Abstract
Gene fusions between CIC and DUX4 define a rare class of soft tissue sarcomas poorly understood at the molecular level. Previous karyotyping and fluorescence in situ hybridization studies support trisomy chromosome 8 as a recurrent alteration, however other driving alterations are largely unknown. Thus, we analyzed eleven formalin fixed paraffin embedded CIC-DUX4 sarcoma tissue samples (including three sample pairs) using targeted Ion Torrent based multiplexed polymerase chain reaction (PCR) next generation sequencing to characterize potential somatic driver alterations in 409 genes. Although we did not identify recurrent somatic mutations (point mutations or indels), copy number analysis showed recurrent, broad, copy number alterations, including gain of chromosome 8 and loss of 1p. In one sample pair (untreated primary and local recurrence resections), we identified similar copy number profiles and a somatic ARID1A R963X nonsense mutation exclusively in the local recurrence sample. In another sample pair (pre- and post-radiation treatment specimens), we observed single copy loss of chromosome 7q exclusively in the post-treatment recurrence sample, supporting it as an acquired event after radiation treatment. In the last sample pair (near concurrent, post-chemotherapy primary and distant metastasis), molecular profiles were highly concordant, consistent with limited inter-tumoral heterogeneity. In summary, next generation sequencing identified limited somatic driver mutations in CIC-DUX4 sarcomas. However, we identified novel, recurrent copy number alterations, including chromosome 1p, which is also the locus of ARID1A. Additional functional work and assessment of larger cohorts is needed to determine the biological and clinical significance of the alterations identified herein.
Keywords: Next-generation sequencing, CIC-DUX4, FFPE tissue, soft tissue sarcoma, ARID1A
1. Introduction
Soft tissue sarcomas are potentially aggressive tumors that are challenging to diagnose and classify due to the morphologic similarities between subgroups [1]. Despite advancements in understanding at the morphologic and genomic level, 5% of sarcomas remain unclassifiable in clinical practice [2–5]. Such tumors have been termed “undifferentiated soft tissue sarcomas” (USTSs) since they do not show histologic or immunohistochemical features characteristic of a specific lineage [2–5]. Although a subset of USTSs have been characterized as “Ewing-like” tumors due to their round cell morphology and immunophenotype [6], undifferentiated round cell sarcomas (URCS) lack characteristic Ewing sarcoma fusions involving EWSR1 and members of the ETS transcription factor family [7]. In 2006, Kawamura-Saito et al. reported that some aggressive URCSs harbored fusions of CIC (a human homolog of Drosophila capicua) to DUX 4 (double homeobox 4), as a result of t(4;19)(q35;q13.1) translocations[8]. CIC-DUX4 sarcomas typically have small round cell morphology, geographic necrosis, coarse chromatin, focal extracellular myxoid matrix, clear cell areas, and mild-moderate nuclear pleomorphism [7–10]. Furthermore, most cells lack a well-defined cell border and contain vesicular nuclei with often enlarged nucleoli [11].
The CIC-DUX4 fusion results in a chimeric protein that includes the majority of the CIC gene but lacks the homeodomains of DUX4 [8]. CIC is a transcription factor member of the HMG box superfamily that is involved in the development of medulloblastoma [10]. DUX4 has primarily been characterized in the context of muscular dystrophy, where aberrant DUX4 expression due to epigenetic changes are thought to cause facioscapulohmueral muscular dystrophy (FSHD) [12]. Previous research has shown CIC-DUX4 fusions expose the DUX4 C-terminus, resulting in increased activation of CIC, even though the DNA binding of the CIC HMG domain is largely not affected [8]. This suggests that CIC downstream targets, such as ETS family members, may be deregulated, supporting CIC-DUX4 as an oncogenic transcription factor [8].
Given the rarity of CIC-DUX4 sarcomas, molecular alterations beyond the defining translocation remain poorly understood and their molecular relationship to Ewing sarcoma [2, 4, 7–10, 13-18]. Previous karyotyping and fluorescence in situ hybridization (FISH) studies support chromosome (chr) 8 trisomy and MYC amplification as recurrent alterations in CIC-DUX4 sarcomas [7, 18], however a more comprehensive analysis of the genomic landscape of CIC-DUX4 sarcomas, including assessment of somatic point mutations, small insertions/deletions (indels), and copy number alterations (CNAs), is lacking. Such an analysis is needed given the aggressive course and rapid chemoresistance of CIC-DUX4 sarcomas and lack of highly efficacious therapeutic strategies [7]. Likewise, it is unclear whether CIC-DUX4 sarcomas are similar to Ewing sarcoma at the genomic level, as Ewing sarcomas have few recurrent point mutations/indels (most frequently involving TP53 and STAG2) but several recurrent, broad, copy number alterations (CNAs). Hence, here we profiled the genomic landscape of eleven formalin fixed paraffin embedded (FFPE) CIC-DUX4 sarcomas (including three pairs of samples) using targeted next generation sequencing (NGS) of the coding sequence from 409 cancer related genes to assess somatic mutations and CNAs.
2. Materials and Methods
2.1 Cohort
We identified eleven CIC-DUX4 sarcoma formalin fixed paraffin embedded (FFPE) tissue samples from the University of Michigan Department of Pathology Archives. IRB approval was obtained to perform targeted next generation sequencing on clinical FFPE tumor material. Clinicopathological information for each sample was obtained from the medical record. Hematoxylin and eosin (H&E) stained slides were reviewed by board-certified Anatomic Pathologists (R.P and S.A.T.) to ensure sufficient tumor content. Of the eleven samples, three represented sequential pairs: Samples 5A and 5B represent a pre-treatment primary tumor and a post radiation therapy pelvic recurrence; Samples 6A and 6B represent a post systemic/adjuvant chemotherapy treated primary tumor and a near concurrent (<1 month) brain metastasis; and Samples 7A and 7B represent an untreated primary tumor resection with no evidence of residual disease and a rapid (<3 months) local recurrence without adjuvant therapy. CIC-DUX4 rearrangement for all samples was confirmed by RT-PCR and/or FISH as described [7] prior to inclusion in our sequencing cohort.
