Abstract
Bacterial pathogens require the iron-containing cofactor heme to cause disease. Heme is essential to the function of hemoproteins, which are involved in energy generation by the electron transport chain, detoxification of host immune effectors, and other processes. During infection, bacterial pathogens must synthesize heme or acquire heme from the host; however, host heme is sequestered in high-affinity hemoproteins. Pathogens have evolved elaborate strategies to acquire heme from host sources, particularly hemoglobin, and both heme acquisition and synthesis are important for pathogenesis. Paradoxically, excess heme is toxic to bacteria and pathogens must rely on heme detoxification strategies. Heme is a key nutrient in the struggle for survival between host and pathogen, and its study has offered significant insight into the molecular mechanisms of bacterial pathogenesis.
Keywords: iron uptake, hemoglobin, Isd system, heme toxicity, NEAT domain
Introduction
Heme and iron are essential for life
The tetrapyrrole cofactor heme is important for the cellular processes of most organisms and essential to many lifeforms across domains of life. Heme, a porphyrin ring complexed with iron, serves as a redox active moiety required for the function of many cellular proteins. Heme functions as an electron shuttle in enzymes of the electron transport chain and is required for cellular respiration. Additionally, cells rely on heme for the function of many widely conserved enzymes including catalase, nitric oxide synthase, hemoglobin (Hb), and others. Heme is also an important molecule involved in diverse cellular processes including signaling, gas sensing, microRNA processing, and cellular differentiation [1–4]. Thus, nearly all organisms must satisfy the requirement for heme through either synthesis or acquisition.
Heme coordinates an iron atom at its center which is vital for heme's electron transfer abilities and redox activity. Like heme, iron is nearly universally required for life, and only a few exceptions have been identified [5,6]. As an inorganic cofactor, iron can act alone or in iron–sulfur clusters as a prosthetic moiety for members of the oxidoreductase, nitrogenase, hydrogenase, dehydrogenase, and hydratase enzyme families [7–12]. Therefore, organisms have evolved elaborate strategies to acquire, store, and regulate intracellular iron for heme-dependent and other iron-dependent enzymes.
Nutritional immunity limits host iron availability
Nutritional immunity, a concept articulated primarily by Eugene Weinberg in the 1970s, describes the processes by which humans and other organisms sequester iron to limit acquisition by bacterial pathogens [13,14]. Nutritional immunity has since been expanded to include the host processes that manipulate local levels of manganese, zinc, and other transition metals in order to metal starve or intoxicate the invading pathogens (reviewed previously in Refs. [15,16–18]). The limited access of pathogens to metals serves as an antimicrobial strategy and limits bacterial replication. For instance, free iron rarely exists in the mammalian host. The solubility of ferric iron in aerobic solution is exceedingly low, and high-affinity iron-binding proteins, including transferrin, lactoferrin, albumin, and ferritin, sequester iron extracellularly and intracellularly. Iron-binding proteins function to transport iron, protect host cells from iron-mediated oxidative damage, and keep iron from pathogens. However, bacterial pathogens have developed exquisite tactics to overcome iron limitation and elaborate high-affinity iron receptors and chelators. In this regard, an evolutionary arms race has developed at the host–pathogen interface involving host iron-binding proteins and the mechanisms bacteria encode to steal iron.
Heme is an important host iron source
Heme makes up the greatest reservoir of iron in the host and serves as an iron source for many bacterial pathogens. Humans and other metazoa synthesize heme through a variety of steps in the mitochondria and cytosol. This pathway, called the Shemin or four-carbon pathway, begins with the condensation of glycine and succinyl-CoA to form the committed precursor δ-aminolevulinic acid (ALA) [19–21]. A series of enzymes produces protoporphyrin IX from ALA and iron is inserted, forming protoheme IX. For the sake of simplicity in this review, heme will refer to ferrous and ferric iron forms of protoheme IX. Heme is then bound by hemoproteins to serve a variety of intracellular and extracellular tasks. Catalase, peroxidase, and myeloperoxidase rely on heme to catalyze the hydrolysis of peroxide molecules. Energy generation by the electron transport chain relies on heme-dependent c- and b-type cytochromes of the ubiquinol–ferricytochrome c oxidoreductase (Complex III) family [22,23]. Hemoproteins involved in tissue oxygen homeostasis include myoglobin and neuroglobin. Perhaps the most well-known hemoprotein is the oxygen transporter Hb. Its abundance and location in erythrocytes make Hb a rich heme source for pathogens. Hb contains about two-thirds of the body's iron, and a single erythrocyte contains more than 280 million molecules of Hb [15,24]. Bacterial pathogens have evolved high-affinity Hb-binding proteins for the acquisition of heme, and these proteins will be described below.
Owing in part to the reactive nature of heme-iron, free heme and Hb are toxic to the human host and bacterial pathogens alike [25,26]. To prevent excess heme toxicity, eukaryotic heme synthesis is highly regulated and heme homeostasis and sequestration are well orchestrated. When Hb is released from erythrocytes or otherwise exists extracellularly, it is rapidly bound by haptoglobin (Hp) [27]. The abundance of cell-free Hb is thought to be very low in healthy adults, but a variety of genetic disorders, infections, and other disease states can increase the concentration of free Hb [28]. Free Hb and its modified forms, in the presence of reactive oxygen species, exhibit cytotoxic effects toward endothelial cells [29]. However, the relevance of these in vivo studies is unclear, and a comprehensive understanding of concentrations to achieve Hb toxicity in healthy humans has not been achieved [25]. On the other hand, in the absence of infection-free heme that has been liberated from its hemoprotein likely only exists transiently in serum or in cells. In serum, heme is immediately bound by the highly abundant albumin (kd ≈ 10 nM) then transferred to hemopexin (kd < 1 pM) [30]. The heme is delivered to cells expressing the hemopexin receptor; these cells then degrade the heme using heme oxygenases [30]. The rapid sequestration and degradation of free heme in the blood is vital to the survival of erythrocytes, as heme in the presence of reactive oxygen species exhibits cytotoxicity and lipid peroxidation at micromolar concentrations [31,32]. During infection of host heme- and Hb-replete niches, bacterial pathogens experience heme toxicity and encode systems to protect from heme toxicity as well [33]. Therefore, heme is at the center of a complex interplay between host and pathogen for survival.
Bacterial Heme Synthesis
Divergent heme synthesis pathways in Gramnegative and Gram-positive organisms
While both humans and bacteria share the early heme precursor ALA, most bacteria (Alphaproteo-bacteria are the exception), archaea, and plants synthesize ALA from charged glutamyl-tRNAGlu via the “C5 pathway” (Fig. 1) [21,34,35]. The gluta-myl-tRNA reductase HemA produces the highly reactive intermediate glutamate-1-semialdehyde, which HemL, a glutamate-1-semialdehyde amino-mutase, converts to ALA [36,37]. The three steps, from ALA to uroporphyrinogen, are well conserved and thought to be the evolutionary core of heme biosynthesis. ALA dehydratase (also called porphobilinogen synthase; annotated as HemB) is responsible for the condensation of two ALA to porphobilinogen (PBG) [38]. The linear tetrapyrrole hydroxymethylbilane (HMB) is produced by a head-to-tail condensation and deamination of four PBG molecules, catalyzed by HMB synthase (alternatively called PBG deaminase, annotated as HemC) [39,40]. Under physiological conditions, HMB will spontaneously cyclize to form the uroporphyrinogen I isomer, a biosynthetic deadend. Therefore, most bacteria utilize uroporphyrinogen III synthase (HemD) to catalyze the cyclization of HMB through a spiro-intermediate to form uroporphyrinogen III [41].
Fig. 1. Bacterial heme biosynthesis.

The heme synthesis pathway of most bacteria begins with charged glutamyl-tRNAGlu to form the universal precursor ALA, and coproporphyrinogen III isformed through a series of conserved enzymatic steps. The classical pathway (blue) forms heme through the protoporphyrinogen IX intermediate; most organisms including Gram-negative bacteria and eukaryotes use this pathway. The noncanonical pathway (green), performed by most Gram-positive bacteria, produces heme through the coproporphyrin III intermediate. Shown for each step is the enzyme name followed by the common protein annotation in bold.
Uroporphyrinogen III can be utilized for the synthesis of several tetrapyrrole-based cofactors. Uroporphyrinogen III decarboxylase (HemE) decarboxylates the four acetate side chains to methyl groups, producing coproporphyrinogen III, the next step in heme synthesis [42]. Additionally, uroporphyrinogen III can be shunted from heme synthesis and converted to precorrin-2 to synthesize vitamin B12, coenzyme F430, and siroheme [43]. The Ahb enzymes of some archaea and sulfur-reducing bacteria can convert siroheme (produced from uroporphyrinogen) to heme [44,45]. The contribution of the Ahb alternative heme pathway has not been demonstrated in bacterial pathogens.
In Gram-negative organisms, as well as eukaryotes, coproporphyrinogen III is converted to proto-porphyrinogen IX by coproporphyrinogen III oxidase. This step is the first of the terminal three steps in the classical heme synthesis pathway (in blue in Fig. 1) and is catalyzed by oxygen dependent HemF or by oxygen independent HemN [46,47]. Protoporphyrinogen IX is subsequently oxidized to form proto-porphyrin IX, by a six-electron oxidation catalyzed by one of three protoporphyrinogen oxidase enzymes. HemG, in Gammaproteobacteria and some Alphaproteobactera and Deltaproteobacteria, uses the respiratory chain as its electron acceptor and is not dependent on oxygen [48]. HemJ is poorly characterized but represents the most common protoporphyrinogen oxidase among Alphaproteobactera and Deltaproteobacteria [49]. The third protoporphyrinogen oxidase is HemY, an FAD- and oxygen-dependent protoporphyrinogen oxidase found in some Proteobacteria as well as eukaryotes [50]. The final step of the classical pathway is the insertion of ferrous iron by protoporphyrin ferrochelatase (HemH) to form protoheme IX, called heme [51]. From ALA to heme, the steps of the classical synthesis pathway are shared by eukaryotes and Gram-negative bacteria.
The terminal steps of the classical pathway were considered universally conserved for all heme synthesizing organisms. However, just in the last few years, the terminal steps of heme synthesis in the Gram-positive phyla Firmicutes and Actinobacteria have been described with genomic and biochemical analysis and termed the non-canonical or transitional pathway [34,52]. Very few HemF or HemN coproporphyrinogen oxidases can be identified in Gram-positive genomes; instead it has been realized that the annotated HemY in these organisms functions as a coproporphyrinogen oxidase to form coproporphyrin III [34,53]. The Gram-positive HemH, a coproporphyrin ferrochelatase, inserts ferrous iron to form coproheme [52]. Finally, coproheme is decarboxylated by HemQ, an enzyme unique to members of the Firmicutes and Actinobacteria to form protoheme IX [54–57]. It is now clear that Grampositive organisms utilize a unique series of terminal steps to synthesize heme (in green in Fig. 1).
Regulation of heme synthesis
Despite the vital role of heme to bacterial physiology, the regulation of heme biosynthesis has not been well studied outside of a few model organisms. In bacteria, regulation has been recognized to occur largely at two steps, abundance of the initial enzyme HemA and transcription of the coproporphyrinogen oxidase enzymes. Regulation of HemA is typically heme-dependent, indicating that bacteria reduce synthesis of heme and all intermediates in heme-replete conditions. This process has been extensively studied in Escherichia coli and Salmonella enterica serovar. Typhimurium. The addition of heme to cell extracts of E. coli reduces total HemA activity, without inhibiting the activity of the purified enzyme [58,59]. This was explained by the observation that excess heme results in the proteolytic degradation of HemA in Salmonella, suggesting that HemA might bind excess heme [60]. The Clp and Lon proteases are responsible for this reduction in HemA levels [61]. Furthermore, mutations in HemA have been described that render HemA resistant to heme- and protease-mediated degradation, indicating that HemA binds excess heme, and holo-HemA but not apo-HemA is a substrate for proteolytic degradation [62,63]. In this manner, cellular levels of heme can regulate the first step of heme synthesis and limit the unnecessary synthesis of heme intermediates as well as the consumption of iron. Recent metabolic engineering efforts to enhance ALA production in E. coli suggest that protoporphyrin IX post-translationally inhibits HemB, an additional example of feedback inhibition [64]. It is likely that for many organisms, heme and terminal heme intermediates can have post-translational regulatory effects on heme synthesis enzymes. Like Salmonella and E. coli, the Gram-positive bacterium Bacillus subtilis regulates levels of HemA. While a mechanistic explanation has not been described, the membrane protein HemX post-transcriptionally regulates HemA abundance in B. subtilis [38,65]. Homologs of B. subtilis HemX exist in multiple Gram-positive pathogens; however, the function of HemX and HemA regulation has yet to be detailed.
In addition to the regulation of HemA enzyme levels, the transcription of hemA is also a point of control for heme biosynthesis. Two promoters exist upstream of hemA in the Gram-negative pathogen Pseudomonas aeruginosa, and these promoters contain binding sites for the regulators Anr (oxygen sensing), Dnr (redox regulator), IHF (integration host factor), and NarL (nitrate regulator) [66,67]. Therefore, hemA expression is induced in the presence of oxygen or when oxygen is lacking but an alternative electron acceptor such as nitrate is present for utilization of heme-dependent respiration. In B. subtilis, hemEHY is induced anaerobically and hemAXCBL is induced by peroxide through de-repression of PerR [38,68]. As in B. subtilis, PerR has been implicated as a regulator of the hemEHY operon in Staphylococcus aureus; yet recent work has demonstrated that major differences exist between B. subtilis and S. aureus PerR orthologs, and therefore, it is difficult to conclude that PerR plays a role in S. aureus heme synthesis [69,70]. Corynebacterium diphtheriae, a member of the Actinobacteria phylum, encodes two heme-responsive two-component systems (TCS). The response regulator HrrA directly binds the promoters of hemA, hemE, and hemH to repress their transcription in heme-replete conditions [71]. Similarly, ChrA can repress transcription of hemA in heme replete conditions [72,73]. These data suggest that in C. diphtheriae, heme utilization is preferred over synthesis when exogenous heme is available. Together, these examples point to the transcriptional and post-translational control of HemA as a central step in heme synthesis regulation.
The expression of coproporphyrinogen oxidase genes is the second major point of heme synthesis regulation. In several species, hemF and hemN are regulated by different oxygen- or anaerobic-responsive regulators to ensure proper expression of oxygen-dependent or oxygen-independent coproporphyrinogen oxidases. OxyR, a global regulator in E. coli, is responsible for the induction of oxygen-dependent hemF expression in hydrogen peroxide stress. It has been suggested that the [Fe-S] cluster in oxygen-independent HemN is vulnerable to peroxide damage, so HemF is produced to take the place of HemN [74]. In B. subtilis, the transcription of coproporphyrinogen III oxidases hemN and hemZ (a second coproporphyrinogen oxidase, not to be confused with oxygen-dependent HemY) is induced anaerobically by the regulatory cascade of ResDE, Fnr, and YwiD to replace the oxygen-dependent HemY [75–78]. Similarly, Pseudomonas hemF and hemN are expressed anaerobically under the control of Anr and Dnr, while Anr induces the expression of only hemN aerobically [79]. It has been suggested, but not validated, that the expression of oxygen-dependent hemF in oxygen limited conditions by Anr and Dnr serves to consume residual oxygen during the transition to anaerobiosis, which would protect other anaerobically induced oxygen-sensitive proteins [79]. Thus, oxygen is a key regulatory of expression of coproporphyrinogen oxidase genes.
