures 6E–F), indicating that K279 acetylation alone is sufficient to override the effect of S83 phosphorylation and turn off Cep76. Considered together, our data suggest that acetylation at K279 negatively regulates Cep76 by limiting protein activity and S83 phosphorylation.
Temporal changes in Cep76 PTMs correlate with protein function
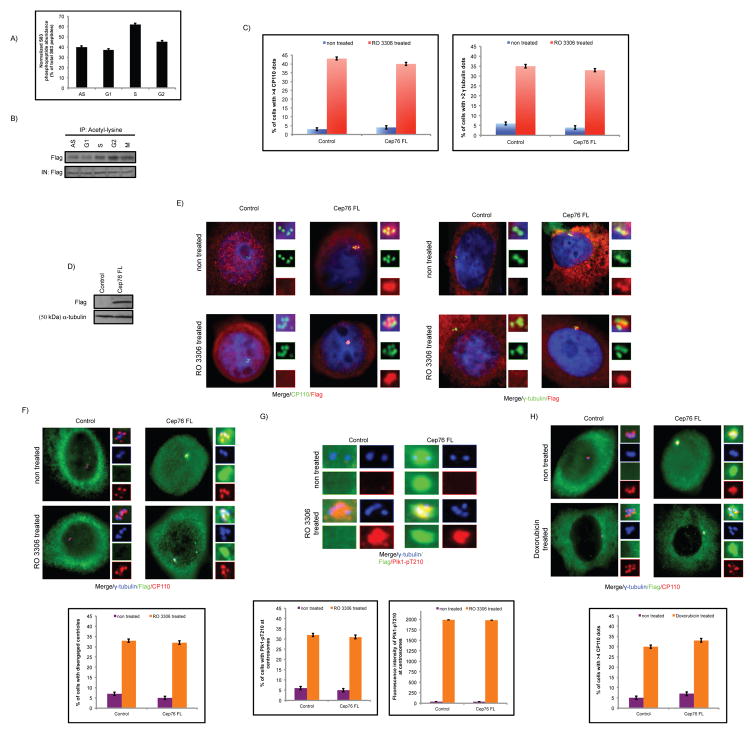
Next, we examined when phosphorylation and acetylation of Cep76 occur in the cell cycle. For analysis of protein phosphorylation, the abundance of S83 phosphopeptide was quantitatively determined by mass spectrometric analysis of Cep76 protein immuno-purified from synchronized cells. These experiments revealed that S83 phosphorylation increases from G1 to S but decreases thereafter (Figure 7A). Likewise, when we immunoprecipitated Flag-Cep76 from synchronized cells and Western blotted the resulting immunoprecipitates with anti-phosphoserine antibody, we found that phosphorylation is highly enriched in S phase (Figure S9). Thus, S83 phosphorylation appears to coincide with Cep76 function during S phase. For analysis of protein acetylation, we expressed Flag-Cep76 in synchronized cells and showed that the protein is more highly acetylated in G2 compared to other cell cycle phases (Figure 7B). Fittingly, Cep76 failed to inhibit centriole amplification induced by RO 3306 (Figures 7C–E), a small molecule inhibitor that triggers prolonged arrest in G2 phase and repeated rounds of centriole duplication (48, 51, 52). In these cells, both centriole disengagement and centrosomal Plk1-pT210 staining were observed (Figures 7F–G). As a further proof that acetylation of Cep76 in G2 phase leads to a loss of protein function, we found that Cep76 could not prevent centriole amplification induced by doxorubicin (DOX) (Figure 7H), a radiomimetic drug that induces G2 arrest (52). Our findings suggest that acetylation of Cep76 in G2 phase renders the protein unable to function.
Figure 7.
Temporal changes in Cep76 PTMs in the cell cycle correlate with protein function. (A) Mass spectrometric quantitation of S83 phosphopeptides from asynchronous (AS) and synchronized U2OS cells expressing Flag-Cep76. (B) Acetylated proteins from asynchronous (AS) and synchronized U2OS cells expressing Flag-tagged Cep76 were immunoprecipitated with an anti-acetyl-lysine antibody, and the resulting immunoprecipitates were Western blotted with an anti-Flag antibody. IN, input. (C) U2OS cells were transfected with plasmid expressing an irrelevant Flag-tagged protein (control) or Flag-Cep76 wild type (Cep76 FL) and either left untreated or treated with RO 3306. The percentages of transfected cells with (left) >4 CP110 or (right) >2 γ-tubulin dots were determined. (D) Western blotting of Flag in U2OS cells expressing the indicated Flag-tagged proteins. α-tubulin was used as a loading control. (E) Cells were stained with DAPI (blue) and antibodies against Flag (red) and CP110 or γ-tubulin (green). (F, G) U2OS cells were transfected with plasmid expressing an irrelevant Flag-tagged protein (control) or Flag-Cep76 wild type (Cep76 FL) and either left untreated or treated with RO 3306. (Top) Cells were stained with antibodies against Flag (green), γ-tubulin (blue) and (F) CP110 or (G) Plk1-pT210 (red). (Bottom) (F) The percentages of transfected cells with disengaged centrioles or (G) Plk1-pT210 at centrosomes were determined and (G) the fluorescence intensities of centrosomal Plk1-pT210 were quantitated. (H) U2OS cells were transfected with plasmid expressing an irrelevant Flag-tagged protein (control) or Flag-Cep76 wild type (Cep76 FL) and either left untreated or treated with DOX. (Top) Cells were stained with antibodies against Flag (green), CP110 (red) and γ-tubulin (blue). (Bottom) The percentages of transfected cells with >4 CP110 dots were determined. In (C, F, G and H), at least 75 transfected cells were scored per condition, and average of three independent experiments is shown. Error bars represent standard errors.
