Abstract
The identification of genes regulating milk secretion by mammary glands is a key-step for exploiting changes in milk composition induced by different diet regimens. Linseed supplementation is a reliable feeding strategy to enhance polyunsaturated fatty acid content in milk fat from sheep, increasing milk quality and nutraceutical value. To investigate the molecular bases of diet induced differences in milk composition, we collected milk from dairy ewes at 90 day of lactation and after 3 weeks of diet supplementation with extruded linseed. The milk of dairy ewes contains milk somatic cells mostly derived by mammary glands. After isolating milk somatic cells by centrifugation, RNAs were purified from these cells, and Illumina RNA sequencing was performed to analyze RNA synthesis. Our data provide a resource (available at Gene Expression Omnibus database under GSE89163) to be employed for comparative analyses of gene expression in milk somatic cells in different breeds and different diets, with the long-term aim of developing strategies to improve sheep milk quality.
| Specifications | |
|---|---|
| Organism/cell line/tissue | Ovis aries/Comisana breed/milk somatic cells |
| Sex | F |
| Sequencer or array type | Illumina HiSeq2000 |
| Data format | Raw data: FASTQ files, analyzed data: txt files |
| Experimental factors | Milk somatic cells before vs. after 3 weeks linseed feeding |
| Experimental features | RNA-Seq dataset for gene expression profiling in sheep milk somatic cells |
| Consent | Full consent |
| Sample source location | Perugia, Italy |
1. Direct link to deposited data
Deposited data can be found here: http://www.ncbi.nlm.nih.gov/geo/info/linking.html.
2. Experimental design, materials and methods
2.1. Sample collection and RNA isolation
To analyze transcriptome in the mammary gland a biopsy would be necessary, but this technique is particularly invasive and it can cause udder damage and infection [1]. Using milk somatic cells (MSC) is preferable for RNA isolation [2]. For transcriptomic analyses on sheep milk somatic cells, five Comisana mid-lactation sheep were used. Starting in February 2015, each sheep was fed grass hay and a commercial concentrate for three weeks, then the same sheep were fed grass hay and a concentrate containing 20% of extruded linseed on dry matter basis for 3 weeks. In both periods, the diets were formulated according to the nutrient requirements of an ewe weighing 65 kg and producing 1 kg of milk at 6.5% fat. Animal health was constantly monitored and since some ewes turned sick, they were excluded from the experiment. Milk from each sheep was collected at the beginning and at the end of the experiment. Total RNA was isolated from MSC, according to the method described by Wickramasinghe et al. [3], with minor modifications [4]. Briefly, MSC were pelleted from 250 ml fresh milk at 1100 g for 10 min at 4 °C. The cell pellet was washed twice in phosphate-buffered saline solution, pH 7.2 with 0.5 mM EDTA and resuspended in 3 ml of Trizol reagent (Life Technologies). RNA extraction continued following the Trizol protocol, except that all centrifugations were performed at 4 °C and precipitation in isopropanol was carried on at − 80 °C for 30 min. Then, a DNAse I (Roche) treatment was performed according to the manufacturer's instructions, to completely remove genomic DNA contamination. Finally, RNA was purified by phenol/chloroform extraction and precipitated following standard procedures.
The handling of the animals was carried out according to the guidelines of the EU Directive 2010/63/EU for animal experiments and to the Institutional Animal Care and Use Committee of the University of Perugia.
2.2. Generation of RNA-Seq data
RNA-Seq libraries were generated using the TruSeq RNA-Seq Sample Prep kit according to the manufacturer's protocol (Illumina Inc., San Diego, CA). Poly-A RNAs were isolated from total RNA and chemically fragmented. First and second strand cDNA synthesis were followed by end-repair and adenosines were added to the 3′ ends. Adapters were ligated to the cDNA and 200 ± 25 bp fragments were gel purified and enriched by PCR. The libraries were quantified using Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA) and run on the Illumina HiSeq2000 (Illumina Inc.) using version 3 reagents. Paired-read sequences,125 nt in length, were collected. Whole RNA-Seq data were submitted to NCBI Sequence Read Archive and Gene Expression Omnibus (series accession number GSE89163).
2.3. Data processing
Raw single reads (in FASTQ format) were subjected to sequence quality control using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). FastQC performed a series of analysis modules on raw data and created a report with statistics for the data analyzed. For each library, FastQC showed high per base sequence quality, exceeding 30 on Phred scale (< 1/1000 chance of a base being wrong) and detected adapter contamination, matching the reads to known adapter sequences. Then, raw reads were trimmed using Trimmomatic [5], version 0.33, with the following parameters: CROP:115 HEADCROP:15 MINLEN:100. After trimming, ribosomal reads were mapped against Ovis aries and Bos taurus ribosomal DNA sequences and rDNA matching reads were removed.
Finally, FastQC was used again to examine the characteristics of the libraries and to verify trimming efficiency, which resulted into removal of a total of 54,860,517 (42.94%) low quality or ribosomal sequences (Table 1).