2.2 DNA/RNA Isolation
For each sample, 5–8 × 10um FFPE sections were cut from a single representative block and macrodissected with a scalpel to enrich for tumor content. DNA was isolated using the Qiagen Allprep FFPE DNA/RNA kit (Qiagen, Valencia, CA) as described[19, 20]. DNA was quantified using the Qubit 2.0 fluorometer (Life Technologies, Foster City, CA).
2.3 Targeted Next Generation Sequencing
We performed targeted, multiplexed PCR based next generation sequencing (NGS) using the Ion Ampliseq Comprehensive Cancer Panel (CCP), which targets 1,688,650 bases from 15,992 amplicons representing 409 cancer genes, essentially as described [19, 20]. Barcoded libraries were generated from 40 ng of DNA per sample using the CCP and the Ion Ampliseq library kit 2.0 (Life Technologies, Foster City, CA) according to manufacturer’s instructions with barcode incorporation. Templates were prepared using the Ion PI Template OT2 200 Kit v3 (Life Technologies, Foster City, CA) on the Ion One Touch 2 according to the manufacturer’s instructions. Sequencing of multiplexed templates was performed using the Ion Torrent Proton Sequencer (Life Technologies, Foster City, CA) on Ion PI chips using the Ion PI Sequencing 200 Kit v3 (Life Technologies, Foster City, CA) according to the manufacturer’s instructions.
2.4 Somatic Variant Identification
Data analysis was performed essentially as described [19, 20] using validated pipelines based on Torrent Suite 4.0.2, with alignment by TMAP using default parameters, and variant calling using the Torrent Variant Caller plugin (version 4.0-r76860) with low-stringency default somatic variant settings. Variants were annotated using Annovar [21]. Called variants were filtered to remove synonymous or non-coding variants, those with flow corrected read depths (FDP) ≤30, flow corrected variant allele containing reads (FAO) ≤6, variant allele frequencies (FAO/FDP) <0.10, extreme skewing of forward/reverse flow corrected reads (FSAF/FSAR <0.2 or >5), FSAF and FSAR >1, or indels within homopolymer runs >4 bases. Variants occurring exclusively in reads with other single nucleotide variants or indels and those occurring in the last mapped base of a read were excluded. Additionally, variants called in >4% of internally sequenced samples using the same panel and not having a cosmic ID were removed. Variants present in ESP6500 or 1000 Genomes (from Annovar) as well as samples with ExAC database (http://exac.broadinstitute.org) at allele frequencies greater than 0.1% were considered germ line variants and removed. Variants reported in ExAC with observed variant allele frequencies in our data between 0.40 and 0.60 or >0.9 were also considered germ line and removed unless occurring at known somatic mutation hotspots. High confidence somatic variants passing the above criteria were then visualized in IGV. From these somatic variants, hotspots (>1 observation at that residue in COSMIC) in oncogenes, or hotspot or deleterious alterations (nonsense/frameshift variants) in tumor suppressors were then considered as prioritized variants.
2.5 Copy number analysis
Copy number analysis was performed as described using a validated approach [19, 22]. Briefly, normalized GC content corrected read counts per amplicon for each sample were divided by those from a composite normal male DNA sample (composed of multiple FFPE and frozen tissue, individual and pooled samples) to identify CNAs through the copy number ratio for each amplicon. Genes with less than four amplicons or a wide distribution of gene-level CN estimates across a large panel of tumor and normal samples internally sequenced on the CCP were removed from all CN analyses.
2.6 Copy Number validated with quantitative PCR
ARID1A copy numbers were assessed by quantitative reverse transcription PCR (qRT-PCR) for a subset of the cohort with sufficient DNA. Primers and probes (5’ FAM; ZEN/Iowa Black FQ dual quenchers) were designed using PrimerQuest (http://www.idtdna.com/Primerquest/Home/Index, hg 19 genome assembly) and obtained from IDT (sequences available upon request). After assay specificity was confirmed using BLAST and BLAT, we excluded primers/probes in areas of SNPs. Each qPCR reaction (15ul) used 5 ng of genomic DNA per reaction, a final concentration of 0.9 uM for each primer and 0.25 uM for each probe in TaqMan Genotyping Master Mix (Applied Biosystems). Triplicate reactions for were performed using 384 well plates on the Quantstudio 12K Flex (Applied Biosystems). Automatic baseline and Ct thresholds were set using QuantStudio 12K Flex Real-Time PCR System Software. Log2 copy number of the genes were determined by the ΔΔCT method using the average Ct of FBXW7, DNMT3A, and IGF1R as the reference (copy number neutral by NGS in all samples) and an unrelated FFPE isolated male genomic DNA sample (copy number neutral by NGS) as the calibrator.
2.7 Sanger sequencing to validate called somatic variants
Bidirectional Sanger sequencing was performed over the prioritized mutations with variant allele frequencies >0.15 on all tumor samples. Genomic DNA (10ng) was used as template in PCR amplifications with Invitrogen Platinum PCR Supermix Hi-Fi (Life-Technologies) with the suggested initial denaturation and cycling conditions. PCR products were subjected to bidirectional Sanger sequencing for both primer pairs by the University of Michigan DNA Sequencing Core after treatment with ExoSAP-OT (GE Healthcare) and sequences were analyzed using SeqMan Pro Software (DNASTAR).
3. Results
3.1 Targeted next generation sequencing (NGS) demonstrates a lack of recurrent driving mutations in CIC-DUX4 sarcomas
To assess the genomic landscape of FISH and/or RT-PCR confirmed CIC-DUX4 sarcomas, we performed targeted NGS on eleven routine FFPE samples from eight patients whose clinical characteristics are presented in Table 1. Representative histology of three CIC-DUX4 sarcomas subjected to sequencing are shown in Figure 1. Amongst the eleven samples, we sequenced two samples from one case (Samples 6A and 6B), representing a primary tumor (located on the calf) and a near concurrent, brain metastasis, both of which had been exposed to prior systemic chemotherapy. We also sequenced two samples from a second case representing a primary pre-treatment tumor (located at the knee) and a post-radiation pelvic metastasis/local recurrence (Samples 5A and 5B). The last sample pair (Samples 7A and 7B) represented a primary, untreated tumor resection from the flank (which obtained pathological confirmed lack of residual disease) and a rapid local recurrence (in the absence of adjuvant therapy).
