Abstract
Massively parallel reporter assays (MPRA) enable nucleotide-resolution dissection of transcriptional regulatory regions, such as enhancers, but only few regions at a time. Here, we present a combined experimental and computational approach, Sharpr-MPRA, that allows high-resolution analysis of thousands of regions simultaneously. Sharpr-MPRA combines dense tiling of overlapping MPRA constructs with a probabilistic graphical model to recognize functional regulatory nucleotides, and to distinguish activating and repressive nucleotides, using their inferred contribution to reporter gene expression. We use Sharpr-MPRA to test 4.6 million nucleotides spanning 15,000 putative regulatory regions tiled at 5-nucleotide resolution in two human cell types. Our results recover known cell type-specific regulatory motifs and evolutionarily-conserved nucleotides, and distinguish known activating and repressive motifs. Our results also show that endogenous chromatin state and DNA accessibility are both predictive of regulatory function in reporter assays, identify retroviral elements with activating roles, and uncover ‘attenuator’ motifs with repressive roles in active chromatin.
Introduction
Epigenome maps predict thousands of putative regulatory regions through their in vivo epigenomic signatures1–8 and are widely used for studying gene regulation9,10 and disease11,12. However, such maps present only indirect evidence of regulatory function, have often limited resolution, and do not distinguish activator from repressor elements5,6,8. DNA motif and sequence pattern analysis can complement epigenome maps5,6,8,13–15, but also provides only indirect evidence and only identifies sequences that match enriched patterns.
Episomal reporter assays1,2 and endogenous modulation11,16–18 provide two complementary approaches to characterize putative regulatory regions. Episomal reporters evaluate sequence function directly, independently of epigenetic effects, whereas endogenous perturbations capture endogenous context effects. Multiplexed endogenous or episomal assays have been used to dissect few regulatory regions at high resolution19–28 or many at low resolution23,28–37.
MPRAs19,30 synthesize DNA sequences on programmable microarrays and integrate them in reporter gene plasmids that are then transfected into cell types of interest. Barcodes placed in reporter gene 3'UTRs (to minimize their effect on pre-transcriptional control) provide a quantitative readout of gene expression levels. The limited number of array spots constrains the number of regions tested and the number of reporter constructs devoted to each region. Due to the short length of synthesized fragments (~145 nucleotides), MPRA requires accurate knowledge of putative regulatory region position and boundaries, which are not generally known.
Here, we overcome these limitations using dense tiling of MPRA constructs and computational analysis to infer activating and repressive nucleotides at high resolution across many regions. We term the combined approach Sharpr-MPRA, for Systematic High-resolution Activation and Repression Profiling with Reporter-tiling using MPRA, and the associated computational method SHARPR. We use Sharpr-MPRA to dissect over 15,000 putative regulatory regions from genome-wide epigenomic maps. We tile each 295-bp region at 5 nucleotide offsets using overlapping 145-nucleotide constructs. We make 4.6 million nucleotide inferences, each in two cell types, and distinguish activating and repressive regulatory functions without use of motifs or other sequence information. Inferred regulatory nucleotides are reproducible, high-resolution, cell type-specific, and supported by evolutionary conservation and regulatory motif evidence. Our strategy enables gene-regulatory insights, including activating motifs lacking well-established regulators, ‘dual-role’ motifs with both activating and repressive roles, strongly-activating repeat elements, and ‘attenuator’ motifs that play repressive roles in active chromatin states.
Results
Pilot design tiling 250 regions at 30-bp resolution
We first developed a low-resolution 'pilot' design, applied to 250 regions showing H3K27ac-marked enhancer chromatin states2 (200 in liver carcinoma HepG2 cells and 50 in leukemia K562 cells). We tiled 385-nucleotide regions at 30-nucleotide offsets using 145-nucleotide constructs, each unique sequence was tested using 24 barcodes (Fig. 1a, Supplementary Fig. 1). We centered our tiling on H3K27ac signal dips known to be indicative of nucleosome displacement due to transcription factor (TF) binding, and thus likely to overlap regulatory nucleotides. For HepG2, we selected 100 regions with strong dip scores, and 100 with wide-ranging dip scores (Supplementary Fig. 1, see Methods). We profiled each region in both K562 and HepG2, each in two replicates (Supplementary Data Files 1–2).
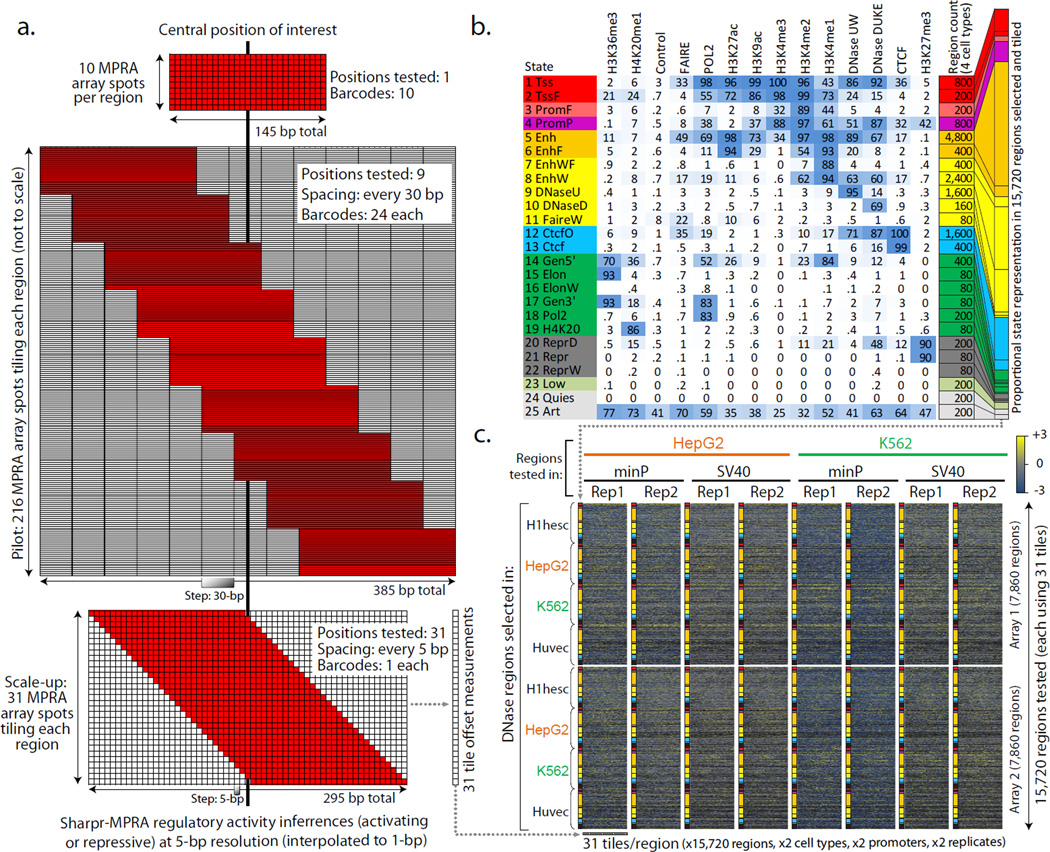
Figure 1. Experimental Design.
(a) Comparison of MPRA strategies for testing regulatory regions. Non-tiling approaches (top, e.g. Ref.30) use multiple barcodes for the same tested sequence. Our pilot design (middle) tests each region using 9 tile offsets, spaced at 30-bp increments, each tested using 24 barcodes (216 MPRA array spots per region). Our scaled-up design (bottom), tests each region using 31 tile offsets spaced at 5-bp increments, each tested using a single barcode per tile offset. The designs are to scale along the horizontal dimension. Only top and bottom are to scale in the vertical dimension. (b) The 25 chromatin states used in selecting regulatory regions for testing in the scale-up design38,39 (Supplementary Fig. 5). Heatmap indicates the emission probabilities (scaled between 0 and 100) for each epigenomic feature (columns) in each chromatin state (rows). Tested regions were restricted to DNase sites in one of four cell types, with the number of regions selected based on a stratified random sampling as indicated. (c) Overview of experiments using the scale-up design (see Supplementary Fig. 1 for the pilot design). We carried out 16 experiments, consisting of two sets of 7860 regions (row groups) across 25 chromatin states (colors), with 31 tiles per region (individual columns), each tested in both HepG2 (orange) and K562 (green), each using both a minimal promoter and an SV40 promoter, each in two replicates. Heatmap shows MPRA reporter gene expression measurements (blue=low, yellow=high, black=missing) (Supplementary Data 3).
Among the nine tile positions, inner tiles (centered on H3K27ac dips) showed the highest level and frequency of activity (Fig. 2a, Supplementary Fig. 2a–c), and the highest variability across regions (Supplementary Fig. 2d), indicative of regulatory nucleotides. Regions with stronger H3K27ac dip scores showed higher activity and the cell type-specificity of epigenomic signals matched the cell type-specificity of reporter expression (Fig. 2a, Supplementary Figs. 1–2), suggesting that endogenous epigenomic information is indicative of reporter assay activity.
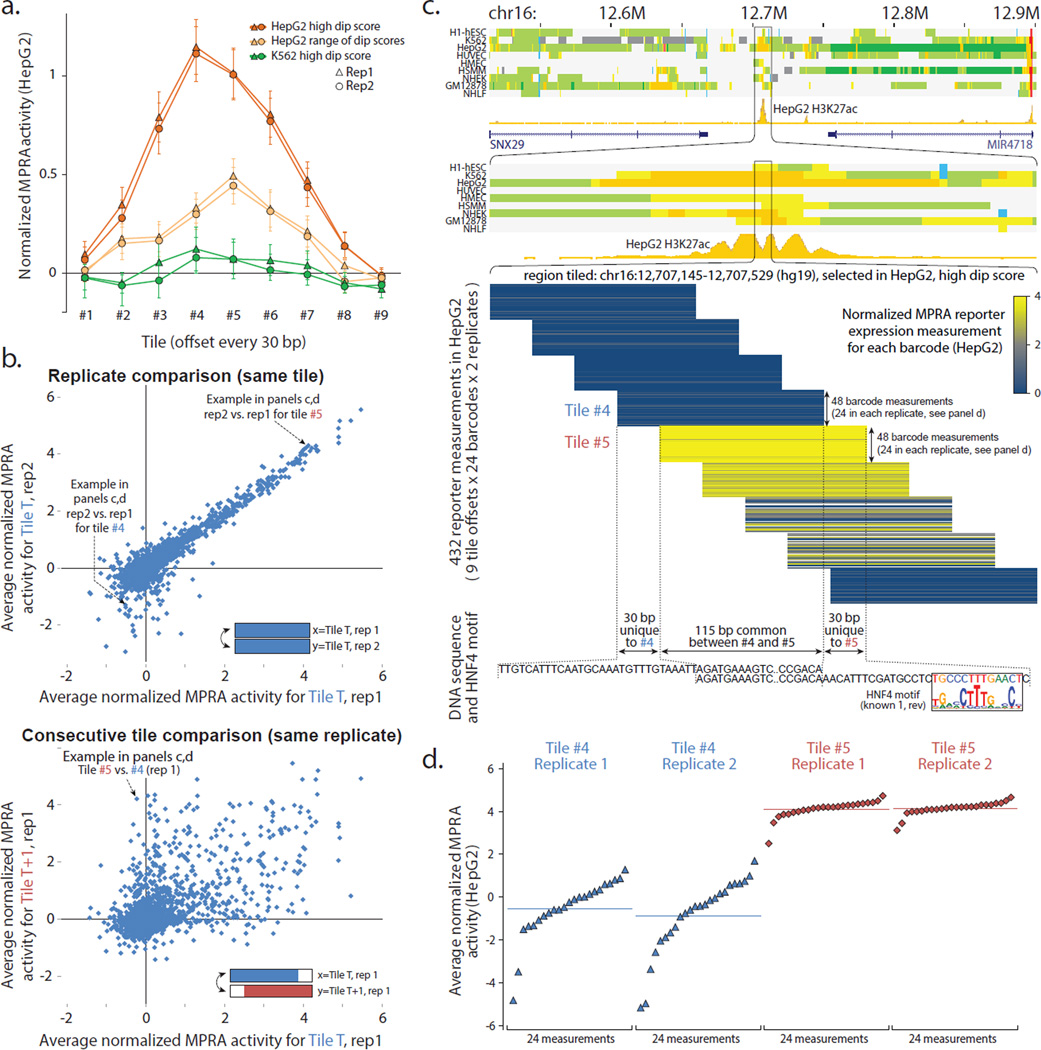
Figure 2. Tiling enhancer regions in pilot design reveals regulatory segments at 30-bp resolution.