Table 1.
Number of raw and high-quality Illumina reads, and number of reads matching the Ovis aries transcriptome for each library (C, control; L, after 3 weeks linseed feeding).
| Library | Total reads | HQ reads | Aligned HQ reads | Aligned HQ reads (%) |
|---|---|---|---|---|
| C1 | 28,098,370 | 25,682,138 | 22,644,050 | 80.59 |
| C2 | 15,834,738 | 12,902,452 | 8,805,705 | 55.61 |
| C3 | 18,805,590 | 14,994,960 | 10,602,801 | 56.38 |
| L1 | 17,380,388 | 5,973,508 | 4,941,935 | 28.43 |
| L2 | 20,894,652 | 8,478,368 | 5,978,026 | 28.61 |
| L3 | 26,732,292 | 24,451,846 | 19,912,996 | 74.49 |
| Total | 127,746,030 | 92,483,272 | 72,885,513 | 57.06 |
2.4. Differential expression quantification
Reads were aligned to the Ovis aries transcriptome (Oar), version 4.0, available at the NCBI site (https://www.ncbi.nlm.nih.gov/genome/?term=ovis%20aries) [6], with a tolerance of up to 2 mismatches, using CLC-BIO Genomic Workbench 8.0.3 (CLC). The percentage of aligned reads per each sample is reported in Table 1.
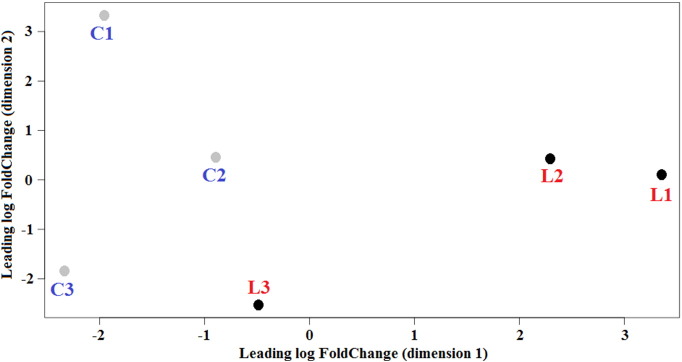
The sample cluster of linseed feeding in determining gene expression differences was verified applying a Multi Dimensional Scaling (MDS) plot to the gene expression data of the whole set of genes (Fig. 1). MSC before and after linseed feeding formed two separate groups.
Fig. 1.
Multidimensional scaling plot of sheep libraries showing cluster of samples. (C, ewes before linseed feeding; L, ewes after linseed feeding). Each ewe is identified by a number.
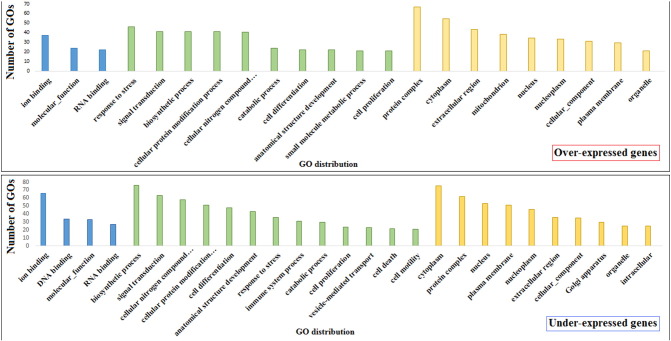
Gene expression level was calculated and expressed as Reads Per Kilobase per Million reads mapped (RPKM [7]). Expression data were evaluated considering RPKM values before and after linseed feeding using likelihood test on edgeR [8]. The log fold changes between treatments were considered as significant when the weight of a sample was at least one-fold higher or lower than another, with an FDR corrected p-value ≤ 0.05. The analysis was limited to genes showing RPKM > 1 in at least one sample, as used in other studies [9]. Of a total of 22,328 analyzed genes, 1421 resulted over-expressed and 1343 under-expressed in ewes after linseed feeding compared to the same sheep before linseed feeding. Genes affected by treatment were annotated by gene ontology analysis using BLAST2GO [10]. The most represented GOs are reported in Fig. 2.
Fig. 2.
The most represented (n > 10) gene ontologies of differentially over- (above) and under-expressed genes (below) in milk somatic cells after 3 weeks of linseed feeding. Columns are separated into molecular functions (blue), biological processes (green), and cellular components (yellow).
3. Discussion
It is known that feeding regimen plays a pivot role in affecting milk nutrient composition. Also milk nutraceutical compounds (products derived from food sources that are purported to provide extra health benefits) can change with animal diet. For example, linseed supplementation in the diet of dairy ewes resulted in an increase of milk polyunsaturated fatty acids, including conjugated linoleic acids (CLA) and alpha-linolenic acid (ALA) [11]. Cheese from sheep milk enriched with CLA and ALA was proven to be effective in reducing plasma cholesterol and endocannabinoids in human [12]. These data prompted us to study mammary gland gene expression of dairy ewes, as affected by linseed-enriched diet.