Table 1.
Clinicopathologic features of CIC-DUX4 sarcomas profiled by next generation sequencing (NGS)
| Sample | Reference | Gender | Age (yrs) |
Size (cm) |
Prior Treatment |
Tumor location |
Metastasis | Tumor Content |
Karyotype | NGS Prioritized mutations |
CIC-DUX4 FISH |
CIC-DUX4 RT-PCR |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | New | F | 21 | 22 | None | Leg | 80% | N/A |
TP53 C238Y; TP53 D208fs |
N/A | Yes | |
| 2 | Choi et al. | F | 20 | 6.0 | Chemo. | Shoulder | - | 70% | N/A | Yes | Yes | |
| 3 | Choi et al. | F | 32 | 14.0 | Chemo. | Pelvis | - | 60% | 17 cells: 46–48, X,t(X;1)(q11.2;p34), del(1)(p21p36), +del(1)(p22p36),t(3;20) (p21;q13.3), t(4;19)(q35;13.1),+8, del(13)(q12.3q14), -14[cp17];3cells:9193, idemx2,+del(13)(q12.3q14) [cp3]93,idemx2,+del(13)(q12.3q14) [cp3] |
Yes | Yes | |
| 4 | New | M | 14 | 17.0 | None | Forearm | - | 70% | 46–47, XY,del(1)(q32q44),t(3;16) (p21;q22), der(4)t(4;19)(q35;q13.1), +17,del(17)(q25q25), der(19)t(19;?22)(p13;q11.2) t(4;19)(q35;q13.1), -20,der(22)t(?19;22)(p13;q11.2) [cp9] |
Yes | ||
| 5A | Choi et al. | M | 43 | 9.8 | None | Knee | - | 70% | N/A | Yes | Yes | |
| 5B | Radiation | - | Pelvis | 50% | N/A | |||||||
| 6A | Choi et al. | F | 25 | 11.0 | Chemo. | Calf | - | 70% | 47,XX,t(4;19)(q35;q13.1), +8[15]* |
CTNNB1 E54K |
Yes | Yes |
| 6B | Chemo. | - | Brain | 70% | Yes | |||||||
| 7A | New | M | 13 | 4 | None | Flank | 60% | N/A | Yes | |||
| 7B | New | M | 14 | 5.5 | None | Flank | 80% | N/A |
ARID1A R693X |
Yes | Yes | |
| 8 | New | M | 17 | 7.4 | Chemo. | Inguinal LN | 70% | N/A | Yes |
Clinicopathologic information for CIC-DUX4 sarcomas profiled by NGS. For each profiled sample, the case number, inclusion in prior published studies (Choi et al. ref 7), gender, age at original diagnosis, tumor size (greatest dimension, cm) by imaging at diagnosis, prior treatment, primary tumor location (if sequenced), metastatic/local recurrence location (if sequenced), estimated tumor content (by histology), and previously determined karyotype (*from primary, pre-chemotherapy specimen). Priotized high confidence somatic mutations (see Methods) identified by NGS in this study are also shown (see Table S2 for details). CIC-DUX4 fusion status for all cases was confirmed as indicated by FISH and/or RT-PCR.
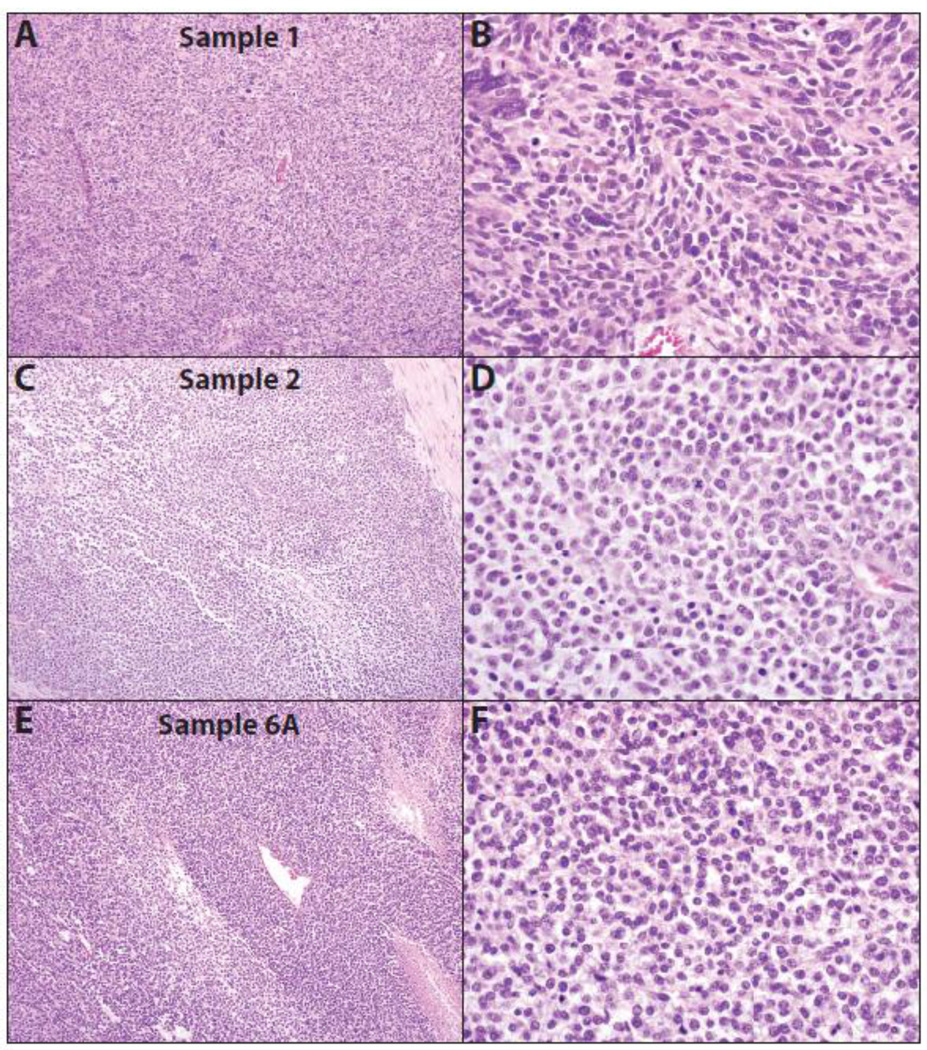
Fig. 1. Histology of CIC-DUX4 sarcomas subjected to next generation sequencing (NGS).
