Abstract
The β-catenin functions as an adhesion molecule and a component of the Wnt signaling pathway. In the absence of the Wnt ligand, β-catenin is constantly phosphorylated, which designates it for degradation by the APC complex. This process is one of the key regulatory mechanisms of β-catenin. The level of β-catenin is also controlled by the E3 ubiquitin protein ligase SIAH-1 via a phosphorylation-independent degradation pathway. Similar to β-catenin, STAT3 is responsible for various cellular processes, such as survival, proliferation, and differentiation. However, little is known about how these molecules work together to regulate diverse cellular processes. In this study, we investigated the regulatory relationship between STAT3 and β-catenin in HEK293T cells. To our knowledge, this is the first study to report that β-catenin-TCF-4 transcriptional activity was suppressed by phosphorylated STAT3; furthermore, STAT3 inactivation abolished this effect and elevated activated β-catenin levels. STAT3 also showed a strong interaction with SIAH-1, a regulator of active β-catenin via degradation, which stabilized SIAH-1 and increased its interaction with β-catenin. These results suggest that activated STAT3 regulates active β-catenin protein levels via stabilization of SIAH-1 and the subsequent ubiquitin-dependent proteasomal degradation of β-catenin in HEK293T cells.
Keywords: β-catenin, proteasomal degradation, SIAH-1, STAT3
INTRODUCTION
The β-catenin functions as an adhesion molecule that associates with E-cadherin and actin filaments at the cell membrane and as a component of the canonical Wnt pathway (Orsulic et al., 1999). In the absence of the Wnt ligand, cytoplasmic β-catenin is constantly degraded by a “destruction complex” consisting of the scaffolding protein axin, tumor suppressor adenomatous polyposis coli gene product (APC), casein kinase 1 (CK1), and glycogen synthase kinase 3β (GSK3β). CK1 and GSK3β sequentially phosphorylate β-catenin, which results in recognition by the E3 ubiquitin protein ligase β-TrCP and subsequent ubiquitin-dependent proteasomal degradation (He et al., 2004). This continual elimination of β-catenin leads to repression of Wnt target genes, such as c-Myc, c-jun, and cyclin D1, by preventing the translocation of β-catenin to the nucleus and formation of complexes with TCF/LEF family of proteins (Liu et al., 2002; Polakis, 2000). Wnt/β-catenin signaling regulates diverse cellular processes, including organ development, cellular proliferation, morphology, motility, stemness maintenance, and fate determination (Cadigan and Nusse, 1997; DALE, 1998).
Control of β-catenin levels via constant degradation is a major regulatory mechanism, and reduced levels of β-catenin prevent its nuclear translocation and the activation of target genes. As mentioned above, phosphorylated β-catenin is recognized by β-TrCP and designated for ubiquitin-dependent proteasomal degradation. In response to the induction of p53, β-catenin also undergoes a phosphorylation-independent degradation pathway via interaction with the E3 ubiquitin protein ligase SIAH-1, which encourages the binding of ubiquitin-conjugating enzymes (E2s) and proteasomal degradation (Liu et al., 2001; Matsuzawa and Reed, 2001). Humans have two SIAH genes encoding Sina-like proteins, SIAH-1 and SIAH-2 (Hu et al., 1997). Sina was initially discovered in Drosophila as a requirement for R7 photoreceptor cell differentiation (Carthew and Rubin, 1990). The SIAH-1 protein plays a key role in many biological processes, such as the cell cycle, programmed cell death, and oncogenesis (Nemani et al., 1996).
STAT3, a member of the STAT family, is a latent transcription factor that mediates cytokine-and growth factor-directed transcription (Levy and Darnell, 2002). In response to the binding of extracellular ligands, receptor and non-receptor protein tyrosine kinases phosphorylate STAT3 at tyrosine residue 705 within the transactivation domain near the carboxy-terminus (Improta et al., 1994). Phosphorylated STAT3, the active form of STAT3, dimerizes with other activated STAT proteins in the cytoplasm to form homo- or hetero-dimers and then translocates to the nucleus to bind to DNA and stimulate the production of target genes (Darnell, 1997). STAT3 participates in a wide variety of cellular processes, including proliferation, postnatal survival, differentiation in the context of growth and development, invasion, angiogenesis, and metastasis in the context of cancer progression (Bromberg et al., 1999; Levy and Lee, 2002).
