Abstract
Interferon Regulatory Factor (IRF)3 is a crucial transcription factor during innate immune responses. Here we show IRF3 also has a role in adaptive T cell immune responses. Expression of IFN-γ, IL-17, and Granzyme B (GrB) during in vitro T cell responses was impaired when either dendritic cells (DCs) or T cells were derived from IRF3KO mice. Unexpectedly, IRF3–dependent NK-activating molecule (INAM), which is an NK cell activating factor of the DC innate immune response, was induced during the T cell response. Additionally, supernatants from responding T cells induced ISG54 in the RAW264.7 macrophage cell line in an IRF3 dependent manner. Moreover, addition of anti-IFN-γ prevented supernatant induction of ISG54 and recombinant IFN-γ stimulated ISG54 expression. Thus, IRF3 in APCs and T cells is required for optimal T-cell effector function and the ability of T cells to influence innate immune function of APCs.
Keywords: Interferon Regulatory Factor-3, Interferon-γ, Interleukin-17, Granzyme-B, T cells, Dendritic Cells, Innate immunity, Interferon stimulated gene-54
1. Introduction
For complete clearance of microbes during infection both innate and adaptive immune responses are necessary. The traditional view is that innate immune responses occur within a day after viral infection by initiating expression of Interferon-stimulated genes (ISGs) and genes for NK cell activation. ISG54 is a critical anti-viral factor induced in cells to initiate apoptosis for innate control of viral replication (1). IRF3–dependent NK-activating molecule (INAM) is an inducible cell surface molecule expressed on dendritic cells (DCs) that stimulates NK cell activation (2). On the other hand, adaptive immune effector functions develop slowly during the first week after viral infection. Adaptive immunity for viral infections requires CD4 T cell responses that produce IFN-γ (3) and CD8 T cell responses that produce Granzyme B (GrB) and IFN-γ(4). GrB is critical to T cell cytotoxicity against virus-infected cells (5) and IFN-γ promotes Th1 differentiation and anti-viral effects (6). In contrast, CD4 T cell expression of IL-17 is linked to viral persistence and pathology during certain viral infections (7). Moreover, inducible Foxp3+ CD4 Tregs exhibit plasticity in the presence of IL-6 from inflammatory macrophages, which induces IL-17 expression but represses Foxp3 expression (8). While it is well-known that the innate immune response can shape the adaptive immune response, T cell factors produced during adaptive immune responses are expected to feedback to cells, such as macrophages, enhancing their innate immune responses (9). Very few studies have examined contributions from adaptive T cell responses that enhance innate immune responses.
Most of the research regarding Interferon Regulatory Factor 3 (IRF3) in immunity has dealt with its role in innate anti-viral responses. However, recent studies have uncovered an unexpected link between IRF3 and T cell immune responses in mice during infection (10, 11) and during responses to antigens (12). We recently reported that mice deficient in IRF3 had impairments in memory T cell expression of GrB and IFN-γ during T cell responses to Influenza A and Theiler’s virus infection (11). This role for IRF3 in T cells responses may be the result of IRF3 activation in APCs that participate in T cell responses, where it transcriptionally regulates expression of APC cytokines governing T cell differentiation during the response. We speculated that impaired T cell responses could be due to inadequate production of IL-12 (13), IL-15 (14), IL-6 (15), and IL-23 (16), all of which rely on IRF3 for expression and which promote T cell expression of IFN-γ, GrB, and IL-17 (17). However, addition of these cytokines to T cell responses of mice deficient in IRF3 failed to restore expression of GrB and IFN-γ. Another possibility is that IRF3 may simply contribute to T cell development at the thymus. However, Taniguchi found that relative to other leukocytes, the percentage of total T cells, CD4 T cells, and CD8 T cells is unaffected by IRF3 gene ablation (1). Still further IRF3 may be activated in the T cells, themselves. Finally, IRF3 may contribute to the manner in which adaptive T cell responses feedback onto APCs to enhance their innate immune responses.
Cytokines produced during T cell responses may indeed feedback to APCs and augment innate immune responses (9). A number of innate immune responses involve activation of IRF3 including expression of IFN-β (18), interferon stimulated genes (ISGs), such as ISG54 (1), and NK-activating factors, such as INAM (2). The experiments here were designed to clarify the role for IRF3 in development of T cell effector functions and production of T cell factors that feedback to stimulate expression of ISGs and INAM by APCs. The results show that IRF3 in T cells and APCs is required for full development of T cell effector function during immune responses. Moreover, we found that IFN-γ from responding T cells was responsible for IRF3 dependent expression of ISG54.
2. Materials and Methods
2.1. Mice and cells
Female C57BL/6 mice were purchased from Harlan Sprague Dawley and used at 10–12 weeks of age. Female IRF3 deficient mice (IRF3KO) on the C57BL/6 background were offspring of breeder pairs obtained from Dr. Karen Mossman (McMaster University), originally produced by Dr. Tadatsugu Taniguchi from the University of Tokyo (19). The absence of IRF3 in IRF3KO mice was periodically verified by western blot (data not shown). Experimental animal procedures using mice were approved by and conducted in accordance with the Institutional Animal Care and Use Committee (IACUC) at the University of Nebraska Medical Center. Total splenic mononuclear cells were isolated by dissociating spleens through 70 µM mesh followed by red blood cell lysis. Purified T cells from spleens were isolated using an EasySep T cell isolation kit from Stem Cell Technologies (Vancouver, BC, Canada). 2.5 × 106 splenic mononuclear cells were mixed with biotinylated antibodies to CD11b, CD19, CD24, CD45R, B220, CD49b, and TER119 before eliminating cells with these cell surface proteins using avidin-magnetic spheres. Bone marrow (BM)-derived DCs were generated as originally described (20) and previously generated by one of us (21), where they were found to be 60–70% CD11c+. Briefly, BM cells from C57Bl/6 or IRF3KO mice were incubated for 8 days with 5 ng/ml GM-CSF in RPMI media containing 10% FBS and 50 µg/ml gentamycin. Fresh GM-CSF was added at day 3, 5, and 7 before harvesting DCs at day 8. RAW264.7 cells (RAW-Lucia™ and RAW-IRF3KO-Lucia (IRF3 gene knockout)) were obtained from InVivogen (San Diego, CA), both of which stably express a plasmid containing a secreted luciferase reporter gene under the control of the ISG54 promoter-5X ISRE. These cells were used as previously described (22).
2.2 T cell stimulation
T cells in 2.5 × 106 primary C57BL/6 and IRF3KO splenic mononuclear cells were stimulated with 1 µg/ml anti-CD3 and/or 1 µg/ml anti-CD28 and/or 25 µg/ml poly I:C. Alternatively mixtures of 3 × 104 DCs were incubated with 3 × 105 purified T cells from C57Bl/6 or IRF3KO mice in culture media were stimulated with these factors. Stimulated cells were incubated at 37°C in 5% CO2 for 24 or 48 h. At those times supernatants were collected for ELISAs and RNA extracted for qRT-PCR.