Low (A, C & E) and high power (B, D & F) hematoxylin and eosin stained (H&E) stained sections from three CIC-DUX4 sarcomas, Sample 1 (A&B), Sample 2 (C&D) and Sample 6A (E&F), subjected to NGS. Tumors show typical small round cell morphology, geographic necrosis, coarse chromatin, and focal extracellular myxoid matrix, with Sample 1 showing more pleomorphic histology. Original magnifications 10× (A, C & E) and 40× (B, D & F).
Targeted NGS from isolated DNA on each sample generated an average of 10,309,255 mapped reads yielding 634× targeted base coverage across the eleven samples (Table S1). An average of 1,347 variants per sample passed standard low stringency default filters, however after stringent filtering to identify high confidence somatic alterations (see Methods), we identified a total of twenty high confidence somatic mutations across the eleven samples (Table S2). Importantly, no genes were recurrently mutated across patients (Samples 7A and 7B harbor the same KMT2D A3318G variant at near 0.50 variant allele frequency, which although passing our somatic filtering is likely to be germline; Sample 1 also harbors a KMT2D mutation that is a known rare germline variant, but the observed variant allele frequency (0.39) is just below our germline filtering threshold (0.40) used herein.
Likewise, across the eleven samples, we identified only four total prioritized variants (Tables 1 & S2). An activating CTNNB1 E54K was identified in Sample 6A; however as this variant was present at low variant allele frequency (0.11) and was not detected in the matched brain metastasis from this case (6B), this alteration likely represents a subclonal alteration. TP53 C238Y and D208fs mutations were identified in Sample 1, with variant allele frequencies of 0.32 and 0.68, respectively, consistent with loss of TP53 function. Lastly, a prioritized ARID1A R693X mutation was identified exclusively in Sample 7B (variant allele frequency 157/417=38% vs. 1/355=0.3% in 7A, Tables 1 & S2). The three prioritized mutations with variant allele frequencies >15% (approximate lower limit for Sanger sequencing) were confirmed by Sanger sequencing (Fig S1). Taken together, these results support a lack of candidate driving somatic mutations in CIC-DUX4 sarcomas.
3.2 Copy number profiling from NGS data demonstrates recurrent copy number alterations (CNAs) in CIC-DUX4 sarcomas
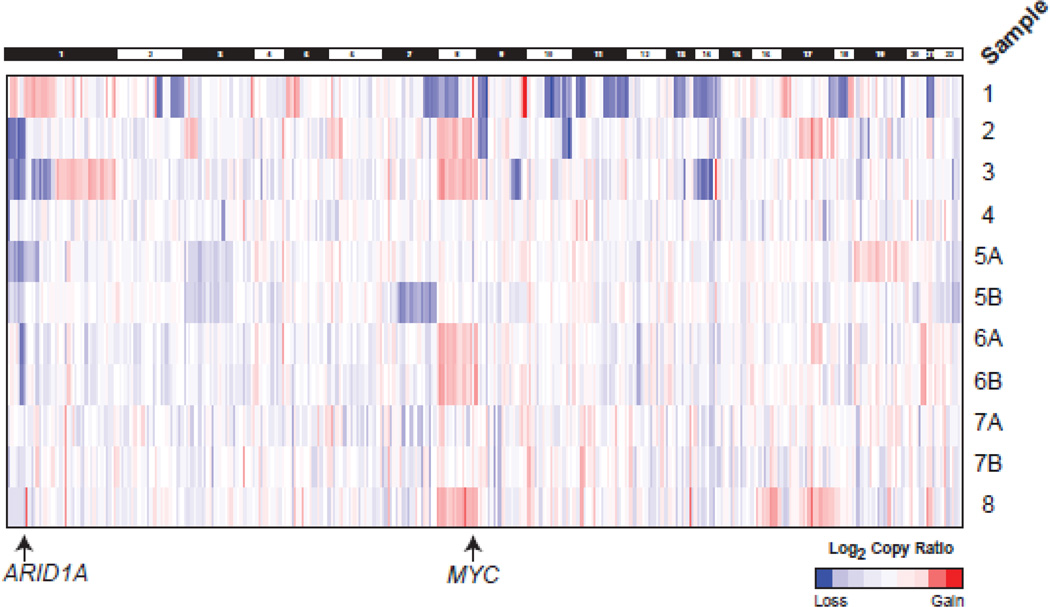
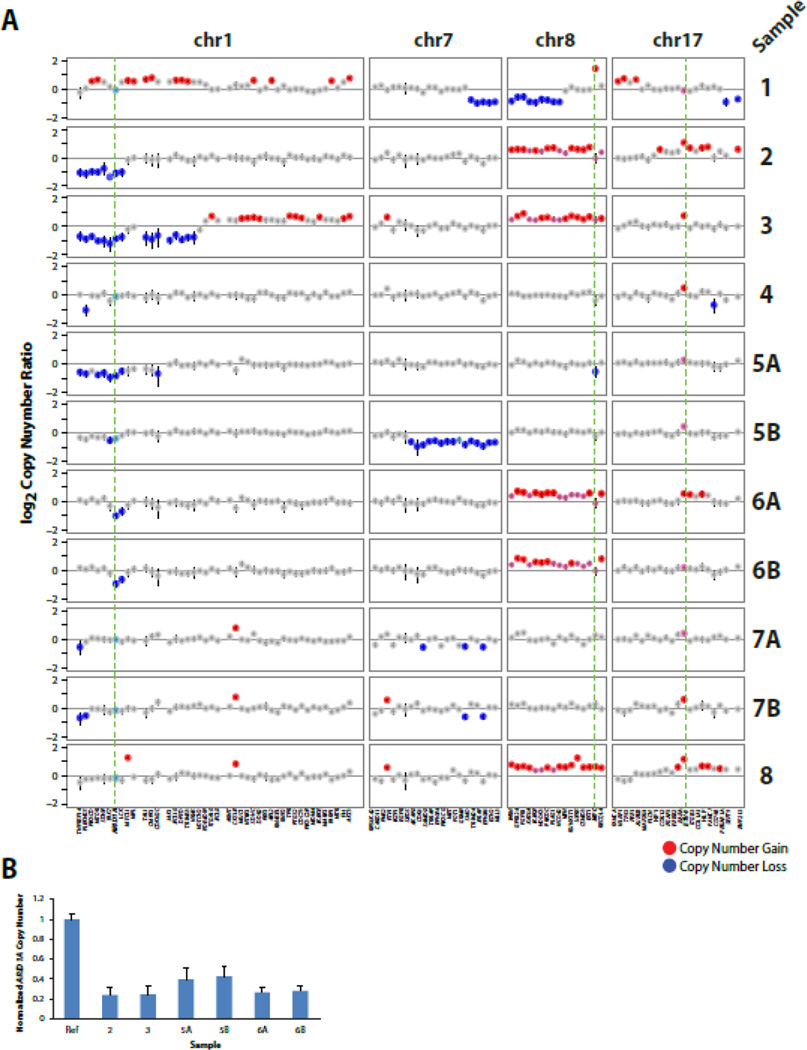
In addition to mutations, we also assessed our NGS data to identify somatic copy number alterations (CNAs) using a validated approach from NGS amplicon read counts. As shown in Figures 2&3A, in contrast to the limited mutational landscape of CIC-DUX4 tumors, we identified CNAs in all samples, including areas of recurrent gain/loss. Consistent with previous karyotyping/FISH studies that demonstrated chromosome (chr) 8 gain and focal MYC (8q24) amplification in CIC-DUX4 sarcomas [7, 18], we observed broad, low-level chr 8 gain in 4 of nine cases (Figs. 2 & 3A); focal high-level MYC amplification was only seen in Sample 1, suggesting that this alteration may be subclonal. Importantly, of the three cases previously karyotyped (3,4 and 6), chr 8 gain by karyotyping (Table 1) and NGS was concordant in each case (Samples 6A and 6B showed concordant chr 8 gain by NGS).
