Abstract
Neisseria meningitidis (N. meningitidis), Streptococcus pneumoniae (S. pneumoniae), and Haemophilus influenzae type b (Hib) are three most common pathogens accounting for most bacterial meningitis, a serious global infectious disease with high fatality, especially in developing nations. Because the treatment and antibiotics differ among each type, the identification of the exact bacteria causing the disease is vital. Herein, we report a polymer/paper hybrid microfluidic biochip integrated with loop-mediated isothermal amplification (LAMP) for multiplexed instrument-free diagnosis of these three major types of bacterial meningitis, with high sensitivity and specificity. Results can be visually observed by the naked eye or imaged by a smartphone camera under a portable UV light source. Without using any specialized laboratory instrument, the limits of detection of a few DNA copies per LAMP zone for N. meningitidis, S. pneumoniae and Hib were achieved within 1 hour. In addition, these three types of microorganisms spiked in artificial cerebrospinal fluid (ACSF) were directly detected simultaneously, avoiding cumbersome sample preparation procedures in conventional methods. Compared with the paper-free non-hybrid microfluidic biochip over a period of three months, the hybrid microfluidic biochip was found to have a much longer shelf life. Hence, this rapid, instrument-free and highly sensitive microfluidic approach has great potential for point-of-care (POC) diagnosis of multiple infectious diseases simultaneously, especially in resource-limited settings.
Keywords: hybrid microfluidic biochip, multiplexed meningitis diagnosis, instrument-free detection, LAMP, low-resource settings
Graphical Abstract
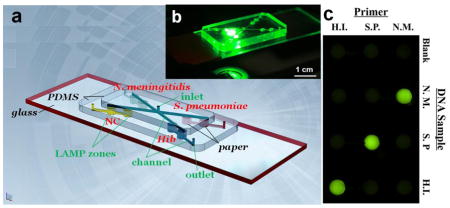
We report a low-cost polymer/paper hybrid microfluidic biochip integrated with loop-mediated isothermal amplification (LAMP) for multiplexed instrument-free meningitis diagnosis with high sensitivity and specificity, especially for resource-limited settings.
1. Introduction
Although great progress has been made to prevent infectious diseases, multiple infectious diseases such as meningitis still cause thousands of deaths every year globally. Meningitis is an inflammation of the lining surrounding the spine and the brain, which can become fatal in as early as 24 hours after symptoms occur (2015). Meningitis can be caused by infection with bacteria, viruses and fungi. Bacterial meningitis remains the most serious form of this disease. As reported by World Health Organization (WHO), without epidemics about 1000000 cases of bacterial meningitis are expected to occur and around 200000 patients die annually worldwide (Castillo 2011). Bacterial meningitis is a more serious issue in developing countries or high poverty regions, with higher reported fatality rates (37–60%) (Castillo 2011). In the ‘Meningitis Belt’ of Africa, bacterial meningitis remains a major unresolved public health problem, where recurrent unpredictable epidemics often occur (Agier et al. 2013).
Neisseria meningitidis (N. meningitidis), Streptococcus pneumoniae (S. pneumoniae) and Haemophilus influenzae type b (Hib) are three major pathogens causing bacterial meningitis. As a medical emergency, the immediate antibiotic therapy is imperative for bacterial meningitis, which must not be postponed by diagnostic delays (Hoffman and Weber 2009). In addition, identification of the exact bacteria causing the disease is vital because treatment and antibiotics differ for each type. Due to the high fatality rate and the serious damaging effect caused by untreated meningitis especially in rural high-poverty areas, a simple, low-cost, rapid and highly sensitive approach for immediate multiplexed bacterial meningitis diagnosis is in great need.
Bacterial culture and gram staining are key approaches to the laboratory diagnosis of bacterial meningitis (Brouwer et al. 2010). However, their limitations are obvious. (1) Their detection sensitivity is low. (2) Their detection rates decrease for patients who have received antibiotic pre-treatment. (3) They have to be done in laboratories with well-trained personnel. (4) Bacterial culture takes up to 72 hours for identification (Dubos et al. 2006). Recently, molecular diagnostic methods such as real-time polymerase chain reaction (qPCR) and loop-mediated isothermal amplification (LAMP) have been increasingly used to provide rapid detection of bacterial meningitis with higher sensitivity and specificity (Lee et al. 2015; Wang et al. 2012). qPCR has become a valuable technique for the detection of bacterial meningitis pathogens (Wang et al. 2012), but it is costly. As a promising isothermal DNA amplification method, LAMP has been developed to amplify the target DNA at a constant temperature in a range of 60~65 °C. The high strand displacement activity from the Bacillus stearothermophilus (Bst) DNA polymerase and identification of 6 distinct regions from 2 or 3 sets of primers in LAMP result in high specificity. There have been several reports on rapid diagnosis of bacterial meningitis (e.g., N. meningitidis, S. pneumoniae, Hib) by using conventional off-chip LAMP (Kim et al. 2012; Kim et al. 2011; McKenna et al. 2011). However, none of these LAMP methods achieved multiplexed bacterial meningitis detection. In addition, specialized equipment is required in laboratories for these DNA amplification-based methods, such as qPCR (e.g., QuantStudio® 6 Flex Real-Time PCR System costs ~$60000), thermal cyclers, centrifuges, and fluorescent microscopes, making them not applicable in low-resource settings.
With various advantages including low sample/reagent consumption, integration, miniaturization and automation, the microfluidic lab-on-a-chip provides a platform for a variety of human health diagnostics with high efficiency (Dou et al. 2016a; Dou et al. 2016b; Dou et al. 2015; Fu et al. 2016; Li et al. 2011; Li et al. 2012; Li and Zhou 2013; Liu et al. 2011; Sanjay et al. 2016a; Sanjay et al. 2015; Shen et al. 2014; Tian et al. 2016; Zhang et al. 2014), as well as the possibility for multiplexed detection (Fang et al. 2012; Zuo et al. 2013). By taking advantage of different substrates, we previously developed a polydimethylsiloxane (PDMS)/paper hybrid microfluidic device integrated with aptamer-functionalized graphene oxide nanosensors for one-step detection of multiple foodborne pathogens simultaneously (Zuo et al. 2013). However, the detection sensitivity is not comparable to DNA amplification-based assays. DNA amplification is usually essential for DNA-based techniques to achieve high detection sensitivity. Therefore, we developed another hybrid microfluidic device integrated with LAMP for sensitive infectious disease diagnosis (Dou et al. 2014). Unlike PCR, LAMP doesn’t need complex microfabrication of heaters and temperature sensors on a microfluidic device, making it more suitable for point-of-care (POC) analysis on a chip. Although high sensitivity was demonstrated, only one pathogen was measured from each assay and pathogen lysis steps were not integrated on the chip. In addition, other microfluidic device-based LAMP methods have been reported for the detection of different pathogens (Borysiak et al. 2015; Chang et al. 2013; Duarte et al. 2013; Fang et al. 2012; Safavieh et al. 2012; Sun et al. 2015; Wang et al. 2011). However, certain limitations hindered their wide applications for low-resource settings, including dependence on bulky detection systems (Safavieh et al. 2012), supporting equipment (Wang et al. 2011), and complicated sample preparation steps (Safavieh et al. 2012).