2.3 ELISAs
ELISA plates were coated with antibodies to mouse IFN-γ or IL-17 (Ebiosciences, Sand Diego, CA). After blocking and washing, supernatants or serial dilutions of recombinant IFN-γ or IL-17 (Ebiosciences) were added. After 2 h, biotinylated antibody to mouse IFN-γ or IL-17 was added to each well. After 1 h, streptavidin-horseradish peroxidase (1:1,000; BD-Pharmingen) was added for 30 min, and then 3,3-,5,5--tetra-methyl-benzindine–hydrogen peroxide solution was added to each well. Cytokine concentrations were estimated by determining optical densities at a 450-nm wavelength (OD450s) with a reference OD570 using an ELISA plate reader.
2.4 RNA preparation and qRT-PCR
RNA was extracted from cells using the Purelink kit from Ambion/Invitrogen (Carlsbad, CA) according to the manufacturer’s specifications. One hundred nanograms to 1 µg of RNA was reverse transcribed as before (23). cDNA samples were incubated with the following primer pairs (Invitrogen): Granzyme B sense 5’ TCCTGCTACT GC TG AC CTTGTC 3’ and antisense 5’ ATGATCTCCCC TG CC TT TGTC 3’, INAM sense 5’ CAACTGCAATGCCACGCTA 3’ and antisense 5’ TCCAACCG AACACCTGAGACT 3’, or GAPDH sense 5’ TTGTCAGCAATGCATCCTGCAC 3’ and antisense 5’ ACAGC TTTCCAGAGGGGCCATC 3’. Quantitative PCRs were run on an ABI Prism 7000 thermal cycler at 50°C for 2 min and 95°C for 10 min, followed by 45 cycles of 95°C for 15 s and 60°C for 30 s. Cycle thresholds (CT) of sample were normalized to the CT of GAPDH for that sample (CT) and then normalized to the average CT of the control samples (CT), after which data were expressed as relative levels of mRNA using 2ΔΔCT.
2.5 Single Color Flow Cytometry
On day 7 of BMDC induction, cells were fixed with 4% paraformaldehyde, washed, and permeabilized in 0.2% Tween 20 in PBS for 15 minutes at 37°C. Following permeabilization, cells were washed with .1% Tween 20 in FACS buffer (PBS with 0.1% Sodium Azide and 2% FBS), and blocked with TruStain fcX™ (Biolegend 101320) for 10 minutes. Cells were then stained with APC-anti-CD11c (Ebioscience, clone N418), PE-anti-TLR3 antibody (Biolegend 141904), or PE-Isotype control (Biolegend 400508) for 30 minutes on ice. Cells were washed twice with 0.1% Tween 20 in FACS buffer, washed twice with FACS buffer, and then analyzed immediately using a FACSCalibur cell analyzer and the data were analyzed with FlowJo software.
2.6 ISG54 promoter activity
Supernatants from T cell / DC cell cultures stimulated with anti-CD3/anti-CD28 without poly I:C were diluted 1:4 in fresh media. Supernatants from T cell/DC cell cultures with anti-CD3/anti-CD28 with poly I:C were diluted 1:8. These diluted supernatants were then incubated with 105 RAW-Lucia or IRF3KO-RAW-Lucia cells in culture media for 24 h. Additionally, serial dilutions of recombinant IFN-γ (Biolegend 575302) were added to 105 RAW-Lucia or IRF3KO RAW-Lucia cells. Recombinant IFN-β (250 pg/ml) (R&D 124001-1) and PolyI:C (25ug/ml) were also used as positive controls for responses of RAW-Lucia and IRF3KO-RAW-Lucia cells. To assess secreted luciferase at 24 h, 10 µl of supernatants were assayed using the QUANTI-Luc assay of InVivogen.
2.7 IFN-γ neutralization assay
Supernatants from T cell / DC cultures were diluted 1:4 in culture media and incubated with either PBS, 250 ng/ml IFN-γ neutralizing antibody (Clone: R4-6A2; Biolegend 505705), or 250 ng/ml isotype control antibody (Biolegend 400413) at 37°C for 90 min prior to incubation with 105 RAW-Lucia cells for 24 hours. 10ul of supernatants were assayed for secreted luciferase using the QUANTI-Luc assay from InVivogen.
2.8 Statistical analysis
Where appropriate, data were subjected to two-way analysis of variance and the Student t test to determine the significance of differences between sample means using the statistical program in Microsoft Excel. P values of <0.05 were considered significant.
3. Results
3.1 IRF3 deficiency modulates T cell responses in vitro
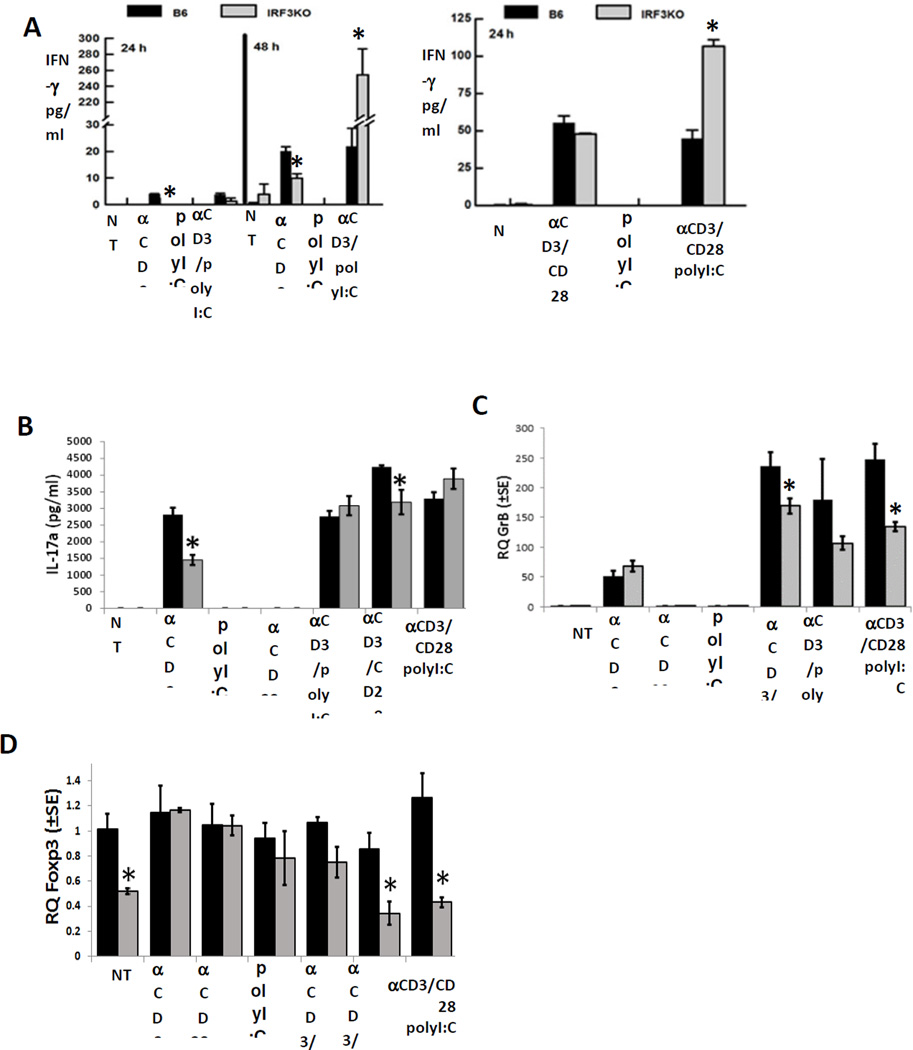
We previously reported that IRF3 deficiency in mice impairs IFN-γ and GrB expression from memory T cells following infections with Influenza A virus or Theiler’s virus (11). To determine if these impairments include other T cell factors, such as IL-17, we stimulated T cells of splenic mononuclear populations from C57Bl/6 or IRF3KO mice with anti-CD3 with or without anti-CD28 in the presence or absence of poly I:C, a TLR3 agonist. As expected, IFN-β expression in response to poly I:C was impaired during stimulation of IRF3KO mononuclear cells with anti-CD3 with or without anti-CD28 (data not shown). Deficiency of IRF3 resulted in significantly decreased production of IFN-γ at 24 and 48 h during the responses (Fig. 1A). Addition of poly I:C during stimulation did not significantly enhance IFN-γ production from T cells of C57Bl/6 mice but did significantly enhance production of IFN-γ from T cells of IRF3KO mice to levels greater than IRF3 sufficient T cells. Similar to IFN-γ, IL-17 production from IRF3 deficient T cells was impaired and poly I:C partially restored IL-17 production (Fig. 1B). In contrast, expression of GrB was only impaired in IRF3 deficient T cells stimulated with anti-CD3 plus anti-CD28, and poly I:C did not restore GrB expression during the response (Fig. 1C). Therefore, deficiency of IRF3 significantly decreases IFN-γ, IL-17 and GrB, but unexpectedly IRF3 deficiency increased production of IFN-γ from T cells responding in the presence of poly I:C.