Fig. 2. Somatic copy-number profiles of CIC-DUX4 sarcomas generated by targeted next generation sequencing (NGS).
Somatic, autosomal copy number profiles are presented for the 11 CIC-DUX4 sarcoma samples from 8 cases assessed by NGS. Gene-level copy number estimates are shown for all target genes with >= 3 amplicons across samples. Colors correspond to log2 copy number ratios (tumor to composite normal) as indicated in legend. Samples 5A&B represent a pre-treatment tumor and a post-radiation metastasis/local recurrence from the same patient. Samples 6A&B represent a primary tumor and near concurrent untreated brain metastasis. Samples 7A&7B are a primary tumor and rapid local recurrence.
Fig. 3. Recurrent copy number alterations (CNAs) identified by next generation sequencing (NGS) of CIC-DUX4 sarcomas.
A. Log2 copy number profiles (tumor to composite normal) for selected chromosomes from the genome wide plots in Figure 2 are shown. Gains and losses are shown in red and blue, respectively, with lighter shades indicating lower level alterations. Genes are indicated below the copy number profiles with ARID1A (chr 1p) MYC (chr 8q) and ETV4 (chr 17q) bolded and indicated by dashed green lines. B. Confirmation of ARID1A copy number loss by quantitative PCR (qPCR). Genomic DNA was assessed by qPCR in triplicate for ARID1A copy number normalized to the average of three reference genes without CNA by NGS (FBXW7, DNMT3A, and IGF1R) from samples with available DNA. Normalized ARID1A copy number ratio was calibrated to an unrelated benign FFPE genomic DNA sample as the calibrator control (Con.). Mean + S.E. are shown.
Both broad and focal low-level gains centered on ETV4 (chr 17) were observed in 6 of 11 samples; however this alteration may also be subclonal given that it was only observed in Samples 5B & 6A and not the matched sample from these patients (Fig 3A). As multiple groups have shown that CIC-DUX4 sarcomas (and the CIC-DUX4 fusion protein more directly) over-express ETS transcription factors [6, 8, 18], including ETV4, our results support low level/subclonal ETV4 gains as contributing to over-expression as well.
Of note, we also identified recurrent, low level deletions centered on the tumor suppressor ARID1A on chr 1p36 (Fig 2 and 3A), which were present in 4 of 8 profiled cases (including concordant loss in Samples 6A & B; loss in case 5A but not 5B). Importantly, case 3, which harbored this chr 1p deletion by NGS, was previously reported to harbor a deletion of chr 1p21-p36 by karyotyping (Table 1), consistent with our NGS results. In addition, qPCR analysis of genomic DNA on Samples 2–6 supported recurrent ARID1A loss across our cohort (Fig 3B). ARID1A protein expression by immunohistochemistry (IHC) was assessed on a tissue microarray (TMA) containing CIC-DUX4 sarcomas and Ewing sarcomas; no consistent differences in ARID1A expression was observed in CIC-DUX4 sarcomas with or without ARID1A loss or deleterious mutation by NGS (data not shown), supporting 1p single copy loss (or subclonal two copy loss) in CIC-DUX4 sarcomas, as well as potential regulation of ARID1A protein expression by mechanisms other than genomic loss and/or mutation.
