Abstract
Extracytoplasmic function (ECF) σ factors have roles related to cell envelope and/or cell membrane functions, in addition to other cellular functions. Without cell-surface stresses, ECF σ factors are sequestered by the cognate anti-σ factor, leading to inactivation and the resultant repression of regulons due to the inhibition of transcription of their own genes. Bacillus subtilis has seven ECF σ factors including σX and σM that transcribe their own structural genes. Here, we report that glucose addition to the medium induced sigX and sigM transcription independent of their anti-σ factors. This induction was dependent on an intracellular acetyl-CoA pool. Transposon mutagenesis searching for the mutants showing no induction of sigX and sigM revealed that the cshA gene encoding DEAD-box RNA helicase is required for gene induction. Global analysis of the acetylome in B. subtilis showed CshA has two acetylated lysine residues. We found that in a cshA mutant with acetylation-abolishing K to R exchange mutations, glucose induction of sigX and sigM was abolished and that glucose addition stimulated acetylation of CshA in the wild type strain. Thus, we present a model wherein glucose addition results in a larger acetyl-CoA pool, probably leading to increased levels of acetylated CshA. CshA is known to associate with RNA polymerase (RNAP), and thus RNAP with acetylated CshA could stimulate the autoregulation of sigX and sigM. This is a unique model showing a functional link between nutritional signals and the basal transcription machinery.
Keywords: protein lysine acetylation, nutritional signal, transposon mutagenesis, sigma factor, RNA polymerase
Introduction
The association of σ factor with RNAP determines the specific binding of the RNAP holoenzyme to target promoters (Helmann and Chamberlin, 1988). The varieties of σ factors enhance the chance for survival in the environment via fine-tuning of the regulation of gene expression. Bacteria have several distinct σ factors including extracytoplasmic function (ECF) σ factors, which are composed of large paralog gene families (Souza et al., 2014; Helmann, 2016). ECF σ factors commonly respond to cell envelope stresses and transcribe genes related to the stress response (Souza et al., 2014; Helmann, 2016). In Bacillus subtilis, seven ECF σ factors, σM, σV, σW, σX, σY, σZ, and σY laC are encoded in the genome (Helmann, 2016). In many cases, these σ factors recognize similar nucleotide sequences. σM, σV, σW, and σX bind to the consensus -35 and -10 elements, (T)GAAACNT and CGT(C/A)T, respectively (Helmann, 2016). As a result, these σ-factor regulons overlap, yet there are genes transcribed by a single ECF σ factor. σX- and σM-RNAPs transcribe their own genes: sigX and sigM, respectively (Asai et al., 2003; Helmann, 2016). The σX regulon contains the genes required for resistance against the antibiotic nisin and the peptidoglycan synthesis inhibitor bacitracin, and the genes encoding enzymes for D-alanylation of teichoic acids, which are required for the resistance against cationic antimicrobial peptides (Huang and Helmann, 1998; Cao and Helmann, 2002, 2004; Asai et al., 2003). In the sigX mutant, levels of heat and oxidative stress resistance are low, but the mechanism responsible for this is not yet known (Huang et al., 1997). The σM regulon includes >60 genes such as core genes for cell wall biogenesis and cell division (rodA, divIC, mreBCDminCD, and murBdivIB), regulatory genes (spx and abh), and the bacitracin resistance gene bcrC (Cao and Helmann, 2002; Thackray and Moir, 2003; Eiamphungporn and Helmann, 2008; Luo and Helmann, 2009).
In most cases, a gene encoding an ECF σ factor is associated with a gene encoding a corresponding anti-σ factor (Ho and Ellermeier, 2012). The anti-σ factor embedded in the cell membrane traps the cognate σ factor through a protein–protein interaction, leading to repressed expression of the regulon. A detailed mechanism for the release of σ factor from anti-σ factor in B. subtilis is well-understood for σW and includes specific proteolysis of its cognate anti-σ factor (Ho and Ellermeier, 2012). Exposure to antibiotics that interfere with cell wall biosynthesis induces ECF σ factors (e.g., the peptidoglycan synthesis inhibitor vancomycin induces sigM) (Thackray and Moir, 2003). In addition, some mutations in the cell envelope biosynthesis pathway induce the sigX and/or sigM regulons (Hashimoto et al., 2013). For example, a mutation in yfhO, encoding a probable flippase for polymers synthesized by the CsbB glucosyltranfserase, enhanced sigM regulon expression (Inoue et al., 2013). A mutation in ugtP, which encodes a UDP glucosyltransferase, induces expression of the σX regulon (Matsuoka et al., 2011). Transposon mutagenesis for mutants that upregulate sigX promoter activity, that is, screening for inhibiting genes of σX activity, resulted in identification of seven mutations including a multidrug efflux pump gene (Turner and Helmann, 2000).
In this study, we identified that protein lysine acetylation is involved in sigX and sigM regulation. Protein lysine acetylation is a well-conserved protein modification in both eukaryotes and prokaryotes (Wang et al., 2010; Thao and Escalante-Semerena, 2011; Bernal et al., 2014). In prokaryotes, including B. subtilis, several global analyses of protein lysine acetylation have been reported, although the functional analysis of each gene was poorly performed (Kim et al., 2013; Kosono et al., 2015; Carabetta et al., 2016).
Spx is a global regulatory protein with a role in the disulfide stress response and the other cellular processes (Zuber, 2004; Rochat et al., 2012). Spx is bound to α-subunit of RNAP. We observed glucose induction (GI) of the spx gene in sporulation medium (Shiwa et al., 2015). Another group has also reported GI of the spx regulon in LB medium supplemented with glucose (Yang et al., 2014). Here, we report that glucose addition to the medium induced sigX and sigM transcription independent of their anti-σ factors. The GI of spx was caused by the GI of sigX and sigM. The induction would be dependent on the cellular acetyl-CoA pool. Transposon mutagenesis for the mutant showing no induction of sigX and sigM revealed that cshA, which encodes DEAD-box RNA helicase, is required for induction (Lehnik-Habrink et al., 2010). According to the global acetylome analysis in B. subtilis, CshA has two lysine residues that can be acetylated (Kosono et al., 2015). We showed that in the cshA mutant with acetylation-abolishing K to R exchange mutations, GI of sigX and sigM was abolished and that glucose addition stimulated acetylation of CshA. Thus, we present a model in which glucose addition results in a larger acetyl-CoA pool, probably leading to an increase in acetylated CshA. CshA is known to be associated with RNAP (Delumeau et al., 2011), and thus RNAP with acetylated CshA could stimulate autoregulation of sigX and sigM.
Materials and Methods
Strains, Media, and β-Galactosidase Analysis
All of the B. subtilis strains used in this study are listed in Supplementary Table S1. One-step competence medium (Kunst et al., 1994) [MC], Schaeffer’s sporulation medium (Schaeffer et al., 1965), and Luria-Bertani (LB) medium (Difco, Lennox) were used. Antibiotic concentrations were described previously (Ogura and Tanaka, 1996; Ogura et al., 1997). Synthetic oligonucleotides were commercially prepared by Tsukuba Oligo Service (Ibaraki, Japan) and are listed in Supplementary Table S2.
Growth Condition
Strains were grown on a LB agar plate (1.5%) containing appropriate antibiotics at 37°C overnight. The cells were scraped from the plate and suspended in the sporulation medium. The suspension was inoculated into 50 ml sporulation medium (with or without glucose) without antibiotics in a 200-ml flask. Klett value was adjusted around 10 units. The flask was gently shaken (110 reciprocation/min) at 37°C. Cell growth was monitored using Klett colorimeter (Klett Mfg., Co., Inc., New York, NY, USA).
