Abstract
Among mitochondrial lipids, cardiolipin occupies a unique place. It is the only phospholipid that is specific to mitochondria and although it is merely a minor component, accounting for 10–20% of the total phospholipid content, cardiolipin plays an important role in the molecular organization, and thus the function of the cristae. This review covers the formation of cardiolipin, a phospholipid dimer containing two phosphatidyl residues, and its assembly into mitochondrial membranes. While a large body of literature exists on this topic, the review focuses on papers that appeared in the past three years.
Introduction
Cardiolipin (CL, diphosphatidylglycerol) is a dimeric phospholipid that is specific to mitochondria and can thus be used to identify and quantify mitochondria in subcellular fractions. CL interacts with various proteins, which plays an important role in the lateral organization of mitochondrial membranes. The presence of CL in mitochondria and its involvement in mitochondrial function, are conserved in all eukaryotes, among them yeast, animals, protozoans, and plants. Even in plants, where energy metabolism does not primarily rely on mitochondria but on chloroplasts, deletion of cardiolipin synthase has detrimental consequences, a phenomenon probably related to increased oxidative stress [1].
The life of a CL molecule can be divided into 3 phases. The first is its biosynthesis in the inner mitochondrial membrane; the second is the remodeling of its fatty acids and its assembly into the inner membrane; and the third is its degradation and oxidative modification and its potential translocation out of the inner membrane. While the first step is relatively well established, the other two are still under scrutiny as their mechanisms are not fully understood and their functional implications are not very clear. A lot of effort has been spent on elucidating the mechanism and the function of the acyl remodeling of CL because deficient CL remodeling causes Barth syndrome. Recently, CL remodeling has also been discovered in bacteria [2]. Since several review articles have been published on this subject, the present paper has the character of an update, focusing mostly on the literature between 2013 and 2016.
Biosynthesis of CL
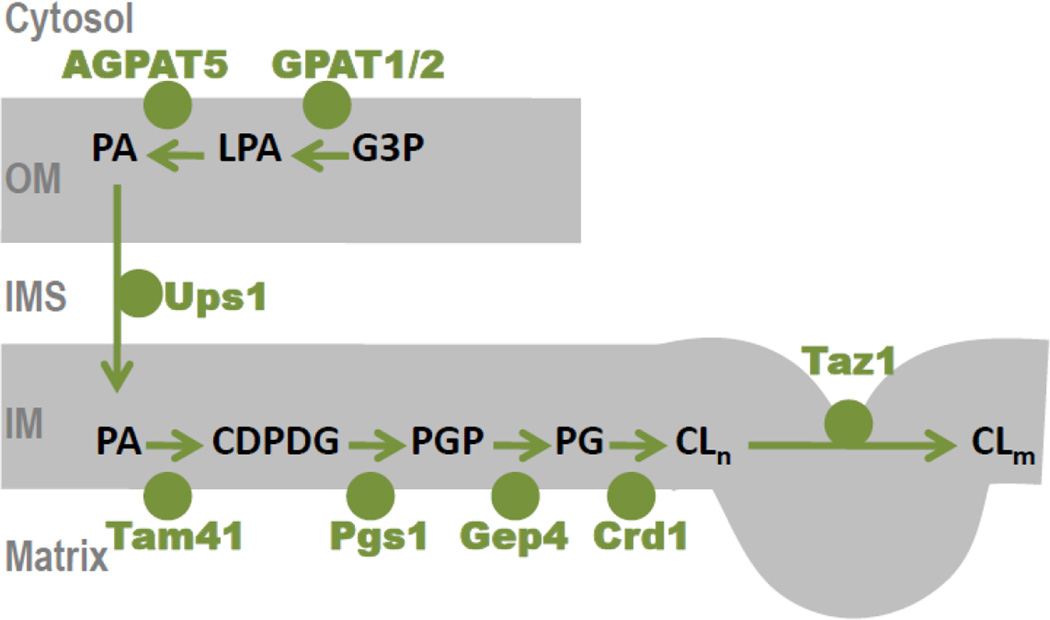
The biosynthetic pathway of CL has been established in the 1970s [3]; it takes place on the matrix face of the inner mitochondrial membrane [4]. The first step of this pathway is catalyzed by Tam41 [5], which was initially described as an activity required for the assembly and the maintenance of protein translocases [6, 7]. Eventually, Tam41 was found to be essential for CL biosynthesis [8] and to have CDP-diacylglycerol synthase activity [5]. The second step is catalyzed by Pgs1 [9], an enzyme that forms phosphatidylglycerophosphate by transferring a phosphatidyl group from CDP-diacylglycerol to the sn-1 hydroxyl group of glycerol-3-phosphate. The conversion of an anhydride bond into an ester bond produces a large drop in free energy, which makes this reaction a suitable target for biological rate control. For instance, Pgs1 can be phosphorylated in yeast, which reduces its activity [10]. The third step on the CL pathway is catalyzed by Gep4 in yeast [11] or PTPMT1 in mammalian cells [12]. These enzymes remove the terminal phosphate group from phosphatidylglycerophosphate to form phosphatidylglycerol. The fourth and final step of CL biosynthesis is catalyzed by CL synthase (Crd1), which uses phosphatidylglycerol and CDP-diacylglycerol, to form CL. CL synthase was discovered in yeast [9, 13, 14] and later identified in humans [15–17] and plants [18, 19]. The CL pathway is illustrated in Figure 1.
Figure 1. Cardiolipin biosynthesis in mitochondria.
Glycero-3-phosphate (G3P) is acylated to lysophosphatidic acid (LPA) and then to phosphatidic acid (PA). PA formation occurs on the outer face of the outer mitochondrial membrane (OM) and in the endoplasmic reticulum. PA is transferred from the outer to the inner mitochondrial membrane (IM) via the intermembrane space (IMS) and is converted to CL on the matrix face of the IM via the intermedates CDPdiacylglycerol (CDP-DG), phosphatidylglycerophosphate (PGP), and phosphatidylglycerol (PG). The final step of the pathway is the remodeling of nascent cardiolipin (CLn) to mature cardiolipin (CLm). Remodeling occurs in the outer leaflet of the inner membrane and requires disturbance of the bilayer packing order. Mammalian genes are shown for the enzymes of PA synthesis and yeast genes are shown for all other enzymes.