(a) Effect of tile offset and H3K27ac dip score on reporter expression. Average HepG2 reporter expression (y-axis) at each of nine offsets (x-axis) for three sets of regions (color): HepG2 candidate enhancers2 with the highest H3K27ac dip scores (orange), candidate enhancers with a range of dip scores (light orange), and regions that are not predicted enhancers in HepG2 but are predicted enhancers with a high dip score in K562 (green). Error bar height is one standard error. (b) Consecutive tiles can differ in reporter expression. Top: Comparison of median reporter activity between biological replicates in HepG2. Only first eight tile offsets shown. Bottom: Comparison of consecutive tiles T (x-axis) and T+1 (y-axis) for the same biological replicate (rep1). (c) Top: Chromatin state annotations2 in nine cell types and H3K27ac signal track in HepG2 over the 500kb and 10kb surrounding the tiled 385-bp region centered in the H3K27ac dip. Middle: Expanded view of tile reporter measurements (yellow blue color) across all nine tiles, 24 barcodes, and two replicates. Bottom: Tiles #4 and #5 share 115-bp in common (abbreviated), and have 30-bp unique to #4 or #5 (shown), indicating the potential presence of activating elements in the sequence unique to #5 and/or repressive elements in the sequence unique to #4. Indeed, the 30-bp segment unique to #5 contains a candidate binding site for HNF4, a known activator of liver-related functions. (d) Expanded view of expression activity measurements for consecutive tiles #4 and #5 for all individual barcodes (points), sorted by their reporter expression levels. For Replicate 1 of Tile #4, 1 of 24 barcode measurements failed. The y-axis coordinates correspond to the ones shown in panel b. See Supplementary Figs. 2–4 for additional results on the pilot design.
Biological replicates of the same tile showed reproducible median reporter activity (Pearson’s correlation 0.92 across replicates for HepG2), but tiles offset by 30 bp sometimes differed substantially (Pearson's correlation 0.57 within the same HepG2 experiment) (Fig. 2b), with greater distances showing greater differences (Supplementary Fig. 3a). K562 showed similar results (0.66 vs. 0.34), with the lower correlation likely reflecting reduced transfection rates for K562 using our experimental protocols, as previously30. To gain insights into the sequences driving these differences, we focused on 637 pairs of neighboring positions in HepG2 and 142 pairs in K562 that showed significant differences in activity (false discovery rate 5%) (Supplementary Table 1, Supplementary Fig. 3b,c), and searched for motif differences in sequence segments distinguishing consecutive tiles (Fig. 2c,d, Supplementary Fig. 4a, see Methods). Segments with increased HepG2 activity were enriched for known liver-function motifs, including HNF4 and HNF1, whereas segments with increased K562 activity were enriched for known hematopoietic motifs including GATA (Supplementary Fig. 4b). This confirms that a tiling approach can reveal nucleotides important for cell type-specific regulatory function.
Scale-up design tiling 15,720 regions at 5-bp resolution
We next scaled-up our MPRA tiling design, increasing resolution, throughput, coverage, and chromatin state diversity. To achieve these goals, we made several modifications:
Resolution: To achieve increased resolution, we positioned reporter sequences at 5-bp increments instead of 30 bp (Fig. 1a).
Throughput: To increase throughput, we used a single reporter construct per position instead of 24 (Fig. 1a), achieving robustness by exploiting the many reporter constructs overlapping each position (15 on average; 25 for the central 105 nucleotides). We also tested smaller 295 bp regions instead of 385 bp, focusing on the most informative positions based on our pilot results (Fig. 2a, Supplementary Fig. 2).
Coverage: To increase coverage, we used two 244K arrays for DNA synthesis (instead of a single 55K array), targeting 15,720 regions for tiling, each profiled in both HepG2 and K562, using both a minimal TATA promoter (minP) and a strong SV40 promoter (SV40P), each in two replicates (Fig. 1c, columns), resulting in a total of 3.9 million measurements (Supplementary Data File 3).
Chromatin state diversity: To enable analyses across diverse chromatin states of a 25-state ChromHMM model38,39(Supplementary Fig. 5), we centered regions on double-cut DNase I peaks instead of H3K27ac dips. To ensure that all states are represented, we used a tiered random sampling approach, which also favored enhancers and other DNase-enriched states (Fig. 1b, counts). To include both active and inactive regulatory regions in the cell types profiled, we selected DNase I regions from HepG2, K562, and two additional cell types, Human umbilical vein endothelial cells (Huvec) and embryonic stem cells (H1hesc) (Fig. 1c, rows).
These design choices, and the SHARPR computational inference method we describe next, allowed us to infer regulatory activity at ~5-nucleotide resolution across 4.6 million nucleotides spanning over 15,000 regions, a 6-fold increase in resolution and 60-fold increase in coverage compared to our pilot design.
Computation inference of activating and repressive nucleotides
We developed a computational method, SHARPR, that scores the relative activating or repressive potential of each 5-bp interval within tiled regions and interpolates these values to make predictions for individual nucleotides (Fig. 3a,b). The inclusion and exclusion of 5-bp nucleotide intervals between consecutive tiles is akin to perturbation experiments, allowing inferences at substantially higher resolution (5-bp) than the original reporter constructs (145-bp) (Fig. 3a). Intuitively, activating intervals (e.g. containing activator motifs) should increase the reporter expression for tiles overlapping them, whereas repressive intervals (e.g. containing repressor motifs) should decrease reporter expression of overlapping tiles, as we show in our pilot experiments (e.g. Fig. 2c). Thus, modeling the relative activity of overlapping tiles should enable inference of activating and repressive nucleotide positions at high resolution (Fig. 3b).
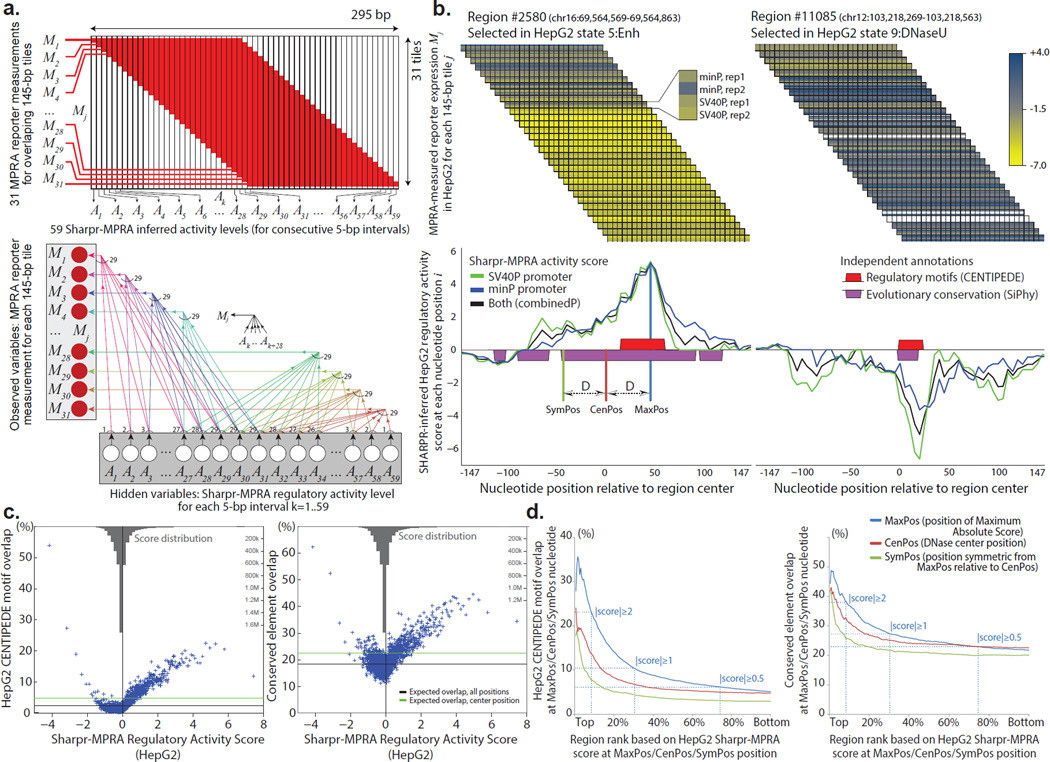
Figure 3. Scale-up design permits dissection of regulatory elements at high-resolution.
