Abstract
Objective
Vascular dysfunction represents a disease initiating event in scleroderma (SSc). Recent data suggest that epigenetic dysregulation impairs normal angiogenesis and can result in abnormal blood vessel growth patterns. Histone deacetylases (HDACs) control endothelial cell (EC) proliferation and regulate EC migration. Specifically, HDAC5 appears to be anti-angiogenic. We hypothesized that HDAC5 contributes to impaired angiogenesis in SSc by repressing pro-angiogenic factors in ECs.
Methods
Dermal ECs were isolated from patients with diffuse cutaneous SSc and healthy controls. Angiogenesis was assessed by an in vitro Matrigel tube formation assay. An assay for transposase-accessible chromatin using sequencing (ATAC-seq) was performed to assess and localize genome-wide effects of HDAC5 knockdown on chromatin accessibility.
Results
The expression of HDAC5 was significantly increased in SSc ECs compared to normal ECs. Silencing of HDAC5 in SSc ECs restored normal angiogenesis. HDAC5 knockdown followed by ATAC-seq in SSc ECs identified key HDAC5-regulated genes involved in angiogenesis and fibrosis, such as CYR61, PVRL2, and FSTL1. Simultaneous knockdown of HDAC5 with either CYR61, PVRL2, or FSTL1 inhibited angiogenesis in SSc ECs while overexpression of these genes individually led to increase in tube formation in Matrigel assay, suggesting that these genes play functional roles in impairing angiogenesis in SSc.
Conclusions
Several novel HDAC5-target genes associated with impaired angiogenesis were identified in SSc ECs by ATAC-seq. This study provides a link between epigenetic regulation and impaired angiogenesis in SSc, and identifies a novel mechanism for dysregulated angiogenesis that characterizes this disease.
Keywords: Scleroderma, epigenetics, HDAC5, vasculopathy, angiogenesis, endothelial cells, chromatin accessibility
Introduction
Systemic sclerosis (scleroderma, SSc) is a poorly understood autoimmune disease characterized by vascular injury and debilitating tissue fibrosis. Activation of endothelial cells (ECs), inflammatory cells, as well as fibroblasts leads to excessive production of extracellular matrices that accumulate in various organs. Widespread vascular damage appears to be an early disease event as morphological changes in the vasculature occur before the onset of tissue fibrosis. This is supported by the early presentation of Raynaud’s phenomenon, a condition that occurs as the first symptom in almost all SSc patients, prior to the occurrence of fibrosis. Other vascular complications include pulmonary arterial hypertension and scleroderma renal crisis, both of which contribute significantly to mortality in SSc (1).
In the skin, SSc vasculopathy leads to loss of dermal capillaries resulting in tissue hypoxia and ischemia, which under normal circumstances, prompts angiogenesis. However, in SSc this compensatory process is impaired and the ECs are incapable of building new blood vessels (2). The mechanism of dysregulated angiogenesis in SSc ECs appears to be multifactorial: the expression of angiogenic-related proteins and transcription factors is altered (2–4), impairment of the urokinase-type plasminogen activator receptor pathway is evident (2), defects in the basic fibroblast growth factor (bFGF) and vascular endothelial growth factor (VEGF) pathway are present (2, 5–7), and changes in chemokine and chemokine receptor expression and signaling occur (2, 4, 8). In addition, SSc ECs can promote fibroblast activation via the CCN2/TGFβ pathway (9). Epigenetic mechanisms in SSc ECs dysfunction is also noted, as the lower expression of bone morphogenetic protein receptor II (BMPR II) in these cells can be modified by inhibitors of DNA methyltransferase and histone deacetylase (HDAC) (10).
Emerging data on the role of epigenetics in angiogenesis are beginning to shed light on abnormal angiogenesis in different diseases. Specifically, HDACs, which enzymatically remove acetyl groups from histones, appear to play significant roles in blood vessel formation. It was reported that non-selective inhibitors of HDACs reduce tube formation of EC in vitro, inhibit postnatal neovascularization in response to hypoxia, and block tumor angiogenesis (11). Moreover, the enzymatic activity of class I and II HDACs is essential for endothelial commitment of progenitor cells (11). Studies that examined individual HDACs in ECs revealed that class IIa HDACs (HDAC5, 7, and 9), class IIb HDACs (HDAC6), as well as class III HDACs (SIRT1) are involved in angiogenesis (11). They have been shown to affect endothelial functions including EC proliferation, migration, and apoptosis. Among them, HDAC5 is anti-angiogenic, as HDAC5 knockdown resulted in EC migration and sprouting (12). It appears that HDAC5 represses a number of angiogenic genes, such as Slit homolog 2 protein (SLIT2) and FGF2 (which encodes fibroblast growth factor 2, also known as bFGF), by binding to their promoter region (12).
In this study, we examined the role of HDAC5 in SSc EC impaired angiogenesis. We first examined the expression of HDAC5 in ECs isolated from healthy volunteers or patients with diffuse cutaneous SSc, and whether knocking it down in SSc ECs altered their angiogenic ability. We then utilized an unbiased approach to assess chromatin accessibility and identify target genes repressed by HDAC5 in ECs, followed by bioinformatics analyses and experimental validation of identified targets. Several HDAC5-regulated genes that play critical role in dysregulated angiogenesis in SSc were identified.
Materials and Methods
Patients
All patients met the American College of Rheumatology/European League Against Rheumatism criteria for the classification of SSc (13). The demographics and clinical characteristics of the enrolled patients are summarized in Table 1. Two 4mm punch biopsies from the distal forearm of healthy volunteers and diffuse cutaneous SSc patients were obtained for EC isolation. All subjects included in this study signed a written informed consent. All procedures in this study were reviewed and approved by the Institutional Review Board of the University of Michigan.
Table 1.
SSc patients and healthy controls characteristics.
| SSc (n=12) | Healthy volunteers (n=8) | |
|---|---|---|
| Age (years) | 50.9 ± 4.2a | 49.8 ± 5.0 |
| Sex | F10/M2 | F6/M2 |
| Diffuse SSc | 12 | N.A.b |
| Disease duration (years) | 3.4 ± 0.7 | N.A. |
| Modified Rodnan Skin Score | 14.3 ± 2.6 | N.A. |
| Raynaud’s phenomenon | 12 | N.A. |
| Early disease (< 5yrs) | 10 | N.A. |
| Digital ulcers | 2 | N.A. |
| Teleangectasias | 7 | N.A. |
| Gastrointestinal disease | 9 | N.A. |
| Interstitial lung disease | 9 | N.A. |
| Pulmonary arterial hypertension | 4 | N.A. |
| Renal involvement | 1 | N.A. |
Mean ± SEM
N.A. = Not applicable
Cell culture
Dermal ECs were isolated and characterized in our laboratory as previously described (4, 6). After digestion ECs were purified using the CD31 MicroBead Kit and a MiniMACS™ Separator with an MS Column (Miltenyi Biotech) and grown in EBM-2 media with growth factors (Lonza). Cells between passage 3 and 6 were used in the experiments.