The CL pathway is different in prokaryotes, where phosphatidyl transfer occurs from one phosphatidylglycerol molecule to another (for a review, see ref [20]) or, as recently discovered, from phosphatidylethanolamine to phosphatidylglycerol [21]. A bacterial-type CL synthase is also present in some unicellular eukaryotes [22]. Not surprisingly, the CL pathway is regulated by PGC-1, the transcription factor that controls the biogenesis of mitochondria [23]. Another regulatory activity may be associated with ATP citrate lyase, a cytosolic enzyme that is placed at the junction between carbohydrate and lipid metabolism. It was proposed that the activation of this enzyme stimulates the formation of CL [24].
Remodeling of CL fatty acids
By definition, remodeling is a structural modification that follows the biosynthesis of CL. In the wake of this process, CL acquires a new set of fatty acids, which is organism and tissue specific. In some tissues, such as mammalian brain, the CL pattern is very diverse [25, 26] but in others, especially in tissues with high energy turnover, CL contains surprisingly few molecular species, and those species are typically dominated by only one or two unsaturated acyl groups [27].
Remodeling is catalyzed by tafazzin, an enzyme that transfers fatty acids from phospholipids to lysophospholipids [28] (Figure 1). However, tafazzin does so non-specifically, which does not provide an explanation for the fatty acid pattern observed in CL; tafazzin is simply a vehicle that allows fatty acids to exchange between phospholipid molecules. In order for tafazzin to produce a specific molecular pattern rich in tetralinoleoyl-CL (the dominant CL molecule of human heart), the free energy of tetralinoleoyl-CL must be lower than the free energy of other molecular species. Such energy differences are dependent on the packing properties of the entire lipid assembly. For instance, we have shown in vitro that negative curvature creates conditions that favor the formation of tetralinoleoyl-CL because the shape of this lipid naturally fits the geometry of negatively curved monolayers [29].
Recently, Abe et al proposed that purified yeast tafazzin catalyzes acyl-specific transacylations in bilayers [30]. However, the claim that the reaction took place in rigid bilayers is contradicted by the fact that the medium contained up to 0.08% (1.28 mM) Triton X-100, which amounts to a 2-fold excess of the detergent over the phospholipids (0.59 mM). Furthermore, the data analysis (Guggenheim plot to determine pseudo-first order rate constants) did not carefully distinguish the properties of the enzyme (rate of catalysis) from the properties of the chemical equilibrium (concentration of substrate and products after completion of the reaction). This is more difficult for tafazzin than for many other enzymes because of the small difference in free energy between substrates and products. As the reaction approaches its equilibrium, the rates of forward and reverse reactions become similar and eventually identical, which causes an underestimation of the actual forward conversion between two time points. Essentially all acyl specificities observed by Abe et al [30] can be explained by differences in the equilibrium composition, which basically confirms that the transacylation specificities are driven by the physical properties of the lipids rather than the enzyme. Finally, transgenic experiments have not supported the idea of intrinsic acyl specificity of tafazzin, because human tafazzin replicates the Drosophila-type CL pattern in flies and the yeast-type CL pattern in yeast [31, 32], although these patterns are different from human CL and from each other.
Thus, when considering the tafazzin reaction in vivo, the question becomes what in the environment of the enzyme gives acyl specificity to the otherwise promiscuous acyl exchange? An unequivocal answer to this question will require further studies, but at this point it seems likely that the process that is driving CL remodeling is the assembly of the large protein complexes and supercomplexes of the inner mitochondrial membrane (see below).
A better understanding of CL remodeling will ultimately hinge on knowing the precise localization and the physical interactions of tafazzin. It has been shown that tafazzin is a nonintegral membrane protein that resides on the outer face of the inner and perhaps also on the inner face of the outer mitochondrial membrane [33, 34]. There it is associated with several protein complexes that range in size from roughly 100 to over 600 kDa [33–36]. Among its interaction partners are the ATP synthase and the ADP-ATP carrier but they hardly account for the entire spectrum [35]. In H9c2 cell, we found the half-life of tafazzin to be shorter than the half-life of other mitochondrial proteins implying that tafazzin is either a temporary resident of protein assemblies or the assemblies themselves are short-lived [36]. However, the half-life of tafazzin varies between cell types and is very sensitive to certain mutations [34]. Given these ambiguities, a simple narrative of the tafazzin function has remained elusive up to now.
Assembly of CL in protein complexes
CL strongly interacts with many different proteins. They constitute a long list (reviewed in ref. [37]) to which recently the uncoupling protein [38], multi-spanning proteins of the outer mitochondrial membrane [39], and the dynamin-related protein 1 [40, 41] were added. Structural features of CL-binding proteins have been defined [42], but high affinity for CL is present in too many proteins to make a convincing argument for true specificity. Rather, the physical basis of this tight association seems to be rooted in the unique shape of CL and its charge state, which together produce a tendency to cluster and a tendency to form strong non-covalent bonds. Crucial herein is the large acyl moiety, which enforces hydrophobic interactions, and the small, motion-restricted head group, which leaves unshielded the two negatively charged phosphates and therefore increases their radius of interaction [43]. Studies in isolated mitochondria have shown that most if not all CL is associated with proteins [44, 45]. In other words, mitochondrial membranes contain virtually no free CL in the bulk lipid phase.
Preliminary evidence has emerged for the involvement of protein complex assembly in the remodeling of CL. This applies specifically to three groups of proteins, namely (i) prohobitins, (ii) the MICOS complex, and (iii) the complexes of oxidative phosphorylation (OXPHOS).
(i) Prohibitins form large complexes in the inner mitochondrial membrane. They interact with other proteins and play a role in phospholipid homeostasis although their function has not been clearly defined [46]. Knockdown of prohibitin 2 in HEK 293 cells led to alterations in the molecular composition of CL [47]. Proteins that interact with prohibitins, such as DNAJC19 and MDM33, also affect CL [47, 48].