(a) Modeling scheme and probabilistic graphical model for the scale-up design. Variables M1,…,M31 represent the observed values of the reporter measurements for the 31 tiles (each 145-bp long), and variables A1,…,A59 represent the unobserved regulatory activity level of each 5-bp interval of the 295-bp covered, which is then normalized into the Sharpr-MPRA regulatory activity score. Bottom: Probabilistic graphical model used for high-resolution inference of activating and repressive intervals, with arrows Ak→Mj illustrating the dependencies between variables when tile Mj overlaps interval Ak, and the direction of information flow in the generative model. Conditional inference allows us to use the observed reporter measurements M1,…,M31 for each tile j in order to infer the unobserved activity levels A1,…,A59 for each 5-bp interval k, which we interpolate to each nucleotide position i, under the modeling assumptions specified in Methods. (b) Observed reporter expression measurements for 145-bp segments (top) and inferred regulatory activity for 5-bp segments, interpolated to individual nucleotides (bottom) for two 295-bp regulatory regions in HepG2 cells. Top: At each offset, the four rows correspond to four measurements of the same tile, using the minP and SV40P promoter, each in two replicates. Measurements for each tile are shown spanning all nucleotide positions the tile covers. White rows represent missing data for a promoter/replicate combination for a given 145-bp tile. Bottom: resulting inference of regulatory activity at each nucleotide i using all four measurements (black), only the two SV40P measurements (green), or only the two minP measurements (blue). Predicted positions of highest activating (positive scores) or repressive (negative scores) activity capture CENTIPEDE8 predicted binding sites (red boxes) and conserved elements identified by the SiPhy-PI method42,65 (purple boxes), even though such information was not used in our inferences. These examples are shown (and boxed) on page 2 of Supplementary Data Files 6a and 6b respectively. (c) Higher activating or repressive Sharpr-MPRA regulation activity score in HepG2 cells (x-axis) results in higher overlap with transcription factor binding sites predicted by CENTIPEDE in HepG2 cells8 (y-axis, left panel), and higher overlap with conserved elements identified by SiPhy-PI42,65 (y-axis, right panel). Each point represents the average of 927 nucleotide positions in each of 5,000 quantiles. Horizontal black line shows the expected overlap averaged across all 295 nucleotide positions of each region, and the green line shows the expected overlap fraction at the center nucleotide position (a stringent control). Reversed grey barplot at the top of each panel shows the density (histogram) of the distribution of Sharpr-MPRA combinedP scores in HepG2 cells. (d) Sharpr-MPRA inferences capture regulatory nucleotides at high resolution. Cumulative overlap (y-axis) with CENTIPEDE predicted transcription factor binding sites in HepG2 (left plot) and evolutionarily conserved elements (right plot) is higher for maximum-absolutely-score nucleotide positions (MaxPos, blue), than for the stringent control of DNase center nucleotide positions (CenPos, red), or for symmetric nucleotide positions (SymPos, green), indicating this is not a positional bias. Each set is ranked from highest (left) to lowest (right) absolute Sharpr-MPRA score in MaxPos/CenPos/SymPos nucleotides (x-axis) in HepG2 cells (see Supplementary Fig. 21 for K562 cells, and for individual promoter types). Dotted lines mark thresholds at absolute score ≥2, ≥1, and ≥0.5. MaxPos, CenPos, and SymPos nucleotide positions are illustrated in the example of panel b.
We constructed a probabilistic graphical model (Fig. 3a, bottom) relating the unobserved regulatory activity of each 5-bp interval (hidden variables A1–A59) to the 145-bp reporter measurements (observed variables M1–M31). We modeled Mj using a normal distribution with mean the average of overlapping Ak and variance the empirical variance of all measurements in the experiment (see Methods). We modeled Ak using a normal distribution with mean the average of all measurements in the experiment and variance a free parameter (smaller values resulting in more smoothed inferences). We inferred the ‘most likely’ values for the regulatory activity variables based on observed reporter measurements and their prior distributions, and standardized these using a z-score to combine results from multiple replicates and inferences (Supplementary Fig. 6). We used a low-variance prior and a high-variance prior and combined these results (see Methods). We carried out piecewise linear interpolation from the 5-bp activity estimates to infer the regulatory activity of each nucleotide in the tiled regions. The computational portion of Sharpr-MPRA is implemented in a software package for which we provide a public software release (http://www.biolchem.ucla.edu/labs/ernst/SHARPR/; Supplementary Code).
We used Sharpr-MPRA to make activating or repressive regulatory activity inferences for 4.6 million nucleotides, each in two cell types, each using two promoter types, each using two replicates. We inferred nucleotide activity for the minP and SV40P promoters both individually (combining two replicate experiments for each), and for their combination (combinedP, using four experiments jointly), resulting in three activity tracks for each cell type (Supplementary Data File 4, Supplementary Figs. S6b, S7a). We also assigned a minP, SV40P and combinedP score to each region (Supplementary Fig. 7b,c), using the signed (activating or repressive) score of the maximum absolute score position (MaxPos) (labeled in Fig. 3b, bottom). We provide visualizations showing all minP, SV40P, and combinedP inferences for HepG2 and K562 in 31,440 figures on our supplementary website, and for several selected subsets (Supplementary Data Files 5–8).
Reproducibility of Sharpr-MPRA results
We evaluated the reproducibility of our results in multiple ways. We first evaluated the agreement between minP and SV40P inferences for each nucleotide position across regions. The central 101 positions showed on average 0.75 Pearson's correlation for HepG2 and 0.66 for K562, which decreased towards outer positions (Supplementary Fig. 8a), attributable to fewer tiles overlapping outer positions and stronger regulatory activity closer to DNase peak centers. Individual nucleotides maintained similar scores between promoter types (Supplementary Fig. 9a), with 83% of scores ≥1.5 in one showing scores ≥1 in the other for HepG2 (71% for K562), and 74% of scores < −0.5 in one showing scores < 0 in the other for HepG2 (73% for K562).
Individual replicates of each promoter type showed strong agreement for increasing absolute scores (Supplementary Fig. 10a) for both cell types and both promoter types (e.g. Pearson correlation 0.7 for |score|≥2, 0.9 for |score|≥3 for minP). Between promoter types, regions with |score|≥2 showed 0.8 correlation on average in HepG2 (0.7 in K562), compared to <0.1 expected by chance (Supplementary Fig. 10b). The MaxPos nucleotide position showed significantly greater concordance between replicates and between promoter types than expected by chance (Supplementary Fig. 11a–c), with the distance decreasing with increasing absolute score. Between minP replicates in HepG2, MaxPos nucleotides were on average within 28-bp, 11-bp and 5-bp respectively for |score|≥2, 4, and 6 for HepG2 (26-bp, 13-bp, and 7-bp respectively for K562), compared to ~60-bp expected by chance when sampling MaxPos positions.
We repeated these comparisons across different sets of barcodes using DNase regions that were independently selected in multiple cell types, and thus tested using multiple barcode sets (Supplementary Data File 5). Forty-four regions overlapped completely and 212 overlapped partially, providing a resource for quantifying potential barcode effects. The position-specific correlation in Sharpr-MPRA scores between SV40P and minP showed a negligible change in HepG2 and a modest reduction in K562 when using the same vs. a different set of barcodes (0.72 vs. 0.71 and 0.69 vs. 0.60 respectively for the central 101 nucleotide positions on average, Supplementary Fig. 8b), indicating a modest barcode effect relative to other sources of biological or technical variability. Individual nucleotide scores showed strong agreement across barcode sets for both partial-overlap and exact-overlap regions (Supplementary Fig. 9c,d), with 90% of combinedP scores ≥1.5 in one showing combinedP score ≥1 in the other for HepG2 (80% in K562) across all 256 multiply-tiled regions. The 44 complete-overlap regions showed high score correlation across barcode sets for regions with high absolute scores (0.96 for HepG2 and 0.77 for K562 for |score|≥2) (Supplementary Fig. 10c). The position of maximum score was within 16-bp between barcode sets for HepG2 (21-bp for K562) for |score|≥2 (Supplementary Fig. 11d). A search for k-mer effects within the barcodes (see Methods) found no influence on inferred activity in HepG2 compared to random expectation, a small effect for shorter k-mers in K562, and no noticeable effect for longer k-mers in either cell type (Supplementary Fig. 12).
Sharpr-MPRA recovers known motifs and conserved regions
To establish whether our inferred regulatory nucleotides are biologically relevant, we compared them to predictions of TF binding sites that were not used to make our inferences, including DNase-based5,6,8,40,41 and DNase-independent13 predictions of TF-bound nucleotides, and both motif-based8,13,41 and motif independent5,6,40 predictions of regulatory nucleotides. For example, CENTIPEDE8 motif annotations showed strong agreement for both activating and repressive scores at the nucleotide level (Fig. 3c, left, Supplementary Fig. 13), at the region level (Fig. 3b, Supplementary Data Files 6–8), and for specific regulators (Supplementary Fig. 14), and CENTIPEDE nucleotides showed reproducible scores (Supplementary Fig. 9b,e,f). Each regulatory annotation set tested showed better agreement with our inferences than with stringent controls (Supplementary Figs. 15–16).
We also compared our results to evolutionarily-conserved elements across 29 mammals42, and found enrichment for both activating and repressive nucleotides (Fig. 3c right, Supplementary Fig. 17), also supporting that Sharpr-MPRA captures functional nucleotides within tiled regions.
K-mer-based DeltaSVM14 predictions of nucleotides expected to have regulatory effects when mutated also agreed with our activating nucleotides (Supplementary Fig. 18, see Methods). However, DeltaSVM predictions failed to capture our repressive nucleotides, even though the latter agreed with both conserved nucleotides and CENTIPEDE motifs (Fig. 3c).
Sharpr-MPRA captures regulatory nucleotides at high resolution
We next evaluated whether our inferences capture high-resolution information within tiled regions. We confirmed that CENTIPEDE motif and conserved element enrichments also held when focusing on maximum-absolute-score positions (MaxPos) nucleotides (Supplementary Fig. 19), a substantial fraction of which disagreed with DNase center locations (66% of MaxPos nucleotides lie outside the 41 central nucleotides, 44% outside the 81 central nucleotides, Supplementary Fig. 20).
We compared the motif and conserved element enrichments of MaxPos nucleotides to those of DNase center position (CenPos) nucleotides, and their symmetric position (SymPos) nucleotides (equidistant from DNase peak centers) (illustrated in Fig. 3b). The CenPos comparison is particularly stringent, given the higher activity expected at central nucleotides and the better power to detect activity with more overlapping constructs. Despite these biases favoring central positions, MaxPos nucleotides were substantially more enriched than CenPos nucleotides for both CENTIPEDE motifs and conserved elements, in both HepG2 and K562, for all ranks of high activation or repression (Fig. 3d, Supplementary Fig. 21). By contrast, their symmetrical (SymPos) nucleotides showed substantially lower enrichments than MaxPos nucleotides for all metrics, indicating that MaxPos enrichments do not stem from distance biases.
We evaluated the effect of tiling density on functional element recovery and replicate correlation, using spaced subsets of our reporter constructs. Higher density led to stronger CENTIPEDE motif and conserved element enrichments (Supplementary Fig. 22) and to higher correlation between replicates (Supplementary Fig. 23). Saturation was not reached at the 5-bp level used here, suggesting that smaller offsets might increase discovery power, at the cost of more constructs per region and thus fewer tiled regions.
Cell type-specific activating and repressing motifs
We used our Sharpr-MPRA results to study a compendium of 1934 known and predicted regulatory motifs13. For each motif, we computed an average motif score (across all central motif positions), and separately an 'activating' and a 'repressive' enrichment score (based on its enrichment in nucleotides with score≥1 and ≤−1 respectively) for each cell type (Supplementary Table 2) (see Methods). As minP and SV40P average motif scores were highly concordant (0.98 correlation in HepG2; 0.95 in K562, Supplementary Fig. 24), we focused on combinedP scores.
Most motifs showed similar average scores between the two cell types (Fig. 4a). For example, motifs for the ETS and NRF1 regulators showed among the strongest activating average scores in both HepG2 and K562, and known repressor REST motif43 showed the most repressive average score in both. Unexpectedly, the most activating average score in both cell types was found for variants of the TCTCGCGAGA palindrome, which was present in our compendium13 based on its de novo discovery in ChIP-seq experiments for diverse regulators (including NR3C1, BRCA1, ETS, CHD2, and ZBTB3344). This motif lacks a well-established regulator in vivo45, despite support for its importance from strong evolutionary conservation46, high nearby gene expression8, and other experimental and bioinformatics evidence44,47,48.