The multiplexed pathogen detection can provide richer information, higher throughput, more convenience, and less sample consumption and less detrimental effects on patients during sampling, compared to multiple times of repeated singleplexed assays. Although LAMP has received increasing attention for applications in low-resource settings, the multiplexed LAMP detection is still challenging because of complexities from multiple primer sets and complicated ladder-patterned amplicons (Iseki et al. 2007; Shao et al. 2011). Most conventional LAMP methods by just mixing different primers for multiple pathogens in a single tube for simultaneous pathogen detection can only identify the occurrence of amplification reactions, but are not efficient to identify the specific target pathogens (Huy et al. 2012; Wenhui et al. 2015). Possibly due to this reason, no LAMP methods were reported for multiplexed meningitis diagnosis yet. Herein, by taking advantage of ease of multiple-compartment fabrication in microfluidics for parallel LAMP reactions, we for the first time developed a PDMS/paper hybrid microfluidic biochip for simultaneous LAMP detection of three most common meningitis-causing pathogens with high sensitivity and specificity, N. meningitidis, S. pneumoniae and Hib. Simple lysis steps were integrated on the chip. Three types of pathogenic microorganisms spiked in ACSF samples were directly analyzed, and detection results were observable to the naked eye within 1 hour, without using laborious sample preparation procedures and any specialized instruments during the entire assay. The limits of detection of a few DNA copies per LAMP zone were achieved.
2. Experimental Section
2.1. Chemicals and materials
LAMP kits and calcein were obtained from Eiken Co. Ltd., Japan. The LAMP reaction mixture was prepared by following the manufacture’s protocol. Table S1 shows the LAMP primers purchased from Integrated DNA Technologies (Coralville, IA) for the target DNA sequences of the N. meningitidis ctrA gene, the S. pneumoniae lytA gene and the Hib capsulation locus region II (Kim et al. 2012; Kim et al. 2011; McKenna et al. 2011).
Microfluidic platform fabrication: Polydimethylsiloxane (PDMS, Sylgard 184) was obtained from Dow Corning (Midland, MI); Whatman #1 chromatography paper and Epoxy glue were purchased from Sigma (St. Louis, MO) and ITW Devcon (Danvers, MA), respectively.
All other chemicals used in preparing the bacterial lysis buffer and the artificial cerebrospinal fluid (ACSF) buffer were purchased from Sigma (St. Louis, MO) and used without further purification. Unless otherwise noted, all solutions were prepared with ultrapure Milli-Q water (18.2 MΩ cm) from a Millipore Milli-Q system (Bedford, MA).
2.2. Microorganism culture and DNA preparation
N. meningitidis (ATCC 13098), S. pneumoniae (ATCC 49619) and Hib (ATCC 33533) were obtained from American Type Culture Collection (ATCC, Rockville, MD). N. meningitidis and Hib were grown on chocolate II agar plates (BD, Sparks, MD). S. pneumoniae was grown in TSA II agar plates supplemented with 5% sheep blood (BD, Sparks, MD). All the microorganisms were incubated at 37 °C for 48 h in an aerobic environment with 5% CO2.
DNA extraction, the concentration measurement and the calculation of copy numbers of template genomic DNA were shown in the Supplementary Material.
2.3. Microfluidic chip design and fabrication
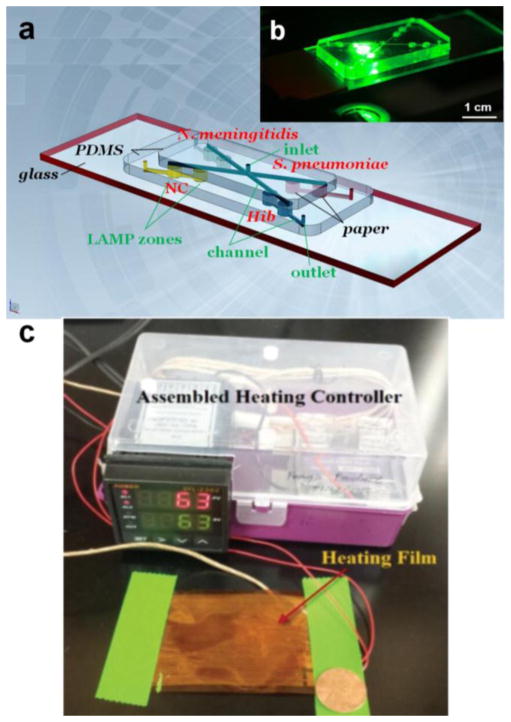
The microfluidic biochip consists of three layers, as shown in Fig. 1a. The top layer is a PDMS layer including a central inlet reservoir with a diameter of 1.0 mm, and 4 microchannels (100 μm in width and depth) for reagent delivery. The middle layer is also a PDMS layer containing 8 LAMP zones (diameter 2.0 mm, and depth 1.0 mm) for LAMP reactions and detection, 4 outlet reservoirs with a diameter of 1.0 mm, and 4 microchannels at the bottom surface of the middle layer. Different LAMP zones were designated for different pathogen detection (N. meningitidis, S. pneumoniae and Hib, respectively) and negative control (NC). A Whatman #1 chromatography paper disk (diameter 2.0 mm) cut by a laser cutter (Epilog Zing 16, Golden, CO) was placed inside each LAMP zone. The bottom plate is a glass slide for structure support. Fig. 1b is a photograph of the assembled biochip on a fluorescence microscope stage. Fig. S1 illustrates the detection principle in the LAMP zone.
Fig. 1.
(a) Schematic of the PDMS/paper hybrid microfluidic biochip for multiplexed bacterial meningitis diagnosis. (b) A photograph of the hybrid microfluidic biochip during fluorescence measurement. (c) A photograph of the portable heater with a heating film controlled by a proportional–integral–derivative (PID)-based temperature controller. A penny was included in this photograph for the size comparison.