mRNA extraction and qRT-PCR
Total RNA from cells was isolated using Direct-zol™ RNA MiniPrep Kit (Zymo Research) or RNeasy Mini kit (Qiagen). Verso cDNA synthesis kit was used to prepare cDNA (Thermo Scientific). Primers for human HDAC4, 5, 6, 7, 9, and 10, VEGF, and β-actin along with Power SYBR Green PCR master mix (Applied Biosystems) were applied for qPCR, which was run by either the ViiA™ 7 Real-Time PCR System or the Applied Biosystems Real-Time PCR System. Primer sequences are as follows: HDAC4 FW:TGTACGACGCCAAAGATGAC; RV: CGGTTCAGAAGCTGTTTTCC; HDAC5 FW: CAGCAGGCGTTCTACAATGA; RV: CGATGCAGAGAGATGTAGAGCA; HDAC6 FW: GAAAGTCACCTCGGCATCAT; RV: TAGTCTGGCCTGGAGTGGAC; HDAC7 FW: ATGGGGGATCCTGAGTACCT; RV: GATGGGCATCACGACTATCC; HDAC9 FW: CTGGAGCCCATCTCACCTT; RV: TCATCATCCTGAGGTCTGTCC; HDAC10 FW: GCCGGATATCACATTGGTTC; RV: GACGCTTCCTGTTGGATGA; VEGF FW: ATGAACTTTCTGCTGTCTTGGGT; RV: TGGCCTTGGTGAGGTTTGATCC; β-actin FW: GTCAGGCAGCTCGTAGCTCT; RV: GCCATGTACGTTGCTATCCA. The rest of the primers were KiCqStart® SYBR® Green Primers from Sigma.
Western blots
Cell lysate was obtained from both healthy subjects and SSc patients. Equal amounts of protein were separated by SDS-PAGE and electroblotted onto nitrocellulose membranes. HDAC5 proteins were detected using anti-human HDAC5 antibodies (Cell Signaling) while β-actin was used as a loading control (anti-β-actin antibodies were from Sigma Aldrich). Band quantification was performed using GelQuant.NET (BiochemLab Solutions).
Gene knockdown experiments
To evaluate the effect of HDAC5 on angiogenesis in SSc ECs, its expression was knocked down using HDAC5 siRNA from Santa Cruz Biotechnology. siRNA from Life Technologies was used as control. We first utilized healthy ECs to establish the transfection condition. ECs were transfected with 25–75 nM of siRNA with TransIT-TKO transfection reagent (Mirus Bio) for 48 hours and mRNA/protein lysate was collected. We then confirmed the knockdown of HDAC5 using SSc ECs. The knockdown condition was also optimized for PVRL2 (50nM), CTNNAL1 (50nM), FSTL1 (25nM), FSTL3 (25nM), (siRNA from Santa Cruz Biotechnology), ID2 (150nM), and CYR61 (25nM), (siRNA from GE Dharmacon).
Gene overexpression experiments
Overexpression of PVRL2, FSTL1, and CYR61 was performed as previously described with some modification (4). ECs were transfected with 0.33 μg of FSTL1, PVRL2 (both from Origene; control vector pCMV6-XL5), or 1.65 μg of CYR61 (Origene; control vector pCMV6-XL4) and lipofectamine 2000 (Invitrogen) for 24 hours in T12.5 flasks. Five hours after transfection, the culture media was changed to allow the cells to grow in EGM supplemented with bovine brain extract (Lonza). Subsequent analysis for qPCR or Matrigel tube formation assay was performed.
Enzyme-linked immunosorbent assay (ELISA)
The levels of VEGF and bFGF in cell culture supernatants were measured using ELISA kits from R&D systems. The absorbance of each well was read using a microplate reader at 450 nm.
Matrigel tube formation assay
The Matrigel tube formation assay was performed to evaluate the effect of HDAC5 on angiogenesis (6). Transfected SSc ECs were suspended in EBM-2 with 0.1% fetal bovine serum and plated in 8-well Lab-Tek chambers coated with growth factor reduced Matrigel (BD Biosciences). The cells were fixed and stained after overnight incubation. Quantitation of the tubes formed by ECs was performed by a blinded researcher. Pictures of each well were taken using EVOS XL Core Cell Imaging System (Life Technologies). In the case of the double knockdown study, SSc ECs were transfected with control siRNA, HDAC5 siRNA, or HDAC5 plus the siRNA of genes of interest for 48 hours, and then plated on Matrigel. For the overexpression experiments, SSc ECs were transfected with control vector, FSTL1, PVRL2, or CYR61 and the Matrigel assay was carried out the next day.
Assay for transposase-accessible chromatin using sequencing (ATAC-seq)
To characterize the impact of HDAC5 knockdown on chromatin accessibility at a genome-wide level, ATAC-seq was performed (14). This utilizes a hyperactive Tn5 transposase that simultaneously cuts and ligates adaptors at regions of open chromatin. Since HDAC5 knockdown alters the chromatin structure, application of ATAC-seq in this study allowed us to assess genome-wide chromatin accessibility in SSc ECs with high degree of accuracy and sensitivity. After transfection, 50,000 ECs were collected and lysed in 50 μL lysing buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% IGEPAL CA-630). After centrifugation the supernatant was discarded and the pellet was used for transposition. The transposition reaction included 25 μL 2×TD buffer (Illumina, San Diego, CA), 2.5 μLTn5 transposase (Illumina, San Diego, CA), and 22.5 μL water. The mixture was incubated at 37°C for 30 min. After purification, DNA libraries were prepared by PCR and purified using a Qiagen MinElute Kit. To assess the quality DNA libraries, gel electrophoresis was performed and the DNA was visualized using Omega Lum C (Aplegen Inc). Libraries that passed quality control measures as previously described (14) were sequenced with 50bp, pair-end reads on the Illumina Hi-Seq 2500 platform (three samples per lane).
ATAC-seq data analysis
Sequencing reads were aligned to the human genome assembly (hg19) using BWA (v0.7.5) (15). Improperly-paired alignment and alignment with mapping quality less than 4 were filtered using SAMtools (v0.1.19) (16). Peak-calling was then performed using MACS2 (v2.0.10) (17), with reads shifted and extended by the parameters “--nomodel --shift -100 --extsize 200 -B --broad”. Blacklisted genomic regions (18) were removed from the peaks. Comparison between HDAC5 knockdown and control was performed using diffReps (v1.55.4) (19), with the three individuals treated as triplicates for each condition. Running diffReps with negative binomial model and 1e-4 p-value threshold identified peaks that have significantly different signal abundance between the two conditions, with a p-value of less than 0.05 after being adjusted by the Benjamini-Hochberg method.