Fig. 1.
IRF3 deficiency reduces T cell cytokine and Granzyme B expression during T cell responses with APCs. Primary splenic mononuclear cells (2.5 × 106) from C57Bl/6 and IRF3KO mice were stimulated with 1 µg/ml anti-CD3 with or without 1 µg/ml anti-CD28 with or without 25 µg/ml poly I:C for 24 or 48 h. Forty eight h supernatant levels of IFN-γ (A) and IL-17 (B) were assayed by ELISA, mRNA levels for GrB (C) and Foxp3 (D) were determined by qRT-PCR. Data are means ± SE, * indicates p< 0.05; NT indicates no treatment.
To determine if the enhanced IFN-γ production in IRF3KO T cells treated with poly I:C is related to diminished Treg activity we evaluated Foxp3 expression. IRF3KO cultures that were not treated or those that were stimulated with anti-CD3 ± anti-CD28 in the presence of poly I:C had significantly less Foxp3 expression compared to stimulated wild-type cells with poly I:C (Fig. 1D). These data suggest that IRF3 contributes to expression of Foxp3 and diminished Foxp3 expression of IRF3KO T cells is could be somewhat responsible for enhanced IFN- γ production from stimulated IRF3KO T cells.
3.2 INAM is induced during T cell responses with APCs
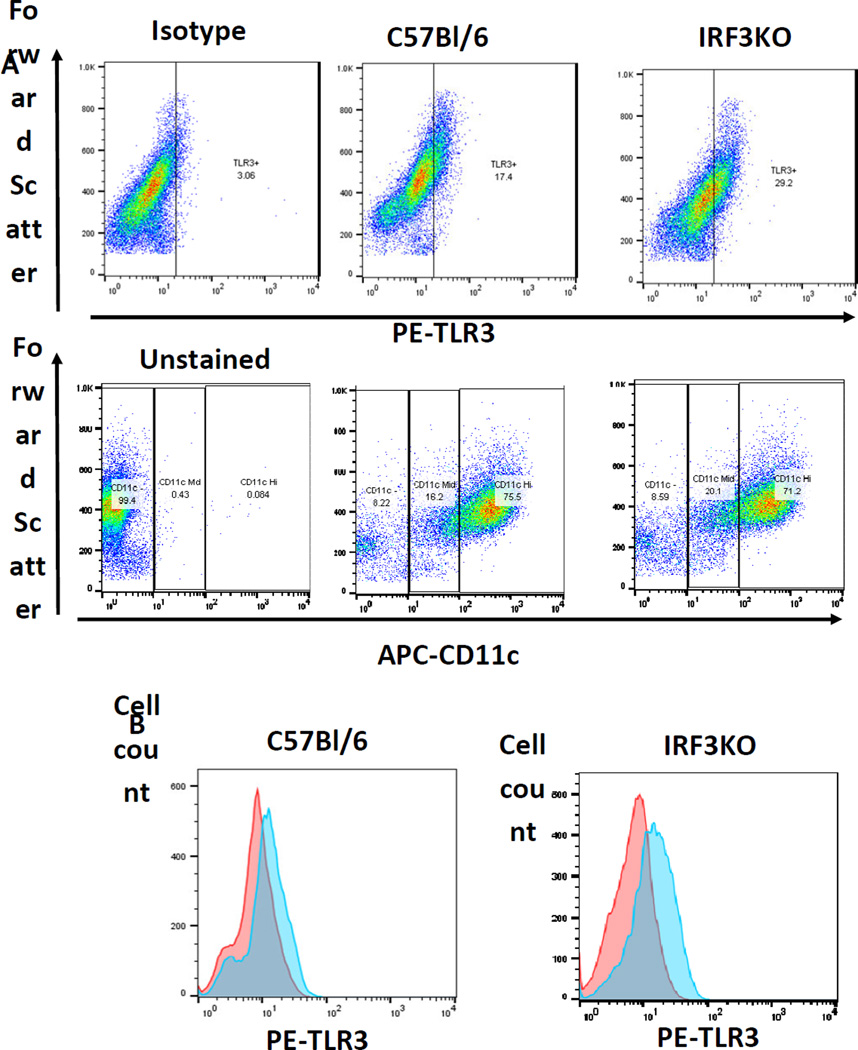
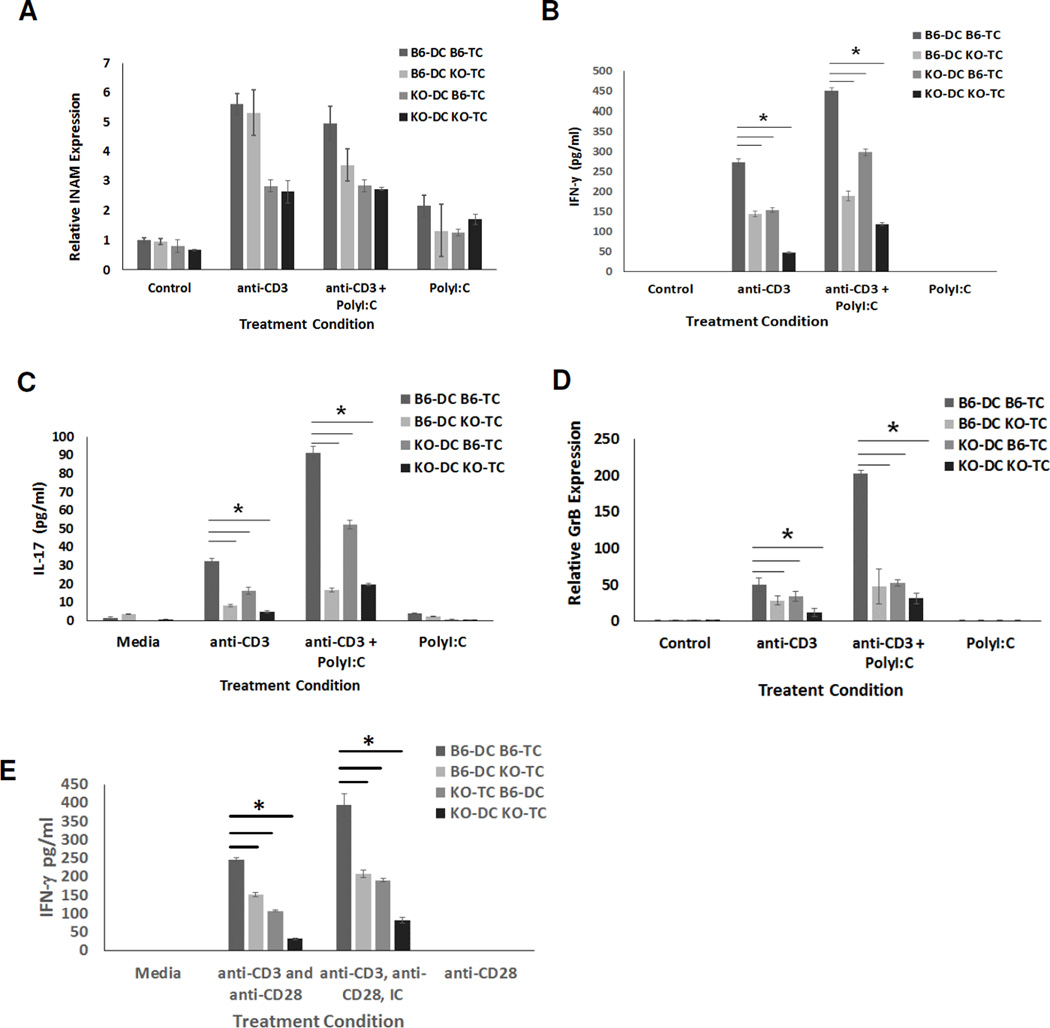
To determine if these impairments in T cell cytokines and GrB were due to intrinsic deficiency of IRF3 in T cells, APCs, or both, we generated bone marrow DCs (BMDCs) from C57Bl/6 and IRF3KO mice. We confirmed that 70–80 % of the BMDCs are CD11c+ (Fig. 2A). These BMDCs were combined at an optimum DC to T cell ratio of 1:10 with enriched T cells from either C57Bl/6 or IRF3KO mice. Cell mixtures were stimulated with anti-CD3 with or without poly I:C. Since we used the TLR3 agonist, poly I:C, we first evaluated TLR3 levels in the DCs. We found that BMDCs from both WT and IRF3KO mice expressed TLR3 (Fig. 2A, B). Because IRF3 activation during the T cell response is transitory and would be difficult estimate when it would occur, we measured expression of INAM as a measure of IRF3 activation because its expression in response to poly I:C in DCs and macrophages is absent during IRF3 deficiency (24). After 48 h of stimulation of T cells with anti-CD3, all DC/TC combinations expressed INAM regardless of IRF3 status (Fig. 3A). Although the highest expression of INAM seemed to occur in combinations with WT DCs, this trend was not statistically significant in each experiment. Therefore as expected, IRF3 contributes significantly to INAM expression and it is likely that activation of IRF3 occurs in BMDCs at some time during T cell responses with APCs.
Fig. 2.
CD11c and TLR3 expression by BM-DC DCs of C57Bl/6 (B6) or IRF3KO mice. DCs were derived by incubating BM cells with GM-CSF for 7 days as described in Methods section. Cells were harvested, stained with PE-anti-TLR3, APC-anti-CD11c, or PE-Ig-Isotype control antibody and then FACS analyzed. Data are scatter plots of cells Forward Scatter vs. PE-anti-TLR3 stained or APC-anti-CD11c stained cells (A) or histograms of PE-anti-TLR3 stained cells in the total population (B).