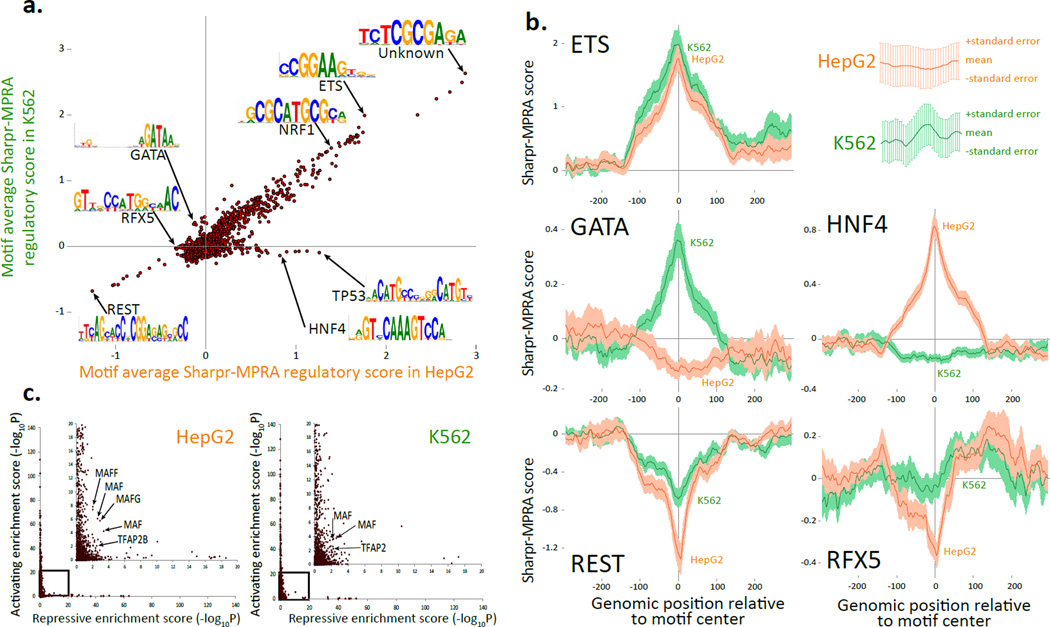
Figure 4. Comparison of Sharpr-MPRA with motif annotations.
(a) Comparison of average Sharpr-MPRA score for regulatory motifs from a previously assembled compendium13 (points) in HepG2 (x-axis) vs. K562 (y-axis), averaged at the center position of all instances for each motif. Arrows highlight motif examples mentioned in the text (Supplementary Table 2). Only motifs with more than 10 instances are shown. (b) Aggregation plots of the regulation score (y-axis) at increasing varying genomic positions relative to the motif center (x-axis) for K562 (green) and HepG2 (orange) for all motif instances, predicted independently of cell type in Ref 13, for ETS_known9, GATA_known14, REST_known2, HNF4_known18, and RFX5_known6 regulatory motifs. Error bar height is one standard error. (c) Activating enrichment score (y-axis) and repressive enrichment score (x-axis) for the regulatory motif compendium13 (points) in HepG2 (left) and K562 (right), based on the statistical significance (−log10P) for the enrichment of the center motif position for nucleotides with Sharpr-MPRA scores ≤−1 (repressive) or ≥1 (activating), using a one-sided binomial test. Inset expands boxed region, and does not cover any points, as no motif was enriched beyond −log10P=20 for both activating and repressing positions. Arrows highlight members of MAF and AP-2 motif families discussed in text. Similar plots using top 5% activating and repressive nucleotides in Supplementary Fig. 29.
A subset of motifs showed significant differences (using a paired t-test) in scores between HepG2 and K562, (Supplementary Fig. 25a). Significantly-different activating motifs include HNF4, RXRA, PPARA, HNF1A, HNF1B, and FOXA in HepG2, consistent with known liver-related roles, and GATA, SP1, and KLF in K562, consistent with K562 roles49 (Supplementary Fig. 26a). Significantly-different repressive motifs included multiple RFX motifs in HepG2 (Supplementary Fig. 26b), consistent with previous evidence for one enhancer50.
Cell type specificity was also reflected in the position-specific aggregated distribution of activity scores surrounding all instances of these motifs (Fig. 4b). Activating motifs (e.g. ETS, GATA, HNF4), showed a positive peak surrounding their instances, and repressor motifs (e.g. REST, RFX) showed a negative peak, in each case matching the expected cell types. These patterns were stronger when motifs occurred in more central positions (Supplementary Fig. 27), as expected given the higher reporter coverage and absolute activity of central positions.
The motif compendium resulted in more activating than repressive motif scores, both based on average scores (Fig. 4a), and based on activating vs. repressive enrichment scores (511 vs. 117 in HepG2; 474 vs. 79 in K562, respectively, at an uncorrected p-value of 0.01) (Fig 4c, Supplementary Fig. 25b, Supplementary Table 2). The higher number of activating motifs also held for average scores of all 7-mer sequences (Supplementary Fig. 28), indicating it is not an ascertainment bias in the compendium used.
A small number of 'dual-role' motifs showed signatures of both activating and repressive function in the same cell type (14 in HepG2 and 15 in K562, of which six were in common). These included several motifs for MAF proteins, consistent with previous reports of both activation and repression51,52 and different motifs of the AP-2 family. Alternative cutoffs (top 5% activating and repressive nucleotides) also resulted in very few dual-role motifs (6 in HepG2 and 9 in K562) (Supplementary Fig. 29).
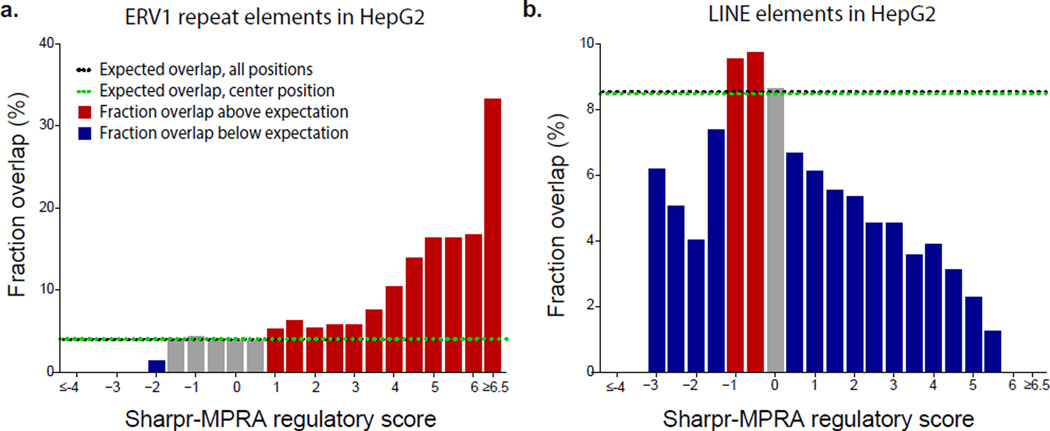
Enrichment of ERV1 repeats in activating nucleotides
The most strongly-activating nucleotides in HepG2 showed substantial enrichment for long terminal repeat (LTR) elements (Fig. 5, Supplementary Fig. 30), consistent with previous reports of their ability to drive gene expression53. This helps explain why conserved elements were less enriched in the most extreme activating scores (Fig. 3c), as no LTRs overlapped conserved elements with the most extreme activating scores.
Figure 5. Regulatory activity of ERV1 and LINE repeats.
For nucleotides of varying Sharpr-MPRA regulatory activity score in HepG2 cells (x-axis bins) the fraction that overlaps with annotated repeat elements (y-axis) shows a strong ERV1 repeat enrichment at the most activating nucleotides (panel a) and a depletion for LINE repeats at the most activating and most repressive nucleotides (panel b). Bins formed by assigning each base to the nearest 0.5 value based on its regulatory score. Extreme bins contain extreme values as indicated. Horizontal lines denote expected overlap based on center position (CenPos, green), and all positions (black). Enrichments and depletions for K562 and for additional repeats shown in Supplementary Fig. 30.
Among LTRs, Endogenous retroviral sequence 1 (ERV1) repeats showed the strongest enrichment in HepG2 activating nucleotides (Fig. 5a), overlapping 33% of the 820 nucleotides with the highest regulatory scores (≥6.5 bin), vs. only 4% expected on average (8-fold enrichment). Regulatory roles were previously hypothesized for ERV1 repeats, based on TF binding and RNAi evidence54–56, and our results indicate they can function autonomously and lead to strong episomal expression.
By contrast, LINE elements were strongly depleted in both activating and repressive nucleotides in HepG2 and K562 (Fig. 5b, Supplementary Fig. 30b), indicating repeat-specific regulatory functions. Moreover, ERV1 and other LTR enrichments were weaker in K562 than HepG2 activating nucleotides (Supplementary Fig. 30a,c), indicating cell type-specific repeat functions.
Endogenous chromatin state is predictive of reporter activity
We analyzed the relationship between regulatory activity scores and endogenous chromatin state, enabled by inclusion of all chromatin states (defined in Supplementary Fig. 5) and both DNase and non-DNase sites in each cell type (by including DNase regions only active in other cell types).
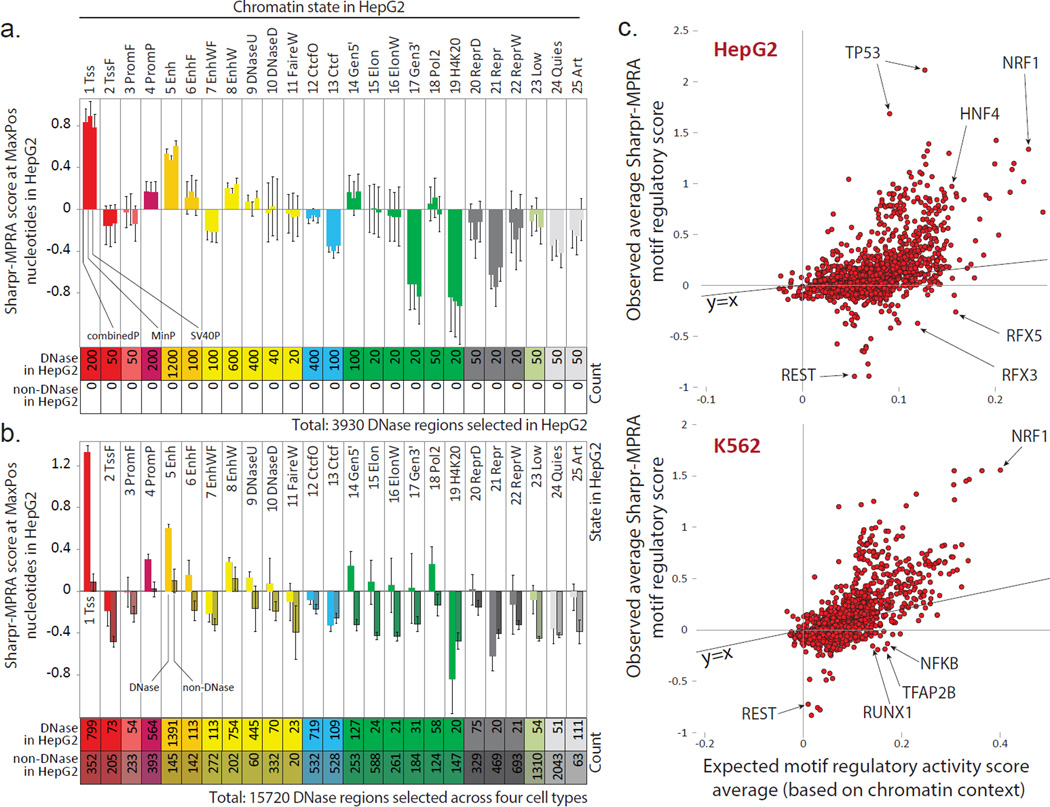
Among DNase regions, endogenous chromatin state was predictive of regulatory function in reporter assays, (quantified for each region by its MaxPos Sharpr-MPRA score) (Fig. 6a, Supplementary Fig. 31a). Regions in active promoter or H3K27ac-marked enhancer chromatin states showed higher Sharpr-MPRA activating scores, regions in weak enhancer states showed intermediate activating scores, and regions in Polycomb-associated states showed repressive Sharpr-MPRA scores, consistent with previous work2,57. Conversely, among genomic locations in the same chromatin state, the DNA accessibility of the region in its endogenous context was predictive of Sharpr-MPRA reporter activity (Fig. 6b, Supplementary Fig. 31b), consistent with previous work in enhancer regions30,31. Together, these results indicate that the endogenous epigenomic signatures of DNA accessibility and chromatin state each capture unique information about regulatory function, and that sequence elements within these regions can have corresponding activating or repressive regulatory functions outside their endogenous context.
