Abstract
Understanding kinetic information is fundamental in understanding biological function. Advanced imaging technologies have fostered the development of kinetic analyses in cells. We have developed Permeabilization Activated Reduction in Fluorescence (PARF) analysis for determination of apparent t1/2 and immobile fraction, describing the dissociation of a protein of interest from intracellular structures. To create conditions where dissociation events are observable, cells expressing a fluorescently-tagged protein are permeabilized with digitonin, diluting the unbound protein into the extracellular media. As the media volume is much larger than the cytosolic volume, the concentration of the unbound pool decreases drastically, shifting the system out of equilibrium--favoring dissociation events. Loss of bound protein is observed as loss of fluorescence from intracellular structures and can be fit to an exponential decay. We compared PARF dissociation kinetics with previously published equilibrium kinetics as determined by FRAP. PARF dissociation rates agreed with the equilibrium-based FRAP analysis predictions of the magnitude of those rates. When used to investigate binding kinetics of a panel of cytoskeletal proteins, PARF analysis revealed that filament stabilization resulted in slower fluorescence loss. Additionally, commonly used “general” F-actin labels display differences in kinetic properties, suggesting that not all fluorescently-tagged actin labels interact with the actin network in the same way. We also observed differential dissociation kinetics for GFP-VASP depending on which cellular structure was being labeled. These results demonstrate that PARF analysis of non-equilibrium systems reveals kinetic information without the infrastructure investment required for other quantitative approaches such as FRAP, photoactivation, or in vitro reconstitution assays.
Key words/phrases: transient kinetic analysis, quantitative fluorescence microscopy, protein-protein interactions, cytoskeleton, digitonin
Introduction
Reversible binding interactions between macromolecules underlie all biology. Life is a dynamic process, and as researchers grapple with uncovering the mechanisms governing life at the subcellular level a great deal of focus has been placed on understanding the kinetics of macromolecular interactions. Investigation of these research questions has been supported by advances in in vitro reconstitution and transient kinetic approaches for ensemble measures of the kinetics of protein-protein interactions [Pollard 2003; Pollard 2010]. The increasing ease in generation of fluorescently tagged proteins and advances in optics have increased the field’s ability to generate quantitative data using microscopy [Murphy and Davidson 2012b].
Fluorescence recovery after photobleaching (FRAP) is one such method that can be used to determine the diffusion of fluorescently labeled proteins in cellular environments [Snapp et al. 2003]. As interaction with other cellular structures alters fluorescence recovery kinetics, by examining the recovery kinetics of a photobleached region of interest, a researcher can determine the apparent exchange rate of the bleached fluor and half-life for recovery under equilibrium conditions [Murphy and Davidson 2012a]. They can also determine the fraction of protein within the region which can exchange and recover fluorescence (the mobile fraction), and the fraction that is not exchanging (the immobile fraction) [Axelrod et al. 1976]. In order for FRAP analysis to be valid, the researcher must make a number of assumptions about the experimental system [Snapp et al. 2003]. First, the bleached region of interest is capable of exchanging material with unbleached pools. Second, the unbleached pool that exchanges with the bleached region of interest has a much larger amount of labeled molecules than the region of interest. Additionally, in order to reliably determine t1/2 of recovery the fluorescence recovery must approach a plateau. This plateau may be at a fluorescence level lower than the initial pre-bleach fluorescence if the region of interest contains a significant immobile fraction which is incapable of exchanging with unbleached pools [White and Stelzer 1999]. A researcher interested in the relative mobility of different proteins of interest can carry out FRAP analysis and compare the relative t1/2 and immobile fractions. A smaller t1/2 indicate faster exchange of bleached fluor with unbleached cellular pools. A larger immobile fraction indicates less exchange of material with other unbleached pools. Similar kinetic analyses can be carried out using photoactivatable or photoswitchable fluors [Lippincott-Schwartz et al. 2003], and all approaches represent the equilibrium condition.
The key to many experiments designed to investigate the kinetics of protein interactions is to establish a condition where association or dissociation events are easily observable [Pollard and De La Cruz 2013]. Like many of the other experimental approaches using biophotonic analysis to gain kinetic insight, FRAP analysis can require a significant investment in specific equipment. Where low intensity illumination can be used to visualize cellular fluorescence, greater doses of photons directed at the sample are required to quickly and efficiently bleach a region of interest. Laser-scanning confocal microscopes are often used in FRAP analysis [Ellenberg and Lippincott-Schwartz 1999], as are wide-field epifluorescence microscopes equipped with an additional laser beam focused to a diffraction-limited spot at the plane of focus [Wadsworth and Salmon 1986; Wang 1985]. These technical advances have expanded the experimental repertoire available to some researchers. However, many researchers who are eager to address experimental questions related to protein dynamics that could be addressed using FRAP and photoactivation techniques may not have access to such an instrument. Additionally, steady-state approaches like FRAP and related techniques allow for the determination of equilibrium constants, but are not useful on their own in characterizing the individual events leading to the equilibrium exchange--association or dissociation rates [Cleland 1990; Pollard 2010]. Transient/pre-steady state kinetic approaches common to in vitro reconstitution experiments, allow for direct characterization of association or dissociation rates under pseudo first-order conditions [De La Cruz and Ostap 2009; Johnson 1992; Pollard and De La Cruz 2013].
Transient kinetic approaches often rely on the manipulation of the concentrations of the components of the system being studied, which can prove difficult with intact cells. Detergents such as digitonin have been used extensively to selectively permeabilize the plasma membrane by removing cholesterol [Schulz 1990] without altering the characteristics of internal membranes lacking cholesterol [Fiskum et al. 1980]. Permeabilization has been used to allow manipulation of cytosolic ion concentrations [Altschuld et al. 1985; Smolen et al. 1986], to allow introduction of large macromolecules [Lorenz et al. 2008], and to allow removal of macromolecules [Lineberger et al. 1989]. In many of these experiments the cell structures that remained following digitonin permeabilization continued to behave similarly to intact cells, allowing researchers to characterize subcellular processes using approaches not available in intact cells or in in vitro reconstitution systems [Marino et al. 2014; Schulz 1990; Subedi et al. 2014].
Here we present a permeabilization-based assay system that, under certain conditions, can be used to characterize the pseudo first-order kinetics of dissociation and association events. Permeabilization-activated reduction in fluorescence (PARF) and permeabilization-activated accumulation of fluorescence (PAAF) analysis, provide an elegant, cost-effective assay system marrying biophotonics with transient kinetics approaches. PARF and PAAF can be used in combination with FRAP and photoactivation techniques to investigate macromolecular interactions within cells. By treating cells with digitonin, the soluble cytosolic contents would be diluted into the incubation media. A fluorescently- tagged protein of interest exists in a cell at equilibrium between bound and unbound states. In transfected cells, sufficient dilution of the unbound label would set the system out of equilibrium, favoring dissociation as the system establishes a new equilibrium where the soluble pool is at a lower concentration [Pollard and De La Cruz 2013]. In untransfected neighbors, the sudden appearance of fluorescently labeled protein in the media could lead to association events, if the on-rate was of sufficient magnitude. Since the protein of interest is fluorescently labeled, dissociation events could be tracked via time-lapse fluorescence imaging, allowing for calculation of t1/2 for dissociation, giving the researcher some indication of the binding kinetics between their protein of interest and intracellular structures. Additionally, identification of an immobile fraction would indicate the relative amounts of exchange between bound and soluble pools overall. If visible, association events could be similarly analyzed. Using PARF and PAAF approaches we have been able to demonstrate that the common F-actin labels, Lifeact, F-tractin, and utrophin, show differences in their F-actin binding kinetics suggesting differences in the what underlying actin structures and dynamics are revealed by their labeling patterns. We were also able to demonstrate that the actin-associated protein, VASP displays differences in PARF kinetics depending on the cellular structure with which it is associating.
Results and Discussion
PARF analysis requires rapid manipulation of the concentration of the protein being studied
In-cell transient kinetic analyses such as PARF take advantage of large-scale dilution of the cytoplasm to reveal dissociation events. Our Rose Chamber [Rose et al. 1958] consists of a 20mm culture area filled with 300µL of buffer. Assuming that the cell growth area was 100% confluent with a monolayer of cells 20µm thick, then the volume of the cell monolayer would be less than 0.1µL. Digitonin-induced mixing with the extracellular buffer would dilute the cytosolic contents approximately 3000-fold. In our experiments, our cells were grown at confluencies of less than 50%, and transfection efficiencies were also less than 50%. Therefore it is likely that the soluble material in the cytoplasm of transfected cells was diluted 6,000- to 12,000-fold by digitonin permeabilization and mixing with extracellular media.
For a cellular system where a GFP-labeled protein of interest was at equilibrium between an unbound cytosolic pool and a bound pool, large scale dilution would result a large drop in the concentration of the unbound pool, disrupting the initial equilibrium condition. Given time, the system would establish a new equilibrium condition dependent on the newly-established concentration of the soluble pool. In previous studies, end-point analyses have been used to demonstrate that particular protein-protein interactions persist after permeabilization [Faire et al. 1999; Svitkina et al. 2003], but the dissociation kinetics of those interactions were not investigated in permeabilized cells. As equilibrium is initially reestablished, the likelihood of association events has decreased dramatically compared to the likelihood of dissociation events (Figure 1). As the protein is fluorescently labeled, a decrease in fluorescence signal would correspond to loss of labeled material from the structure of interest. Decrease in fluorescence signal would also follow pseudo first-order kinetics, and fitting the decrease in the fluorescence signal for an intracellular structure to an exponential could be used to determine a t1/2 for fluorescence loss and an immobile fraction [Pollard and De La Cruz 2013]. We hypothesized that comparing t1/2 and immobile fractions for different fluorescently tagged proteins of interest allows for the comparison of the binding characteristics relative to each other. If the same protein interacts differently with different cellular structures, then permeabilization-based kinetic analyses may reveal differences in association and dissociation rates as well.