Discussion
Centriole duplication is a highly orchestrated process that occurs once per cell cycle. Deregulated duplication often leads to amplification, a hallmark of cancer cells (5). Because centriole amplification is believed to be an early event in tumorigenesis (5, 7–9), it is essential to identify the players that control the duplication process and examine their relevance to cancer development. CDK2 has emerged as a master regulator of duplication. However, much remains to be studied about how CDK2 controls this process, in part because a complete set of centrosomal CDK2 substrates have not been identified. Here, we uncovered a link between CDK2 and Cep76, a protein that limits centriole duplication to once per cell cycle (29), and demonstrated that the latter is a bona fide substrate of the former. To our knowledge, Cep76 is the first CDK2 substrate known to negatively regulate centriole amplification, since a small handful of substrates identified to date promote duplication and amplification in a positive manner.
Several centrosomal proteins, including Cep76, origin recognition complex 1 (ORC1), minichromosome maintenance 5 (MCM5), RNA-binding motif protein 14 (RBM14) and CDC14 are reported to suppress centriole amplification without affecting normal duplication (29, 53–57). Their mechanisms of action, however, have not been fully deciphered. While MCM5 and ORC1 associate with both cyclin E and cyclin A, only ORC1 inhibits cyclin/CDK2 activity. CDC14 dephosphorylates certain CDK substrates and may counteract CDK-mediated phosphorylation of centrosomal substrates (58, 59). RBM14 has no connection to CDK2; it suppresses the assembly of centriolar protein complexes that would lead to the formation of extra centrioles. Unlike the aforementioned proteins, Cep76 is a CDK2 substrate, and its phosphorylation by CDK2 is required to suppress centriole amplification. Although CDK2 can impinge on Cep76 function, the converse is not true, since Cep76 does not substantially affect the activity or localization of the kinase. Instead, our data revealed that Cep76 phosphorylation blocks Plk1 activity, thereby preventing premature centriole disengagement and amplification. Thus, the mechanism by which Cep76 suppresses centriole amplification appears to be unique.
Centriole disengagement is known to require the activities of Plk1 and the anaphase-promoting complex (APC) (52, 60, 61). Although either activity alone is sufficient to disengage centrioles, the extent to which they contribute to this intricate process remains unclear. We showed that Cep76 can prevent centriole disengagement and amplification in S phase but not in G2 phase. Thus, it is possible that HU-induced centriole disengagement is more highly dependent on Plk1 which can be easily overcome by Cep76 expression, whereas RO 3306- and DOX-induced disengagement is less dependent on Plk1. In support of this, we found that expression of a constitutively active Cep76 mutant S83EK279R, which retains the ability to block Plk1 activation at the centrosome, cannot suppress centriole amplification in RO 3306-treated cells (Figure S10). Another possibility is that Cep76 could specifically function in S phase and not in G2 phase. The two possibilities are not mutually exclusive.
Our data suggest that the function of Cep76 is modulated in a temporal manner during the cell cycle by two different types of PTM. Since Cep76 protein levels and cyclin/CDK2 activity peak in S phase and coincides with centriole duplication (29, 60, 61), we envision that this protein is phosphorylated and becomes activated by cyclin A/CDK2 in S phase to control the fidelity of duplication. Disruption of S83 phosphorylation, such as the phosphorylation site mutation S83A and the S83C mutation found in human cancer, compromises protein function. Once duplication is completed, Cep76 becomes acetylated at K279 in G2 phase. Acetylation of Cep76 serves to block S83 phosphorylation and to directly inhibit protein function possibly by relieving Plk1 from inhibition (Figures 6 and S10). It is uncertain how acetylation prevents protein phosphorylation. The K279Q mutant can still bind cyclin A/CDK2 (data not shown), suggesting that acetylation does not affect the interaction between the substrate and the kinase. Instead, acetylation of Cep76 may enhance its binding to a phosphatase which can promote dephosphorylation at S83, in a manner similar to the effect of acetylation on glycogen phosphorylase phosphorylation (62). As several phosphatases were identified in our proteomic screen for Cep76-interacting partners (data not shown), experiments are currently underway to determine if any of these could interact with Cep76.
In conclusion, we established that CDK2-mediated phosphorylation of a novel centrosomal substrate Cep76 is required to block centriole amplification in S phase. We also demonstrated the significance of a functional crosstalk between phosphorylation and acetylation. These results raise the intriguing possibility that deregulation of Cep76 might be linked to cancer risk.
Materials and Methods
Cell culture and plasmids
U2OS, Saos-2 and HEK293 cells (American Type Culture Collection) were grown in DMEM supplemented with 5% FBS at 37°C in a humidified 5% CO2 atmosphere. A plasmid expressing human recombinant Flag-Cep76 protein, pCDEF3-Flag-Cep76, was described previously (29). The following Cep76 mutations (S83C, S83A, S83E, S83AT335AS558AS615A, K279R, K279Q, S83AK279R, S83EK279Q, S83EK279R, AXA12-14, AXA54-56, AXA67-69, AXA106-108, AXA325-327, AXA331-333, AXA448-450, AXA533-535, AXA541-543) were introduced into full-length cDNA by employing a two-step PCR mutagenesis strategy and sub-cloned into pCDEF3-Flag. All constructs were verified by DNA sequencing. Myc-cyclin A and CDK2-HA plasmids were obtained from J. Archambault, and myc-Plk1-T210D was obtained from Addgene.