The standard soft lithography procedures were followed to produce PDMS films. Firstly, the liquid PDMS base and the curing agent mixture were prepared at a weight ratio of 10:1. Then the PDMS precursor mixture was poured into a petri dish, which was degassed in a vacuum desiccator for about 30 min and incubated in an oven at 85 °C for 2.5 hours. Microchannels on the surface of PDMS films were directly created by using a laser cutter (Epilog Zing 16, Golden, CO). Inlet reservoir, outlet reservoirs and LAMP zones on PDMS films were drilled by using biopsy punches. PDMS films and the glass slide were face-to-face sandwiched to bond irreversibly by exposure in oxidizing air Plasma Cleaner (Ithaca, NY) for 30 seconds. After biochip assembly, the specific primers for N. meningitidis, S. pneumoniae and Hib DNA sequences were pre-loaded on paper disks in LAMP zones. The concentrations and loading amount of the pre-loaded primers are shown in Table S2. No primers were pre-loaded in LAMP zones for NC for the convenience of subsequent reagent deliveries, which was confirmed to provide consistent results with another type of negative control in the presence of primers but not the template DNA (see Fig. S2). Thus, the biochip became ready for use.
2.4. On-chip LAMP process and confirmation tests
A 26 μL LAMP mixture was prepared following the manufacturer’s protocol and introduced into the LMAP zones of the microfluidic biochip from the inlet. The inlet and outlets were sealed with Epoxy glue afterward. The microfluidic biochip was heated by using a portable heater (or in a water bath) at 63 °C for 1 hour for LAMP reactions, and was further heated at 95 °C for 2 min to terminate the LAMP reactions. The portable heater devised by our laboratory includes a heating film and an inexpensive portable proportional–integral–derivative (PID)-based temperature controller (Fig. 1c). The material cost of this whole heating unit was about $60.
After LAMP reactions, a cellphone camera (e.g., iPhone 5) was applied to capture the generated fluorescence images under a portable UV light pen. The cellphone camera captured images were analyzed by the NIH software ImageJ to obtain gray values. The fluorescence intensities were measured by using a Nikon Ti-E fluorescence microscope (Melville, NY) equipped with a motorized stage and a cooled CCD camera to confirm instrument-free detection results, using appropriate FITC optical filters. LAMP products were also analysed using the conventional gel electrophoresis (Sub-Cell GT, Bio-Rad, CA) and Nanodrop (Nanodrop 1000, Thermo Scientific, MA). During gel electrophoresis, amplified products were resolved by applying 90 V for 1 hour in 1.5% agarose gel.
2.5. Multiplexed instrument-free detection of microorganisms
The bacteria lysis buffer is composed of 50 mM Tris buffer (pH 7.5), 4 M urea and 0.1% triton. The artificial cerebrospinal fluid (ACSF) buffer was prepared by following a published protocol (Cold Spring 2011). The ACSF buffer contained 119 mM NaCl, 26.2 mM NaHCO3, 2.5 mM KCl, 1 mM NaH2PO4, 1.3 mM MgCl2, 10 mM glucose. It was bubbled with 5% CO2/95% O2 for 10–15 min, before CaCl2 was added to the buffer to reach a final concentration of 2.5 mM CaCl2. The prepared ACSF buffer was filtered with a 0.20 μm filter apparatus, and stored at 4 °C before use.
Microorganisms were spiked in ACSF to prepare mock clinical samples. We swabbed a tiny amount of bacterial colonies from each microorganism culture plate of N. meningitidis, S. pneumoniae and Hib, and suspended them in 1 mL ACSF buffer to form an ACSF sample for bacterial cell lysis by adjusting the turbidity to McFarland Standard 0.5 (Key Scientific Products, TX) with an approximate bacterial density of 1.5 × 108 CFU/mL (de Almeida Gomes et al. 2006).
To test microorganism in ACSF samples, only 2 μL of the ACSF sample was mixed with 18 μL of the bacterial lysis buffer to prepare the bacterial lysis buffer/ACSF sample mixture. Then 16 μL of this mixture was introduced into the microfluidic biochip from the common inlet. The on-chip lysis step could be simply completed by just incubating the lysis buffer/ACSF sample mixture in the microfluidic biochip for about 10 min at room temperature. After the lysis buffer/ACSF sample mixture dried to avoid dilution of LAMP solutions, the LAMP reagent mix was introduced into the microfluidic biochip to complete the subsequent LAMP reactions for multiplexed detection of meningitis microorganisms from ACSF samples. The bacterial cell lysis and incubation process could also be performed off the chip, with no differences observed in LAMP results when compared with on-chip lysis procedures (see Fig. S2 for more information). If lysis incubation was performed off-chip, 2 μL of the lysate was directly used to prepare the LAMP mix.
3. Results and discussion
3.1. Singleplexed pathogen detection by using extracted DNA
By using the template DNA extracted from bacterial culture, we first tested the feasibility of the on-chip LAMP detection of each pathogen separately.
Paper is a highly porous material with 3D structures and a high surface-to-volume ratio (Martinez 2011). It is believed that paper is able to protect DNA in harsh conditions, so paper has been advantageously used in collecting, storing and processing DNA samples (Chaisomchit et al. 2005; Moscoso et al. 2004; Smith and Burgoyne 2004). Therefore, in this work, we placed a paper disk in each of the LAMP zone, forming a PDMS/paper hybrid microfluidic biochip. In this way, paper functions as a 3D substrate where DNA primers were pre-loaded for subsequent on-chip LAMP reactions. Previously, we also developed a few other paper/polymer hybrid microfluidic devices for rapid disease diagnosis (Dou et al. 2014; Sanjay et al. 2016b; Zuo et al. 2013). Hybrid devices can draw more benefits from multiple device substrates (Dou et al. 2015; Sanjay et al. 2015).