(ii) MICOS stands for Mitochondrial Contact site and Cristae Organizing System. It comprises a large heterogeneous complex that localizes to cristae junctions and connects the inner with the outer mitochondrial membrane [49]. The MICOS complex plays a central role in both the formation of cristae and the formation of intermembrane contact sites. In yeast, MICOS consists of two sub-complexes, Mic60/Mic19 and Mic27/Mic10/Mic12, the latter of which requires the presence of CL [50]. While this fact itself does not imply any role in CL remodeling, the stability of the MICOS complex seems to control the level of tafazzin and therefore may indirectly affect the CL species composition [51, 52].
(iii) OXPHOS complexes I to V and proteins of the solute carrier family have all been shown to bind tightly to CL. Together they account for the majority of proteins in the mitochondrial inner membrane. CL not only binds to the surface of these proteins, it also promotes their assembly to supercomplexes [53–55] and is found in high concentration within these supercomplexes [56, 57]. Thus, the available evidence strongly supports that CL asserts a stabilizing effect on various supercomplex assemblies. However, the reverse seems to be true as well. Specifically, we have shown that the sequestration of CL within protein supercomplexes is responsible for the extremely slow turnover of CL. In contrast, when supercomplexes are forced to dissociate, CL is rapidly degraded [45]. Resveratrol, a drug that promotes supercomplex association [58], prolongs the half-life of CL but not the half-life of other phospholipids [45]. It is known that resveratrol activates sirtuins, which in turn produces protein deacetylation and desuccinylation. New evidence suggests that these modifications may directly increase the binding affinity for CL [59]. Together the data suggest mutual stabilization between supercomplexes and CL.
However, it remains to be determined whether supercomplex assembly actually plays a role in the acyl specificity of CL remodeling. In favor of this idea, it has been shown that deficient CL remodeling impairs supercomplex assembly [45, 60–63] and in one example, defective supercomplex assembly was associated with subtle changes in the CL composition [64]. In the latter experiment, a new CL-binding protein was identified, C11orf83, that is part of complex III and that stabilizes respiratory supercomplexes. Another protein that stabilizes supercomplexes, SLP-2 also seems to interact with CL [65].
Turnover of CL
The half- life of CL is several-fold longer than the half- life of other phospholipids. This was first observed in rats labeled with 32P phosphate [66] or 2H2O [67] and later confirmed in cell cultures and flies [45, 68]. For instance, there is no measurable CL turnover in Drosophila over a period of 9 days, which is remarkable given that these animals only live 60 days and show plenty of turnover in all other phospholipids during the 9 day period [45]. The high stability of CL applies to both the glycerol groups and the acyl groups. The sequestration of CL in protein complexes is critical for the long half- life of CL because conditions that promote supercomplex association increase CL stability whereas conditions that dissociate supercomplexes have the opposite effect [45]. Thus, there is mutual stabilization between CL and proteins. While CL promotes protein association, the large supramolecular assemblies that form as a result, protect CL from degradation. The idea that the inner membrane contains long-lived structures is also supported by mitochondrial fusion experiments, in which mixing of inner membrane proteins took much longer than mixing of matrix or outer membrane proteins [69].
Despite its high stability and firm integration in protein complexes, CL is not completely immune to chemical modification and re-localization, events that are probably involved in mitophagy and apoptosis [70]. CL can be transferred from the inner to the outer mitochondrial membrane by NDPK-D, a protein of the intermembrane space [71]. This translocation is part of a program that makes CL accessible to cytosolic factors at the outer surface of mitochondria; these factors then trigger mitophagy [72]. If CL is not only translocated but also oxidized, it may trigger another program, which leads to apoptosis [26]. The mitochondrial phospholipase iPLA2β shows preference for oxidized CL molecules, which can protect cells against apoptotic death [73]. Monolyso-cardiolipin (MLCL), the product of this reaction can be re-acylated to CL by the enzyme ALCAT, which is located in the mitochondria-associated endoplasmic reticulum. However, the actual function of ALCAT has remained obscure because deletion of this enzyme does not seem to have any obvious physiologic effects [74]. Furthermore, ALCAT is not specific for MLCL but carries broad lysophospholipid:acyl-CoA activity [75].
Abnormal CL metabolism in Barth syndrome
Mutations in the human tafazzin gene cause Barth syndrome (BTHS), a disease that presents with cardiomyopathy, skeletal muscle weakness, fatigue, neutropenia, and abnormal growth. Patients have low CL levels and aberrant CL species composition and they accumulate MLCL, a lipid that is not detectable in normal tissues [37]. The MLCL/CL ratio of blood, measured in platelets, leukocytes, or blood spots, has become an important diagnostic marker for BTHS [76– 81].
Because tetralinoleoyl-CL, the predominant CL species in normal heart, is virtually absent from BTHS patients’ cells [76], many studies have suggested that the pathology in BTHS results from this deficiency. The inability to completely block CL deacylation in mammalian cells complicates experiments to distinguish between the effects of MLCL/CL vs. abnormal CL species composition. However, this question was addressed in yeast cells using CL remodeling mutants [82, 83]. Mutant cells lacking the single CL-specific phospholipase (Cld1), which exhibit wild type levels of CL and MLCL but lack remodeled CL, were found to be normal with respect to growth, oxidative phosphorylation, and mitochondrial morphology. Importantly, the cld1Δ mutant rescued the growth, lifespan, and respiratory defects of the tafazzin mutant. These experiments indicate that the deleterious effects of defective tafazzin in yeast are caused by CL deficiency or MLCL accumulation rather than by abnormal CL molecular species.
If the inability to remodel CL is not deleterious to yeast cells, what is the function of the remodeling enzymes? Some insight into the function of CLD1 comes from the finding that expression of the CLD1 gene in yeast cells is highest when the demand for respiration is increased [82, 84]. One possible explanation for increased CLD1 expression is that CL remodeling is needed for optimal oxidative phosphorylation; however, as mentioned, oxidative phosphorylation is not defective in the cld1Δ mutant. A more likely possibility is that increased CLD1 expression is a damage control mechanism to deal with the deleterious effects of increased respiration, such as increased generation of reactive oxygen species. In this light, remodeling may function to eliminate oxidized CL species. A third and intriguing possibility is that the enzymes that remodel CL have cellular functions in addition to CL remodeling. Thus, as a non-specific transacylase, tafazzin may regulate levels of other phospholipids and lysolipids, in addition to CL and MLCL, which may affect other functions than those attributed to CL.