Antibodies
Antibodies used in this study included anti-HA (sc-7392), anti-myc (sc-40), anti-CDK2 (sc-163), anti-CDK1 (sc-954), anti-CDK4 (sc-4601), anti-CDK6 (sc-177), anti-cyclin A (sc-751), anti-cyclin E (sc-481), anti-C-Nap1 (sc-1358) (Santa Cruz); anti-α-tubulin (T5168), anti-Flag (F7425), anti-γ-tubulin (T3559) (Sigma); anti-acetyl lysine (#9441; Cell Signaling); anti-phosphoserine (05-1000X; Millipore); anti-Plk1-pT210 (ab39068; Abcam) and anti-CP110 (A301-344A) and anti-Cep76 (A302-326A) (Bethyl Laboratories)
Mass spectrometric identification of Cep76 associated proteins and phosphorylation sites
To identify Cep76-interacting proteins, Flag-Cep76 was expressed in HEK293 and U2OS cells and immunoprecipitated with anti-Flag agarose beads (Sigma) for 2 hours at 4°C. Bounded proteins were eluted with Flag peptide for 30 minutes, and the resultant eluates were precipitated with trichloroacetic acid and fractionated by SDS-PAGE. Six gel slices containing polypeptides were excised after Coomassie staining and subjected to proteolytic digestion mass spectrometric analysis. Analyses were performed at the Harvard Taplin Mass Spectrometry facility and the mass spectrometry core facility from IRCM by micro-capillary LC/MS/MS. For the identification of phosphorylation sites, cells were treated with 100 nM calyculin A for 30 min before lysis, and the lysis buffer contained 10 mM sodium fluoride, 0.2 mM sodium orthovanadate and 50mM β-glycerophosphate. A gel slice corresponding to Flag-Cep76 was excised. LC-MS peptide quantitation was performed by manual integration of the extracted ion chromatogram of each peptide using Qual Browser/Xcalibur version 2.2 (ThermoFisher Scientific).
Immunoprecipitation, immunoblotting, and immunofluorescence microscopy
Immunoprecipitation, immunoblotting and immunofluorescence were performed as described (63, 64). Densitometric analyses were performed with the ImageJ software (v1.43m, US National Institutes of Health) using the gel analysis and label peaks tools. ImageJ was also used to quantify the Plk1-pT210 signal at centrosomes.
Centrosome overduplication assay, cell cycle synchronization and FACS analysis
Cells were treated with 2mM HU (Sigma), 15 μM RO 3306 (Sigma) or 0.1 μM DOX (Santa Cruz) for 48 hours. The drug concentration and treatment time were optimized to minimize toxicity and to obtain the largest number of cells with overduplicated centrosomes. For cell cycle synchronization, G1, S, G2 and M phase cells were obtained by treating cells for 24 hours with 0.4 mM mimosine, 2 mM HU and release for 4 hours, 15 μM RO 3306, and 40ng/ml nocodazole, respectively. Cell cycle distribution was confirmed by fluorescence-activated cell sorting as described previously (64).
RNA interference
Transfection of siRNAs was performed using siImporter (Millipore) as per manufacturer’s instructions. The 21-nucleotide siRNA sequence for the non-specific control was 5′-AATTCTCCGAACGTGTCACGT-3′. The 21-nucleotide siRNA sequences for Cep76 were 5′-GAGCGTACAACAAGTATATTT-3′ and 5′-AATCACAATCTGGCTTGAATGTT-3′ (for targeting the 3′UTR region) (Dharmacon).
Production of recombinant proteins
Purified and active cyclin A/CDK2 and cyclin E/CDK2 were obtained from Millipore. Flag-Cep76 wild type or mutant, or Flag-NPHP5, was expressed in U2OS cells, lysed and incubated with anti-Flag beads for 2 hours. Beads were washed twice with ELB buffer containing 500mM NaCl and proteins were eluted with Flag peptide. A small sample was run on a gel and Coomassie stained to assess protein purity. We determined that our Flag-Cep76 or Flag-NPHP5 protein was at least 90% pure.
In vitro binding assay
1 μg of purified Flag-Cep76 protein was mixed with 1 μg of purified cyclin A/CDK2 or cyclin E/CDK2 at 4°C for 1h, followed by incubation with anti-Flag agarose beads at 4°C for 2h. After extensive washing with ELB buffer, bound proteins were analyzed by SDS-PAGE and immunoblotting.
In vitro kinase assay
The ADP-Glo™ kinase assay (Promega) was used for the in vitro kinase assay. The assay was carried out as per manufacturer’s instructions in a 96-well plate in kinase buffer (40 mM Tris pH 7.5, 20 mM MgCl2, 0.1% BSA) containing 100 ng of active cyclin A/CDK2 or cyclin E/CDK2, 1 μg of purified Flag-Cep76, histone H1 or Flag-NPHP5, and 1.25 μl of 1 mM ATP (Promega). The signal was quantitated using the GloMax 96 Microplate Luminometer (Promega). For the kinase inhibition assay, histone H1 was used as a substrate for cyclin A/CDK2 and increasing amounts of purified Flag-76 mutant protein S83A or S83E were then added to the reaction.
Statistical analysis
Experiments were performed three times. Graphs were generated in Microsoft Excel. Error bars represent standard error of the mean.
