Abstract
Neorickettsia (Rickettsiales, Anaplasmataceae) is a genus of obligate intracellular bacterial endosymbionts of digeneans (Platyhelminthes, Digenea). Some Neorickettsia are able to invade cells of the digenean's vertebrate host and are known to cause diseases of domestic animals, wildlife, and humans. In this study we report the results of screening digenean samples for Neorickettsia collected from bats in Egypt and Mindoro Island, Philippines, snails and fishes from Thailand, and fishes from Vietnam and the USA. Neorickettsiae were detected using a real-time PCR protocol targeting a 152 bp fragment of the heat shock protein coding gene, GroEL, and verified with nested PCR and sequencing of a 1853 bp long region of the GroESL operon and a 1371 bp long region of 16S rRNA. Eight unique genotypes of Neorickettsia were obtained from digenean samples. Neorickettsia sp. 8 obtained from Lecithodendrium sp. from Egypt; Neorickettsia sp. 9 and 10 obtained from two species of Paralecithodendrium from Mindoro, Philippines; Neorickettsia sp. 11 from Lecithodendrium sp. and Neorickettsia sp. 4 (previously identified from Saccocoelioides lizae, from China) from Thailand; Neorickettsia sp. 12 from Dicrogaster sp. Florida, USA; Neorickettsia sp. 13 and SF agent from Vietnam. Sequence comparison and phylogenetic analysis demonstrated that the forms, provisionally named Neorickettsia sp. 8-13, represent new genotypes. We have for the first time detected Neorickettsia in a digenean from Egypt (and the African continent as a whole), the Philippines, Thailand and Vietnam based on PCR and sequencing evidence. Our findings suggest that further surveys from the African continent, SE Asia, and the Island countries are likely to reveal new Neorickettsia lineages as well as new digenean host associations.
Keywords: Neorickettsia, Southeast Asia, Africa, North America, real-time PCR, molecular phylogeny, Digenea
Graphical Abstract
1. Introduction
Obligate intracellular bacteria in the genus Neorickettsia (Rickettsiales, Anaplasmataceae) are endosymbionts of digeneans. Neorickettsia are vertically transmitted through all stages of complex digenean life cycles. Additionally, they are capable of being horizontally transmitted to the vertebrate hosts of the digenean, both human and wildlife, where they can cause disease [1]. These diseases are potentially debilitating, e.g., Sennetsu fever in humans (Neorickettsia sennetsu), or even fatal, e.g., salmon dog poisoning (Neorickettsia helminthoeca) and Potomac horse fever (Neorickettsia risticii)[1].
Greiman et al. [2] conducted the first large scale molecular screening of digeneans for Neorickettsia from multiple countries and continents. Their screening revealed 7 new genotypes of Neorickettsia, bringing the known number of species/genotypes of the bacteria to 20. Additionally, they detected Neorickettsia from digeneans for the first time from the Australian continent and China. Records resulting from PCR detection of neorickettsial DNA are known only from North and South America, eastern Asia, Antarctica, and Australia [2], [3] and [4]. Africa and Europe are the last continents where Neorickettsia has yet to be found.
In this study, we screened DNA extracts of lecithodendriid digeneans from bats collected in Egypt and the Philippines, and other digenean samples collected from fishes and snails in the United States, Thailand, and Vietnam. As a result we found 6 new genotypes of Neorickettsia and two previously identified genotypes of Neorickettsia. We have for the first time detected Neorickettsia in digeneans from the African continent, the Philippines, Vietnam, and Thailand using PCR-based detection and DNA sequencing. Also, we have conducted molecular phylogenetic analyses in order to reveal the interrelationships among the newly discovered genotypes with previously known species/genotypes of Neorickettsia.