Despite their common cellular functions, such as proliferation and fate determination, research on the relationship between STAT3 and β-catenin is limited. A previous study reported that STAT3 cooperates with β-catenin to exert oncogenic effects in breast cancer cells (Armanious et al., 2010). In contrast, another study showed that treatment with siSTAT3 in HCC increased β-catenin levels (Wang et al., 2010). To study the precise regulatory relationship between STAT3 and β-catenin, we created an artificial system by transfecting STAT3 and β-catenin in HEK293T cells. Here we report that STAT3 activation regulates the protein levels of β-catenin. Specifically, STAT3 stabilizes SIAH-1, which enhances the interaction between SIAH-1 and active β-catenin and results in the ubiquitin-dependent proteasomal degradation of β-catenin in HEK293T cells.
MATERIAL AND METHODS
Cell lines
Human embryonic kidney cells, HEK293T was maintained in DMEM (Life Technologies, Inc., USA) containing 10% FBS (Life Technologies, Inc.) and 1% antibiotics (Life Technologies, Inc.). The cells were maintained in a humidified incubator at 37°C in the presence of 5% CO2.
Plasmids
To generate expression vector for STAT3 cDNA (GenBankTM accession number NM_213662, purchased from Origene Technologies Inc.), the corresponding STAT3 cDNA was cloned in-frame into pLL3.7 vector (Enzynomics Inc.). A point mutant plasmids of the tyrosine 705 residue, the serine 727 residue, and the tyrosine 705 and serine 727 residues of STAT3 was made using pLL3.7-STAT3 (Enzynomics Inc.). The plasmid encoding constitutively activated STAT3 used in the present study, namely pCMV-caSTAT3, was purchased from Addgene (USA). HA-β-catenin was kindly provided by Dr. Jong-Wan Park.
Transfection, siRNA and MG132 treatment
The transient plasmid DNA transfection was done using Lipofectamine 2000 (Invitrogen, UK) according to the manufacturer’s instructions. siRNAs against STAT3 and negative control were used at 100 nM on HEK293T cells transfected using Qiagen HiPerfect according to the manufacturer’s instructions (Catalog No. 301705). MG132 (10 μM) (PeproTech) was used to treat HEK293T cells for 10 h before harvest.
Western blot analysis
Cells were washed with cold PBS and lysed in a cold Radio ImmunoPrecipitation Assay (RIPA) buffer containing protease inhibitor (2 mM PhenylMethylSulphonylFluoried (PMSF), 10 μg/ml leupeptin and 2 mM EthyleneDiamineTetraAcetic acid (EDTA)). The lysates were collected and centrifuged for 20 min at 13,000 rpm at 4°C, and the supernatants were collected. Equal amounts of proteins from the supernatants were separated by SDS-PAGE and transferred on to nitrocellulose (NC) membrane. The membranes were blocked in a TBS-T containing 5% non-fat dried milk for at least one hour and subsequently incubated with specific primary antibodies overnight at 4°C. After washing with TBS-T for 30 min at room temperature, the membranes were further incubated with a HRP-conjugated secondary antibody for 1 h. After washing with TBS-T, the signals were detected using SuperSignal West Femto (Thermoscientific, USA). The following antibodies were used: phosphotyrosine STAT3, total STAT3, HA-tag, and ubiquitin (Cell signaling technology, USA), and active β-catenin, Myc-tag (Millipore), and total β-catenin (Santa Cruz Biotechnologies, USA) and α-tubulin (Thermoscientific).
Immunoprecipitation
Cell extracts were precleared with anti-Myc antibody or anti-HA antibody overnight at 4°C and incubated with protein G sepharose beads for 1 h at 4°C. The beads were then washed five times with ice-cold lysis buffer and suspended in SDS sample loading buffer. Western blotting was then performed.
Luciferase assay
Cells were cotransfected with both 0.25 to 2 μg each of reporter construct and CMV-β-gal plasmid. Luciferase activity was measured using Lumat-LB960 luminometer (Berthold), and divided by β-gal activity to normalize the transfection efficiency.
Statistical analysis
Data are presented as average ± Standard Deviation (SD) and statistical significance was determined by unpaired two-tailed Student’s t test implemented in Microsoft Excel software. **p < 0.001, *p < 0.01.