Bioinformatics analysis
To identify genes that are relevant to SSc, genes with differential chromatin accessibility with HDAC5 knockdown in ECs were subjected to bioinformatics analysis using IRIDESCENT (20). This analysis searches for co-occurrence of the genes and the key words (i.e. fibrosis or angiogenesis) within the scientific literature. A statistical method was used to score the relevance of the co-occurrence by comparing the observed frequencies to ones randomly occurring by chance (20).
Statistical analysis
Results were expressed as mean ± S.D. To determine the differences between the groups in Figure 1, Mann-Whitney U test was performed using GraphPad Prism version 6 (GraphPad Software, Inc). To compare the biomarkers before and after HDAC5 knockdown in Table 3, a paired t-test was performed. P-values of less than 0.05 were considered statistically significant.
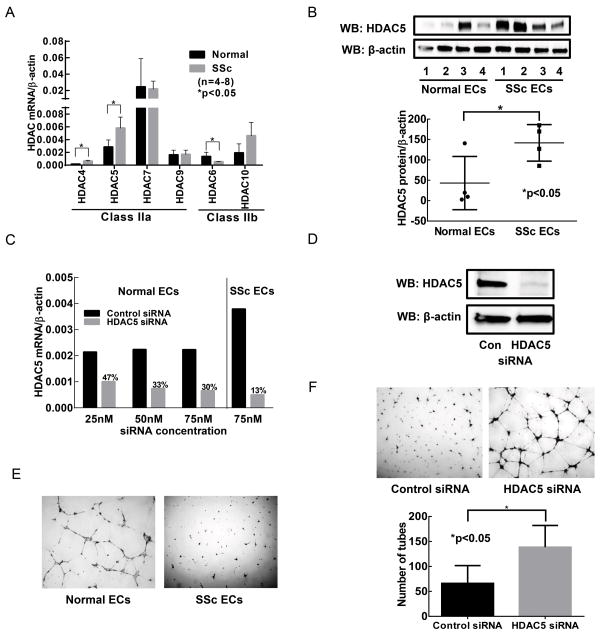
Figure 1. The effect of HDAC5 expression on the angiogenic ability of SSc ECs.
(A) The expression of HDAC4 and HDAC5, both class IIa HDACs, was significantly elevated in SSc ECs compared to healthy ECs, while class IIb HDAC6 was down-regulated (p<0.05). (B) HDAC5 protein levels were upregulated in SSc patients compared to healthy controls. (C) HDAC5 was knocked down in both healthy and SSc ECs. The use of 75 nM of HDAC5 siRNA resulted in more than 70% knockdown in both cell types. (D) HDAC5 knockdown was confirmed at the protein level. (E) SSc ECs were unable to form tubes on Matrigel while normal ECs spontaneously form tubes, showing the anti-angiogenic nature of SSc ECs. (F) Knockdown of HDAC5 in SSc ECs led to increased tube formation on Matrigel. Quantification of the tubes was done by counting the tubes by a blinded investigator. Significant increase in tube formation after HDAC5 knockdown was observed (p<0.05). n=3 patient lines. Results are expressed as mean +/− SD and p<0.05 was considered significant.
Table 3.
mRNA expression levels of candidate genes after HDAC5 knockdown in SSc ECs compared to control siRNA-treated cells (mean +/− S.D).
| Gene | Expression fold change vs. control (n=7 patient pairs) | p-value |
|---|---|---|
| ID2 | 2.188 ± 1.002 | 0.02 |
| CTNNAL1 | 2.064 ± 0.880 | 0.03 |
| MBP | 4.073 ± 5.550 | 0.03 |
| PVRL2 | 3.091 ± 1.600 | 0.02 |
| CYR61 | 2.423 ± 1.562 | 0.03 |
| FSTL1 | 2.542 ± 1.514 | 0.04 |
| FSTL3 | 4.586 ± 4.358 | 0.03 |
| SQSTM1 | 2.080 ± 1.419 | 0.04 |
| KRAS | 1.884 ± 1.250 | 0.08 |
| SOD2 | 3.446 ± 2.591 | 0.08 |
| NINJ1 | 1.849 ± 1.469 | 0.55 |
| THSD1 | 1.762 ± 1.106 | 0.43 |
| FGF18 | 1.023 ± 1.576 | 0.35 |
| FAM198 | 1.086 ± 0.793 | 0.86 |
| TAB2 | 1.532 ± 1.017 | 0.75 |
| TMEM18 | 1.339 ± 0.688 | 0.99 |
| IQSEC1 | 1.301 ± 0.661 | 0.22 |
| NFYA | 1.757 ± 1.622 | 0.19 |
| ETV5 | 2.453 ± 2.228 | 0.83 |
| EDEM1 | 1.342 ± 0.685 | 0.35 |
| HSF2 | 1.807 ± 1.062 | 0.14 |
| MAP3K3 | 1.417 ± 0.898 | 0.55 |
| PRDX3 | 1.365 ± 0.664 | 0.31 |
| HSPA2 | 1.152 ± 0.720 | 0.31 |
| MATN1 | N.D. |
N.D.=Not Detected
Results
Aberrant expression of class II HDACs in SSc ECs
We examined the expression of class IIa and IIb HDACs using qPCR. As shown in Figure 1A, HDAC4 and 5, both class IIa HDACs, were upregulated in SSc ECs compared to control ECs, while HDAC6, a class IIb HDAC, was downregulated (p<0.05). We further confirmed increased HDAC5 expression in SSc ECs at the protein level (Figure 1B). Although HDAC5 expression was variable among SSc patients, there was a significant increase in HDAC5 expression in SSc ECs (p<0.05, n=4). Since HDAC5 is anti-angiogenic, we proceeded to knockdown HDAC5 and examined whether the decrease in HDAC5 altered the angiogenic ability of SSc ECs. We first established the HDAC5 knockdown condition (Figure 1C). In healthy ECs, 75 nM of HDAC5 siRNA resulted in approximately 70% knockdown of HDAC5 compared to cells transfected with control siRNA. We used this condition in SSc ECs and achieved approximately 87% knockdown. Under this condition, we confirmed that at the protein level, HDAC5 was indeed negligible (Figure 1D). Therefore in subsequent experiments the condition we used for HDAC5 knockdown was to use 75 nM siRNA for 48 hours.
HDAC5 knockdown increased tube formation in SSc ECs
We then tested whether a decrease in HDAC5 expression altered the angiogenic ability of SSc ECs using the Matrigel tube formation assay. We first used ECs isolated from healthy subjects and SSc patients and performed Matrigel tube formation assay. Similar to previous reports (3, 5, 7), in the absence of an angiogenic stimulus SSc ECs were unable to form tube-like structures on Matrigel, while healthy ECs could (Figure 1E). SSc ECs transfected with control siRNA also demonstrated a diminished capability to form tubes (Figure 1F). In contrast, knocking down HDAC5 restored the ability of SSc EC to form tubes and resulted in approximately a two fold increase in the number of tubes formed (p<0.05).