2. Materials and methods
2.1 Sample collections
Digenean samples were collected from bats, fishes and snails from the United States (Salt Springs, Florida, 29°21′01″N, 81°43′57″ W), Vietnam (Cat-Ba Island) 20°43′27.86″N, 107°2′58.61″E), Egypt (30°1′39″N, 31°12′37″E), Philippines (Mindoro Island, 12°47′14.99″N, 120°54′57.96″E), and Thailand (Aranyaprathet, Sa Kae, 13°36′37″N, 102°34′14″E and Talay Thai Seafood market, Sumut Sakhon (13°32′56.29″N, 100°15′26.11″E). Fishes from Vietnam and Thailand were purchased from local and commercial fish markets.
2.2 Sample processing
Live adult worms were rinsed in saline, examined briefly, killed with hot water, and fixed in 80% ethanol that allowed for both morphological examination and molecular study. For morphological identifications, fixed worms were stained in aqueous alum carmine or Mayer's hematoxylin; dehydrated in a graded ethanol series; cleared in clove oil (carmine) or methyl salicylate (Mayer's); and mounted permanently in Damar Gum.
Genomic DNA was extracted one of two ways. Samples collected from Florida were extracted using the Qiagen DNAeasy tissue kit (Qiagen, Inc., Valencia, California) following the manufacturer's instructions. Samples collected from Egypt, Thailand, Vietnam, and Philippines were first homogenized by direct sonication using a UP100H compact ultrasonic processor (Hielscher USA, Inc., Ringwood, New Jersey) at an amplitude of 90% for 20 seconds and further DNA extracted using the guanidine thiocyanate method according to Tkach and Pawlowski [5].
2.3 Molecular screening
DNA extracts were first tested for the presence of Neorickettsia using a real-time PCR protocol as described by Greiman et al. [2]. Five microliters of each DNA extract were used. The real-time PCR amplified a 152-bp portion of the 3' end of the heat shock protein coding gene, GroEL. Samples that tested positive with real-time PCR were verified using a substantially modified nested PCR protocol as described by Greiman et al. [6]. Five microliters of each DNA extract were used for the first PCR reaction and 1 μl of the first PCR product was used for the nested PCR. A 1470-bp long fragment of the 16S rRNA gene was first amplified, followed by the nested PCR step which amplified a 1371-bp fragment using internal primers. The same nested PCR primers were used in sequencing reactions along with internal forward and internal reverse primers [6]. DNA of Neorickettsia sp. from the digenean Plagiorchis elegans, obtained from a laboratory life cycle, was used as a positive control [7]. Pure water was used for negative controls in both real-time and nested PCRs.
A nested PCR amplifying a 1940-bp fragment of the GroESL operon was carried out for members of the “Neorickettsia risticii” clade (Fig. 1, subclade B). Five microliters of each DNA extract were used for the first PCR reaction and 2 μl of the first PCR product was used for the nested PCR. The primers used for the initial PCR were; hs10F 5′-CTCAAATGAAACAAT-CCGTTTGTTTGTAGC-3′ and hs2090R 5′-CATTCCACCCATGCCA-CCACCAGGCAT-3′. The primers used for the nested PCR were hs90F 5′-GTAGGTCTTGAAAAATATCACAGCG-3′ and hs2030R 5′-GTAGTCACTAGAACACTAGCAACAGA-3′. The same nested PCR primers were used for sequencing along with internal forward primers; hs120F 5′- TACGATATTTGATTCTGTAGGTCATTAG -3′and hs910F 5′- TGGTTCAATTTCTGCTAACGGCAAT-3′ and internal reverse primers; hs700R 5′ GCTTTTTCATTCGCCTGTGAGGTAGCCT-3′ and hs1620R 5′- CTTTAACCTCAACTTCTGTAGCACCAC-3′ designed by SEG.
Fig 1.