The expression of 22,328 genes included in the O. aries transcriptome database was studied in milk somatic cells to infer the effect of linseed feeding on gene expression in mammary glands. Linseed feeding determined deep variation in the overall expression of genes, as indicated by MDS plot. Most detected genes were expressed before and after linseed feeding, however a number of genes were significantly induced or repressed by linseed, similar to results obtained in the liver of cows fed with CLA [13].
Expressed genes were characterised by gene ontology. Considering the three principal gene ontologies, molecular functions, biological processes, and cellular components, the most represented ontologies of genes induced by linseed feeding were Ion Binding, Response to Stress, and Protein Complex, respectively. On the other hand, the most common ontologies of repressed genes were Ion Binding, Biosynthetic Process, and Cytoplasm, respectively.
The biological significance of this finding remains to be elucidated in the future. The dataset of expression data will help improving our understanding of molecular mechanisms regulating sheep milk quality as affected by different diets.
Conflict of interest
Authors declare no conflict of interest.
Acknowledgements
This research work was supported by University of Pisa, Italy, project “Progetto di Ricerca di Ateneo: Metodi innovativi per l'analisi del trascrittoma mammario di pecore alimentate con semi di lino”. Thanks are due to Prof. Mariano Pauselli (University of Perugia) for ensuring availability of sheep and to Dr. Giulia Pasqualetto (University of Pisa) for help in RNA isolation, respectively.
References
- 1.Murrieta C.M., Hess B.W., Scholljegerdes E.J., Engle T.E., Hossner K.L., Mossand G.E., Rule D.C. Evaluation of milk somatic cells as a source of mRNA for study of lipogenesis in the mammary gland of lactating beef cows supplemented with dietary high-linoleate safflower seeds. J. Anim. Sci. 2006;84:2399–2405. doi: 10.2527/jas.2005-677. [DOI] [PubMed] [Google Scholar]
- 2.Mura M.C., Daga C., Bodano S., Paludo M., Luridiana S., Pazzola M., Dettori M.L., Vacca G.M., Carcangiu V. Development of a RNA extraction method from milk for gene expression study in the mammary gland of sheep. Mol. Biol. Rep. 2013;40:2169–2173. doi: 10.1007/s11033-012-2276-6. [DOI] [PubMed] [Google Scholar]
- 3.Wickramasinghe S., Rincon G., Islas-Trejo A., Medrano J.F. Transcriptional profiling of bovine milk using RNA sequencing. BMC Genomics. 2012;13:45. doi: 10.1186/1471-2164-13-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vangelisti A., Giordani T., Natali L., Serra A., Conte G., Mele M., Ranieri A., Cavallini A. RNA isolation from the milk of dairy ewes for transcriptome analysis. Agrochimica. 2016;60:320–324. [Google Scholar]
- 5.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The International Sheep Genomic Consortium The sheep genome reference sequence: a work in progress. Anim. Genet. 2010;5:449–453. doi: 10.1111/j.1365-2052.2010.02100.x. [DOI] [PubMed] [Google Scholar]
- 7.Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 8.Robinson M.D., McCarthy D.J., Smith G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cossu R.M., Giordani T., Cavallini A., Natali L. High-throughput analysis of transcriptome variation during water deficit in a poplar hybrid: a general overview. Tree Genet. Genomes. 2014;10:53–66. [Google Scholar]
- 10.Conesa A., Götz S., Garcia-Gomez J.M., Terol J., Talon M., Robles M. Blast2GO: a universal tool for annotation, visualization, and analysis in functional genomics research. Bioinformatics. 2005;10:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 11.Mele M., Contarini G., Cercaci L., Serra A., Buccioni A., Povolo M., Conte G., Funaro A., Banni S., Lercker G., Secchiari P. Enrichment of Pecorino cheese with conjugated linoleic acid by feeding dairy ewes with extruded linseed: effect on fatty acid and triglycerides composition and on oxidative stability. Int. Dairy J. 2011;21:365–372. [Google Scholar]
- 12.Pintus S., Murru E., Carta G., Cordeddu L., Batetta B., Accossu S., Pistis D., Uda S., Elena G., Ghiani M., Mele M., Secchiari P., Almerighi G., Pintus P., Banni S. Sheep cheese naturally enriched in α-linolenic, conjugated linoleic and vaccenic acids improves the lipid profile and reduces anandamide in the plasma of hypercholesterolaemic subjects. British J. Nutrit. 2013;109:1453–1462. doi: 10.1017/S0007114512003224. [DOI] [PubMed] [Google Scholar]
- 13.Ringseis R., Windisch W., Eder K. Transcript profiling in the liver of early-lactating dairy cows fed conjugated linoleic acid. Genomics Data. 2016;10:101–103. doi: 10.1016/j.gdata.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]