Figure 6. Endogenous chromatin state is predictive of reporter activity.
(a,b) Average HepG2 Sharpr-MPRA regulatory score (y-axis) and standard error (vertical error bars) for each chromatin state (columns) for (a) all 3930 DNase regions selected in HepG2 and (b) all 15,720 regions selected in all four cell types, evaluated at nucleotide positions of maximum absolute activity (MaxPos). In panel a, each group of consecutive bars shows the combinedP, minP, and SV40P results. All 3930 regions correspond to DNase sites in HepG2, as they were selected in HepG2. In panel b, the combinedP score is shown separately for regions corresponding to DNase sites in HepG2 (light shading) and non-DNase sites in HepG2 (darker shading). Some DNase sites selected in other cell types were also DNase in HepG2, leading to an increased DNase count compared to panel a. All non-DNase regions in HepG2 were DNase regions in the cell type in which they were selected. The chromatin state of the center position is shown. K562 plots in Supplementary Fig. 31. (c) For all motifs (Ref 13) (circles) in HepG2-selected regions (top) and K562-selected regions (bottom), relationship between their average combinedP Sharpr-MPRA score in the corresponding cell type (y-axis) and their expected score based on the chromatin states in which the motif occurs (x-axis), quantified as the median of randomized motif occurrences that preserve positional and chromatin state distributions (see Methods). Only motifs with 20 or more evaluated instances in selected regions are shown. Diagonal line shows y=x line. Randomization 95th percentile confidence intervals shown in Supplementary Fig. 39.
The fraction of regions that show activating and repressive MaxPos scores varied greatly between chromatin states and DNase classes (Supplementary Fig. 32). In HepG2, activating regions with scores ≥1 included 36% of HepG2-selected DNase regions in an active promoter state (Tss) and 29% in an H3K27ac-marked enhancer state (Enh) (Supplementary Fig. 32a), compared to only 6% for non-DNase sites in the quiescent states (Quies) (Supplementary Fig. 32d) (41% and 32% vs. 5% respectively in K562). Repressive regions with MaxPos scores ≤−1 in HepG2 included 29% of HepG2-selected DNase regions in Polycomb repressed states ReprD and Repr (21% for K562) (Supplementary Fig. 32a), compared to only 6% of all DNase regions in the active promoter state (10% in K562) (Supplementary Fig. 32c). These comparisons allow us to estimate false discovery rates for both activating regions (e.g. 6% for HepG2, 5% for K562) and for repressive regions (e.g. 6% and 10% respectively) relative to their respective backgrounds. These estimates are likely conservative, as all regions tested were in DNase sites in at least one cell type, and thus more likely to contain activating or repressive elements than random background nucleotides.
Beyond these thresholds, the full distribution of MaxPos scores across chromatin states and DNA accessibility (Supplementary Fig. 33), confirmed consistently higher activation scores for active promoter and H3K27ac-marked enhancer states across a broad range of ranked positions. For non-DNase regions, the strongest repressive scores were found for chromatin state DNaseD (associated with single-cut DNase58 and lack of double-cut DNase7), indicating that it contains repressive elements, consistent with a previously-hypothesized repressive role38 (Supplementary Fig. 33c).
The role of H3K27ac as a signature of active enhancer regions is well-established2,4,30,59 and agrees with our results here, but was recently questioned in an isolated study31 using a similar reporter assay (CRE-seq), which suggested that H3K27ac-marked regions show weaker reporter activity. That study31 used a 7-state segmentation39 that merged ChromHMM60 and Segway61 results and tested smaller segments (130-bp) without a tiling approach and without anchoring on DNase sites, making its results dependent on the positioning of tested segments, and specifically whether DNase sites or their flanking elements were captured. Mapping their tested segments31 on the 25-state ChromHMM annotations considered here, we find that the H3K27ac-marked enhancers selected by the study preferentially lay outside DNase regions, and the non-H3K27ac enhancers selected preferentially lay in DNase regions. Correcting for this bias by analyzing DNase and non-DNase regions separately, we find that H3K27ac enhancers have increased CRE-Seq activity compared with non-H3K27ac enhancers (Supplementary Fig. 34), which is fully consistent with our results and the previous literature.
Similar to other studies30,31,62,63, many predicted enhancer and promoter regions did not have reporter activity (Supplementary Fig. 7b). These regions showed distinct levels and patterns of endogenous TF binding and DNA accessibility, including: less frequent endogenous TF binding within the tested regions; more frequent endogenous TF binding in the surrounding 2-kb; and proximity to other DNase sites (Supplementary Figs. 35–38). We interpret these findings to indicate that their endogenous activation may arise at least in part from TF binding in nearby regions, consistent with their lower reporter gene expression when tested in isolation.
A Subset of Repressive Motifs in Active Chromatin
For each motif, we analyzed the relationship between its observed average regulatory score and the average regulatory score that would be expected based on the chromatin states where the motif occurs, quantified as the median of randomized motif occurrences that preserve positional and chromatin state distributions (see Methods). Overall, the observed average score of a motif was correlated with its expected average score (0.54 in HepG2 and 0.68 in K562 for motifs with ≥20 instances, Fig. 6c, Supplementary Fig. 39, Supplementary Table 2). For example, NRF1 showed both a high average regulation score and a high expected score in both HepG2 and K562, indicating that it acts as an activator in active chromatin states.
Several motifs showed only moderate expected scores but very strong activating or repressive motif scores, suggesting they maintain their functions regardless of their genomic context. In HepG2 for example, TP53 showed only a moderate expected score, but the highest score among all evaluated motifs, consistent with its proposed role as a pioneer factor64. At the other end of the spectrum, REST showed only an intermediate expected score, but had the most repressive motif score, indicating strong repressor functions irrespective of context.
A small number of motifs showed repressive motif scores, but among the highest expected scores, suggesting they play ‘attenuator’ repressive roles in activating chromatin contexts. RFX family motifs had among the most repressive motif scores in HepG2, but among the most activating expected scores (Fig. 6c). Consistent with 'attenuator' roles, they showed repressive (negative) activity in our positional activity analysis, but they were flanked by activating (positive) scores (Fig. 4b). Moreover, in our pilot analysis in HepG2, RFX family motifs were discovered as enriched in segments inferred to be repressive, but found in active enhancer regions (Supplementary Fig. 4). Indeed, a repressive role was experimentally confirmed in an enhancer of the Cdx2 gene for a single RFX1 motif instance in HepG2 cells50. Our results indicate a broader repressive role in active regions, a discovery that stems directly from our ability to distinguish activating vs. repressive nucleotides using our tiling approach.
Discussion
We presented Sharpr-MPRA, a combined experimental and computational approach for high-resolution mapping of activating and repressive nucleotides across thousands of genomic regions. We used dense tiling of MPRA constructs spanning 4.6 million nucleotides targeting 15,720 regions at a resolution typically not afforded without perturbation experiments, which are traditionally not applicable at this scale. Sharpr-MPRA distinguishes activating from repressive nucleotides, and directly assesses regulatory function in a reporter assay, thus complementing the endogenous epigenomic signatures surveyed by ENCODE1, Roadmap Epigenomics4 and related projects.
Nucleotides with stronger activating or repressive Sharpr-MPRA scores were enriched in evolutionarily-conserved elements and predicted cell type-specific TF binding sites. Surprisingly however, both enrichments were weaker for the most highly-active nucleotides in HepG2 cells. Instead, the strongest reporter activity overlapped ERV1 endogenous retroviral repeat elements, which might speak to a potential role in regulatory turnover across even closely-related species.
Endogenous epigenomic signatures were predictive of reporter gene expression, with chromatin state and DNA accessibility each providing relevant information. Segments with endogenous active promoter and H3K27ac-marked enhancer signatures drove the strongest reporter gene activation, and segments showing endogenous Polycomb-associated signatures showed among the strongest reporter gene repression. These results indicate that even when tested outside their endogenous context, DNA sequence elements maintain the activating and repressive functions reflected in their endogenous epigenomic signatures.
Aggregation of activity scores at the motif level revealed cell type-specific motifs, and distinguished activator and repressor motifs. Motif activity typically correlated with chromatin context, with activating motifs found in active chromatin states, and repressive motifs in repressive states. Notable exceptions included putative 'attenuator' motifs that showed repressive roles but were found in active chromatin states (e.g. RFX motifs in HepG2) and putative ‘pioneer’ motifs, which showed strong activity regardless of their chromatin state context (e.g. activator TP53 and repressor NRSF), although directed experimentation and endogenous modulation will be needed to confirm these predictions. Most motifs showed activating-only or repressive-only signatures, but a small number of 'dual-role' motifs showed both activating and repressive signatures (e.g. members of the MAF and AP-2 protein families). Surprisingly, the sequence pattern with the strongest average activity in both cell types, TCTCGCGAGA, is not associated with a well-established regulator, highlighting our still incomplete understanding of regulatory elements, and the importance of unbiased dissection of regulatory regions.
Limitations include a need for longer sequences to show reporter activity in some regions, which might be overcome by improved DNA synthesis, and limited transfection efficiency in some cell types, which may require alternative delivery approaches (e.g. viral transduction). Additionally, we assumed additive effects in our analysis, which may miss interactions between different nucleotide positions. Barcode effects and other factors may cause experimental noise, which could be overcome by higher density tiling or different experimental bar-coding strategies63. We only tested elements in episomal assays, which provides direct information on regulatory activity, but does not capture potential effects of the endogenous chromatin context, and we only transfected unstimulated cells, although some sites may only function after specific stimulations.