Although the pre- and post-radiation samples from case 5 (Samples 5A and 5B, respectively) showed broad low level chromosome 6p gain (Fig 2), consistent with clonality, the pre-radiation sample (5A) uniquely showed a broad chr 19 gain and chr 1p loss, while the post-radiation sample (5B) showed a broad chr 7 loss (Fig 2 & 3A), suggesting that this alteration may be associated with post-treatment recurrence and supporting heterogeneity between pre and post-treatment samples. In contrast, other than the low level gain of chr 17 (involving ETV4) exclusively in the primary tumor (Sample 6A), the paired post-chemotherapy primary tumor and nearly concurrent brain metastasis from case 6 showed nearly identical copy number profiles (including gain of chr 8 and loss of chr 18), supporting limited intertumoral heterogeneity (Fig 2 & 3A). The paired primary and untreated local recurrence in case 7 (Samples 7A and 7B, respectively) showed similar copy number profiles, with both samples showing CKS1B gain on chromosome 1 (also observed in Sample 8), PAX5 and SYK loss on chromosome 9, IRS2 gain on chr 13 (also observed in Sample 8), loss of the X chromosome and evidence of chromothripsis on chromosome 7. The recurrence sample (7B) showed unique SOX2 gain on chromosome 3, in addition to the ARID1A non-sense mutation described above.
4. Discussion
Although morphologically similar to Ewing sarcomas, CIC-DUX4 sarcomas are driven by a distinct gene fusion. Given the rarity of CIC-DUX4 sarcomas, they are much more poorly characterized at the molecular level than conventional Ewing sarcomas. Hence, here we performed targeted NGS on eleven FFPE CIC-DUX4 samples (from eight patients) to identify somatic mutations and CNAs in CIC-DUX4 sarcomas as well as assess the molecular relationship to Ewing sarcomas.
Overall, we did not identify any genes with recurrent driving point mutations. However, one sample (6A) harbored a prioritized potentially driving activating oncogenic mutation (CTNNB1 E54K, likely subclonal), while Sample 1 harbored TP53 C238Y and D208fs mutations, and Sample 7B (local recurrence) harbored an ARID1A R693X nonsense mutation. Our cohort included six previously treated samples (five post-chemotherapy and one post-radiation), suggesting that point mutations/indels are not major drivers of treatment resistance in CIC-DUX4 sarcomas. Additionally, our data is consistent with Ewing sarcomas, which shows a very low rate of somatic mutations; of note, our panel did not target STAG2, which has been shown to be recurrently mutated in Ewing sarcomas [23–25], however case 7 showed a single copy loss of the entire X chromosome (location of STAG2, not shown in heatmap), which would result in complete STAG2 loss in this male patient.
Although we identified limited focal, high level gains or losses in CIC-DUX4 sarcomas by copy number profiling analysis of NGS data (nearly exclusively in cases 1& 8), we identified areas of broad, low level CNAs, including recurrent gains of chr 8, as we have previously reported in CIC-DUX4 sarcomas [18] and has been observed in Ewing sarcomas [25]. Of note, although MYC has been nominated as the target of chr 8 gain (and was focally amplified in Sample 1), Sample 8 harbored a gain of chromosome 8 with a focal high level gain exclusively of UBR5 (chr 8q22), centromeric to MYC. We also observed low level recurrent gains of ETV4, which were focal in some cases, which may contribute to ETV4 over-expression as has been observed in CIC-DUX4 sarcomas[6, 8, 18]. Likewise, we identified recurrent low level chr 1 p deletions that included the frequently mutated tumor suppressor AR1D1A [26] (at 1p36), in 4 of 8 cases, in addition to the ARID1A R693X nonsense mutation exclusively in the local recurrence sample of case 7 (Sample 7B). Of interest, chr 1p, and specifically 1p36, has been identified as recurrently deleted in 6–22% of FISH/karyotyping based studies of Ewing sarcomas [27–30]. Results from our small series suggest that this alteration may be more frequent in CIC-DUX4 sarcomas, and the identification of both ARID1A copy loss in multiple samples and a deleterious mutation in Sample 7B is intriguing. Of note, however, ARID1A protein expression was not correlated with copy number/mutation status, and the ARID1A nonsense mutation is estimated as heterozygous (variant allele frequency 0.38 and estimated tumor content of 80%). Likewise, 4 of the 5 samples (from 3 of 4 cases) with chr 1p loss (including ARID1A) were obtained after neo-adjuvant chemotherapy, although the local recurrence sample harboring the ARID1A nonsense mutation (Sample 7B) was treatment naïve. Hence, although ARID1A alterations have shown to be subclonal in other tumors through NGS[26], whether ARID1A has a role in CIC-DUX4 sarcoma development or progression is unclear and requires validation in larger cohorts and through functional studies.
Notably, other recurrent copy number alterations seen in Ewing sarcomas (1q gain, 16q loss, 12q gain, and TP53 (chr 17) deletion [29], were not recurrent in our limited cohort. However, case 3 showed broad 1q gain, while case 1—which showed atypical morphology—harbored deleterious TP53 mutations and CDKN2A two copy loss. Larger CIC-DUX4 cohorts will need to be assessed to identify specific CNAs that may occur at different frequencies in Ewing sarcomas and CIC-DUX4 sarcomas.
Our cohort included three sets of paired specimens, one representing near concurrent, post-chemotherapy primary tumor resection and a brain metastasis (Samples 6A&B), one representing a pre-treatment primary tumor and a post-radiation therapy pelvic recurrence (Samples 5A&B), and one representing a treatment-naïve primary tumor resection and a rapid local recurrence (Samples 7A&B). Samples 6A and 6B showed clonal copy number alterations (including chr 8 gain and 18 loss), however the primary tumor exclusively harbored a CTNNB1 E54K mutation (at subclonal variant frequency) and low level chr 17q gain (involving ETV4), supporting potentially relevant intertumoral heterogeneity. Likewise, although Samples 5A and B both showed chr 6 gain, both samples had unique CNAs, including broad loss of chr 7q in the post-radiation recurrence, supporting intertumoral heterogeneity and potential chr 7q loss as an adaptive response in the radio-resistant clone. Lastly, Samples 7A and B had similar copy number profiles, with the recurrence exclusively showing SOX2 gain and the ARID1A non-sense mutation.