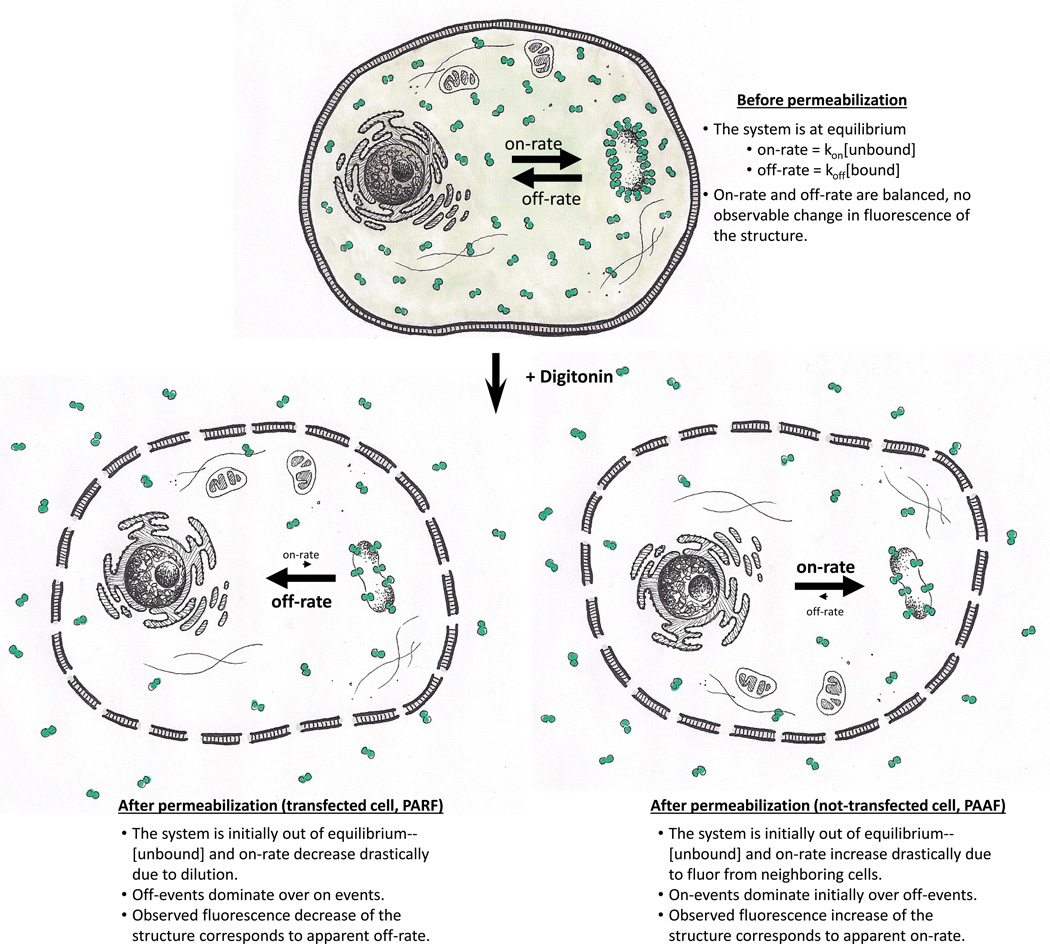
Figure 1. Diagram of PARF and PAAF method.
Before permeabilization of transfected cells, unbound labeled protein is in equilibrium with bound labeled protein. After permeabilization, the unbound labeled protein undergoes a very large dilution into the extracellular fluid, drastically decreasing the rate of label associating with the structure to such an extent that the dissociation rate becomes apparent (PARF). The permeabilization also allows access of soluble label from transfected cells to intracellular structures of untransfected cells, drastically increasing the rate of label associating with the structure to such an extent that the association rate becomes apparent (PAAF).
We first established an appropriate digitonin dose optimized for our cells, balancing speed of permeabilization with minimal disruption of cellular structures of interest. Using mcherry as a cytosolic marker, we treated transfected HeLa cells with increasing doses of digitonin and tracked the loss in fluorescence (Figure 2A). At doses lower than 25µM, the loss in mcherry signal was minimal over the time course observed, indicating that permeabilization was incomplete. We noticed a transient increase in fluorescence for the cells treated with 25µM digitonin that we attribute to the delayed loss of fluorescence from the nucleus traversing the region of interest (ROI) as it leaves the nucleus and diffuses out of the cell. The concentration of digitonin chosen for these experiments was similar to or lower than concentrations previously shown to permeabilize the membrane with minimal effects on intracellular structures [Altschuld et al. 1985; Fiskum et al. 1980; Lineberger et al. 1989; Manor et al. 2015].
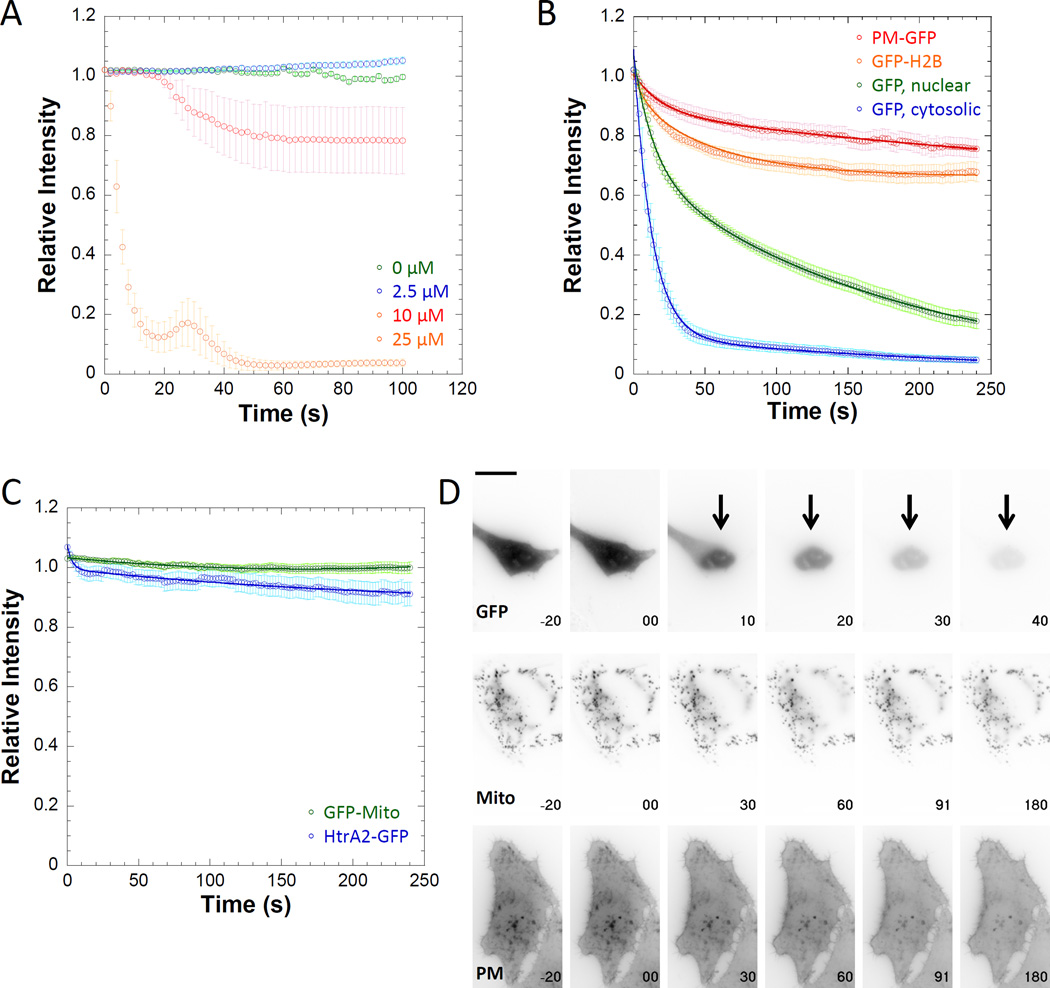
Figure 2. Digitonin permeabilization reveals the apparent off-rates of fluors.
A) HeLa cells transfected with mcherry were treated with varying concentrations of digitonin. Permeabilization was not rapid or complete at concentrations less than 25 µM. B) GFP-labeled proteins in different cellular compartments displayed different PARF kinetics after 25µM digitonin treatment, indicating that digitonin permeabilization did not completely disrupt all cell structures. C) Fluorescent proteins localization to the mitochondrial matrix (GFP-Mito) and the mitochondrial inner membrane space (HtrA2-GFP) was not disrupted by digitonin treatment, indicating that some internal membranes are resistant to digitonin permeabilization. D) Individual frames from movies with time in seconds on each frame for GFP, GFP-Mito, and PM-GFP. (t = 0 is when permeabilization occurred) and scale bar is 20 µm. Data points represent the sample mean ± SEM. For panel A), n0µM = 18 cells, n2.5µM = 14 cells, n10µM = 13 cells, and n25µM = 15 cells. For panel B), nPM-GFP = 17 cells, nGFP-H2B= 32 cells, nGFP, nuclear= 18 nuclei, and nGFP, cytosol= 16 cells. For panel C), nGFP-Mito= 29 cells, nHtrA2-GFP = 27 cells.
We next examined the fluorescence loss of GFP-tagged proteins associated with particular cellular structures. We hypothesized that a plasma membrane label PM-GFP [Teruel et al. 1999] and a histone subunit inside the nucleus (GFP-H2B) would display slower fluorescence loss than GFP [Cormack et al. 1996], a soluble cytoplasmic label (Figure 2B and 2D, Supplemental Movie 1). We observed that both PM-GFP and GFP-H2B showed minimal fluorescence loss, indicating that digitonin treatment did not lead to total disruption of the cellular structures with which they were associated. GFP signal was lost quickly from the cytosol. It is also worth noting that in the soluble GFP timelapses there is a clear delay in GFP leaving the nucleus likely through nuclear pore complexes [Wang and Brattain 2007], suggesting that digitonin treatment is not having as significant an impact on nuclear membrane permeability as it is on plasma membrane permeability.
According to our model, we predicted that cytosolic GFP would have the smallest t1/2 and immobile fraction, and fluors labeling more stable structures would have larger t1/2 and immobile fractions. In all cases the data analysis requires correction to account for fluorescence contributions from unincorporated label, background signal, and fluorescence photofading due to image acquisition. First, we chose to analyze regions of interest that primarily consisted of the structure we wished to analyze. As these data were collected using wide-field epifluorescence microscopy, careful choice of region of interest limited the contribution of unbound fluorescence signal to the total fluorescence signal. Next, we subtracted the extracellular background fluorescence, and corrected the fluorescence signal for photofading due to acquisition [Applewhite et al. 2007]. We calculated a correction equation by fitting the observed photofading to a single exponential decay for each fluorescently-labeled protein over the same experimental time course used for PARF. Correcting the data in this manner ensured that the observed loss in fluorescence most strongly correlated to dissociation events.
In all cases the corrected data fit well to double-exponential functions, and the calculated fits agreed with our predictions. The t1/2 for cytosolic GFP (11.7±3.0s) was shorter than for nuclear GFP (58.5±6.8s), and both displayed minimal immobile fractions (0.027±0.042 and 0.004±0.005, respectively). PM-GFP and GFP-H2B had large immobile fractions (0.67±0.08 and 0.67±0.03, respectively), indicating that a large portion of the fluor was not dissociating over the 4-minute time course of the experiment. As relative intensity never reached 0.5 for either PM-GFP or GFP-H2B, a t1/2 could not be calculated (Table 1).