Contribution of heme synthesis to pathogenesis
With a few notable exceptions including Bartonella hensaela, Enterococcus faecalis, Haemophilus influenzae, and Streptococcus spp., most human pathogens encode complete heme biosynthetic pathways [80–83]. However, the contribution of heme synthesis to the pathogenesis of bacterial pathogens is largely understudied. For S. aureus, whose reliance on heme acquisition during infection has been well established, it is now clear that heme biosynthesis is vital to cause disease in murine models of infection [84–86]. Inactivation of hemA, which renders S. aureus heme deficient, causes the small-colony variant (SCV) phenotype [87]. During systemic infection, this mutant is highly defective at colonizing the murine heart and liver relative to wildtype S. aureus [87]. A mutant lacking hemB, also a heme-deficient SCV, demonstrates reduced colonization and bone destruction in a murine model of osteomyelitis [88,89]. These data demonstrate that for S. aureus, heme acquisition is insufficient to support organ colonization and therefore heme biosynthesis is critical to pathogenesis. Importantly, the SCV phenotype is encountered clinically. Despite their reduced virulence, SCVs are generally more resistant to antibiotics and oxidative stress, more equipped to evade the immune system by living intracellularly, and are likely the etiological agent of persistent staphylococcal infections [89–92] (reviewed in Ref. [93]).
Less evidence for the role of heme synthesis during infection is available for other pathogens. For the intracellular pathogen Brucella abortus, hemH is required for virulence in a murine model of brucellosis [94]. Therefore, like S. aureus, host heme utilization is insufficient and synthesis is required for full virulence. In addition to B. abortus and S. aureus, the advent of whole genome in vivo analysis of mutants using techniques such as transposon-sequencing and signature tagged mutagenesis has highlighted the role of heme synthesis. In these infections, genes with marked mutations that are recovered at a lower frequency from the infected tissue relative to growth in vitro are considered important to infection. These types of experiments have demonstrated a role for different heme synthesis genes during infection. Transposon mutantsdisrupted in hemY were found to be defective for P. aeruginosa colonization of the murine gastrointestinal tract [95]. hemN was found to be important for Yersinia pestis infection of deep tissue [96]. Transposon mutants lacking hemE in Acinetobacter baumannii were less effective at colonizing the murine lung [97]. Finally, hemG was found to be important for Listeria mono-cytogenes oral infection [98]. Based on these trans-poson library infections, heme synthesis is vital to the fitness of a variety of pathogens.
Current challenges and opportunities
The divergence between the terminal steps of Gram-positive heme synthesis and the classical pathway utilized by Gram-negative organisms as well as humans presents the opportunity for targeted small molecule interventions to inhibit or activate Gram-positive heme synthesis. The terminal Gram-positive enzymes HemQ, which exists only in Actinobacteria and Firmicutes, as well as HemY and HemH, which recognize different substrates than the eukaryotic host enzymes, present three potential targets. Small molecules have been described that modulate heme synthesis in vivo, while in vitro inhibitors of S. aureus HemY have recently been reported, suggesting that Gram-positive heme synthesis is an attractive drug target [52,99,100].
Outside of a few model pathogens, very little is understood regarding the regulation of heme synthesis, particularly during pathogenesis. Regulation is a central question in understanding the role of heme synthesis in infection. Considering that in some niches host heme is available and can reach toxic levels, pathogens with the capacity to both steal and synthesize heme must regulate both pathways. For S. aureus, in which heme synthesis and acquisition are vital during infection, the regulation of heme synthesis is unknown. This is despite the observation over half a century ago that the rate of staphylococcal heme synthesis is modulated by exogenous heme [101]. For other pathogens, the contribution of heme synthesis to disease is still unclear, but whole-genome in vivo fitness experiments like transposon-sequencing suggest many bacterial pathogens rely on heme biosynthesis to cause disease, and this field of research provides ample opportunity for further exploration.
Gram-Positive Heme Acquisition Strategies
Bacterial pathogens utilize a variety of heme acquisition strategies during infection, ranging from surface receptors to secreted proteins that bind either heme or hemoproteins. Heme acquired from the host is used fully intact or degraded to liberate heme-iron and both processes are important during bacterial pathogenesis. Gram-positive pathogens, including S. aureus, Bacillus anthracis, and C. diphtheriae rely on heme acquisition during infection. The heme uptake pathways of these three pathogens will be presented as models for theGram-positive processes, along with the regulation of the pathway and evidence for the role of heme uptake during pathogenesis.
The S. aureus Isd paradigm
The Iron-regulated surface determinant system (Isd), first described in S. aureus, is the paradigm for Gram-positive heme acquisition [102]. During infection, S. aureus utilizes the leukocidins HlgAB and LukED to lyse erythrocytes and liberate Hb into the bloodstream [103]. This results in accessible free heme, heme bound by hemopexin (Hx), free Hb (Hb), and Hb bound by Hp to form the Hp–Hbcomplex. The Isd system enables utilization of free heme, or heme bound to Hb and Hp–Hb complexes. Isd proteins bind heme and Hb at the cell wall surface with conserved near transporter (NEAT) domains. The NEAT domains are 120–125 aa domains that constitute a conserved eight-stranded β-sandwich fold [104,105]. Heme is bound in a hydrophobic pocket with critical coordination by tyrosine residues in a YXXXY motif. These NEAT-containing surface proteins (IsdB, IsdH, IsdA in S. aureus) shuttle heme to NEAT-containing IsdC. IsdC transfers heme to the membrane-associated transporter IsdDEF for transit across the membrane. To access host heme and hemoproteins, IsdB, IsdH, and IsdA are covalently attached to the peptidoglycan by the standard Sortase A cysteine transpeptidase [106]. IsdB contains two NEAT domains, NEAT1 (N1) binds Hb and Hb–Hp, but not Hp and N2 binds heme; as such IsdB is believed to be the primary Hb-binding protein [85,107,108]. IsdH contains three NEAT domains, N1 and N2 bind both Hb and Hp, andN3bindsheme [109,110]. IsdA, which is partially surface exposed, contains a single heme-binding NEAT domain [102]. The current model (Fig. 2), supported by strong structural evidence, suggests that IsdB-N1 binds Hb, and IsdB-N2 extracts heme [111]. Similarly, IsdHN1 and N2 bind Hb and Hp, and IsdH-N3 extracts the heme. The heme is then transferred either directly to IsdC or shuttled via IsdA to IsdC.
Fig. 2. Gram-positive heme uptake systems.
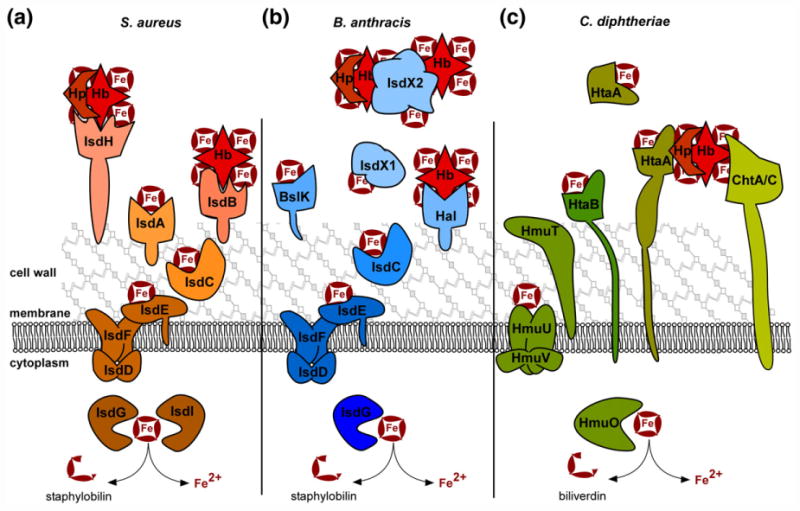
The iron-regulated surface determinant (Isd) systems for heme acquisition in S. aureus and B. anthracis, as well as the non-Isd systems of C. diphtheriae are diagrammed. Host Hb, Hp-bound Hb, and free heme (Fe-containing ring) can serve as heme sources during infection. (a) In S. aureus, IsdH is the primary Hp–Hb receptor and IsdB is the principle Hb receptor. Both are sortase-linked on the surface of the cell wall, bind host hemoproteins with NEAT domains, and extract heme using additional NEAT domains. IsdA can bind free heme or accept heme from IsdB and IsdH. Heme is transferred to IsdC, which is embedded in the cell wall and transits heme to the membrane complex IsdDEF. IsdDEF transports heme to the cytoplasm for utilization intact or for degradation by the heme oxygenases IsdG/I. (b) Similarly, B. anthracis uses Isd proteins to acquire heme. IsdX1 and IsdX2 are secreted hemophores that bind Hb, Hp–Hb, or free heme as depicted. IsdX2, which has five NEAT domains, may also serve as a heme storage protein. Additionally, the sortase anchored Hal serves as a Hb receptor on the cell surface and uses its NEAT and leucine-rich repeat domains to acquire heme. BslK is cell wall associated and binds heme via its NEAT domain. IsdC transports heme to the IsdDEF membrane importer for utilization or degradation by IsdG. (c) C. diphtheriae utilizes a unique set of heme uptake proteins for heme utilization. HtaA is a cell wall spanning lipoprotein that can acquire heme from Hp–Hb in conjunction with ChtA or ChtC. HtaB can bind free heme or accept heme transfer from HtaA and transfers heme to the HmuTUV membrane transporter. A portion of HtaA may also serve as a secreted hemophore. C. diphtheriae HmuO heme oxygenase can liberate iron from imported heme.
S. aureus encodes an iron-regulated Sortase B (SrtB) for which IsdC is the only substrate, and SrtB attaches IsdC to peptidoglycan in such a way that IsdC is not surface exposed but rather buried in the cell wall, which is 15–30 nm thick [112,113]. This organization allows heme transferred from surface Isd proteins to pass through the cell wall to the membrane by IsdC's single heme-binding NEAT domain. IsdC alone transfers heme to the IsdE of the IsdDEF transporter [114]. At the membrane, IsdDEF transit heme across the membrane and into the cytosol.
Upon import, heme is incorporated into staphylococcal proteins or degraded. Exogenous heme accumulates in the membrane and is also capable of complementing the growth of heme-deficient mutants [84]. Alternatively, the heme oxygenases IsdG and IsdI degrade heme to release iron [115] (reviewed in Ref. [116]). IsdG and IsdI are structurally similar and are the first described members of the Isd heme oxygenase family, which catabolizes heme to staphylobilin instead of biliverdin [117–119]. IsdG and IsdI are required for growth using heme as a sole iron source and are expressed during infection [115,120].
The widely conserved ferric uptake regulator (Fur) is the principle regulator of the expression of heme acquisition systems in S. aureus. In iron-deplete conditions, Fur no longer represses its regulon, allowing the transcription of the isdB, isdA, isdC-DEFsrtBisdG, and isdI loci [102]. During infection of iron-deplete niches, the heme acquisition system and associated iron-liberating heme oxygenases are expressed. Further regulation of the heme oxygenases exists; IsdG abundance increases in the presence of heme and IsdG half-life is increased when heme-bound [120]. Also, the Clp proteases have a role in Hb acquisition by modulating IsdB levels [121]. Additional heme-dependent regulation likely exists but has not been described.
Isd-mediated heme acquisition is vital to the virulence of S. aureus. Heme is the preferred iron-source during systemic infection, in part because a heme-responsive transcriptional regulator activates iron siderophore synthesis only when heme-iron is unavailable [84,122]. The role of the Isd system has been extensively demonstrated in murine infection models. Mutants lacking components of the Isd system are highly defective inpathogenesis, highlighting the importance of heme acquisition to staphylo-coccal disease [84–86,108,120,123,124].
Isd-dependent heme uptake by B. anthracis
B. anthracis encodes a heme uptake system that shares the core of the S. aureus Isd, but with additional unique proteins. B. anthracis encodes two secreted hemophores termed IsdX1 and IsdX2 [125]. These are the first described Gram-positive hemophores and bind heme, Hb, and methemoglobin [125– 129]. IsdX1 contains one NEAT domain, while IsdX2 contains five NEAT domains; both are secreted past the cell wall as they lack sortase signals or membrane spanning domains [125]. B. anthracis also encodes other NEAT contain proteins; Hal contains a single NEAT domain and leucine-rich repeats, which extract heme from Hb [130]. Unlike IsdX1/2, Hal is sortase anchored to the cell wall [131]. A second, recently described NEAT protein is BslK, which is non-covalently attached to the cell wall and transfers heme to IsdC [132]. The current proposed model (Fig. 2) is that IsdX1 is secreted, binds heme, and transfers heme to wall-anchored IsdC. IsdX2 can bind free heme, accept heme from IsdX1, and transfer heme to IsdC. The multiple NEAT domains ofIsdX2 have been proposed to be important for these multiple functions, and it has been suggested that IsdX2 can serve as a heme storage protein. IsdDEF transports heme across the membrane for utilization by IsdG, an orthologue of the S. aureus heme oxygenase [133]. The diversity of heme and Hb-binding proteins relative to S. aureus may be the result of the greater variety of environmental niches that germinant and sporulent B. anthracis inhabits.
The role of B. anthracis heme acquisition during infection is not clear. A guinea pig infection model demonstrated that ΔisdCX1X2 was as virulent as wild type, yet these proteins are expressed during infection [134]. Also, a mutant of B. anthracis lacking Hal demonstrated reduced virulence in a model of inhalational anthrax [135]. It is likely that the IsdX1/X2 hemophores, BslK, and Hal are partially redundant, and a mutant lacking all four proteins would be highly defective in causing anthrax.
In addition to S. aureus and B. anthracis, many other pathogens have evolved NEAT-containing heme acquisition systems, including Staphylococcus lugdunensis, L. monocytogenes, and Streptococcus pyogenes [136–143]. The conservation of NEAT-mediated heme uptake highlights the contribution of host heme to bacterial infection.
C. diphtheriae heme uptake
C. diphtheriae utilizes non-NEAT-mediated heme uptake systems for heme-iron acquisition, termed HmuTUV, HtaABC, and ChtABC/CirA. The Hmu (hemin-uptake) system was the first heme acquisition system described in Gram-positive organisms. The associated heme oxygenase, HmuO, was discovered and described first, and then HmuTUV was discovered for the ability of a plasmid encoding hmuTUV to complement a Corynebacterium ulcerans strain that cannot grow on Hb as a sole iron source [144,145]. Sequence analysis suggests that HmuTUV acts asan ABC transporter that shuttles heme across the cell membrane [146]. It was later discovered that an additional gene is encoded within the hmuTUV operon, termed htaA (heme-transport associated) [147]. Adjacent to this locus are the genes htaB and htaC. Unlike the sortase anchoring of other Gram-positive uptake systems, HtaA and HtaB contain N-terminal secretion signals as well as C-terminal intermembrane domains. This results in surface exposure of HtaA and HtaB, which both bind heme. Interestingly, a portion of HtaA is secreted and not anchored to the cell envelope. HtaA isolated from cell culture is unable to complement the growth of an htaA mutant, suggesting that surface bound HtaA may serve as a heme receptor and secreted HtaA may serve as a hemophore [147,148]. However, heme transfer between HtaA molecules and further description of the function of HtaA on the surface have not been reported. In addition to heme, HtaA can acquire heme from Hb and transfer heme to HtaB, suggesting a heme shuttle from HtaA toHtaB toHmuT; HmuT isa surface-anchored lipoprotein, which then transfers heme to the cognate ABC transporter HmuUV [148]. While the Isd NEAT domains rely on tyrosine alone as the axial ligand for heme binding, HmuT relies on an N-terminal histidine and a C-terminal tyrosine to coordinate heme [149].