RESULTS
Activated STAT3 suppresses β-catenin-TCF-4 transcriptional activity
To determine if constitutively activated STAT3 (caSTAT3) influences β-catenin transcriptional activity, we analyzed TOP-flash reporter activity in human embryonic kidney (HEK293T) cells overexpressing both caSTAT3 and β-catenin. The protein levels of phosphorylated STAT3 at tyrosine residue 705 and total STAT3 showed a concentration-dependent elevation based on the amount of transfected DNA (Fig. 1A). The β-catenin/TCF-4-dependent transcriptional activity showed a concentration-dependent decrease in response to the amount of transfected caSTAT3. To confirm that STAT3 activation is associated with decreased β-catenin transcriptional activity, we completed cotransfections of several STAT3 mutants with β-catenin (Fig. 1B). The decreased TOP-flash activity induced by STAT3 was restored to control levels in cells transfected with STAT3 mutants. These results contradicted previous reports that STAT3 aids in β-catenin activation in cancer cells (Armanious et al., 2010). Thus, we confirmed that activated STAT3 supports the transcriptional activity of β-catenin in breast cancer cells (Supplementary Figs. 1A and 1B) but not in human embryonic kidney cells. These findings indicate that phosphorylated STAT3 suppresses the transcriptional activity of β-catenin in non-tumorous cells.
Fig. 1.
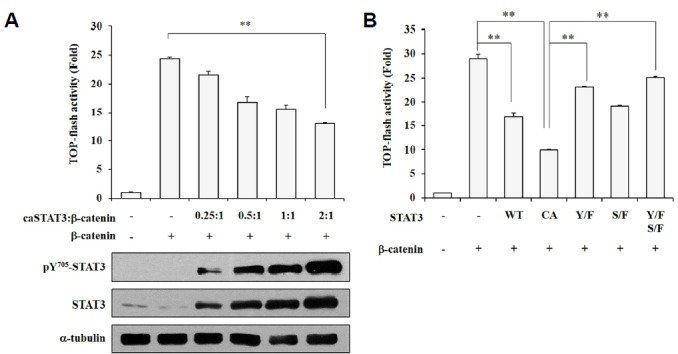
Activated STAT3 suppresses β-catenin-TCF-4 transcriptional activity. (A) HEK293T cells were transfected with caSTAT3 and β-catenin. After a 24-h incubation, cells were subjected to a luciferase reporter assay. HEK293T cells were transfected with caSTAT3 and β-catenin for 24 h and then harvested for Western blot analysis. (B) HEK293T cells were transfected with wild-type STAT3 (WT), caSTAT3 (CA), dominant-negative Y705F STAT3 mutant (Y/F), dominant-negative S727F STAT3 mutant (S/F), dominant-negative double STAT3 mutant (Y/F-S/F), or β-catenin. The luciferase reporter activity was measured. (A, B) β-gal activity was used to normalize transfection efficiency. Data are shown as the mean ± SD, **P < 0.001.
Inhibition of STAT3 increases active β-catenin levels
To examine the role of STAT3 on the protein levels of β-catenin, we performed a siRNA-mediated STAT3 knockdown with or without a GSK inhibitor treatment in HEK293T cells. Western blot analysis showed that the GSK inhibitor increased the protein levels of endogenous β-catenin. STAT3 knockdown combined with a GSK inhibitor treatment further enhanced the protein levels of active β-catenin (Fig. 2A). To confirm if the inhibition of STAT3 activity also increased non-phosphorylatable mutant β-catenin (active-β-catenin), we incubated cells with AG490 (40 μM) or a STAT3 inhibitor (10 μM) overnight. HA-β-catenin is a mutant form of β-catenin, also termed active β-catenin because the phosphorylation of the serine 33 residue of β-catenin is mutated and thus cannot be degraded by the β-TrCP-dependent pathway. As shown in Fig. 2B, treatment with either AG490 or a STAT3 inhibitor increased the protein levels of active β-catenin. These results suggest that STAT3 is an important regulator of active β-catenin levels.
Fig. 2.