Supplementary Material
CDK inhibition attenuates S83 phosphorylation. Mass spectrometric quantitation of S83 phosphopeptides from Flag-Cep76 expressing U2OS cells grown asynchronously (AS) or synchronized with HU and released into S phase in the absence (S) or presence of roscovitine (S+roscovitine).
Cep76 AXA mutants bind cyclin A and are functional. (A) Schematic representation of AXA mutants of Cep76. (B) Flag (control), Flag-Cep76 wild type (FL) or AXA mutants were expressed in U2OS cells and immunoprecipitated from lysates with anti-Flag beads. The resulting immunoprecipitates were Western blotted with anti-Flag and cyclin A antibodies. IN, input. (C) The percentages of transfected U2OS cells with (top) >4 CP110 or (bottom) >2 γ-tubulin dots were determined. At least 75 transfected cells were scored per condition, and average of three independent experiments is shown. Error bars represent standard errors.
Cep76 AXA mutants suppress HU-induced centriole amplification. U2OS cells transfected with the indicated plasmids and either left untreated or treated with HU were stained with DAPI (blue) and antibodies against Flag (red) and γ-tubulin (green). Control denotes expression of an irrelevant Flag-tagged protein.
A ~1.5 fold overexpression of Flag-Cep76 is sufficient to rescue centriole amplification induced by loss of Cep76 or HU treatment. (A) U2OS cells were transfected with control siRNA (siNSp) or siRNA targeting Cep76 3′UTR and the indicated amounts of plasmid expressing Flag-Cep76. (Top) Western blotting of endogenous and recombinant Cep76 using an anti-Cep76 antibody. α-tubulin was used as a loading control. (Bottom) The percentages of transfected cells with >4 CP110 dots were determined. (B) U2OS cells expressing increasing amounts of Flag-Cep76 were either left untreated or treated with HU. (Top) Western blotting of endogenous and recombinant Cep76 using an anti-Cep76 antibody. α-tubulin was used as a loading control. (Bottom) The percentages of transfected cells with >4 CP110 dots were determined. In (A and B), at least 75 transfected cells were scored per condition, and average of three independent experiments is shown. Error bars represent standard errors. Densitometric analyses were performed with ImageJ.
A cancer-associated Cep76 S83C mutation is unable to suppress centriole amplification. U2OS cells transfected with control siRNA (siNSp) or siRNA targeting Cep76 3′UTR (siCep76) and plasmid expressing an irrelevant Flag-tagged protein (control), Flag-Cep76 wild type (Cep76 FL) or mutant (Cep76 S83C). (A) The percentages of transfected cells with >4 CP110 dots were determined. At least 75 transfected cells were scored per condition, and average of three independent experiments is shown. Error bars represent standard errors. (B) Cells were stained with DAPI (blue) and antibodies against Flag (red) and CP110 (green).
Cep76 slightly but not dramatically affects CDK2 and Cyclin A localization to the centrosome. (A) Fluorescence intensity of endogenous CDK2 at the centrosome was measured in U2OS cells transfected with control siRNA (siNSp) or Cep76 siRNA. (B) Fluorescence intensity of endogenous cyclin A at the centrosome was measured in U2OS cells transfected with control siRNA (siNSp) or Cep76 siRNA. In (A and B), at least 75 transfected cells were scored per condition, and average of three independent experiments is shown. Error bars represent standard errors.
Cep76 antagonizes Plk1 function to suppress centriole amplification. (A) (Left) U2OS cells were transfected with control siRNA (siNSp) or Cep76 siRNA and either left untreated or treated with BI 2536. The percentages of transfected cells with >4 centrin dots were determined. (Right) Cells were stained with DAPI (blue) and antibodies against Cep76 (red) and centrin (green). (B) U2OS cells transfected with plasmids expressing Flag and Myc, Flag-Cep76 and Myc, or Flag-Cep76 and constitutively active Plk1 (Myc-Plk1-T210D), were either left untreated or treated with HU. The percentages of transfected cells with >4 CP110 dots were determined. (A–B) At least 75 transfected cells were scored per condition, and average of three independent experiments is shown. Error bars represent standard errors.
Flag-Cep76 is acetylated in vivo. (A) U2OS cells expressing Flag-tagged Cep76 were either left untreated or treated with TSA. Recombinant proteins were immunoprecipitated with anti-Flag beads, and the resulting immunoprecipitates were Western blotted with anti-acetyl-lysine and Flag antibodies. IN, input. (B) Same as (A), except that proteins were immunoprecipitated with an anti-acetyl-lysine antibody. The resulting immunoprecipitates were Western blotted with an anti-Flag antibody. IN, input. Densitometric analyses were performed with ImageJ.
Cep76 is highly phosphorylated in S phase. Flag-Cep76 expressed in asynchronous (AS) or synchronized U2OS cells were immunoprecipitated with an anti-Flag antibody, and the resulting immunoprecipitates were Western blotted with anti-Flag and anti-phosphoserine antibodies. IN, input.
Acetylation of Cep76 relieves Plk1 from inhibition but is insufficient to prevent centriole amplification induced by RO 3306. U2OS cells were transfected with plasmid expressing an irrelevant Flag-tagged protein (control), Flag-Cep76 wild type (Cep76 FL) or mutant (S83E, S83EK279Q and S83EK279R) and either left untreated or treated with RO 3306. The percentages of transfected cells with (top) >4 CP110 dots or (bottom) Plk1-pT210 at centrosomes were determined. At least 75 transfected cells were scored per condition, and average of two independent experiments is shown. Error bars represent standard errors.