What are the consequences of low CL levels in mitochondria? It has been established that CL deficiency can be responsible for the instability of cristae membrane supercomplexes [85]. Indeed, the abundance of such supercomplexes was found to be low in BTHS [45, 86, 87]. However, new data from our laboratory suggest that supercomplex instability may be the cause rather than the effect of CL deficiency in BTHS. According to this idea, the lack of CL remodeling causes the dissociation of supercomplexes, which then reduces the amount of proteinassociated CL and thus exposes CL to phopholipases [45]. Of course, this scenario implies that CL remodeling by tafazzin is intrinsically linked to the assembly of protein complexes. Surely, this idea requires further confirmation. Nevertheless, we have demonstrated in different experimental models, including yeast, mammalian cells, and flies, that CL loses its stability in the absence of tafazzin and gets rapidly degraded to MLCL [45]. This challenges the traditional view according to which MLCL accumulates because it is an intermediate of CL remodeling. More work is necessary though to understand the mechanism of CL remodeling and its potential connection to the assembly of protein complexes.
Conclusions
The biosynthesis of CL occurs in the inner leaflet of the inner mitochondrial membrane and is followed by the remodeling of the fatty acids of CL and its assembly into the inner membrane. While the biosynthetic pathway has been established, controversy continues to exist about the mechanism and the functional implications of CL remodeling.
Arguably, the most important function of CL in mitochondria is the effect it has on the association of protein complexes and supercomplexes. The significance of such assemblies that can reach a mass of several million daltons, has increasingly been recognized. Protein complexes entrap a large proportion of mitochondrial CL and the sequestration of CL in these complexes is responsible for its very long half-life. Thus, in the future it will be necessary to dissect the mechanism by which CL is incorporated into protein complexes. There is good reason to believe that the remodeling of CL acyl groups and the assembly of CL into protein complexes are linked. Mechanistic insight into this process will be crucial to understand the molecular pathogenesis of BTHS.
Highlights.
-
-
Cardiolipin is the specific phospholipid of mitochondria
-
-
The review covers biosynthesis, membrane assembly, and turnover of cardiolipin
-
-
Aberrant cardiolipin metabolism causes Barth syndrome
Acknowledgments
We acknowledge support from the National Institutes of Health, including grant GM115593 (to MS) and grant HL117880 (to MLG).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pineau B, Bourge M, Marion J, Mauve C, Gilard F, Maneta-Peyret L, Moreau P, Satiat-Jeunemaitre B, Brown SC, De Paepe R, Danon A. The importance of cardiolipin synthase for mitochondrial ultrastructure, respiratory function, plant development, and stress responses in Arabidopsis. The Plant cell. 2013;25:4195–4208. doi: 10.1105/tpc.113.118018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin Y, Bogdanov M, Tong S, Guan Z, Zheng L. Substrate Selectivity of Lysophospholipid Transporter LplT Involved in Membrane Phospholipid Remodeling in Escherichia coli. The Journal of biological chemistry. 2016;291:2136–2149. doi: 10.1074/jbc.M115.700419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hostetler KY, Van den Bosch H, Van Deenen LL. Biosynthesis of cardiolipin in liver mitochondria. Biochimica et biophysica acta. 1971;239:113–119. doi: 10.1016/0005-2760(71)90201-3. [DOI] [PubMed] [Google Scholar]
- 4.Schlame M, Haldar D. Cardiolipin is synthesized on the matrix side of the inner membrane in rat liver mitochondria. The Journal of biological chemistry. 1993;268:74–79. [PubMed] [Google Scholar]
- 5.Tamura Y, Harada Y, Nishikawa S, Yamano K, Kamiya M, Shiota T, Kuroda T, Kuge O, Sesaki H, Imai K, Tomii K, Endo T. Tam41 is a CDP-diacylglycerol synthase required for cardiolipin biosynthesis in mitochondria. Cell metabolism. 2013;17:709–718. doi: 10.1016/j.cmet.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tamura Y, Harada Y, Yamano K, Watanabe K, Ishikawa D, Ohshima C, Nishikawa S, Yamamoto H, Endo T. Identification of Tam41 maintaining integrity of the TIM23 protein translocator complex in mitochondria. The Journal of cell biology. 2006;174:631–637. doi: 10.1083/jcb.200603087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gallas MR, Dienhart MK, Stuart RA, Long RM. Characterization of Mmp37p, a Saccharomyces cerevisiae mitochondrial matrix protein with a role in mitochondrial protein import. Molecular biology of the cell. 2006;17:4051–4062. doi: 10.1091/mbc.E06-04-0366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kutik S, Rissler M, Guan XL, Guiard B, Shui G, Gebert N, Heacock PN, Rehling P, Dowhan W, Wenk MR, Pfanner N, Wiedemann N. The translocator maintenance protein Tam41 is required for mitochondrial cardiolipin biosynthesis. The Journal of cell biology. 2008;183:1213–1221. doi: 10.1083/jcb.200806048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang SC, Heacock PN, Clancey CJ, Dowhan W. The PEL1 gene (renamed PGS1) encodes the phosphatidylglycero-phosphate synthase of Saccharomyces cerevisiae. The Journal of biological chemistry. 1998;273:9829–9836. doi: 10.1074/jbc.273.16.9829. [DOI] [PubMed] [Google Scholar]
- 10.He Q, Greenberg ML. Post-translational regulation of phosphatidylglycerolphosphate synthase in response to inositol. Molecular microbiology. 2004;53:1243–1249. doi: 10.1111/j.1365-2958.2004.04202.x. [DOI] [PubMed] [Google Scholar]