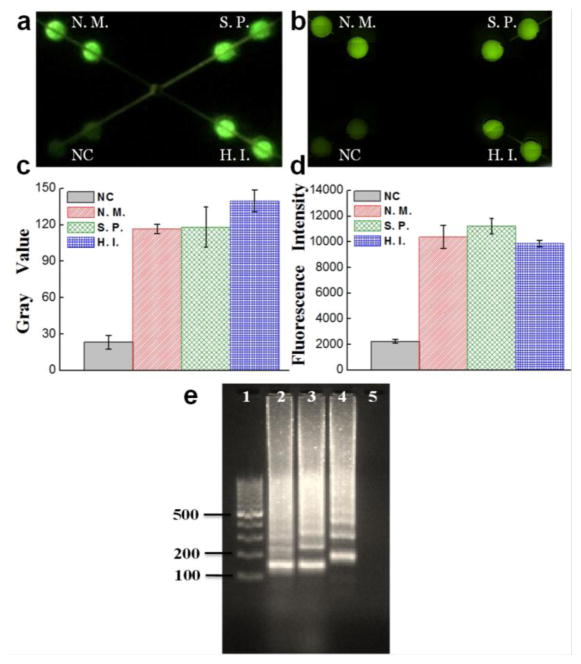
Before LAMP reactions, the fluorescence of calcein, the detection reagent, is quenched by manganese ions. After LAMP reactions, the byproduct pyrophosphate ion forms a complex with manganese ions. As a result, calcein is free to combine magnesium ions, producing bright fluorescence (see Fig. S1) (Mori and Notomi 2009; Tomita et al. 2008). Hence, the instrument-free determination can be achieved by the naked eye or a smartphone camera, based on fluorescence from the LAMP zones under a UV light pen. Fig. 2a showed a photograph taken by a smartphone camera for detection of Hib by using extracted DNA under a UV light pen, in which only the LAMP zones for Hib detection were lit up with bright green fluorescence but not for NC LAMP zones. Similar results were obtained from fluorescence microscopy (see Fig. 2b) to confirm the results from a smartphone camera. Similarly, the on-chip LAMP pathogen detection of N. meningitidis and S. pneumoniae was performed by using a smartphone camera and confirmed by a fluorescence microscope, respectively. As shown in Fig. 2c–d and Fig. S3, LAMP zones for N. meningitidis and S. pneumoniae detection exhibited strong fluorescence, while very weak fluorescence was exhibited in LAMP zones for NC. In addition, the hybrid microfluidic chip was designed to offer versatile functions. Each LAMP products can be extracted separately from each outlet for confirmatory tests such as by gel electrophoresis (see Fig. 2e). The LAMP reaction generates a mixture of stem-loop DNA products with various sizes (Tomita et al. 2008). As can be seen from Fig. 2e, the specific ladder-pattern bands of LAMP products from each pathogen but not from NC represented a mixture of DNA amplicons with different sizes due to loop-mediated reactions, which confirmed that the on-chip LAMP reactions for singleplexed pathogen detection were successful. As also can be seen from Fig. 2e, the complicated ladder-pattern bands make it challenging for multiplexed LAMP detection in a single microwell or tube.
Fig. 2.
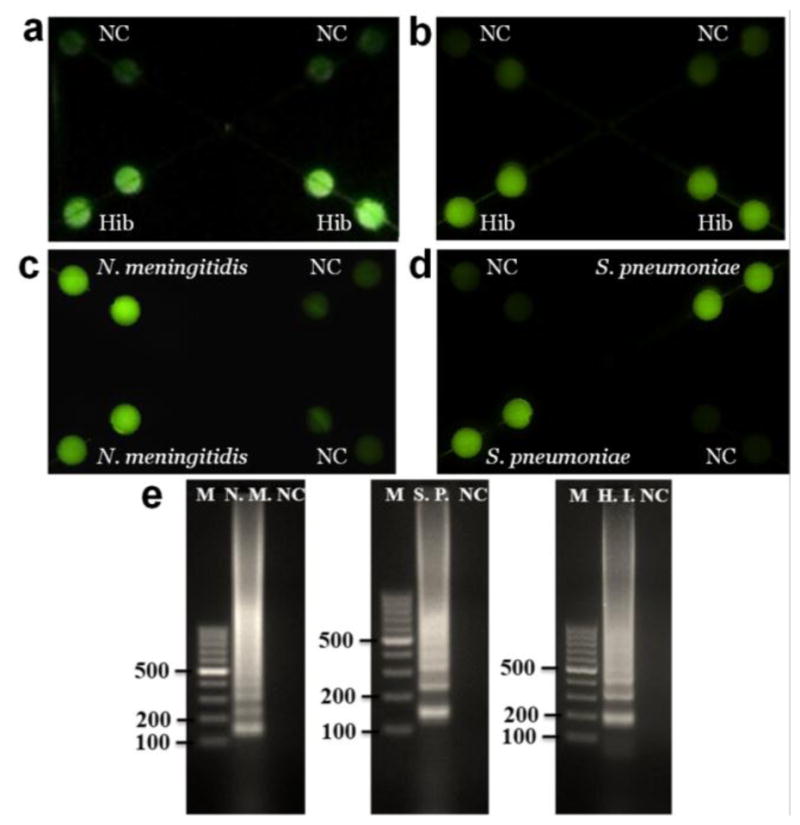
On-chip LAMP singleplexed meningitis detection of Hib, N. meningitidis and S. pneumoniae using extracted DNA by a smartphone camera (a) and a fluorescence microscopy (b–d). (e) Gel electrophoresis analysis of collected LAMP products for confirmatory analysis. Lanes N.M., S.P., H.I. were LAMP products from N. meningitidis, S. pneumoniae and Hib respectively; Lane M, 100 bp ladder; Lane NC, negative control. The extracted DNA templates used were 2 μL 29.6 ng/μL for N. meningitidis, 2 μL 56.1 ng/μL for S. pneumoniae and 2 μL 39.7 ng/μL for Hib, which corresponded to 3 × 106, 6 × 106 and 5 × 106 copies per LAMP zone, respectively.
3.2. Multiplexed meningitis detection by using extracted DNA
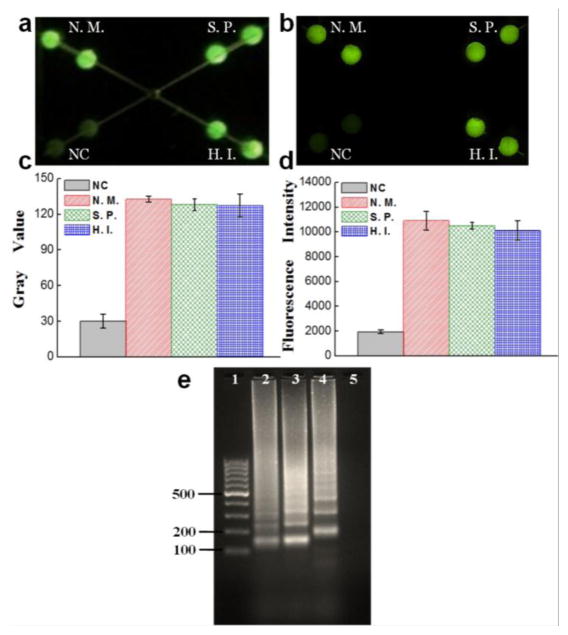
Multiple pathogens can coexist. Identification of the exact bacteria causing meningitis is vital because treatment and antibiotics differ for each type. Multiplexed pathogen detection provides a more efficient way for rapid diagnostics from the real world through a single assay, avoiding inconvenience and delays from multiple repeated singleplexed assays. Therefore, upon the success of the singleplexed detection of these three pathogens separately, we investigated the feasibility of our microfluidic approach for multiplexed detection of three meningitis-causing pathogens using extracted DNA. Different primers targeting different pathogens were pre-loaded in different LAMP zones in a ready-to-use device. During the multiplexed assay, a LAMP mix including template DNA from all the three pathogens was introduced from the common inlet. The LAMP zones in the middle PDMS layer are not directly connected on the same plane, which can effectively prevent the cross-talk and the risk of cross-contamination among these different LAMP zones. This allows us to perform parallel LAMP reactions in independent compartments for multiplexed bacterial meningitis diagnosis. As shown in Fig. 3a, end point detection results could be visually determined from the green fluorescence of the LAMP zones under UV light. All the LAMP zones targeting these three pathogens (i.e. N. meningitidis, S. pneumoniae and Hib) were lit up with strong fluorescence, but not for NC LAMP zones. Their average gray values were measured by the ImageJ software (see Fig. 3c), quantitatively indicating the brightness difference between pathogen detection zones and NC zones. In addition, fluorescence image and intensities from Fig. 3b and 3d show similar results. Fluorescence intensities of the LAMP products from the three pathogens were about five times higher than that of the NC. Furthermore, LAMP products from these three pathogens were separately collected from each outlet to run gel electrophoresis. As shown in Fig. 3e, the ladder-pattern bands of LAMP products from these three pathogens further verified that the multiplexed on-chip meningitis detection was successful.