Inactivation of the Hmu/Hta systems does not completely eliminate growth with heme as a sole iron source, suggesting the involvement of an additional heme uptake system [147]. This led to the characterization of the ChtAB and CirAChrC operons, which are regulated by iron levels via DtxR. DtxR is the Diphtheria Toxin regulator which activates the expression of Diphtheria Toxin as well as HmuTUV and HtaABC [150,151]. ChtAB and ChtC appear to be the result of gene duplication of HtaAB, as all three groups of proteins have sequence similarity, N-terminal secretion signals, and C-terminal transmembrane domains, and contain the same heme-binding domain [152]. Like HtaAB, ChtAB and CirAChtC are surface exposed and ChtAB and ChtC bind heme and Hb. It appears that these heme-binding proteins serve redundant functions, and as such, a mutant lacking both HtaB and ChtB is deficient at utilizing Hb as an iron source [152]. Recently, it has been shown that ChtA and ChtC are both capable of binding Hp–Hb for heme extraction, and acquisition of heme from Hp–Hb requires HtaA [153]. The current model (Fig. 2) for Hp–Hb heme acquisition involves binding of Hp–Hb by a combination of HtaA and ChtA or ChtC, heme extraction either actively or passively, and transfer to HtaB, HmuT, and HmuUV [153].
Gram-Negative Heme Acquisition Strategies
The outer membrane of the cellular envelope of Gram-negative organisms presents an additional barrier to heme acquisition. Therefore, Gram-negative heme uptake systems consist of outer-membrane receptors that either bind heme and hemoproteins directly, or bind heme-bound secreted hemophores. Heme then transits the periplasm and is brought into the cell via ABC transporters at the inner membrane. The versatile opportunistic pathogen P. aeruginosa encodes direct heme uptake and hemophore systems at the outer membrane, H. influenzae uses a hemophore uptake system, and Neisseria meningitidis uses a unique bipartite receptor for heme acquisition from host hemoproteins . These pathogens are presented as models for Gram-negative heme uptake systems.
P. aeruginosa
P. aeruginosa encodes direct and indirect systems for heme uptake. The Phu (Pseudomonas heme uptake) consists of a TonB-dependent PhuR which binds heme and transports it to the periplasm. PhuR activity is representative of Gram-negative TonB-dependent outer-membrane receptors. These β-barrel proteins bind substrates (often iron containing molecules) with high affinity, and rely on proton motive force and TonB for transport across the outer membrane [154]. TonB is an inner-membrane protein with a substantial periplasmic portion for direct interaction with periplasmic domains of the outer membrane proteins. Upon PhuR translocation of heme into the periplasm, the soluble periplasmic protein PhuT binds heme and brings it to PhuUV, an ABC transporter at the inner membrane.
In addition, HasA/HasR (heme assimilation system) is utilized for heme uptake. HasA is a secreted hemophore that binds heme and transfers it to a second TonB-dependent transporter, HasR. Like other Gram-negative heme-binding motifs, HasA coordinates heme using histidine and tyrosine residues with picomolar affinity. Data from the orthologous HasA hemophore of Serratia marcescens suggest that HasA binds Hb and extracts heme, then HasA transfers heme to HasR [155,156]. The present model (Fig. 3) for these two heme uptake systems suggests that Phu is the principle heme acquisition system but full heme utilization requires HasA/HasR. HasA/HasR may be more relevant as a heme sensing system; under low heme conditions, the inner-membrane HasS binds the sigma factor inhibitor HasI. When heme is available, HasS instead binds HasR, and HasI is free to recruit RNA polymerase to activate the transcription of hasAR, hasSI, phuSTUV, and phuR [157]. The P. aeruginosa heme uptake system PhuSTUV/PhuR is regulated by Fur in addition to the HasI sigma factor detailed above. Recently, small regulatory RNAs have been described that impact phuS mRNA levels, suggesting another layer of heme-responsive regulation [158,159]
Fig. 3. Gram-negative heme acquisition.

The heme uptake systems as described in the text are depicted. (a) P. aeruginosa PhuR binds heme at the outer membrane and imports heme into the periplasm in a TonB-dependent manner. Heme is transferred to PhuT, which subsequently transfers heme to the PhuUV inner-membrane transporter for transit into the cytoplasm. There, PhuS binds and stores heme or transfers heme to the heme oxygenase HemO for iron utilization. P. aeruginosa also secretes the hemophore HasA which binds Hb or free heme, and transfers heme to the TonB-dependent outer-membrane receptor HasR. The fate of HasR imported heme is not fully understood, but may be trafficked to PhuTUV for import. HasS serves as an inner-membrane sensor and regulates expression of the has and phu systems through the sigma factor HasI (not shown). (b) H. influenzae can utilize a variety of host heme sources. Secreted HxuA specifically binds hemopexin (Hx), and heme from Hx is transferred into the periplasm when HxuA interacts with HxuBC at the outer membrane. Independent of HxuA, HxuC can also import heme from serum albumin (Alb). HgpA, HgpB, and HgpC are highly similar outer-membrane receptors for heme acquisition from Hb complexed with Hp, free Hb, and Hp bound to myoglobin (not shown). The inner-membrane heme transporter has not been clearly defined, but the Hip system has been implicated for heme transit into the cytoplasm. Interestingly, all imported heme may be utilized intact, as no heme oxygenase has been identified yet. (c) The N. meningitidis outer-membrane, TonB-dependent complex of HpuAB can acquire heme from Hb and Hp–Hb and bring heme into the periplasm. Additionally, the HmbR outer-membrane receptor specifically extracts heme from Hb for transport. The identity of the inner-membrane heme transporter is unclear at this time, but heme somehow enters the cytoplasm where it can be utilized or degraded by the HemO heme oxygenase.
In contrast to many other organisms, Pseudomonas encodes a soluble cytoplasmic heme-binding protein that is not a heme oxygenase. This protein, PhuS, transfers heme to the heme-oxygenase HemO for iron liberation. PhuS, unlike many hemoproteins, binds ferric-iron heme and subsequently transfers it to HemOunder iron-deplete conditions [160]. The dissociation constant of the heme–PhuS–HemO complex is in the nanomolar range, suggesting that PhuS transfers heme to HemO specifically and not to the second Pseudomonas heme oxygense, BphO [160]. While the PhuS heme transfer has not been described completely, PhuS has been shown to bind heme as a monomer utilizing one of two histidine residues (His209 and His212), and a third binding site exists when PhuS is in dimeric form [161]. Further in vitro characterization and structural analysis has led to a model whereby heme coordination occurs primarily at the His212 ligand and induces a conformational change required for interaction with HemO [162,163]. Additionally, in vitro heme oxygenase activity has been attributed to PhuS; however, the in vivo relevance of this function is unclear as no biliverdin-β (the product of HemO heme catabolism) is detected in a mutant lacking hemO [164,165].
A recent clinical evaluation of genetic changes to P. aeruginosa during infection of cystic fibrosis lungs revealed the importance of heme acquisition during infection [166]. Long-term infection led to the selection of mutations in the promoters of the phuSTUWV and phuR loci, resulting in greater Phu expression. These changes to phu transcription confer a growth advantage enabling the utilization of heme from Hb as the sole iron source and suggest that heme is an important iron source during chronic Pseudomonas infection. The infections also selected for mutants that demonstrate enhanced expression of the feo ferrous-iron acquisition genes, indicating that ferrous iron is also a source of bioavailable iron. These clinical data confirm experimental findings suggesting that P. aeruginosa heme acquisition contributes to chronic infection.
H. influenzae
H. influenzae is a notable exception to the other pathogens outlined here, as it is incapable of synthesizing heme and therefore requires heme uptake for aerobic respiration [167]. It is capable of acquiring heme from diverse host sources (Fig. 3), including hemopexin, free heme, albumin-bound heme, myoglobin, and Hb; the variety of heme sources is in accordance with its absolute reliance on exogenous heme [168]. H. influenzae has evolved a variety of heme uptake systems important for growth in vitro using various host heme sources. While some systems are well described, less is known about others, and a global understanding of the utilization of these heme uptake systems during infection is lacking.
The HxuCBA system, described primarily in H. influenzae type B, is capable of heme acquisition from free heme and heme-hemopexin (Hx). HxuA is a secreted hemophore that is released from the outer membrane by its transporter HxuB [169–171]. HxuA exhibits no heme-binding motif but rather demonstrates high-affinity binding specifically to Hx with little distinction between apo- and holo-Hx [172]. HxuC is a TonB-dependent transporter that binds heme after release from the Hx–heme–HxuA complex and imports it into the periplasm [173]. Additionally, HxuC is capable of acquiring heme from serum albumin (Alb) independent of HxuA [174]. HpbA is another heme acquisition protein identified in nontypeable and type B H. influenzae. A lipoprotein, HbpA is important for growth using Hb, Hp–Hb, and heme-human serum albumin as heme sources [175,176]. The inner-membrane heme transporter has not been definitively identified, but the Hip proteins have been implicated [174].
Additionally, H. influenzae encodes three receptors, HgpA, HgpB, and HgpC, that can acquire heme from Hp–Hb and Hp bound myoglobin, albeit it at greater concentrations than thought to be physiologically relevant [177,178]. While the contribution of the Hgps seems redundant, HgpB has been demonstrated to be most important for utilization of Hp–Hb and Hp–myoglobin.
There are many outstanding questions regarding H. influenzae heme uptake. Many proteins have been attributed to be involved in heme uptake, but their function requires further investigation [179–183]. The regulation of the heme uptake system expression is not well described, except that hxuCBA and the hgp genes are expressed under in vitro iron/heme deplete conditions during experimental infection of the chinchilla ear [184,185]. Lastly, a heme oxygenase of Haemophilus has not been described, suggesting that acquired heme is utilized intact and that other iron acquisition pathways, from transferrin and lactoferrin sources, are sufficient for cellular iron needs. However, it is also possible that a heme oxygenase exists and has not yet been identified.
Genetic evidence from clinical isolates suggests that heme uptake is vital to pathogenic strains of H. influenzae. Isolates from otitis media infection in children relative to commensal throat isolates exhibit greater rates of hxuA, hxuB, hxuC, and hgpB gene prevalence, indicating that heme uptake may be a virulence determinant [186,187]. Several animal models have been used to demonstrate the role of heme uptake during H. influenzae infection. In a model of H. influenzae bacteremia, infant rats infected with a mutant lacking HbpA completely clear the infection after one week while rats infected with wildtype remain infected [176]. Likewise, a mutant lacking both HxuC and HgpABC uptake proteins is unable to cause bacteremia in the same rat model [188]. Additionally, the Hgp proteins are required to cause otitis media in a chinchilla model [189]. It is clear that for H. influenzae pathogenesis, heme uptake is a critical virulence determinant.
N. meningitidis
N. meningitidis encodes a bipartite heme uptake system consisting of HpuAB and HmbR (Fig. 3). HpuAB is expressed from an iron-repressed operon and consists of the HpuA lipoprotein and HpuB, the TonB-dependent receptor capable of binding Hb, apo-Hp, and Hp–Hb [190,191]. Upon heme transport into the cytoplasm, the HemO heme oxygenase degrades heme to biliverdin and liberates iron. As such, HemO is required for survival using heme, Hb, or Hp–Hb as a sole iron source [192,193]. Heme is extracted from these hemoproteins and is imported intact, as Hb can complement the deficiencies of a heme synthesis mutant in an HpuAB-dependent manner [194]. The inner-membrane transporter has not yet been identified, but a zinc transporter has been implicated [195].
Initial studies of the individual function of HpuA and HpuB failed to describe the role of HpuA in heme acquisition. HpuB is sufficient to bind Hb, but a high-affinity HpuB-Hb complex requires the presence of HpuA, even though HpuA-Hb binding was not detected by a flow cytometry assay [196,197]. Additionally, HpuA is required for growth with Hb as a sole iron source and heme import [198]. However, a recent structural characterization has described a direct, albeit weak, interaction between HpuA and Hb, and a co-crystal structure of Hb and an HpuA homolog from Kingella denitrificans has been solved [199]. While these data are not conclusive, they suggest that HpuA and HpuA homologs interact with Hb, and this interaction is required for HpuAB-mediate heme uptake.
HmbR (Hb receptor) is an additional N. meningitidis heme uptake protein that binds host Hb with species specificity, exhibiting a greater utilization of human Hb but is unable to bind the Hp-Hb complex and therefore likely binds free Hb only [200,201]. Like HpuAB, it is subject to phase variation [202]. HmbR, based on spectroscopy and mutational analysis, also coordinates heme with a Tyr residue, which further confirms that diverse heme-binding domains have evolved to utilize tyrosine as the axial ligand [203]. The mechanism of heme extraction by HmbR, the associated inner-membrane heme transporter that partners with HmbR extraction, and structural descriptions of ligand binding are still undescribed for HmbR heme uptake.
In N. meningitidis, expression of hemO and hmbR is regulated by Fur as well as the MisRS TCS [204,205]. MisRS activates the expression of hemO and hmbR independent of Hb and iron concentration, which suggests an additional layer of regulation for Hb acquisition. However, the activating signal of MisRS has not yet been described.
The genetic diversity of N. meningitidis clinical isolates has highlighted the importance of heme uptake to meningococcal virulence. While not all N. meningitidis strains express both the HmbR and HpuAB systems, most express at least one. Most pathogenic isolates express at least HmbR, but HpuAB expression is equally associated with disease and carriage isolates, which indicates that HmbR is an indicator of pathogenesis [206,207]. N. meningitidis serotype B isolates associated with disease also exhibit “on” phase variation of HmbR, correlating virulence with the expression of HmbR [208]. Additionally, HmbR is required for virulence in an infant rat model of meningitis [200]. These data implicate heme uptake, particularly HmbR, as an important component of Neisseria infection.
Current challenges and opportunities
Study of heme uptake strategies has offered great insight into bacterial pathogenesis and nutrient acquisition. There is still great opportunity for discovery. For most bacterial heme-binding motifs, the transfer from host hemoprotein has not been demonstrated as either passive dissociation or active extraction. The redundancy of heme uptake systems in pathogens like B. anthracis, P. aeruginosa, and H. influenzae is well appreciated, but the role of each system during infection of various niches or commensal environments has yet to be fully elucidated. The relative contribution of host heme to iron acquisition by bacterial pathogens during infection is understudied. It is unclear if pathogens rely on heme for iron in unique spatiotemporal niches and rely on ferrous iron and siderophore acquisition systems in other niches. Opportunity abounds to understand the role of heme-iron utilization across time and tissues during infection. Finally, while global abundance of heme and Hb in the host has been measured, the local availability of heme and hemoproteins during infection has not been described and presents an opportunity to understand the microenvironment of an infectious niche as well as the host response to infection.
In terms of clinical application, heme uptake systems may be attractive therapeutic targets. S. aureus Isd proteins have been the target of vaccine development with mixed success and monoclonal antibodies against IsdB have been studied for therapeutic use [209–213]. Considering the importance of heme acquisition to infection, using surface-exposed heme uptake proteins as targets for vaccine and antibodies should continue to be investigated. Additionally, the Mycobacterium tuberculosis heme uptake system, which comprises three unique proteins and is sufficient to rescue the growth of a heme auxotroph, has been proposed as a new mycobacteria-specific antimicrobial target to be explored [214–216]
The interactions between host hemoproteins and bacterial hemoprotein binding proteins offer an excellent opportunity to study host–pathogen co-evolution. It has been recently demonstrated that the human and primate iron-binding protein transferrin has undergone positive selection at the interface of binding by bacterial transferrin receptors, suggesting that the co-evolution of humans and pathogens has produced an evolutionary arms race in the context of nutritional immunity [217,218]. In the same vein, the Hb-binding IsdB of S. aureus exhibits species specificity and more efficiently utilizes human Hb relative to mouse Hb [86]. In keeping with this, transgenic mice expressing human Hb are more susceptible to S. aureus disease [86]. The contribution of bacterial heme acquisition to human evolution presents ample opportunity to further investigate co-evolution and nutritional immunity.
Heme Toxicity and Tolerance
Bacterial pathogens dedicate extensive cellular machinery to the synthesis and acquisition of heme. Paradoxically, excess heme is toxic and thus during infection, invading pathogens must contend with heme toxicity as a component of pathogenesis. While heme toxicity is well studied in eukaryotes, less is known in bacteria [25,219,220]. A brief description of heme toxicity in bacteria and strategies utilized to combat toxicity follow.