- 11.Osman C, Haag M, Wieland FT, Brugger B, Langer T. A mitochondrial phosphatase required for cardiolipin biosynthesis: the PGP phosphatase Gep4. The EMBO journal. 2010;29:1976–1987. doi: 10.1038/emboj.2010.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J, Guan Z, Murphy AN, Wiley SE, Perkins GA, Worby CA, Engel JL, Heacock P, Nguyen OK, Wang JH, Raetz CR, Dowhan W, Dixon JE. Mitochondrial phosphatase PTPMT1 is essential for cardiolipin biosynthesis. Cell metabolism. 2011;13:690–700. doi: 10.1016/j.cmet.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang F, Rizavi HS, Greenberg ML. Cardiolipin is not essential for the growth of Saccharomyces cerevisiae on fermentable or non-fermentable carbon sources. Molecular microbiology. 1997;26:481–491. doi: 10.1046/j.1365-2958.1997.5841950.x. [DOI] [PubMed] [Google Scholar]
- 14.Tuller G, Hrastnik C, Achleitner G, Schiefthaler U, Klein F, Daum G. YDL142c encodes cardiolipin synthase (Cls1p) and is non-essential for aerobic growth of Saccharomyces cerevisiae. FEBS letters. 1998;421:15–18. doi: 10.1016/s0014-5793(97)01525-1. [DOI] [PubMed] [Google Scholar]
- 15.Chen D, Zhang XY, Shi Y. Identification and functional characterization of hCLS1, a human cardiolipin synthase localized in mitochondria. The Biochemical journal. 2006;398:169–176. doi: 10.1042/BJ20060303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu B, Xu FY, Jiang YJ, Choy PC, Hatch GM, Grunfeld C, Feingold KR. Cloning and characterization of a cDNA encoding human cardiolipin synthase (hCLS1) Journal of lipid research. 2006;47:1140–1145. doi: 10.1194/jlr.C600004-JLR200. [DOI] [PubMed] [Google Scholar]
- 17.Houtkooper RH, Akbari H, van Lenthe H, Kulik W, Wanders RJ, Frentzen M, Vaz FM. Identification and characterization of human cardiolipin synthase. FEBS letters. 2006;580:3059–3064. doi: 10.1016/j.febslet.2006.04.054. [DOI] [PubMed] [Google Scholar]
- 18.Katayama K, Sakurai I, Wada H. Identification of an Arabidopsis thaliana gene for cardiolipin synthase located in mitochondria. FEBS letters. 2004;577:193–198. doi: 10.1016/j.febslet.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 19.Nowicki M, Muller F, Frentzen M. Cardiolipin synthase of Arabidopsis thaliana. FEBS letters. 2005;579:2161–2165. doi: 10.1016/j.febslet.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 20.Schlame M. Cardiolipin synthesis for the assembly of bacterial and mitochondrial membranes. Journal of lipid research. 2008;49:1607–1620. doi: 10.1194/jlr.R700018-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan BK, Bogdanov M, Zhao J, Dowhan W, Raetz CR, Guan Z. Discovery of a cardiolipin synthase utilizing phosphatidylethanolamine and phosphatidylglycerol as substrates. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:16504–16509. doi: 10.1073/pnas.1212797109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Serricchio M, Butikofer P. An essential bacterial-type cardiolipin synthase mediates cardiolipin formation in a eukaryote. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E954–E961. doi: 10.1073/pnas.1121528109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai L, Wang M, Martin OJ, Leone TC, Vega RB, Han X, Kelly DP. A role for peroxisome proliferator-activated receptor gamma coactivator 1 (PGC-1) in the regulation of cardiac mitochondrial phospholipid biosynthesis. The Journal of biological chemistry. 2014;289:2250–2259. doi: 10.1074/jbc.M113.523654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Das S, Morvan F, Jourde B, Meier V, Kahle P, Brebbia P, Toussaint G, Glass DJ, Fornaro M. ATP citrate lyase improves mitochondrial function in skeletal muscle. Cell metabolism. 2015;21:868–876. doi: 10.1016/j.cmet.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 25.Kiebish MA, Han X, Cheng H, Chuang JH, Seyfried TN. Cardiolipin and electron transport chain abnormalities in mouse brain tumor mitochondria: lipidomic evidence supporting the Warburg theory of cancer. Journal of lipid research. 2008;49:2545–2556. doi: 10.1194/jlr.M800319-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tyurina YY, Poloyac SM, Tyurin VA, Kapralov AA, Jiang J, Anthonymuthu TS, Kapralova VI, Vikulina AS, Jung MY, Epperly MW, Mohammadyani D, Klein-Seetharaman J, Jackson TC, Kochanek PM, Pitt BR, Greenberger JS, Vladimirov YA, Bayir H, Kagan VE. A mitochondrial pathway for biosynthesis of lipid mediators. Nature chemistry. 2014;6:542–552. doi: 10.1038/nchem.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schlame M, Ren M, Xu Y, Greenberg ML, Haller I. Molecular symmetry in mitochondrial cardiolipins. Chemistry and physics of lipids. 2005;138:38–49. doi: 10.1016/j.chemphyslip.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 28.Xu Y, Malhotra A, Ren M, Schlame M. The enzymatic function of tafazzin. The Journal of biological chemistry. 2006;281:39217–39224. doi: 10.1074/jbc.M606100200. [DOI] [PubMed] [Google Scholar]
- 29.Schlame M, Acehan D, Berno B, Xu Y, Valvo S, Ren M, Stokes DL, Epand RM. The physical state of lipid substrates provides transacylation specificity for tafazzin. Nature chemical biology. 2012;8:862–869. doi: 10.1038/nchembio.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abe M, Hasegawa Y, Oku M, Sawada Y, Tanaka E, Sakai Y, Miyoshi H. Mechanism for Remodeling of the Acyl Chain Composition of Cardiolipin Catalyzed by Saccharomyces cerevisiae Tafazzin. The Journal of biological chemistry. 2016;291:15491–15502. doi: 10.1074/jbc.M116.718510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaz FM, Houtkooper RH, Valianpour F, Barth PG, Wanders RJ. Only one splice variant of the human TAZ gene encodes a functional protein with a role in cardiolipin metabolism. The Journal of biological chemistry. 2003;278:43089–43094. doi: 10.1074/jbc.M305956200. [DOI] [PubMed] [Google Scholar]