Acknowledgments
We thank all members of the Tsang laboratory for constructive advice, E. Gomes and J. Song for assistance with cloning, and Ross Tomaino and Denis Faubert for assistance with mass spectrometry. W.Y.T. is a Canadian Institutes of Health Research New Investigator and a Fonds de recherche Santé Junior 1 Research Scholar. This work was supported by the Canadian Institutes of Health Research (MOP-115033 to W.Y.T.) and the Natural Sciences and Engineering Research Council of Canada.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Bornens M. The centrosome in cells and organisms. Science. 2012;335(6067):422–6. doi: 10.1126/science.1209037. [DOI] [PubMed] [Google Scholar]
- 2.Nigg EA, Stearns T. The centrosome cycle: Centriole biogenesis, duplication and inherent asymmetries. Nat Cell Biol. 2011;13(10):1154–60. doi: 10.1038/ncb2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nigg EA, Raff JW. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139(4):663–78. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 4.Lingle WL, Lutz WH, Ingle JN, Maihle NJ, Salisbury JL. Centrosome hypertrophy in human breast tumors: implications for genomic stability and cell polarity. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(6):2950–5. doi: 10.1073/pnas.95.6.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan JY. A clinical overview of centrosome amplification in human cancers. International journal of biological sciences. 2011;7(8):1122–44. doi: 10.7150/ijbs.7.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pihan GA, Purohit A, Wallace J, Knecht H, Woda B, Quesenberry P, et al. Centrosome defects and genetic instability in malignant tumors. Cancer Res. 1998;58(17):3974–85. [PubMed] [Google Scholar]
- 7.Duensing S, Lee LY, Duensing A, Basile J, Piboonniyom S, Gonzalez S, et al. The human papillomavirus type 16 E6 and E7 oncoproteins cooperate to induce mitotic defects and genomic instability by uncoupling centrosome duplication from the cell division cycle. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(18):10002–7. doi: 10.1073/pnas.170093297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lingle WL, Barrett SL, Negron VC, D’Assoro AB, Boeneman K, Liu W, et al. Centrosome amplification drives chromosomal instability in breast tumor development. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(4):1978–83. doi: 10.1073/pnas.032479999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pihan GA, Wallace J, Zhou Y, Doxsey SJ. Centrosome abnormalities and chromosome instability occur together in pre-invasive carcinomas. Cancer Res. 2003;63(6):1398–404. [PubMed] [Google Scholar]
- 10.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460(7252):278–82. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Godinho SA, Picone R, Burute M, Dagher R, Su Y, Leung CT, et al. Oncogene-like induction of cellular invasion from centrosome amplification. Nature. 2014;510(7503):167–71. doi: 10.1038/nature13277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berthet C, Aleem E, Coppola V, Tessarollo L, Kaldis P. Cdk2 knockout mice are viable. Current biology: CB. 2003;13(20):1775–85. doi: 10.1016/j.cub.2003.09.024. [DOI] [PubMed] [Google Scholar]
- 13.Ortega S, Prieto I, Odajima J, Martin A, Dubus P, Sotillo R, et al. Cyclin-dependent kinase 2 is essential for meiosis but not for mitotic cell division in mice. Nature genetics. 2003;35(1):25–31. doi: 10.1038/ng1232. [DOI] [PubMed] [Google Scholar]
- 14.Santamaria D, Barriere C, Cerqueira A, Hunt S, Tardy C, Newton K, et al. Cdk1 is sufficient to drive the mammalian cell cycle. Nature. 2007;448(7155):811–5. doi: 10.1038/nature06046. [DOI] [PubMed] [Google Scholar]
- 15.Merrick KA, Wohlbold L, Zhang C, Allen JJ, Horiuchi D, Huskey NE, et al. Switching Cdk2 on or off with small molecules to reveal requirements in human cell proliferation. Mol Cell. 2011;42(5):624–36. doi: 10.1016/j.molcel.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hinchcliffe EH, Li C, Thompson EA, Maller JL, Sluder G. Requirement of Cdk2-cyclin E activity for repeated centrosome reproduction in Xenopus egg extracts. Science. 1999;283(5403):851–4. doi: 10.1126/science.283.5403.851. [DOI] [PubMed] [Google Scholar]
- 17.Lacey KR, Jackson PK, Stearns T. Cyclin-dependent kinase control of centrosome duplication. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(6):2817–22. doi: 10.1073/pnas.96.6.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meraldi P, Lukas J, Fry AM, Bartek J, Nigg EA. Centrosome duplication in mammalian somatic cells requires E2F and Cdk2-cyclin A. Nat Cell Biol. 1999;1(2):88–93. doi: 10.1038/10054. [DOI] [PubMed] [Google Scholar]
- 19.Matsumoto Y, Hayashi K, Nishida E. Cyclin-dependent kinase 2 (Cdk2) is required for centrosome duplication in mammalian cells. Current biology: CB. 1999;9(8):429–32. doi: 10.1016/s0960-9822(99)80191-2. [DOI] [PubMed] [Google Scholar]
- 20.Okuda M, Horn HF, Tarapore P, Tokuyama Y, Smulian AG, Chan PK, et al. Nucleophosmin/B23 is a target of CDK2/cyclin E in centrosome duplication. Cell. 2000;103(1):127–40. doi: 10.1016/s0092-8674(00)00093-3. [DOI] [PubMed] [Google Scholar]