Multi-faceted mechanism of heme toxicity
The reactive nature of heme that makes it such a versatile cofactor also results in toxicity at excess concentrations. While the toxicity of heme toward bacteria has been observed for over 60 years, a complete understanding of the mechanisms of heme toxicity is lacking [26,221]. Free heme is rapidly bactericidal toward various Gram-positive and Gram-negative pathogens in low- to mid-micromolar concentrations [33,222-225]. However, investigation of heme toxicity in a variety of bacterial species has led to a model of heme inducing iron- and non-iron-related damage to the cell.
The accumulation of heme results in excess iron by one of two mechanisms, both of which are likely at play under aerobic conditions. First, a portion of iron is freed by the heme oxygenases. Second, iron itself may be liberated from the porphyrin ring upon reaction with reactive oxygen species (ROS). Irrespective of the source, iron can cycle between ferrous and ferric states via Fenton chemistry and the Haber-Weiss reaction (reaction 1), yielding a regenerating supply of ROS.
| (1) |
| (2) |
Iron-mediated production of ROS can damage DNA, lipids, and proteins [226,227]. Further evidence for the contribution of oxidative stress to heme toxicity comes from S. aureus. In conditions of excess heme toxicity, membrane proteins are highly oxidized and superoxide is formed by redox cycling of heme-iron through membrane menaquinone [228]. Superoxide production is a separate source of oxidative damage from ferrous iron-mediated ROS and is a major component of heme damage in S. aureus [228]. In addition to experimentally validating that heme-mediated ROS is a key to heme toxicity, this work also localized heme toxicity primarily to the membrane. The lipophilic nature of heme suggests that it partitions to the membrane of bacteria, and this has been demonstrated in S. aureus, likely resulting in damage to membrane proteins and lipids [84].
Further evidence suggests that iron-mediated ROS production and subsequent membrane damage are an insufficient description of heme toxicity. First, heme is toxic in anaerobic conditions, and second, non-iron protoporphyrins are toxic to bacteria and activate the cellular response to heme toxicity [229–231]. Also, porphyrins cause significant damage to bacterial DNA [232]. Finally, resistance to heme toxicity is in part mediated in N. meningitidis by Ght (gene of hydrophobic agent tolerance), suggesting that damage by heme is similar to other hydrophobic molecules and may disrupt the Gram-negative outer membrane [224,233]. The toxicity of heme is likely the result of a combination of membrane disruption, membrane protein and lipid oxidation, and DNA damage. However, a total understanding of heme-mediated damage is far from complete.
Strategies to overcome heme toxicity
While the direct result of excess heme is unclear, it is evident that bacteria must contend with heme damage and have evolved a variety of strategies to overcome heme toxicity (Fig. 4). These systems consist primarily of efflux and sequestration. Additionally, the heme oxygenase outlined as part of heme acquisition strategies may contribute to the reduction of heme toxicity by cleaving the porphyrin ring and liberating iron for use.
Fig. 4. Strategies to avoid heme toxicity.
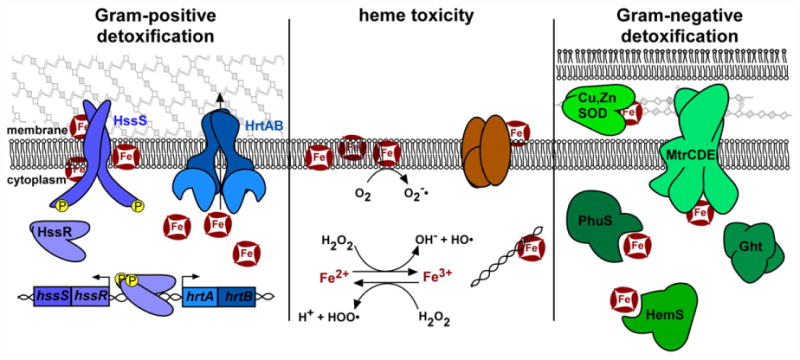
Heme toxicity (center) is a combination of heme damage to membrane lipids, membrane proteins, and DNA, and oxidative damage. Oxidative damage is mediated by the production of superoxide dismutase ( ), hydroxyl radical (HO•), and hydroperoxyl radical (HOO•). To reduce heme damage, many Gram-positive organisms (the S. aureus system is diagrammed here) encode the HrtAB efflux pump. The HssRS two-component system responds to excess heme and activates the transcription of the hrtAB system, thus preventing the accumulation of toxic levels of heme. Alternatively, Gram-negative organisms rely on intracellular heme sequestration proteins (PhuS of P. aeruginosa, HemS of Yersinia), the periplasmic heme-binding, copper and zinc dependent superoxide dismutase (Cu,Zn SOD, of H. ducreyi), and systems that respond to hydrophobic molecules, including heme (MtrCDE efflux and Ght of Neisseria).
Heme efflux strategies have been primarily characterized in Gram-positive organisms, potentially because efflux across a single-membrane barrier is simpler to achieve than in Gram-negative pathogens. Three systems have been described, HrtAB, PefAB/CD, and MtrCDE. The S. aureus heme-regulated transporter HrtAB is required for survival in toxic concentrations of heme. hrtAB expression is activated by the HssRS heme sensing TCS [222,234,235]. While the ligand of the HssS histidine kinase has remained elusive, excess exogenous or endogenous heme leads to activation, either directly or indirectly [99]. HrtA is an ATPase that drives efflux by HrtB permease of its ligand, likely heme. Orthologues of HrtAB have been described in B. anthracis and Lactococcus lactis and are required for resistance to heme toxicity in these organisms [236]. When the Hrt efflux pump is inactivated in both S. aureus and L. lactis, levels of intracellular heme increase, suggesting that heme is the substrate of HrtAB export [229,237]. In B. anthracis, an HssRS orthologue controls the expression of HrtAB and cross-talks with a second TCS that responds to cellular envelope stresses, further implicating membrane damage as a component of heme stress [238]. HrtAB is actively expressed during murine anthrax, suggesting that organisms that replicate in the bloodstream must tolerate heme toxicity [33].
Additional efflux systems exist, suggesting that this strategy is well conserved. Streptococcus agalactiae encodes an orthologue of HrtAB, as well as a dual efflux system PefAB and PefRCD [223]. In heme stress, hrtAB and pefAB/RCD are expressed at high levels, and the Pef systems are required for resistance to heme toxicity [223]. The Gram-negative N. gonorrhoeae encodes an efflux pump, MtrCDE, for hydrophobic molecules that is required for resistance to heme stress [239].
Heme sequestration and storage is a second theme in strategies to resist heme toxicity. The conserved HemS family has been described in Yersinia enterocolitica, Y. pestis, Shigella dysenteriae (termed ShuS), P. aeruginosa (called PhuS, detailed above), and E. coli (ChuS, which also has heme oxygenase activity) [146,160,225,240–244]. While a variety of heme storage, transfer, and degradation properties have been assigned to these proteins, their involvement in resisting heme toxicity is clear. Additionally, non-HemS family proteins have been found to bind heme and play a role in heme homeostasis, including the small outer-membrane Protein E of H. influenzae and the Cu,Zn superoxide dismutase of Haemophilus ducreyi [245,246].
Current challenges and opportunities
While numerous systems are involved in detoxifying heme, there are many outstanding questions. The efflux systems have been described genetically, but a complete understanding of the ligands exported is still murky. For Gram-positive pathogens, the efflux systems may provide an additional therapeutic target for infection. Inhibition of efflux may offer a treatment option for bloodstream infections by S. aureus and B. anthracis; presumably the effects of heme toxicity would be deadly to the bacterium if the HrtAB pump were pharmacologically inactivated. This strategy could also pair well with small molecule activation of heme synthesis, which has been developed [99]. In terms of heme sequestration proteins, it has been difficult to fully interpret the contribution of heme sequestration because additional properties like oxygenase (PhuS and ChuS) and DNA binding (ShuS) have been observed. Finally, the role of heme oxygenases in resisting heme stress has not been well studied.
Concluding Remarks
Heme synthesis, uptake, utilization, and toxicity have been an area of intense investigation in bacterial pathogenesis. As outlined throughout, there are many additional questions in this field. Some of the most fundamental aspects of heme homeostasis have not been studied in detail. For most pathogens, regulation of heme synthesis is unclear and the contribution of heme synthesis to infection has not been investigated. For organisms that acquire and synthesize heme, a full model of preference between exogenous and endogenous sources is unknown. Based on limited evidence, exogenous heme is preferred when available, but is there a division between exogenous and endogenous heme in the partitioning to hemoproteins and heme oxygenases? Also, when heme enters the cell or is synthesized, does it exist in the free state or are there heme chaperones analogous to metallochaperones?
As the heme uptake, synthesis, and toxicity processes are well conserved and vital to bacterial pathogenesis, they present an opportunity for therapeutic intervention. As the field gains further insight into these processes, hopefully academia and industry will pursue small molecule interventions and vaccine candidates for the treatment of the bacterial pathogens outlined in this review, many of which are recalcitrant current treatment options.
Acknowledgments
We thank members of the Skaar laboratory for critical review of this manuscript. J.E.C. is supported by NIH T32 GM065086. Heme acquisition, synthesis, and toxicity research in the Skaar laboratory is supported by NIH grants AI069233 and AI073843.
Abbreviations
- Hb
hemoglobin
- Hp
haptoglobin
- ALA
δ-aminolevulinic acid
- PBG
porphobilinogen
- HMB
hydroxymethylbilane
- Hx
hemopexin
- SCV
small colony variant
- PPIX
protoporphyrin IX
- NEAT
near-transporter domain
- Isd
iron-regulated surface determinants
- TCS
two-component system
References
- 1.Chen JJ, London IM. Hemin enhances the differentiation of mouse 3T3 cells to adipocytes. Cell. 1981;26:117–122. doi: 10.1016/0092-8674(81)90039-8. [DOI] [PubMed] [Google Scholar]
- 2.Shelver D, Kerby RL, He Y, Roberts GP. CooA, a CO-sensing transcription factor from Rhodospirillum rubrum, is a CO-binding heme protein. Proc Natl Acad Sci USA. 1997;94:11216–11220. doi: 10.1073/pnas.94.21.11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Faller M, Matsunaga M, Yin S, Loo JA, Guo F. Heme is involved in microRNA processing. Nat Struct Mol Biol. 2007;14:23–29. doi: 10.1038/nsmb1182. [DOI] [PubMed] [Google Scholar]
- 4.Bonyhady RE, Hendry IA, Hill CE, McLennan IS. Effects of haemin on neurones derived from the neural crest. Dev Neurosci. 1982;5:125–129. doi: 10.1159/000112669. [DOI] [PubMed] [Google Scholar]
- 5.Posey JE, Gherardini FC. Lack of a role for iron in the Lyme disease pathogen. Science. 2000;288:1651–1654. doi: 10.1126/science.288.5471.1651. [DOI] [PubMed] [Google Scholar]
- 6.Archibald F. Lactobacillus plantarum, an organism not requiring iron. FEMS Microbiol Lett. 1983;19:29–32. [Google Scholar]
- 7.Kessler D, Leibrecht I, Knappe J. Pyruvate-formate-lyasedeactivase and acetyl-CoA reductase activities of Escherichia coli reside on a polymeric protein particle encoded by adhE. FEBS Lett. 1991;281:59–63. doi: 10.1016/0014-5793(91)80358-a. [DOI] [PubMed] [Google Scholar]
- 8.Yamamoto I, Saiki T, Liu SM, Ljungdahl LG. Purification and properties of NADP-dependent formate dehydrogenase from Clostridium thermoaceticum, a tungsten–selenium–iron protein. J Biol Chem. 1983;258:1826–1832. [PubMed] [Google Scholar]
- 9.Peters JW, Lanzilotta WN, Lemon BJ, Seefeldt LC. X-ray crystal structure of the Fe-only hydrogenase (CpI) from Clostridium pasteurianum to 1.8 Angstrom resolution. Science. 1998;282:1853–1858. doi: 10.1126/science.282.5395.1853. [DOI] [PubMed] [Google Scholar]
- 10.Nagashima S, Nakasako M, Dohmae N, Tsujimura M, Takio K, Odaka M, et al. Novel non-heme iron center of nitrile hydratase with a claw setting of oxygen atoms. Nat Struct Biol. 1998;5:347–351. doi: 10.1038/nsb0598-347. [DOI] [PubMed] [Google Scholar]
- 11.Johnson DC, Dean DR, Smith AD, Johnson MK. Structure, function, and formation of biological iron–sulfur clusters. Annu Rev Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- 12.Shomura Y, Yoon KS, Nishihara H, Higuchi Y. Structural basis for a [4Fe–3S] cluster in the oxygen-tolerant membrane-bound [NiFe]-hydrogenase. Nature. 2011;479:253–256. doi: 10.1038/nature10504. [DOI] [PubMed] [Google Scholar]
- 13.Weinberg ED. Iron and susceptibility to infectious disease. Science. 1974;184:952–956. doi: 10.1126/science.184.4140.952. [DOI] [PubMed] [Google Scholar]
- 14.Weinberg ED. Nutritional immunity: host's attempt to withold iron from microbial invaders. JAMA. 1975;231:39–41. doi: 10.1001/jama.231.1.39. [DOI] [PubMed] [Google Scholar]
- 15.Cassat JE, Skaar EP. Iron in infection and immunity. Cell Host Microbe. 2013;13:509–519. doi: 10.1016/j.chom.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hood MI, Skaar EP. Nutritional immunity: transition metals at the pathogen-host interface. Nat Rev Microbiol. 2012;10:525–537. doi: 10.1038/nrmicro2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zackular JP, Chazin WJ, Skaar EP. Nutritional immunity: S100 proteins at the host–pathogen interface. J Biol Chem. 2015;290:18991–18998. doi: 10.1074/jbc.R115.645085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Juttukonda LJ, Skaar EP. Manganese homeostasis and utilization in pathogenic bacteria. Mol Microbiol. 2015;97:216–228. doi: 10.1111/mmi.13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wriston JC, Jr, Lack L, Shemin D. The mechanism of porphyrin formation; further evidence on the relationship of the citric acid cycle and porphyrin formation. J Biol Chem. 1955;215:603–611. [PubMed] [Google Scholar]
- 20.Kikuchi G, Shemin D, Bachmann BJ. The enzymic synthesis of delta-aminolevulinic acid. Biochim Biophys Acta. 1958;28:219–220. doi: 10.1016/0006-3002(58)90461-x. [DOI] [PubMed] [Google Scholar]
- 21.Hamza I, Dailey HA. One ring to rule them all: trafficking of heme and heme synthesis intermediates in the metazoans. Biochim Biophys Acta. 2012;(1823):1617–1632. doi: 10.1016/j.bbamcr.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rieske JS. Composition, structure and function of complex III of the respiratory chain. Biochim Biophys Acta. 1976;456:195–247. doi: 10.1016/0304-4173(76)90012-4. [DOI] [PubMed] [Google Scholar]
- 23.Omura T, Sato R. The carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J Biol Chem. 1964;239:2370–2378. [PubMed] [Google Scholar]
- 24.Bridges KR, Seligman PA. Disorders of Iron Metabolism. J.B Lippincott Company; New York: 1995. [Google Scholar]
- 25.Everse J, Hsia N. The toxicities of native and modified hemoglobins. Free Radic Biol Med. 1997;22:1075–1099. doi: 10.1016/s0891-5849(96)00499-6. [DOI] [PubMed] [Google Scholar]