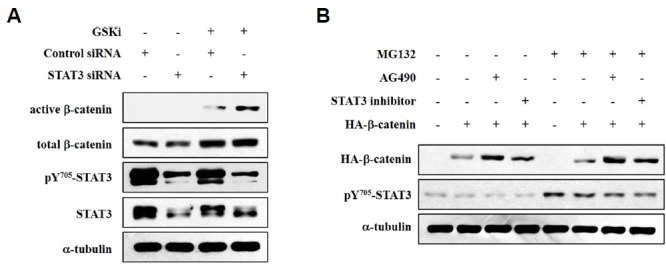
Inhibition of STAT3 elevates active β-catenin levels. (A) HEK293T cells were transfected with STAT3 siRNA (100 nM) or a scrambled control siRNA (100 nM, nonspecific siRNA used as a negative control). After treatment with a GSK inhibitor at a concentration of 1 μM for 4 h, cells were harvested, and the proteins were extracted in RIPA buffer for Western blotting. Anti-α-tubulin was used as a loading control. (B) HEK293T cells were transfected with β-catenin and then treated with 10 μM MG132 for 10 h before harvesting. Cells were incubated with 40 μM AG490 or a 10 μM STAT3 inhibitor overnight before harvesting. Cells were harvested, and the proteins were extracted in RIPA buffer for Western blotting. Anti-α-tubulin was used as the loading control.
STAT3 stabilizes SIAH-1 protein via a direct interaction
STAT3 knockdown increased the levels of non-phosphorylatable mutant β-catenin, which suggests that SIAH-1 mediates the degradation of active β-catenin. To determine if STAT3 regulates SIAH-1, we examined the effect of STAT3 on the RNA and protein levels of SIAH-1 in HEK293T cells using RT-PCR and Western blotting, respectively. As shown in Fig. 3A, the RNA expression of SIAH-1 was not effected by STAT3 transfection; however, STAT3 activation increased the protein levels of SIAH-1. This result indicates that SIAH-1 stability is regulated by STAT3 activation. Next, we confirmed the STAT3-dependence of SIAH-1 protein levels using STAT3 siRNA and a STAT3 expression plasmid (Fig. 3B). We speculated that STAT3 interacts with SIAH-1 to improve protein stability; thus, we assessed the protein–protein interaction between STAT3 and SIAH-1 using immunoprecipitation. Either myc-tagged wild type STAT3 (wtSTAT3-myc) or myc-tagged STAT3 mutated at tyrosine residue 705 (STAT3-Y705F-myc) was transfected in HEK293T cells, and immunoprecipitation was performed with an anti-myc antibody (Fig. 3C). Ectopic STAT3 co-precipitated with endogenous SIAH-1, and the interaction between STAT3 and SIAH-1 was reduced when STAT3 activation was decreased via the STAT3 mutation at tyrosine residue 705. These results indicate that STAT3 activation is essential for its interaction with SIAH-1. To evaluate if β-catenin affects the interaction between STAT3 and SIAH-1, wtSTAT3-myc and HA-β-catenin was co-transfected in HEK293T cells. Next, we performed Western blotting and co-immunoprecipitation with an anti-myc antibody (Fig. 3D). The data show that STAT3 interacted with SIAH-1, despite the overexpression of HA-β-catenin. These results suggest that STAT3 interacts with SIAH-1 and that active β-catenin does not influence the interaction between STAT3 and SIAH-1.
Fig. 3.
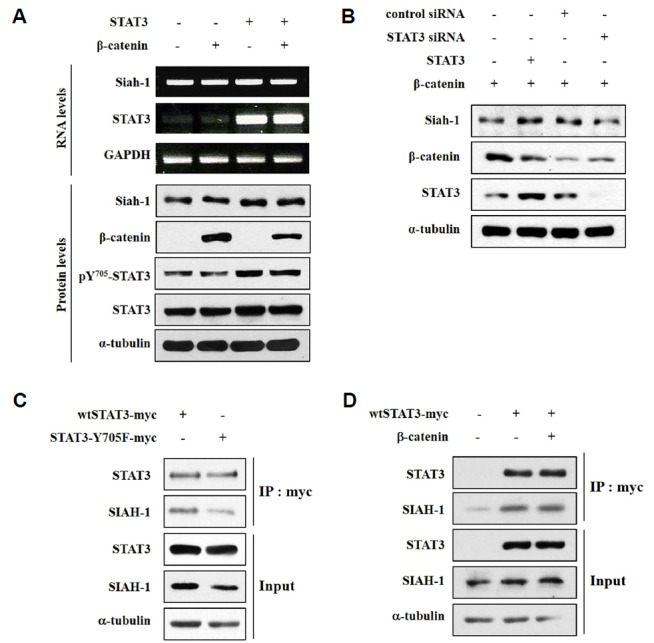
STAT3 stabilizes SIAH-1 via a direct interaction. (A) HEK293T cells were transfected with wtSTAT3 and β-catenin. After 24 h, cells were harvested and the mRNA and proteins were extracted for RT-PCR and Western blotting, respectively. (B) HEK293T cells were transfected with siSTAT3 (100 nM) or scrambled siRNA (100 nM, nonspecific siRNA used as a negative control) and with wtSTAT3, β-catenin, or a control pCMV vector. After culturing for 24 h, cells were subjected to Western blotting. (C) HEK293T cells were transfected with wtSTAT3-myc or STAT3-Y705F-myc. After 24 h, an immunoprecipitation assay was performed with an anti-myc-antibody, and Western blotting was completed with the indicated antibodies. (D) HEK293T cells were transfected with wtSTAT3-myc and β-catenin. After 24 h, an immunoprecipitation assay was performed with an anti-myc-antibody and Western blotting was completed with the indicated antibodies.