- 21.Fisk HA, Winey M. The mouse Mps1p-like kinase regulates centrosome duplication. Cell. 2001;106(1):95–104. doi: 10.1016/s0092-8674(01)00411-1. [DOI] [PubMed] [Google Scholar]
- 22.Chen Z, Indjeian VB, McManus M, Wang L, Dynlacht BD. CP110, a cell cycle-dependent CDK substrate, regulates centrosome duplication in human cells. Developmental cell. 2002;3(3):339–50. doi: 10.1016/s1534-5807(02)00258-7. [DOI] [PubMed] [Google Scholar]
- 23.Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA. The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol. 2005;7(11):1140–6. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- 24.Korzeniewski N, Zheng L, Cuevas R, Parry J, Chatterjee P, Anderton B, et al. Cullin 1 functions as a centrosomal suppressor of centriole multiplication by regulating polo-like kinase 4 protein levels. Cancer Res. 2009;69(16):6668–75. doi: 10.1158/0008-5472.CAN-09-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duensing A, Liu Y, Tseng M, Malumbres M, Barbacid M, Duensing S. Cyclin-dependent kinase 2 is dispensable for normal centrosome duplication but required for oncogene-induced centrosome overduplication. Oncogene. 2006;25(20):2943–9. doi: 10.1038/sj.onc.1209310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adon AM, Zeng X, Harrison MK, Sannem S, Kiyokawa H, Kaldis P, et al. Cdk2 and Cdk4 regulate the centrosome cycle and are critical mediators of centrosome amplification in p53-null cells. Molecular and cellular biology. 2010;30(3):694–710. doi: 10.1128/MCB.00253-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Errico A, Deshmukh K, Tanaka Y, Pozniakovsky A, Hunt T. Identification of substrates for cyclin dependent kinases. Adv Enzyme Regul. 2010;50(1):375–99. doi: 10.1016/j.advenzreg.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 28.Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426(6966):570–4. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- 29.Tsang WY, Spektor A, Vijayakumar S, Bista BR, Li J, Sanchez I, et al. Cep76, a centrosomal protein that specifically restrains centriole reduplication. Developmental cell. 2009;16(5):649–60. doi: 10.1016/j.devcel.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balczon R, Bao L, Zimmer WE, Brown K, Zinkowski RP, Brinkley BR. Dissociation of centrosome replication events from cycles of DNA synthesis and mitotic division in hydroxyurea-arrested Chinese hamster ovary cells. The Journal of cell biology. 1995;130(1):105–15. doi: 10.1083/jcb.130.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosenblatt J, Gu Y, Morgan DO. Human cyclin-dependent kinase 2 is activated during the S and G2 phases of the cell cycle and associates with cyclin A. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(7):2824–8. doi: 10.1073/pnas.89.7.2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harper JW, Adams PD. Cyclin-dependent kinases. Chem Rev. 2001;101(8):2511–26. doi: 10.1021/cr0001030. [DOI] [PubMed] [Google Scholar]
- 33.Malumbres M. Cyclin-dependent kinases. Genome Biol. 2014;15(6):122. doi: 10.1186/gb4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nigg EA. The substrates of the cdc2 kinase. Semin Cell Biol. 1991;2(4):261–70. [PubMed] [Google Scholar]
- 35.Moreno S, Nurse P. Substrates for p34cdc2: in vivo veritas? Cell. 1990;61(4):549–51. doi: 10.1016/0092-8674(90)90463-o. [DOI] [PubMed] [Google Scholar]
- 36.Nigg EA. Cellular substrates of p34(cdc2) and its companion cyclin-dependent kinases. Trends in cell biology. 1993;3(9):296–301. doi: 10.1016/0962-8924(93)90011-o. [DOI] [PubMed] [Google Scholar]
- 37.Adams PD, Sellers WR, Sharma SK, Wu AD, Nalin CM, Kaelin WG., Jr Identification of a cyclin-cdk2 recognition motif present in substrates and p21-like cyclin-dependent kinase inhibitors. Molecular and cellular biology. 1996;16(12):6623–33. doi: 10.1128/mcb.16.12.6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen J, Saha P, Kornbluth S, Dynlacht BD, Dutta A. Cyclin-binding motifs are essential for the function of p21CIP1. Molecular and cellular biology. 1996;16(9):4673–82. doi: 10.1128/mcb.16.9.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu L, Harlow E, Dynlacht BD. p107 uses a p21CIP1-related domain to bind cyclin/cdk2 and regulate interactions with E2F. Genes & development. 1995;9(14):1740–52. doi: 10.1101/gad.9.14.1740. [DOI] [PubMed] [Google Scholar]
- 40.Sheaff RJ, Groudine M, Gordon M, Roberts JM, Clurman BE. Cyclin E-CDK2 is a regulator of p27Kip1. Genes & development. 1997;11(11):1464–78. doi: 10.1101/gad.11.11.1464. [DOI] [PubMed] [Google Scholar]
- 41.Tsang WY, Wang L, Chen Z, Sanchez I, Dynlacht BD. SCAPER, a novel cyclin A-interacting protein that regulates cell cycle progression. The Journal of cell biology. 2007;178(4):621–33. doi: 10.1083/jcb.200701166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsang WY, Dynlacht BD. Double identity of SCAPER: a substrate and regulator of cyclin A/Cdk2. Cell cycle. 2008;7(6):702–5. doi: 10.4161/cc.7.6.5611. [DOI] [PubMed] [Google Scholar]