- 26.Anzaldi LL, Skaar EP. Overcoming the heme paradox: heme toxicity and tolerance in bacterial pathogens. Infect Immun. 2010;78:4977–4989. doi: 10.1128/IAI.00613-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith A, McCulloh RJ. Hemopexin and haptoglobin: allies against heme toxicity from hemoglobin not contenders. Front Physiol. 2015;6:187. doi: 10.3389/fphys.2015.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schaer DJ, Buehler PW. Cell-free hemoglobin and its scavenger proteins: new disease models leading the way to targeted therapies. Cold Spring Harb Perspect Med. 2013;3 doi: 10.1101/cshperspect.a013433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeney V, Balla J, Yachie A, Varga Z, Vercellotti GM, Eaton JW, et al. Pro-oxidant and cytotoxic effects of circulating heme. Blood. 2002;100:879–887. doi: 10.1182/blood.v100.3.879. [DOI] [PubMed] [Google Scholar]
- 30.Tolosano E, Altruda F. Hemopexin: structure, function, and regulation. DNA Cell Biol. 2002;21:297–306. doi: 10.1089/104454902753759717. [DOI] [PubMed] [Google Scholar]
- 31.Lin H, McFaul SJ, Brady JC, Everse J. The mechanism of peroxidase-mediated cytotoxicity. II. Role of the heme moiety. Proc Soc Exp Biol Med. 1988;187:7–13. doi: 10.3181/00379727-187-42629. [DOI] [PubMed] [Google Scholar]
- 32.Balla G, Jacob HS, Eaton JW, Belcher JD, Vercellotti GM. Hemin: a possible physiological mediator of low density lipoprotein oxidation and endothelial injury. Arter-ioscler Thromb. 1991;11:1700–1711. doi: 10.1161/01.atv.11.6.1700. [DOI] [PubMed] [Google Scholar]
- 33.Stauff DL, Skaar EP. Bacillus anthracis HssRS signalling to HrtAB regulates haem resistance during infection. Mol Microbiol. 2009;72:763–778. doi: 10.1111/j.1365-2958.2009.06684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dailey HA, Gerdes S, Dailey TA, Burch JS, Phillips JD. Noncanonical coproporphyrin-dependent bacterial heme biosynthesis pathway that does not use protoporphyrin. Proc Natl Acad Sci USA. 2015;112:2210–2215. doi: 10.1073/pnas.1416285112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beale SI, Gough SP, Granick S. Biosynthesis of delta-aminolevulinic acid from the intact carbon skeleton of glutamic acid in greening barley. Proc Natl Acad Sci USA. 1975;72:2719–2723. doi: 10.1073/pnas.72.7.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoober JK, Kahn A, Ash DE, Gough S, Kannangara CG. Biosynthesis of delta-aminolevulinate in greening barley leaves. IX. Structure of the substrate, mode of gabaculine inhibition, and the catalytic mechanism of glutamate 1-semialdehyde aminotransferase. Carlsb Res Commun. 1988;53 doi: 10.1007/BF02908411. [DOI] [PubMed] [Google Scholar]
- 37.Kannangara CG, Gough SP, Bruyant P, Hoober JK, Kahn A, von Wettstein D. tRNA(Glu) as a cofactor in delta-aminolevulinate biosynthesis: steps that regulate chlorophyll synthesis. Trends Biochem Sci. 1988;13:139–143. doi: 10.1016/0968-0004(88)90071-0. [DOI] [PubMed] [Google Scholar]
- 38.Hansson M, Rutberg L, Schroder I, Hederstedt L. The Bacillus subtilis hemAXCDBL gene cluster, which encodes enzymes of the biosynthetic pathway from glutamate to uroporphyrinogen III. J Bacteriol. 1991;173:2590–2599. doi: 10.1128/jb.173.8.2590-2599.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Louie GV, Brownlie PD, Lambert R, Cooper JB, Blundell TL, Wood SP, et al. Structure of porphobilinogen deaminase reveals a flexible multidomain polymerase with a single catalytic site. Nature. 1992;359:33–39. doi: 10.1038/359033a0. [DOI] [PubMed] [Google Scholar]
- 40.Bung N, Pradhan M, Srinivasan H, Bulusu G. Structural insights into E. coli porphobilinogen deaminase during synthesis and exit of 1-hydroxymethylbilane. PLoS Comput Biol. 2014;10:e1003484. doi: 10.1371/journal.pcbi.1003484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jordan PM. The biosynthesis of tetrapyrroles. Elsevier; New York, NY: 1991. [Google Scholar]
- 42.Hansson M, Hederstedt L. Cloning and characterization of the Bacillus subtilis hemEHY gene cluster, which encodes protoheme IX biosynthetic enzymes. J Bacteriol. 1992;174:8081–8093. doi: 10.1128/jb.174.24.8081-8093.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leeper FJ. The biosynthesis of porphyrins, chlorophylls, and vitamin B12. Nat Prod Rep. 1987;4:441–469. doi: 10.1039/np9870400441. [DOI] [PubMed] [Google Scholar]
- 44.Bali S, Lawrence AD, Lobo SA, Saraiva LM, Golding BT, Palmer DJ, et al. Molecular hijacking of siroheme for the synthesis of heme and d1 heme. Proc Natl Acad Sci USA. 2011;108:18260–18265. doi: 10.1073/pnas.1108228108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bali S, Palmer DJ, Schroeder S, Ferguson SJ, Warren MJ. Recent advances in the biosynthesis of modified tetrapyrroles: the discovery of an alternative pathway for the formation of heme and heme d 1. Cell Mol Life Sci. 2014;71:2837–2863. doi: 10.1007/s00018-014-1563-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Troup B, Hungerer C, Jahn D. Cloning and characterization of the Escherichia coli hemN gene encoding the oxygen-independent coproporphyrinogen III oxidase. J Bacteriol. 1995;177:3326–3331. doi: 10.1128/jb.177.11.3326-3331.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seehra JS, Jordan PM, Akhtar M. Anaerobic and aerobic coproporphyrinogen III oxidases of Rhodopseudomonas spheroides. Mechanism and stereochemistry of vinyl group formation. Biochem J. 1983;209:709–718. doi: 10.1042/bj2090709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boynton TO, Daugherty LE, Dailey TA, Dailey HA. Identification of Escherichia coli HemG as a novel, menadione-dependent flavodoxin with protoporphyrinogen oxidase activity. Biochemistry. 2009;48:6705–6711. doi: 10.1021/bi900850y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kobayashi K, Masuda T, Tajima N, Wada H, Sato N. Molecular phylogeny and intricate evolutionary history of the three isofunctional enzymes involved in the oxidation of protoporphyrinogen IX. Genome Biol Evol. 2014;6:2141–2155. doi: 10.1093/gbe/evu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Porra RJ, Falk JE. The enzymic conversion of coproporphyrinogen III into protoporphyrin IX. Biochem J. 1964;90:69–75. doi: 10.1042/bj0900069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Camadro JM, Labbe P. Purification and properties of ferrochelatase from the yeast Saccharomyces cerevisiae. Evidence for a precursor form of the protein. J Biol Chem. 1988;263:11675–11682. [PubMed] [Google Scholar]
- 52.Lobo SA, Scott A, Videira MA, Winpenny D, Gardner M, Palmer MJ, et al. Staphylococcus aureus haem biosynthesis: characterisation of the enzymes involved in final steps of the pathway. Mol Microbiol. 2015;97:472–487. doi: 10.1111/mmi.13041. [DOI] [PubMed] [Google Scholar]
- 53.Hansson M, Gustafsson MC, Kannangara CG, Hederstedt L. Isolated Bacillus subtilis HemY has copropor-phyrinogen III to coproporphyrin III oxidase activity. Biochim Biophys Acta. 1997;1340:97–104. doi: 10.1016/s0167-4838(97)00030-7. [DOI] [PubMed] [Google Scholar]
- 54.Mayfield JA, Hammer ND, Kurker RC, Chen TK, Ojha S, Skaar EP, et al. The chlorite dismutase (HemQ) from Staphylococcus aureus has a redox-sensitive heme and is associated with the small colony variant phenotype. J Biol Chem. 2013;288:23488–23504. doi: 10.1074/jbc.M112.442335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dailey HA, Gerdes S. HemQ: an iron-coproporphyrin oxidative decarboxylase for protoheme synthesis in Firmicutes and Actinobacteria. Arch Biochem Biophys. 2015;574:27–35. doi: 10.1016/j.abb.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dailey TA, Boynton TO, Albetel AN, Gerdes S, Johnson MK, Dailey HA. Discovery and characterization of HemQ: an essential heme biosynthetic pathway component. J Biol Chem. 2010;285:25978–25986. doi: 10.1074/jbc.M110.142604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Celis AI, Streit BR, Moraski GC, Kant R, Lash TD, Lukat-Rodgers GS, et al. Unusual peroxide-dependent, heme-transforming reaction catalyzed by HemQ. Biochemistry. 2015;54:4022–4032. doi: 10.1021/acs.biochem.5b00492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jahn D, Michelsen U, Soll D. Two glutamyl-tRNA reductase activities in Escherichia coli. J Biol Chem. 1991;266:2542–2548. [PubMed] [Google Scholar]
- 59.Javor GT, Febre EF. Enzymatic basis of thiol-stimulated secretion of porphyrins by Escherichia coli. J Bacteriol. 1992;174:1072–1075. doi: 10.1128/jb.174.3.1072-1075.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang LY, Brown L, Elliott M, Elliott T. Regulation of heme biosynthesis in Salmonella typhimurium: activity of glutamyl-tRNA reductase (HemA) is greatly elevated during heme limitation by a mechanism which increases abundance of the protein. J Bacteriol. 1997;179:2907–2914. doi: 10.1128/jb.179.9.2907-2914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang L, Elliott M, Elliott T. Conditional stability of the HemA protein (glutamyl-tRNA reductase) regulates heme biosynthesis in Salmonella typhimurium. J Bacteriol. 1999;181:1211–1219. doi: 10.1128/jb.181.4.1211-1219.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jones AM, Elliott T. A purified mutant HemA protein from Salmonella enterica serovar Typhimurium lacks bound heme and is defective for heme-mediated regulation in vivo. FEMS Microbiol Lett. 2010;307:41–47. doi: 10.1111/j.1574-6968.2010.01967.x. [DOI] [PubMed] [Google Scholar]
- 63.Wang L, Wilson S, Elliott T. A mutant HemA protein with positive charge close to the N terminus is stabilized against heme-regulated proteolysis in Salmonella typhimurium. J Bacteriol. 1999;181:6033–6041. doi: 10.1128/jb.181.19.6033-6041.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang J, Kang Z, Chen J, Du G. Optimization of the heme biosynthesis pathway for the production of 5-aminolevulinic acid in Escherichia coli. Sci Rep. 2015;5:8584. doi: 10.1038/srep08584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schroder I, Johansson P, Rutberg L, Hederstedt L. The hemX gene of the Bacillus subtilis hemAXCDBL operon encodes a membrane protein, negatively affecting the steady-state cellular concentration of HemA (glutamyl-tRNA reductase) Microbiology. 1994;140:731–740. doi: 10.1099/00221287-140-4-731. [DOI] [PubMed] [Google Scholar]
- 66.Krieger R, Rompf A, Schobert M, Jahn D. The Pseudomonas aeruginosa hemA promoterisregulatedby Anr, Dnr, NarL and Integration Host Factor. Mol Gen Genomics. 2002;267:409–417. doi: 10.1007/s00438-002-0672-7. [DOI] [PubMed] [Google Scholar]
- 67.Hungerer C, Troup B, Romling U, Jahn D. Regulation of the hemA gene during 5-aminolevulinic acid formation in Pseudomonas aeruginosa. J Bacteriol. 1995;177:1435–1443. doi: 10.1128/jb.177.6.1435-1443.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ye RW, Tao W, Bedzyk L, Young T, Chen M, Li L. Global gene expression profiles of Bacillus subtilis grown under anaerobic conditions. J Bacteriol. 2000;182:4458–4465. doi: 10.1128/jb.182.16.4458-4465.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ji CJ, Kim JH, Won YB, Lee YE, Choi TW, Ju SY, et al. Staphylococcus aureus PerR Is a hypersensitive hydrogen peroxide sensor using iron-mediated histidine oxidation. J Biol Chem. 2015;290:20374–20386. doi: 10.1074/jbc.M115.664961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ravcheev DA, Best AA, Tintle N, Dejongh M, Osterman AL, Novichkov PS, et al. Inference of the transcriptional regulatory network in Staphylococcus aureus by integration of experimental and genomics-based evidence. J Bacteriol. 2011;193:3228–3240. doi: 10.1128/JB.00350-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Frunzke J, Gatgens C, Brocker M, Bott M. Control of heme homeostasis in Corynebacterium glutamicum by the two-component system HrrSA. J Bacteriol. 2011;193:1212–1221. doi: 10.1128/JB.01130-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bibb LA, Kunkle CA, Schmitt MP. The ChrA–ChrS and HrrA–HrrS signal transduction systems are required for activation of the hmuO promoter and repression of the hemA promoter in Corynebacterium diphtheriae. Infect Immun. 2007;75:2421–2431. doi: 10.1128/IAI.01821-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bibb LA, Schmitt MP. The ABC transporter HrtAB confers resistance to hemin toxicity and is regulated in a hemin-dependent manner by the ChrAS two-component system in Corynebacterium diphtheriae. J Bacteriol. 2010;192:4606–4617. doi: 10.1128/JB.00525-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mancini S, Imlay JA. The induction of two biosynthetic enzymes helps Escherichia coli sustain heme synthesis and activate catalase during hydrogen peroxide stress. Mol Microbiol. 2015;96:744–763. doi: 10.1111/mmi.12967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Marino M, Ramos HC, Hoffmann T, Glaser P, Jahn D. Modulation of anaerobic energy metabolism of Bacillus subtilis by arfM (ywiD) J Bacteriol. 2001;183:6815–6821. doi: 10.1128/JB.183.23.6815-6821.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hippler B, Homuth G, Hoffmann T, Hungerer C, Schumann W, Jahn D. Characterization of Bacillus subtilis hemN. J Bacteriol. 1997;179:7181–7185. doi: 10.1128/jb.179.22.7181-7185.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Homuth G, Rompf A, Schumann W, Jahn D. Transcriptional control of Bacillus subtilis hemN and hemZ. J Bacteriol. 1999;181:5922–5929. doi: 10.1128/jb.181.19.5922-5929.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen L, Keramati L, Helmann JD. Coordinate regulation of Bacillus subtilis peroxide stress genes by hydrogen peroxide and metal ions. Proc Natl Acad Sci USA. 1995;92:8190–8194. doi: 10.1073/pnas.92.18.8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rompf A, Hungerer C, Hoffmann T, Lindenmeyer M, Romling U, Gross U, et al. Regulation of Pseudomonas aeruginosa hemF and hemN by the dual action of the redox response regulators Anr and Dnr. Mol Microbiol. 1998;29:985–997. doi: 10.1046/j.1365-2958.1998.00980.x. [DOI] [PubMed] [Google Scholar]
- 80.Sander A, Kretzer S, Bredt W, Oberle K, Bereswill S. Hemin-dependent growth and hemin binding of Bartonella henselae. FEMS Microbiol Lett. 2000;189:55–59. doi: 10.1111/j.1574-6968.2000.tb09205.x. [DOI] [PubMed] [Google Scholar]
- 81.Frankenberg L, Brugna M, Hederstedt L. Enterococcus faecalis heme-dependent catalase. J Bacteriol. 2002;184:6351–6356. doi: 10.1128/JB.184.22.6351-6356.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Biberstein EL, Mini PD, Gills MG. Action of Haemophilus cultures on δ-aminolevulinic acid. J Bacteriol. 1963;86:814–819. doi: 10.1128/jb.86.4.814-819.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Baureder M, Hederstedt L. Heme proteins in lactic acid bacteria. Adv Microb Physiol. 2013;62:1–43. doi: 10.1016/B978-0-12-410515-7.00001-9. [DOI] [PubMed] [Google Scholar]