Plasmid Construction
The plasmids used in this study are listed in Supplementary Table S1. pIS284-sigM-del1 and pDG1663-sigX-del2 were constructed by cloning of the double-stranded oligonucleotides PsigM-F/PsigM-R and PsigX-F/PsigX-R into pIS284 and pDG1663 which were treated with EcoRI/BamHI, respectively (Guérout-Fleury et al., 1996; Tsukahara and Ogura, 2008). To construct pDG1663-sigX-Wt and pDL2-sigX-del1, PCR products amplified by using the oligonucleotide pair SigX-F/SigX-R and SigX-F/SigX-R2, respectively, were digested with EcoRI/BamHI and cloned into pDG1663 and pDL2 treated with the same enzymes (Yuan and Wong, 1995). To construct pDG1729-PcshA, PCR products amplified by using the oligonucleotide pair, pDG1729-PcshA-E/pDG1729-PcshA-H, were digested with EcoRI/HindIII and cloned into pDG1729 treated with the same enzymes (Guérout-Fleury et al., 1996). To construct pMUTIN3DZ, PCR products amplified by using the oligonucleotide pair, ypuN-F/ypuN-R, were digested with HindIII/BamHI and cloned into pMUTIN3DZ treated with the same enzymes (Yoshimura et al., 2004).
Construction of Strains
The bkdB::Pxyl-cshA unit was constructed using PCR (Supplementary Figure S1). The bkdB-up linked to Kmr and xylR-Pxyl units were amplified from the total DNA of the strain containing bkdB::sinR and the pX plasmid, respectively (Hori et al., 2002; Ogura, 2016). The other fragments were amplified from the wild type derived total DNA. PrimeSTAR MAX DNA polymerase was used (TaKaRa). Four DNA fragments were assembled by using Gibson Assembly Master Mix (New England Biolabs). The sequences of the cshA ORF in the resultant strains were confirmed by sequencing using the primers pX-cshA-seq-F and pX-cshA-seq-R. The cshA::Tcr unit was constructed using PCR. Briefly the upstream and downstream regions and Tcr from pBEST304 (Itaya, 1992) were amplified using the indicated primers (Supplementary Table S2) and then combined by PCR.
Sublancin Production Assay
Spot-on-lawn assays were performed as previously reported (Luo and Helmann, 2009). To prepare the lawn, overnight culture of the Sublancin-sensitive Y13 cells in liquid LB medium was inoculated into 4 ml of melted 0.7% LB agar (2%), and poured onto the solid 1.5% LB agar plate. Both media were supplemented with or without 1% glucose. Plates were dried for 30 min in an incubator (37°C), and 3 μl of each overnight culture in liquid LB medium was spotted on plates and incubated at 37°C.
Transposon Mutagenesis
The transposon delivery vector pMarA (Le Breton et al., 2006) was introduced into the strain OAM709. The resultant strain was grown in liquid LB medium containing kanamycin at 30°C overnight. The cells were diluted and plated onto sporulation medium with 1.5% agar plates containing 2% glucose, X-gal (100 μg/ml), kanamycin, and erythromycin. The plates were incubated at 42°C and the pale colonies were chosen. Otherwise, the diluted cells were plated onto LB medium with 1.5% agar plates containing kanamycin and erythromycin, and incubated at 42°C. Colonies were then transferred onto sporulation medium with 1.5% agar plates containing 2% glucose, X-gal (100 μg/ml), kanamycin, and erythromycin. The plates were incubated at 37°C to choose pale colonies. The insertion mutations were backcrossed into the parental strain and the resultant strains were used for the Lac-assay. Total DNA was taken from the candidate strain, SauIIIA1-digested, self-ligated, and subjected to inverse PCR using oligonucleotides 695 and 696 as described previously (Chan et al., 2014, Supplementary Table S2). The PCR products were sequenced using the oligonucleotide 696.
Purification of CshA-His
CshA-His was purified using a Ni-affinity column from B. subtilis OAM730 (cshA bkdB::Pxyl-cshA-His) cells as described previously (Ogura et al., 2014).
Western Blot Analysis
Western blot analysis was performed by a method described previously (Hata et al., 2001). Monoclonal anti-acetylated lysine rabbit antibody was purchased from Cell Signaling Technology (Danvers, MA, USA). This antibody was diluted (1/1000) in 1x TBS with 5% BSA and 0.1% Tween20, and the solution was incubated with a protein-blotted nitrocellulose filter overnight at 4°C. To enhance signal, Can Get Signal solution 2 (ToYoBo) was used for secondary antibody.
Results
Abolishment of GI of spx in the sigX and sigM Mutants
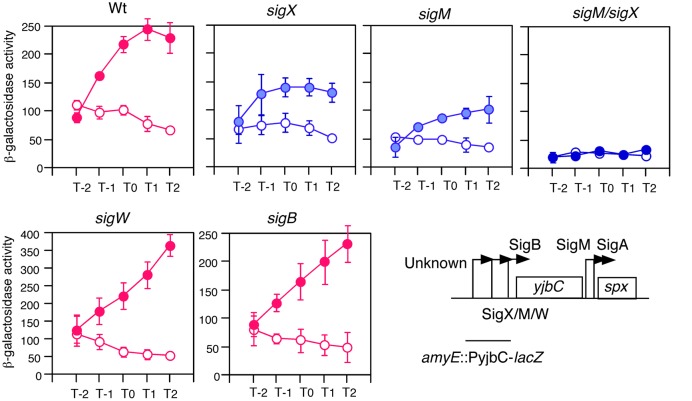
We observed GI of the spx gene in sporulation medium (Shiwa et al., 2015). The transcription of the yjbC-spx operon is complex. Five promoters and five σ factors have been reported to play roles in transcription of the operon (Antelmann et al., 2000; Thackray and Moir, 2003; Cao and Helmann, 2004; Jervis et al., 2007, Figure 1). To clarify which σ factors other than σA are responsible for GI, we introduced several mutations in the genes encoding σ factors and examined expression of the promoter for yjbC fused to lacZ. The typical cell growth in the sporulation medium with or without 2% glucose is shown (Supplementary Figure S2A). In the sigX and sigM mutants GI was reduced (Figure 1). In a double mutant of sigX and sigM, GI was completely lost, while in the sigB and sigW mutants, GI was still observed (Figure 1). Similar results were obtained using an spx-lacZ fusion (Supplementary Figure S3). This raised the possibility that sigX and sigM themselves might be induced by glucose.
FIGURE 1.
GI of PyjbC promoter and effects of σ gene disruption. Cells were grown in sporulation medium with (closed symbols) or without (open symbols) 2% glucose. Data sets showing GI and not showing GI are shown in red and blue, respectively. Averages from at least three independent experiments and standard deviations (error bar) are shown. Cells were sampled hourly. β-galactosidase activities are shown in Miller units. The X-axis represents the growth time in hours relative to the end of vegetative growth (T0, see Supplementary Figure S2). The chromosomal structure of PyjbC-lacZ in OAM702 is shown. Each introduced gene disruption into OAM702 is indicated above the panel. Boxes and bent arrows show open-reading frames and promoters, respectively. Text along with the bent arrow show the σ factors responsible for the promoter activity.