Limitations of our study include the small cohort size, requiring validation of our findings in additional cohorts. Such studies will require intra-institutional collaborations given the rarity of CIC-DUX4 sarcomas. Likewise, although our NGS panel was designed to assess over 400 known cancer genes and is capable of detecting both mutations and CNAs, more comprehensive platforms will be needed to assess the existence of chromosomal rearrangements or recurrent mutations/focal CNAs in genes not targeted herein (e.g. STAG2). Lastly, future studies will be needed to evaluate any potential clinical implications as well as any biological links between these alterations and CIC-DUX4 sarcoma development/progression, given the general lack of relevant cell line and animal models.
In summary, using NGS we report the somatic mutation and CNA landscape of eleven routine FFPE CIC-DUX4 specimens from 8 patients, including three paired samples. Like Ewing sarcomas, we identify a very low mutational rate amongst a large panel of cancer related genes in CIC-DUX4 sarcomas. Additionally, we identified known (e.g. chr 8 gain) and novel alterations in CIC-DUX4 sarcomas, including copy number loss and a deleterious mutation in ARID1A (chr 1p36). Additional studies are needed to confirm these findings.
Supplementary Material
Acknowledgments
S.A.T. has received travel support from Thermo Fisher Scientific, who produces the CCP, and had a separate sponsored research agreement with Life Technologies/Thermo Fisher Scientific that was not used to support the research performed herein. S.A.T. has consulted for Ventana Medical Systems, Abbvie, Janssen, Astellas/Medivation, and has a sponsored research agreement with Astellas. S.A.T. is a co-founder, equity holder, and consultant for Strata Oncology.
The authors thank Angela Fullen, Mandy Davis and Javed Siddiqui for histology. This work was supported by the National Institutes of Health (SARC Sarcoma S.P.O.R.E. grant U54 CA168512 to the University of Michigan). S.A.T. is supported by the A. Alfred Taubman Medical Research Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures/Conflicts of Interest
The remaining authors have no disclosures.
References
- 1.Davicioni E, Wai DH, Anderson MJ. Diagnostic and prognostic sarcoma signatures. Mol Diagn Ther. 2008;12:359–374. doi: 10.1007/BF03256302. [DOI] [PubMed] [Google Scholar]
- 2.Somers GR, Gupta AA, Doria AS, Ho M, Pereira C, Shago M, et al. Pediatric undifferentiated sarcoma of the soft tissues: a clinicopathologic study. Pediatr Dev Pathol. 2006;9:132–142. doi: 10.2350/08-05-0098.1. [DOI] [PubMed] [Google Scholar]
- 3.Qualman SJ, Coffin CM, Newton WA, Hojo H, Triche TJ, Parham DM, et al. Intergroup Rhabdomyosarcoma Study: update for pathologists. Pediatr Dev Pathol. 1998;1:550–561. doi: 10.1007/s100249900076. [DOI] [PubMed] [Google Scholar]
- 4.Yoshimoto M, Graham C, Chilton-MacNeill S, Lee E, Shago M, Squire J, et al. Detailed cytogenetic and array analysis of pediatric primitive sarcomas reveals a recurrent CIC-DUX4 fusion gene event. Cancer Genet Cytogenet. 2009;195:1–11. doi: 10.1016/j.cancergencyto.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 5.Kreiger PA, Judkins AR, Russo PA, Biegel JA, Lestini BJ, Assanasen C, et al. Loss of INI1 expression defines a unique subset of pediatric undifferentiated soft tissue sarcomas. Mod Pathol. 2009;22:142–150. doi: 10.1038/modpathol.2008.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Specht K, Sung YS, Zhang L, Richter GH, Fletcher CD, Antonescu CR. Distinct transcriptional signature and immunoprofile of CIC-DUX4 fusion-positive round cell tumors compared to EWSR1-rearranged Ewing sarcomas: further evidence toward distinct pathologic entities. Genes Chromosomes Cancer. 2014;53:622–633. doi: 10.1002/gcc.22172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi EY, Thomas DG, McHugh JB, Patel RM, Roulston D, Schuetze SM, et al. Undifferentiated small round cell sarcoma with t(4;19)(q35;q13.1) CIC-DUX4 fusion: a novel highly aggressive soft tissue tumor with distinctive histopathology. Am J Surg Pathol. 2013;37:1379–1386. doi: 10.1097/PAS.0b013e318297a57d. [DOI] [PubMed] [Google Scholar]
- 8.Kawamura-Saito M, Yamazaki Y, Kaneko K, Kawaguchi N, Kanda H, Mukai H, et al. Fusion between CIC and DUX4 up-regulates PEA3 family genes in Ewing-like sarcomas with t(4;19)(q35;q13) translocation. Hum Mol Genet. 2006;15:2125–2137. doi: 10.1093/hmg/ddl136. [DOI] [PubMed] [Google Scholar]
- 9.Rakheja D, Goldman S, Wilson KS, Lenarsky C, Weinthal J, Schultz RA. Translocation (4;19)(q35;q13.1)-associated primitive round cell sarcoma: report of a case and review of the literature. Pediatr Dev Pathol. 2008;11:239–244. doi: 10.2350/07-06-0296.1. [DOI] [PubMed] [Google Scholar]
- 10.Italiano A, Sung YS, Zhang L, Singer S, Maki RG, Coindre JM, et al. High prevalence of CIC fusion with double-homeobox (DUX4) transcription factors in EWSR1-negative undifferentiated small blue round cell sarcomas. Genes Chromosomes Cancer. 2012;51:207–218. doi: 10.1002/gcc.20945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Antonescu C. Round cell sarcomas beyond Ewing: emerging entities. Histopathology. 2014;64:26–37. doi: 10.1111/his.12281. [DOI] [PubMed] [Google Scholar]