- 32.Xu Y, Zhang S, Malhotra A, Edelman-Novemsky I, Ma J, Kruppa A, Cernicica C, Blais S, Neubert TA, Ren M, Schlame M. Characterization of tafazzin splice variants from humans and fruit flies. The Journal of biological chemistry. 2009;284:29230–29239. doi: 10.1074/jbc.M109.016642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Claypool SM, McCaffery JM, Koehler CM. Mitochondrial mislocalization and altered assembly of a cluster of Barth syndrome mutant tafazzins. The Journal of cell biology. 2006;174:379–390. doi: 10.1083/jcb.200605043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu YW, Galbraith L, Herndon JD, Lu YL, Pras-Raves M, Vervaart M, Van Kampen A, Luyf A, Koehler CM, McCaffery JM, Gottlieb E, Vaz FM, Claypool SM. Defining functional classes of Barth syndrome mutation in humans. Human molecular genetics. 2016;25:1754–1770. doi: 10.1093/hmg/ddw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Claypool SM, Boontheung P, McCaffery JM, Loo JA, Koehler CM. The cardiolipin transacylase, tafazzin, associates with two distinct respiratory components providing insight into Barth syndrome. Molecular biology of the cell. 2008;19:5143–5155. doi: 10.1091/mbc.E08-09-0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu Y, Malhotra A, Claypool SM, Ren M, Schlame M. Tafazzins from Drosophila and mammalian cells assemble in large protein complexes with a short half-life. Mitochondrion. 2015;21:27–32. doi: 10.1016/j.mito.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ren M, Phoon CK, Schlame M. Metabolism and function of mitochondrial cardiolipin. Progress in lipid research. 2014;55:1–16. doi: 10.1016/j.plipres.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 38.Lee Y, Willers C, Kunji ER, Crichton PG. Uncoupling protein 1 binds one nucleotide per monomer and is stabilized by tightly bound cardiolipin. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:6973–6978. doi: 10.1073/pnas.1503833112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sauerwald J, Jores T, Eisenberg-Bord M, Chuartzman SG, Schuldiner M, Rapaport D. Genome-Wide Screens in Saccharomyces cerevisiae Highlight a Role for Cardiolipin in Biogenesis of Mitochondrial Outer Membrane Multispan Proteins. Molecular and cellular biology. 2015;35:3200–3211. doi: 10.1128/MCB.00107-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bustillo-Zabalbeitia I, Montessuit S, Raemy E, Basanez G, Terrones O, Martinou JC. Specific interaction with cardiolipin triggers functional activation of Dynamin-Related Protein 1. PloS one. 2014;9:e102738. doi: 10.1371/journal.pone.0102738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Macdonald PJ, Stepanyants N, Mehrotra N, Mears JA, Qi X, Sesaki H, Ramachandran R. A dimeric equilibrium intermediate nucleates Drp1 reassembly on mitochondrial membranes for fission. Molecular biology of the cell. 2014;25:1905–1915. doi: 10.1091/mbc.E14-02-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Planas-Iglesias J, Dwarakanath H, Mohammadyani D, Yanamala N, Kagan VE, Klein-Seetharaman J. Cardiolipin Interactions with Proteins. Biophysical journal. 2015;109:1282–1294. doi: 10.1016/j.bpj.2015.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lewis RN, McElhaney RN. The physicochemical properties of cardiolipin bilayers and cardiolipin-containing lipid membranes. Biochimica et biophysica acta. 2009;1788:2069–2079. doi: 10.1016/j.bbamem.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 44.Schlame M, Horvath L, Vigh L. Relationship between lipid saturation and lipid-protein interaction in liver mitochondria modified by catalytic hydrogenation with reference to cardiolipin molecular species. The Biochemical journal. 1990;265:79–85. doi: 10.1042/bj2650079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu Y, Phoon CK, Berno B, D'Souza K, Hoedt E, Zhang G, Neubert TA, Epand RM, Ren M, Schlame M. Loss of protein association causes cardiolipin degradation in Barth syndrome. Nature chemical biology. 2016;12:641–647. doi: 10.1038/nchembio.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Osman C, Merkwirth C, Langer T. Prohibitins and the functional compartmentalization of mitochondrial membranes. Journal of cell science. 2009;122:3823–3830. doi: 10.1242/jcs.037655. [DOI] [PubMed] [Google Scholar]
- 47.Richter-Dennerlein R, Korwitz A, Haag M, Tatsuta T, Dargazanli S, Baker M, Decker T, Lamkemeyer T, Rugarli EI, Langer T. DNAJC19, a mitochondrial cochaperone associated with cardiomyopathy, forms a complex with prohibitins to regulate cardiolipin remodeling. Cell metabolism. 2014;20:158–171. doi: 10.1016/j.cmet.2014.04.016. [DOI] [PubMed] [Google Scholar]
- 48.Klecker T, Wemmer M, Haag M, Weig A, Bockler S, Langer T, Nunnari J, Westermann B. Interaction of MDM33 with mitochondrial inner membrane homeostasis pathways in yeast. Scientific reports. 2015;5:18344. doi: 10.1038/srep18344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pfanner N, van der Laan M, Amati P, Capaldi RA, Caudy AA, Chacinska A, Darshi M, Deckers M, Hoppins S, Icho T, Jakobs S, Ji J, Kozjak-Pavlovic V, Meisinger C, Odgren PR, Park SK, Rehling P, Reichert AS, Sheikh MS, Taylor SS, Tsuchida N, van der Bliek AM, van der Klei IJ, Weissman JS, Westermann B, Zha J, Neupert W, Nunnari J. Uniform nomenclature for the mitochondrial contact site and cristae organizing system. The Journal of cell biology. 2014;204:1083–1086. doi: 10.1083/jcb.201401006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Friedman JR, Mourier A, Yamada J, McCaffery JM, Nunnari J. MICOS coordinates with respiratory complexes and lipids to establish mitochondrial inner membrane architecture. eLife. 2015;4 doi: 10.7554/eLife.07739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harner ME, Unger AK, Izawa T, Walther DM, Ozbalci C, Geimer S, Reggiori F, Brugger B, Mann M, Westermann B, Neupert W. Aim24 and MICOS modulate respiratory function, tafazzin-related cardiolipin modification and mitochondrial architecture. eLife. 2014;3:e01684. doi: 10.7554/eLife.01684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koob S, Barrera M, Anand R, Reichert AS. The non-glycosylated isoform of MIC26 is a constituent of the mammalian MICOS complex and promotes formation of crista junctions. Biochimica et biophysica acta. 2015;1853:1551–1563. doi: 10.1016/j.bbamcr.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 53.Zhang M, Mileykovskaya E, Dowhan W. Gluing the respiratory chain together. Cardiolipin is required for supercomplex formation in the inner mitochondrial membrane. The Journal of biological chemistry. 2002;277:43553–43556. doi: 10.1074/jbc.C200551200. [DOI] [PubMed] [Google Scholar]