Table 1.
PARF kinetic analysis for GFP-tagged proteins used in this study
| Protein and condition | t1/2 | Immobile fraction |
R | n | ||
|---|---|---|---|---|---|---|
| Cells | Structures | Experiments | ||||
| GFP in cytosol | 11.7 ± 3.0 | 0.027 ± 0.042 | 0.998 | 16 | - | 5 |
| GFP in nucleus | 58.5 ± 6.8 | 0.004 ± 0.005 | 0.999 | 18 | - | 5 |
| GFP-H2B | - | 0.67 ± 0.03 | 0.998 | 32 | - | 6 |
| PM-GFP | - | 0.67 ± 0.08 | 0.998 | 17 | - | 6 |
| GFP-αtubulin | 29.5 ± 8.0 | 0.35 ± 0.06 | 0.995 | 22 | - | 8 |
| GFP-αtubulin + taxol | - | 0.75 ± 0.02 | 0.998 | 26 | - | 5 |
| 3xGFP-EMTB | - | 0.82 ± 0.03 | 0.943 | 23 | - | 6 |
| 3xGFP-EMTB + taxol | - | 0.85 ± 0.03 | 0.936 | 12 | - | 3 |
| GFP-actin | 26.1 ± 6.2 | 0.36 ± 0.02 | 0.991 | 36 | - | 7 |
| GFP-actin + jas | - | 0.62 ± 0.02 | 0.994 | 24 | - | 5 |
| GFP-LifeAct | 17.8 ± 3.9 | 0.06 ± 0.01 | 1.000 | 25 | - | 6 |
| GFP-LifeAct + jas | 11.8 ± 1.9 | 0.10 ± 0.13 | 0.989 | 29 | - | 6 |
| GFP-Ftractin | 16.2 ± 2.3 | 0.06 ± 0.01 | 1.000 | 42 | - | 9 |
| GFP-Ftractin + jas | 26.1 ± 4.7 | 0.09 ± 0.01 | 0.999 | 32 | - | 5 |
| GFP-Utrophin | 52.4 ± 10.5 | 0.31 ± 0.04 | 1.000 | 29 | - | 7 |
| GFP-Utrophin + jas | - | 0.63 ± 0.07 | 0.997 | 43 | - | 6 |
| GFP-VASP, whole cell | 20.7 ± 5.0 | 0.11 ± 0.02 | 0.998 | 23 | - | 8 |
| GFP-VASP, focal adhesions |
39.2 ± 3.3 | 0.21 ± 0.03 | 0.973 | 23 | 145 | 8 |
| GFP-VASP, filopodial tips | - | 0.71 ± 0.07 | 0.943 | 23 | 31 | 6 |
Digitonin permeabilization minimally disrupts some subcellular compartments and leads to disassembly of others
To assay for digitonin-induced disruption of internal structures, we undertook PARF analysis of GFP-Mito, a mitochondrial matrix label, and HtrA2-GFP, a serine protease found in the inner membrane space [Martins et al. 2002] (Figure 2C, Supplemental Movie 1). Neither GFP-Mito nor HtrA2-GFP lost more than ~10% of their initial fluorescence over the course of the experiment. We previously used FRAP to determine that the t1/2 for exchange of GFP-Mito is slow under equilibrium conditions (~180s) [Quintero et al. 2009], and FRAP analysis of mitochondrial matrix-targeted DsRed1 also showed that recovery after photobleaching is slow [Collins and Bootman 2003], in agreement with the minimal loss of fluorescence that we observed by PARF. These data suggest that over short timescales digitonin treatment is not impacting the mitochondrial membranes to the same extent as the plasma membrane.
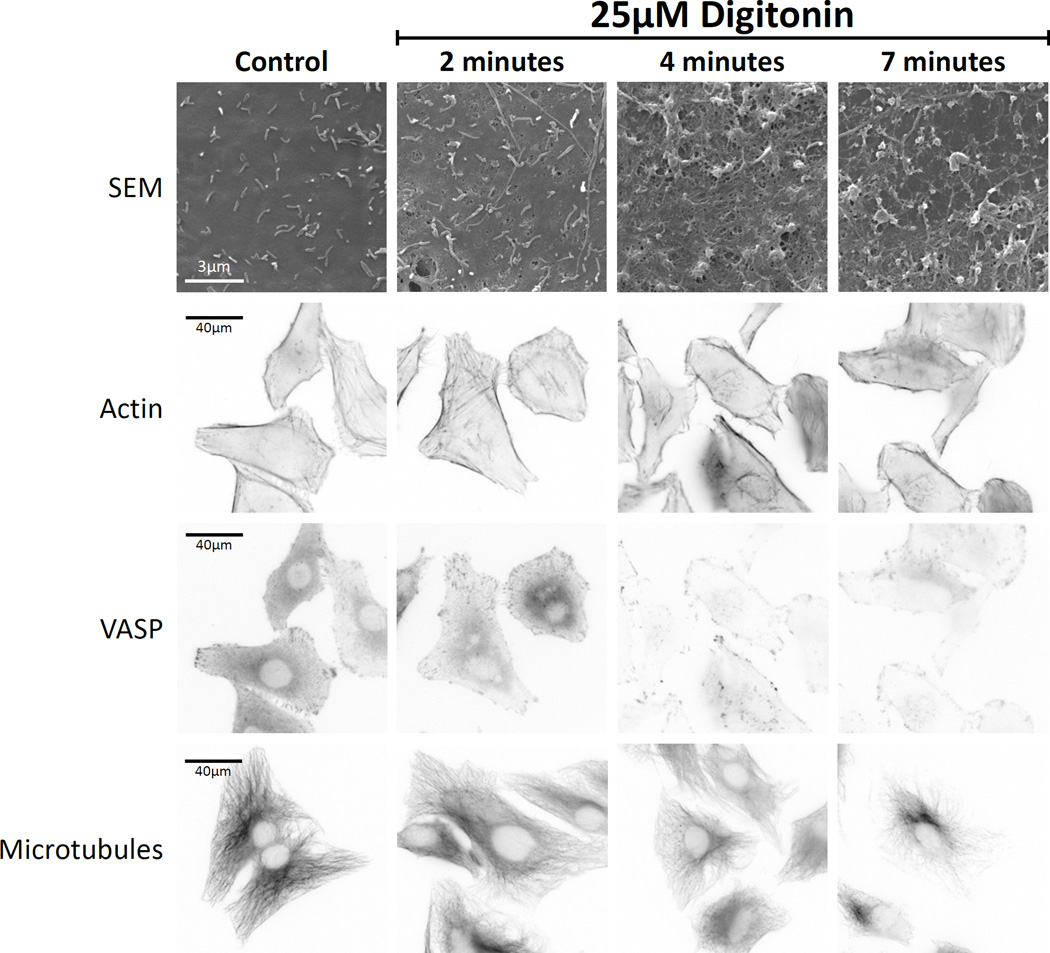
We used digitonin treatment at fixed time points to determine the impact of permeabilization on endogenous cellular structures (Figure 3). Scanning electron microscopy showed that permeabilization disrupted the plasma membrane but did not remove it completely. Cytoskeletal structures such as the actin filament network, VASP-labeled focal adhesions, and the microtubule network were all sensitive to digitonin permeabilization, as fluorescent signal associated with those structures decreased over time. However, in all cases fluorescence signal persisted and the cellular structures were evident even seven minutes after permeabilization. These data indicate that the structures in which we were interested remain intact over the time course of PARF analysis (approximately 4 minutes).
Figure 3. Cellular structures continue to persist following 25µM digitonin permeabilization.
Scanning-electron microscopy of the surface of digitonin-treated HeLa cells revealed increasing appearance of pores in the plasma membrane over 7 minutes of treatment. Cytoskeletal structures such as actin filaments (visualized by phalloidin-staining), focal adhesions (visualized by VASP immunostaining), and microtubules (visualized by immunostaining), were reduced in intensity but persisted throughout the 7-minute time course of the experiment.
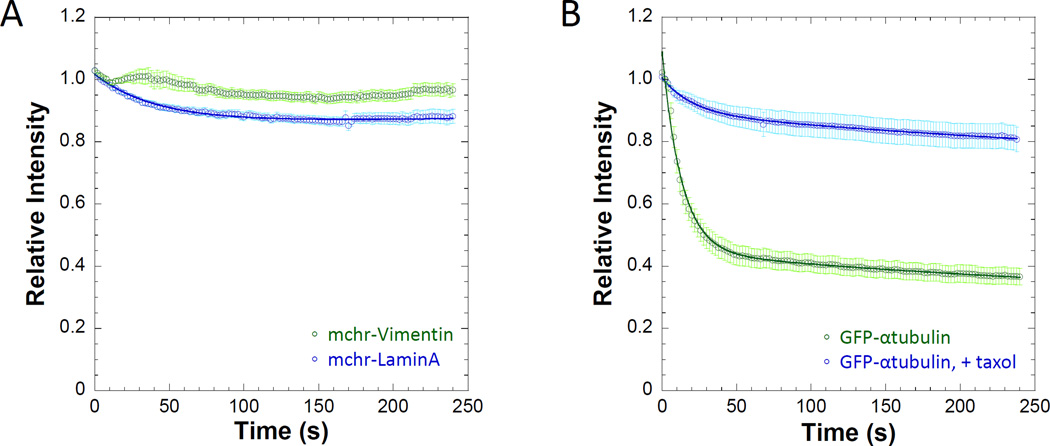
Unlike FRAP where it is assumed that the bleaching event does not disrupt the cellular structure being observed, digitonin permeabilization impacts every other equilibrium present in the cytosol, including the processes that may help to maintain a structure of interest. Digitonin permeabilization will impact the soluble concentration of cytoskeletal elements such as actin [Fiskum et al. 1980] or microtubules [Bloom and Vallee 1983]. We carried out PARF analysis of two cytoskeletal filament systems which undergo exchange with soluble pools: intermediate filaments (Figure 4A) and microtubules (Figure 4B). The two intermediate filament proteins assayed, lamin A and vimentin, both displayed very little fluorescence loss over the time course of the experiment, in agreement with steady-state analyses indicating that both vimentin (t1/2 for FRAP ~ 5 minutes) [Yoon et al. 1998], and lamin A (t1/2 for FRAP ~90 minutes) [Moir et al. 2000] display slow exchange with cellular pools. Drastically decreasing the concentration of soluble tubulin will impact microtubule polymerization dynamics. PARF experiments using GFP-α-tubulin in the presence and absence of 10µM taxol (Figure 4B) demonstrate that stabilizing interactions lead to a much larger immobile fraction (0.35±0.06 without taxol versus 0.75±0.02 with taxol), indicating less filament disassembly in the presence of taxol. Additionally, as GFP-labeled tubulin subunits within the microtubule lattice do not have an equal likelihood of dissociation as compared to subunits at the ends of microtubules, the observed kinetics in these experiments describes the conditions at the filament ends, not of the structure as a whole where any labeled subunit has an equal likelihood of dissociation. We observed similar results for GFP-actin (t1/2=26.1±6.2s, immobile fraction = 0.36±0.02) when stabilized with jasplakinolide (immobile fraction = 0.62±0.02) as well (Figure 5A).