Fig. 3.
INAM is induced during T cell responses and IRF3 contributes to T cell expression of cytokines and GrB. BM-derived DCs from C57Bl/6 (B6) or IRF3KO mice were incubated in cell culture with T cells (TC) from C57Bl/6 (B6) or IRF3KO mice (3 × 104 DCs with 3 × 105 TCs) and left unstimulated or stimulated with anti-CD3 with or without poly I:C (A–D) or stimulated with anti-CD3/anti-CD28 with or without poly I:C (E). After for 48 h supernatants were collected for IFN-γ (B,E) and IL-17 (C) ELISAs or total RNA isolated for INAM (A) and GrB (D) qRT-PCR. Data are means ± SE, * indicates p< 0.05 using two-tailed Student t test. Results shown are representative of two independent experiments (n = 3)
3.3 IRF3 deficiency in both APCs and T cells impairs T cell responses
Production of IFN-γ (Fig. 3B) and IL-17 (Fig. 3C) and expression of GrB (Fig. 3D) declined during the T cell responses when IRF3 was absent in either T cells or DCs. The least amount of IFN-γ, IL-17, and GrB was produced and expressed during T cell responses when both T cells and DCs were deficient in IRF3. In each circumstance, with or without poly I:C, IRF3 deficiency in T cells or DCs independently lowered cytokine and GrB production to a significant degree (Table 1, 2). Stimulation of T cell/DC cultures with anti-CD3 plus anti-CD28 resulted in similar effects on IFN-γ production (Fig. 3B, E), as well as on expression of INAM, IL-17 and GrB (data not shown). As expected, anti-CD28 alone did not stimulate any production of IFN-γ (Fig. 3E), or production of IL-17, GrB, or INAM (data not shown). Similarly, T cells alone stimulated with anti-CD3 with or without anti-CD28 did not produce detectable IFN-γ by 48 h (data not shown). Therefore, the results indicate that highest induction of IFN-γ, IL-17, and GrB during T cell responses depends on IRF3 activity in both T cells and DCs.
Table 1.
ANOVA: Two Factor Statistics for IL-17, IFN-γ, and Granzyme B from T cell – Dendritic cell cultures stimulated with anti-CD3 antibody
| IL-17 | |||||
| Stimulant | Cell type | Experiment 1 | Experiment 2 | ||
| F | p | F | p | ||
| Anti-CD3 | Dendritic Cell | 47.05 | .0001 | 15.16 | 0.0046 |
| T Cell | 158.40 | 1.49E-06 | 11.50 | .0095 | |
| Interaction | 20.85 | .0018 | 0.145 | 0.713 | |
| IFN-y | |||||
| Stimulant | Cell type | Experiment 1 | Experiment 2 | ||
| F | p | F | p | ||
| Anti-CD3 | Dendritic Cell | 265.65 | 2.02E-07 | 60.82 | 5.24E-05 |
| T Cell | 314.67 | 1.04E-07 | 151.72 | 1.76E-06 | |
| Interaction | 2.595 | 0.1459 | 34.39 | 0.0004 | |
| Granzyme B | |||||
| Stimulant | Cell type | Experiment 1 | Experiment 2 | ||
| F | p | F | p | ||
| Anti-CD3 | Dendritic Cell | 6.1341 | .0383 | 6.858 | .0307 |
| T Cell | 11.193 | .0102 | 32.895 | .0004 | |
| Interaction | .00108 | .9746 | 9.9166 | .0136 | |
Table 2.