We envision diverse uses for the results and methodology presented here. On one hand, the annotation of activating or repressive function for 4.6 million nucleotides can be useful to ask biological questions beyond the ones addressed here, and to train new computational models (e.g. for predicting activating and repressive nucleotides outside the regions surveyed here, or predicting the effect of non-coding variation on regulatory function62,63). On the other hand, Sharpr-MPRA's combined experimental and computational strategy can be broadly useful for dissecting regulatory regions across individuals, species, cell types, conditions, and disease state.
Online Methods
Pilot Large-step Massively Parallel Reporter Assay Design
As a pilot design we selected 250 regulatory regions to test. Each selected regulatory region was tiled by nine sequence tiles of 145bp in length placed in 30bp offsets so that adjacent sequences would have 115bp in common. Each tile was associated with 24 unique barcodes. In this design we used 216 barcodes per putative regulatory region.
Of the 250 regions we selected 200 of them so that their center position came from a HepG2 H3K27ac-marked "strong enhancer" chromatin states from the 15-state chromatin state model in Ref. 2 (Supplementary Fig. 1b). In order to define the specific locations to test we first defined a dip score based on the ENCODE hg18 HepG2 H3K27ac signal2 on chr1–22 and chrX. We defined the dip score to be the sum of the signal from positions 200bp away in both directions minus twice the signal at the dip center. We then ranked all positions at a 25-nucleotide resolution based on their dip score excluding from consideration positions that either (i) did not have the minimum HepG2 H3K27ac signal within 200 nucleotides, or (ii) were tied for the minimum H3K27ac signal with another position within 200 nucleotides and did not have a strictly greater dip score than the tied position. This requirement often enabled us to center our tiles within nucleosome depleted regions. We excluded positions if they were within 2kb of an annotated transcription start site based on the GENCODE v2b annotations. We also excluded from testing those sequences that contained a GGTACC, TCTAGA, or GGCCNNNNNGGCC as these were recognition sequences of the restriction enzymes. We selected 100 positions to be the highest ranked non-excluded positions. We selected an additional 100 positions to cover a range of dip scores among the non-excluded positions. Let m denote the dip-score of the 101st ranked position. We define an interval width w to be (ln(m)−ln(10))/99. We selected as the ith range selection where i=1,…,100 the region which had the greatest dip score, v, such that ln(ν) ≤ m − (i − 1)×w. The center of the selected 25-bp position interval was used as the center of the center tile.
An additional 50 sequences were selected based on the same procedure to select the top 100 sequences for HepG2, but based on the K562 data, limited to the top 50, with the additional constraint that they were in low-activity states in HepG2 (‘weak transcribed’ or ‘heterochromatin;low signal’) (Supplementary Fig. 1b).
For the motif enrichment analysis, we converted the coordinates from this design to hg19 using the UCSC Genome Browser liftover tool.
Identification of Significant Adjacent Changes in the Pilot Large-step Data
Among all pairs of adjacent tiles we identified a set of pairs that had a significant difference in reporter expression level at a 5% False Discovery Rate. Our procedure for doing this was as follows. Let pi denote the p-value for the ith adjacent pair (where i=1,…,2000 in our case) that there is a significant difference in the reporter expression between the first and second tiles of the pair. We let pi,L denote the p-value for the one sided test that the second tile has lower expression than the first and pi,G the p-value for a one sided test that the second tile has greater expression than the first. We further let pi,L,r and pi,G,r for r=1,…,R, where R=2 in our case, denote the p-value for that in the ith pair and the rth replicate that the second tile had lower or greater expression than the first respectively.
We compute the individual pi,L,r and pi,G,r p-values based on a one-sided Mann-Whitney Test on all the individual barcoded expression values for a tile, which was up to 24 in our case. To compute pi,L,r we first obtained a two-sided p-value, v, for a Mann-Whitney Test using Apache Commons Math 3.3 (https://commons.apache.org/). We then assigned the p-value v/2 if the second tile had average lower or equal ranks than the first and the p-value 1-v/2 otherwise; we made the opposite assignments for pi,G,r. The pi,L p-value was computed based on Fisher’s method combining the pi,L,r for r=1,…,R p-values, and likewise for the pi,G p-values. The p-value pi we defined to be
We multiply by two here to correct for having testing both p-values separately as one-sided tests. We obtained a false discovery rate for the set of p-values pi for i=1,…,2000 using a Benjamini-Hochberg procedure.
Scale-up Sharpr-MPRA Assay Design
We targeted 15,720 DNase regions, consisting of 3,930 tiled regions in each of four cell types: HepG2, H1hesc, K562, and Huvec. The DNase peaks were generated by the University of Washington ENCODE Group7 and specifically we used the peaks contained in the hg19 files: wgEncodeUwDnaseHepg2PkRep1.narrowPeak.gz, wgEncodeUwDnaseH1hescPkRep1.narrowPeak.gz, wgEncodeUwDnaseK562PkRep1.narrowPeak.gz, and wgEncodeUwDnaseHuvecPkRep1.narrowPeak.gz for the HepG2, H1hesc, K562, and Huvec cell types respectively. The selection of the subset of 3,930 regulatory regions based on each cell type was conducted independently. To increase chromatin state diversity, we selected regions using a richer 25-state chromatin state model (the ChromHMM60 model from Refs 38,39), which was based on 14 input tracks, consisting of 8 histone modification marks, CTCF, POL2, DNase (single-cut, generated by Duke58; and double-cut, generated by University of Washington7), FAIRE58, and input. The counts of each state were manually specified to ensure some coverage of each chromatin state, greater coverage in states more associated with DNase, and deeper coverage of enhancer chromatin states (specified in Fig. 1b). The regions were then randomly selected given the counts for each state. As with the Pilot design we excluded from testing those sequences that contained a GGTACC, TCTAGA, or GGCCNNNNNGGCC as these were recognition sequences of the restriction enzymes, but we had no restriction in this design with respect to position relative to annotated genes. The tiled regions based on each cell type and each chromatin state were randomly and evenly divided between the two array designs, which led to each design having 7860 tiled regions. If the same or overlapping regions were selected based on different cell types we retained both tiled regions and considered them separately except in forming the browser tracks in which case we averaged all regulatory scores for a given nucleotide. In total, we targeted 15,720 regions, some of which overlapped, resulting in 15,455 unique non-overlapping regions.
Experimental Procedure
The experimental methods for a Massively Parallel Reporter Assay are described in (Melinkov et al., 2014)66. Oligo nucleotide library synthesis was performed by Agilent Inc.67, and the cell culture, transfection and plasmid construction were done as in (Kheradpour et al, 2013)30. The MPRA vector backbone and promoter-reporter cassettes are available from Addgene (Plasmid # 49349, 49353 and 49354). The K562 and HepG2 cell lines were obtained directly from ATCC (CCL-243 and HB-8065).
For the pilot design, we used a single 55K-spot array for synthesis and designed sequences for 54,000 of them (2,250 unique sequences with 24 barcodes each). Transfection and barcode sequencing experiments were conducted in replicate in both K562 and HepG2 cells using the SV40 promoter (Supplementary Fig. 1a). The plasmid pools were amplified and sequenced in replicate and shared among the K562 and HepG2 experiments.
For the scale-up design, we used two 244K-spot arrays for synthesis and designed sequences for 243,660 of them of which 243,573 and 243,564 (99.96%) were included in the synthesis for the two arrays, leading a total of 487,137 probed tiles. Transfection and barcode sequencing experiments were conducted for each array in both HepG2 and K562, each using both a minimal and SV40P promoter, each in two replicates (16 experiments total). For these experiments we amplified and sequenced four plasmid pools, one for each combination of array design and promoter type (one for minP in HepG2, one for SV40P in HepG2, one for minP in K562, and one for SV40P in K562), with the two RNA replicate experiments normalized to the same DNA pool.
Data Normalization
The initial data processing for data from one experiment was as follows. We generated DNA and RNA counts based on the procedure described in (Kheradpour et al, 2013)30. And added a pseudo-count of 1 to these counts. We then divided all RNA values by the sum of all RNA-values and divided the DNA values by the sum of all the DNA values. For the readout corresponding to a given barcode we computed the log base two ratio of the RNA to DNA counts. We treated as missing those barcodes that had less than 20 for the original DNA counts associated with them. For the pilot design, we then normalized the values by taking the difference with the average value for the expression of the tiles in the positions furthest from the H3K27ac dip centers (tiles #1 and #9) as an approximation for background in these experiment. For the scale-up design, additional normalization was conducted on the inferred activity values (see below).
Sharpr-MPRA Regulatory Activity Scores
To compute Sharpr-MPRA regulatory activity scores from tiled reporter data we assume each reporter sequence has length L, which was 145bp in our case, and a consistent step size, s, between adjacent reporter sequences, which was 5 in our case. We assume that L is divisible by s, and let N=L/s denote the number of intervals of the step size overlapped by a reporter sequence, which was 29 in our case. We let J denote the number of overlapping reporter tiles covering a tiled region, which was 31 in our case. We let K denote the total number of non-overlapping intervals of step size s intervals that have coverage by at least one reporter sequence which is N + (J − 1), or 59 in our case. We let W = s×K denote the total number of nucleotides covered by at least one reporter sequence, which was 295 in our case, and we index them using i=1,…,295. We let T denote the total number of tiled regions tested in a single design, which was 7860 in our case. We let R denote the number of experiments for the design that we are combining, where R = 2 when we considered the SV40P and minP experiments separately and R = 4 when we combined all experiments for a design in the same cell type.
We let Ar,t,k denote a random variable for the unobserved regulatory activity for the kth s=5-bp interval where k = 1, …, K = 59 in the tth tiled region in the rth experiment being combined. We let Mr,t,j denote a random variable for the normalized expression value for the reporter sequence at the jth tile offset, where j = 1, …, J = 31, of the tth tiled region in the rth experiment being combined (Fig. 3a). We let mr,t,j denote the corresponding observed value, and if there was no observed value it is set to null. Our objective is to infer the maximum a posteriori values of the Ar,t,k variables conditioned on the observed values for the Mr,t,j variables.
We assume that each Ar,t,k is normally distributed with mean μar and variance , that is
Let denote the set of all observed reporter values. μar is set to the empirical mean of the observed reporter values in the rth experiment, that is
is a free parameter. We performed the inference with set both to 1 and 50 and combined the inferences using a procedure described below.
We assume that each Mr,t,j is normally distributed with mean μmr,t,j and variance , that is
We assume μmr,t,j to be the mean of all the unobserved regulatory activity variables corresponding to intervals for which the jth reporter tile overlaps, that is
We set to the empirical variance of the observed reporter values in rth experiment, that is
The vector Xr,t comprised of the Ar,t,k and Mr,t,j variables in the rth experiment for tth tiled region, can be expressed as a multivariate normal distribution
where Ar,t = [Ar,t,1 … Ar,t,K]T and Mr,t = [Mr,t,1 … Mr,t,J]T. If a mr,t,j is considered missing, that is has a null value, the corresponding Mr,t,j variable is omitted, but we do not re-index the remaining Mr,t,j variables. We let
Both μAr,t and μMr,t are column vectors with all values equal to μar.