STAT3 facilitates the proteasomal degradation of β-catenin
Activated STAT3 increases the protein levels of SIAH-1 via a direct interaction; thus, it is possible that STAT3 promotes the interaction between SIAH-1 and leads to proteosomal degradation of active β-catenin. The results showed that co-expression of STAT3 and HA-β-catenin increased the binding of endogenous SIAH-1 and HA-β-catenin (Fig. 4A). These findings indicate that HA-β-catenin easily undergoes proteasomal degradation through SIAH-1. To examine if STAT3 enhances polyubiquitination of HA-β-catenin, STAT3 and HA-β-catenin were co-expressed in HEK293T cells in the presence or absence of the proteasome inhibitor MG132 (10 μM) to check for the ubiquitination of HA-β-catenin. As shown in Fig. 4B, the polyubiquitination of HA-β-catenin was increased in the presence of both MG132 and a STAT3 inhibitor. Taken together, our results suggest distinct role of phosphorylated STAT3 in β-catenin degradation in non-tumorous cells. STAT3 enhances the protein expression and transcriptional activity of b-catenin in many type of tumor. In contrast, STAT3 activation leads to proteosomal degradation of active β-catenin by stabilization of SIAH-1 in the developmental process (Fig. 4C).
Fig. 4.
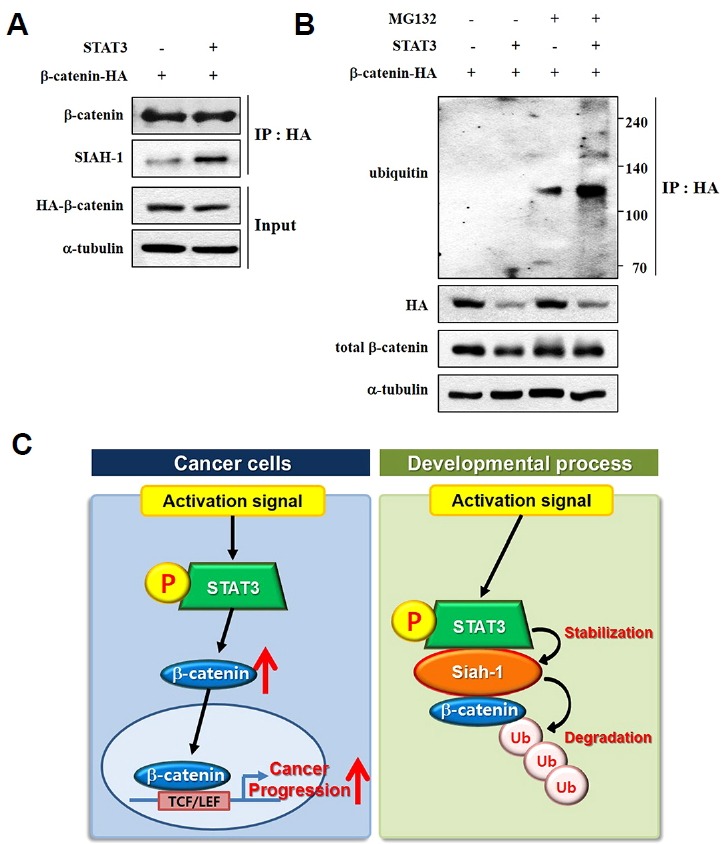
STAT3 facilitates the proteasomal degradation of β-catenin. (A) HEK293T cells were transfected with wtSTAT3-myc and β-catenin-HA. After 24 h, cells were harvested. We performed an immunoprecipitation assay with an anti-HA-antibody, and Western blotting was completed with the indicated antibodies. (B) HEK293T cells were transfected with wtSTAT3-myc and β-catenin-HA and then treated with 10 μM MG132 for 10 h before harvest. After 24 h of transfection, cells were harvested. An immunoprecipitation assay was performed with an anti-HA-antibody, and Western blotting was completed with the indicated antibodies. (C) Graphical summarization. Two distinct mechanisms of β-catenin degradation by activated STAT3 is described.