ANOVA: Two Factor Statistics for IL-17, IFN-γ, and Granzyme B from T cell – Dendritic cell cultures stimulated with anti-CD3 antibody and poly I:C
| IL-17 | |||||
| Stimulant | Cell Type | Experiment 1 | Experiment 2 | ||
| F | P | F | P | ||
| Anti-CD3 +polyI:C |
Dendritic Cell | 65.93 | 3.90E-05 | 28.07 | 0.0007 |
| T Cell | 595.5 | 8.49E-09 | 228.9 | 3.61E-07 | |
| Interaction | 90.85 | 1.21E-05 | 1.80 | 0.217 | |
| IFN-y | |||||
| Stimulant | Cell Type | Experiment 1 | Experiment 2 | ||
| F | P | F | P | ||
|
Anti-CD3 +polyI:C |
Dendritic Cell | 170.8099 | 1.12E-06 | 100.765 | 8.25E-06 |
| T Cell | 671.9407 | 5.26E-09 | 274.891 | 1.77E-07 | |
| Interaction | 22.66285 | 0.0014 | 29.182 | 0.0006 | |
| Granzyme B | |||||
| Stimulant | Cell Type | Experiment 1 | Experiment 2 | ||
| F | P | F | P | ||
|
Anti-CD3 +polyI:C |
Dendritic Cell | 39.67 | .0003 | 4.155 | .0759 |
| T Cell | 45.02 | .0002 | 10.475 | .0119 | |
| Interaction | 25.467 | .001 | 1.87 | .2085 | |
3.4 Factors produced during T cell responses with APCs induce ISG54 promoter activity
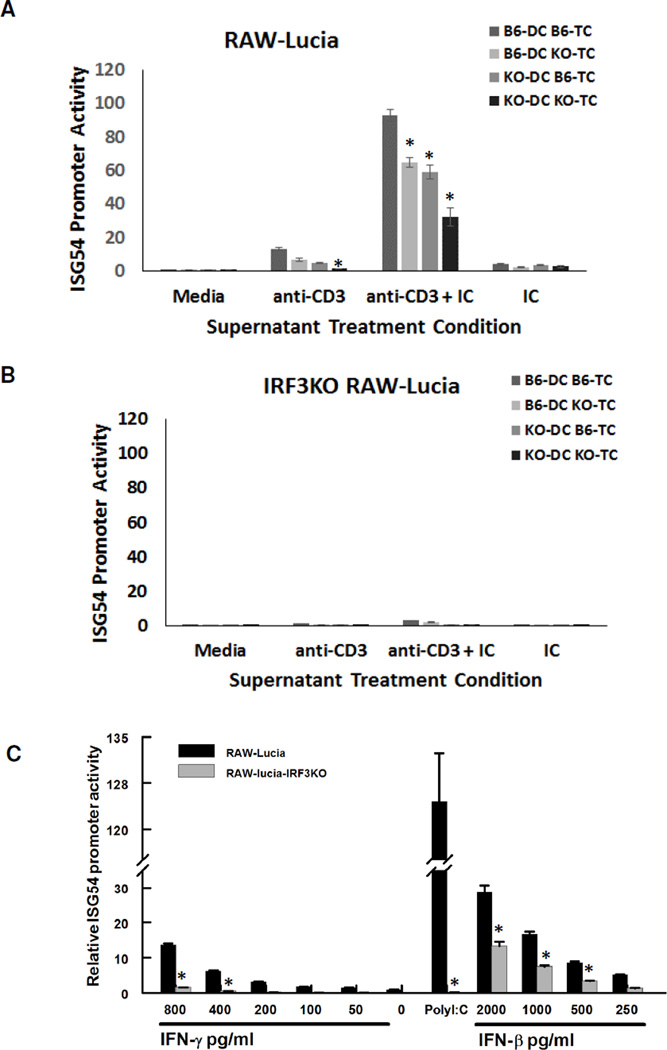
As suggested by INAM expression, stimulated T cells could produce factor(s) that in turn cause APC expression of ISGs in an IRF3 dependent manner (1). To see if ISG expression could be induced during T cell responses, we determined the ability of supernatants from T cell/DC cultures to induce ISG54 promoter activity in RAW264.7 cells (RAW-Lucia) compared with RAW264.7 cells in which the IRF3 gene is deleted (RAW-Lucia-IRF3KO). The supernatants from T cell/DC cultures stimulated with anti-CD3 with or without poly I:C induced ISG54 promoter activity in RAW-Lucia cells but not in RAW-Lucia-IRF3KO cells (Fig. 4A,B). Moreover, the T cell factor(s) that induce ISG54 was dependent on IRF3, as supernatants from IRF3KO DCs or IRF3KO T cells induced significantly less ISG54 compared to those from C57BL/6 DCs or C57BL/6 T cells (Fig. 4A). The supernatants from the combination IRF3KO DC and TC induced the lowest amount of ISG54 promoter activity. Moreover, supernatants from T cell/DC cultures stimulated with poly I:C alone (Fig. 4A) or with anti-CD28 alone (data not shown) induced little ISG54 promoter activity. Therefore, soluble factors produced during T cell responses induce ISG54 expression in APCs in an IRF3 dependent fashion.
Fig. 4.
Soluble factors from T cell responses with APCs induce ISG54 promoter activity in an IRF3 dependent fashion. Supernatants from Fig. 3 (A, B) or recombinant IFN-γ, poly I:C, or recombinant IFN-β (C) were incubated with RAW-Lucia (105) or IRF3KO RAW-Lucia (105) cells. Secreted luciferase from RAW-Lucia or IRF3KO RAW-Lucia was measured 24 h after incubation of these cells with T cell supernatants or recombinant IFN-γ, poly I:C, or recombinant IFN-β. Data are mean of relative luciferase activity ± SE, * indicates p< 0.05 using two-tailed Student’s t test. Results shown are representative of two independent repeats (n = 3)
3.5 IFN-γ is essential for T cell mediated induction of ISG54 promoter activity in APCs
One of the cytokines produced during stimulation of T cell/APC cultures is IFN-γ, which has been shown to induce ISG54 from APCs (25). Therefore, we stimulated RAW-Lucia and RAW-Lucia-IRF3KO cells with recombinant IFN-γ. In a dose dependent fashion, IFN-γ stimulated ISG54 promoter activity in RAW-Lucia cells with IRF3 but not in RAW-Lucia-IRF3KO cells (Fig. 4C). We also stimulated RAW-Lucia cells and RAW-Lucia-IRF3KO cells with several concentrations of IFN-β, which is known to induce expression of ISGs through an ISGF3 complex composed of STAT1, STAT2, and IRF9 (26). IFN-β stimulated ISG54 promoter activity in both the WT RAW-Lucia cells and the IRF3KO RAW-Lucia cells. However, the IRF3 deficiency decreased expression of ISG54 induction in response to IFN-β. Altogether, these data demonstrate that IFN-γ is sufficient to induce ISG54 in APCs in an IRF3 dependent fashion.