ATAC-seq revealed differential open-chromatin regions after HDAC5 knockdown in SSc ECs
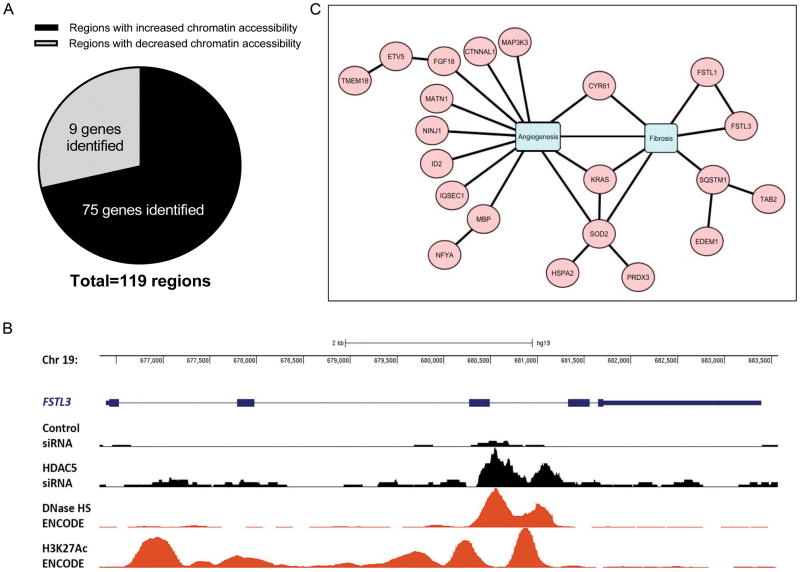
HDAC5 knockdown showed increased chromatin accessibility compared to control siRNA-treated cells (Figure 2A). A total of 119 regions were identified with at least a 20% fold difference (≥1.2 fold or ≤0.80 fold) in chromatin accessibility, represented by a change in sequencing peak intensity (adjusted p-value <0.05). Of those, there were 85 regions with increased chromatin accessibility, and 34 regions with decreased chromatin accessibility. In the regions of increased chromatin accessibility, we identified a total of 75 genes located in these sites (Figure 2A). In contrast, only 9 genes were located in less-accessible regions. The genes that were located in the more accessible regions that had a 1.5 fold increase in peak intensity between HDAC5 knockdown and control were listed in Table 2. Figure 2B shows genome tracks of the FSTL3 locus. Knockdown of HDAC5 increased chromatin accessibility as indicated by the increase in peak intensity compared to control-siRNA treated cells. For validation we compared our ATAC-seq data with chromatin-accessibility data sets such as DNase-seq. The ATAC-seq peaks correlated with DNases hypersensitivity regions generated in human umbilical ECs (HUVECs, generated by University of Washington ENCODE group). In addition, we observed that ATAC-seq data aligned with ChIP-seq peaks from the ENCODE project (21) for histone marks associated with active chromatin (H3K27ac).
Figure 2. ATAC-seq revealed differential open-chromatin regions after HDAC5 knockdown in SSc ECs.
(A) HDAC5 knockdown increased chromatin accessibility compared to control siRNA transfected cells. With a cut-off of 20% change with a p-value adjusted for multiple testing less than 0.05, we identified a total of 119 regions: 85 of them were regions with increased chromatin accessibility, and 34 were regions with decreased chromatin accessibility. In the regions of increased chromatin accessibility, 75 genes were located in these sites and only 9 genes were located in less-accessible regions. (B) UCSC genome browser tracks of the FSTL3 locus with data of ATAC-seq signals from control or HDAC5 siRNA treated samples, DNase I hypersensitivity mapping from human umbilical endothelial cells (HUVECs, generated by the ENCODE project), as well as H3K27ac ChIP (HUVECs, ENCODE). HDAC5 knockdown resulted in increase in peak intensity compared to control siRNA. The ATAC-seq signals were very similar compared to DNase I hypersensitivity sequencing and overlapped H3K27ac enrichment in HUVECs. (C) Genes located in the more-accessible chromatin regions after HDAC5 knockdown were analyzed using IRIDESCENT to identify genes that were related to angiogenesis or fibrosis.
Table 2.
Genes located in regions with at least 1.5 fold increase in chromatin accessibility following HDAC5 knockdown.
| Chr | Positions | Gene | Fold change (HDAC5 vs. control) | p-value | Adjusted p-value |
|---|---|---|---|---|---|
| chr19 | 680601-681600 | FSTL3 | 2.17 | 9.32E-06 | 8.14E-04 |
| chr7 | 156930501-156931700 | UBE3C | 1.93 | 2.96E-06 | 3.57E-04 |
| chr13 | 52979801-52980900 | THSD1 | 1.84 | 1.54E-06 | 2.48E-04 |
| chr21 | 47604401-47605600 | C21orf56 | 1.83 | 2.43E-06 | 3.32E-04 |
| chr1 | 156697701-156698700 | RRNAD1 | 1.77 | 8.04E-05 | 3.91E-03 |
| chr12 | 9066701-9067700 | PHC1 | 1.75 | 4.40E-06 | 4.95E-04 |
| chr11 | 134122401-134124000 | THYN1 | 1.73 | 3.93E-06 | 4.49E-04 |
| chr17 | 77070501-77071500 | ENGASE | 1.73 | 6.03E-05 | 3.26E-03 |
| chr14 | 96570401-98571900 | - | 1.72 | 1.61E-06 | 2.48E-04 |
| chr3 | 120169201-120170200 | FSTL1 | 1.65 | 1.09E-05 | 9.27E-04 |
| chr3 | 49058601-49059800 | NDUFAF3 | 1.64 | 2.04E-06 | 3.08E-04 |
| chr6 | 74363001-74364300 | SLC17A5 | 1.62 | 2.79E-05 | 1.93E-03 |
| chr2 | 238600001-238601100 | LRRFIP1 | 1.61 | 1.13E-05 | 9.53E-04 |
| chr5 | 179233401-179234700 | SQSTM1 | 1.61 | 2.70E-06 | 3.49E-04 |