Fig. 3.
On-chip LAMP multiplexed pathogen detection of N. meningitidis (N.M.), S. pneumoniae (S.P.) and Hib (H.I.) using extracted DNA by a smartphone camera (a) and fluorescence microscopy (b). (c) Gray values of the LAMP products measured by ImageJ; (d) Fluorescent intensities of the LAMP products measured by a fluorescence microscope. (e) Gel electrophoresis analysis of collected LAMP products for confirmatory analysis. Lanes 1–5: 100 bp ladder, LAMP products from N. meningitidis, S. pneumoniae and Hib, NC. The information of the extracted DNA templates is mentioned in Fig. 2.
3.3. Specificity test
To investigate the specificity of the approach for N. meningitidis, S. pneumoniae and Hib detection, we designed a microfluidic array to investigate the cross-reactions among these three bacteria DNA with their corresponding and non-corresponding LAMP primers (see Fig. S4).
As shown in Fig. 4a, specific LAMP primers for N. meningitidis, S. pneumoniae and Hib were pre-loaded in lateral rows from the top to the bottom, respectively. Blank and three DNA samples of N. meningitidis, S. pneumoniae and Hib were introduced to columns from left to right, respectively. It can be seen from Fig. 4 that only LAMP zones with corresponding primers and DNA samples showed bright green fluorescence. For instance, for the column with the N. meningitidis DNA sample, only the LAMP zone with pre-loaded N. meningitidis LAMP primers produced bright fluorescence, the intensity of which was about 5 times higher than that of the blank (see Fig. 4b). Other LAMP zones were as dim as the blank. Similar phenomena were observed with S. pneumoniae and Hib, indicating the high specificity of the approach for multiplexed meningitis detection.
Fig. 4.
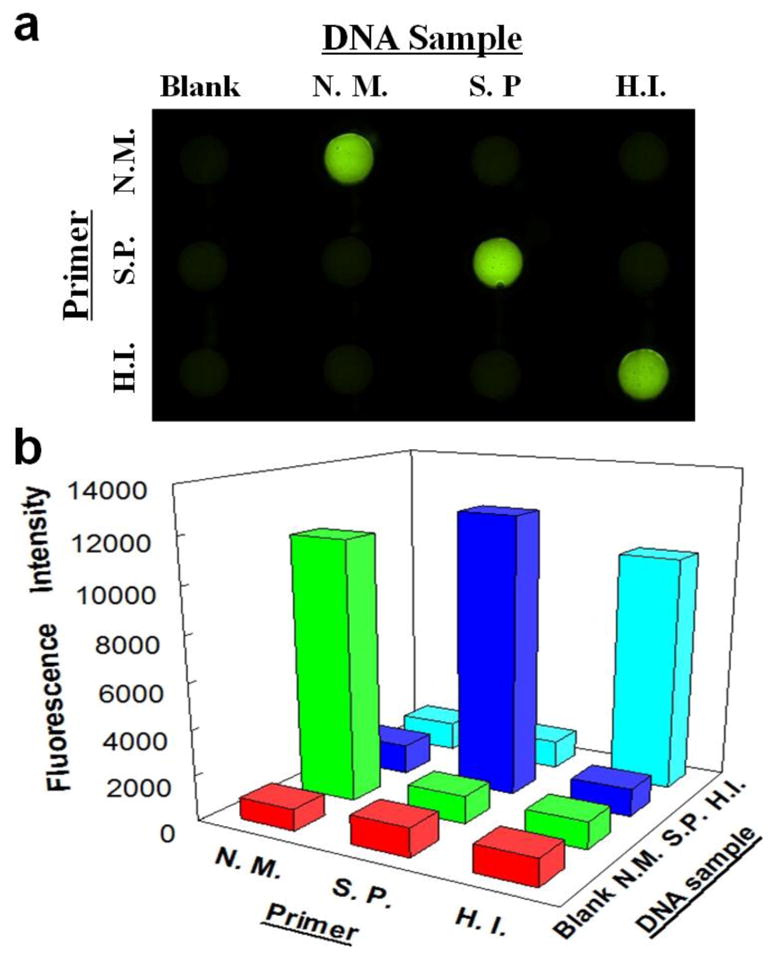
Specificity investigation among N. meningitidis, S. pneumoniae and Hib with their corresponding and non-corresponding LAMP primers by a fluorescence image (a) and fluorescence intensities (b). Lateral rows from the top to the bottom were pre-loaded with LAMP primers for N. meningitidis (N.M.), S. pneumoniae (S.P.) and Hib (H.I.) respectively; columns from left to right were introduced with Blank, DNA samples of N. meningitidis (N.M.), S. pneumoniae (S.P.) and Hib (H.I.), respectively. The RSDs for N. meningitidis, S. pneumoniae and Hib are 3.6%, 3.8% and 12.0% (n=3) on a different microfluidic device designed for the specificity test (see Fig. S4).
3.4. Multiplexed instrument-free detection of microorganisms from ACSF samples
The multiplexed meningitis detection discussed above was performed by using extracted pathogen DNA. However, to test real clinical samples, such as cerebrospinal fluid (CSF) or blood, extra complicated sample preparing steps including DNA extraction and purification before the amplification are required. Traditional clinical sample preparation procedures not only are time-consuming, but also rely on specialized equipment such as a centrifuge in laboratory settings (Safavieh et al. 2012). These limitations make them not feasible for POC pathogen detection.