Figure 4. Cytoskeletal structures containing fluorescently labeled subunits continue to persist following 25µM digitonin permeabilization.
A) Nuclear (mchr-lamin A) and cytosolic (mchr-vimentin) intermediate filaments are both display a large insoluble fraction following permeabilization, indicating extremely slow exchange with soluble pools. B) The dissociation kinetics of GFP-labeled α-tubulin were slowed by stabilization with 10 µM taxol, where relative intensity levels never reach 50% of initial intensity due to a large immobile fraction (0.75±0.02) as a result of stabilization. Data points represent the sample mean ± SEM. For A), nlamin= 38 cells and nvimentin= 42 cells. For B), ntubulin=22 cells and n+taxol= 26 cells.
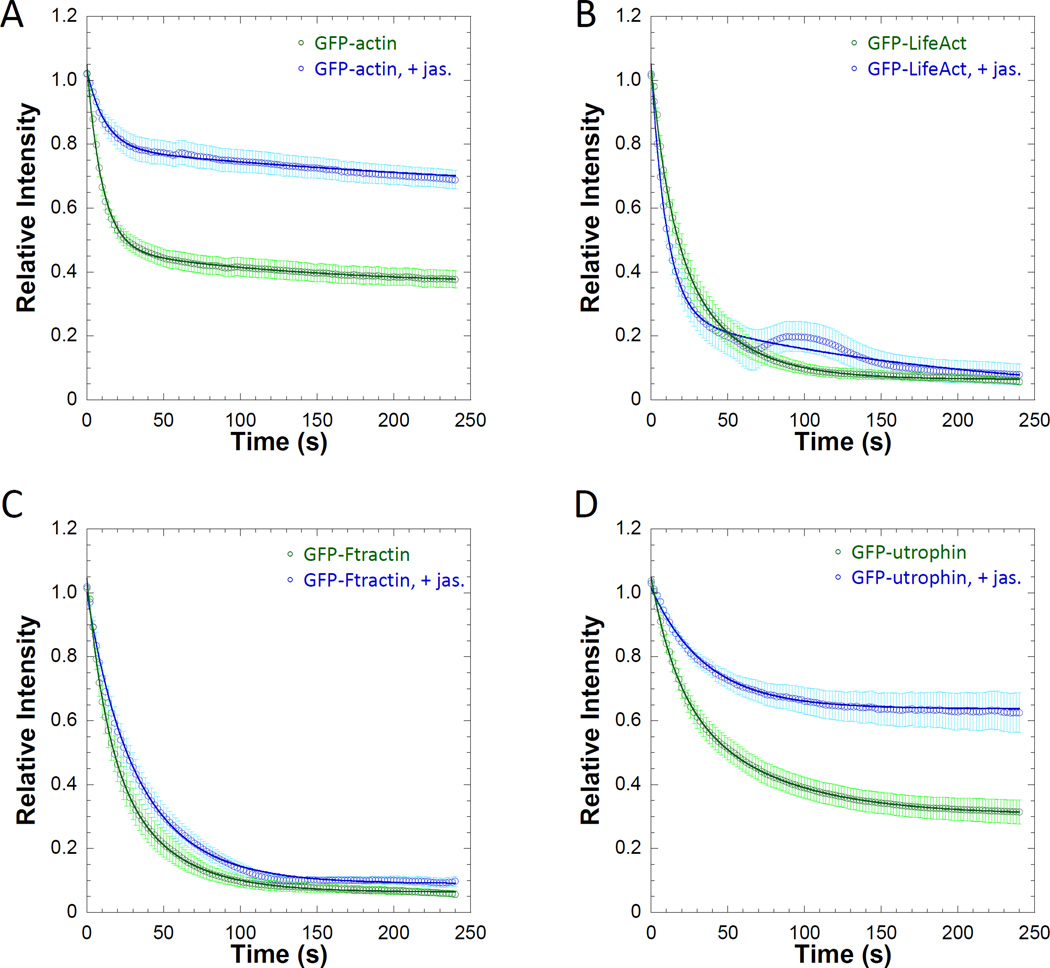
Figure 5. Jasplakinolide stabilizes the actin network and alters the PARF kinetics of actin-associated proteins.
Relative intensity plots of regions of interest of A) GFP-actin, B) GFP-Lifeact, C) GFP-F-tractin, and D) GFP-utrophin transfected cells in the presence or absence of 0.5µM jasplakinolide demonstrate that stabilization of filamentous actin decreases the apparent dissociation rate of subunits of the filament network as well as the dissociation rate of actin-binding proteins. The curves and calculated half-lives also show that GFP-Lifeact (t1/2= 17.8±3.9s versus t1/2 = 11.8±1.9s with jasplakinolide) and GFP-F-tractin (t1/2= 16.2±2.3s versus t1/2 = 26.1±4.7s with jasplakinolide) had similar fluorescence whether actin was stabilized or not. GFP-utrophin (t1/2= 54.2±10.5s) and GFP-actin (t1/2= 26.1±6.2s) PARF was slower than that of GFP-Lifeact and GFP-F-tractin in the absence jasplakinolide. Fluorescence loss was further slowed by actin stabilization as in both cases the relative fluorescence of the jasplakinolide-treated cells never reached 0.50 as demonstrated by the immobile fractions of GFP-actin (0.62±0.02) and GFP-utrophin (0.63±0.07). Data points on the plots represent the sample mean ± SEM. For A), nactin=36 cells and nact+jas=24 cells. For B), nLifeact=25 cells and nLife+jas=29 cells. For C), nF-tractin=42 cells and nF-tract+jas=32 cells. For D), nutrophin=29cells and nutro+jas=43 cells.
PARF analysis reveals differences in dissociation kinetics of commonly used F-actin labels
Altering the dynamics of the structure may also reveal characteristics of the labeled protein of interest. Lifeact [Riedl et al. 2008], F-tractin [Johnson and Schell 2009], and utrophin [Burkel et al. 2007] are commonly used labels for the actin network. Lifeact consists of the first 17 amino acids of actin-binding protein 140 (Abp140) from Saccharomyces cerevisiae, and is the smallest of the labels examined. The N-terminal 66 amino acids of Rattus Norvegicus inositol triphosphate make up the actin-binding domain of F-tractin, and utrophin labels contain the first 261 amino acids of the calponin-homology domains from the Homo sapiens protein. Previous reports indicate that these proteins may label different subsets of actin [Belin et al. 2015; Burkel et al. 2007; Lemieux et al. 2014], and that their overexpression may differentially impact other cellular activities [Spracklen et al. 2014]. PARF analysis (Figure 4B–D, Table 1) reveals that Lifeact (t1/2= 17.8±3.9s) and F-tractin (t1/2= 16.2±2.3s) had a 2 to 3-fold faster t1/2 compared to utrophin (t1/2= 52.4±10.5s), in agreement with previous reports where actin interactions were assayed via FRAP and showed faster recovery for Lifeact and F-tractin than for utrophin in actin structures with slow dynamics, such as stress fibers [Belin et al. 2015; Burkel et al. 2007; Johnson and Schell 2009]. In the presence of the actin stabilizer jasplakinolide, utrophin PARF kinetics were more greatly impacted than Lifeact or F-tractin (Figure 4B–D). In the presence of 0.5µM jasplakinolide, the utrophin immobile fraction rose to 0.63±0.07. Our observations with respect to utrophin dynamics in the presence of jasplakinolide are in agreement with a previous report by Burkel et al. indicating slower FRAP kinetics for labeled utropin and slower fluorescence decay for photo-activatable utrophin in the presence of jasplakinolide [Burkel et al. 2007]. Taken together these data lend more evidence to the growing body of literature suggesting that the common fluorescent actin labels are not equivalent and may not label actin in the same way. Labels with faster dynamics than the actin network itself may not be useful labels for representing the dynamics of the underlying structure [Belin et al. 2015].
PAAF analysis reveals differences in association kinetics of commonly used F-actin labels
Just as PARF reveals the apparent dissociation kinetics of a labeled protein in a transfected cell where permeabilization disrupts equilibrium, it may also be possible to observe association events under permissible experimental conditions. Inefficient transfection results in a population of cells with a mix of transfected and untransfected cells. Permeabilization allows for the mixing of the cytoplasm of transfected cells with the extracellular buffer, and the same is true of non-transfected cells. Fluorescent label released from transfected cells now has access to binding sites in untransfected cells made accessible by permeabilization. If a labeled protein of interest is at a high enough concentration in the extracellular fluid and has a sufficiently fast association rate to be visible within the timescale of the experiment, then association events may be visible in the untransfected cells following PARF.