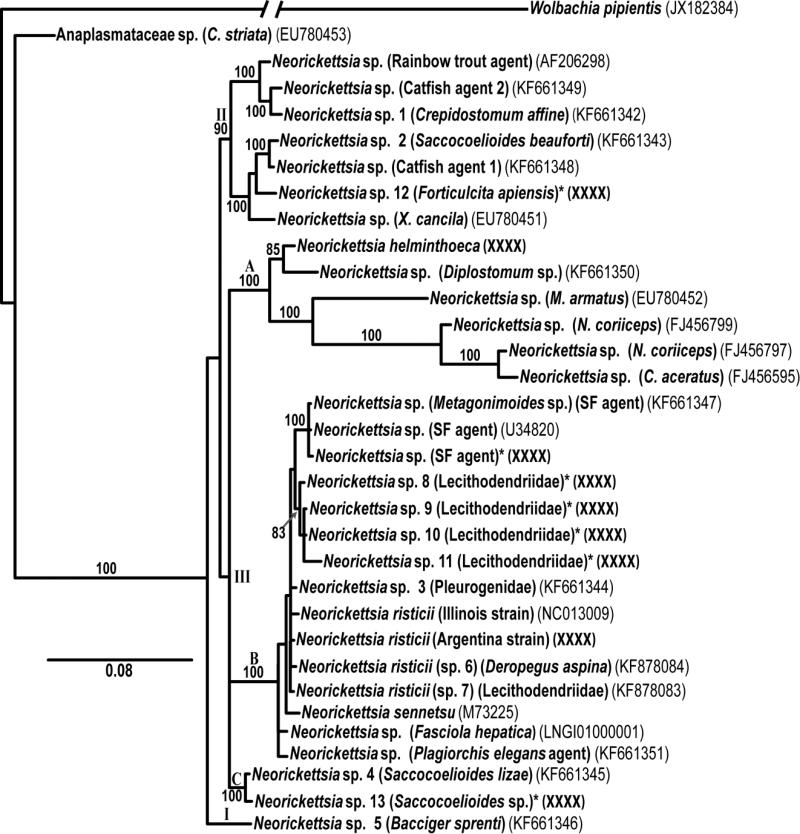
Phylogenetic interrelationships among 30 genetic lineages of Neorickettsia based on Bayesian analysis of partial 16S rRNA sequences. Numbers above internodes indicate posterior probabilities. An asterisk (*) at the end of a taxon name indicates a new sequence obtained in this study. GenBank numbers are given here for all taxa.
2.4 Digenean host identification
The digenean hosts were identified based on morphology using stained whole mounts and using partial sequences of the nuclear large ribosomal subunit gene (28S). Digenean DNA was amplified from the extracted DNA by the PCR protocol described by Greiman et al. [2].
2.5 DNA sequencing
PCR amplicons of both Neorickettsia and digeneans were purified using the Zymo DNA Clean & Concentrator™-5 (Zymo Research, Irvine, CA) or ExoSap PCR clean-up enzymatic kit from Affimetrix (Santa Clara, CA) according to the manufacturer's instructions. The PCR products were cycle-sequenced using ABI BigDye™ chemistry, ethanol precipitated, and run on an ABI Prism 3100™ automated capillary sequencer. Contiguous sequences of Neorickettsia were assembled using Sequencher™ ver. 4.2 (GeneCodes Corp., Ann Arbor, MI) and submitted to GenBank under accession numbers XXXX.
2.6 Phylogenetic analysis
Newly obtained sequences of neorickettsiae from GenBank were used in the phylogenetic analyses (Figs. 1, 2). The sequences were initially aligned with the aid of ClustalW as implemented in the BioEdit program, version 7.0.1 [8]. Two distinct alignments were prepared and two analyses were done. The alignments were manually refined in MacClade, version 4 [9].
Fig. 2.