In this work, we further integrated simple on-chip lysis procedures on the hybrid biochip for instrument-free pathogen detection. To mimic the real clinical samples, we prepared ACSF samples containing pathogenic microorganisms of N. meningitidis, S. pneumoniae and Hib. The mixture of the ACSF sample with the bacterial lysis buffer that we developed was simply introduced into the device from the common inlet, and was further incubated for about 10 min at room temperature to complete the lysis step before the introduction of the LAMP mix. This on-chip lysis procedure was not only simple, without the use of any centrifuges, but also fully compatible with the subsequent LAMP reaction. No noticeable inhibition to LAMP was observed. As shown in Fig. 5a and 5b, LAMP products from the ACSF sample produced bright fluorescence under a UV light pen as well as under a fluorescence microscope. The fluorescence difference between pathogen LAMP zones and NC zones can be readily distinguished by the naked eye, as indicated in the photograph taken by a smartphone camera (see Fig. 5a). Their gray values and fluorescence intensities shown in Fig. 5c and 5d respectively indicate their difference quantitatively. As shown in Fig. 5c, the RSDs for the detection of N. meningitidis, S. pneumoniae, and Hib were 4.9%, 3.6%, and 6.3% (n=6), respectively. In addition, as shown in Fig. 5e, the ladder-pattern bands from conventional gel electrophoresis of these three pathogens’ amplicons further confirmed that the on-chip LAMP reaction was successful. No significant difference was found in the instrument-free detection between the ACSF sample (Fig. 5) and the extracted DNA (Fig. 3). Therefore, the instrument-free multiplexed meningitis diagnosis directly from microorganism samples was readily achieved without relying on any special equipment.
Fig. 5.
Multiplexed detection of microorganisms of N. meningitidis (N.M.), S. pneumoniae (S.P.) and Hib (H.I.) spiked in ACSF samples by a smartphone camera (a) and a fluorescence microscopy (b). (c) Gray values of different LAMP zones measured by ImageJ from a photograph taken by a smartphone camera. (d) Fluorescent intensities of different LAMP zones measured by a fluorescence microscope. (e) Gel electrophoresis analysis of LAMP products from N. meningitidis, S. pneumoniae and Hib in ACSF. Lanes 1–5: 100 bp ladder, LAMP products of N. meningitidis, S. pneumoniae and Hib in ACSF, NC.
3.5. Limits of detection (LODs)
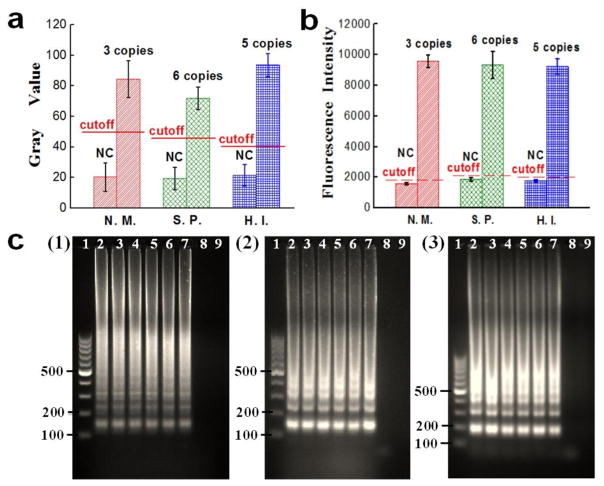
By preparing a serial of 10-fold diluted template DNA samples, the LOD for each pathogen was studied. The gray values and fluorescence intensities of different LAMP zones for N. meningitidis, S. pneumoniae and Hib are shown in Fig. 6a and 6b respectively. It indicated that strong fluorescence was still observed even when the template DNA was only a few copies per LAMP zone (see Fig. S5 for fluorescence images). On the contrary, no obvious fluorescence was generated when the template DNA was lower than 3 copies. Based on the negative control’s mean gray value plus its 3-fold standard deviations, the gray values of the cutoff lines for N. meningitidis, S. pneumoniae and Hib were calculated to be 48.1, 41.3 and 42.5 respectively, as shown by the cutoff lines in Fig. 6a. The gray values of 3 copies N. meningitidis, 6 copies S. pneumoniae and 5 copies Hib DNA templates were much higher than their cutoff lines. Similar results were achieved by measuring the fluorescence intensities (see Fig. 6b). Gel electrophoresis results from Fig. 6c further confirmed the results. The ladder-pattern bands of the LAMP products produced with more than 3 copies template DNA indicated that these on-chip LAMP reactions were successful (see Fig. 6c). On the contrary, when the template DNA was below 1 copy, no DNA bands were observed. Therefore, we concluded that the LODs of N. meningitidis, S. pneumoniae and Hib were 3 copies, 6 copies and 5 copies per LAMP zone, respectively. The LODs corresponded to 2 μL of each original template DNA samples of N. meningitidis (29.6 fg/μL), S. pneumoniae (56.1 fg/μL) and Hib (39.7 fg/μL), respectively. This indicates that although no expensive instrument was used, the sensitivity of our microfluidic method is comparable to or higher than qPCR (9 copies for N. meningitidis (Wang et al. 2012), 14 copies of S. pneumoniae (Park et al. 2010), and 10 copies for Hib per reaction (Chiba et al. 2009).
Fig. 6.
LOD determination. (a–b) Gray values and fluorescence intensities of LAMP products from 3 copies N. meningitidis (N. M.) DNA template, 6 copies S. pneumoniae (S. P.) DNA template and 5 copies Hib (H. I.) DNA template as well as their corresponding NC respectively. The dash lines were the cutoff gray values and fluorescence intensities for their corresponding pathogen’s LOD. (c) Gel electrophoresis analysis. Lane 1: 100 bp marker; Lanes 2–8: LAMP products using a series of 10-fold diluted DNA samples of (1) N. meningitidis 3 × 105, 3 × 104, …, 3 × 100, 3 × 10−1 copies per LAMP zone; (2) S. pneumoniae 6 × 105, 6 × 104, …, 6 × 100, 6 × 10−1 copies per LAMP zone; (3) Hib 5 × 105, 5 × 104, …, 5 × 100, 5 × 10−1 copies per LAMP zone; Lane 9: NC;
3.6. Shelf life test
Using N. meningitidis as an example, the LAMP performance of the hybrid biochip with paper inside was investigated and compared with the paper-free non-hybrid biochip over a period of three months. We pre-loaded DNA primers of N. meningitidis in a series of these two types of microfluidic biochips, and stored those ready-to-use biochips in dark at room temperature (25 °C). To investigate the on-chip LAMP performance, LAMP products from different hybrid and non-hybrid biochips were measured by using Nanodrop for each reaction after different time periods (in half a month, one month, one and a half months, two months and three months, respectively). As shown in Fig. S6, the LAMP performance of the non-hybrid biochip decreased sharply within the first half month (by ~22%), and continued decreasing over the three months, with a total decline of ~40%. It indicated that without paper inside, the performance of the non-hybrid biochip was not stable. Fresh biochips should be prepared right before each LAMP assay, making them user-unfriendly and inappropriate for in-field and POC applications. However, the hybrid biochip exhibited stable LAMP performance over the experimental period. Only a slight decrease (~6%) was observed within the first two months. And the on-chip LAMP performance remained stable even after three months (>85%). Therefore, our hybrid biochip with paper inside warranted more stable performance of LAMP reactions than the paper-free non-hybrid system for a long period time. This is probably mainly because that the paper with highly interwoven fibers provides a 3D matrix for protection of the DNA primers from harsh environmental conditions and also prevent the primers forming aerosols resulting in primer loss in the air. Owens et al. reported a simple and economical filter paper-based method to store and preserve insect DNA as a template for PCR analysis, which showed successful PCR amplification after 7 months (Owens and Szalanski 2005). We expect that our hybrid microfluidic biochip with paper inside for storage and preservation of the pre-loaded DNA primers should have a much longer shelf life than the tested period of 3 months.