- 12.Himeda CL, Debarnot C, Homma S, Beermann ML, Miller JB, Jones PL, et al. Myogenic enhancers regulate expression of the facioscapulohumeral muscular dystrophy-associated DUX4 gene. Mol Cell Biol. 2014;34:1942–1955. doi: 10.1128/MCB.00149-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Machado I, Cruz J, Lavernia J, Rubio L, Campos J, Barrios M, et al. Superficial EWSR1-negative undifferentiated small round cell sarcoma with CIC/DUX4 gene fusion: a new variant of Ewing-like tumors with locoregional lymph node metastasis. Virchows Arch. 2013;463:837–842. doi: 10.1007/s00428-013-1499-9. [DOI] [PubMed] [Google Scholar]
- 14.Bielle F, Zanello M, Guillemot D, Gil-Delgado M, Bertrand A, Boch AL, et al. Unusual primary cerebral localization of a CIC-DUX4 translocation tumor of the Ewing sarcoma family. Acta Neuropathol. 2014;128:309–311. doi: 10.1007/s00401-014-1312-0. [DOI] [PubMed] [Google Scholar]
- 15.Graham C, Chilton-MacNeill S, Zielenska M, Somers GR. The CIC-DUX4 fusion transcript is present in a subgroup of pediatric primitive round cell sarcomas. Hum Pathol. 2012;43:180–189. doi: 10.1016/j.humpath.2011.04.023. [DOI] [PubMed] [Google Scholar]
- 16.Richkind KE, Romansky SG, Finklestein JZ. t(4;19)(q35;q13.1): a recurrent change in primitive mesenchymal tumors? Cancer Genet Cytogenet. 1996;87:71–74. doi: 10.1016/0165-4608(95)00240-5. [DOI] [PubMed] [Google Scholar]
- 17.Panagopoulos I, Gorunova L, Bjerkehagen B, Heim S. The "grep" command but not FusionMap, FusionFinder or ChimeraScan captures the CIC-DUX4 fusion gene from whole transcriptome sequencing data on a small round cell tumor with t(4;19)(q35;q13) PLoS One. 2014;9:e99439. doi: 10.1371/journal.pone.0099439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith SC, Buehler D, Choi EY, McHugh JB, Rubin BP, Billings SD, et al. CIC-DUX sarcomas demonstrate frequent MYC amplification and ETS-family transcription factor expression. Mod Pathol. 2015;28:57–68. doi: 10.1038/modpathol.2014.83. [DOI] [PubMed] [Google Scholar]
- 19.Kadakia KC, Tomlins SA, Sanghvi SK, Cani AK, Omata K, Hovelson DH, et al. Comprehensive serial molecular profiling of an "N of 1" exceptional non-responder with metastatic prostate cancer progressing to small cell carcinoma on treatment. J Hematol Oncol. 2015;8:109. doi: 10.1186/s13045-015-0204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warrick JI, Hovelson DH, Amin A, Liu CJ, Cani AK, McDaniel AS, et al. Tumor evolution and progression in multifocal and paired non-invasive/invasive urothelial carcinoma. Virchows Arch. 2015;466:297–311. doi: 10.1007/s00428-014-1699-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang X, Wang K. wANNOVAR: annotating genetic variants for personal genomes via the web. J Med Genet. 2012;49:433–436. doi: 10.1136/jmedgenet-2012-100918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grasso C, Butler T, Rhodes K, Quist M, Neff TL, Moore S, et al. Assessing copy number alterations in targeted, amplicon-based next-generation sequencing data. J Mol Diagn. 2015;17:53–63. doi: 10.1016/j.jmoldx.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brohl AS, Solomon DA, Chang W, Wang J, Song Y, Sindiri S, et al. The genomic landscape of the Ewing Sarcoma family of tumors reveals recurrent STAG2 mutation. PLoS Genet. 2014;10:e1004475. doi: 10.1371/journal.pgen.1004475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crompton BD, Stewart C, Taylor-Weiner A, Alexe G, Kurek KC, Calicchio ML, et al. The genomic landscape of pediatric Ewing sarcoma. Cancer Discovery. 2014;4:1326–1341. doi: 10.1158/2159-8290.CD-13-1037. [DOI] [PubMed] [Google Scholar]
- 25.Tirode F, Surdez D, Ma X, Parker M, Le Deley MC, Bahrami A, et al. Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer Discov. 2014;4:1342–1353. doi: 10.1158/2159-8290.CD-14-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones S, Li M, Parsons DW, Zhang X, Wesseling J, Kristel P, et al. Somatic mutations in the chromatin remodeling gene ARID1A occur in several tumor types. Hum Mutat. 2012;33:100–103. doi: 10.1002/humu.21633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roberts P, Burchill SA, Brownhill S, Cullinane CJ, Johnston C, Griffiths MJ, et al. Ploidy and karyotype complexity are powerful prognostic indicators in the Ewing's sarcoma family of tumors: a study by the United Kingdom Cancer Cytogenetics and the Children's Cancer and Leukaemia Group. Genes Chromosomes Cancer. 2008;47:207–220. doi: 10.1002/gcc.20523. [DOI] [PubMed] [Google Scholar]
- 28.Neale G, Su X, Morton CL, Phelps D, Gorlick R, Lock RB, et al. Molecular characterization of the pediatric preclinical testing panel. Clin Cancer Res. 2008;14:4572–4583. doi: 10.1158/1078-0432.CCR-07-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jahromi MS, Jones KB, Schiffman JD. Copy Number Alterations and Methylation in Ewing's Sarcoma. Sarcoma. 2011;2011:362173. doi: 10.1155/2011/362173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hattinger CM, Rumpler S, Strehl S, Ambros IM, Zoubek A, Potschger U, et al. Prognostic impact of deletions at 1p36 and numerical aberrations in Ewing tumors. Genes Chromosomes Cancer. 1999;24:243–254. doi: 10.1002/(sici)1098-2264(199903)24:3<243::aid-gcc10>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.