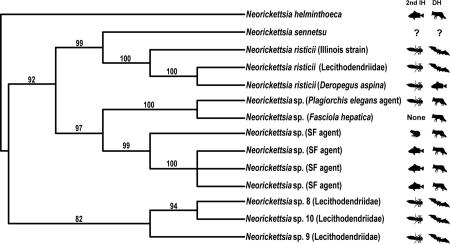
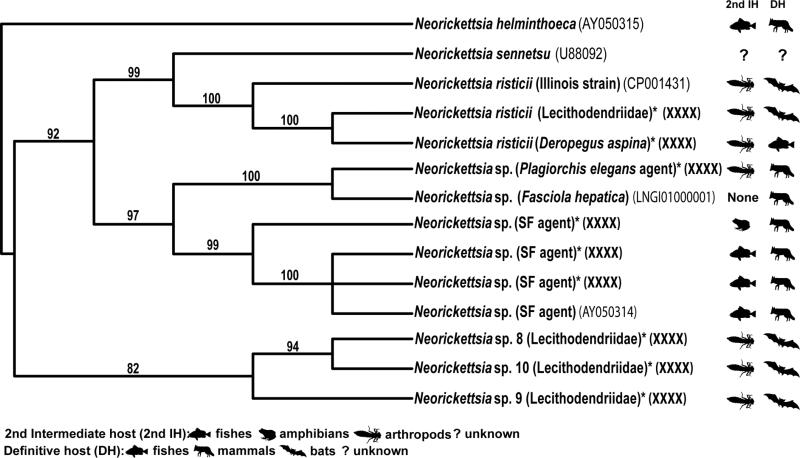
Cladogram based on Bayesian analysis of partial GroESL sequences depicting phylogenetic interrelationships among 14 genetic lineages of N. risticii and N. risticii-like neorickettsiae. Posterior probabilities greater than 80% are shown. Digenean second intermediate host groups and vertebrate definitive host groups are indicated by symbols. An asterisk (*) at the end of a taxon name indicates a new sequence obtained in this study. GenBank numbers are given here for all taxa.
The first phylogenetic analysis was run using a larger dataset including all new and previously available 16S sequences of Neorickettsia (a total of 30 sequences), one sequence of closely related Anaplasmataceae sp., and Wolbachia pipientis as the outgroup (Fig. 1). The analysis was carried out using Bayesian inference as implemented in the MrBayes program, version 2.01 [10] with the following nucleotide substitution parameters: lset nst=6, rates=invgamma, prset (prior assumptions) pinvarpr=fixed (0.6170) shapepr=fixed (0.4360), that correspond to a “three-parameter model (TPM3) [11] model including estimates of the proportion of invariant sites (I) and rate variation among sites with a number of rate categories (G). Posterior probabilities were approximated over 1,500,000 generations, log-likelihood scores plotted, and only the final 75% of trees were used to produce the consensus trees by setting the “burnin” parameters at 375,000 generations. The TPM3+I+G model was used for the analysis based off of the results obtained from jModelTest, version 0.1.1 [12], [13].
The second phylogenetic analysis was run using a much smaller dataset based on the partial sequences of the GroESL operon, including only species/genotypes of Neorickettsia closely related to Neorickettsia risticii (Fig. 2), a total of 13 sequences. Neorickettsia helminthoeca was used as an outgroup based on the outcome of the 16S analysis (Fig. 1). The analysis was carried out using Bayesian inference (BI) as implemented in the MrBayes program, version 2.01 [10] with the following nucleotide substitution parameters: lset nst=6, rates=propinv, prset (prior assumptions) pinvarpr=fixed (0.6380), that correspond to a Tamura-Nei (TrN) model including estimates of the proportion of invariant sites (I). Posterior probabilities were approximated over 1,000,000 generations, log-likelihood scores plotted, and only the final 75% of trees were used to produce the consensus trees by setting the “burnin” parameters at 250,000 generations. The TrN+I model was used for the analysis based off of the results obtained from jModelTest, version 0.1.1 [12], [13].