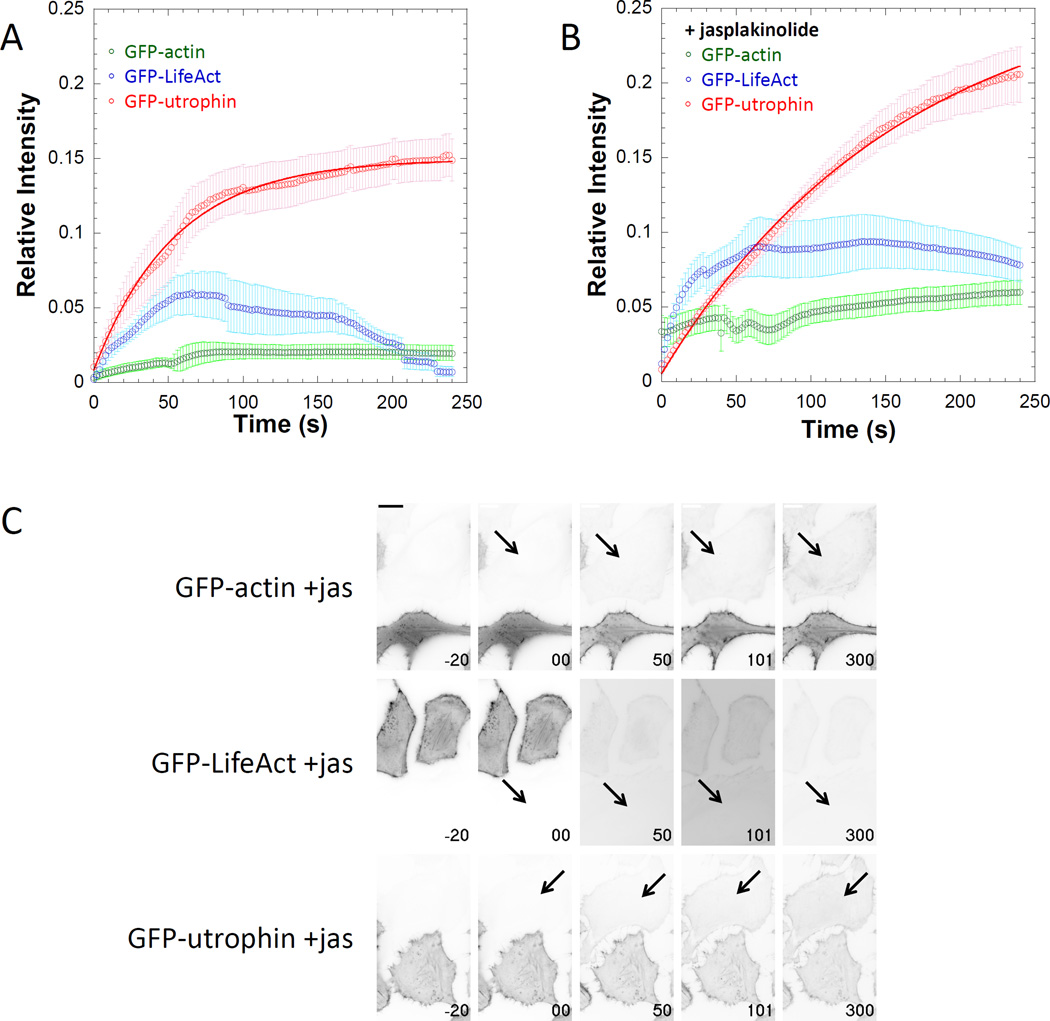
We chose to use this permeabilization associated accumulation of fluorescence (PAAF) as a qualitative comparison of relative apparent association rates between fluors interacting with the same structure. Because the contribution of dissociation events to the apparent on-rate would become more apparent over time (depending on the magnitude of the dissociation rate), it would be difficult to calculate an association rate from these data. Since both PARF and PAAF are relative measures we thought it important to calculate both measures on similar brightness scales. To do so, we calculated the PAAF increase in fluorescence relative to the average brightness of the transfected cells in the same field of view. Binding events were detectable for some of the actin labels (Figure 6A), and magnitude of those events was increased by jasplakinolide stabilization of the actin cytoskeleton (Figure 6B–C, Supplemental Movie 2). Although binding events were easily detectable for Lifeact and utrophin, utrophin PAAF was longer lived. This can be explained by the difference in the apparent off-rate kinetics for the two fluors. Since the t1/2 for utrophin dissociation is approximately 3-fold slower than for Lifeact, the PAAF fluorescence signal would persist longer for utrophin than for Lifeact. We suspect that for labels where PAAF association kinetics are rapid but measurable and PARF dissociation kinetics are rapid, the PAAF curve will rise initially and then fall. Additionally, evidence of rebinding events may become apparent as a “PAAF bump” in the PARF dissociation curves of the transfected cells, as is the case for the GFP-Lifeact in the presence of jasplakinolide (Figure 5B).
Figure 6. Under certain circumstances binding can become apparent after permeabilization.
Relative intensity plots of regions of interest of GFP-actin, GFP-Lifeact, and GFP-utrophin entering untransfected neighboring cells following digitonin permeabilization A) in the absence or B) in the presence of 0.5µM jasplakinolide demonstrate that the apparent on-rates are detectible for some fluorescence labels. C) Individual frames from movies demonstrating PAAF for GFP-actin, GFP-utrophin, and GFP-Lifeact in the presence of jasplakinolide with time in seconds on each frame (t = 0 is when permeabilization occurred) and scale bar is 20 µm. Data points represent the sample mean ± SEM. For panel A), nactin= 10 cells, nLifeact= 7 cells, and nutrophin= 10 cells. For panel B), nactin= 11 cells, nLifeact= 12 cells, and nutrophin= 22 cells. Note that the signal intensities for images in panel C are inverted from those used in the supplemental movies.
PARF analysis reveals differences in dissociation kinetics of GFP-VASP between focal adhesions and filopodia
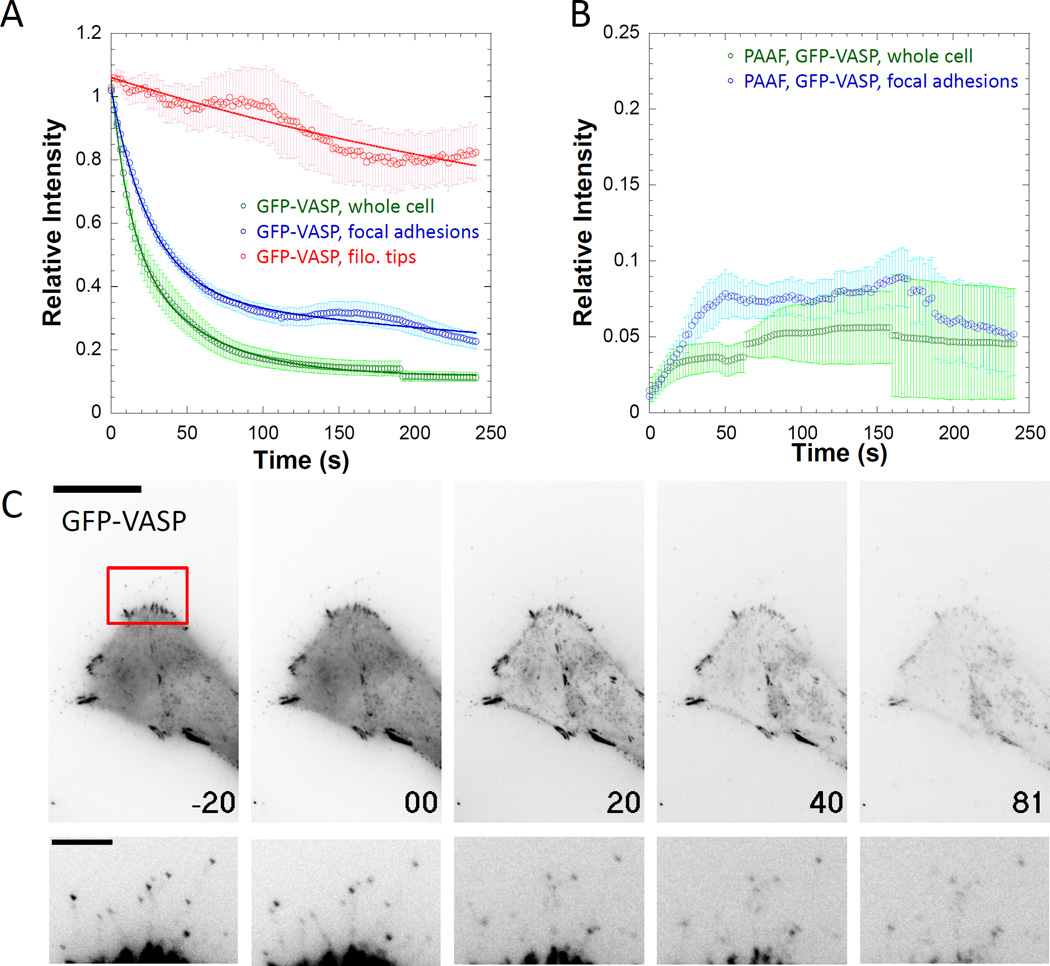
Just as these permeabilization approaches can be used to compare the binding kinetics of different proteins for the same intracellular structure, PARF and PAAF can be used to compare the binding kinetics for a particular protein to different intracellular structures. VASP localizes to multiple cellular structures including the leading edge of migrating cells, intercellular adhesions, focal adhesions, and filopodial tips [Han et al. 2002; Rottner et al. 1999; Svitkina et al. 2003; Vasioukhin et al. 2000]. As HeLa cells do not generate VASP-labeled lamellopodia or intracellular adhesions, we chose to analyze the kinetic properties of VASP within focal adhesions and at filopodial tips. Steady-state analysis of FRAP kinetics indicate that VASP exchange rates differ depending on the structure being assayed. Applewhite and colleagues demonstrated that in B16F1 cells VASP localized to filopodial tips showed little to no exchange over the approximately 2-minute time course of their experiment, while leading-edge VASP showed a half-time of recovery of approximately 10 seconds [Applewhite et al. 2007]. We demonstrated in HeLa cells that VASP recovery at the filopodial tip was minimal, while exchange in focal adhesions showed a half-time of recovery of approximately 10–20 seconds [Quintero et al. 2003]; Hoffman and colleagues also demonstrated similar t1/2 for VASP in focal adhesions in REF52 cells [Hoffmann et al. 2014]. Differential steady-state exchange rates would suggest faster PARF dissociation kinetics for focal adhesions compared to filopodial tips. Our PARF analysis (Figure 7A, Supplemental Movie 3, Supplemental Movie 4) indicated slow dissociation of VASP the filopodial tip (immobile fraction = 0.71±0.07) while the apparent dissociation from focal adhesions was greater (t1/2= 39.2±3.3s, immobile fraction = 0.21±0.03). It is worth noting that the intensity of the VASP signal at filopodial tips was much lower than that of focal adhesions within the same cells, leading to more variability in measurements as the signal-to-noise ratio of the filopodial tip measurements was smaller than that of the focal adhesion measurements. Additionally, rapid steady-state exchange kinetics at focal adhesions suggested a fast apparent on-rate. PAAF events were visible for VASP interacting with focal adhesions, but their magnitude was less than 10% of the initial brightness of neighboring cells, likely due the rapid apparent dissociation rates observed in the transfected cells (Figure 7B). We were unable to locate PAAF events at filopodial tips which could be due to slower association kinetics than we could detect, a high overall signal-to-noise ratio for filopodial tip measurements making it difficult to detect association events, or a combination of both. The dissociation curve for GFP-VASP at focal adhesions displayed less pronounced “PAAF bump” than for Lifeact with jasplakinolide (Figure 7A). Taken together, these data suggest that the association and dissociation kinetics for a protein can vary depending on the particular cellular structure with which it is interacting, and that these differences can be revealed by PARF and PAAF approaches.
Figure 7. PARF kinetics for GFP-VASP vary depending on the subcellular localization of labeled protein.