- 84.Skaar EP, Humayun M, Bae T, DeBord KL, Schneewind O. Iron-source preference of Staphylococcus aureus infections. Science. 2004;305:1626–1628. doi: 10.1126/science.1099930. [DOI] [PubMed] [Google Scholar]
- 85.Pishchany G, Sheldon JR, Dickson CF, Alam MT, Read TD, Gell DA, et al. IsdB-dependent hemoglobin binding is required for acquisition of heme by Staphylococcus aureus. J Infect Dis. 2014;209:1764–1772. doi: 10.1093/infdis/jit817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pishchany G, McCoy AL, Torres VJ, Krause JC, Crowe JE, Jr, Fabry ME, et al. Specificity for human hemoglobin enhances Staphylococcus aureus infection. Cell Host Microbe. 2010;8:544–550. doi: 10.1016/j.chom.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hammer ND, Reniere ML, Cassat JE, Zhang Y, Hirsch AO, Hood MI, et al. Two heme-dependent terminal oxidases power Staphylococcus aureus organ-specific colonization of the vertebrate host. mBio. 2013;4 doi: 10.1128/mBio.00241-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hammer ND, Cassat JE, Noto MJ, Lojek LJ, Chadha AD, Schmitz JE, et al. Inter- and intraspecies metabolite exchange promotes virulence of antibiotic-resistant Staphylococcus aureus. Cell Host Microbe. 2014;16:531–537. doi: 10.1016/j.chom.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.von Eiff C, Heilmann C, Proctor RA, Woltz C, Peters G, Gotz F. A site-directed Staphylococcus aureus hemB mutant is a small-colony variant which persists intracellularly. J Bacteriol. 1997;179:4706–4712. doi: 10.1128/jb.179.15.4706-4712.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Balwit JM, van Langevelde P, Vann JM, Proctor RA. Gentamicin-resistant menadione and hemin auxotrophic Staphylococcus aureus persist within cultured endothelial cells. J Infect Dis. 1994;170:1033–1037. doi: 10.1093/infdis/170.4.1033. [DOI] [PubMed] [Google Scholar]
- 91.Painter KL, Strange E, Parkhill J, Bamford KB, Armstrong-James D, Edwards AM. Staphylococcus aureus adapts to oxidative stress by producing H2O2-resistant small-colony variants via the SOS response. Infect Immun. 2015;83:1830–1844. doi: 10.1128/IAI.03016-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sadowska B, Bonar A, von Eiff C, Proctor RA, Chmiela M, Rudnicka W, et al. Characteristics of Staphylococcus aureus, isolated from airways of cystic fibrosis patients, and their small colony variants. FEMS Immunol Med Microbiol. 2002;32:191–197. doi: 10.1111/j.1574-695X.2002.tb00553.x. [DOI] [PubMed] [Google Scholar]
- 93.Proctor RA, von Eiff C, Kahl BC, Becker K, McNamara P, Herrmann M, et al. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat Rev Microbiol. 2006;4:295–305. doi: 10.1038/nrmicro1384. [DOI] [PubMed] [Google Scholar]
- 94.Almiron M, Martinez M, Sanjuan N, Ugalde RA. Ferrochelatase is present in Brucella abortus and is critical for its intracellular survival and virulence. Infect Immun. 2001;69:6225–6230. doi: 10.1128/IAI.69.10.6225-6230.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Skurnik D, Roux D, Aschard H, Cattoir V, Yoder-Himes D, Lory S, et al. A comprehensive analysis of in vitro and in vivo genetic fitness of Pseudomonas aeruginosa using high-throughput sequencing of transposon libraries. PLoS Pathog. 2013;9:e1003582. doi: 10.1371/journal.ppat.1003582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Palace SG, Proulx MK, Lu S, Baker RE, Goguen JD. Genome-wide mutant fitness profiling identifies nutritional requirements for optimal growth of Yersinia pestis in deep tissue. mBio. 2014:5. doi: 10.1128/mBio.01385-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang N, Ozer EA, Mandel MJ, Hauser AR. Genomewide identification of Acinetobacter baumannii genes necessary for persistence in the lung. mBio. 2014;5:e01163–14. doi: 10.1128/mBio.01163-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cummins J, Casey PG, Joyce SA, Gahan CG. A mariner transposon-based signature-tagged mutagenesis system for the analysis of oral infection by Listeria monocytogenes. PLoS ONE. 2013;8:e75437. doi: 10.1371/journal.pone.0075437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mike LA, Dutter BF, Stauff DL, Moore JL, Vitko NP, Aranmolate O, et al. Activation of heme biosynthesis by a small molecule that is toxic to fermenting Staphylococcus aureus. Proc Natl Acad Sci USA. 2013;110:9206–8211. doi: 10.1073/pnas.1303674110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dutter BF, Mike LA, Reid PR, Chong KM, Ramos-Hunter SJ, Skaar EP, et al. Decoupling activation of heme biosynthesis from anaerobic toxicity in a molecule active in Staphylococcus aureus. ACS Chem Biol. 2016 doi: 10.1021/acschembio.5b00934. PMID: 26890615, Epub ahead of print http://dx.doi.org/10.1021/acschembio.5b00934. [DOI] [PMC free article] [PubMed]
- 101.Jensen J. The effect of heme on tetrapyrrol synthesis in a heme requiring Staphylococcus aureus. Biochem Biophys Res Commun. 1962;8:271–277. doi: 10.1016/0006-291x(62)90276-0. [DOI] [PubMed] [Google Scholar]
- 102.Mazmanian SK, Skaar EP, Gaspar AH, Humayun M, Gornicki P, Jelenska J, et al. Passage of heme-iron across the envelope of Staphylococcus aureus. Science. 2003;299:906–909. doi: 10.1126/science.1081147. [DOI] [PubMed] [Google Scholar]
- 103.Spaan AN, Reyes-Robles T, Badiou C, Cochet S, Boguslawski KM, Yoong P, et al. Staphylococcus aureus targets the Duffy Antigen Receptor for Chemokines (DARC) to lyse erythrocytes. Cell Host Microbe. 2015;18:363–370. doi: 10.1016/j.chom.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Grigg JC, Vermeiren CL, Heinrichs DE, Murphy ME. Haem recognition by a Staphylococcus aureus NEAT domain. Mol Microbiol. 2007;63:139–149. doi: 10.1111/j.1365-2958.2006.05502.x. [DOI] [PubMed] [Google Scholar]
- 105.Sharp KH, Schneider S, Cockayne A, Paoli M. Crystal structure of the heme-IsdC complex, the central conduit of the Isd iron/heme uptake system in Staphylococcus aureus. J Biol Chem. 2007;282:10625–10631. doi: 10.1074/jbc.M700234200. [DOI] [PubMed] [Google Scholar]
- 106.Mazmanian SK, Liu G, Ton-That H, Schneewind O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science. 1999;285:760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- 107.Fonner BA, Tripet BP, Eilers BJ, Stanisich J, Sullivan-Springhetti RK, Moore R, et al. Solution structure and molecular determinants of hemoglobin binding of the first NEAT domain of IsdB in Staphylococcus aureus. Biochemistry. 2014;53:3922–3933. doi: 10.1021/bi5005188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Torres VJ, Pishchany G, Humayun M, Schneewind O, Skaar EP. Staphylococcus aureus IsdB is a hemoglobin receptor required for heme iron utilization. J Bacteriol. 2006;188:8421–8429. doi: 10.1128/JB.01335-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dryla A, Gelbmann D, Von Gabain A, Nagy E. Identification of a novel iron regulated staphylococcal surface protein with haptoglobin-haemoglobin binding activity. Mol Microbiol. 2003;49:37–53. doi: 10.1046/j.1365-2958.2003.03542.x. [DOI] [PubMed] [Google Scholar]
- 110.Pilpa RM, Robson SA, Villareal VA, Wong ML, Phillips M, Clubb RT. Functionally distinct NEAT (NEAr Transporter) domains within the Staphylococcus aureus IsdH/HarA protein extract heme from methemoglobin. J Biol Chem. 2009;284:1166–1176. doi: 10.1074/jbc.M806007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Muryoi N, Tiedemann MT, Pluym M, Cheung J, Heinrichs DE, Stillman MJ. Demonstration of the iron-regulated surface determinant (Isd) heme transfer pathway in Staphylococcus aureus. J Biol Chem. 2008;283:28125–28136. doi: 10.1074/jbc.M802171200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mazmanian SK, Ton-That H, Su K, Schneewind O. An iron-regulated sortase anchors a class of surface protein during Staphylococcus aureus pathogenesis. Proc Natl Acad Sci USA. 2002;99:2293–2298. doi: 10.1073/pnas.032523999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Beveridge J. Ultrastructure of Gram-positive cell walls. In: Fishetti VA, Novick RP, Feretti JJ, Portnoy DA, Rood JI, editors. Gram-Positive Pathogens. second. ASM Press; Washington, DC: 2000. pp. 3–10. [Google Scholar]
- 114.Zhu H, Xie G, Liu M, Olson JS, Fabian M, Dooley DM, et al. Pathway for heme uptake from human methemoglobin by the iron-regulated surface determinants system of Staphylococcus aureus. J Biol Chem. 2008;283:18450–18460. doi: 10.1074/jbc.M801466200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Skaar EP, Gaspar AH, Schneewind O. IsdG and IsdI, heme-degrading enzymes in the cytoplasm of Staphylococcus aureus. J Biol Chem. 2004;279:436–443. doi: 10.1074/jbc.M307952200. [DOI] [PubMed] [Google Scholar]
- 116.Reniere ML, Torres VJ, Skaar EP. Intracellular metalloporphyrin metabolism in Staphylococcus aureus. Biometals. 2007;20:333–345. doi: 10.1007/s10534-006-9032-0. [DOI] [PubMed] [Google Scholar]
- 117.Reniere ML, Ukpabi GN, Harry SR, Stec DF, Krull R, Wright DW, et al. The IsdG-family of haem oxygenases degrades haem to a novel chromophore. Mol Microbiol. 2010;75:1529–1538. doi: 10.1111/j.1365-2958.2010.07076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lee WC, Reniere ML, Skaar EP, Murphy ME. Ruffling of metalloporphyrins bound to IsdG and IsdI, two hemed-egrading enzymes in Staphylococcus aureus. J Biol Chem. 2008;283:30957–30963. doi: 10.1074/jbc.M709486200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wu R, Skaar EP, Zhang R, Joachimiak G, Gornicki P, Schneewind O, et al. Staphylococcus aureus IsdG and IsdI, heme-degrading enzymes with structural similarity to monooxygenases. J Biol Chem. 2005;280:2840–2846. doi: 10.1074/jbc.M409526200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Reniere ML, Skaar EP. Staphylococcus aureus haem oxygenases are differentially regulated by iron and haem. Mol Microbiol. 2008;69:1304–1315. doi: 10.1111/j.1365-2958.2008.06363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Farrand AJ, Reniere ML, Ingmer H, Frees D, Skaar EP. Regulation of host hemoglobin binding by the Staphylococcus aureus Clp proteolytic system. J Bacteriol. 2013;195:5041–5050. doi: 10.1128/JB.00505-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Laakso HA, Marolda CL, Pinter TB, Stillman MJ, Heinrichs DE. A heme-responsive regulator controls synthesis of Staphyloferrin B in Staphylococcus aureus. J Biol Chem. 2016;291:29–40. doi: 10.1074/jbc.M115.696625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Visai L, Yanagisawa N, Josefsson E, Tarkowski A, Pezzali I, Rooijakkers SH, et al. Immune evasion by Staphylococcus aureus conferred by iron-regulated surface determinant protein IsdH. Microbiology. 2009;155:667–679. doi: 10.1099/mic.0.025684-0. [DOI] [PubMed] [Google Scholar]
- 124.Cheng AG, Kim HK, Burts ML, Krausz T, Schneewind O, Missiakas DM. Genetic requirements for Staphylococcus aureus abscess formation and persistence in host tissues. FASEB J. 2009;23:3393–3404. doi: 10.1096/fj.09-135467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Maresso AW, Garufi G, Schneewind O. Bacillus anthracis secretes proteins that mediate heme acquisition from hemoglobin. PLoS Pathog. 2008;4:e1000132. doi: 10.1371/journal.ppat.1000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Fabian M, Solomaha E, Olson JS, Maresso AW. Heme transfer to the bacterial cell envelope occurs via a secreted hemophore in the Gram-positive pathogen Bacillus anthracis. J Biol Chem. 2009;284:32138–32146. doi: 10.1074/jbc.M109.040915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Honsa ES, Fabian M, Cardenas AM, Olson JS, Maresso AW. The five near-iron transporter (NEAT) domain anthrax hemophore, IsdX2, scavenges heme from hemoglobin and transfers heme to the surface protein IsdC. J Biol Chem. 2011;286:33652–33660. doi: 10.1074/jbc.M111.241687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Maresso AW, Chapa TJ, Schneewind O. Surface protein IsdC and Sortase B are required for heme-iron scavenging of Bacillus anthracis. J Bacteriol. 2006;188:8145–8152. doi: 10.1128/JB.01011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Honsa ES, Owens CP, Goulding CW, Maresso AW. The near-iron transporter (NEAT) domains of the anthrax hemophore IsdX2 require a critical glutamine to extract heme from methemoglobin. J Biol Chem. 2013;288:8479–8490. doi: 10.1074/jbc.M112.430009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Balderas MA, Nobles CL, Honsa ES, Alicki ER, Maresso AW. Hal Is a Bacillus anthracis heme acquisition protein. J Bacteriol. 2012;194:5513–5521. doi: 10.1128/JB.00685-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gaspar AH, Marraffini LA, Glass EM, Debord KL, Ton-That H, Schneewind O. Bacillus anthracis sortase A (SrtA) anchors LPXTG motif-containing surface proteins to the cell wall envelope. J Bacteriol. 2005;187:4646–4655. doi: 10.1128/JB.187.13.4646-4655.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tarlovsky Y, Fabian M, Solomaha E, Honsa E, Olson JS, Maresso AW. A Bacillus anthracis S-layer homology protein that binds heme and mediates heme delivery to IsdC. J Bacteriol. 2010;192:3503–3511. doi: 10.1128/JB.00054-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Skaar EP, Gaspar AH, Schneewind O. Bacillus anthracis IsdG, a heme-degrading monooxygenase. J Bacteriol. 2006;188:1071–1080. doi: 10.1128/JB.188.3.1071-1080.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gat O, Zaide G, Inbar I, Grosfeld H, Chitlaru T, Levy H, et al. Characterization of Bacillus anthracis iron-regulated surface determinant (Isd) proteins containing NEAT domains. Mol Microbiol. 2008;70:983–999. doi: 10.1111/j.1365-2958.2008.06460.x. [DOI] [PubMed] [Google Scholar]