- 54.Claypool SM, Oktay Y, Boontheung P, Loo JA, Koehler CM. Cardiolipin defines the interactome of the major ADP/ATP carrier protein of the mitochondrial inner membrane. The Journal of cell biology. 2008;182:937–950. doi: 10.1083/jcb.200801152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pfeiffer K, Gohil V, Stuart RA, Hunte C, Brandt U, Greenberg ML, Schagger H. Cardiolipin stabilizes respiratory chain supercomplexes. The Journal of biological chemistry. 2003;278:52873–52880. doi: 10.1074/jbc.M308366200. [DOI] [PubMed] [Google Scholar]
- 56.Althoff T, Mills DJ, Popot JL, Kuhlbrandt W. Arrangement of electron transport chain components in bovine mitochondrial supercomplex I1III2IV1. The EMBO journal. 2011;30:4652–4664. doi: 10.1038/emboj.2011.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mileykovskaya E, Penczek PA, Fang J, Mallampalli VK, Sparagna GC, Dowhan W. Arrangement of the respiratory chain complexes in Saccharomyces cerevisiae supercomplex III2IV2 revealed by single particle cryo-electron microscopy. The Journal of biological chemistry. 2012;287:23095–23103. doi: 10.1074/jbc.M112.367888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hofer A, Noe N, Tischner C, Kladt N, Lellek V, Schauss A, Wenz T. Defining the action spectrum of potential PGC-1alpha activators on a mitochondrial and cellular level in vivo. Human molecular genetics. 2014;23:2400–2415. doi: 10.1093/hmg/ddt631. [DOI] [PubMed] [Google Scholar]
- 59.Zhang Y, Bharathi SS, Rardin MJ, Uppala R, Verdin E, Gibson BW, Goetzman ES. SIRT3 and SIRT5 regulate the enzyme activity and cardiolipin binding of very long-chain acyl-CoA dehydrogenase. PloS one. 2015;10:e0122297. doi: 10.1371/journal.pone.0122297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brandner K, Mick DU, Frazier AE, Taylor RD, Meisinger C, Rehling P. Taz1, an outer mitochondrial membrane protein, affects stability and assembly of inner membrane protein complexes: implications for Barth Syndrome. Molecular biology of the cell. 2005;16:5202–5214. doi: 10.1091/mbc.E05-03-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McKenzie M, Lazarou M, Thorburn DR, Ryan MT. Mitochondrial respiratory chain supercomplexes are destabilized in Barth Syndrome patients. Journal of molecular biology. 2006;361:462–469. doi: 10.1016/j.jmb.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 62.Gonzalvez F, D'Aurelio M, Boutant M, Moustapha A, Puech JP, Landes T, Arnaune-Pelloquin L, Vial G, Taleux N, Slomianny C, Wanders RJ, Houtkooper RH, Bellenguer P, Moller IM, Gottlieb E, Vaz FM, Manfredi G, Petit PX. Barth syndrome: cellular compensation of mitochondrial dysfunction and apoptosis inhibition due to changes in cardiolipin remodeling linked to tafazzin (TAZ) gene mutation. Biochimica et biophysica acta. 2013;1832:1194–1206. doi: 10.1016/j.bbadis.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 63.Dudek J, Cheng IF, Balleininger M, Vaz FM, Streckfuss-Bomeke K, Hubscher D, Vukotic M, Wanders RJ, Rehling P, Guan K. Cardiolipin deficiency affects respiratory chain function and organization in an induced pluripotent stem cell model of Barth syndrome. Stem cell research. 2013;11:806–819. doi: 10.1016/j.scr.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 64.Desmurs M, Foti M, Raemy E, Vaz FM, Martinou JC, Bairoch A, Lane L. C11orf83, a mitochondrial cardiolipin-binding protein involved in bc1 complex assembly and supercomplex stabilization. Molecular and cellular biology. 2015;35:1139–1156. doi: 10.1128/MCB.01047-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mitsopoulos P, Chang YH, Wai T, Konig T, Dunn SD, Langer T, Madrenas J. Stomatin-like protein 2 is required for in vivo mitochondrial respiratory chain supercomplex formation and optimal cell function. Molecular and cellular biology. 2015;35:1838–1847. doi: 10.1128/MCB.00047-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Landriscina C, Megli FM, Quagliariello E. Turnover of fatty acids in rat liver cardiolipin: comparison with other mitochondrial phospholipids. Lipids. 1976;11:61–66. doi: 10.1007/BF02532585. [DOI] [PubMed] [Google Scholar]
- 67.Wahjudi PN, J KY, Martinez SR, Zhang J, Teitell M, Nikolaenko L, Swerdloff R, Wang C, Lee WN. Turnover of nonessential fatty acids in cardiolipin from the rat heart. Journal of lipid research. 2011;52:2226–2233. doi: 10.1194/jlr.M015966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu Y, Schlame M. The turnover of glycerol and acyl moieties of cardiolipin. Chemistry and physics of lipids. 2014;179:17–24. doi: 10.1016/j.chemphyslip.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 69.Wilkens V, Kohl W, Busch K. Restricted diffusion of OXPHOS complexes in dynamic mitochondria delays their exchange between cristae and engenders a transitory mosaic distribution. Journal of cell science. 2013;126:103–116. doi: 10.1242/jcs.108852. [DOI] [PubMed] [Google Scholar]