4. Conclusions
We have developed a low-cost polymer/paper hybrid microfluidic biochip for multiplexed instrument-free detection of three major meningitis-causing pathogens. Microorganisms in ACSF samples can be directly analysed due to the integration of simple centrifuge-free lysis on the chip, thus avoiding cumbersome sample preparation procedures in conventional pathogen methods. The visual detection can be readily achieved by the naked eye within one hour based on the recovered fluorescence under a portable UV light, without relying on any specialized instruments. The LOD of our approach for N. meningitidis, S. pneumoniae and Hib are 3 copies, 6 copies and 5 copies per LAMP zone, comparable to those of real-time PCR.
The polymer/paper hybrid microfluidic biochip fuses the merits of different low-cost materials, such as high liquid manipulation performance and transparency from PDMS and 3D porous structures for preserving DNA primers from cellulose paper. The shelf life experiment showed that the hybrid microfluidic biochip has a stable performance over a longer time period at room temperature (25 °C). Storage in a refrigerator is expected to further prolong the shelf life time.
With all of these characteristics, the hybrid microfluidic biochip is able to achieve POC detection of multiplexed meningitis, especially in resource-limited settings. In addition, by employing other DNA primers targeting to different pathogens, this hybrid microfluidic biochip should have broad applications in instrument-free multiplexed detection of a variety of other pathogens (e.g., Zika virus). In this study, the data analysis still relies on the software ImageJ installed on a computer. A smartphone application which is under development would further increase convenience and portability of our method.
Supplementary Material
Highlights.
This is the first time to report such a microfluidic device for simultaneous instrument-free detection of three major meningitis-causing pathogens with high specificity (N. meningitidis, S. pneumoniae and Hib).
The method is highly rapid and sensitive. The limits of detection of a few DNA copies (e.g. 3 copies) per LAMP zone for N. meningitidis, S. pneumoniae and Hib were achieved within one hour without using any specialized laboratory instrument, which are comparable or even better than costly real-time PCR.
Taking advantages of both chip substrates, PDMS and paper, the hybrid microfluidic biochip had a much longer shelf life with stable on-chip LAMP performance compared with the paper-free non-hybrid microfluidic biochip over a period of three months.
Simple lysis procedures are integrated on the chip, and athogenic microorganisms can be directly analyzed, avoiding laborious sample preparation procedures and any specialized instruments such as centrifuges.
The low-cost, rapid, instrument-free and high-sensitivity microfluidic approach that we developed has great potential for point-of-care (POC) diagnosis of multiplexed infectious disease diagnosis, especially in resource-limited settings.
Acknowledgments
The authors would like to acknowledge the financial support of the NIH/NIGMS under award number SC2GM105584 and the NIH/NIAID under award number R21AI107415. Financial support from the IDR Program at the UTEP and the NIH RCMI Pilot Grant is also gratefully acknowledged. We would like to express special thanks to the staff of the Genomic Analysis Core Facility of UTEP. This core is supported by Grant G12MD007592 from the National Institute on Minority Health and Health Disparities (NIMHD).
Appendix A. Supplementary material
Supplementary data associated with this article can be found in the online version at:
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Cold Spring Harbor Protocols. 2011;(9) pdb.rec065730. [Google Scholar]
- World Health Organization. Fact sheet N 141. 2015 http://www.who.int/mediacentre/factsheets/fs141/en/
- Agier L, Broutin H, Bertherat E, Djingarey MH, Lingani C, Perea W, Hugonnet S. Transactions of The Royal Society of Tropical Medicine and Hygiene. 2013;107(1):30–36. doi: 10.1093/trstmh/trs010. [DOI] [PubMed] [Google Scholar]
- Borysiak MD, Kimura KW, Posner JD. Lab on a Chip. 2015;15(7):1697–1707. doi: 10.1039/c4lc01479k. [DOI] [PubMed] [Google Scholar]
- Brouwer MC, Tunkel AR, van de Beek D. Clinical Microbiology Reviews. 2010;23(3):467–492. doi: 10.1128/CMR.00070-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo D. WHO Manual. 2 2011. [Google Scholar]
- Chaisomchit S, Wichajarn R, Janejai N, Chareonsiriwatana W. Southeast Asian J Trop Med Public Health. 2005;36(1):270–273. [PubMed] [Google Scholar]
- Chang WH, Yang SY, Wang CH, Tsai MA, Wang PC, Chen TY, Chen SC, Lee GB. Sensors and Actuators B: Chemical. 2013;180:96–106. [Google Scholar]
- Chiba N, Murayama SY, Morozumi M, Nakayama E, Okada T, Iwata S, Sunakawa K, Ubukata K. Journal of Infection and Chemotherapy. 2009;15(2):92–98. doi: 10.1007/s10156-009-0670-3. [DOI] [PubMed] [Google Scholar]
- de Almeida Gomes BPF, Vianna ME, Sena NT, Zaia AA, Ferraz CCR, de Souza Filho FJ. Oral Surgery, Oral Medicine, Oral Pathology, Oral Radiology, and Endodontology. 2006;102(4):544–550. doi: 10.1016/j.tripleo.2006.04.010. [DOI] [PubMed] [Google Scholar]
- Dou M, Dominguez DC, Li X, Sanchez J, Scott G. Analytical Chemistry. 2014;86(15):7978–7986. doi: 10.1021/ac5021694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou M, García JM, Zhan S, Li X. Chemical Communications. 2016a;52:3470–3473. doi: 10.1039/c5cc09066k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou M, Lopez J, Rios M, Garcia O, Xiao C, Eastman M, Li X. Analyst. 2016b;141(12):3898–3903. doi: 10.1039/c6an00370b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou M, Sanjay ST, Benhabib M, Xu F, Li X. Talanta. 2015;145:43–54. doi: 10.1016/j.talanta.2015.04.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte C, Salm E, Dorvel B, Reddy B, Jr, Bashir R. Biomed Microdevices. 2013;15(5):821–830. doi: 10.1007/s10544-013-9769-5. [DOI] [PubMed] [Google Scholar]