GI of sigX and sigM Expression
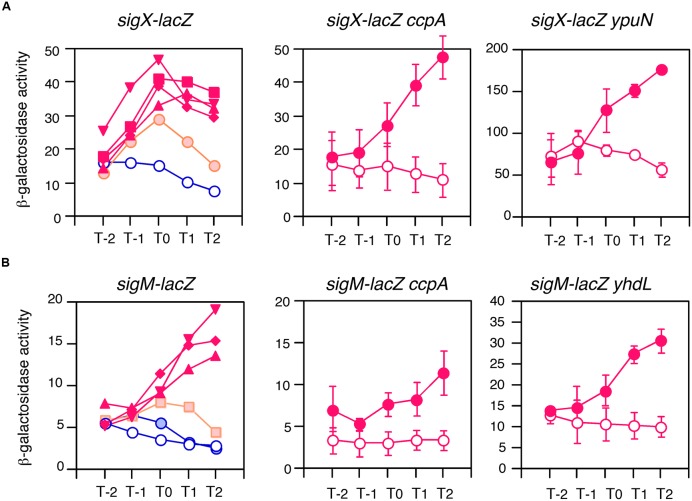
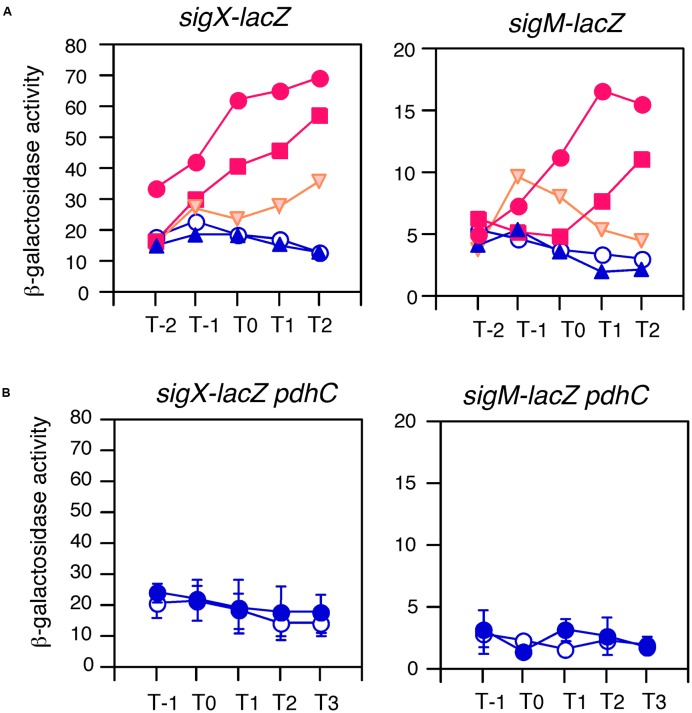
To examine this possibility, we tested the expression of two genes in glucose-containing medium. As expected, sigX and sigM were induced by glucose in sporulation medium (Figure 2). This induction was very sensitive to low concentrations of glucose, 0.05% for sigX and 0.1% for sigM. In B. subtilis the ccpA gene has been known to play a major role in glucose-dependent gene induction and repression (Fujita, 2009). Thus, to test whether CcpA is involved in GI of sig genes, we examined the expression of sigX and sigM in a ccpA disruptant in the glucose-containing medium and observed normal GI (middle panels in Figure 2). This demonstrated that CcpA is not involved in GI.
FIGURE 2.
Anti-σ factor-independent GI of sigX and sigM. Cells were grown in sporulation medium and sampled hourly for β-galactosidase activities shown in Miller units. The X-axis is the same as that in Figure 1. Averages from at least three independent experiments are shown. (A,B) Show expression profiles of sigX-lacZ and sigM-lacZ, respectively. Each gene fusion is “Wt” in Figure 4 and expressed only by their ECF σ-dependent promoters. Left panels in (A,B) show the concentrations of glucose-induced expression of the lacZ fusions. Graphs showing GI and absence of GI are shown in red and blue, respectively. Graphs showing mild GI are shown in orange. Open and closed symbols indicate no glucose and glucose-supplemented culture, respectively. Glucose concentrations: closed circles, 0.05%; squares, 0.1%; diamonds, 0.5%; triangles, 1%; reverse triangles, 2%. Standard deviations of all the points are less than 20%. In the middle and right panels in (A,B), closed and open circles show expression with and without 2% glucose, respectively. Data sets showing GI are shown in red. Standard deviations (error bar) are shown. Middle panels show expression in the ccpA strain. Right panels show expression in the disruptant or depleted strain of anti-σ factor genes, ypuN for (A) and yhdL for (B). The growth of the yhdL depleted strain is dependent on IPTG (yhdL expression is dependent on an IPTG-inducible promoter). Thus, pre-culture and culture for the expression test contained 1 mM and 0.05 mM IPTG, respectively.
The expression of sigX and sigM has been shown to be downregulated by their respective anti-σ factors (Brutsche and Braun, 1997; Horsburgh and Moir, 1999). To investigate the involvement of the anti-σ factors in the GI phenomenon regulating sigX and sigM, mutations in anti-σ factor genes were introduced into each strain with the lacZ fusion. It should be noted that a strain with an IPTG-dependent anti-σM factor gene was constructed and the expression was tested in medium containing 0.05 mM IPTG, since the anti-σM factor gene is essential (Horsburgh and Moir, 1999). We observed enhanced expression of both genes as expected, because the mutations increased the free forms of the σ factors. The increase was 3.5-fold for sigX and threefold for sigM (right panels in Figure 2). As shown in these panels, GI was observed in both mutants, indicating no involvement of anti-σ factors in GI of two σ-regulated genes. Among the seven ECF σ genes, only these two genes were subject to GI (Supplementary Figure S4). It should be noted that GI of sigX was observed in cells grown in LB medium (Supplementary Figure S5).
GI of Sublancin Production
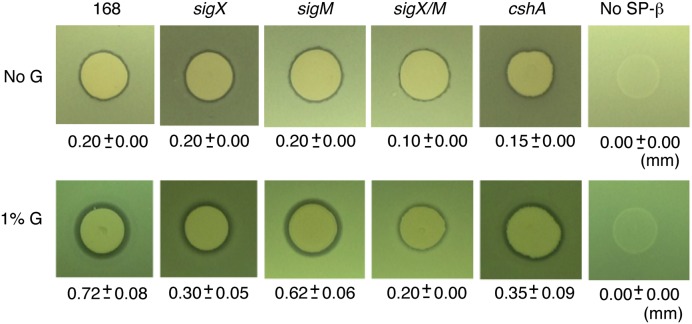
Bacillus subtilis has been shown to produce Sublancin, an antimicrobial glycopeptide (Luo and Helmann, 2009). The precursor peptide-encoding gene and the self-resistance gene for Sublancin (sunA and sunI, respectively) are located on the SP-β prophage region in the chromosome (Luo and Helmann, 2009). The sunA, sunT (Sublancin transporter) and sunS (Sublancin glycosyltransferase) genes are activated by the transition regulator Abh, whose transcription requires σX in addition to σM, although σM has a minor role (Luo and Helmann, 2009). Thus, GI of sigX might enhance Sublancin production through the σX/Abh/SunA pathway. To test this possibility, we spotted Sublancin-producing wild type 168 cells on the lawn of Y13 cells lacking SP-β, which have defects in the production and immunity for Sublancin. On the 1% glucose-containing LB agar media, a larger inhibition zone of Y13 growth was observed as compared to that observed in LB medium (Figure 3). This indicates that glucose addition induces sigX, leading to enhanced transcription of abh and the operon containing sunA. Recently, glucose addition was shown to enhance Sublancin resistance in LB medium (Garcia De Gonzalo et al., 2015). Thus, the resistance levels of the lawn would be higher, and the apparent enhancement of inhibition zone would also be underestimated. As reported previously, we also observed a smaller inhibition zone in the sigX mutant (Luo and Helmann, 2009).
FIGURE 3.
Enhancement of sublancin production by glucose addition. “1% G” and “No G” indicate the lawn cell plate supplemented with or without 1% glucose. The phenotypes of the strains are listed above the photographs. The strains were spotted onto Y13 lawn. The radii of the zones of inhibition produced by various strains were measured as the total diameter of the zone of clearing minus the diameter of the colony spot, divided by a factor of 2. Three independent experiments were performed and averages with standard deviations are shown. Strains; sigX, ASK4705; sigM, ASK4706; sigX/M, ASK4710; cshA, YDBRd; No SP-b, Y13.