- 135.Carlson PE, Jr, Carr KA, Janes BK, Anderson EC, Hanna PC. Transcriptional profiling of Bacillus anthracis Sterne (34F2) during iron starvation. PLoS One. 2009;4:e6988. doi: 10.1371/journal.pone.0006988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Farrand AJ, Haley KP, Lareau NM, Heilbronner S, McLean JA, Foster T, et al. An iron-regulated autolysin remodels the cell wall to facilitate heme acquisition in Staphylococcus lugdunensis. Infect Immun. 2015;83:3578–3589. doi: 10.1128/IAI.00397-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zapotoczna M, Heilbronner S, Speziale P, Foster TJ. Iron-regulated surface determinant (Isd) proteins of Staphylococcus lugdunensis. J Bacteriol. 2012;194:6453–6467. doi: 10.1128/JB.01195-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Haley KP, Janson EM, Heilbronner S, Foster TJ, Skaar EP. Staphylococcus lugdunensis IsdG liberates iron from host heme. J Bacteriol. 2011;193:4749–4757. doi: 10.1128/JB.00436-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Malmirchegini GR, Sjodt M, Shnitkind S, Sawaya MR, Rosinski J, Newton SM. Novel mechanism of hemin capture by Hbp2, the hemoglobin-binding hemophore from Listeria monocytogenes. J Biol Chem. 2014;289:34886–34899. doi: 10.1074/jbc.M114.583013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Xiao Q, Jiang X, Moore KJ, Shao Y, Pi H, Dubail I, et al. Sortase independent and dependent systems for acquisition of haem and haemoglobin in Listeria monocytogenes. Mol Microbiol. 2011;80:1581–1597. doi: 10.1111/j.1365-2958.2011.07667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Dahesh S, Nizet V, Cole JN. Study of streptococcal hemoprotein receptor (Shr) in iron acquisition and virulence of M1T1 group A streptococcus. Virulence. 2012;3:566–575. doi: 10.4161/viru.21933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ouattara M, Cunha EB, Li X, Huang YS, Dixon D, Eichenbaum Z. Shr of group A streptococcus is a new type of composite NEAT protein involved in sequestering haem from methaemoglobin. Mol Microbiol. 2010;78:739–756. doi: 10.1111/j.1365-2958.2010.07367.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ouattara M, Pennati A, Devlin DJ, Huang YS, Gadda G, Eichenbaum Z. Kinetics of heme transfer by the Shr NEAT domains of Group A Streptococcus. Arch Biochem Biophys. 2013;538:71–79. doi: 10.1016/j.abb.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 144.Schmitt MP. Utilization of host iron sources by Coryne-bacterium diphtheriae: identification of a gene whose product is homologous to eukaryotic heme oxygenases and is required for acquisition of iron from heme and hemoglobin. J Bacteriol. 1997;179:838–845. doi: 10.1128/jb.179.3.838-845.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Drazek ES, Hammack CA, Schmitt MP. Corynebacterium diphtheriae genes required for acquisition of iron from haemin and haemoglobin are homologous to ABC haemin transporters. Mol Microbiol. 2000;36:68–84. doi: 10.1046/j.1365-2958.2000.01818.x. [DOI] [PubMed] [Google Scholar]
- 146.Thompson JM, Jones HA, Perry RD. Molecular characterization of the hemin uptake locus (hmu) from Yersinia pestis and analysis of hmu mutants for hemin and hemoprotein utilization. Infect Immun. 1999;67:3879–3892. doi: 10.1128/iai.67.8.3879-3892.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Allen CE, Schmitt MP. HtaA is an iron-regulated hemin binding protein involved in the utilization of heme iron in Corynebacterium diphtheriae. J Bacteriol. 2009;191:2638–2648. doi: 10.1128/JB.01784-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Allen CE, Schmitt MP. Novel hemin binding domains in the Corynebacterium diphtheriae HtaA protein interact with hemoglobin and are critical for heme iron utilization by HtaA. J Bacteriol. 2011;193:5374–5385. doi: 10.1128/JB.05508-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Draganova EB, Akbas N, Adrian SA, Lukat-Rodgers GS, Collins DP, Dawson JH, et al. Heme binding by Corynebacterium diphtheriae HmuT: function and heme environment. Biochemistry. 2015;54:6598–6609. doi: 10.1021/acs.biochem.5b00666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kunkle CA, Schmitt MP. Analysis of a DtxR-regulated iron transport and siderophore biosynthesis gene cluster in Corynebacterium diphtheriae. J Bacteriol. 2005;187:422–433. doi: 10.1128/JB.187.2.422-433.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kunkle CA, Schmitt MP. Analysis of the Corynebacterium diphtheriae DtxR regulon: identification of a putative siderophore synthesis and transport system that is similar to the Yersinia high-pathogenicity island-encoded yersiniabactin synthesis and uptake system. J Bacteriol. 2003;185:6826–6840. doi: 10.1128/JB.185.23.6826-6840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Allen CE, Burgos JM, Schmitt MP. Analysis of novel iron-regulated, surface-anchored hemin-binding proteins in Corynebacterium diphtheriae. J Bacteriol. 2013;195:2852–2863. doi: 10.1128/JB.00244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Allen CE, Schmitt MP. Utilization of host iron sources by Corynebacterium diphtheriae: multiple hemoglobin-binding proteins are essential for the use of iron from the hemoglobin– haptoglobin complex. J Bacteriol. 2015;197:553–562. doi: 10.1128/JB.02413-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Noinaj N, Guillier M, Barnard TJ, Buchanan SK. TonB-dependent transporters: regulation, structure, and function. Annu Rev Microbiol. 2010;64:43–60. doi: 10.1146/annurev.micro.112408.134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Letoffe S, Nato F, Goldberg ME, Wandersman C. Interactions of HasA, a bacterial haemophore, with haemoglobin and with its outer membrane receptor HasR. Mol Microbiol. 1999;33:546–555. doi: 10.1046/j.1365-2958.1999.01499.x. [DOI] [PubMed] [Google Scholar]
- 156.Letoffe S, Deniau C, Wolff N, Dassa E, Delepelaire P, Lecroisey A, et al. Haemophore-mediated bacterial haem transport: evidence for a common or overlapping site for haem-free and haem-loaded haemophore on its specific outer membrane receptor. Mol Microbiol. 2001;41:439–450. doi: 10.1046/j.1365-2958.2001.02530.x. [DOI] [PubMed] [Google Scholar]
- 157.Smith AD, Wilks A. Differential contributions of the outer membrane receptors PhuR and HasR to heme acquisition in Pseudomonas aeruginosa. J Biol Chem. 2015;290:7756–7766. doi: 10.1074/jbc.M114.633495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Oglesby-Sherrouse AG, Vasil ML. Characterization of a heme-regulated non-coding RNA encoded by the prrF locus of Pseudomonas aeruginosa. PLoS One. 2010;5:e9930. doi: 10.1371/journal.pone.0009930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Reinhart AA, Powell DA, Nguyen AT, O'Neill M, Djapgne L, Wilks A, et al. The prrF-encoded small regulatory RNAs are required for iron homeostasis and virulence of Pseudomonas aeruginosa. Infect Immun. 2015;83:863–875. doi: 10.1128/IAI.02707-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lansky IB, Lukat-Rodgers GS, Block D, Rodgers KR, Ratliff M, Wilks A. The cytoplasmic heme-binding protein (PhuS) from the heme uptake system of Pseudomonas aeruginosa is an intracellular heme-trafficking protein to the delta-regioselective heme oxygenase. J Biol Chem. 2006;281:13652–13662. doi: 10.1074/jbc.M600824200. [DOI] [PubMed] [Google Scholar]
- 161.Block DR, Lukat-Rodgers GS, Rodgers KR, Wilks A, Bhakta MN, Lansky IB. Identification of two heme-binding sites in the cytoplasmic heme-trafficking protein PhuS from Pseudomonas aeruginosa and their relevance to function. Biochemistry. 2007;46:14391–14402. doi: 10.1021/bi701509n. [DOI] [PubMed] [Google Scholar]
- 162.Tripathi S, O'Neill MJ, Wilks A, Poulos TL. Crystal structure of the Pseudomonas aeruginosa cytoplasmic heme binding protein, Apo-PhuS. J Inorg Biochem. 2013;128:131–136. doi: 10.1016/j.jinorgbio.2013.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.O'Neill MJ, Bhakta MN, Fleming KG, Wilks A. Induced fit on heme binding to the Pseudomonas aeruginosa cytoplasmic protein (PhuS) drives interaction with heme oxygenase (HemO) Proc Natl Acad Sci USA. 2012;109:5639–5644. doi: 10.1073/pnas.1121549109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lee MJ, Schep D, McLaughlin B, Kaufmann M, Jia Z. Structural analysis and identification of PhuS as a heme-degrading enzyme from Pseudomonas aeruginosa. J Mol Biol. 2014;426:1936–1946. doi: 10.1016/j.jmb.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 165.O'Neill MJ, Wilks A. The P. aeruginosa heme binding protein PhuS is a heme oxygenase titratable regulator of heme uptake. ACS Chem Biol. 2013;8:1794–1802. doi: 10.1021/cb400165b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Marvig RL, Damkiaer S, Khademi SM, Markussen TM, Molin S, Jelsbak L. Within-host evolution of Pseudomonas aeruginosa reveals adaptation toward iron acquisition from hemoglobin. mBio. 2014;5:e00966–14. doi: 10.1128/mBio.00966-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Granick S, Gilder H. The porphyrin requirements of Haemophilus influenzae and some functions of the vinyl and propionic acid side chains of heme. J Gen Physiol. 1946;30:1–13. [PubMed] [Google Scholar]
- 168.Stull TL. Protein sources of heme for Haemophilus influenzae. Infect Immun. 1987;55:148–153. doi: 10.1128/iai.55.1.148-153.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Cope LD, Thomas SE, Hrkal Z, Hansen EJ. Binding of heme-hemopexin complexes by soluble HxuA protein allows utilization of this complexed heme by Haemophilus influenzae. Infect Immun. 1998;66:4511–4516. doi: 10.1128/iai.66.9.4511-4516.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Cope LD, Yogev R, Muller-Eberhard U, Hansen EJ. A gene cluster involved in the utilization of both free heme and heme: hemopexin by Haemophilus influenzae type b. J Bacteriol. 1995;177:2644–2653. doi: 10.1128/jb.177.10.2644-2653.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Baelen S, Dewitte F, Clantin B, Villeret V. Structure of the secretion domain of HxuA from Haemophilus influenzae. Acta Crystallogr Sect F: Struct Biol Cryst Commun. 2013;69:1322–1327. doi: 10.1107/S174430911302962X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Fournier C, Smith A, Delepelaire P. Haem release from haemopexin by HxuA allows Haemophilus influenzae to escape host nutritional immunity. Mol Microbiol. 2011;80:133–148. doi: 10.1111/j.1365-2958.2011.07562.x. [DOI] [PubMed] [Google Scholar]
- 173.Cope LD, Love RP, Guinn SE, Gilep A, Usanov S, Estabrook RW, et al. Involvement of HxuC outer membrane protein in utilization of hemoglobin by Haemophilus influenzae. Infect Immun. 2001;69:2353–2363. doi: 10.1128/IAI.69.4.2353-2363.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Morton DJ, Seale TW, Madore LL, VanWagoner TM, Whitby PW, Stull TL. The haem-haemopexin utilization gene cluster (hxuCBA) as a virulence factor of Haemophilus influenzae. Microbiology. 2007;153:215–224. doi: 10.1099/mic.0.2006/000190-0. [DOI] [PubMed] [Google Scholar]
- 175.Morton DJ, Madore LL, Smith A, Vanwagoner TM, Seale TW, Whitby PW, et al. The heme-binding lipoprotein (HbpA) of Haemophilus influenzae: role in heme utilization. FEMS Microbiol Lett. 2005;253:193–199. doi: 10.1016/j.femsle.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 176.Morton DJ, Seale TW, Bakaletz LO, Jurcisek JA, Smith A, VanWagoner TM, et al. The heme-binding protein (HbpA) of Haemophilus influenzae as a virulence determinant. Int J Med Microbiol. 2009;299:479–488. doi: 10.1016/j.ijmm.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Morton DJ, Van Wagoner TM, Seale TW, Whitby PW, Stull TL. Utilization of myoglobin as a heme source by Haemophilus influenzae requires binding of myoglobin to haptoglobin. FEMS Microbiol Lett. 2006;258:235–240. doi: 10.1111/j.1574-6968.2006.00230.x. [DOI] [PubMed] [Google Scholar]
- 178.Morton DJ, VanWagoner TM, Seale TW, Whitby PW, Stull TL. Differential utilization by Haemophilus influenzae of haemoglobin complexed to the three human haptoglobin phenotypes. FEMS Immunol Med Microbiol. 2006;46:426–432. doi: 10.1111/j.1574-695X.2006.00052.x. [DOI] [PubMed] [Google Scholar]
- 179.Morton DJ, Smith A, Ren Z, Madore LL, VanWagoner TM, Seale TW, et al. Identification of a haem-utilization protein (Hup) in Haemophilus influenzae. Microbiology. 2004;150:3923–3933. doi: 10.1099/mic.0.27238-0. [DOI] [PubMed] [Google Scholar]
- 180.Morton DJ, Seale TW, Vanwagoner TM, Whitby PW, Stull TL. The dppBCDF gene cluster of Haemophilus influenzae: role in heme utilization. BMC Res Notes. 2009;2:166. doi: 10.1186/1756-0500-2-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Morton DJ, Smith A, VanWagoner TM, Seale TW, Whitby PW, Stull TL. Lipoprotein e (P4) of Haemophilus influenzae: role in heme utilization and pathogenesis. Microbes Infect. 2007;9:932–939. doi: 10.1016/j.micinf.2007.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.LaCross NC, Marrs CF, Gilsdorf JR. Otitis media associated polymorphisms in the hemin receptor HemR of nontypeable Haemophilus influenzae. Infect Genet Evol. 2014;26:47–57. doi: 10.1016/j.meegid.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Vogel AR, Szelestey BR, Raffel FK, Sharpe SW, Gearinger RL, Justice SS, et al. SapF-mediated heme-iron utilization enhances persistence and coordinates biofilm architecture of Haemophilus. Front Cell Infect Microbiol. 2012;2:42. doi: 10.3389/fcimb.2012.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Whitby PW, VanWagoner TM, Seale TW, Morton DJ, Stull TL. Comparison of transcription of the Haemophilus influenzae iron/heme modulon genes in vitro and in vivo in the chinchilla middle ear. BMC Genomics. 2013;14:925. doi: 10.1186/1471-2164-14-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Whitby PW, Seale TW, VanWagoner TM, Morton DJ, Stull TL. The iron/heme regulated genes of Haemophilus influenzae: comparative transcriptional profiling as a tool to define the species core modulon. BMC Genomics. 2009;10:6. doi: 10.1186/1471-2164-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Xie J, Juliao PC, Gilsdorf JR, Ghosh D, Patel M, Marrs CF. Identification of new genetic regions more prevalent in nontypeable Haemophilus influenzae otitis media strains than in throat strains. J Clin Microbiol. 2006;44:4316–4325. doi: 10.1128/JCM.01331-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Hariadi NI, Zhang L, Patel M, Sandstedt SA, Davis GS, Marrs CF, et al. comparative profile of heme acquisition genes in disease-causing and colonizing nontypeable Haemophilus influenzae and Haemophilus haemolyticus. J Clin Microbiol. 2015;53:2132–2137. doi: 10.1128/JCM.00345-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Seale TW, Morton DJ, Whitby PW, Wolf R, Kosanke SD, VanWagoner TM, et al. Complex role of hemoglobin and hemoglobin–haptoglobin binding proteins in Haemophilus influenzae virulence in the infant rat model of invasive infection. Infect Immun. 2006;74:6213–6225. doi: 10.1128/IAI.00744-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Morton DJ, Bakaletz LO, Jurcisek JA, VanWagoner TM, Seale TW, Whitby PW, et al. Reduced severity of middle ear infection caused by nontypeable Haemophilus influenzae lacking the hemoglobin/hemoglobin–haptoglobin binding proteins (Hgp) in a chinchilla model of otitis media. Microb Pathog. 2004;36:25–33. doi: 10.1016/j.micpath.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 190.Lewis LA, Gray E, Wang YP, Roe BA, Dyer DW. Molecular characterization of hpuAB, the haemoglobin–hapto-globin-utilization operon of Neisseria meningitidis. Mol Microbiol. 1997;23:737–749. doi: 10.1046/j.1365-2958.1997.2501619.x. [DOI] [PubMed] [Google Scholar]