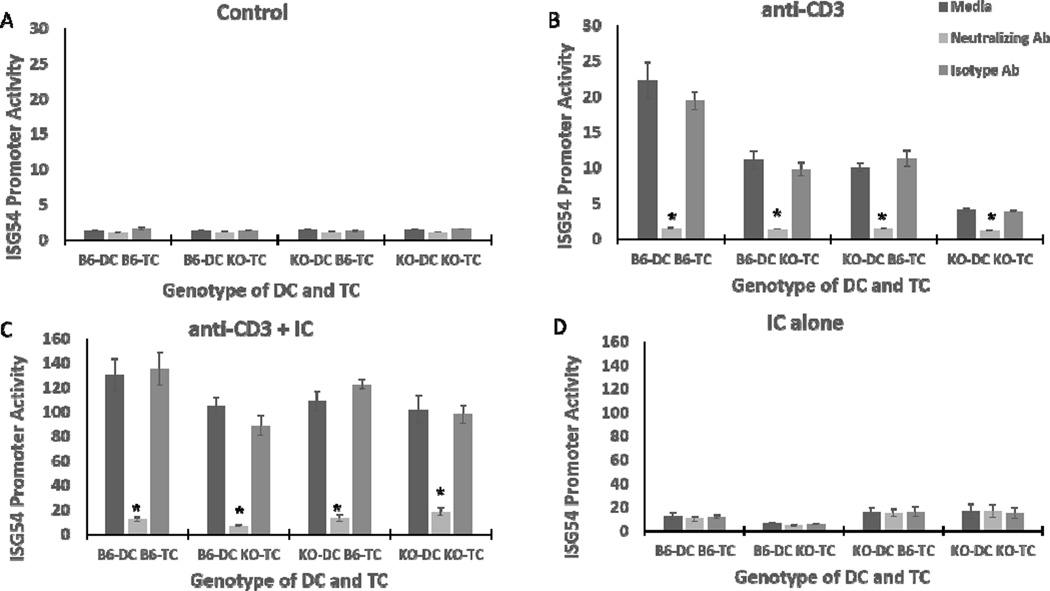
We next wanted to see if IFN-γ was the responsible soluble factor in the responding T cell/DC supernatants for induction of ISG54 promoter activity. T cell/DC supernatants were treated with a neutralizing antibody targeting IFN-γ, prior to incubation with RAW-Lucia cells. ISG54 activity was almost completely abolished when neutralizing antibody was added to the anti-CD3 or anti-CD3/poly I:C T cell/DC supernatants compared to supernatants alone, or supernatants with an isotype control antibody (Fig. 5B, C). In contrast, control supernatants from unstimulated T cell DC cultures failed to induce ISG54 promoter activity, which was unaffected by IFN-γ neutralizing antibody (Fig. 5A). Supernatants from poly I:C alone T cell/DC cultures stimulated a small level of ISG54 promoter activity, which was not affected by IFN-γ neutralizing antibody (Fig. 5D). Therefore, IFN-γ is the essential IRF3 dependent factor produced by responding T cells that induces ISG54 promoter activity in APCs.
Fig. 5.
IFN-γ from T cell responses with APCs induces ISG54 promoter activity. Supernatants from Fig. 3 were incubated with IFN-γ neutralizing antibody, isotype control immunoglobulin, or PBS for 90 minutes at 37°C. Control supernatants (A), supernatants from cells stimulated with anti-CD3 (B), anti-CD3 plus poly I:C (C), or poly I:C (D) were then incubated with 105 RAW-Lucia cells for 24 hours. After 24 hours secreted luciferase in supernatants was measured. Data are mean of relative luciferase activity ± SE, * indicates p< 0.05 using two-tailed Student’s t test. Results shown are representative of two independent repeats (n = 3)
4. Discussion
The appropriate differentiation of T cells responding to pathogens is central to adaptive immunity and the establishment of correct T cell effector functions to resist the microbial infection at hand. The results here confirm that IRF3 is required for induction of cytokines and GrB during T cell responses. Moreover, the data indicate that there are independent roles for IRF3 in both T cells and DCs for induction of IFN-γ, IL-17, and GrB. IRF3 is a transcription factor that is constitutively expressed by most cells and post-translationally activated in response to viral infection (27), intracellular bacterial infection (28), TLR4 signaling (29), HMGB1 (30), DNA damage (31), or endoplasmic reticulum stress (32). Therefore, it is not entirely surprising that IRF3 plays a significant role in development of T cell effector functions. During viral infections, TLR3 binding to dsRNA of viruses in endosomes is linked to signaling pathways that activate cytoplasmic TBK1/IKKε, which ultimately activate IRF3 (33). However, IRF3 activation can occur if cells become infected with viruses as well (34). In this case, cytoplasmic helicase Retinoic Acid Inducible gene-I (RIG-I) recognition of uncapped 5’-triphosphorylated-RNA (35) and Melanoma Differentiation-associated gene (MDA)-5 recognition of RNA from picornaviruses that are triphosphorylated and capped with viral protein (VPg) induces RIG-I and MDA5 interaction with mitochondrial MAVS (also known as, IPS-1, CARDIF, VISA) that then activates TBK1/IKKε for IRF3 phosphorylation (36). TBK1/IKKε activation of IRF3 can also occur upon host recognition of viral DNA via the STING pathway (37). Ultimately, activation of IRF3 is a common feature of macrophage and DC innate anti-viral responses (38, 39). The unique aspect of the data presented is the indication that IRF3 dependent genes are activated during T cell responses with DCs and this activation occurs without the addition of exogenous RNA or DNA.
IRF3 is a key transcription factor for IFN-β expression and innate anti-viral immune responses (40). We and others showed that IRF3 also contributes to production of IL-6 (41), IL-15 (24), IL-12 (13), and IL-23 (16) during innate immune responses. These APC cytokines are also known to be important for expression of GrB (42–44), IFN-γ (45), and IL-17 (17) by T cells in adaptive immune responses. Several other transcription factors have been linked to development of specific T cell responses. The transcription factor Eomesodermin (Eomes) is associated with cytolytic T cell differentiation (46), T-bet is associated with IFN-γ from T cells (47), and ROR-γT with IL-17 from T cells (48). A more recent paper showed that IRF3 is activated during responses of purified naïve T cell to anti-CD3/anti-CD28 (12). Moreover, CD4 and CD8 T cells that were differentiated under Th17 and Tc17 conditions express ROR-γt and activated IRF3 together and both of these transcription factors interacted together in the cytoplasm of responding T cells (12). Binding of IRF3 to ROR-γt intracellularly in Tc17 cell dampened IL-17 production from responding Tc17 cells but not from responding naïve T cells. It is still unclear if IRF3 interacts with or influences Eomes and T-bet in T cells. However, association of IRF3 with other transcription factors is not unexpected. Activated IRF3 interacts with several additional transcription factors including IRF7 or CBP/p300 (49). The data in the present report suggest that IRF3 association with Eomes and T-bet is a possibility.
In the present report during responses of T cells with DCs, one of the ISGs, ISG54, and an NK cell activating factors, INAM, are expressed in an IRF3 dependent fashion. Previously we showed that activation of IRF3 was required for expression of INAM by macrophages in response to poly I:C nucleic acid (24). However here, even in the absence of exogenous nucleic acids, expression of ISG54 and INAM was induced during T cell responses with DCs. We show that T cell IFN-γ stimulates ISG54 promoter activity in an IRF3 dependent manner. It is also likely that other ISGs are induced during T cell responses. ISG54, also called IFN-induced protein with tetratricopeptide repeat 2 (IFIT2), is one of 3 related murine ISGs (ISG56/ IFIT1, ISG54/ IFIT2, and ISG49/IFIT3). In humans, ISG54 is one of four related proteins, with ISG56/IFIT1, ISG58 /IFIT5, and ISG60/IFIT3 being the other three (50). Previous reports showed ISG54 expression in response to IFN-γ (25). This under-investigated effect of IFN-γ on ISG54 expression suggests that this is related to its potent anti-viral effects and may also be related to its potent anti-cancer effector function. Taken together the results here demonstrate that the transcription factor, IRF3, is involved in T cell responses. IRF3 contributes to expression of several T cell effector molecules but is also involved in responses of APCs to T cell IFN-γ.