The Minimum Promoter Region Required for GI
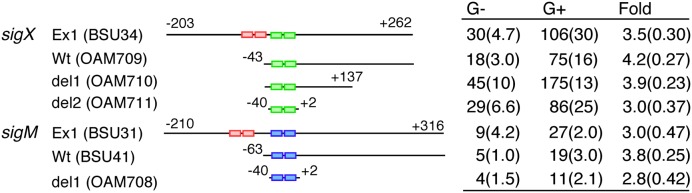
We determined a minimum promoter region responsible for GI in sigX and sigM by testing GI in lacZ fusions carrying various promoter regions. GI is expected to be observed in all constructs because the strains bear endogenous sigX or sigM genes. In both genes, similar levels of GI were observed in all fusions including the core promoter region fusions (-40 to +2 relative to the transcription start site, Figure 4). Moreover, between the core promoter regions of sigX and sigM there is no common nucleotide sequence except for the -35 and -10 elements for σX-RNAP and σM-RNAP. Thus, the similar levels of GI in all constructs suggested no involvement of some unknown trans-acting factors in GI.
FIGURE 4.
Minimum promoter regions for GI. Expression of various promoter lacZ transcriptional fusions is shown. The names of the lacZ fusion and the corresponding strain are shown. The pair of small rectangles on the horizontal line show the -35 and -10 elements of the promoter. Red, green and blue indicate σA-, σX-, and σM-dependent promoters. The numbers indicate the nucleotide positions relative to the transcription start sites by ECF σ factors. The “G-” and “G+” columns show the β-galactosidase activities (Miller units) of each fusion grown in sporulation medium without or with 2% glucose, respectively. The activities were assayed at five time-points (log phase to early stationary phase), and the experiments were performed independently three times. Averages of the peak values with standard deviations are shown. “Fold” with calculated standard deviations through propagation of errors means extent of GI.
Dependency of GI of sigX and sigM on the Cellular Acetyl-CoA Pool
We were interested in which carbon sources could induce sigX and sigM expression. Thus, three carbon sources were added, and we examined the expression levels of both sig genes. Glycerol or pyruvate addition induced gene expression to some extent, while succinate did not (Figure 5A). We note that the effects of pyruvate were modest. Glycerol and pyruvate have been shown to be incorporated into the glycolysis pathway and succinate is not (Wang et al., 2010). Thus, this pattern is reminiscent of acetyl-CoA production. Recently, protein-lysine acetylation has been studied in B. subtilis (Kim et al., 2013; Kosono et al., 2015; Carabetta et al., 2016). This modification may change protein function. In Escherichia coli, glucose addition changed and enhanced global acetylation profiles (Schilling et al., 2015). Thus, we hypothesized that glucose addition might result in a higher cellular pool of acetyl-CoA, leading to enhancement of acetylation of proteins regulating sigX and sigM. To examine the involvement of acetyl-CoA in GI of sigX and sigM, a disruption mutation of pdhC, which encodes the E2 subunit of pyruvate dehydrogenase and whose reaction product is acetyl-CoA (Gao et al., 2002), was introduced into the lacZ fusion strain. The rationale for the experiment is that in the pdhC mutant an acetyl-CoA pool would be reduced, although the cells still have other acetyl-CoA synthesis pathway involving acetyl-CoA synthetase. It should be noted that the gene encoding acetyl-CoA synthetase has been known to be repressed by glucose (Grundy et al., 1994). In the resultant strains, GI of sigX and sigM was completely abolished (Figure 5B). We observed slow growth and poor cell mass of the mutants as previously reported (Gao et al., 2002, Supplementary Figure S2B). These results strongly suggested acetyl-CoA dependent GI of sigX and sigM.
FIGURE 5.
Effects of various carbon sources and pdhC disruption on GI. Cells were grown in sporulation medium and sampled hourly. β-galactosidase activities are shown in Miller units. The X-axis is the same as that in Figure 1. Averages from at least three independent experiments are shown. Each gene fusion (for sigX-lacZ, BSU43 and for sigM-lacZ, BSU41 are used) is indicated above the panels. (A) Graphs showing GI and absence of GI are shown in red and blue, respectively. Those showing mild GI are shown in orange. Open circles, control (no glucose); closed circles, 1% glucose; squares, 0.8% glycerol; reverse triangles, 0.8% pyruvate; triangles, 0.4% succinate. Standard deviations of all the points are less than 25%. (B) Data sets not showing GI are shown in blue. The same fusions were examined in the pdhC background in sporulation medium without (open symbols) or with (closed symbols) 2% glucose. Standard deviations (error bar) are shown.
Identification of cshA as a Positive Regulator by Transposon Mutagenesis
To further gain insights into the mechanism of GI, we performed transposon mutagenesis for GI-deficient mutants by using a transposon delivery vector, pMarA (Le Breton et al., 2006). Mutants with sigX-lacZ showing pale blue on LB or sporulation agar plate containing X-Gal and 2% glucose were chosen among about 12,000 colonies. Several mutants were obtained and one was a disruptant of cshA, which encodes DEAD-box RNA helicase (Lehnik-Habrink et al., 2010). Transposon was inserted into the 342nd codon of the cshA ORF. In this mutant, sigX and sigM expression was not induced by glucose and basal expression was reduced but not abolished (Supplementary Figure S6). The decrease in sigX expression resulted in a decrease in Sublancin production in the cshA mutant (Figure 3). Among many candidates of the Tn mutagenesis, only additional four mutations negatively affected GI of both sigX and sigM genes (data not shown). Two carried a disruption of a gene of which function was not yet identified (ylxR and yqfO) and the rests were ptsH and gcp mutants. Probably in the ptsH mutant efficient glucose transport would be inhibited, leading to abolishment of GI. The detailed analysis of these mutants will be reported elsewhere.
Operon Structure and Expression of cshA
The chromosomal region surrounding cshA is shown in Supplementary Figure S7A, cshA seems to be a monocistronic operon with its own promoter and terminator (Nicolas et al., 2012). Thus, we created a promoter-lacZ fusion of cshA and examined its expression. The expression was observed and found to be unaffected by glucose addition (Supplementary Figure S7B, left). The σX/M/V-driven promoter within the ydbO ORF has been reported to co-transcribe ydbO-(ydbP)-ddlA-murF (Eiamphungporn and Helmann, 2008). Between murF and cshA there is no terminator, and thus this promoter may transcribe cshA. In addition, there is a promoter upstream of ddlA (Nicolas et al., 2012). These suggested a read-through mechanism from the upstream promoters. To confirm this, we examined activities of a lacZ-fusion located at the original locus in the chromosome. This lacZ fusion showed higher activity as compared to its own promoter fused to lacZ. Glucose addition also did not affect the activity (Supplementary Figure S7B, right). It should be noted that the strain with this fusion at the original locus carries the cshA disruption. These observations showed that read-through transcription contributed to cshA expression. It was previously reported that the cshA expression is constitutive at the protein level (Lehnik-Habrink et al., 2010).
Complementation of the cshA Mutation by cshA in Trans
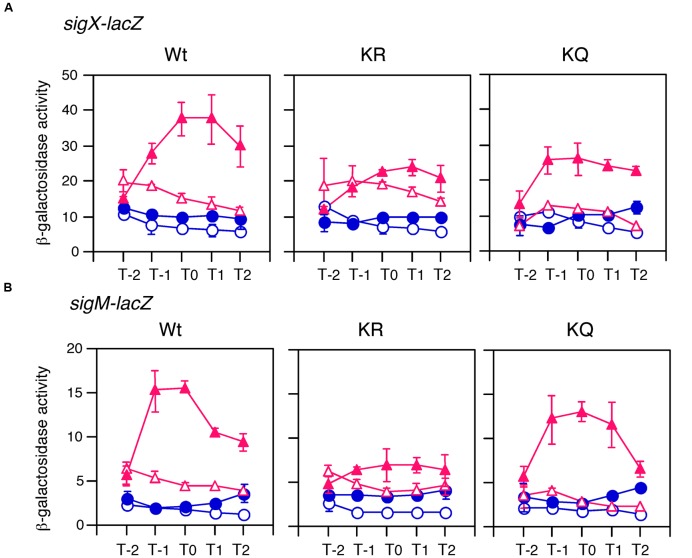
We created a xylose-inducible cshA cassette at the bkdB locus in the cshA mutant with sigX-lacZ or sigM-lacZ (Supplementary Figure S1A). Xylose addition to the culture of these strains without glucose recovered the basal expression levels of sigX-lacZ and sigM-lacZ (left panels in Figure 6). Xylose addition to the culture with glucose also rescued GI of these strains, demonstrating that the cshA gene is responsible for GI. We noted that xylose addition did not have any effect on GI in the control strain with sigX-lacZ nor sigM-lacZ (data not shown).