- 70.Kagan VE, Chu CT, Tyurina YY, Cheikhi A, Bayir H. Cardiolipin asymmetry, oxidation and signaling. Chemistry and physics of lipids. 2014;179:64–69. doi: 10.1016/j.chemphyslip.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kagan VE, Jiang J, Huang Z, Tyurina YY, Desbourdes C, Cottet-Rousselle C, Dar HH, Verma M, Tyurin VA, Kapralov AA, Cheikhi A, Mao G, Stolz D, St Croix CM, Watkins S, Shen Z, Li Y, Greenberg ML, Tokarska-Schlattner M, Boissan M, Lacombe ML, Epand RM, Chu CT, Mallampalli RK, Bayir H, Schlattner U. NDPK-D (NM23-H4)-mediated externalization of cardiolipin enables elimination of depolarized mitochondria by mitophagy. Cell death and differentiation. 2016;23:1140–1151. doi: 10.1038/cdd.2015.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chu CT, Ji J, Dagda RK, Jiang JF, Tyurina YY, Kapralov AA, Tyurin VA, Yanamala N, Shrivastava IH, Mohammadyani D, Qiang Wang KZ, Zhu J, Klein-Seetharaman J, Balasubramanian K, Amoscato AA, Borisenko G, Huang Z, Gusdon AM, Cheikhi A, Steer EK, Wang R, Baty C, Watkins S, Bahar I, Bayir H, Kagan VE. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nature cell biology. 2013;15:1197–1205. doi: 10.1038/ncb2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Song H, Wohltmann M, Tan M, Ladenson JH, Turk J. Group VIA phospholipase A2 mitigates palmitate-induced beta-cell mitochondrial injury and apoptosis. The Journal of biological chemistry. 2014;289:14194–14210. doi: 10.1074/jbc.M114.561910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li J, Romestaing C, Han X, Li Y, Hao X, Wu Y, Sun C, Liu X, Jefferson LS, Xiong J, Lanoue KF, Chang Z, Lynch CJ, Wang H, Shi Y. Cardiolipin remodeling by ALCAT1 links oxidative stress and mitochondrial dysfunction to obesity. Cell metabolism. 2010;12:154–165. doi: 10.1016/j.cmet.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhao Y, Chen YQ, Li S, Konrad RJ, Cao G. The microsomal cardiolipin remodeling enzyme acyl-CoA lysocardiolipin acyltransferase is an acyltransferase of multiple anionic lysophospholipids. Journal of lipid research. 2009;50:945–956. doi: 10.1194/jlr.M800567-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schlame M, Kelley RI, Feigenbaum A, Towbin JA, Heerdt PM, Schieble T, Wanders RJ, DiMauro S, Blanck TJ. Phospholipid abnormalities in children with Barth syndrome. Journal of the American College of Cardiology. 2003;42:1994–1999. doi: 10.1016/j.jacc.2003.06.015. [DOI] [PubMed] [Google Scholar]
- 77.Kulik W, van Lenthe H, Stet FS, Houtkooper RH, Kemp H, Stone JE, Steward CG, Wanders RJ, Vaz FM. Bloodspot assay using HPLC-tandem mass spectrometry for detection of Barth syndrome. Clin Chem. 2008;54:371–378. doi: 10.1373/clinchem.2007.095711. [DOI] [PubMed] [Google Scholar]
- 78.Houtkooper RH, Rodenburg RJ, Thiels C, van Lenthe H, Stet F, Bwee TPT, Stone JE, Steward CG, Wanders RJ, Smeitink J, Kulik W, Vaz FM. Cardiolipin and monolysocardiolipin analysis in fibroblasts, lymphocytes, and tissues using high-performance liquid chromatography-mass spectrometry as a diagnostic test for Barth syndrome. Analytical biochemistry. 2009;387:230–237. doi: 10.1016/j.ab.2009.01.032. [DOI] [PubMed] [Google Scholar]
- 79.Valianpour F, Wanders RJ, Barth PG, Overmars H, van Gennip AH. Quantitative and compositional study of cardiolipin in platelets by electrospray ionization mass spectrometry: application for the identification of Barth syndrome patients. Clin Chem. 2002;48:1390–1397. [PubMed] [Google Scholar]
- 80.Bowron A, Frost R, Powers VE, Thomas PH, Heales SJ, Steward CG. Diagnosis of Barth syndrome using a novel LC-MS/MS method for leukocyte cardiolipin analysis. Journal of inherited metabolic disease. 2013;36:741–746. doi: 10.1007/s10545-012-9552-4. [DOI] [PubMed] [Google Scholar]
- 81.Angelini R, Lobasso S, Gorgoglione R, Bowron A, Steward CG, Corcelli A. Cardiolipin fingerprinting of leukocytes by MALDI-TOF/MS as a screening tool for Barth syndrome. Journal of lipid research. 2015;56:1787–1794. doi: 10.1194/jlr.D059824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ye C, Lou W, Li Y, Chatzispyrou IA, Huttemann M, Lee I, Houtkooper RH, Vaz FM, Chen S, Greenberg ML. Deletion of the cardiolipin-specific phospholipase Cld1 rescues growth and life span defects in the tafazzin mutant: implications for Barth syndrome. The Journal of biological chemistry. 2014;289:3114–3125. doi: 10.1074/jbc.M113.529487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Baile MG, Sathappa M, Lu YW, Pryce E, Whited K, McCaffery JM, Han X, Alder NN, Claypool SM. Unremodeled and remodeled cardiolipin are functionally indistinguishable in yeast. The Journal of biological chemistry. 2014;289:1768–1778. doi: 10.1074/jbc.M113.525733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Baile MG, Whited K, Claypool SM. Deacylation on the matrix side of the mitochondrial inner membrane regulates cardiolipin remodeling. Molecular biology of the cell. 2013;24:2008–2020. doi: 10.1091/mbc.E13-03-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mileykovskaya E, Dowhan W. Cardiolipin-dependent formation of mitochondrial respiratory supercomplexes. Chemistry and physics of lipids. 2014;179:42–48. doi: 10.1016/j.chemphyslip.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Huang Y, Powers C, Madala SK, Greis KD, Haffey WD, Towbin JA, Purevjav E, Javadov S, Strauss AW, Khuchua Z. Cardiac metabolic pathways affected in the mouse model of Barth syndrome. PloS one. 2015;10:e0128561. doi: 10.1371/journal.pone.0128561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dudek J, Cheng IF, Chowdhury A, Wozny K, Balleininger M, Reinhold R, Grunau S, Callegari S, Toischer K, Wanders RJ, Hasenfuss G, Brugger B, Guan K, Rehling P. Cardiac-specific succinate dehydrogenase deficiency in Barth syndrome. EMBO molecular medicine. 2016;8:139–154. doi: 10.15252/emmm.201505644. [DOI] [PMC free article] [PubMed] [Google Scholar]