- Dubos F, Moulin F, Gajdos V, De Suremain N, Biscardi S, Lebon P, Raymond J, Breart G, Gendrel D, Chalumeau M. The Journal of Pediatrics. 2006;149(1):72–76. doi: 10.1016/j.jpeds.2006.02.034. [DOI] [PubMed] [Google Scholar]
- Fang X, Chen H, Xu L, Jiang X, Wu W, Kong J. Lab on a Chip. 2012;12(8):1495–1499. doi: 10.1039/c2lc40055c. [DOI] [PubMed] [Google Scholar]
- Fu G, Sanjay ST, Dou M, Li XJ. Nanoscale. 2016;8:5422–5427. doi: 10.1039/c5nr09051b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman O, Weber JR. Therapeutic Advances in Neurological Disorders. 2009;2(6):401–412. doi: 10.1177/1756285609337975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huy NT, Boamah D, Lan NTP, Van Thanh P, Watanabe K, Huong VTT, Kikuchi M, Ariyoshi K, Morita K, Hirayama K. FEMS Microbiology Letters. 2012;337(1):25–30. doi: 10.1111/1574-6968.12002. [DOI] [PubMed] [Google Scholar]
- Iseki H, Alhassan A, Ohta N, Thekisoe OM, Yokoyama N, Inoue N, Nambota A, Yasuda J, Igarashi I. Journal of Microbiological Methods. 2007;71(3):281–287. doi: 10.1016/j.mimet.2007.09.019. [DOI] [PubMed] [Google Scholar]
- Kim DW, Kilgore PE, Kim EJ, Kim SA, Anh DD, Dong BQ, Kim JS, Seki M. PLoS One. 2012;7(8):e42954. doi: 10.1371/journal.pone.0042954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DW, Kilgore PE, Kim EJ, Kim SA, Anh DD, Seki M. Journal of Clinical Microbiology. 2011;49(10):3621–3626. doi: 10.1128/JCM.00515-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Kim EJ, Kilgore PE, Kim SA, Takahashi H, Ohnishi M, Anh DD, Dong BQ, Kim JS, Tomono J. PloS One. 2015 doi: 10.1371/journal.pone.0122922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Chen Y, Li PC. Lab on a Chip. 2011;11(7):1378–1384. doi: 10.1039/c0lc00626b. [DOI] [PubMed] [Google Scholar]
- Li X, Valadez AV, Zuo P, Nie Z. Bioanalysis. 2012;4(12):1509–1525. doi: 10.4155/bio.12.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XJ, Zhou Y. Elsevier; 2013. [Google Scholar]
- Liu P, Li X, Greenspoon SA, Scherer JR, Mathies RA. Lab on a Chip. 2011;11(6):1041–1048. doi: 10.1039/c0lc00533a. [DOI] [PubMed] [Google Scholar]
- Martinez AW. Bioanalysis. 2011;3(23):2589–2592. doi: 10.4155/bio.11.258. [DOI] [PubMed] [Google Scholar]
- McKenna JP, Fairley DJ, Shields MD, Cosby SL, Wyatt DE, McCaughey C, Coyle PV. Diagnostic Microbiology and Infectious Disease. 2011;69(2):137–144. doi: 10.1016/j.diagmicrobio.2010.10.008. [DOI] [PubMed] [Google Scholar]
- Mori Y, Notomi T. Journal of Infection and Chemotherapy. 2009;15(2):62–69. doi: 10.1007/s10156-009-0669-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscoso H, Thayer SG, Hofacre CL, Kleven SH. Avian Diseases. 2004;48(4):841–850. doi: 10.1637/7215-060104. [DOI] [PubMed] [Google Scholar]
- Owens CB, Szalanski AL. Journal of Medical Entomology. 2005;42(4):709–711. doi: 10.1093/jmedent/42.4.709. [DOI] [PubMed] [Google Scholar]
- Park HK, Lee HJ, Kim W. FEMS Microbiology Letters. 2010;310(1):48–53. doi: 10.1111/j.1574-6968.2010.02044.x. [DOI] [PubMed] [Google Scholar]
- Safavieh M, Ahmed MU, Tolba M, Zourob M. Biosensors and Bioelectronics. 2012;31(1):523–528. doi: 10.1016/j.bios.2011.11.032. [DOI] [PubMed] [Google Scholar]
- Sanjay ST, Dou M, Fu G, Xu F, Li X. Current Pharmaceutical Biotechnology. 2016a;17(9):772–787. doi: 10.2174/1389201017666160127110440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjay ST, Dou M, Sun J, Li X. Sci Rep. 2016b;6:30474. doi: 10.1038/srep30474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjay ST, Fu G, Dou M, Xu F, Liu R, Qi H, Li X. Analyst. 2015;140(21):7062–7081. doi: 10.1039/c5an00780a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Zhu S, Jin C, Chen F. International Journal of Food Microbiology. 2011;148(2):75–79. doi: 10.1016/j.ijfoodmicro.2011.05.004. [DOI] [PubMed] [Google Scholar]
- Shen F, Li X, Li PC. Biomicrofluidics. 2014;8(1):014109. doi: 10.1063/1.4866358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L, Burgoyne LA. BMC ecology. 2004;4(1):4. doi: 10.1186/1472-6785-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staroscik A. URI Genomics & Sequencing Center. 2004 http://cels.uri.edu/gsc/cndna.html.
- Sun Y, Quyen TL, Hung TQ, Chin WH, Wolff A, Bang DD. Lab on a Chip. 2015;15(8):1898–1904. doi: 10.1039/c4lc01459f. [DOI] [PubMed] [Google Scholar]
- Tian T, Li J, Song Y, Zhou L, Zhu Z, Yang CJ. Lab on a Chip. 2016;16(7):1139–1151. doi: 10.1039/c5lc01562f. [DOI] [PubMed] [Google Scholar]
- Tomita N, Mori Y, Kanda H, Notomi T. Nature Protocols. 2008;3(5):877–882. doi: 10.1038/nprot.2008.57. [DOI] [PubMed] [Google Scholar]
- Wang CH, Lien KY, Wu JJ, Lee GB. Lab on a Chip. 2011;11(8):1521–1531. doi: 10.1039/c0lc00430h. [DOI] [PubMed] [Google Scholar]
- Wang X, Theodore MJ, Mair R, Trujillo-Lopez E, du Plessis M, Wolter N, Baughman AL, Hatcher C, Vuong J, Lott L. Journal of Clinical Microbiology. 2012;50(3):702–708. doi: 10.1128/JCM.06087-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenhui L, Bingjie Z, Qinxin S, Guohua Z. Yi Chuan. 2015;37(9):899. [Google Scholar]
- Zhang J, Sheng W, Fan ZH. Chemical Communications. 2014;50(51):6722–6725. doi: 10.1039/c4cc02002b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo P, Li X, Dominguez DC, Ye BC. Lab on a Chip. 2013;13(19):3921–3928. doi: 10.1039/c3lc50654a. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.