FIGURE 6.
Complementation test of cshA for GI and the effects of substitutions of acetylation residues in CshA on GI of sigX and sigM. Cells were grown in sporulation medium with (closed symbols) or without (open symbols) 1 and 0.5% glucose for (A,B), respectively. Triangles and circles indicate the culture with or without 2% xylose, respectively. Cells were sampled hourly and β-galactosidase activities are shown in Miller units. The X-axis is the same as that in Figure 1. Data sets with and without xylose are shown in red and blue, respectively. Averages from at least three independent experiments and standard deviations (error bar) are shown. Structures for the xylose-inducible cshA cassette are shown in Supplementary Figure S1. “Wt,” “KR,” and “KQ” mean strains with the genes encoding wild type CshA, CshA with two K to R substitutions, and CshA with two K to Q substitutions, respectively. (A) For Wt, KR, and KQ, OAM725, OAM727, and OAM729 were used. (B) For Wt, KR, and KQ, OAM724, OAM726, and OAM728 were used.
Stimulation of the Acetylation of CshA by Glucose Addition
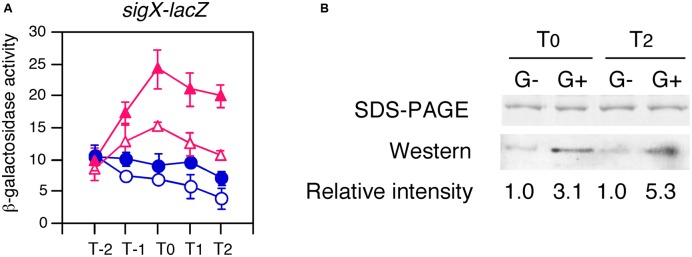
We added His-tag to the CshA protein encoded by the cassette at bkdB using PCR (Supplementary Figure S1B). The His-CshA protein was confirmed to be functional by a complementation test (Figure 7A). Xylose addition to the culture with or without glucose caused expression of His-CshA, and the two protein fractions containing His-CshA from the two time points (T0 and T2), were purified by Ni-affinity column (Figure 7B). A global analysis of the acetylome in B. subtilis revealed that CshA was acetylated at K244 and K296 (Kosono et al., 2015). Thus, we examined the acetylation status of purified His-CshA by Western blot analysis using an anti-acetyl lysine antibody. The acetyl lysine signals were enhanced in His-CshA purified from the cells with glucose, indicating that glucose addition stimulates CshA acetylation (Figure 7B). A similar result was reproducibly observed from independent culture (data not shown).
FIGURE 7.
Enhancement of acetylation of CshA by glucose addition. (A) Confirmation that His-CshA is functional. OAM731 cells with the xylose-inducible gene encoding His-tagged CshA were grown in sporulation medium with (closed symbols) or without (open symbols) 1% glucose. Triangles and circles indicate the culture with or without 2% xylose, respectively. Cells were sampled hourly. β-galactosidase activities are shown in Miller units. The X-axis is the same as that in Figure 1. Data sets with and without xylose are shown in red and blue, respectively. Averages from at least three independent experiments and standard deviations (error bar) are shown. (B) Western blot using anti-acetylated lysine residue antibody. OAM730 cells were grown in 800 ml sporulation medium containing 2% xylose with or without 1% glucose and harvested at T0 and T2. Each His-CshA was purified using Ni-affinity column, analyzed using 15% SDS-PAGE, and stained with Coomassie blue (top panel). Equal amount of the protein (0.1 μg) was electrophoresed in 15% SDS-PAGE and blotted onto a nitrocellulose membrane. Western blot analysis with monoclonal anti-acetylated lysine antibody is shown in the bottom panel. The “G-” and “G+” show the protein purified from the culture without or with 1% glucose, respectively.
GI of sigX and sigM in the rny Mutant
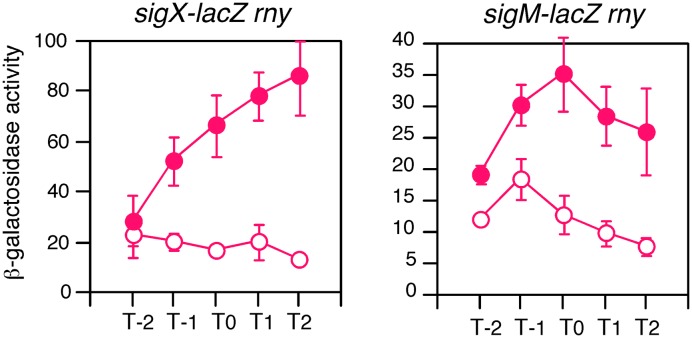
CshA has been known to be involved in RNA degradosome containing endoribonuclease RNase Y (Lehnik-Habrink et al., 2010). RNase Y has global impact on the gene expression through mRNA metabolism (Lehnik-Habrink et al., 2011; Durand et al., 2012; Laalami et al., 2013). Thus, CshA may cause GI of sigX and sigM due to the inhibition of RNase Y activity through acetylated CshA-dependent mechanism, leading to the longer half-life of mRNAs of sigX and sigM. Otherwise RNase Y may regulate sigX and sigM indirectly. Indeed, sigM expression was enhanced in an RNase Y-depleted mutant, rny, according to the three analyses (Lehnik-Habrink et al., 2011; Durand et al., 2012; Laalami et al., 2013). To examine this, we introduced rny depletion into the strains bearing sigX-lacZ or sigM-lacZ and tested whether GI was observed. GI of sigX was clearly observed in the rny mutant (Figure 8, left). With respect to sigM, we observed enhancement of sigM-lacZ expression especially in the log phase as expected (Figure 8, right). Moreover, GI of sigM was also clearly observed. These results demonstrated that CshA would act on expression of sigX and sigM independent of influence to RNase Y due to interaction of CshA with RNA degradosome.
FIGURE 8.
GI in the RNase Y-depleted mutant. Cells were grown in sporulation medium with (closed symbols) or without (open symbols) 2% glucose. Left, sigX-lacZ with rny (OAM735); right, sigM-lacZ with rny (OAM734). Cells were sampled hourly and β-galactosidase activities are shown in Miller units. The X-axis is the same as that in Figure 1. Data sets showing GI are shown in red. Averages from at least three independent experiments and standard deviations (error bar) are shown. The mutant of rny disruption is associated with Pxyl -rny, and thus the strains were maintained in the presence of 1% xylose and the preculture for the experiments lacked xylose.
The Effects of K–R and K–Q Mutations on GI
K–R and K–Q mutations have been shown to mimic non-acetylation and acetylation states, respectively (Kamieniarz and Schneider, 2009). Hence, we also introduced these mutations into the xylose-inducible cshA cassette system (Supplementary Figure S1A). Introduction of these mutations resulted in similar effects on the expression of sigX-lacZ and sigM-lacZ (Figure 6). In the strain with CshA bearing two K–R substitutions, basal expression was recovered to that of wild type CshA, whereas GI was completely abolished. In the strain with CshA bearing two K–Q substitutions, basal expression was not recovered, whereas GI was observed. These results strongly suggested that acetylation of two lysine residues in CshA is required for GI of sigX and sigM. Moreover, the results also indicated that basal expression and GI of sigX and sigM were mutually distinguishable in a genetic context. We note that the strains bearing wild type and mutant CshA proteins did not grow in culture with 2% glucose in the presence of xylose, and the extent of the resistance against glucose changed in the strains bearing sigX-lacZ and sigM-lacZ due to unknown reasons (data not shown). Thus, different glucose concentrations were used for the experiments for sigX-lacZ and sigM-lacZ.