Conditioning on observing Mr,t = mr,t the maximum a posteriori values for values in Ar,t is determined as the mean in a conditional multivariate normal distribution which is given by68
For ΣMr,t,Mr,t we have:
For ΣAr,t,Mr,t we have:
The above expressions are derived in the Supplementary Note.
We denote with ar,t,k the inferred value for the kth interval in the tth tiled region in the rth experiment. We note that the modeling to infer these values can also be viewed as a specific instance of Bayesian linear regression.
We then standardize all inferred values within an experiment by subtracting the mean and dividing by the standard deviation. Formally we denote zr,t,k the standardized regulatory score for the kth interval in the tth tiled region in the rth experiment. We then define:
We also define as our merged regulatory score ẑt,k for the kth interval in the tth tiled region which averages multiple replicates as:
When conducting inference with two different values for , denoted and , we denote the merged regulatory scores for kth interval in the tth tiled region for these two parameter settings, and , respectively. We combined them to obtain the value denoted as follows
This procedure takes the more conservative score when the signs of the values agree and 0 otherwise. Note that if , then . We inferred activity values using both a low-variance prior and a high-variance prior . We found this strategy for combining the results using two different variance prior parameters was more robust compared to using one specific parameter setting as it would reduce overfitting in cases when a single variance parameter was set to be too large, which sometimes led to unlikely high activating and repressive inferences in the same small region, while also reducing underfitting when a single variance parameter was set to be too small (Supplementary Fig. 6). We evaluated the enrichments of inferred highly activating and repressive nucleotides for conserved elements as a function of the variance prior parameters. We found them to be relatively robust across a substantial range of parameter settings, and in particular when using this strategy of using the more conservative inference from two substantially different settings for the variance prior (Supplementary Fig. 40). We also verified for the values of these parameters we used that using the higher variance parameter in conjunction with the lower variance parameter had little overall effect on the motif analyses as compared to just using the higher variance parameter (Supplementary Fig. 41).
To obtain nucleotide regulatory scores, bt,i, for each nucleotide i = 0, …, W − 1 in the tth tiled region we conducted piecewise linear interpolations between the merged regulatory scores. Formally we define bt,i as
where
The inference on the two designs was conducted separately, but they were combined for conducting the downstream analysis. The matrix operations were implemented using the library Apache Commons Math (https://commons.apache.org/proper/commons-math/) library v3.3. For the analysis on step sizes greater than 5 (Supplementary Figs. 22–23), we effectively applied the method here with a step size of 5, but treated entire positions of reporter tiles as missing.
Region and Chromatin State Scores
The region score for a tiled region t, denoted et is defined as bt,i (see above) where i is selected to maximize |bt,i| for i = 0, …, W − 1. The average region score for a chromatin state u, denoted su, based on matched data in a cell type, is the average value of et for all tiled regions t selected based on chromatin state u in the cell type.
Footprint, Motif, Transcription Factor ChIP-Seq, Conservation and Repeat Data
The HepG2 and K562 CENTIPEDE elements were from Ref. 8 obtained from http://centipede.uchicago.edu/. The footprints from Ref. 6 were obtained from ftp://ftp.ebi.ac.uk/pub/datanucleotides/ensembl/encode/integration_data_jan2011/byDataType/footprints/jan2011/. The Wellington footprints in K562 were obtained from the supplementary data of Ref. 40. The footprints of Ref. 5 were the hg19 footprints obtained from http://fureylab.web.unc.edu/datasets/footprints/. The PIQ footprints41 were the version 1 footprints obtained from http://piq.csail.mit.edu/ including both forward and reverse footprints. The motif instances were those provided by Ref. 13. The ENCODE transcription factor binding peak call datasets used in the analyses of Supplementary Figs. 35 and 36 were the ENCODE21,69 uniform peak calls downloaded from http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeAwgTfbsUniform which included 150 files in K562 cells and 77 for HepG2 cells. The conserved elements were the hg19 liftover of the SiPhy-PI65 conserved elements from Ref. 42. The repeats were based on RepeatMasker70 obtained through the UCSC genome browser71.
The Motif Analysis in the Scale-up Experiments Data
The motif analysis comparing K562 and HepG2 cells (Fig. 4, Supplementary Figure 25, Supplementary Table 2) was computed based on averaging the bt,i values at the center of all instances for a motif. The p-values were computed using a paired t-test over all instances tested using the Apache Commons Math library v3.3 implementation. The average motif score for the analysis in Fig. 6c was based on just motif instances overlapping a tiled region selected based on HepG2 chromatin data when testing in HepG2 cells and likewise for K562 restricted to motifs with at least 20 such instances. The expected motif scores for these same set of instances were computed by permuting among tiled regions assigned to the same chromatin state and selected by the cell type of the measurements, which set of reporter values were assigned to which tiled regions. These permutations would preserve the same set of rows in a matrix where the rows correspond to reporter expression values and the columns the tile offsets. This was done for 1000 permutations. For each motif the median average motif score across all permutations as well as the value of the 2.5% and 97.5% quantiles were recorded to form the expected motif values and 95% confidence intervals.
The p-values for motif enrichment as an activator or repressor in Fig. 4c and Supplementary Figs. 25–26, 29 were computed based on one-side binomial tests where the probability of success in the binomial distribution is the fraction of total nucleotides tested that had a regulatory score greater than or equal to the activation threshold for activators or less than or equal to the repression threshold for repressors. The number of trials is the number of instances of a motif with a center position overlapping a nucleotide tested. The number of successes is the number of instances of the motif with a center position having a regulatory score equal to or greater than the activation threshold for activators or less than or equal to the repression threshold for repressors. The p-value threshold for defining activator and repressor motifs was an uncorrected p-value of 0.01. In total 1934 motifs were tested. Motif instances that appeared on both strands at the same position were only counted once in the analysis.
Motif Analysis in the Pilot Large-step Experiment Data
We defined four sets of sequences to analyze for motifs based on the pilot data. Two sets were defined based on adjacent pairs of tiles with significant differences at a 5% FDR in the HepG2 data, with one set corresponding to the sequences that on average had higher expression as determined based on the average ranks and the other set lower expression. The other two sets were based on the K562 data defined in the same way as for the HepG2 data. For each set of sequences we conducted motif analysis on the 30-bp that were unique to each sequence in the set compared to its corresponding adjacent tile plus 10 additional bp into the common sequence. The motif enrichments with known motifs were computed using the program of Ref. 13 modified so that the background set of motifs only included those overlapping sequences part of the array design. We ran de novo motif discovery using MEME72 through the MEME suite with its default settings except requesting 10 motifs. The motifs were matched to a known motif using TOMTOM73.
DeltaSVM Comparison
For the comparison with important regulatory mutations predicted by the DeltaSVM approach in Supplementary Fig. 18a we identified a top 1% set of nucleotides associated with the maximum decrease in the sequence predicted to be regulatory when mutating the reference sequence. Specifically we obtained the gkm-SVM 10-mer weights based on Human ENCODE UW DHS from the website http://www.beerlab.org/deltasvm/ and used those in the files tup2_UwDnaseHepg2Aln_500_nc30_np_top10k_nsr1×1_gkm_1_10_6_3_weights.out and tup2_UwDnaseK562Aln_500_nc30_np_top10k_nsr1×1_gkm_1_10_6_3_weights.out for Hepg2 and K562 cells respectively. For each nucleotide tested we computed the sum of the k-mer weights for the k-mers overlapping the nucleotide, which would be ten weights or fewer if the nucleotide was within the first or last nine nucleotides of the 295-bp region. We denote this sum as sREF. We also computed this sum for each of the three possible nucleotide substitutions to the reference sequence at the position denoted by sM1, sM2, sM3. We ranked nucleotide position based on the extent to which they minimized min(sM1 –sREF, sM2 –sREF, sM3 –sREF). The focus on the top 1% nucleotides was consistent with a percentage threshold used previously with DeltaSVM scores14.
We also identified a top 1% set of nucleotides associated with the maximum increase in the sequence predicted to be regulatory when mutating the reference sequence (Supplementary Fig. 18b) using the same procedure as above except ranking nucleotides based on the extent to which they maximized the value of max(sM1 –sREF,sM2 – sREF, sM3 –sREF).
Barcode k-mer Analysis
For the analysis of barcode k-mer effect on inferred activity (Supplementary Fig. 12) we first define et,j to be the barcode sequence for the tth tiled region where t=1,…15720, for the jth reporter tile offset where j=1,…,31, and et,j,p the nucleotide at the pth position in the barcode for p=1,…,10.
For considering the occurrence of a k-mer sequence regardless of position within the barcode we then define for a k-mer sequence s the set Us,j={(t,p) | s=et,j,p… et,j,p+k−1} which gives all pairs of regions and barcode positions containing the k-mer sequence in the jth reporter tile offset. We computed average inferred regulatory activity scores for all tuples (s, j, i) such that s is a sequence of length k found within at least one barcode sequence, j=1,…,31 corresponds to one of the 31 reporter tile offsets, and i=1,…,295 corresponds to one of the inferred activity positions. The average inferred regulatory activity average is then defined as
where u. t denotes the tiled region of u. We then ranked these averages to determine the cumulative distributions. To determine the expected distribution of these averages for this analysis we randomly reassigned each barcode sequence to reporter sequences, but still preserving the activity inferences based on the real assignments. We repeated the same analysis based on the randomized assignments. We did this for 400 randomizations of barcode assignments to reporter sequences and obtained 400 separate rankings of averages. For each rank in the ranking we determined the median value over the 400 randomizations and the 2.5 and 97.5th percentiles. This analysis was done separately for each value of k=1,…,6.
Availability of Data, Software, and Sharpr-MPRA Scores
Raw data is available through Gene Expression Omnibus Accession (GEO) GSE71279. Count data is available in Supplementary Data Files 1 and 3, from GEO, and the supplementary website (http://www.biolchem.ucla.edu/labs/ernst/SHARPR/). The Sharpr-MPRA scores are available in text file, image, and browser track formats from the supplementary website and also in text format in Supplementary Data File 4. The SHARPR software is also available from the supplementary website and the source code is maintained at http://github.com/jernst98/SHARPR.
Supplementary Material
Acknowledgments
We thank Pouya Kheradpour and Jean-Philippe Vert for useful discussions related to this work. This work was supported by NIH grants R01ES024995, U01HG007912, and U01MH105578 (J.E.), R01HG006785 (T.S.M.), R01GM113708, U01HG007610, R01HG004037, U54HG006991, U41HG007000 (M.K), an NSF CAREER Award #1254200, and an Alfred P. Sloan Fellowship (J.E.).
Footnotes
Accession Codes
Gene Expression Omnibus GSE71279
Author Contributions
J.E. and M.K. designed the sequences, developed the computational methods, and analyzed the results. A.M., X.Z., L.W., P.R., and T.S.M. conducted the experimental work. T.S.M. oversaw the experimental work. J.E. and M.K. wrote the paper with substantial input from T.S.M.
Competing Financial Interests Statement
The Broad Institute has filed patents on the original MPRA technology. Patent protection for Sharpr-MPRA is currently being pursued.