DISCUSSION
STAT3 is responsible for various cellular processes, such as survival, proliferation, and differentiation. The β-catenin, as a co-activator of canonical Wnt signaling, is also involved in diverse cellular procedures. Nevertheless, little is known about whether these molecules work with or regulate each other. In this report, we found that STAT3 controls active β-catenin protein levels through regulation of SIAH-1, leading to the ubiquitin-dependent proteasomal degradation of active β-catenin in nontumorous HEK293T cells.
In cancer, the synergistic role between phosphorylated STAT3 and active β-catenin to potentiate tumorigenic progression has been reported (Armanious et al., 2010; Bromberg et al., 1999; Polakis, 2000; Wang et al., 2010). Thus, targeting STAT3 phosphorylation and active β-catenin gives therapeutic benefits in malignant tumors. On the other hand, STAT3 and β-catenin also have critical role such as embryonic stem cell pluripotency in non-tumorous embryonic cells (Hao et al., 2006; Kielman et al., 2002). During developmental processes, both STAT3 and β-catenin play important roles in cell survival, maintenance of pluripotency, and specific cellular differentiation. The Wnt/β-catenin pathway acts to prevent ES cell differentiation through convergence on the LIF/JAK-STAT3 pathway (Hao et al., 2006). However, genetic and molecular evidence shows that the ability and sensitivity of embryonic stem cells to differentiate into the three germ layers is inhibited by increased doses of β-catenin by specific Apc mutations (Kielman et al., 2002). Sokol and his colleagues suggest that Wnt/β-catenin signaling pathway is recognized as a potential mechanism to reinforce embryonic cell fate decision (Sokol, 2011). For this reason, the knowledge of the mechanism to regulate the balance of β-catenin protein levels is important in embryonic developmental stage.
In this study, we found that phosphorylated STAT3 inhibits the transcriptional activity of β-catenin in non-tumorous HEK293T cells (Fig. 1). Interestingly, phosphorylated STAT3 repress only non-phosphorylatable β-catenin (active β-catenin) and these data suggested that phosphorylated STAT3 regulates non-canonical β-catenin degradation pathway not β-TrCP-dependent degradation pathway (Fig. 2). Expectedly, SIAH-1, the major component of non-canonical β-catenin degradation pathway, stabilized by protein-protein interaction with activated STAT3 (Fig. 3). Through the stabilization of SIAH-1 protein, STAT3 facilitates the proteasomal degradation of β-catenin (Fig. 4). From these results, we suggest that STAT3 activation is involved in the degradation of active β-catenin via SIAH-1 in non-tumorous cells.
Our findings provide not only a comprehensive understanding of the interaction between STAT3 and SIAH-1 but also a novel mechanism of β-catenin degradation by STAT3 phosphorylation might be one of the strategy to orchestrate developmental process of embryonic cells.
Supplementary Information
ACKNOWLEDGMENTS
This work was supported by grants from the National R&D program for Cancer Control, Ministry of Health & Welfare (0720540), the National Research Foundation of Korea (NRF) funded by the Korea government (MESF; 2014R1A2A1A11053203), Seoul National University Hospital (SNUH) Research Fund, Republic of Korea (3420130270 and 0320140100), This work was supported by the Brain Korea 21 PLUS program.