- 191.Lewis LA, Dyer DW. Identification of an iron-regulated outer membrane protein of Neisseria meningitidis involved in the utilization of hemoglobin complexed to haptoglobin. J Bacteriol. 1995;177:1299–1306. doi: 10.1128/jb.177.5.1299-1306.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Zhu W, Wilks A, Stojiljkovic I. Degradation of heme in gram-negative bacteria: the product of the hemO gene of Neisseriae is a heme oxygenase. J Bacteriol. 2000;182:6783–6790. doi: 10.1128/jb.182.23.6783-6790.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Zhu W, Hunt DJ, Richardson AR, Stojiljkovic I. Use of heme compounds as iron sources by pathogenic neisseriae requires the product of the hemO gene. J Bacteriol. 2000;182:439–447. doi: 10.1128/jb.182.2.439-447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Lewis LA, Sung MH, Gipson M, Hartman K, Dyer DW. Transport of intact porphyrin by HpuAB, the hemoglobin– haptoglobin utilization system of Neisseria meningitidis. J Bacteriol. 1998;180:6043–6047. doi: 10.1128/jb.180.22.6043-6047.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Kumar P, Sannigrahi S, Tzeng YL. The Neisseria meningitidis ZnuD zinc receptor contributes to interactions with epithelial cells and supports heme utilization when expressed in Escherichia coli. Infect Immun. 2012;80:657–667. doi: 10.1128/IAI.05208-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Chen CJ, Elkins C, Sparling PF. Phase variation of hemoglobin utilization in Neisseria gonorrhoeae. Infect Immun. 1998;66:987–993. doi: 10.1128/iai.66.3.987-993.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Rohde KH, Dyer DW. Analysis of haptoglobin and hemoglobin –haptoglobin interactions with the Neisseria meningitidis TonB-dependent receptor HpuAB by flow cytometry. Infect Immun. 2004;72:2494–2506. doi: 10.1128/IAI.72.5.2494-2506.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Rohde KH, Gillaspy AF, Hatfield MD, Lewis LA, Dyer DW. Interactions of haemoglobin with the Neisseria meningitidis receptor HpuAB: the role of TonB and an intact proton motive force. Mol Microbiol. 2002;43:335–354. doi: 10.1046/j.1365-2958.2002.02745.x. [DOI] [PubMed] [Google Scholar]
- 199.Wong CT, Xu Y, Gupta A, Garnett JA, Matthews SJ, Hare SA. Structural analysis of haemoglobin binding by HpuA from the Neisseriaceae family. Nat Commun. 2015;6:10172. doi: 10.1038/ncomms10172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Stojiljkovic I, Hwa V, de Saint Martin L, O'Gaora P, Nassif X, Heffron F, et al. The Neisseria meningitidis haemoglobin receptor: its role in iron utilization and virulence. Mol Microbiol. 1995;15:531–541. doi: 10.1111/j.1365-2958.1995.tb02266.x. [DOI] [PubMed] [Google Scholar]
- 201.Stojiljkovic I, Larson J, Hwa V, Anic S, So M. HmbR outer membrane receptors of pathogenic Neisseria spp.: iron-regulated, hemoglobin-binding proteins with a high level of primary structure conservation. J Bacteriology. 1996;178:4670–4678. doi: 10.1128/jb.178.15.4670-4678.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Richardson AR, Stojiljkovic I. HmbR, a hemoglobin-binding outer membrane protein of Neisseria meningitidis, undergoes phase variation. J Bacteriol. 1999;181:2067–2074. doi: 10.1128/jb.181.7.2067-2074.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Mokry DZ, Nadia-Albete A, Johnson MK, Lukat-Rodgers GS, Rodgers KR, Lanzilotta WN. Spectroscopic evidence for a 5-coordinate oxygenic ligated high spin ferric heme moiety in the Neisseria meningitidis hemoglobin binding receptor. Biochim Biophys Acta. 2014;(1840):3058–3066. doi: 10.1016/j.bbagen.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Sebastian S, Agarwal S, Murphy JR, Genco CA. The gonococcal Fur regulon: identification of additional genes involved in major catabolic, recombination, and secretory pathways. J Bacteriol. 2002;184:3965–3974. doi: 10.1128/JB.184.14.3965-3974.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Zhao S, Montanez GE, Kumar P, Sannigrahi S, Tzeng YL. Regulatory role of the MisR/S two-component system in hemoglobin utilization in Neisseria meningitidis. Infect Immun. 2010;78:1109–1122. doi: 10.1128/IAI.00363-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Harrison OB, Evans NJ, Blair JM, Grimes HS, Tinsley CR, Nassif X, et al. Epidemiological evidence for the role of the hemoglobin receptor, MmbR, in meningococcal virulence. J Infect Dis. 2009;200:94–98. doi: 10.1086/599377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Harrison OB, Bennett JS, Derrick JP, Maiden MC, Bayliss CD. Distribution and diversity of the haemoglobin-haptoglobin iron-acquisition systems in pathogenic and non-pathogenic Neisseria. Microbiology. 2013;159:1920–1930. doi: 10.1099/mic.0.068874-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Lucidarme J, Findlow J, Chan H, Feavers IM, Gray SJ, Kaczmarski EB, et al. The distribution and ‘in vivo’ phase variation status of haemoglobin receptors in invasive meningococcal serogroup B disease: genotypic and phenotypic analysis. PLoS One. 2013;8:e76932. doi: 10.1371/journal.pone.0076932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Fowler VG, Jr, Proctor RA. Where does a Staphylococcus aureus vaccine stand? Clin Microbiol Infect. 2014;20(5):66–75. doi: 10.1111/1469-0691.12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Kuklin NA, Clark DJ, Secore S, Cook J, Cope LD, McNeely T, et al. A novel Staphylococcus aureus vaccine: iron surface determinant B induces rapid antibody responses in rhesus macaques and specific increased survival in a murine S. aureus sepsis model. Infect Immun. 2006;74:2215–2223. doi: 10.1128/IAI.74.4.2215-2223.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.McNeely TB, Shah NA, Fridman A, Joshi A, Hartzel JS, Keshari RS, et al. Mortality among recipients of the Merck V710 Staphylococcus aureus vaccine after postoperative S. aureus infections: an analysis of possible contributing host factors. Hum Vaccin Immunother. 2014;10:3513–3516. doi: 10.4161/hv.34407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Ebert T, Smith S, Pancari G, Clark D, Hampton R, Secore S, et al. A fully human monoclonal antibody to Staphylococcus aureus iron regulated surface determinant B (IsdB) with functional activity in vitro and in vivo. Hum Antibodies. 2010;19:113–128. doi: 10.3233/HAB-2010-0235. [DOI] [PubMed] [Google Scholar]
- 213.Pancari G, Fan H, Smith S, Joshi A, Haimbach R, Clark D, et al. Characterization of the mechanism of protection mediated by CS-D7, a monoclonal antibody to Staphylococcus aureus iron regulated surface determinant B (IsdB) Front Cell Infect Microbiol. 2012;2:36. doi: 10.3389/fcimb.2012.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Parish T, Schaeffer M, Roberts G, Duncan K. HemZ is essential for heme biosynthesis in Mycobacterium tuberculosis. Tuberculosis (Edinb) 2005;85:197–204. doi: 10.1016/j.tube.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 215.Tullius MV, Harmston CA, Owens CP, Chim N, Morse RP, McMath LM, et al. Discovery and characterization of a unique mycobacterial heme acquisition system. Proc Natl Acad Sci USA. 2011;108:5051–5056. doi: 10.1073/pnas.1009516108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Owens CP, Chim N, Goulding CW. Insights on how the Mycobacterium tuberculosis heme uptake pathway can be used as a drug target. Future Med Chem. 2013;5:1391–1403. doi: 10.4155/fmc.13.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Barber MF, Elde NC. Nutritional immunity. Escape from bacterial iron piracy through rapid evolution of transferrin. Science. 2014;346:1362–1366. doi: 10.1126/science.1259329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Barber MF, Elde NC. Buried treasure: evolutionary perspectives on microbial iron piracy. Trends Genet. 2015;31:627–636. doi: 10.1016/j.tig.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Chou AC, Fitch CD. Mechanism of hemolysis induced by ferriprotoporphyrin IX. J Clin Invest. 1981;68:672–677. doi: 10.1172/JCI110302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Schmitt TH, Frezzatti WA, Jr, Schreier S. Hemin-induced lipid membrane disorder and increased permeability: a molecular model for the mechanism of cell lysis. Arch Biochem Biophys. 1993;307:96–103. doi: 10.1006/abbi.1993.1566. [DOI] [PubMed] [Google Scholar]
- 221.Van Heyningen WE. Inhibition of aerobic sporing bacilli by haematin. Nature. 1948;162:114. doi: 10.1038/162114b0. [DOI] [PubMed] [Google Scholar]
- 222.Stauff DL, Torres VJ, Skaar EP. Signaling and DNA-binding activities of the Staphylococcus aureus HssR–HssS two-component system required for heme sensing. J Biol Chem. 2007;282:26111–26121. doi: 10.1074/jbc.M703797200. [DOI] [PubMed] [Google Scholar]
- 223.Fernandez A, Lechardeur D, Derre-Bobillot A, Couve E, Gaudu P, Gruss A. Two coregulated efflux transporters modulate intracellular heme and protoporphyrin IX availability in Streptococcus agalactiae. PLoS Pathog. 2010;6:e1000860. doi: 10.1371/journal.ppat.1000860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Rasmussen AW, Alexander HL, Perkins-Balding D, Shafer WM, Stojiljkovic I. Resistance of Neisseria meningitidis to the toxic effects of heme iron and other hydrophobic agents requires expression of ght. J Bacteriol. 2005;187:5214–5223. doi: 10.1128/JB.187.15.5214-5223.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Wyckoff EE, Lopreato GF, Tipton KA, Payne SM. Shigella dysenteriae ShuS promotes utilization of heme as an iron source and protects against heme toxicity. J Bacteriol. 2005;187:5658–5664. doi: 10.1128/JB.187.16.5658-5664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Imlay JA, Chin SM, Linn S. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science. 1988;240:640–642. doi: 10.1126/science.2834821. [DOI] [PubMed] [Google Scholar]
- 227.Imlay JA, Linn S. DNA damage and oxygen radical toxicity. Science. 1988;240:1302–1309. doi: 10.1126/science.3287616. [DOI] [PubMed] [Google Scholar]
- 228.Wakeman CA, Hammer ND, Stauff DL, Attia AS, Anzaldi LL, Dikalov SI, et al. Menaquinone biosynthesis potentiates haem toxicity in Staphylococcus aureus. Mol Microbiol. 2012;86:1376–1392. doi: 10.1111/mmi.12063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Wakeman CA, Stauff DL, Zhang Y, Skaar EP. Differential activation of Staphylococcus aureus heme detoxification machinery by heme analogues. J Bacteriol. 2014;196:1335–1342. doi: 10.1128/JB.01067-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Stojiljkovic I, Kumar V, Srinivasan N. Non-iron metalloporphyrins: potent antibacterial compounds that exploit haem/Hb uptake systems of pathogenic bacteria. Mol Microbiol. 1999;31:429–442. doi: 10.1046/j.1365-2958.1999.01175.x. [DOI] [PubMed] [Google Scholar]
- 231.Nitzan Y, Wexler HM, Finegold SM. Inactivation of anaerobic bacteria by various photosensitized porphyrins or by hemin. Curr Microbiol. 1994;29:125–131. doi: 10.1007/BF01570752. [DOI] [PubMed] [Google Scholar]
- 232.Nir U, Ladan H, Malik Z, Nitzan Y. In vivo effects of porphyrins on bacterial DNA. J Photochem Photobiol B. 1991;11:295–306. doi: 10.1016/1011-1344(91)80035-g. [DOI] [PubMed] [Google Scholar]
- 233.Putker F, Grutsch A, Tommassen J, Bos MP. Ght protein of Neisseria meningitidis is involved in the regulation of lipopoly-saccharide biosynthesis. J Bacteriol. 2014;196:780–789. doi: 10.1128/JB.00943-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Stauff DL, Bagaley D, Torres VJ, Joyce R, Anderson KL, Kuechenmeister L, et al. Staphylococcus aureus HrtA is an ATPase required for protection against heme toxicity and prevention of a transcriptional heme stress response. J Bacteriol. 2008;190:3588–3596. doi: 10.1128/JB.01921-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Torres VJ, Stauff DL, Pishchany G, Bezbradica JS, Gordy LE, Iturregui J, et al. A Staphylococcus aureus regulatory system that responds to host heme and modulates virulence. Cell Host Microbe. 2007;1:109–119. doi: 10.1016/j.chom.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Lechardeur D, Cesselin B, Liebl U, Vos MH, Fernandez A, Brun C, et al. Discovery of intracellular heme-binding protein HrtR, which controls heme efflux by the conserved HrtB–HrtA transporter in Lactococcus lactis. J Biol Chem. 2012;287:4752–4758. doi: 10.1074/jbc.M111.297531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Joubert L, Derre-Bobillot A, Gaudu P, Gruss A, Lechardeur D. HrtBA and menaquinones control haem homeostasis in Lactococcus lactis. Mol Microbiol. 2014;93:823–833. doi: 10.1111/mmi.12705. [DOI] [PubMed] [Google Scholar]
- 238.Mike LA, Choby JE, Brinkman PR, Olive LQ, Dutter BF, Ivan SJ, et al. Two-component system cross-regulation integrates Bacillus anthracis response to heme and cell envelope stress. PLoS Pathog. 2014;10:e1004044. doi: 10.1371/journal.ppat.1004044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Hagman KE, Pan W, Spratt BG, Balthazar JT, Judd RC, Shafer WM. Resistance of Neisseria gonorrhoeae to antimicrobial hydrophobic agents is modulated by the mtrRCDE efflux system. Microbiology. 1995;141(Pt 3):611–622. doi: 10.1099/13500872-141-3-611. [DOI] [PubMed] [Google Scholar]
- 240.Stojiljkovic I, Hantke K. Transport of haemin across the cytoplasmic membrane through a haemin-specific periplasmic binding-protein-dependent transport system in Yersinia enter-ocolitica. Mol Microbiol. 1994;13:719–732. doi: 10.1111/j.1365-2958.1994.tb00465.x. [DOI] [PubMed] [Google Scholar]
- 241.Suits MD, Pal GP, Nakatsu K, Matte A, Cygler M, Jia Z. Identification of an Escherichia coli O157:H7 heme oxygenase with tandem functional repeats. Proc Natl Acad Sci USA. 2005;102:16955–16960. doi: 10.1073/pnas.0504289102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Ouellet YH, Ndiaye CT, Gagne SM, Sebilo A, Suits MD, Jubinville E, et al. An alternative reaction for heme degradation catalyzed by the Escherichia coli O157:H7 ChuS protein: release of hematinic acid, tripyrrole and Fe(III) J Inorg Biochem. 2016;154:103–113. doi: 10.1016/j.jinorgbio.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 243.Kaur AP, Lansky IB, Wilks A. The role of the cytoplasmic heme-binding protein (PhuS) of Pseudomonas aeruginosa in intracellular heme trafficking and iron homeostasis. J Biol Chem. 2009;284:56–66. doi: 10.1074/jbc.M806068200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Kaur AP, Wilks A. Heme inhibits the DNA binding properties of the cytoplasmic heme binding protein of Shigella dysenteriae (ShuS) Biochemistry. 2007;46:2994–3000. doi: 10.1021/bi061722r. [DOI] [PubMed] [Google Scholar]
- 245.Al Jubair T, Singh B, Fleury C, Blom AM, Morgelin M, Thunnissen MM, et al. Haemophilus influenzae stores and distributes hemin by using protein E. Int J Med Microbiol. 2014;304:662–668. doi: 10.1016/j.ijmm.2014.04.015. [DOI] [PubMed] [Google Scholar]
- 246.Negari S, Sulpher J, Pacello F, Ingrey K, Battistoni A, Lee BC. A role for Haemophilus ducreyi Cu, ZnSOD in resistance to heme toxicity. Biometals. 2008;21:249–258. doi: 10.1007/s10534-007-9113-8. [DOI] [PubMed] [Google Scholar]