3. Results
In northeastern Egypt, during the fall of 2012, authors RE and AIK collected a total of 50 bats (Pipistrellus kuhli marginatus) infected with adult digeneans. In northern Vietnam, during the fall of 2013, authors SEG and NVH dissected a total of 75 fishes, representing at least 6 fish species. From those dissections, a total of 30 fishes were infected with either larval or adult digeneans. In southeastern Thailand, during the winter of 2014, authors SEG, JAV, PA, and VVT dissected a total of 114 fishes. From those dissections, 50 fishes were infected with either larval or adult digeneans. In spring of 2013 (March), TJF obtained 8 Mugil cephalus via Hawaiian sling from Salt Springs, Florida. Two-thirds of these hosts harbored coinfections of F. apiensis and Saccocoelioides sp. and one individual harbored a single mature specimen of Intromugil alachuaensis.
Digenean samples were screened for Neorickettsia. Screening revealed eight unique genotypes of Neorickettsia from digenean samples: Neorickettsia sp. 8 obtained from Lecithodendrium sp. from Egypt; Neorickettsia sp. 9 and 10 obtained from two species of Paralecithodendrium from Mindoro, Philippines; Neorickettsia sp. 11 from Lecithodendrium sp. and Neorickettsia sp. 4 (previously identified from Saccocoelioides lizae, from China) from Thailand; Neorickettsia sp. 12 from Florida, USA; Neorickettsia sp. 13 and SF agent from Vietnam (Table 1). Genotype numbering is the continuation of that proposed by Greiman et al. [2].
Table 1.
Neorickettsia genotypes found in this study, their digenean host associations and corresponding digenean life cycle information.
| Neorickettsia species | Digenean family | Digenean genus and species | Digenean life cycle stage | Vertebrate host (this study) | Definitive host | 1st/2nd intermediate hosts | Origin |
|---|---|---|---|---|---|---|---|
| Neorickettsia sp. 8 (XXXX) | Lecithodendriidae | Lecithodendrium sp. | Adult | ________ | Bats | Aquatic snail / aquatic arthropod | Egypt |
| Neorickettsia sp. 9 (XXXX) | Lecithodendriidae | Paralecithodendrium sp. | Adult | Horsfield's bat (Myotis horsfieldii) | Bats | Aquatic snail / aquatic arthropod | Philippines |
| Neorickettsia sp. 10 (XXXX) | Lecithodendriidae | Paralecithodendrium sp. | Adult | Large-eared horseshoe bat (Rhinolophus philippinensis) | Bats | Aquatic snail / aquatic arthropod | Philippines |
| Neorickettsia sp. 11 (XXXX) | Lecithodendriidae | Lecithodendrium sp. | Cercariae | ________ | Bats | Aquatic snail / aquatic arthropod | Thailand |
| Neorickettsia sp. 4 (XXXX) | Haploporidae | Saccocoelioides lizae | Adult | Striped mullet (Mugil cephalus) | Fishes | Unknown | Thailand |
| Neorickettsia sp. 12 (XXXX) | Haploporidae | Forticulcita apiensis | Adult | Striped mullet (Mugil cephalus) | Fishes | Unknown | USA |
| Neorickettsia sp. 13 (XXXX) | Haploporidae | Saccocoelioides sp. | Adult | Striped mullet (Mugil cephalus) | Fishes | Unknown | Vietnam |
| Neorickettsia sp. SF agent (XXXX) | Heterophyidae | Stellantchasmus falcatus | metacercariae | Striped mullet (Mugil cephalus) | Mammals | Aquatic snail/fish | Vietnam |
The 16S alignment included a total of 1,258 sites, of which 1,249 could be aligned unambiguously. Positions of ambiguous homology were excluded from the analysis. Bayesian analysis produced a tree where all Neorickettsia sequences clearly fall into a well-defined clade, with 100% support (Fig. 1), composed of three subclades – subclade I (1 genotype), subclade II (7 genotypes), and subclade III (22 genotypes). Subclade IIIA included N. helminthoeca, agent of salmon dog poisoning and subclade IIIB included N. risticii, agent of Potomac horse fever and N. sennetsu, agent of Sennetsu fever in humans The topology of the consensus tree generated by this analysis is essentially identical to the phylogeny published by Greiman et al. [2], with the only exception being the inclusion of the new genotypes of Neorickettsia discovered in this study.