A) Relative intensity plots of regions of interest of GFP-VASP in the whole cell, at focal adhesions, or at filopodial tips indicate that the apparent dissociation kinetics depend on the cellular structure being observed, as dissociation from focal adhesions (t1/2=39.2±3.3s), was slower than dissociation from the whole cell (t1/2 =20.7±5.0s), but faster than dissociation from filopodial tips (immobile fraction = 0.52±0.23). B) PAAF kinetics for GFP-VASP at focal adhesions are rare but observable. C) Individual frames from movies demonstrating PARF for GFP-VASP with time in seconds on each frame (t = 0 is when permeabilization occurred) and scale bar is 20 µm. The second row of images is an inset from the region indicated by the red box scaled to make filopodial tips visible; scale bar is 5µm. Data points represent the sample mean ± SEM. For panel A), ncell=23 cells, nfocal adh.= 142 adhesions from 23 cells, and nfilopodia= 31 filopodial tips from 31 cells. For panel B), n = 16 focal adhesions from 4 cells. Note that the signal intensities for images in panel C are inverted from those used in the supplemental movies.
PARF is a flexible and useful technique capable of addressing a subset of protein interaction questions
Here we present a novel method for investigating the kinetic parameters underlying biochemical reactions within a cellular context. This “in vivtro” method [Narang et al. 1986] falls somewhere between truly in vitro methods where all the constituents of a system are known, and in vivo methods where the system being assayed is living and consists solely of endogenous components. Similar to Xenopus egg extract systems, it is clear that some cellular activities will be disrupted by establishment of the assay system while other activities, such as nuclear import [Turgay et al. 2010], calcium signaling [Subedi et al. 2014], cardiomyocyte contractility [Altschuld et al. 1985], and platelet function[Lineberger et al. 1989], remain intact. Although PARF may be applicable to the study of a specific subset of cellular processes, the technical capabilities required for PARF are less than those required for other quantitative imaging techniques (such as photoactivation or FRAP), nor does PARF require the technical expertise in protein expression and purification necessary for reconstitution approaches. It does require an understanding of basic chemical kinetics and the limitations of the model system being assayed. We were able to generate useful data with an inverted wide-field epifluorescence microscope and without having to express of purify multiple components of an intracellular complex. The epifluorescence illumination was from a mixed gas lamp with a computer-controlled shutter, and images were acquired digitally. Careful design of such a work station, taking advantage of the newer generations of affordable digital cameras and open-source microscopy control software [Edelstein et al. 2010] are not nearly as expensive as the laser-illuminated confocal systems often required for FRAP and/or photoactivation analysis.
As PARF analysis disrupts the stoichiometry of intracellular processes that may impact the structure being observed, such analyses may be better suited for the investigation of interactions with particular cellular structures less likely to be disrupted by plasma membrane permeabilization. PARF may be particularly well-suited for characterizing the binding interactions of proteins associated with long-lived intracellular structures, which may include some cytoskeletal assemblies. Additionally, stabilizing the structures using agents that prevent depolymerization may be applicable, assuming that the structure-stabilizing agent does not alter the binding kinetics of the protein of interest. In other experimental approaches, stabilizing drugs have been used both in vivo [Shannon et al. 2005] and in vitro [Kron and Spudich 1986] to gain detailed understanding of the functions of cytoskeletal assemblies. Examining the binding interactions of proteins that localize to the cytosolic face of internal membranes may also be appropriate [Hawthorne et al. 2016]. It may also be possible to minimize changes to the concentrations of biomolecules of interest through the careful selection of the composition of the extracellular buffer, such as carrying out the experiments in Xenopus egg extract [Marino et al. 2014].
Like other techniques designed to investigate the kinetic properties of macromolecular interactions, proper application of PARF depends on a number of assumptions holding true for interpretation to be valid [Snapp et al. 2003]. For one of the simplest situations where PARF is a valid approach we assume that: a soluble pool of label exists in equilibrium with a bound pool, that the soluble pool is diluted quickly upon permeabilization, and that all molecules in the bound pool are described by the same dissociation kinetics. Any of these assumptions may be false, but can be accounted for by carefully observing the data for indications that the output does not completely fit the model, and then modifying the exponential fitting used to better fit the actual cellular situation. Suppose that two subpopulations of bound molecules exist within the bound pool, each with different dissociation kinetics—this situation can also be described by a multi-exponential fit. The extreme case also exists where a fraction of the bound pool is immobile and incapable of dissociation after permeabilization, which we observed for multiple fluors, and accounted for in a manner similar to how immobile fractions are accounted for in FRAP analysis [Lippincott-Schwartz et al. 2001]. It may be the case that over a longer time course, the mobility of this “immobile” fraction may become apparent.
Like with other approaches that provide quantitative understanding of biological processes, the usefulness of PARF is primarily limited by the user’s depth of understanding of an experimental system, their creativity, and their resourcefulness. Variations on this approach might include a system where the extracellular buffer is something more akin to cytosol so that permeabilization is less disruptive to the stoichiometry of the intracellular machinery. It may also be possible to observe association events under equilibrium conditions if said extracellular buffer was similar to cytosol, contained a labeled protein of interest at a known concentration, and the cells being permeabilized did not contain labeled protein. With careful a thorough understanding of the limitations of the technique and the system, careful optimization, and appropriate controls, an investigator is well positioned to make sense of unexpected results and can have greater confidence in their data interpretation.
Materials and methods
Plasmids
mcherry, eGFP, eGFP-α-tubulin, eGFP-βactin, and eGFP-Mito were obtained from Clontech. eGFP-VASP [Svitkina et al. 2003] was a gift from the Borisy lab. eGFP-Utrophin [Burkel et al. 2007] was a gift from the Bement lab. eGFP-Lifeact [Riedl et al. 2008] and eGFP-F-tractin [Johnson and Schell 2009] were gifts from the Baum lab. EMTB-3xGFP [Miller and Bement 2009] was a gift from William Bement (Addgene plasmid #26741). PM-GFP [Teruel et al. 1999] was a gift from Tobias Meyer (Addgene plasmid # 21213). EGFP-H2B-6 (Addgene plasmid # 56436), mcherry-LaminA-C-18 (Addgene plasmid # 55068), and mcherry-Vimentin-7 (Addgene plasmid # 55156) [Shaner et al. 2007] were a gift from Michael Davidson. pcDNA3-HtrA2-EGFP [Martins et al. 2002] was a gift from L. Miguel Martins (Addgene plasmid # 14121). For simplicity, all text references referring to “GFP” are describing enhanced-GFP constructs.
Cell culture and transfection
HeLa cells [Scherer et al. 1953] were maintained in DMEM high-glucose media supplemented with 10% FBS, 50 units/mL penicillin, and 50µg/mL streptomycin (growth media) at 37°C in a humidified atmosphere of 5% CO2. Cells were passaged using 0.25% trypsin-EDTA. For imaging, HeLa cells were grown on acid-washed, 22mm #1.5 glass coverslips. Cells were transfected with Lipofectamine 2000 (Life Technologies) using the manufacturer’s protocol. DNA was diluted to 6.7 µg/ml or less in Optimem without serum or antibiotics in a final volume of 150µL. 4µL of Lipofectamine was diluted into 150µL of Optimem without serum or antibiotics and then mixed with the DNA dilution. Complexes were allowed to form for 5 minutes at room temperature. The entirety of the DNA/reagent mix was added drop-wise to a well of a 6-well plate. Cells were imaged and permeabilized 20–30 hours after transfection.
Microscopy and data acquisition
Cover slips were assembled in open-top Rose chambers [Rose et al. 1958] with 300µL KHM buffer (20 mM HEPES, pH 7.4, 110 mM potassium acetate, and 2 mM MgCl2) [Lorenz et al. 2008]. Images were acquired using an Olympus IX-83 microscope outfitted with a PLAN APON 60×/1.42NA DIC objective, an EXFO mixed gas light source, Sutter filter wheels and shutters, Sedat Quad filterset (Chroma), a Hamamatsu ORCA-Flash 4.0 V2 sCMOS camera, and Metamorph imaging software. Time-lapse images were captured at an interval of one frame every two seconds with a constant exposure time for 200 frames. For PARF, cells were permeabilized after the first 20 frames by addition of digitonin to a final concentration of 25 µM. In experiments where cells were treated with taxol or jasplakinolide, the drugs were included in the KHM buffer before permeabilization. For photofading due to acquisition (PDA) control, cells were imaged under the same conditions but without digitonin treatment.
Scanning Electron Microscopy of fixed time point digitonin-treated cells
HeLa cells were plated on acid-washed, 18mm #1.5 coverslips and allowed to adhere overnight. One day following plating, the cells were washed with KHM buffer, and then treated with 25µM digitonin dissolved in KHM buffer for the appropriate amount of time. Cells were immediately fixed by adding an equal volume of PBS with 5% gluteraldehyde to the sample well and allowed to fix at a final concentration of 2.5% glutaraldehyde for approximately 20 minutes. The coverslips were then washed in PBS, and then in increasing concentrations of ethanol for 5 minutes at each step (30%, 50%, 70%, 90%, and 100%). Following ethanol washes, the cells were critical-point dried and sputter coated as described previously [Applewhite et al. 2015]. Cells were imaged using a JEOL 6360 LV scanning electron microscope.
Immunofluorescence of fixed time point digitonin-treated cells
HeLa cells were plated on acid-washed, 22mm #1.5 coverslips and allowed to adhere overnight. One day following plating, the cells were washed with KHM buffer, and then treated with 25µM digitonin dissolved in KHM buffer for the appropriate amount of time. Cells were immediately fixed by adding an equal volume of PBS with 8% paraformaldehyde to the sample well and allowed to fix at a final concentration of 4% paraformaldehyde for approximately 10 minutes. The coverslips were then washed in PBS, permeabilized with 0.5% TritonX100 in PBS for 5 minutes. Cells were blocked in PBS with 5% donkey serum and 0.1% TritonX100 for 1h. Primary antibodies, 0.2 µg/ml α-tubulin DM1A (Sigma) or 1µg/ml α-VASP (BD Biosciences) were diluted in blocking reagent and incubated with the cells for 30 minutes. Following 4 five-minute PBS wash steps, ALEXA488-conjugated donkey α-mouse secondary antibody (Jackson Immunochemicals) was diluted to 0.2µg/ml in blocking reagent containing 6.6nM ALEXA568-phalloidin (Life Technologies) and 75nM DAPI (Life Technologies), and incubated on with the cells for 30 minutes. After 4 five-minute PBS washes, the coverslips were mounted in PBS containing 0.5% N-propyl gallate and 80% glycerol.