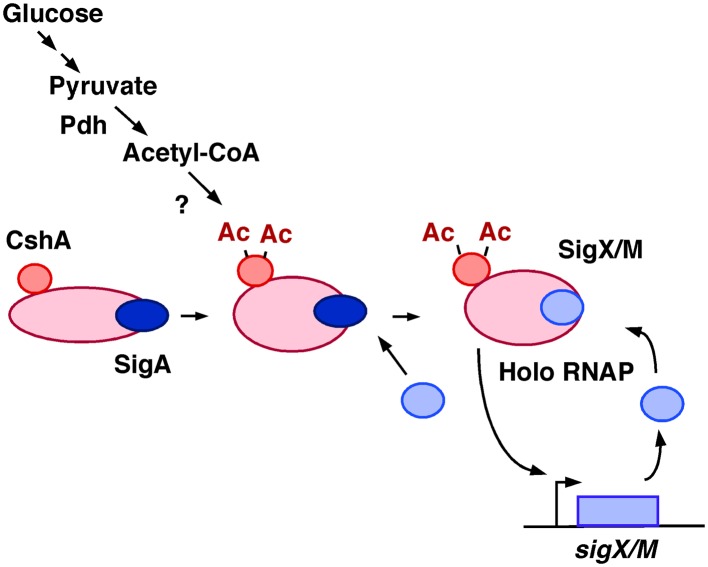
Recent analysis using mass spectrometry of B. subtilis RNAP reported that half of the RNAP pool was associated with CshA (Delumeau et al., 2011). Thus, we present a model for the mechanism by which GI of sigX and sigM occurs as shown in Figure 9.
FIGURE 9.
Model of GI of sigX and sigM. Glucose addition would result in an increase of the cellular acetyl-CoA pool, leading to more acetylated CshA. CshA has been shown to associate with RNAP. RNAP with acetylated CshA may stimulate replacement of σX/M for σA in the RNAP holoenzyme, although the mode of action is not known. Moreover, the acetyl transfer enzyme that acetylates CshA is also not known. Ac, acetyl moiety; Pdh, pyruvate dehydrogenase.
Discussion
Extracytoplasmic function σ factor genes have been shown to be induced mainly by cell envelope and cell membrane stresses through a mechanism involving anti-σ factor embedded in the cell membrane. As an exception, Caulobacter crescentus ECF σ factors play roles in many other stresses (for example, heat, glucose starvation, and oxidative stresses), because stress-responsive sigma factor homologs of σB in B. subtilis and σS in E. coli are lacking in C. crescentus. Metabolic stress such as carbon-starvation induced ECF σ factor σT perhaps in an anti-σ factor dependent manner in this bacterium (Britos et al., 2011). Our study finds that glucose addition to the medium induces the sigX and sigM genes independent of their anti-σ factors, probably through a CshA-dependent functional modification of RNAP. This is the first report for GI of ECF σ genes to our knowledge. The addition of glucose to the medium affects σX- and σM-dependent cellular functions, such as Sublancin production and Spx overproduction.
RNA polymerase with acetylated CshA may somehow stimulate the autoregulatory loop of sigX and sigM. Our model shown in Figure 9 encompasses all of the experimental data: involvement of pdhC, the different effects of carbon sources on GI, the effects of non-acetylation mimicking mutations, and the enhancement of CshA acetylation by glucose, yet some key elements in this model remain unknown. It is of interest whether CshA is acetylated before it binds to RNAP or if acetylation of CshA stimulates RNAP-binding of CshA. In addition, there are several outstanding questions, for example, what enzyme acetylates CshA, or how σX/M replaces σA in the RNAP holoenzyme, during GI. According to an analysis of the RNAP holoenzyme, the core RNAP with no σ factor was barely detected (Delumeau et al., 2011). Thus, replacement of σX/M for σA is a likely event. Generally, competition between housekeeping σA and alternative σ factors for the core RNAP is favorable for σA-binding due to the higher affinity of σA for RNAP (Österberg et al., 2011). To change the balance of σ binding, various mechanisms have been reported (Österberg et al., 2011). For example, the small subunit of RNAP, the ω- subunit, is involved in σ factor recruitment (Weiss and Shaw, 2015). This demonstrates that a protein associated with RNAP can modulate its affinity to a specific σ factor perhaps through structural change as we may see with CshA.
Thousands of protein acetylation sites have been reported in B. subtilis, but with respect to transcription factors, a link between acetylation and a functional change has only been elucidated in very rare cases (Thao et al., 2010; Lima et al., 2011; Hu et al., 2013; Hentchel et al., 2015). Generally, the link to function was shown by introducing acetylation-mimicking and -abolishing mutations into the protein. In our study, the functional modulation of CshA by acetylation was also clarified using this method.
CshA has been characterized as a multi-functional protein and required for growth in low temperatures (Lehnik-Habrink et al., 2013). CshA exists in an RNA degradosome complex and affects the expression of many genes through its interaction with RNase Y (Lehnik-Habrink et al., 2013). In the case of sigX and sigM, GI was observed in rny-depleted mutant. This indicates that the mechanism of CshA-dependent GI does not occur via mRNA metabolism by RNase Y. Moreover, CshA is required for ribosome biogenesis, and thus CshA has a role in translation (Lehnik-Habrink et al., 2013). A possibility that translational control of sigX and sigM by CshA cannot be completely excluded. However, it is difficult to speculate how the changes in ribosome biogenesis caused by the cshA depletion results in the specific regulation of sigX and sigM expression. Thus, we favored the model shown in Figure 9, although further experiments will be required to prove the model.
Author Contributions
MO and KA designed research, performed experiments and analyzed results. MO wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank A. I. Alonson (Purdue University), S. Kosono (Tokyo University), T. Morimoto (Kao Co.), and BGSC for kindly supplying the bacterial strains and plasmids used in this study.