The GroESL alignment included a total of 1,822 sites, of which all could be aligned unambiguously. Bayesian analysis produced a well-structured consensus tree with strong support of nearly all topologies (Fig. 2). Based on this analysis, Neorickettsia sp. 8 from Egypt and Neorickettsia spp. 9 and 10 from the Philippines appeared in a cluster that formed a sister clade to all other genotypes of Neorickettsia (N. sennetsu, N. risticii, Neorickettsia sp. from Plagiorchis elegans, Neorickettsia sp. from Fasciola hepatica, and SF agent).
4. Discussion
Greiman et al. [2] emphasized that the known distribution of Neorickettsia is very geographically uneven, with no well-documented records supported by PCR/sequence data from Africa, Europe, most of South America, most of Asia, and nearly all island countries. In this study we have further expanded taxonomic and geographic coverage of screening of digeneans for Neorickettsia. As suggested by Greiman et al. [2] it resulted in discoveries of novel forms of Neorickettsia from additional digenean taxa and geographic regions including the first record from Africa.
Digeneans in the family Lecithodendriidae parasitic in bats appear to be common hosts of Neorickettsia, with a total of 7 species of lecithodendriids known to harbor the bacteria including 4 added by the present study [1], [2]. This is a relatively large digenean family comprising at least 12 genera parasitizing mammals (mostly bats) and occasionally birds, on all continents [14]. Currently, lecithodendriids have been found to harbor the bacterial endosymbiont in Egypt, Thailand, North America (USA), South America (Argentina), and the Philippines, with those in the Americas known to harbor N. risticii, the etiological agent of Potomac horse fever.
Mugil cephalus (grey mullet) is the 2nd intermediate or definitive host of four digenean species harboring Neorickettsia in this study (Saccocoelioides lizae, Saccocoelioides sp., Forticulcita apiensis, and Stellantchasmis falcatus (host of SF agent)) and two digenean species (Saccocoelioides beauforti and S. lizae) harboring Neorickettsia in the study by Greiman et al. [2]. Although this is a globally distributed marine fish, its digenean species appear to be much more restricted in their distribution. Saccocoelioides lizae has only been recorded off the cost of southeastern China [15]; however, we have now found it off the coast of Thailand. The distribution of Saccocoelioides sp. from northern Vietnam is unknown, however, the sequence of Neorickettsia sp. 13 differs from Neorickettsia sp. 4 identified from S. lizae from Thailand (this study) and China [2] by only 3 bp, likely indicating that its distribution is limited to the marine habitats of the region. Based on the high sequence similarity, Neorickettsia sp. 13 from Vietnam is likely the same species as Neorickettsia identified from China and Thailand. The last Neorickettsia hosting digenean species from mullet, F. apiensis was recently described from Salt Springs, Florida [16]. Salt Springs is an artesian spring that discharges slightly saline water in Lake George. This haploporid is likely endemic to the region, and therefore the genotype/species of Neorickettsia (sp. 12) is likely also unique to this area.