Quantitative Image Analysis
Using FIJI [Schindelin et al. 2012] image analysis software, regions of interest (ROI) were selected for the structure with bound labeled-protein to measure mean intensity. Measurements underwent corrections prior to analysis. First, regions of background outside of the cells were selected and value of average relative background intensity at each time point was subtracted from the ROI mean intensity for each time point. Second, the frame at which the ROI was permeabilized based on the visual cues of structural changes noted during imaging was set to t = 0. For all subsequent frames, intensity relative to t = 0 was calculated. Third, the rate of photofading due to acquisition (PDA) was calculated from unpermeabilized cells and used to correct permeabilized samples for PDA [Applewhite et al. 2007]. For the PDA control the relative mean intensity versus time was curve-fit with the single exponential, y = a*e−bt where y is the mean relative intensity remaining, t is time elapsed. Correction curves were calculated for each individual fluorescently-tagged protein on each image-acquisition day. PARF trials were corrected for PDA by dividing the relative intensity of the ROI by the fraction of remaining fluorescence intensity calculated for that particular time-point and fluor on that day.
Data from multiple experimental trials were averaged together, and exponential fits for analysis were performed using Kaleidagraph. PARF fluorescence reduction data were then fit to y = a*e−bt + c*e−dt + f (double exponential with immobile fraction). PAAF fluorescence accumulation data were then fit to y = a*(1 − e−bt) + c, where the maximum relative intensity of 1 was set to the average fluorescence intensity of all the transfected cells within the same field of view at a time point prior to permeabilization.
Measured data, such as data points on curves, are expressed as mean ± standard error of the mean (SEM). Error estimates for values calculated from curve fits (such as t1/2 and immobile fractions) were determined by generating curve fits of the mean + SEM and mean − SEM, and reporting the error as the difference between the value calculated from the mean fit and the value calculated from the SEM-adjusted fit.
All images were prepared for publication using FIJI and Metamorph. Images in figures and data videos were not corrected for photofading due to acquisition, and image brightness was scaled identically for all planes within a time series.
Supplementary Material
HeLa cells transfected with GFP (left), GFP-Mito (center), or PM-GFP (right) were permeabilized with 25µM digitonin at t =0. In the GFP cell, cytoplasmic signal dissipated more quickly than nuclear fluorescence, while mitochondria-associated fluorescence and plasma membrane-associated fluorescence decreased slowly from the GFP-Mito and PM-GFP transfected cells, respectively. Time is indicated in seconds, scale bar represents 20µm. Movies represent raw data not corrected for photofading due to acquisition.
HeLa cells transfected with GFP-actin, GFP-Lifeact, or GFP-utrophin permeabilized with 25µM digitonin in the presence of 0.5µM jasplakinolide at t =0. Fluorescence decreased in the transfected cells, while it increased in untransfected cells as a result of GFP-tagged protein release into the permeabilization media and subsequent binding to actin structures in untransfected, permeabilized cells. Note that GFP-Lifeact signal dissipates quickly from untransfected cells while GFP-utrophin does not. Movies represent raw data not corrected for photofading due to acquisition. Arrows indicate untransfected cells. Time is indicated in seconds, scale bar represents 20µm.
HeLa cells transfected with GFP-VASP were permeabilized with 25µM digitonin at t =0. As determined by PARF analysis, cytoplasmic signal dissipated more quickly than focal adhesion fluorescence. Time is indicated in seconds, scale bar represents 20µm. Movies represent raw data not corrected for photofading due to acquisition. Regions of interest used in focal adhesion PARF calculation indicated in red.
HeLa cells transfected with GFP-VASP were permeabilized with 25µM digitonin at t =0. As determined by PARF analysis, filopodial tip fluorescence dissipated more slowly than focal adhesion fluorescence. Time is indicated in seconds, scale bar represents 5µm. Movies represent raw data not corrected for photofading due to acquisition, but scaled differently than Supplemental Movie 3 to reveal the dimmer relative fluorescence of filopodial tips.
Acknowledgments
This work would have been impossible without the teaching and research environment fostered by the “Integrated Quantitative Sciences” undergraduate curriculum at University of Richmond, which was funded by the Howard Hughes Medical Institute Undergraduate Science Education Award (HHMI 52007567). PPS and JLH were supported in this work in part through the HHMI award, and the Robert F. Smart Award in Biology. OAQ was supported by a grant from the National Cancer Institute at the NIH (K01CA160667) and by funding from the University of Richmond School of Arts and Sciences. We would like to thank Michael Kerckhove, Derek Applewhite, Uri Manor, and Rebecca Adikes for critical reading and discussions related to this manuscript. We also thank Melisa Kortan for early work in piloting the PARF system, Edward Salmon for the gift of Rose chambers, and Sarah Friday for her artful rendition of PARF and PAAF (Figure 1). Hypotheses and experiments in this study were conceived by OAQ.
Footnotes
Author contributions
Experiments were performed by PPS, JLH, CAD, and OAQ. The data analysis workflow was conceived by PPS and JLH. The manuscript was written by PPS, JLH, CAD, and OAQ.
References
- Altschuld RA, Wenger WC, Lamka KG, Kindig OR, Capen CC, Mizuhira V, Vander Heide RS, Brierley GP. Structural and functional properties of adult rat heart myocytes lysed with digitonin. J Biol Chem. 1985;260(26):14325–14334. [PubMed] [Google Scholar]
- Applewhite DA, Barzik M, Kojima S, Svitkina TM, Gertler FB, Borisy GG. Ena/VASP proteins have an anti-capping independent function in filopodia formation. Mol Biol Cell. 2007;18(7):2579–2591. doi: 10.1091/mbc.E06-11-0990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Applewhite DA, Davis CA, Griffis ER, Quintero OA. Imaging of the cytoskeleton using live and fixed Drosophila tissue culture cells. In: Gavin RH, editor. Cytoskeleton methods and protocols. third. Springer; 2015. p in press. [DOI] [PubMed] [Google Scholar]
- Axelrod D, Ravdin P, Koppel DE, Schlessinger J, Webb WW, Elson EL, Podleski TR. Lateral motion of fluorescently labeled acetylcholine receptors in membranes of developing muscle fibers. Proc Natl Acad Sci U S A. 1976;73(12):4594–4598. doi: 10.1073/pnas.73.12.4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin BJ, Goins LM, Mullins RD. Comparative analysis of tools for live cell imaging of actin network architecture. BioArchitecture. 2015;4(6):1–14. doi: 10.1080/19490992.2014.1047714. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom GS, Vallee RB. Association of microtubule-associated protein 2 (MAP 2) with microtubules and intermediate filaments in cultured brain cells. J Cell Biol. 1983;96(6):1523–1531. doi: 10.1083/jcb.96.6.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkel BM, von Dassow G, Bement WM. Versatile fluorescent probes for actin filaments based on the actin-binding domain of utrophin. Cell Motil Cytoskeleton. 2007;64(11):822–832. doi: 10.1002/cm.20226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleland WW. 3 Steady-State Kinetics. The enzymes (Amsterdam) 1990;19:99–158. [Google Scholar]
- Collins TJ, Bootman MD. Mitochondria are morphologically heterogeneous within cells. J Exp Biol. 2003;206(Pt 12):1993–2000. doi: 10.1242/jeb.00244. [DOI] [PubMed] [Google Scholar]
- Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173(1 Spec No):33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- De La Cruz EM, Ostap EM. Kinetic and equilibrium analysis of the myosin ATPase. Methods Enzymol. 2009;455:157–192. doi: 10.1016/S0076-6879(08)04206-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Computer Control of Microscopes Using µManager. John Wiley & Sons, Inc; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenberg J, Lippincott-Schwartz J. Dynamics and mobility of nuclear envelope proteins in interphase and mitotic cells revealed by green fluorescent protein chimeras. Methods. 1999;19(3):362–372. doi: 10.1006/meth.1999.0872. [DOI] [PubMed] [Google Scholar]
- Faire K, Waterman-Storer CM, Gruber D, Masson D, Salmon ED, Bulinski JC. E-MAP-115 (ensconsin) associates dynamically with microtubules in vivo and is not a physiological modulator of microtubule dynamics. J Cell Sci. 1999;112(Pt 23):4243–4255. doi: 10.1242/jcs.112.23.4243. [DOI] [PubMed] [Google Scholar]
- Fiskum G, Craig SW, Decker GL, Lehninger AL. The cytoskeleton of digitonin-treated rat hepatocytes. Proc Natl Acad Sci U S A. 1980;77(6):3430–3434. doi: 10.1073/pnas.77.6.3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han YH, Chung CY, Wessels D, Stephens S, Titus MA, Soll DR, Firtel RA. Requirement of a vasodilator-stimulated phosphoprotein family member for cell adhesion, the formation of filopodia, and chemotaxis in dictyostelium. J Biol Chem. 2002;277(51):49877–49887. doi: 10.1074/jbc.M209107200. [DOI] [PubMed] [Google Scholar]
- Hawthorne JL, Mehta PR, Singh PP, Wong N, Quintero OA. Positively charged residues within the MYO19 MyMOMA domain are essential for proper localization of MYO19 to the mitochondrial outer membrane. Cytoskeleton. 2016 doi: 10.1002/cm.21305. submitted and under review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann JE, Fermin Y, Stricker RL, Ickstadt K, Zamir E. Symmetric exchange of multi-protein building blocks between stationary focal adhesions and the cytosol. Elife. 2014;3:e02257. doi: 10.7554/eLife.02257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson HW, Schell MJ. Neuronal IP3 3-kinase is an F-actin-bundling protein: role in dendritic targeting and regulation of spine morphology. Mol Biol Cell. 2009;20(24):5166–5180. doi: 10.1091/mbc.E09-01-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KA. 1 Transient-State Kinetic Analysis of Enzyme Reaction Pathways. The enzymes (Amsterdam) 1992;20:1–61. [Google Scholar]
- Kron SJ, Spudich JA. Fluorescent actin filaments move on myosin fixed to a glass surface. Proc Natl Acad Sci U S A. 1986;83(17):6272–6276. doi: 10.1073/pnas.83.17.6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux MG, Janzen D, Hwang R, Roldan J, Jarchum I, Knecht DA. Visualization of the actin cytoskeleton: different F-actin-binding probes tell different stories. Cytoskeleton (Hoboken) 2014;71(3):157–169. doi: 10.1002/cm.21160. [DOI] [PubMed] [Google Scholar]