Highlights.
IRF3 is required in both T cells and APCs for optimal T cell effector function
IRF3 dependent innate immune factors of APCs are induced during T cell responses
IFN-γ from T cells induces ISG54 in macrophages in an IRF3 dependent manner.
Acknowledgments
The authors convey their thanks to Marian Schmid for her excellent animal care and to Alex Vogel for his outstanding review of this manuscript and helpful suggestions. We also acknowledge of the help and support of the Nebraska Center for Virology flow cytometry core facility. This work was supported by funding from the University of Nebraska Medical Center College of Dentistry and University of Nebraska Lincoln, School of Biological Sciences, and supported by Award Number P30GM10350903 and P20GM103489 from the National Institute of General Medicine, a component of the National Institutes of Health (NIH). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Research Resources or the National Institutes of Health.
Abbreviations
- IRF
Interferon Regulatory Factor
- IFN
Interferon
- GrB
Granzyme B
- ISG
Interferon stimulated gene
- DCs
Dendritic cells
- INAM
IRF3–dependent NK-activating molecule
- Eomes
Eomesodermin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nakaya T, Sato M, Hata N, Asagiri M, Suemori H, Noguchi S, Tanaka N, Taniguchi T. Gene induction pathways mediated by distinct IRFs during viral infection. Biochem Biophys Res Commun. 2001;283:1150–1156. doi: 10.1006/bbrc.2001.4913. [DOI] [PubMed] [Google Scholar]
- 2.Ebihara T, Azuma M, Oshiumi H, Kasamatsu J, Iwabuchi K, Matsumoto K, Saito H, Taniguchi T, Matsumoto M, Seya T. Identification of a polyI:C-inducible membrane protein that participates in dendritic cell-mediated natural killer cell activation. J Exp Med. 2010;207:2675–2687. doi: 10.1084/jem.20091573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lucey DR, Clerici M, Shearer GM. Type 1 and type 2 cytokine dysregulation in human infectious, neoplastic, and inflammatory diseases. Clin Microbiol Rev. 1996;9:532–562. doi: 10.1128/cmr.9.4.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bannard O, Kraman M, Fearon D. Pathways of memory CD8+ T-cell development. Eur J Immunol. 2009;39:2083–2087. doi: 10.1002/eji.200939555. [DOI] [PubMed] [Google Scholar]
- 5.Andrade F, Roy S, Nicholson D, Thornberry N, Rosen A, Casciola-Rosen L. Granzyme B directly and efficiently cleaves several downstream caspase substrates: implications for CTL-induced apoptosis. Immunity. 1998;8:451–460. doi: 10.1016/s1074-7613(00)80550-6. [DOI] [PubMed] [Google Scholar]
- 6.Trinchieri G. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat Rev Immunol. 2003;3:133–146. doi: 10.1038/nri1001. [DOI] [PubMed] [Google Scholar]
- 7.Hou W, Kang HS, Kim BS. Th17 cells enhance viral persistence and inhibit T cell cytotoxicity in a model of chronic virus infection. J Exp Med. 2009;206:313–328. doi: 10.1084/jem.20082030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Josefowicz SZ, Rudensky A. Control of regulatory T cell lineage commitment and maintenance. Immunity. 2009;30:616–625. doi: 10.1016/j.immuni.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strutt TM, McKinstry KK, Swain SL. Control of innate immunity by memory CD4 T cells. Adv Exp Med Biol. 2011;780:57–68. doi: 10.1007/978-1-4419-5632-3_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharma S, tenOever BR, Grandvaux N, Zhou GP, Lin R, Hiscott J. Triggering the interferon antiviral response through an IKK-related pathway. Science. 2003;300:1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- 11.Moore TC, Vogel AJ, Petro TM, Brown DM. IRF3 deficiency impacts granzyme B expression and maintenance of memory T cell function in response to viral infection. Microbes Infect. 2015;17:426–439. doi: 10.1016/j.micinf.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ysebrant de Lendonck L, Tonon S, Nguyen M, Vandevenne P, Welsby I, Martinet V, Molle C, Charbonnier LM, Leo O, Goriely S. Interferon regulatory factor 3 controls interleukin-17 expression in CD8 T lymphocytes. Proc Natl Acad Sci U S A. 2013;110:3189–3197. doi: 10.1073/pnas.1219221110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dahlberg A, Auble MR, Petro TM. Reduced expression of IL-12 p35 by SJL/J macrophages responding to Theiler's virus infection is associated with constitutive activation of IRF-3. Virology. 2006;353:422–432. doi: 10.1016/j.virol.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 14.Soudja SM, Ruiz AL, Marie JC, Lauvau G. Inflammatory Monocytes Activate Memory CD8(+) T and Innate NK Lymphocytes Independent of Cognate Antigen during Microbial Pathogen Invasion. Immunity. 2012;37:549–562. doi: 10.1016/j.immuni.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sweeney SE, Kimbler TB, Firestein GS. Synoviocyte innate immune responses: II. Pivotal role of IFN regulatory factor 3. J Immunol. 2010;184:7162–7168. doi: 10.4049/jimmunol.0903944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Al-Salleeh F, Petro TM. Promoter Analysis Reveals Critical Roles for SMAD-3 and ATF-2 in Expression of IL-23 p19 in Macrophages. J Immunol. 2008;181:4523–4533. doi: 10.4049/jimmunol.181.7.4523. [DOI] [PubMed] [Google Scholar]
- 17.Harrington LE, Hatton RD, Mangan PR, Turner H, Murphy TL, Murphy KM, Weaver CT. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol. 2005;6:1123–1132. doi: 10.1038/ni1254. [DOI] [PubMed] [Google Scholar]
- 18.Wathelet MG, Lin CH, Parekh BS, Ronco LV, Howley PM, Maniatis T. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-beta enhancer in vivo. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 19.Sato M, Suemori H, Hata N, Asagiri M, Ogasawara K, Nakao K, Nakaya T, Katsuki M, Noguchi S, Tanaka N, Taniguchi T. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity. 2000;13:539–548. doi: 10.1016/s1074-7613(00)00053-4. [DOI] [PubMed] [Google Scholar]
- 20.Datta SK, Redecke V, Prilliman KR, Takabayashi K, Corr M, Tallant T, DiDonato J, Dziarski R, Akira S, Schoenberger SP, Raz E. A Subset of Toll-Like Receptor Ligands Induces Cross-presentation by Bone Marrow-Derived Dendritic Cells. J Immunol. 2003;170:4102–4110. doi: 10.4049/jimmunol.170.8.4102. [DOI] [PubMed] [Google Scholar]