- 43.Pascreau G, Eckerdt F, Churchill ME, Maller JL. Discovery of a distinct domain in cyclin A sufficient for centrosomal localization independently of Cdk binding. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(7):2932–7. doi: 10.1073/pnas.0914874107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pagano M, Pepperkok R, Verde F, Ansorge W, Draetta G. Cyclin A is required at two points in the human cell cycle. The EMBO journal. 1992;11(3):961–71. doi: 10.1002/j.1460-2075.1992.tb05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Boer L, Oakes V, Beamish H, Giles N, Stevens F, Somodevilla-Torres M, et al. Cyclin A/cdk2 coordinates centrosomal and nuclear mitotic events. Oncogene. 2008;27(31):4261–8. doi: 10.1038/onc.2008.74. [DOI] [PubMed] [Google Scholar]
- 46.Nishimura T, Takahashi M, Kim HS, Mukai H, Ono Y. Centrosome-targeting region of CG-NAP causes centrosome amplification by recruiting cyclin E-cdk2 complex. Genes Cells. 2005;10(1):75–86. doi: 10.1111/j.1365-2443.2005.00816.x. [DOI] [PubMed] [Google Scholar]
- 47.Tsou MF, Stearns T. Mechanism limiting centrosome duplication to once per cell cycle. Nature. 2006;442(7105):947–51. doi: 10.1038/nature04985. [DOI] [PubMed] [Google Scholar]
- 48.Loncarek J, Hergert P, Khodjakov A. Centriole reduplication during prolonged interphase requires procentriole maturation governed by Plk1. Current biology: CB. 2010;20(14):1277–82. doi: 10.1016/j.cub.2010.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang XJ, Seto E. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol Cell. 2008;31(4):449–61. doi: 10.1016/j.molcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327(5968):1000–4. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vassilev LT, Tovar C, Chen S, Knezevic D, Zhao X, Sun H, et al. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(28):10660–5. doi: 10.1073/pnas.0600447103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Douthwright S, Sluder G. Link between DNA damage and centriole disengagement/reduplication in untransformed human cells. J Cell Physiol. 2014;229(10):1427–36. doi: 10.1002/jcp.24579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu J, Cho HP, Rhee DB, Johnson DK, Dunlap J, Liu Y, et al. Cdc14B depletion leads to centriole amplification, and its overexpression prevents unscheduled centriole duplication. The Journal of cell biology. 2008;181(3):475–83. doi: 10.1083/jcb.200710127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hemerly AS, Prasanth SG, Siddiqui K, Stillman B. Orc1 controls centriole and centrosome copy number in human cells. Science. 2009;323(5915):789–93. doi: 10.1126/science.1166745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ferguson RL, Maller JL. Cyclin E-dependent localization of MCM5 regulates centrosome duplication. Journal of cell science. 2008;121(Pt 19):3224–32. doi: 10.1242/jcs.034702. [DOI] [PubMed] [Google Scholar]
- 56.Ferguson RL, Pascreau G, Maller JL. The cyclin A centrosomal localization sequence recruits MCM5 and Orc1 to regulate centrosome reduplication. Journal of cell science. 2010;123(Pt 16):2743–9. doi: 10.1242/jcs.073098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shiratsuchi G, Takaoka K, Ashikawa T, Hamada H, Kitagawa D. RBM14 prevents assembly of centriolar protein complexes and maintains mitotic spindle integrity. The EMBO journal. 2015;34(1):97–114. doi: 10.15252/embj.201488979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li L, Ljungman M, Dixon JE. The human Cdc14 phosphatases interact with and dephosphorylate the tumor suppressor protein p53. The Journal of biological chemistry. 2000;275(4):2410–4. doi: 10.1074/jbc.275.4.2410. [DOI] [PubMed] [Google Scholar]
- 59.Ovejero S, Ayala P, Bueno A, Sacristan MP. Human Cdc14A regulates Wee1 stability by counteracting CDK-mediated phosphorylation. Mol Biol Cell. 2012;23(23):4515–25. doi: 10.1091/mbc.E12-04-0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pagano M, Pepperkok R, Lukas J, Baldin V, Ansorge W, Bartek J, et al. Regulation of the cell cycle by the cdk2 protein kinase in cultured human fibroblasts. The Journal of cell biology. 1993;121(1):101–11. doi: 10.1083/jcb.121.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsai LH, Lees E, Faha B, Harlow E, Riabowol K. The cdk2 kinase is required for the G1-to-S transition in mammalian cells. Oncogene. 1993;8(6):1593–602. [PubMed] [Google Scholar]
- 62.Zhang T, Wang S, Lin Y, Xu W, Ye D, Xiong Y, et al. Acetylation negatively regulates glycogen phosphorylase by recruiting protein phosphatase 1. Cell Metab. 2012;15(1):75–87. doi: 10.1016/j.cmet.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barbelanne M, Hossain D, Chan DP, Peranen J, Tsang WY. Nephrocystin proteins NPHP5 and Cep290 regulate BBSome integrity, ciliary trafficking and cargo delivery. Hum Mol Genet. 2014 doi: 10.1093/hmg/ddu738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barbelanne M, Song J, Ahmadzai M, Tsang WY. Pathogenic NPHP5 mutations impair protein interaction with Cep290, a prerequisite for ciliogenesis. Hum Mol Genet. 2013;22(12):2482–94. doi: 10.1093/hmg/ddt100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
CDK inhibition attenuates S83 phosphorylation. Mass spectrometric quantitation of S83 phosphopeptides from Flag-Cep76 expressing U2OS cells grown asynchronously (AS) or synchronized with HU and released into S phase in the absence (S) or presence of roscovitine (S+roscovitine).