- Lineberger B, Dawicki DD, Agarwal KC, Kessimian N, Steiner M. Permeabilization of platelets: an investigation of biochemical, ultrastructural and functional aspects. Biochim Biophys Acta. 1989;1012(1):36–45. doi: 10.1016/0167-4889(89)90008-6. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Altan-Bonnet N, Patterson GH. Photobleaching and photoactivation: following protein dynamics in living cells. Nat Cell Biol Suppl. 2003:S7–S14. [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Snapp E, Kenworthy A. Studying protein dynamics in living cells. Nat Rev Mol Cell Biol. 2001;2(6):444–456. doi: 10.1038/35073068. [DOI] [PubMed] [Google Scholar]
- Lorenz H, Hailey DW, Lippincott-Schwartz J. Addressing membrane protein topology using the fluorescence protease protection (FPP) assay. Methods Mol Biol. 2008;440:227–233. doi: 10.1007/978-1-59745-178-9_17. [DOI] [PubMed] [Google Scholar]
- Manor U, Bartholomew S, Golani G, Christenson E, Kozlov M, Higgs H, Spudich J, Lippincott-Schwartz J. A mitochondria-anchored isoform of the actin-nucleating Spire protein regulates mitochondrial division. eLife. 2015 doi: 10.7554/eLife.08828. 10.7554/eLife.08828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino J, Champion L, Wandke C, Horvath P, Mayr MI, Kutay U. An in vitro system to study nuclear envelope breakdown. Methods Cell Biol. 2014;122:255–276. doi: 10.1016/B978-0-12-417160-2.00012-6. [DOI] [PubMed] [Google Scholar]
- Martins LM, Iaccarino I, Tenev T, Gschmeissner S, Totty NF, Lemoine NR, Savopoulos J, Gray CW, Creasy CL, Dingwall C, et al. The serine protease Omi/HtrA2 regulates apoptosis by binding XIAP through a reaper-like motif. J Biol Chem. 2002;277(1):439–444. doi: 10.1074/jbc.M109784200. [DOI] [PubMed] [Google Scholar]
- Miller AL, Bement WM. Regulation of cytokinesis by Rho GTPase flux. Nat Cell Biol. 2009;11(1):71–77. doi: 10.1038/ncb1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir RD, Yoon M, Khuon S, Goldman RD. Nuclear lamins A and B1: different pathways of assembly during nuclear envelope formation in living cells. J Cell Biol. 2000;151(6):1155–1168. doi: 10.1083/jcb.151.6.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy D, Davidson MW. Fundamentals of Light Microscopy and Electronic Imaging. 2nd. Hoboken, NJ: Wiley-Blackwell; 2012a. FLuroescence Imaging of Dynamic Molecular Processes; pp. 233–263. 2nd ed. [Google Scholar]
- Murphy D, Davidson MW. Fundamentals of Light Microscopy and Electronic Imaging. 2nd. Hoboken, NJ: Wiley-Blackwell; 2012b. p. 522. [Google Scholar]
- Narang SA, Dubuc G, Yao FL, Michniewicz JJ. "In vivtro" method of assembling a synthetic gene. Biochem Biophys Res Commun. 1986;134(1):407–411. doi: 10.1016/0006-291x(86)90578-4. [DOI] [PubMed] [Google Scholar]
- Pollard TD. The cytoskeleton, cellular motility and the reductionist agenda. Nature. 2003;422(6933):741–745. doi: 10.1038/nature01598. [DOI] [PubMed] [Google Scholar]
- Pollard TD. A guide to simple and informative binding assays. Mol Biol Cell. 2010;21(23):4061–4067. doi: 10.1091/mbc.E10-08-0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, De La Cruz EM. Take advantage of time in your experiments: a guide to simple, informative kinetics assays. Mol Biol Cell. 2013;24(8):1103–1110. doi: 10.1091/mbc.E13-01-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero OA, DiVito MM, Adikes RC, Kortan MB, Case LB, Lier AJ, Panaretos NS, Slater SQ, Rengarajan M, Feliu M, et al. Human Myo19 is a novel myosin that associates with mitochondria. Curr Biol. 2009;19(23):2008–2013. doi: 10.1016/j.cub.2009.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero OA, Svitkina TM, Chaga OY, Bhaskar A, Borisy GG, Cheney RE. Dynamics of myosin-X (Myo10) and VASP at the filopodial tip. Mol Biol Cell. 2003;14s:1010. [Google Scholar]
- Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, et al. Lifeact: a versatile marker to visualize F-actin. Nat Methods. 2008;5(7):605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose GG, Pomerat CM, Shindler TO, Trunnell JB. A cellophane-strip technique for culturing tissue in multipurpose culture chambers. J Biophys Biochem Cytol. 1958;4(6):761–764. doi: 10.1083/jcb.4.6.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottner K, Behrendt B, Small JV, Wehland J. VASP dynamics during lamellipodia protrusion. Nat Cell Biol. 1999;1(5):321–322. doi: 10.1038/13040. [DOI] [PubMed] [Google Scholar]
- Scherer WF, Syverton JT, Gey GO. Studies on the propagation in vitro of poliomyelitis viruses. IV. Viral multiplication in a stable strain of human malignant epithelial cells (strain HeLa) derived from an epidermoid carcinoma of the cervix. J Exp Med. 1953;97(5):695–710. doi: 10.1084/jem.97.5.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz I. Permeabilizing cells: some methods and applications for the study of intracellular processes. Methods Enzymol. 1990;192:280–300. doi: 10.1016/0076-6879(90)92077-q. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Patterson GH, Davidson MW. Advances in fluorescent protein technology. J Cell Sci. 2007;120(Pt 24):4247–4260. doi: 10.1242/jcs.005801. [DOI] [PubMed] [Google Scholar]
- Shannon KB, Canman JC, Ben Moree C, Tirnauer JS, Salmon ED. Taxol-stabilized microtubules can position the cytokinetic furrow in mammalian cells. Mol Biol Cell. 2005;16(9):4423–4436. doi: 10.1091/mbc.E04-11-0974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolen JE, Stoehr SJ, Boxer LA. Human neutrophils permeabilized with digitonin respond with lysosomal enzyme release when exposed to micromolar levels of free calcium. Biochim Biophys Acta. 1986;886(1):1–17. doi: 10.1016/0167-4889(86)90205-3. [DOI] [PubMed] [Google Scholar]
- Snapp EL, Altan N, Lippincott-Schwartz J. Measuring protein mobility by photobleaching GFP chimeras in living cells. Current Protocols in Cell Biology [Electronic Resource] 2003;Chapter 21(Unit 21):1. doi: 10.1002/0471143030.cb2101s19. [DOI] [PubMed] [Google Scholar]
- Spracklen AJ, Fagan TN, Lovander KE, Tootle TL. The pros and cons of common actin labeling tools for visualizing actin dynamics during Drosophila oogenesis. Dev Biol. 2014;393(2):209–226. doi: 10.1016/j.ydbio.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subedi KP, Paudel O, Sham JS. Detection of differentially regulated subsarcolemmal calcium signals activated by vasoactive agonists in rat pulmonary artery smooth muscle cells. Am J Physiol Cell Physiol. 2014;306(7):C659–C669. doi: 10.1152/ajpcell.00341.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina TM, Bulanova EA, Chaga OY, Vignjevic DM, Kojima S, Vasiliev JM, Borisy GG. Mechanism of filopodia initiation by reorganization of a dendritic network. J Cell Biol. 2003;160(3):409–421. doi: 10.1083/jcb.200210174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teruel MN, Blanpied TA, Shen K, Augustine GJ, Meyer T. A versatile microporation technique for the transfection of cultured CNS neurons. J Neurosci Methods. 1999;93(1):37–48. doi: 10.1016/s0165-0270(99)00112-0. [DOI] [PubMed] [Google Scholar]
- Turgay Y, Ungricht R, Rothballer A, Kiss A, Csucs G, Horvath P, Kutay U. A classical NLS and the SUN domain contribute to the targeting of SUN2 to the inner nuclear membrane. EMBO J. 2010;29(14):2262–2275. doi: 10.1038/emboj.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasioukhin V, Bauer C, Yin M, Fuchs E. Directed actin polymerization is the driving force for epithelial cell-cell adhesion. Cell. 2000;100(2):209–219. doi: 10.1016/s0092-8674(00)81559-7. [DOI] [PubMed] [Google Scholar]
- Wadsworth P, Salmon ED. Analysis of the treadmilling model during metaphase of mitosis using fluorescence redistribution after photobleaching. J Cell Biol. 1986;102(3):1032–1038. doi: 10.1083/jcb.102.3.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Brattain MG. The maximal size of protein to diffuse through the nuclear pore is larger than 60kDa. FEBS Lett. 2007;581(17):3164–3170. doi: 10.1016/j.febslet.2007.05.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YL. Exchange of actin subunits at the leading edge of living fibroblasts: possible role of treadmilling. J Cell Biol. 1985;101(2):597–602. doi: 10.1083/jcb.101.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J, Stelzer E. Photobleaching GFP reveals protein dynamics inside live cells. Trends in Cell Biology. 1999;9(2):61–65. doi: 10.1016/s0962-8924(98)01433-0. [DOI] [PubMed] [Google Scholar]
- Yoon M, Moir RD, Prahlad V, Goldman RD. Motile properties of vimentin intermediate filament networks in living cells. J Cell Biol. 1998;143(1):147–157. doi: 10.1083/jcb.143.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HeLa cells transfected with GFP (left), GFP-Mito (center), or PM-GFP (right) were permeabilized with 25µM digitonin at t =0. In the GFP cell, cytoplasmic signal dissipated more quickly than nuclear fluorescence, while mitochondria-associated fluorescence and plasma membrane-associated fluorescence decreased slowly from the GFP-Mito and PM-GFP transfected cells, respectively. Time is indicated in seconds, scale bar represents 20µm. Movies represent raw data not corrected for photofading due to acquisition.
HeLa cells transfected with GFP-actin, GFP-Lifeact, or GFP-utrophin permeabilized with 25µM digitonin in the presence of 0.5µM jasplakinolide at t =0. Fluorescence decreased in the transfected cells, while it increased in untransfected cells as a result of GFP-tagged protein release into the permeabilization media and subsequent binding to actin structures in untransfected, permeabilized cells. Note that GFP-Lifeact signal dissipates quickly from untransfected cells while GFP-utrophin does not. Movies represent raw data not corrected for photofading due to acquisition. Arrows indicate untransfected cells. Time is indicated in seconds, scale bar represents 20µm.
HeLa cells transfected with GFP-VASP were permeabilized with 25µM digitonin at t =0. As determined by PARF analysis, cytoplasmic signal dissipated more quickly than focal adhesion fluorescence. Time is indicated in seconds, scale bar represents 20µm. Movies represent raw data not corrected for photofading due to acquisition. Regions of interest used in focal adhesion PARF calculation indicated in red.
HeLa cells transfected with GFP-VASP were permeabilized with 25µM digitonin at t =0. As determined by PARF analysis, filopodial tip fluorescence dissipated more slowly than focal adhesion fluorescence. Time is indicated in seconds, scale bar represents 5µm. Movies represent raw data not corrected for photofading due to acquisition, but scaled differently than Supplemental Movie 3 to reveal the dimmer relative fluorescence of filopodial tips.