| chr1 | 86045601-86047200 | CYR61 | 1.60 | 2.31E-06 | 3.27E-04 |
| chr17 | 61698601-61699700 | MAP3K3 | 1.60 | 8.41E-06 | 7.55E-04 |
| chr17 | 38108801-38109900 | - | 1.59 | 9.42E-05 | 4.43E-03 |
| chr19 | 45348901-45349900 | PVRL2 | 1.57 | 2.79E-06 | 3.49E-04 |
| chr2 | 26256501-26257700 | RAB10 | 1.56 | 9.68E-05 | 4.45E-03 |
| chr5 | 40834901-40835900 | RPL37 | 1.56 | 8.95E-05 | 4.25E-03 |
| chr12 | 109219701-109220800 | SSH1 | 1.55 | 3.23E-05 | 2.14E-03 |
| chr2 | 112811101-112812900 | TMEM87B | 1.54 | 2.56E-05 | 1.85E-03 |
| chr7 | 105752501-105753600 | SYPL1 | 1.54 | 1.19E-05 | 9.77E-04 |
| chr9 | 95895401-95896500 | NINJ1 | 1.54 | 2.45E-06 | 3.32E-04 |
| chr6 | 160114401-160115600 | SOD2 | 1.53 | 2.74E-06 | 3.49E-04 |
| chr9 | 77702101-77703100 | C9orf95 | 1.53 | 2.28E-06 | 3.27E-04 |
| chr11 | 112096901-112098000 | PTS | 1.52 | 8.11E-06 | 7.37E-04 |
| chr12 | 22696601-22697600 | KIAA0528 | 1.52 | 8.08E-05 | 3.91E-03 |
| chr20 | 57267501-57268600 | NPEPL1 | 1.52 | 2.22E-04 | 9.74E-03 |
| chr1 | 171710501-171711500 | VAMP4 | 1.51 | 3.49E-05 | 2.27E-03 |
Up-regulation of genes involved in angiogenesis or fibrosis after HDAC5 knockdown
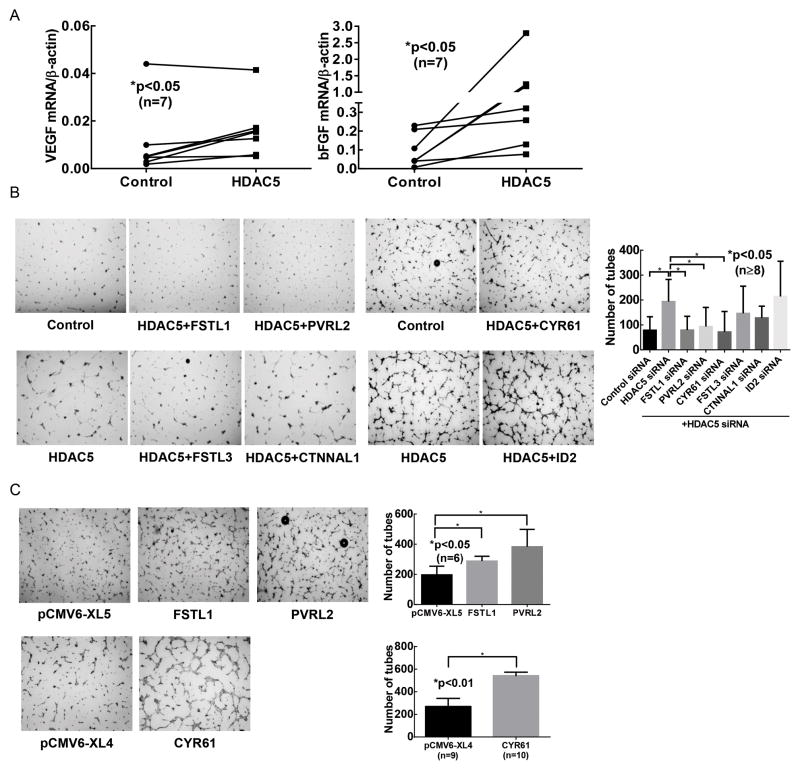
Bioinformatics analysis using IRIDESCENT revealed that among genes that are located in the regions of increased chromatin accessibility in SSc ECs after HDAC5 knockdown, at least 16 genes might be involved in angiogenesis and a few involved in fibrosis (Figure 2C). Three genes were identified as possibly involved in both angiogenesis and fibrosis (CYR61, KRAS, and SOD2). We then examined the expression of the genes listed in Figure 2C in SSc ECs before and after HDAC5 knockdown. We found 8 genes to be significantly upregulated (Table 3, p<0.05). These include ID2, MBP, PVRL2, CTNNAL1, FSTL3, SQSTM1, CYR61, and FSTL1. In addition to the genes identified by ATAC-seq, we also examined the expression of two pro-angiogenic factors, VEGF and bFGF. As shown in Figure 3A, both VEGF and bFGF mRNA levels were significantly upregulated after HDAC5 knockdown. We also observed 1.5 to 2-fold increase in protein levels of both VEGF and bFGF in SSc ECs culture supernatants after HDAC5 knockdown (data not shown).
Figure 3. Several upregulated pro-angiogenic factors promote tube formation after HDAC5 knockdown.
(A) The expression of VEGF and bFGF was quantified by qPCR. Increased expression was observed in both cases after HDAC5 knockdown. (B) Matrigel tube formation was performed after diffuse cutaneous SSc ECs were transfected with siRNA of control, HDAC5, or HDAC5 plus genes of interest. PVRL2, FSTL1, and CYR61 appeared to play key role in promoting angiogenesis after HDAC5 knockdown, while FSTL3, ID2, and CTNNAL1 did not. n=4 patient lines. (C) Overexpression of PVRL2, FSTL1, and CYR61 resulted in increase in tube formation by SSc ECs. Results are expressed as mean +/− SD and p<0.05 was considered significant.
Identification of genes that play functional roles in HDAC5-mediated angiogenesis
We performed double knockdown experiments to identify genes that led to increase in tube formation after HDAC5 knockdown. We chose angiogenic genes that were significantly upregulated after HDAC5 knockdown, as listed in Table 3. This included ID2, PVRL2, CYR61, and CTNNAL1. In addition, FSTL1 and FSTL3 are also included since upon comprehensive literature search, they appear to be pro-angiogenic. MBP was excluded as we were unable to identify its role in angiogenesis. As shown in Figure 3B, in SSc ECs, when FSTL1, PVRL2, or CYR61 were knocked down together with HDAC5, the number of tubes on Matrigel decreased, while transfecting the cells with ID2, FSTL3 or CTNNAL1 siRNA had minimal effect, suggesting that HDAC5 mediated its anti-angiogenic effect at least in part through changing chromatin accessibility of FSTL1, PVRL2, and CYR61. To further confirm their roles, we overexpressed FSTL1, PVRL2, or CYR61 individually in SSc ECs and showed that increase expression of these genes led to increased angiogenic ability (Figure 3C).