- 21.Brown DM, Kamperschroer C, Dilzer AM, Roberts DM, Swain SL. IL-2 and antigen dose differentially regulate perforin- and FasL-mediated cytolytic activity in antigen specific CD4+ T cells. Cell Immunol. 2009;257:69–79. doi: 10.1016/j.cellimm.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stein SC, Lam E, Falck-Pedersen E. Cell-specific regulation of nucleic acid sensor cascades: a controlling interest in the antiviral response. J Virol. 2012;86:13303–13312. doi: 10.1128/JVI.02296-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore TC, Bush KL, Cody L, Brown DM, Petro TM. Interleukin-6 control of early Theiler's Murine Encephalomyelitis Virus replication in macrophages occurs in conjunction with STAT1 activation and nitric oxide production. J Virol. 2012;86:10841–10851. doi: 10.1128/JVI.01402-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore TC, Kumm PM, Brown DM, Petro TM. Interferon response factor 3 is crucial to poly-I:C induced NK cell activity and control of B16 melanoma growth. Cancer Lett. 2014;346:122–128. doi: 10.1016/j.canlet.2013.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ito S, Ansari P, Sakatsume M, Dickensheets H, Vazquez N, Donnelly RP, Larner AC, Finbloom DS. Interleukin-10 inhibits expression of both interferon alpha- and interferon gamma- induced genes by suppressing tyrosine phosphorylation of STAT1. Blood. 1999;93:1456–1463. [PubMed] [Google Scholar]
- 26.Fu XY, Kessler DS, Veals SA, Levy DE, Darnell JE., Jr ISGF3, the transcriptional activator induced by interferon alpha, consists of multiple interacting polypeptide chains. Proc Natl Acad Sci U S A. 1990;87:8555–8559. doi: 10.1073/pnas.87.21.8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Navarro L, David M. p38-dependent activation of interferon regulatory factor 3 by lipopolysaccharide. J Biol Chem. 1999;274:35535–35538. doi: 10.1074/jbc.274.50.35535. [DOI] [PubMed] [Google Scholar]
- 28.Woodward JJ, Iavarone AT, Portnoy DA. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science. 2010;328:1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 30.Bauer EM, Shapiro R, Billiar TR, Bauer PM. High mobility group Box 1 inhibits human pulmonary artery endothelial cell migration via a Toll-like receptor 4- and interferon response factor 3-dependent mechanism(s) J Biol Chem. 2013;288:1365–1373. doi: 10.1074/jbc.M112.434142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim T, Kim TY, Song Y-H, Min IM, Yim J, Kim TK. Activation of Interferon Regulatory Factor 3 in Response to DNA-damaging Agents. J Biol Chem. 1999;274:30686–30689. doi: 10.1074/jbc.274.43.30686. [DOI] [PubMed] [Google Scholar]
- 32.Hu F, Yu X, Wang H, Zuo D, Guo C, Yi H, Tirosh B, Subjeck JR, Qiu X, Wang XY. ER stress and its regulator X-box-binding protein-1 enhance polyIC-induced innate immune response in dendritic cells. Eur J Immunol. 2011;41:1086–1097. doi: 10.1002/eji.201040831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 34.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 35.Seth RB, Sun L, Ea C-K, Chen ZJ. Identification and Characterization of MAVS, a Mitochondrial Antiviral Signaling Protein that Activates NF-kB and IRF3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 36.Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo Y-M, Gale M, Akira S, Yonehara S, Kato A, Fujita T. Shared and Unique Functions of the DExD/H-Box Helicases RIG-I, MDA5, and LGP2 in Antiviral Innate Immunity. J Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 37.Goubau D, Deddouche S, Reis e Sousa C. Cytosolic sensing of viruses. Immunity. 2013;38:855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gautier G, Humbert M, Deauvieau F, Scuiller M, Hiscott J, Bates EE, Trinchieri G, Caux C, Garrone P. A type I interferon autocrine-paracrine loop is involved in Toll-like receptor-induced interleukin-12p70 secretion by dendritic cells. J Exp Med. 2005;201:1435–1446. doi: 10.1084/jem.20041964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moore TC, Petro TM. IRF3 and ERK MAP-kinases control nitric oxide production from macrophages in response to poly-I:C. FEBS Lett. 2013;587:3014–3020. doi: 10.1016/j.febslet.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fan CM, Maniatis T. Two different virus-inducible elements are required for human beta-interferon gene regulation. Embo J. 1989;8:101–110. doi: 10.1002/j.1460-2075.1989.tb03353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moore TC, Cody L, Kumm PM, Brown DM, Petro TM. IRF3 helps control acute TMEV infection through IL-6 expression but contributes to acute hippocampus damage following TMEV infection. Virus Res. 2013;178:226–233. doi: 10.1016/j.virusres.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marshall HD, Prince AL, Berg LJ, Welsh RM. IFN-alpha beta and self-MHC divert CD8 T cells into a distinct differentiation pathway characterized by rapid acquisition of effector functions. J Immunol. 2010;185:1419–1428. doi: 10.4049/jimmunol.1001140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kilinc MO, Aulakh KS, Nair RE, Jones SA, Alard P, Kosiewicz MM, Egilmez NK. Reversing tumor immune suppression with intratumoral IL-12: activation of tumor-associated T effector/memory cells, induction of T suppressor apoptosis, and infiltration of CD8+ T effectors. J Immunol. 2006;177:6962–6973. doi: 10.4049/jimmunol.177.10.6962. [DOI] [PubMed] [Google Scholar]
- 44.MacLeod MK, McKee AS, David A, Wang J, Mason R, Kappler JW, Marrack P. Vaccine adjuvants aluminum and monophosphoryl lipid A provide distinct signals to generate protective cytotoxic memory CD8 T cells. Proc Natl Acad Sci U S A. 2011;108:7914–7919. doi: 10.1073/pnas.1104588108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hsieh CS, Macatonia SE, Tripp CS, Wolf SF, O'Garra A, Murphy KM. Development of TH1 CD4+ T cells through IL-12 produced by Listeria-induced macrophages. Science. 1993;260:547–549. doi: 10.1126/science.8097338. [DOI] [PubMed] [Google Scholar]
- 46.Pearce EL, Mullen AC, Martins GA, Krawczyk CM, Hutchins AS, Zediak VP, Banica M, DiCioccio CB, Gross DA, Mao CA, Shen H, Cereb N, Yang SY, Lindsten T, Rossant J, Hunter CA, Reiner SL. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science. 2003;302:1041–1043. doi: 10.1126/science.1090148. [DOI] [PubMed] [Google Scholar]
- 47.Lighvani AA, Frucht DM, Jankovic D, Yamane H, Aliberti J, Hissong BD, Nguyen BV, Gadina M, Sher A, Paul WE, O'Shea JJ. T-bet is rapidly induced by interferon-γ in lymphoid and myeloid cells. Proc Natl Acad Sci U S A. 2001;98:15137–15142. doi: 10.1073/pnas.261570598. %R 10.1073/pnas.261570598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Annunziato F, Cosmi L, Santarlasci V, Maggi L, Liotta F, Mazzinghi B, Parente E, Filì L, Ferri S, Frosali F, Giudici F, Romagnani P, Parronchi P, Tonelli F, Maggi E, Romagnani S. Phenotypic and functional features of human Th17 cells. J Exp Med. 2007;204:1849–1861. doi: 10.1084/jem.20070663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weaver BK, Kumar KP, Reich NC. Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1. Mol Cell Biol. 1998;18:1359–1368. doi: 10.1128/mcb.18.3.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou X, Michal JJ, Zhang L, Ding B, Lunney JK, Liu B, Jiang Z. Interferon induced IFIT family genes in host antiviral defense. Int J Biol Sci. 2013;9:200–208. doi: 10.7150/ijbs.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]