The least resolved part of the 16S tree is the polytomy in the sub-clade B of clade III that includes several genotypes occupying a derived position in relation to N. sennetsu (Fig. 1; also in [2]). The GroESL phylogenetic analysis helped resolve the relationships among a majority of these taxa and has produced a tree with strong branch support for all topologies (Fig. 2). The GroESL operon was used for the second phylogenetic analysis following its use by Dumler et al. [17] for the members of the family Anaplasmataceae. The new genotypes (Neorickettsia spp. 8, 9, and 10) (from Egypt and the Philippines, respectively) identified in our study, fall into a clade that is sister to the well-supported clade including N. risticii, N. sennetsu, Neorickettsia (P. elegans agent), Neorickettsia sp. (from F. hepatica) and SF agent. This brings up the potential question regarding the pathogenicity of the new genotypes of Neorickettsia from Egypt and the Philippines. Neorickettsia risticii, N. sennetsu, and SF agent are all known to cause potentially debilitating or even fatal diseases in the vertebrate hosts of digeneans [1]. Neorickettsia spp. 8, 9, and 10 from lecithodendriids from Egypt and the Philippines, are closely related to these three species of Neorickettsia and therefore have the potential of causing yet unknown disease in wildlife or domestic animals. Further studies of these three genotypes may reveal their potential as pathogens. The GroESL phylogeny also indicates that the genotype of Neorickettsia from Deropegus aspina, originally called Neorickettsia sp. by Greiman et al. [2], is in fact N. risticii. Additionally, Greiman et al. [6] identified the species of Neorickettsia from P. elegans as N. risticii. However, our GroESL phylogeny (Fig. 2) corroborates the suggestion by Greiman et al. [2] regarding the status of this genotype as a separate species related to N. risticii.
Lastly, Mitreva et al. (unpublished; GenBank LNGI01000001) sequenced the complete genome of a species of Neorickettsia from Fasciola hepatica. Based on our GroESL phylogeny the species falls into the same clade as our species from P. elegans. However, due to the high 16S sequence similarity (99.6%) between these two forms, it remains unknown whether they represent two distinct species or different strains of one species.
The GroESL phylogenetic tree shows close associations among Neorickettsia and their digenean hosts' 2nd intermediate or definitive hosts (Fig. 2). In the sub-clade including N. sennetsu and N. risticii, the 2nd intermediate host of all digenean hosts (at least where the digenean host is known) is an arthropod, and the definitive hosts for all but one are mammals (bats) and potentially birds. The digenean host of N. sennetsu, the causative agent of the human disease Sennetsu fever in Southeast Asia, is currently unknown. Greiman et al. [2] hypothesized a mammal or fish as a definitive host. The topology of the GroESL phylogenetic tree obtained in this study strongly suggests that it also should be a digenean that uses an arthropod as a second intermediate host. This is in contrast with the previous notion that the digenan host of N. sennetsu utilizes a fish second intermediate host. This previous hypothesis was based on the discovery that people became ill with N. sennetsu after consuming raw mullet infected with digenean metacercariae in Japan [18]. However, based on our GroESL phylogeny, and the transmission biology of the closely related species N. risticii, we hypothesize that the tissue of the fish may become infected with the bacteria in a similar manner to how horses become infected with N. risticii. Horses are dead-end hosts to the digeneans infected with N. risticii, and therefore, the mullet may also be dead end hosts to the digenean harboring N. sennetsu, and become infected when the fish ingests an arthropod infected with the metacercariae of the digenean harboring the bacteria. Further studies of trematodes from different host taxa are needed in the regions endemic for Sennetsu in order to answer this intriguing question of public health importance.
Highlights.
Eight unique isolates of Neorickettsia discovered in SE Asia, Africa and North America.
Seven isolates represent new species level genetic lineages.
This is the first report of Neorickettsia from digeneans in Africa and three SE Asian countries.
Phylogenetic relationships Neorickettsia has been studied 16S and GroESL genes.
Host associations of digeneans harboring Neorickettsia are discussed.