Discussion
In this study, we identified a novel epigenetic mechanism that contributes to the inhibition of angiogenesis in SSc. HDAC5, which was overexpressed in SSc ECs, inhibited angiogenesis by repressing pro-angiogenic genes. This was achieved by applying ATAC-seq to assess changes in the chromatin state after HDAC5 knockdown, thereby identifying key genes located in regions with increased chromatin accessibility. Several angiogenic and fibrotic-related genes were pinpointed, as their expression increased significantly after HDAC5 knockdown. Among the pro-angiogenic genes identified by ATAC-seq, we showed that PVRL2, FSTL1, and CYR61 played critical roles in promoting angiogenesis in SSc ECs, as overexpression of these genes resulted in increase in tube formation by SSc ECs, while knocking down these genes together with HDAC5 led to decrease in tube formation.
It is apparent that epigenetic changes are involved in SSc vasculopathy, as in this study, we showed that HDAC5 was involved in impaired angiogenesis in SSc ECs. Indeed our results complement the work in fibrosis, where it was shown that histone acetylation patterns differed in SSc patients and that the use of HDAC inhibitors showed promising anti-fibrotic effect both in vivo and in vitro (22, 23). However, global inactivation of HDACs by nonspecific inhibitors may affect angiogenesis by modifying both pro- and anti-angiogenic HDACs. These pan-HDAC inhibitors target class I, II and/or IV HDACs that govern cell proliferation, differentiation, migration, and angiogenesis. They inhibit angiogenesis through VEGF and hypoxia-inducible factor (24, 25), affect class IIb HDAC-mediated cell migration, and induce class I HDAC-mediated cell apoptosis (26). Therefore one should be cautious when using HDAC inhibitors to treat fibrosis in SSc, as this may be detrimental to the vasculopathic component of this disease.
Our results agree with the study by Ulbich et al. in that HDAC5 knockdown led to increase in tube formation by ECs on Matrigel (12). They showed that target genes regulated by HDAC5 include FGF2, SLIT2, and EPHB4. We showed that bFGF (encoded by FGF2) was upregulated after HDAC5 knockdown (Figure 3A), and also identified additional genes that are repressed by HDAC5, including PVRL2, FSTL1 and CYR61 (Table 3). The discrepancies between the two studies might be due to the different type of ECs used. The methods that were applied to identify genes were also different (ATAC-seq vs. oligonucleotide array). Of note, the discrepancy between bFGF and VEGF expression and the ATAC-seq results may seem contradictory; however, it is not uncommon for chromatin accessibility and gene expression not to correlate with each other. As shown in Table 3, only 8 of the 25 genes that showed significant increase in chromatin accessibility had increased mRNA expression. These results point to the importance of studying the epigenome and transcriptome simultaneously, as the integration of data obtained from both approaches would greatly facilitate target gene identification.
We identified several pro-angiogenic factors controlled by HDAC5 (Table 3). CYR61 (also known as CCN1) is a member of the CCN protein family that binds to the extracellular matrix and supports angiogenesis (27). In contrast to CCN2 (also known as CTGF, connective tissue growth factor) which is pro-fibrotic, CYR61 is antifibrotic through activation of an integrin/oxidative stress-mediated pathway (28). Saigusa et al showed that CYR61 is downregulated in dermal small blood vessels of SSc patients compared to controls (29). We postulate that the anti-fibrotic nature of this molecule should also play a significant role in inhibiting the fibrotic process in SSc, since it has been shown that CYR61 restricted fibrosis in several models of fibrosis including the skin (28, 30, 31). However the role of CYR61 in SSc fibrosis awaits further investigation.
The role of FSTL1 in fibrosis is shown mainly in pulmonary fibrosis; depletion or blockade of FSTL1 in mice attenuated bleomycin-induced lung fibrosis (32). However, contradicting results were obtained in a renal fibrosis mouse model (33). Although our bioinformatics analysis showed that FSTL1 played a role solely in fibrosis (Figure 2C), upon a more detailed literature search, we found that it is also involved in EC proliferation and tube formation (34). Although its role in SSc pathogenesis remains to be explored, it has been shown that FSTL1 is elevated in SSc serum (35). In this study, we showed that the expression of FSTL1 increased after HDAC5 knockdown, agreeing with a previous study showing that FSTL1 is regulated by histone acetylation (36).
PVRL2 is a cell adhesion molecule that connects to the actin cytoskeleton by binding to afadin (37). The mechanism of how nectin-2 promotes angiogenesis is not known. It is possible that it is mediated directly by its cell adhesion properties, but also indirectly through afadin, which promotes angiogenesis (38), or through interacting with other members of adhesion molecules (39). As adhesion molecules have been suggested to play a role in SSc pathogenesis (2), our finding that PVRL2 improves the angiogenic ability of SSc ECs offers new insights as how these adhesion molecules are involved in this disease.
It appears that HDAC5 also controls fibrosis through SSc ECs, as a few fibrotic genes, including FSTL1, FSTL3, and SQSTM1, were identified to be upregulated after HDAC5 knockdown in our study (Table 3). These genes are involved in the TGFβ (FSTL1 and FSTL3) and autophagy pathways (SQSTM1) (40, 41). FSTL3 also plays a role in angiogenesis through inhibiting activin A (42, 43). However in our study, simultaneous knockdown of HDAC5 and FSTL3 did not reverse the angiogenic phenotype of SSc ECs. Although autophagy has been shown to play a role in various fibrotic disorders including SSc (44, 45), whether SQSTM1 is critical warrants additional studies.
In addition to its role in angiogenesis, HDAC5 appears to be involved in inflammation and immunity. It controls the inflammatory responses of macrophages and T cell functions (46, 47). In addition, HDAC5 negatively regulates chemokine and cytokine production in fibroblast-like synoviocytes in rheumatoid arthritis (48). Therefore, it is also possible that HDAC5 has functional roles in the inflammatory and autoimmune aspect of SSc pathogenesis.
In conclusion, our results clearly show that epigenetic mechanisms contribute significantly to impaired angiogenesis in SSc. Increased expression of anti-angiogenic HDAC5 repressed several pro-angiogenic genes resulting in dysregulated angiogenesis in SSc ECs. Coupling ATAC-seq with functional assays, we identified novel angiogenic and fibrotic genes that warrant additional studies to delineate their roles in SSc pathogenesis.
Acknowledgments
We thank Paul Renauer for his assistance in preparing figures for this manuscript.
Funding: This work was supported by National Institutes of Health/National Institute of Arthritis and Musculoskeletal and Skin Diseases grant number T32AR007080 and the Scleroderma Foundation to Dr. Tsou. Dr. Sawalha is supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health grants number R01AI097134 and U19AI110502. Dr. Khanna is supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases grant number K24AR063120 and the National Institute of Allergy and Infectious Diseases grant number UM1AI110557. Dr. Fox is supported by the National Institute of Allergy and Infectious Diseases grant number UM1AI110557. Dr. Wren is supported by the National Institutes of Health grant number P20GM103636.