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Armanious H., Gelebart P., Mackey J., Ma Y., Lai R. STAT3 upregulates the protein expression and transcriptional activity of beta-catenin in breast cancer. Int J Clin Exp Pathol. 2010;3:654–664. [PMC free article] [PubMed] [Google Scholar]
- Bromberg J.F., Wrzeszczynska M.H., Devgan G., Zhao Y., Pestell R.G., Albanese C., Darnell J.E. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- Cadigan K.M., Nusse R. Wnt signaling: a common theme in animal development. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- Carthew R.W., Rubin G.M. seven in absentia, a gene required for specification of R7 cell fate in the Drosophila eye. Cell. 1990;63:561–577. doi: 10.1016/0092-8674(90)90452-k. [DOI] [PubMed] [Google Scholar]
- DALE C.T. Signal transduction by the Wnt family of ligands. Biochem J. 1998;329:209–223. doi: 10.1042/bj3290209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell J.E. STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- Hao J., Li T.-G., Qi X., Zhao D.-F., Zhao G.-Q. WNT/β-catenin pathway up-regulates Stat3 and converges on LIF to prevent differentiation of mouse embryonic stem cells. Dev Biol. 2006;290:81–91. doi: 10.1016/j.ydbio.2005.11.011. [DOI] [PubMed] [Google Scholar]
- He X., Semenov M., Tamai K., Zeng X. LDL receptor-related proteins 5 and 6 in Wnt/β-catenin signaling: arrows point the way. Development. 2004;131:1663–1677. doi: 10.1242/dev.01117. [DOI] [PubMed] [Google Scholar]
- Hu G., Chung Y.-L., Glover T., Valentine V., Look A.T., Fearon E.R. Characterization of Human Homologs of the Drosophila seven in absentia (sina) gene. Genomics. 1997;46:103–111. doi: 10.1006/geno.1997.4997. [DOI] [PubMed] [Google Scholar]
- Improta T., Schindler C., Horvath C.M., Kerr I.M., Stark G., Darnell J. Transcription factor ISGF-3 formation requires phosphorylated Stat91 protein, but Stat113 protein is phosphorylated independently of Stat91 protein. Proc Natl Acad Sci USA. 1994;91:4776–4780. doi: 10.1073/pnas.91.11.4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielman M.F., Rindapää M., Gaspar C., van Poppel N., Breukel C., van Leeuwen S., Taketo M.M., Roberts S., Smits R., Fodde R. Apc modulates embryonic stem-cell differentiation by controlling the dosage of β-catenin signaling. Nat Genet. 2002;32:594–605. doi: 10.1038/ng1045. [DOI] [PubMed] [Google Scholar]
- Levy D.E., Darnell J. Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- Levy D.E., Lee C.-k. What does Stat3 do? J Clin Invest. 2002;109:1143–1148. doi: 10.1172/JCI15650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Stevens J., Rote C.A., Yost H.J., Hu Y., Neufeld K.L., White R.L., Matsunami N. Siah-1 mediates a novel β-catenin degradation pathway linking p53 to the adenomatous polyposis coli protein. Mol Cell. 2001;7:927–936. doi: 10.1016/s1097-2765(01)00241-6. [DOI] [PubMed] [Google Scholar]
- Liu C., Li Y., Semenov M., Han C., Baeg G.-H., Tan Y., Zhang Z., Lin X., He X. Control of β-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell. 2002;108:837–847. doi: 10.1016/s0092-8674(02)00685-2. [DOI] [PubMed] [Google Scholar]
- Matsuzawa S.-i., Reed J.C. Siah-1, SIP, and Ebi collaborate in a novel pathway for β-catenin degradation linked to p53 responses. Mol Cell. 2001;7:915–926. doi: 10.1016/s1097-2765(01)00242-8. [DOI] [PubMed] [Google Scholar]
- Nemani M., Linares-Cruz G., Bruzzoni-Giovanelli H., Roperch J.- P., Tuynder M., Bougueleret L., Cherif D., Medhioub M., Pasturaud P., Alvaro V. Activation of the human homologue of the Drosophila sina gene in apoptosis and tumor suppression. Proc Natl Acad Sci USA. 1996;93:9039–9042. doi: 10.1073/pnas.93.17.9039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsulic S., Huber O., Aberle H., Arnold S., Kemler R. E-cadherin binding prevents beta-catenin nuclear localization and beta-catenin/LEF-1-mediated transactivation. J Cell Sci. 1999;112:1237–1245. doi: 10.1242/jcs.112.8.1237. [DOI] [PubMed] [Google Scholar]
- Polakis P. Wnt signaling and cancer. Genes Dev. 2000;14:1837–1851. [PubMed] [Google Scholar]
- Sokol S.Y. Maintaining embryonic stem cell pluripotency with Wnt signaling. Development. 2011;138:4341–4350. doi: 10.1242/dev.066209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.-H., Meng X.-W., Xing H., Qu B., Han M.-Z., Chen J., Fan Y.-J., Lu C.-Q., Lu Z.-W. STAT3 and betacatenin signaling pathway may affect GSK-3beta expression in hepatocellular carcinoma. Hepato-gastroenterology. 2010;58:487–491. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