References
- 1.ENCODE Project Consortium et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heintzman ND, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roadmap Epigenomics Consortium et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyle AP, et al. High-resolution genome-wide in vivo footprinting of diverse transcription factors in human cells. Genome Res. 2011;21:456–464. doi: 10.1101/gr.112656.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neph S, et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;489:83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pique-Regi R, et al. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res. 2011;21:447–455. doi: 10.1101/gr.112623.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu J, et al. Genome-wide Chromatin State Transitions Associated with Developmental and Environmental Cues. Cell. 2013;152:642–654. doi: 10.1016/j.cell.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Claussnitzer M, et al. FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. N. Engl. J. Med. 2015;373:895–907. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suvà ML, Riggi N, Bernstein BE. Epigenetic reprogramming in cancer. Science. 2013;339:1567–1570. doi: 10.1126/science.1230184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kheradpour P, Kellis M. Systematic discovery and characterization of regulatory motifs in ENCODE TF binding experiments. Nucleic Acids Res. 2014;42:2976–2987. doi: 10.1093/nar/gkt1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee D, et al. A method to predict the impact of regulatory variants from DNA sequence. Nat. Genet. 2015;47:955–961. doi: 10.1038/ng.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou J, Troyanskaya OG. Predicting effects of noncoding variants with deep learning-based sequence model. Nat. Methods. 2015;12:931–934. doi: 10.1038/nmeth.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gröschel S, et al. A Single Oncogenic Enhancer Rearrangement Causes Concomitant EVI1 and GATA2 Deregulation in Leukemia. Cell. 2014;157:369–381. doi: 10.1016/j.cell.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, et al. CRISPR Reveals a Distal Super-Enhancer Required for Sox2 Expression in Mouse Embryonic Stem Cells. PLoS ONE. 2014;9:e114485. doi: 10.1371/journal.pone.0114485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mendenhall EM, et al. Locus-specific editing of histone modifications at endogenous enhancers. Nat. Biotechnol. 2013;31:1133–1136. doi: 10.1038/nbt.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Melnikov A, et al. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat. Biotechnol. 2012;30:271–277. doi: 10.1038/nbt.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patwardhan RP, et al. Massively parallel functional dissection of mammalian enhancers in vivo. Nat. Biotechnol. 2012;30:265–270. doi: 10.1038/nbt.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patwardhan RP, et al. High-resolution analysis of DNA regulatory elements by synthetic saturation mutagenesis. Nat. Biotechnol. 2009;27:1173–1175. doi: 10.1038/nbt.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwasnieski JC, Mogno I, Myers CA, Corbo JC, Cohen BA. Complex effects of nucleotide variants in a mammalian cis-regulatory element. Proc. Natl. Acad. Sci. U. S. A. 2012;109:19498–19503. doi: 10.1073/pnas.1210678109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen SQ, et al. Massively parallel cis-regulatory analysis in the mammalian central nervous system. Genome Res. 2016;26:238–255. doi: 10.1101/gr.193789.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rajagopal N, et al. High-throughput mapping of regulatory DNA. Nat. Biotechnol. 2016;34:167–174. doi: 10.1038/nbt.3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Canver MC, et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527:192–197. doi: 10.1038/nature15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vierstra J, et al. Functional footprinting of regulatory DNA. Nat. Methods. 2015;12:927–930. doi: 10.1038/nmeth.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Findlay GM, Boyle EA, Hause RJ, Klein JC, Shendure J. Saturation editing of genomic regions by multiplex homology-directed repair. Nature. 2014;513:120–123. doi: 10.1038/nature13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korkmaz G, et al. Functional genetic screens for enhancer elements in the human genome using CRISPR-Cas9. Nat. Biotechnol. 2016;34:192–198. doi: 10.1038/nbt.3450. [DOI] [PubMed] [Google Scholar]
- 29.White MA, Myers CA, Corbo JC, Cohen BA. Massively parallel in vivo enhancer assay reveals that highly local features determine the cis-regulatory function of ChIP-seq peaks. Proc. Natl. Acad. Sci. 2013 doi: 10.1073/pnas.1307449110. 201307449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kheradpour P, et al. Systematic dissection of regulatory motifs in 2000 predicted human enhancers using a massively parallel reporter assay. Genome Res. 2013;23:800–811. doi: 10.1101/gr.144899.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwasnieski JC, Fiore C, Chaudhari HG, Cohen BA. High-throughput functional testing of ENCODE segmentation predictions. Genome Res. 2014 doi: 10.1101/gr.173518.114. gr.173518.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gisselbrecht SS, et al. Highly parallel assays of tissue-specific enhancers in whole Drosophila embryos. Nat. Methods. 2013;10:774–780. doi: 10.1038/nmeth.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vanhille L, et al. High-throughput and quantitative assessment of enhancer activity in mammals by CapStarr-seq. Nat. Commun. 2015;6 doi: 10.1038/ncomms7905. [DOI] [PubMed] [Google Scholar]
- 34.Arnold CD, et al. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science. 2013;339:1074–1077. doi: 10.1126/science.1232542. [DOI] [PubMed] [Google Scholar]
- 35.Dickel DE, et al. Function-based identification of mammalian enhancers using site-specific integration. Nat. Methods. 2014;11:566–571. doi: 10.1038/nmeth.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murtha M, et al. FIREWACh: high-throughput functional detection of transcriptional regulatory modules in mammalian cells. Nat. Methods. 2014;11:559–565. doi: 10.1038/nmeth.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Diao Y, et al. A new class of temporarily phenotypic enhancers identified by CRISPR/Cas9 mediated genetic screening. Genome Res. 2016 doi: 10.1101/gr.197152.115. gr.197152.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ernst J, Kellis M. Interplay between chromatin state, regulator binding, and regulatory motifs in six human cell types. Genome Res. 2013;23:1142–1154. doi: 10.1101/gr.144840.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoffman MM, et al. Integrative annotation of chromatin elements from ENCODE data. Nucleic Acids Res. 2013;41:827–841. doi: 10.1093/nar/gks1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piper J, et al. Wellington: a novel method for the accurate identification of digital genomic footprints from DNase-seq data. Nucleic Acids Res. 2013;41:e201. doi: 10.1093/nar/gkt850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sherwood RI, et al. Discovery of directional and nondirectional pioneer transcription factors by modeling DNase profile magnitude and shape. Nat. Biotechnol. 2014;32:171–178. doi: 10.1038/nbt.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lindblad-Toh K, et al. A high-resolution map of human evolutionary constraint using 29 mammals. Nature. 2011;478:476–482. doi: 10.1038/nature10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schoenherr CJ, Anderson DJ. The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science. 1995;267:1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- 44.Raghav SK, et al. Integrative Genomics Identifies the Corepressor SMRT as a Gatekeeper of Adipogenesis through the Transcription Factors C/EBPβ and KAISO. Mol. Cell. 2012;46:335–350. doi: 10.1016/j.molcel.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 45.Blattler A, et al. ZBTB33 binds unmethylated regions of the genome associated with actively expressed genes. Epigenetics Chromatin. 2013;6:13. doi: 10.1186/1756-8935-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xie X, et al. Systematic discovery of regulatory motifs in human promoters and 3’ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mikula M, et al. Comprehensive Analysis of the Palindromic Motif TCTCGCGAGA: A Regulatory Element of the HNRNPK Promoter. DNA Res. 2010;17:245–260. doi: 10.1093/dnares/dsq016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Di Vona C, et al. Chromatin-wide Profiling of DYRK1A Reveals a Role as a Gene-Specific RNA Polymerase II CTD Kinase. Mol. Cell. 2015;57:506–520. doi: 10.1016/j.molcel.2014.12.026. [DOI] [PubMed] [Google Scholar]
- 49.Hu JH, Navas P, Cao H, Stamatoyannopoulos G, Song C-Z. Systematic RNAi Studies on the Role of Sp/KLF Factors in Globin Gene Expression and Erythroid Differentiation. J. Mol. Biol. 2007;366:1064–1073. doi: 10.1016/j.jmb.2006.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Watts JA, et al. Study of FoxA Pioneer Factor at Silent Genes Reveals Rfx-Repressed Enhancer at Cdx2 and a Potential Indicator of Esophageal Adenocarcinoma Development. PLoS Genet. 2011;7:e1002277. doi: 10.1371/journal.pgen.1002277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang Y, Cvekl A. Large Maf Transcription Factors: Cousins of AP-1 Proteins and Important Regulators of Cellular Differentiation. Einstein J. Biol. Med. EJBM. 2007;23:2–11. doi: 10.23861/ejbm20072347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yoshida S, et al. Expression profiling of the developing and mature Nrl−/− mouse retina: identification of retinal disease candidates and transcriptional regulatory targets of Nrl. Hum. Mol. Genet. 2004;13:1487–1503. doi: 10.1093/hmg/ddh160. [DOI] [PubMed] [Google Scholar]
- 53.Bannert N, Kurth R. Retroelements and the human genome: New perspectives on an old relation. Proc. Natl. Acad. Sci. 2004;101:14572–14579. doi: 10.1073/pnas.0404838101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bourque G, et al. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 2008 doi: 10.1101/gr.080663.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kunarso G, et al. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat. Genet. 2010;42:631–634. doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- 56.Wang T, et al. Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc. Natl. Acad. Sci. 2007;104:18613–18618. doi: 10.1073/pnas.0703637104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat. Biotechnol. 2010;28:817–825. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Song L, et al. Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res. 2011;21:1757–1767. doi: 10.1101/gr.121541.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U. S. A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hoffman MM, et al. Unsupervised pattern discovery in human chromatin structure through genomic segmentation. Nat. Methods. 2012;9:473–476. doi: 10.1038/nmeth.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ulirsch JC, et al. Systematic Functional Dissection of Common Genetic Variation Affecting Red Blood Cell Traits. Cell. 2016;165:1530–1545. doi: 10.1016/j.cell.2016.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tewhey R, et al. Direct Identification of Hundreds of Expression-Modulating Variants using a Multiplexed Reporter Assay. Cell. 2016;165:1519–1529. doi: 10.1016/j.cell.2016.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sammons MA, Zhu J, Drake AM, Berger SL. TP53 engagement with the genome occurs in distinct local chromatin environments via pioneer factor activity. Genome Res. 2015;25:179–188. doi: 10.1101/gr.181883.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Garber M, et al. Identifying novel constrained elements by exploiting biased substitution patterns. Bioinformatics. 2009;25:i54–i62. doi: 10.1093/bioinformatics/btp190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Melnikov A, Zhang X, Rogov P, Wang L, Mikkelsen TS. Massively parallel reporter assays in cultured mammalian cells. J. Vis. Exp. JoVE. 2014 doi: 10.3791/51719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.LeProust EM, et al. Synthesis of high-quality libraries of long (150mer) oligonucleotides by a novel depurination controlled process. Nucleic Acids Res. 2010;38:2522–2540. doi: 10.1093/nar/gkq163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bickel PJ, Doksum KA. Mathematical Statistics: Basic Ideas and Selected Topics, Volume I. Second. CRC Press; 2015. [Google Scholar]
- 69.Gerstein MB, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smit A, Hubley R, Green P. RepeatMasker Open-30. 1996 [Google Scholar]
- 71.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. ISMB Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 73.Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS. Quantifying similarity between motifs. Genome Biol. 2007;8:R24. doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.