Footnotes
Conflict of interest: None of the authors has any financial conflict of interest with the work presented
References
- 1.Steen VD, Medsger TA. Changes in causes of death in systemic sclerosis, 1972–2002. Ann Rheum Dis. 2007;66(7):940–4. doi: 10.1136/ard.2006.066068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rabquer BJ, Koch AE. Angiogenesis and vasculopathy in systemic sclerosis: evolving concepts. Current rheumatology reports. 2012;14(1):56–63. doi: 10.1007/s11926-011-0219-1. [DOI] [PubMed] [Google Scholar]
- 3.Mazzotta C, Romano E, Bruni C, Manetti M, Lepri G, Bellando-Randone S, et al. Plexin-D1/Semaphorin 3E pathway may contribute to dysregulation of vascular tone control and defective angiogenesis in systemic sclerosis. Arthritis Res Ther. 2015;17(1):221. doi: 10.1186/s13075-015-0749-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsou PS, Rabquer BJ, Ohara RA, Stinson WA, Campbell PL, Amin MA, et al. Scleroderma dermal microvascular endothelial cells exhibit defective response to pro-angiogenic chemokines. Rheumatology (Oxford) 2015 doi: 10.1093/rheumatology/kev399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Romano E, Chora I, Manetti M, Mazzotta C, Rosa I, Bellando-Randone S, et al. Decreased expression of neuropilin-1 as a novel key factor contributing to peripheral microvasculopathy and defective angiogenesis in systemic sclerosis. Annals of the Rheumatic Diseases. 2015 doi: 10.1136/annrheumdis-2015-207483. [DOI] [PubMed] [Google Scholar]
- 6.Tsou PS, Amin MA, Campbell P, Zakhem G, Balogh B, Edhayan G, et al. Activation of the Thromboxane A2 Receptor by 8-Isoprostane Inhibits the Pro-Angiogenic Effect of Vascular Endothelial Growth Factor in Scleroderma. J Invest Dermatol. 2015 doi: 10.1038/jid.2015.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manetti M, Guiducci S, Romano E, Ceccarelli C, Bellando-Randone S, Conforti ML, et al. Overexpression of VEGF165b, an inhibitory splice variant of vascular endothelial growth factor, leads to insufficient angiogenesis in patients with systemic sclerosis. Circ Res. 2011;109(3):e14–26. doi: 10.1161/CIRCRESAHA.111.242057. [DOI] [PubMed] [Google Scholar]
- 8.Ichimura Y, Asano Y, Akamata K, Takahashi T, Noda S, Taniguchi T, et al. Fli1 deficiency contributes to the suppression of endothelial CXCL5 expression in systemic sclerosis. Arch Dermatol Res. 2014;306(4):331–8. doi: 10.1007/s00403-013-1431-9. [DOI] [PubMed] [Google Scholar]
- 9.Serratì S, Chillà A, Laurenzana A, Margheri F, Giannoni E, Magnelli L, et al. Systemic sclerosis endothelial cells recruit and activate dermal fibroblasts by induction of a connective tissue growth factor (CCN2)/transforming growth factor β–dependent mesenchymal-to-mesenchymal transition. Arthritis & Rheumatism. 2013;65(1):258–69. doi: 10.1002/art.37705. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, Kahaleh B. Epigenetic repression of bone morphogenetic protein receptor II expression in scleroderma. J Cell Mol Med. 2013 doi: 10.1111/jcmm.12105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turtoi A, Peixoto P, Castronovo V, Bellahcene A. Histone deacetylases and cancer-associated angiogenesis: current understanding of the biology and clinical perspectives. Critical reviews in oncogenesis. 2015;20(1–2):119–37. doi: 10.1615/critrevoncog.2014012423. [DOI] [PubMed] [Google Scholar]
- 12.Urbich C, Rossig L, Kaluza D, Potente M, Boeckel JN, Knau A, et al. HDAC5 is a repressor of angiogenesis and determines the angiogenic gene expression pattern of endothelial cells. Blood. 2009;113(22):5669–79. doi: 10.1182/blood-2009-01-196485. [DOI] [PubMed] [Google Scholar]
- 13.van den Hoogen F, Khanna D, Fransen J, Johnson SR, Baron M, Tyndall A, et al. 2013 classification criteria for systemic sclerosis: an American college of rheumatology/European league against rheumatism collaborative initiative. Ann Rheum Dis. 2013;72(11):1747–55. doi: 10.1136/annrheumdis-2013-204424. [DOI] [PubMed] [Google Scholar]
- 14.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10(12):1213–8. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS) Genome biology. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen L, Shao NY, Liu X, Maze I, Feng J, Nestler EJ. diffReps: detecting differential chromatin modification sites from ChIP-seq data with biological replicates. PloS one. 2013;8(6):e65598. doi: 10.1371/journal.pone.0065598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wren JD, Garner HR. Shared relationship analysis: ranking set cohesion and commonalities within a literature-derived relationship network. Bioinformatics. 2004;20(2):191–8. doi: 10.1093/bioinformatics/btg390. [DOI] [PubMed] [Google Scholar]
- 21.Ram O, Goren A, Amit I, Shoresh N, Yosef N, Ernst J, et al. Combinatorial patterning of chromatin regulators uncovered by genome-wide location analysis in human cells. Cell. 2011;147(7):1628–39. doi: 10.1016/j.cell.2011.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huber LC, Distler JH, Moritz F, Hemmatazad H, Hauser T, Michel BA, et al. Trichostatin A prevents the accumulation of extracellular matrix in a mouse model of bleomycin-induced skin fibrosis. Arthritis Rheum. 2007;56(8):2755–64. doi: 10.1002/art.22759. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Fan PS, Kahaleh B. Association between enhanced type I collagen expression and epigenetic repression of the FLI1 gene in scleroderma fibroblasts. Arthritis Rheum. 2006;54(7):2271–9. doi: 10.1002/art.21948. [DOI] [PubMed] [Google Scholar]
- 24.Deroanne CF, Bonjean K, Servotte S, Devy L, Colige A, Clausse N, et al. Histone deacetylases inhibitors as anti-angiogenic agents altering vascular endothelial growth factor signaling. Oncogene. 2002;21(3):427–36. doi: 10.1038/sj.onc.1205108. [DOI] [PubMed] [Google Scholar]
- 25.Kim SH, Jeong JW, Park JA, Lee JW, Seo JH, Jung BK, et al. Regulation of the HIF-1alpha stability by histone deacetylases. Oncology reports. 2007;17(3):647–51. [PubMed] [Google Scholar]
- 26.Khan O, La Thangue NB. HDAC inhibitors in cancer biology: emerging mechanisms and clinical applications. Immunology and cell biology. 2012;90(1):85–94. doi: 10.1038/icb.2011.100. [DOI] [PubMed] [Google Scholar]