Acknowledgements
This project was funded by the grant R15AI092622 from the National Institutes of Health, USA to Vasyl V. Tkach and Jefferson A. Vaughan. We are grateful for the Florida Fish and Wildlife Conservation Commission for issuing the FNW-13-05 (renewal) that allowed for the collection of freshwater fish in Florida. The authors are also grateful to Dr. Richard Heard for the provision of microscopy and sampling equipment. Lastly, SEG was partially funded through a National Science foundation Postdoctoral Research Fellowship in Biology (1523410).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Vaughan JA, Tkach VV, Greiman SE. Neorickettsial endosymbionts of the Digenea: diversity, transmission and distribution. Adv. Parasitol. 2012;79:253–297. doi: 10.1016/B978-0-12-398457-9.00003-2. [DOI] [PubMed] [Google Scholar]
- 2.Greiman SE, Tkach VV, Pulis E, Fayton TJ, Curran SS. Large scale screening of Digeneans for Neorickettsia endosymbionts using real-time PCR reveals new Neorickettsia genotypes, host associations and geographic records. PLoS ONE. 2014;9:e98453. doi: 10.1371/journal.pone.0098453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ward NL, Steven B, Penn K, Methe BA, Detrich WH. Characterization of the intestinal microbiota of two Antarctic notothenioid fish species. Extremophiles. 2009;13:679–685. doi: 10.1007/s00792-009-0252-4. [DOI] [PubMed] [Google Scholar]
- 4.Tkach VV, Schroeder JA, Greiman SE, Vaughan JA. New genetic lineages, host associations and circulation pathways of Neorickettsia endosymbionts of digeneans. Acta Parasitol. 2012;57:285–292. doi: 10.2478/s11686-012-0043-4. [DOI] [PubMed] [Google Scholar]
- 5.Tkach VV, Pawlowski J. A new method of DNA extraction from the ethanol-fixed parasitic worms. Acta Parasitol. 1999;44:147–148. [Google Scholar]
- 6.Greiman SE, Tkach VV, Vaughan JA. Transmission rates of the bacterial endosymbiont, Neorickettsia risticii, during the asexual phase of its digenean host, Plagiorchis elegans, within naturally infected lymnaeid snails. Parasit. Vectors. 2013;6:303–309. doi: 10.1186/1756-3305-6-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greiman SE, Tkach M, Vaughan JA, Tkach VV. Laboratory maintenance of the bacterial endosymbiont, Neorickettsia sp., through the life cycle of a digenean, Plagiorchis elegans. Exp. Parasitol. 2015;157:78–83. doi: 10.1016/j.exppara.2015.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 9.Maddison DR, Maddison WP. MacClade 4: Analysis of phylogeny and character evolution. Version 4.08a. 2005 doi: 10.1159/000156416. Available: http://macclade.org. [DOI] [PubMed]
- 10.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 11.Kimura M. Estimation of evolutionary distances between homologous nucleotide sequences. Proc. Natl. Acad. Sci. U.S.A. 1981;78:454–458. doi: 10.1073/pnas.78.1.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 13.Posada D. jModelTest: Phylogenetic Model Averaging. Mol. Biol. Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 14.Lotz JM, Font WF. Family Lecithodendriidae Luhe, 1901. In: Bray RA, Gibson DI, Jones A, editors. Keys to the Trematoda. Vol. 3. CABI Publishing Wallingford; U.K.: 2008. pp. 527–536. [Google Scholar]
- 15.Liu SF. One new species of Saccocoelioides (Digenea: Haploporidae) from Liza carinatus in Taiwan Strait. J. Oceanogr. 2002;21:37–44. [Google Scholar]
- 16.Andres MJ, Curran SS, Fayton TJ, Pulis EE, Overtsreet RM. An additional genus and two additional species of Forticulcitinae (Digenea: Haploporidae) Folia Parasitol. 2015;62 doi: 10.14411/fp.2015.025. [DOI] [PubMed] [Google Scholar]
- 17.Dumler JS, Barbet AF, Bekker CPJ, Dasch GA, Palmer GH, Ray SC, Rikihisa Y, Rurangirwa FR. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ‘HE agent’ as subjective synonyms of Ehrlichia phagocytophila. Int. J. Syst. Evol. Microbiol. 2001;51:2145–2165. doi: 10.1099/00207713-51-6-2145. [DOI] [PubMed] [Google Scholar]
- 18.Fukuda T. Rickettsial mononucleosis (synonyms: Hyuganetsu, kagaminetsu) Kitasato Arch. Exp. Med. 1958;31:51–56. [Google Scholar]