Footnotes
Funding. This work was supported by JSPS KAKENHI Grant Number 15K07367, and the Research Program of the Institute of Oceanic Research and Development. The author has no conflict of interest to declare.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01918/full#supplementary-material
References
- Antelmann H., Scharf C., Hecker M. (2000). Phosphate starvation-inducible proteins of Bacillus subtilis: proteomics and transcriptional analysis. J. Bacteriol. 182 4478–4490. 10.1128/JB.182.16.4478-4490.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai K., Yamaguchi H., Kang C. M., Yoshida K., Fujita Y., Sadaie Y. (2003). DNA microarray analysis of Bacillus subtilis sigma factors of extracytoplasmic function family. FEMS Microbiol. Lett. 220 155–160. 10.1016/S0378-1097(03)00093-4 [DOI] [PubMed] [Google Scholar]
- Bernal V., Castaño-Cerezo S., Gallego-Jara J., Écija-Conesa A., de Diego T., Iborra J. L., et al. (2014). Regulation of bacterial physiology by lysine acetylation of proteins. N. Biotechnol. 31 586–595. 10.1016/j.nbt.2014.03.002 [DOI] [PubMed] [Google Scholar]
- Britos L., Abeliuk E., Taverner T., Lipton M., McAdams H., Shapiro L. (2011). Regulatory response to carbon starvation in Caulobacter crescentus. PLoS ONE 6:e18179 10.1371/journal.pone.0018179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutsche S., Braun V. (1997). SigX of Bacillus subtilis replaces the ECF sigma factor fecI of Escherichia coli and is inhibited by RsiX. Mol. Gen. Genet. 256 416–425. 10.1007/s004380050585 [DOI] [PubMed] [Google Scholar]
- Cao M., Helmann J. D. (2002). Regulation of the Bacillus subtilis bcrC bacitracin resistance gene by two extracytoplasmic function sigma factors. J. Bacteriol. 184 6123–6129. 10.1128/JB.184.22.6123-6129.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M., Helmann J. D. (2004). The Bacillus subtilis extracytoplasmic-function sigmaX factor regulates modification of the cell envelope and resistance to cationic antimicrobial peptides. J. Bacteriol. 186 1136–1146. 10.1128/JB.186.4.1136-1146.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabetta V. J., Greco T. M., Tanner A. W., Cristea I. M., Dubnau D. (2016). Temporal regulation of the Bacillus subtilis acetylome and evidence for a role of MreB acetylation in cell wall growth. mSystems 1:e00005-16 10.1128/mSystems.00005-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J. M., Guttenplan S. B., Kearns D. B. (2014). Defects in the flagellar motor increase synthesis of poly-γ-glutamate in Bacillus subtilis. J. Bacteriol. 196 740–753. 10.1128/JB.01217-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia De Gonzalo C. V., Denham E. L., Mars R. A., Stülke J., van der Donk W. A., van Dijl J. M. (2015). The phosphoenolpyruvate:sugar phosphotransferase system is involved in sensitivity to the glucosylated bacteriocin sublancin. Antimicrob. Agents Chemother. 59 6844–6854. 10.1128/AAC.01519-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delumeau O., Lecointe F., Muntel J., Guillot A., Guédon E., Monnet V., et al. (2011). The dynamic protein partnership of RNA polymerase in Bacillus subtilis. Proteomics 11 2992–3001. 10.1002/pmic.201000790 [DOI] [PubMed] [Google Scholar]
- Durand S., Gilet L., Bessières P., Nicolas P., Condon C. (2012). Three essential ribonucleases-RNase Y, J1, and III-control the abundance of a majority of Bacillus subtilis mRNAs. PLoS Genet. 8:e1002520 10.1371/journal.pgen.1002520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiamphungporn W., Helmann J. D. (2008). The Bacillus subtilis sigma(M) regulon and its contribution to cell envelope stress responses. Mol. Microbiol. 67 830–848. 10.1111/j.1365-2958.2007.06090.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y. (2009). Carbon catabolite control of the metabolic network in Bacillus subtilis. Biosci. Biotechnol. Biochem. 73 245–259. 10.1271/bbb.80479 [DOI] [PubMed] [Google Scholar]
- Gao H., Jiang X., Pogliano K., Aronson A. I. (2002). The E1beta and E2 subunits of the Bacillus subtilis pyruvate dehydrogenase complex are involved in regulation of sporulation. J. Bacteriol. 184 2780–2788. 10.1128/JB.184.10.2780-2788.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy F. J., Turinsky A. J., Henkin T. M. (1994). Catabolite regulation of Bacillus subtilis acetate and acetoin utilization genes by CcpA. J. Bacteriol. 176 4527–4533. 10.1128/jb.176.15.4527-4533.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guérout-Fleury A. M., Frandsen N., Stragier P. (1996). Plasmids for ectopic integration in Bacillus subtilis. Gene 180 57–61. 10.1016/S0378-1119(96)00404-0 [DOI] [PubMed] [Google Scholar]
- Hashimoto M., Seki T., Matsuoka S., Hara H., Asai K., Sadaie Y., et al. (2013). Induction of extracytoplasmic function sigma factors in Bacillus subtilis cells with defects in lipoteichoic acid synthesis. Microbiology 159 23–35. 10.1099/mic.0.063420-0 [DOI] [PubMed] [Google Scholar]
- Hata M., Ogura M., Tanaka T. (2001). Involvement of stringent factor RelA in expression of the alkaline protease gene aprE in Bacillus subtilis. J. Bacteriol. 183 4648–4651. 10.1128/JB.183.15.4648-4651.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmann J. D. (2016). Bacillus subtilis extracytoplasmic function (ECF) sigma factors and defense of the cell envelope. Curr. Opin. Microbiol. 30 122–132. 10.1016/j.mib.2016.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmann J. D., Chamberlin M. J. (1988). Structure and function of bacterial sigma factors. Ann. Rev. Biochem. 57 839–872. 10.1146/annurev.bi.57.070188.004203 [DOI] [PubMed] [Google Scholar]
- Hentchel K. L., Thao S., Intile P. J., Escalante-Semerena J. C. (2015). Deciphering the regulatory circuitry that controls reversible lysine acetylation in Salmonella enterica. mBio 6:e00891 10.1128/mBio.00891-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho T. D., Ellermeier C. D. (2012). Extra cytoplasmic function σ factor activation. Curr. Opin. Microbiol. 5 182–188. 10.1016/j.mib.2012.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K., Kaneko M., Tanji Y., Xing X. H., Unno H. (2002). Construction of self-disruptive Bacillus megaterium in response to substrate exhaustion for polyhydroxybutyrate production. Appl. Microbiol. Biotechnol. 59 211–216. 10.1007/s00253-002-0986-8 [DOI] [PubMed] [Google Scholar]
- Horsburgh M. J., Moir A. (1999). Sigma M, an ECF RNA polymerase sigma factor of Bacillus subtilis 168, is essential for growth and survival in high concentrations of salt. Mol. Microbiol. 32 41–50. 10.1046/j.1365-2958.1999.01323.x [DOI] [PubMed] [Google Scholar]
- Hu L. I., Chi B. K., Kuhn M. L., Filippova E. V., Walker-Peddakotla A. J., Bäsell K., et al. (2013). Acetylation of the response regulator RcsB controls transcription from a small RNA promoter. J. Bacteriol. 195 4174–4186. 10.1128/JB.00383-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Decatur A., Sorokin A., Helmann J. D. (1997). The Bacillus subtilis sigma(X) protein is an extracytoplasmic function sigma factor contributing to survival at high temperature. J. Bacteriol. 179 2915–2921. 10.1128/jb.179.9.2915-2921.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Helmann J. D. (1998). Identification of target promoters for the Bacillus subtilis sigma X factor using a consensus-directed search. J. Mol. Biol. 279 165–173. 10.1006/jmbi.1998.1765 [DOI] [PubMed] [Google Scholar]
- Inoue H., Suzuki D., Asai K. (2013). A putative bactoprenol glycosyltransferase, CsbB, in Bacillus subtilis activates SigM in the absence of co-transcribed YfhO. Biochem. Biophys. Res. Commun. 436 6–11. 10.1016/j.bbrc.2013.04.064 [DOI] [PubMed] [Google Scholar]
- Itaya M. (1992). Construction of a novel tetracycline resistance gene cassette useful as a marker on the Bacillus subtilis chromosome. Biosci. Biotechnol. Biochem. 56 685–686. 10.1271/bbb.56.685 [DOI] [PubMed] [Google Scholar]
- Jervis A. J., Thackray P. D., Houston C. W., Horsburgh M. J., Moir A. (2007). SigM-responsive genes of Bacillus subtilis and their promoters. J. Bacteriol. 189 4534–4538. 10.1128/JB.00130-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamieniarz K., Schneider R. (2009). Tools to tackle protein acetylation. Chem. Biol. 16 1027–1029. 10.1016/j.chembiol.2009.10.002 [DOI] [PubMed] [Google Scholar]