- 27.Jun JI, Lau LF. Taking aim at the extracellular matrix: CCN proteins as emerging therapeutic targets. Nature reviews Drug discovery. 2011;10(12):945–63. doi: 10.1038/nrd3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jun JI, Lau LF. The matricellular protein CCN1 induces fibroblast senescence and restricts fibrosis in cutaneous wound healing. Nature cell biology. 2010;12(7):676–85. doi: 10.1038/ncb2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saigusa R, Asano Y, Taniguchi T, Yamashita T, Takahashi T, Ichimura Y, et al. A possible contribution of endothelial CCN1 downregulation due to Fli1 deficiency to the development of digital ulcers in systemic sclerosis. Experimental dermatology. 2015;24(2):127–32. doi: 10.1111/exd.12602. [DOI] [PubMed] [Google Scholar]
- 30.Kim KH, Chen CC, Monzon RI, Lau LF. Matricellular protein CCN1 promotes regression of liver fibrosis through induction of cellular senescence in hepatic myofibroblasts. Mol Cell Biol. 2013;33(10):2078–90. doi: 10.1128/MCB.00049-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qin Z, Fisher GJ, Quan T. Cysteine-rich protein 61 (CCN1) domain-specific stimulation of matrix metalloproteinase-1 expression through alphaVbeta3 integrin in human skin fibroblasts. J Biol Chem. 2013;288(17):12386–94. doi: 10.1074/jbc.M112.424358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dong Y, Geng Y, Li L, Li X, Yan X, Fang Y, et al. Blocking follistatin-like 1 attenuates bleomycin-induced pulmonary fibrosis in mice. J Exp Med. 2015;212(2):235–52. doi: 10.1084/jem.20121878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hayakawa S, Ohashi K, Shibata R, Kataoka Y, Miyabe M, Enomoto T, et al. Cardiac myocyte-derived follistatin-like 1 prevents renal injury in a subtotal nephrectomy model. Journal of the American Society of Nephrology : JASN. 2015;26(3):636–46. doi: 10.1681/ASN.2014020210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ouchi N, Asaumi Y, Ohashi K, Higuchi A, Sono-Romanelli S, Oshima Y, et al. DIP2A functions as a FSTL1 receptor. J Biol Chem. 2010;285(10):7127–34. doi: 10.1074/jbc.M109.069468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li D, Wang Y, Xu N, Wei Q, Wu M, Li X, et al. Follistatin-like protein 1 is elevated in systemic autoimmune diseases and correlated with disease activity in patients with rheumatoid arthritis. Arthritis Res Ther. 2011;13(1):R17. doi: 10.1186/ar3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao W, Han HB, Zhang ZQ. Suppression of lung cancer cell invasion and metastasis by connexin43 involves the secretion of follistatin-like 1 mediated via histone acetylation. Int J Biochem Cell Biol. 2011;43(10):1459–68. doi: 10.1016/j.biocel.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 37.Takai Y, Irie K, Shimizu K, Sakisaka T, Ikeda W. Nectins and nectin-like molecules: roles in cell adhesion, migration, and polarization. Cancer science. 2003;94(8):655–67. doi: 10.1111/j.1349-7006.2003.tb01499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tawa H, Rikitake Y, Takahashi M, Amano H, Miyata M, Satomi-Kobayashi S, et al. Role of afadin in vascular endothelial growth factor- and sphingosine 1-phosphate-induced angiogenesis. Circ Res. 2010;106(11):1731–42. doi: 10.1161/CIRCRESAHA.110.216747. [DOI] [PubMed] [Google Scholar]
- 39.Fukuhara A, Irie K, Nakanishi H, Takekuni K, Kawakatsu T, Ikeda W, et al. Involvement of nectin in the localization of junctional adhesion molecule at tight junctions. Oncogene. 2002;21(50):7642–55. doi: 10.1038/sj.onc.1205875. [DOI] [PubMed] [Google Scholar]
- 40.Sidis Y, Tortoriello DV, Holmes WE, Pan Y, Keutmann HT, Schneyer AL. Follistatin-related protein and follistatin differentially neutralize endogenous vs. exogenous activin. Endocrinology. 2002;143(5):1613–24. doi: 10.1210/endo.143.5.8805. [DOI] [PubMed] [Google Scholar]
- 41.Panse KD, Felkin LE, Lopez-Olaneta MM, Gomez-Salinero J, Villalba M, Munoz L, et al. Follistatin-like 3 mediates paracrine fibroblast activation by cardiomyocytes. Journal of cardiovascular translational research. 2012;5(6):814–26. doi: 10.1007/s12265-012-9400-9. [DOI] [PubMed] [Google Scholar]
- 42.Wankell M, Kaesler S, Zhang YQ, Florence C, Werner S, Duan R. The activin binding proteins follistatin and follistatin-related protein are differentially regulated in vitro and during cutaneous wound repair. The Journal of endocrinology. 2001;171(3):385–95. doi: 10.1677/joe.0.1710385. [DOI] [PubMed] [Google Scholar]
- 43.Robertson R, Mahmood W, McGonnell I, Mukherjee A. Society for Endocrinology BES 2013. Harrogate, UK: 2013. Follistatin-like 3 (FSTL3), a transforming growth factor β ligand inhibitor, is essential for placental development in mice; p. OC3.6. [Google Scholar]
- 44.Del Principe D, Lista P, Malorni W, Giammarioli AM. Fibroblast autophagy in fibrotic disorders. J Pathol. 2013;229(2):208–20. doi: 10.1002/path.4115. [DOI] [PubMed] [Google Scholar]
- 45.Dumit VI, Kuttner V, Kappler J, Piera-Velazquez S, Jimenez SA, Bruckner-Tuderman L, et al. Altered MCM protein levels and autophagic flux in aged and systemic sclerosis dermal fibroblasts. J Invest Dermatol. 2014;134(9):2321–30. doi: 10.1038/jid.2014.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiao H, Jiao J, Wang L, O’Brien S, Newick K, Wang LC, et al. HDAC5 controls the functions of Foxp3(+) T-regulatory and CD8(+) T cells. International journal of cancer. 2016;138(10):2477–86. doi: 10.1002/ijc.29979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poralla L, Stroh T, Erben U, Sittig M, Liebig S, Siegmund B, et al. Histone deacetylase 5 regulates the inflammatory response of macrophages. J Cell Mol Med. 2015;19(9):2162–71. doi: 10.1111/jcmm.12595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Angiolilli C, Grabiec AM, Ferguson BS, Ospelt C, Malvar Fernandez B, van Es IE, et al. Inflammatory cytokines epigenetically regulate rheumatoid arthritis fibroblast-like synoviocyte activation by suppressing HDAC5 expression. Ann Rheum Dis. 2016;75(2):430–8. doi: 10.1136/annrheumdis-2014-205635. [DOI] [PMC free article] [PubMed] [Google Scholar]