Cep76 AXA mutants bind cyclin A and are functional. (A) Schematic representation of AXA mutants of Cep76. (B) Flag (control), Flag-Cep76 wild type (FL) or AXA mutants were expressed in U2OS cells and immunoprecipitated from lysates with anti-Flag beads. The resulting immunoprecipitates were Western blotted with anti-Flag and cyclin A antibodies. IN, input. (C) The percentages of transfected U2OS cells with (top) >4 CP110 or (bottom) >2 γ-tubulin dots were determined. At least 75 transfected cells were scored per condition, and average of three independent experiments is shown. Error bars represent standard errors.
Cep76 AXA mutants suppress HU-induced centriole amplification. U2OS cells transfected with the indicated plasmids and either left untreated or treated with HU were stained with DAPI (blue) and antibodies against Flag (red) and γ-tubulin (green). Control denotes expression of an irrelevant Flag-tagged protein.
A ~1.5 fold overexpression of Flag-Cep76 is sufficient to rescue centriole amplification induced by loss of Cep76 or HU treatment. (A) U2OS cells were transfected with control siRNA (siNSp) or siRNA targeting Cep76 3′UTR and the indicated amounts of plasmid expressing Flag-Cep76. (Top) Western blotting of endogenous and recombinant Cep76 using an anti-Cep76 antibody. α-tubulin was used as a loading control. (Bottom) The percentages of transfected cells with >4 CP110 dots were determined. (B) U2OS cells expressing increasing amounts of Flag-Cep76 were either left untreated or treated with HU. (Top) Western blotting of endogenous and recombinant Cep76 using an anti-Cep76 antibody. α-tubulin was used as a loading control. (Bottom) The percentages of transfected cells with >4 CP110 dots were determined. In (A and B), at least 75 transfected cells were scored per condition, and average of three independent experiments is shown. Error bars represent standard errors. Densitometric analyses were performed with ImageJ.
A cancer-associated Cep76 S83C mutation is unable to suppress centriole amplification. U2OS cells transfected with control siRNA (siNSp) or siRNA targeting Cep76 3′UTR (siCep76) and plasmid expressing an irrelevant Flag-tagged protein (control), Flag-Cep76 wild type (Cep76 FL) or mutant (Cep76 S83C). (A) The percentages of transfected cells with >4 CP110 dots were determined. At least 75 transfected cells were scored per condition, and average of three independent experiments is shown. Error bars represent standard errors. (B) Cells were stained with DAPI (blue) and antibodies against Flag (red) and CP110 (green).
Cep76 slightly but not dramatically affects CDK2 and Cyclin A localization to the centrosome. (A) Fluorescence intensity of endogenous CDK2 at the centrosome was measured in U2OS cells transfected with control siRNA (siNSp) or Cep76 siRNA. (B) Fluorescence intensity of endogenous cyclin A at the centrosome was measured in U2OS cells transfected with control siRNA (siNSp) or Cep76 siRNA. In (A and B), at least 75 transfected cells were scored per condition, and average of three independent experiments is shown. Error bars represent standard errors.
Cep76 antagonizes Plk1 function to suppress centriole amplification. (A) (Left) U2OS cells were transfected with control siRNA (siNSp) or Cep76 siRNA and either left untreated or treated with BI 2536. The percentages of transfected cells with >4 centrin dots were determined. (Right) Cells were stained with DAPI (blue) and antibodies against Cep76 (red) and centrin (green). (B) U2OS cells transfected with plasmids expressing Flag and Myc, Flag-Cep76 and Myc, or Flag-Cep76 and constitutively active Plk1 (Myc-Plk1-T210D), were either left untreated or treated with HU. The percentages of transfected cells with >4 CP110 dots were determined. (A–B) At least 75 transfected cells were scored per condition, and average of three independent experiments is shown. Error bars represent standard errors.
Flag-Cep76 is acetylated in vivo. (A) U2OS cells expressing Flag-tagged Cep76 were either left untreated or treated with TSA. Recombinant proteins were immunoprecipitated with anti-Flag beads, and the resulting immunoprecipitates were Western blotted with anti-acetyl-lysine and Flag antibodies. IN, input. (B) Same as (A), except that proteins were immunoprecipitated with an anti-acetyl-lysine antibody. The resulting immunoprecipitates were Western blotted with an anti-Flag antibody. IN, input. Densitometric analyses were performed with ImageJ.
Cep76 is highly phosphorylated in S phase. Flag-Cep76 expressed in asynchronous (AS) or synchronized U2OS cells were immunoprecipitated with an anti-Flag antibody, and the resulting immunoprecipitates were Western blotted with anti-Flag and anti-phosphoserine antibodies. IN, input.
Acetylation of Cep76 relieves Plk1 from inhibition but is insufficient to prevent centriole amplification induced by RO 3306. U2OS cells were transfected with plasmid expressing an irrelevant Flag-tagged protein (control), Flag-Cep76 wild type (Cep76 FL) or mutant (S83E, S83EK279Q and S83EK279R) and either left untreated or treated with RO 3306. The percentages of transfected cells with (top) >4 CP110 dots or (bottom) Plk1-pT210 at centrosomes were determined. At least 75 transfected cells were scored per condition, and average of two independent experiments is shown. Error bars represent standard errors.