- Kim D., Yu B. J., Kim J. A., Lee Y. J., Choi S. G., Kang S., et al. (2013). The acetylproteome of Gram-positive model bacterium Bacillus subtilis. Proteomics 13 1726–1736. 10.1002/pmic.201200001 [DOI] [PubMed] [Google Scholar]
- Kosono S., Tamura M., Suzuki S., Kawamura Y., Yoshida A., Nishiyama M., et al. (2015). Changes in the acetylome and succinylome of Bacillus subtilis in response to carbon source. PLoS ONE 10:e0131169 10.1371/journal.pone.0131169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst F., Msadek T., Rapoport G. (1994). “Signal transduction network controlling degradative enzyme synthesis and competence,” in Bacillus subtilis in Regulation of Bacterial Differentiation, eds Piggot P. J., Moran C. P., Jr., Youngman P. (Washington, DC: ASM Press; ), 1–20. [Google Scholar]
- Laalami S., Bessières P., Rocca A., Zig L., Nicolas P., Putzer H. (2013). Bacillus subtilis RNase Y activity in vivo analysed by tiling microarrays. PLoS ONE 8:e54062 10.1371/journal.pone.0054062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Breton Y., Mohapatra N. P., Haldenwang W. G. (2006). In vivo random mutagenesis of Bacillus subtilis by use of TnYLB-1, a mariner-based transposon. Appl. Environ. Microbiol. 72 327–333. 10.1128/AEM.72.1.327-333.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnik-Habrink M., Pförtner H., Rempeters L., Pietack N., Herzberg C., Stülke J. (2010). The RNA degradosome in Bacillus subtilis: identification of CshA as the major RNA helicase in the multiprotein complex. Mol. Microbiol. 77 958–971. 10.1111/j.1365-2958.2010.07264.x [DOI] [PubMed] [Google Scholar]
- Lehnik-Habrink M., Rempeters L., Kovács Á. T., Wrede C., Baierlein C., Krebber H., et al. (2013). DEAD-Box RNA helicases in Bacillus subtilis have multiple functions and act independently from each other. J. Bacteriol. 195 534–544. 10.1128/JB.01475-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnik-Habrink M., Schaffer M., Mäder U., Diethmaier C., Herzberg C., Stülke J. (2011). RNA processing in Bacillus subtilis: identification of targets of the essential RNase Y. Mol. Microbiol. 81 1459–1473. 10.1111/j.1365-2958.2011.07777.x [DOI] [PubMed] [Google Scholar]
- Lima B. P., Antelmann H., Gronau K., Chi B. K., Becher D., Brinsmade S. R., et al. (2011). Involvement of protein acetylation in glucose-induced transcription of a stress-responsive promoter. Mol. Microbiol. 81 1190–1204. 10.1111/j.1365-2958.2011.07742.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Helmann J. D. (2009). Extracytoplasmic function sigma factors with overlapping promoter specificity regulate sublancin production in Bacillus subtilis. J. Bacteriol. 191 4951–4958. 10.1128/JB.00549-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S., Chiba M., Tanimura Y., Hashimoto M., Hara H., Matsumoto K. (2011). Abnormal morphology of Bacillus subtilis ugtP mutant cells lacking glucolipids. Genes Genet. Syst. 86 295–304. 10.1266/ggs.86.295 [DOI] [PubMed] [Google Scholar]
- Nicolas P., Mäder U., Dervyn E., Rochat T., Leduc A., Pigeonneau N., et al. (2012). Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 335 1103–1106. 10.1126/science.1206848 [DOI] [PubMed] [Google Scholar]
- Ogura M. (2016). Post-transcriptionally generated cell heterogeneity regulates biofilm formation in Bacillus subtilis. Genes Cells 21 335–349. 10.1111/gtc.12343 [DOI] [PubMed] [Google Scholar]
- Ogura M., Ohshiro Y., Hirao S., Tanaka T. (1997). A new Bacillus subtilis gene, med, encodes a positive regulator of comK. J. Bacteriol. 179 6244–6253. 10.1128/jb.179.20.6244-6253.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura M., Tanaka T. (1996). Transcription of Bacillus subtilis degR is σD-dependent and suppressed by multicopy proB through σD. J. Bacteriol. 178 216–222. 10.1128/jb.178.1.216-222.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura M., Yoshikawa H., Chibazakura T. (2014). Regulation of the response regulator gene degU through the binding of SinR/SlrR and exclusion of SinR/SlrR by DegU in Bacillus subtilis. J. Bacteriol. 196 873–881. 10.1128/JB.01321-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Österberg S., del Peso-Santos T., Shingler V. (2011). Regulation of alternative sigma factor use. Ann. Rev. Microbiol. 65 37–55. 10.1146/annurev.micro.112408.134219 [DOI] [PubMed] [Google Scholar]
- Rochat T., Nicolas P., Delumeau O., Rabatinová A., Korelusová J., Leduc A., et al. (2012). Genome-wide identification of genes directly regulated by the pleiotropic transcription factor Spx in Bacillus subtilis. Nucleic Acids Res. 40 9571–9583. 10.1093/nar/gks755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer P., Millet J., Aubert J. (1965). Catabolite repression of bacterial sporulation. Proc. Natl. Acad. Sci. U.S.A. 54 704–711. 10.1073/pnas.54.3.704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling B., Christensen D., Davis R., Sahu A. K., Hu L. I., Walker-Peddakotla A., et al. (2015). Protein acetylation dynamics in response to carbon overflow in Escherichia coli. Mol. Microbiol. 98 847–863. 10.1111/mmi.13161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiwa Y., Yoshikawa H., Tanaka T., Ogura M. (2015). Bacillus subtilis degSU operon is regulated by the ClpXP-Spx regulated proteolysis system. J. Biochem. 157 321–330. 10.1093/jb/mvu076 [DOI] [PubMed] [Google Scholar]
- Souza B. M., Castro T. L., Carvalho R. D., Seyffert N., Silva A., Miyoshi A., et al. (2014). σ(ECF) factors of gram-positive bacteria: a focus on Bacillus subtilis and the CMNR group. Virulence 5 587–600. 10.4161/viru.29514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thackray P. D., Moir A. (2003). SigM, an extracytoplasmic function sigma, factor of Bacillus subtilis, is activated in response to cell wall antibiotics, ethanol, heat, acid, and superoxide stress. J. Bacteriol. 185 3491–3498. 10.1128/JB.185.12.3491-3498.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thao S., Chen C. S., Zhu H., Escalante-Semerena J. C. (2010). N𝜀-lysine acetylation of a bacterial transcription factor inhibits its DNA-binding activity. PLoS ONE 5:e15123 10.1371/journal.pone.0015123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thao S., Escalante-Semerena J. C. (2011). Control of protein function by reversible N𝜀-lysine acetylation in bacteria. Curr. Opin. Microbiol. 14 200–204. 10.1016/j.mib.2010.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukahara K., Ogura M. (2008). Promoter selectivity of the Bacillus subtilis responseregulator DegU, a positive regulator of the fla/che operon and sacB. BMC Microbiol. 8:8 10.1186/1471-2180-8-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner M. S., Helmann J. D. (2000). Mutations in multidrug efflux homologs, sugar isomerases, and antimicrobial biosynthesis genes differentially elevate activity of the sigma(X) and sigma(W) factors in Bacillus subtilis. J. Bacteriol. 182 5202–5210. 10.1128/JB.182.18.5202-5210.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang Y., Yang C., Xiong H., Lin Y., Yao J., et al. (2010). Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science 327 1004–1007. 10.1126/science.1179687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss A., Shaw L. N. (2015). Small things considered: the small accessory subunits of RNA polymerase in Gram-positive bacteria. FEMS Microbiol. Rev. 39 541–554. 10.1093/femsre/fuv005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C. K., Tai P. C., Lu C. D. (2014). Time-related transcriptome analysis of B. subtilis 168 during growth with glucose. Curr. Microbiol. 68 12–20. 10.1007/s00284-013-0432-4 [DOI] [PubMed] [Google Scholar]
- Yoshimura M., Asai K., Sadaie Y., Yoshikawa H. (2004). Interaction of Bacillus subtilis extracytoplasmic function (ECF) sigma factors with the N-terminal regions of their potential anti-sigma factors. Microbiology 150 591–599. 10.1099/mic.0.26712-0 [DOI] [PubMed] [Google Scholar]
- Yuan G., Wong S. L. (1995). Regulation of groE expression in Bacillus subtilis: the involvement of the sigma A-like promoter and the roles of the inverted repeat sequence (CIRCE). J. Bacteriol. 177 5427–5433. 10.1128/jb.177.19.5427-5433.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber P. (2004). Spx-RNA polymerase interaction and global transcriptional control during oxidative stress. J. Bacteriol. 186 1911–1918. 10.1128/JB.186.7.1911-1918.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.